Abstract
Background
The diverse cellular constituents of childhood brain tumor ependymoma, recently revealed by single cell RNA-sequencing, may underly therapeutic resistance. Here we use spatial transcriptomics to further advance our understanding of the tumor microenvironment, mapping cellular subpopulations to the tumor architecture of ependymoma posterior fossa subgroup A (PFA), the commonest and most deadly childhood ependymoma variant.
Methods
Spatial transcriptomics data from intact PFA sections was deconvoluted to resolve the histological arrangement of neoplastic and non-neoplastic cell types. Key findings were validated using immunohistochemistry, in vitro functional assays and outcome analysis in clinically-annotated PFA bulk transcriptomic data.
Results
PFA are comprised of epithelial and mesenchymal histological zones containing a diversity of cellular states, each zone including co-existing and spatially distinct undifferentiated progenitor-like cells; a quiescent mesenchymal zone population, and a second highly mitotic progenitor population that is restricted to hypercellular epithelial zones and that is more abundant in progressive tumors. We show that myeloid cell interaction is the leading cause of mesenchymal transition in PFA, occurring in zones spatially distinct from hypoxia-induced mesenchymal transition, and these distinct EMT-initiating processes were replicated using in vitro models of PFA.
Conclusions
These insights demonstrate the utility of spatial transcriptomics to advance our understanding of ependymoma biology, revealing a clearer picture of the cellular constituents of PFA, their interactions and influence on tumor progression.
Keywords: ependymoma, spatial transcriptomics, tumor microenvironment
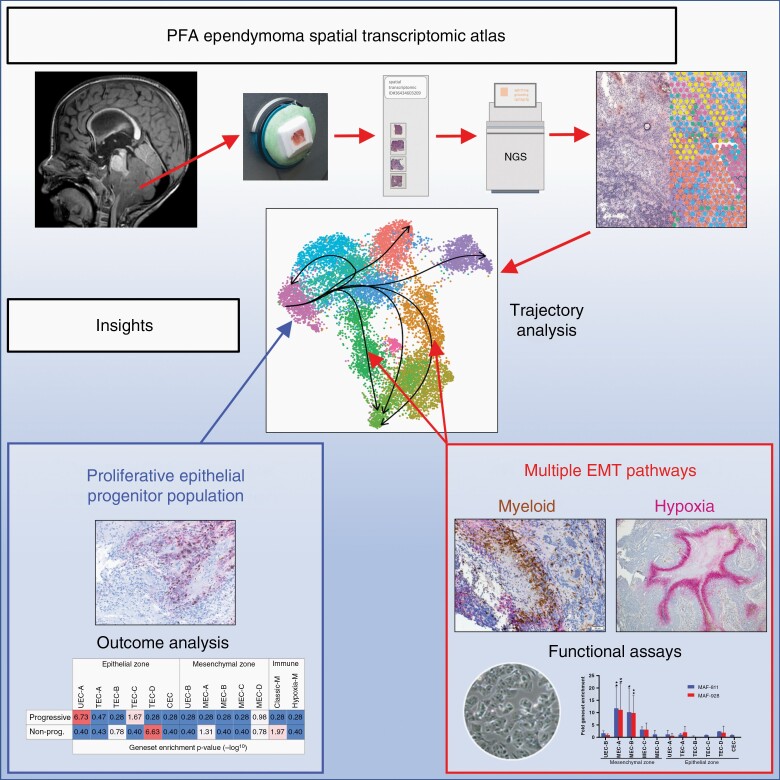
Graphical Abstract
Graphical Abstract.
Key Points.
Spatial transcriptomics advances understanding of ependymoma tumor microenvironment.
Comprehensive delineation of ependymoma cellular constituents and their interactions.
Importance of the Study.
Application of spatial transcriptomic analysis to PFA ependymoma identifies a higher level of cellular diversity than previous scRNAseq analyses. We identify major epithelial and mesenchymal transcriptional zones within all samples, each containing a variety of cellular subtypes. Analysis of the spatial arrangement of mesenchymal cell types revealed that EMT is initiated by both myeloid cell interaction as well as hypoxia in all samples, further highlighting the role of immune cells on ependymoma biology with immunotherapeutic implications. Discrete mesenchymal and epithelial progenitor-like populations were shown to co-exist in all samples, with distinct niches and cellular states. Importantly, the epithelial zone progenitor showed a high proliferation index and was proportionally higher at presentation in those tumors that later recurred, providing a novel cellular therapeutic target.
Our understanding of childhood ependymoma (EPN) has been driven by advances in genomic, transcriptomic and methylomic techniques. This has led to delineation of two major EPN subgroups defined by anatomic location—supratentorial and posterior fossa—the latter being more common in childhood and largely comprised of posterior fossa subgroup A (PFA). In recent years, the cellular heterogeneity of the EPN tumor microenvironment (TME) has been revealed using single cell RNA-sequencing (scRNAseq), identifying multiple neoplastic cell types that include undifferentiated progenitors, ependymal- and mesenchymal-differentiated progeny.1,2 Our previous scRNAseq analysis of PFA identified four major neoplastic subpopulations.1 Two ependymal-differentiated cell types were delineated—ciliated EPN cells (CEC) and transportive EPN cells (TEC)—the names representative of their respective gene and protein expression profiles that are related to normal ependymal functions such as cilia function or trans-membrane transport of molecules, respectively. A mesenchymal subpopulation (MEC; mesenchymal EPN cells) was distinguished by mesenchyme-associated gene and protein expression and are shown by immunohistochemistry (IHC) to border necrotic zones. A progenitor-like population, termed undifferentiated EPN cell (UEC), was seen to colocalize with MEC. The existence of these diverse PFA cell states was corroborated in a parallel scRNAseq study.2
In the present study, we utilize spatial transcriptomics (ST), that combines microscopy, molecular, and single cell techniques, to advance our understanding of PFA by mapping different cell types within the TME, thereby elucidating novel mechanisms that drive PFA EPN progression.
Methods
Tumor Acquisition and Processing
Surgical material was collected at our institution at the time of surgery with informed consent (COMIRB#95-500) and snap frozen (n = 14) (Supplementary Table 1) or formalin-fixed/paraffin-embedded for spatial transcriptomic (ST) analysis and IHC respectively (see Supplementary Methods).
Spatial Transcriptomic Analysis
Frozen samples were OCT embedded and 10 μm sections were mounted on ST capture slides (Visium, 10× Genomics). Following H&E image capture, sections were permeabilized and processed to generate RNA libraries, which were sequenced at 70 000 read pairs per spot (Novaseq6000, Illumina). Sequencing data were processed, filtered, integrated, and clustered using Space Ranger (10× Genomics), Seurat,3 Harmony,4 and Clustree5 packages. Differential ST cluster gene expression was defined by Wilcoxon test. Cell types were defined by (1) manual inspection of ST cluster markers, (2) Jaccard index calculation of ST cluster marker gene overlap with scRNAseq markers,1 and (3) ontological analysis. ST cluster overlays on H&E image were produced in Loupe Browser (10× Genomics). Cell cycle phases were inferred by Seurat function CellCycleScoring. Pseudotime trajectories were inferred with the R package Slingshot.6 Distance calculations between spots and clusters were performed with clustifyr function calc_distance.
Immunohistochemistry
Dual staining immunohistochemistry (IHC) was performed for COL9A2/Ki-67 and Iba1/CA9 was performed using antibodies previously selected and optimized for EPN subpopulation specificity.1,7,8
Perturbation Studies of PFA EPN in Vitro
PFA EPN cell lines MAF-811 and MAF-928 were co-cultured with CD14+ myeloid cells in vitro for 3 days. Cell lines RNA (RNeasy, Qiagen) was sequenced using the Illumina Novaseq6000 and quantified (FPKM) using by Cufflinks. Hypoxia treated EPN cell line data from a prior study,1 was reanalyzed in the present study. Genes upregulated in treated cells compared to controls for each experiment were identified and subjected to hypergeometric analysis to quantify enrichment of the ST cluster marker genesets (Supplementary data 1).
Bulk-Tumor Transcriptome Outcome Analyses
We reanalyzed a bulk transcriptomic PFA patient sample dataset (n = 40) (GSE125861)1 with samples categorized as progressive (n = 27) or non-progressive at 5 years (n = 13). Differentially expressed genes between the two categories were then subject to hypergeometric analysis to quantify enrichment of the ST cluster marker genesets as described above.
Statistical Analyses
Statistical analyses were performed using R bioinformatics, Prism (GraphPad), and Excel (Microsoft) software. For all tests, statistical significance was defined as P < .05.
Data Availability
ST data have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are publicly accessible through GEO accession number GSE195661. A browsable internet resource of the ST data is available at https://www.pneuroonccellatlas.org.
Results
Spatial Transcriptomics Reveals that the PFA TME is Predominantly Comprised of Epithelial and Mesenchymal Zones
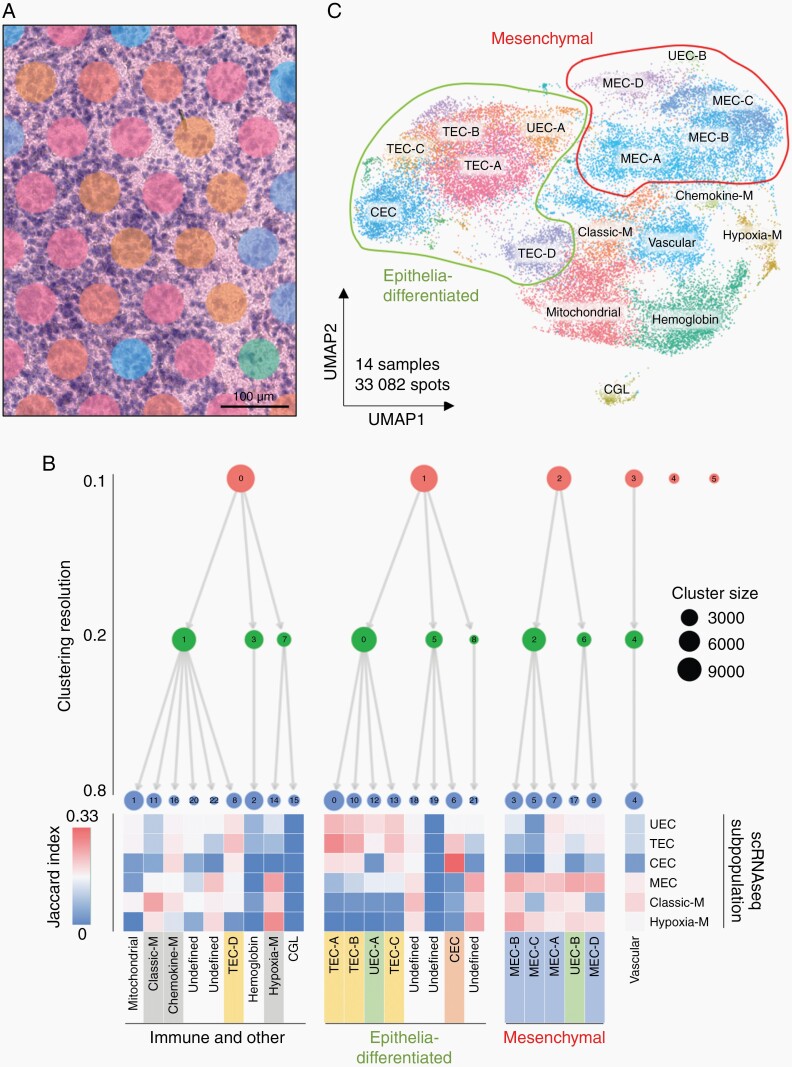
Spatial transcriptomics (ST) was used to map transcriptomic data onto tumor cellular architecture in PFA (Visium, 10× Genomics). We analyzed 14 (11 primary; 3 matched recurrence) snap frozen surgical sections, representing a range of PFA molecular and clinical variables (Supplementary Table 1). ST sequencing data was processed and filtered (Supplementary Figure 1) resulting in 33 082 spots (14 samples; ~2400 spots per sample). Spots from each sample were clustered with batch correction/alignment which identified 23 ST clusters, 17 of which were present in all 14 samples.
Transcriptomic profiles from each spot cluster were examined using three methods to characterize the identities of underlying cells: (1) Jaccard index comparison of ST cluster marker genes with previously identified PFA scRNAseq subpopulation marker genes1; (2) Clustree analysis5 of ST cluster similarity; and (3) ontological analysis of ST marker genes. Spots (diameter 55 µm) were comprised of multiple cells, ranging from 1 to 30 cells depending on the underlying cellularity of the histological region. Spots are spaced at 100 μm intervals so cells between spots are not captured by this technique (Figure 1A). Thus, ST spot clusters represent a sub-sampling of conserved composites of cells rather than single cell transcriptomes and are not therefore perfectly comparable with transcriptomic profiles of single cells identified using scRNAseq. The exact identify of individual cells congregated within each spot is therefore uncertain. Despite this caveat, the aggregated results of our characterization methods allowed us to identify a predominant previously identified scRNAseq neoplastic or immune cell type in each major ST cluster. The four major scRNAseq neoplastic cell subpopulations—transportive (TEC), ciliated (CEC), mesenchymal (MEC), and undifferentiated EPN cells (UEC)—were differentially enriched in ST clusters (Jaccard index analysis) which were annotated accordingly (Figure 1B, C). Clustering hierarchies dichotomized into two major neoplastic branches, predominated either by TEC/CEC or MEC subpopulations (Figure 1B) that represent distinct histological states. CEC and TEC are ependymal-differentiated cell types, corresponding to cilia function, and trans-membrane transport of molecules, respectively. Mesenchyme-differentiated MEC are thought to arise in response to cellular stress such as hypoxia through a process of epithelial-mesenchymal transition (EMT) that has been identified in PFA.1,9,10 Epithelial and mesenchymal ST clusters were seen to segregate by UMAP 2D projection of all 14 samples (Figure 1C).
Fig. 1.
PFA ST spot clusters correspond to PFA neoplastic subpopulations and tumor-infiltrating immune cells. (A) Representative ST sample snapshot highlighting varying numbers of underlying cells per spot (range ~5 to ~30). (B) Sequencing data from 33 082 spots in 14 PFA samples was aggregated and processed to identify 23 conserved spot clusters using Seurat with Harmony alignment. ST spot clusters were annotated according to cell type deconvolution using Jaccard index analysis, which calculates marker gene overlap (displayed as heatmap) with previously published scRNAseq subpopulation markers.1 ST spot clusters were visualized at different resolutions using Clustree analysis which identified four major neoplastic and non-neoplastic spot cluster groups: (1) epithelial-differentiated, (2) mesenchymal, (3) immune and other, and (4) vascular endothelium. (C) UMAP clustering of spatial transcriptomic data demonstrates that neoplastic spot clusters congregate into epithelial or mesenchymal groups. CEC, ciliated EPN cells; TEC, transportive EPN cells; UEC, undifferentiated EPN cells; MEC, mesenchymal EPN cells; CGL, cerebellar granular layer.
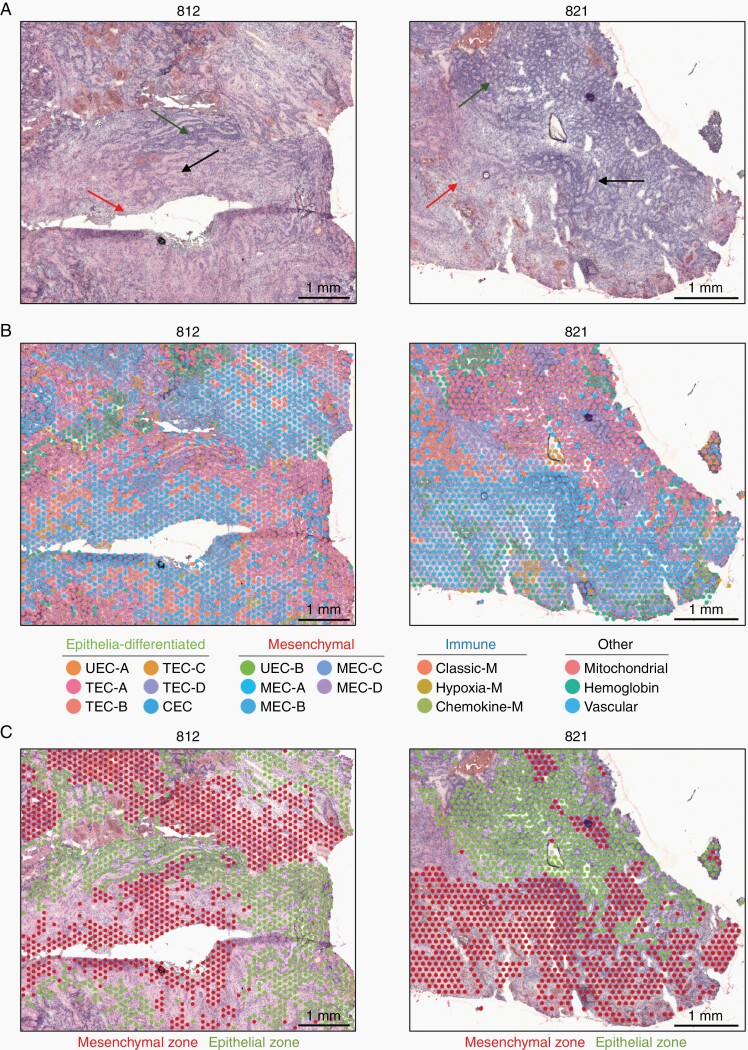
Morphologically, PFA EPN are typified by areas of varying cell density, occasionally arranged in rosettes (Figure 2A). When overlayed onto H&E tumor sections, ST clusters mirrored this morphological diversity, showing significant heterogeneity across the cellular landscape of PFA (Figure 2B). To lessen the visual complexity inherent in attempting to visualize the spatial distribution of 23 ST clusters, we visualized ST clusters at a lower resolution, revealing that most samples are comprised of widespread zones of either epithelial or mesenchymal ST cluster spots (Figure 2C and Supplementary Figure 2). As expected, epithelial zones associated with cell morphologies displaying the rosette-rich and highly cellular areas. Conversely, mesenchymal zones were often moderately or sparsely cellular and associated with areas of necrosis. ST cluster proportions provide an opportunity to quantify subpopulation proportions more accurately than data from scRNAseq analysis, as ST is a direct measure of relatively unmanipulated tissue whereas scRNAseq involves numerous tissue processing steps that are likely to skew cellular proportions. On average, epithelial zone spots were more abundant than mesenchymal in PFA at presentation (average 1.7:1, n = 11), but with a high level of variability (range 3.0:1 to 1:5.6). Both epithelial and mesenchymal zones contained multiple ST subclusters with significant spatial and molecular distinctions, details of which are elaborated in subsequent sections.
Fig. 2.
The PFA TME is comprised of major epithelial and mesenchymal transcriptomic zones. (A) Representative low magnification images of 2 PFA EPN showing classic EPN histology by H&E staining, including regions of hypercellularity with prominent perivascular pseudorosettes (black arrow), areas of increased true ependymal rosettes and epithelial differentiation (green arrow), and paucicellular regions with a lesser degree of differentiation (red arrow). (B) H&E images with overlayed ST spot clusters revealing extensive heterogeneity of transcriptomic signatures across the TME. (C) ST spot clusters congregate into two major classes (epithelial-differentiated and mesenchymal), which when overlayed onto H&E images are arranged into cohesive epithelial and mesenchymal zones. Epithelial and mesenchymal zones are associated with hypercellular differentiated and paucicellular less differentiated regions, respectively. CEC, ciliated EPN cells; TEC, transportive EPN cells; UEC, undifferentiated EPN cells; MEC, mesenchymal EPN cells.
The remaining ST clusters were largely associated with non-neoplastic cell types. A singular and relatively abundant ST cluster was associated with vascular endothelium (VE) gene expression including VWF and PECAM1 (Figure 1B, C and Supplementary Data 1, 2), forming a solitary Clustree branch (Figure 1B). This finding highlights an additional strength of ST, as this cell type was not identified in prior scRNAseq analyses, likely due to the cohesive nature of VE preventing mechanical disaggregation into single cells. A number of ST clusters were predominated by myeloid immune cell gene expression (Figure 1B, C and Supplementary Data 1, 2) which we examine in a subsequent section. Two other ST clusters were predominated either by hemoglobin gene expression, in areas with histological evidence of bleeding (hemosiderin), or by high expression of mitochondrial genes. A single sample (928_2) contained a significant area of normal brain adjacent to tumor, predominated by cerebellar granular layer (CGL) cells, forming a discrete ST cluster (Figure 1C). This cluster, along with the 6 least abundant ST clusters that were only present in 1 or 2 samples each, were excluded from the remainder of this study.
In the following sections, we individually characterize the epithelial, mesenchymal and immune compartments and their interactions.
Identification of a Proliferative Progenitor Subpopulation in PFA Epithelial Zones
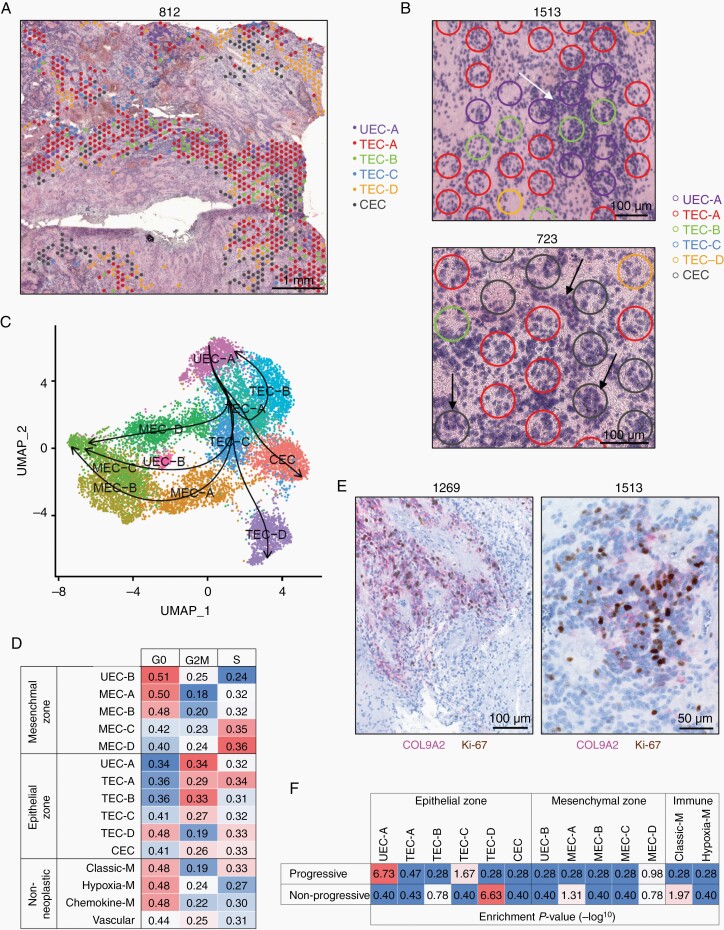
ST epithelial zones were comprised of six ST clusters that were present in all 14 PFA samples (Figure 3A and Supplementary Figure 3). This includes four ST clusters with similarity to the TEC scRNAseq subpopulation that were accordingly labeled TEC-A, TEC-B, TEC-C and TEC-D (Figure 1B). TEC ST clusters showed subtle distinctions in transcriptomic profiles (Supplementary Data 1, 2) and were largely associated with denser cellular areas with classic EPN histological features, apart from TEC-D that was associated with hypocellular areas (Figure 3A, B and Supplementary Figure 3). A single ST cluster (labeled CEC) corresponding to the scRNAseq CEC subpopulation was accordingly enriched for genes related to cilia formation (Supplementary Data 1, 2) and often associated with ependymal rosettes (Figure 3B).
Fig. 3.
PFA epithelial zones are comprised of progenitor-like and ependyma-differentiated cells. (A) Representative section showing spatial distribution of PFA epithelial zone subclusters. (B) Representative sections indicating colocalization of UEC-A, TEC and CEC subclusters with PFA histological features (white arrow = hypercellularity; true ependymal rosettes and epithelial differentiation = black arrow). (C) Pseudotime trajectories of PFA neoplastic cells with directionality designated by setting UEC-A as the starting cluster. (D) Cell cycle phase fractions in PFA neoplastic and non-neoplastic subclusters. (E) Representative dual IHC staining showing colocalization of UEC-A marker COL9A2 and mitosis marker Ki-67 in PFA. (F) Association of ST spot clusters with progression in PFA patients. Bulk-tumor gene expression profiles from 40 PFA at initial presentation were categorized into those that were either non-progressive (n = 13) or progressive (n = 27) at 5 years follow-up. Differentially expressed genes between progressive and non-progressive (P < .05) were subjected to enrichment analysis for ST spot cluster markers (top 50). Values shown are hypergeometric analysis marker geneset enrichment P-values (−log10). CEC, ciliated EPN cells; TEC, transportive EPN cells; UEC, undifferentiated EPN cells; MEC, mesenchymal EPN cells.
A single epithelial zone ST cluster, UEC-A, expressed a subset of genes restricted to the undifferentiated EPN cell (UEC) scRNAseq subpopulation, confirmed by reanalysis of existing scRNAseq data for expression of ELN, MFAP2, MDK and COL9A2 (Supplementary Figure 4 and Supplementary Data 1). Our prior scRNAseq analyses identified UEC as a potential progenitor population in PFA, findings that were mirrored by a parallel study that labeled this subpopulation as “PF-Neural-Stem-Cell-like”.2 In the present study, application of pseudotime trajectory analysis ST spots revealed a cellular hierarchy initiating at UEC-A, in keeping with both prior scRNAseq analyses, then following a trajectory traversing TEC spot clusters then splitting into either CEC, TEC-D or mesenchymal zone spot clusters (Figure 3C). UEC-A was distinguished from other epithelial zone clusters by enrichment of ribosome gene expression (FC = 60.1, P < .0001) (Supplementary Data 2), indicating a relatively high level of protein synthesis. Analysis of cell cycle phase based on gene expression revealed that epithelial zone ST clusters in general were more proliferative than mesenchymal zone or non-neoplastic clusters (Figure 3D). Notably, UEC-A was the most proliferative ST cluster, concordant with the predicted high protein synthesis inferred by ribosome gene expression. Histologically, UEC-A ST spot clusters were associated with denser cellular areas (Figure 3A, B). To further examine the histology of this putative progenitor population, we selected COL9A2 as an IHC marker of UEC-A by examination of UEC-A subpopulation-specific marker genes (Supplementary Data 1). We performed dual IHC staining for COL9A2 and Ki-67 to confirm the high level of proliferation associated with UEC-A, which showed colocalization of UEC-A marker COL9A2 with mitotic marker Ki-67 in hypercellular regions (Figure 3E). Scoring of Ki-67 mitotic cells in COL9A2 positive versus negative zones in PFA from initial presentation (n = 7) confirmed a significantly higher mitotic index (P < .05) each individual sample in the UEC-A subpopulation [average 9.3-fold (range 2.2–67.1); one-tailed, paired t-test P = .0087].
Given that proliferation index is well established as a prognostic factor for poor survival in EPN11 we examined whether this proliferative UEC-A or other ST clusters were associated with risk of tumor recurrence using a clinically-annotated bulk gene expression microarray dataset (GSE125861) of PFA tumor samples at initial presentation (n = 40). Samples were categorized into those from patients that were progressive (n = 27) and non-progressive (n = 13) at 5 years. Genes differentially expressed between progressive and non-progressive (P < .05) were then measured for enrichment of ST cluster marker genesets (hypergeometric analysis). UEC-A was the most enriched ST cluster marker geneset in progressive sample signature genes, and conversely the relatively hypocellular TEC-D cluster markers were enriched in the non-progressive samples (Figure 3F). Collectively, these data identify UEC-A as a highly proliferative epithelial zone progenitor cell that is associated with progression in PFA.
PFA Mesenchymal Zones are Comprised of Spatially Restricted EMT Stages
The process of EMT has been observed in PFA9,10 and was corroborated by our recent scRNAseq study that identified a mesenchymal subpopulation (MEC; mesenchymal EPN cell), showing that this subpopulation could be induced in PFA cell lines under hypoxic culture conditions.1 However, initiation of EMT in situ can potentially be triggered by a number of extrinsic and intrinsic stimuli in addition to hypoxia, including nutrient deprivation, cellular stress, and tissue damage.12 In the present ST study, the mesenchymal zone is comprised of 5 ST clusters, 4 of which are enriched for MEC scRNAseq markers, which were labeled MEC-A, MEC-B, MEC-C and MEC-D in order of abundance (Figure 1B, C). These distinct MEC ST clusters may reflect different EMT stimuli, or alternatively discrete transition states in the EMT process. To test these hypotheses, we examined differences between mesenchymal zone ST clusters with respect to gene expression (Supplementary Data 1, 2) and spatial arrangement (Figure 4A and Supplementary Figure 5).
Fig. 4.
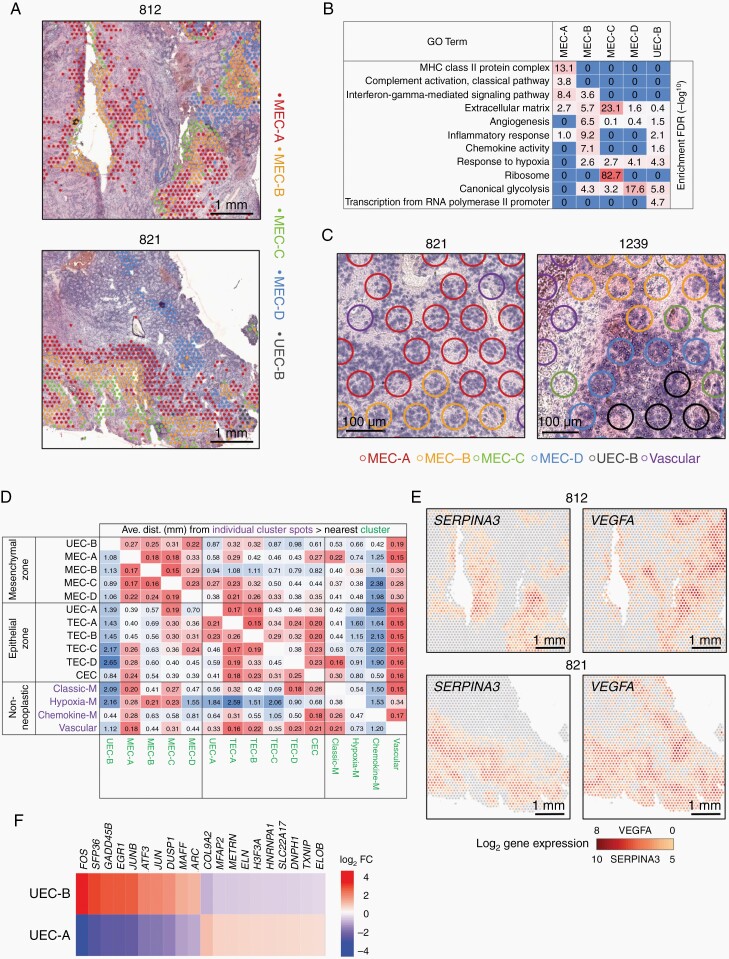
Spatial transcriptomics delineates distinct stages of EMT in PFA. (A) Representative spatial arrangement of mesenchymal zone subclusters in PFA samples. (B) Top GO term geneset enrichments [hypergeometric test FDR (−log10)] for mesenchymal zone subclusters. (C) Representative histology of mesenchymal zone subclusters. (D) Heatmap of average individual cluster spot (y-axis) distances (mm) to nearest other cluster (x-axis). (E) Representative spatial gene expression of key markers of MEC-A (SERPINA3) and MEC-D (VEGFA). (F) Heatmap of differentially expressed genes between UEC-A and UEC-B clusters. CEC, ciliated EPN cells; TEC, transportive EPN cells; UEC, undifferentiated EPN cells; MEC, mesenchymal EPN cells.
The most abundant mesenchymal zone subcluster MEC-A is distinguished by enrichment of immune genes related to MHC-class II and complement (Figure 4B and Supplementary Data 1, 2) that could potentially be derived from immune cells contained within MEC-A spots. However, reanalysis of existing scRNAseq data showed that MEC-A cluster complement-related marker genes (C1S, C1R and CFB) were restricted to the neoplastic MEC subpopulation rather than immune cells (Supplementary Figure 6), suggesting a neoplastic cell-intrinsic activation of the classic complement cascade. Serine protease inhibitor SERPINA3 is the top marker in MEC-A (Supplementary Data 1) and was also restricted to neoplastic cells (Supplementary Figure 6), playing a potentially cytoprotective role against complement component 1 factors that are serine proteases. Conversely MEC-A MHC class-II genes (HLA-DRA, HLA-DRB1) and complement cascade component C3 marker genes were more restricted to myeloid cells in scRNAseq data (Supplementary Figure 6), confirming that MEC-A ST subclusters contain a measurable proportion of myeloid cells, as indicated by Jaccard index analysis (Figure 1B).
MEC-D ST cluster spots showed a spatial arrangement distinct from MEC-A, commonly arising as islands within hypercellular epithelial zones (Figure 4A and Supplementary Figure 5) and MEC-D were microscopically distinguished from other MEC ST subclusters by a more hypercellular histology (Figure 4A, C). Glycolysis-associated genes, including CA9, GAPDH, TPI1 and LDHA, were enriched in MEC-D (Figure 4B and Supplementary Data 1, 2). Upregulation of glycolytic metabolism is a hallmark of cells responding to hypoxia, and hypoxia-related genes, in particular angiogenic VEGFA, are significantly enriched in MEC-D. This suggests that MEC-D arises as a result of hypoxic and/or metabolite depleted conditions occurring in hypercellular, hyperproliferative, epithelial zones.
MEC-C is equivalent to MEC-D with respect to enrichment of hypoxia-related genes such as VEGFA and CA9 (Figure 4B), but in contrast has a predominantly low cell density, and is often seen in necrotic zones (Figure 4A, C and Supplementary Figure 5).
MEC-B is distinguished by genes related to extracellular matrix remodeling, including NFKB components (NFKB1, NFKB2, NFKBIA, NFKBIZ), cytoprotective protease inhibitors (SERPINs and TIMP1), chitinases (CHI3L1, CHI3L2), metalloproteases (MMP7, MMP9), and extracellular matrix genes (Figure 4B and Supplementary Data 1, 2). Additionally, MEC-B was distinguished by an enrichment of genes involved with the inflammatory response characterized by chemokines CXCL1, CXCL8 (IL8) and CCL2. MEC-B is predominately located in moderately cellular areas with less evidence of residual ependymal-differentiated architecture than MEC-A or MEC-D (Figure 4A, C and Supplementary Figure 5). Based on gene expression characteristics of tissue remodeling and less epithelial-differentiated, paucicellular and necrotic histology, we hypothesized that MEC-B and MEC-C zones represent later EMT stages subsequent to MEC-A and MEC-D. Pseudotime trajectory analysis supported this hypothesis, showing MEC-B and MEC-C being the terminal clusters in the mesenchymal trajectory (Figure 3C).
To further elucidate the biology of different MEC subclusters, we examined their spatial relationships. MEC subclusters typically reside in neighboring histological zones (Figure 4A and Supplementary Figure 5) with distinct tissue morphology, supporting the hypothesis that the MEC subclusters represent discrete stages in mesenchymal differentiation. MEC cluster zones are often arranged in a conserved order of consecutive waves radiating away from epithelial zones. This striking spatial arrangement suggests a spatial lineage trajectory, potentially representing stages in the EMT with a sequence commonly starting with MEC-A, followed by MEC-B and finally MEC-C (Figure 4A and Supplementary Figure 5). This spatial evidence was corroborated by pseudotime trajectory analysis, that is agnostic to spatial data, which identified distinct trajectories for MEC-A and MEC-D arising from TEC clusters (Figure 3C). Spatial proximity between cell types is an important factor in their biological roles within the TME. We therefore performed a proximity analysis that systematically calculated inter-spot distances across all 18 major ST clusters in all 14 samples to provide a more quantitative measure of proximity (Figure 4D). Proximity analysis indicated that MEC-A is significantly more distant from MEC-D than MEC-B (P = 8.4 × 10−9) or MEC-C (P = 4.7 × 10−5). This is further illustrated by spatially distinct expression of MEC-A and MEC-D marker genes SERPINA3 and VEGFA (Figure 4E and Supplementary Data 1). Collectively these data suggest that MEC-A and MEC-D subpopulations represent independent roles in initiation of EMT in PFA.
A Second Potential Undifferentiated Progenitor Subpopulation Resides in PFA Mesenchymal Zones
A single ST mesenchymal zone cluster was distinguished by high expression of early response genes such as FOS and JUN that were previously identified as markers of the scRNAseq UEC progenitor subpopulation (Supplementary Data 1 and Supplementary Figure 4). This second potential progenitor undifferentiated subpopulation was therefore labeled UEC-B, being less abundant than the epithelial zone progenitor subpopulation UEC-A. Despite both UEC-A and UEC-B ST clusters expressing subsets of scRNAseq UEC subpopulation markers, these subpopulations were biologically discrete by transcriptomic and spatial analysis. UEC-B and UEC-A harbored exclusive gene expression signatures (Figure 4F) despite these markers being largely restricted to the scRNAseq UEC subpopulation (Supplementary Figure 4). Our prior scRNAseq study used FOS IHC to map the spatial location of UEC cells in relation to other neoplastic subpopulations, revealing an association of UEC with MEC in perinecrotic and perivascular areas.1 Accordingly, UEC-B ST spots were seen to reside in proximity to mesenchymal zone clusters (Figure 4A and Supplementary Figure 3). Proximity analysis confirmed a significantly closer spatial relationship of UEC-B with mesenchymal zone clusters than with epithelial zone clusters (one-tailed t-test P = .014) (Figure 4D).
Ontological analysis revealed that UEC-B is distinguished from other mesenchymal zone subclusters by gene expression related to transcriptional activation of the Pol-II promoter, but shares “response to hypoxia” and “canonical glycolysis” with specific MEC subclusters, notably MEC-D (Figure 4B and Supplementary Data 2), consistent with the commonalities between UEC-B and MEC-D seen by clustering analysis (Figure 1B). Cell cycle phase analysis showed mesenchymal zone clusters in general had a higher proportion of cells in G0, with the highest G0 proportion in UEC-B, consistent with the quiescent phenotype seen in some progenitors (Figure 2C), and in contrast to the highly proliferative UEC-A progenitor. The collective spatial and molecular characteristics of UEC-B suggest that this subpopulation is specifically a progenitor, or transition state of the EMT process in PFA. This is supported by pseudotime trajectory analysis that places UEC-B between TEC clusters and the later mesenchymal stage MEC-B and MEC-C clusters (Figure 3C). UEC-A and UEC-B together may therefore be critical in epithelial and mesenchymal cell transitions, similar to the co-existing proliferative epithelial and quiescent mesenchymal cancer stem cells (CSCs) identified in breast cancer that underly the plasticity of epithelial and mesenchymal states.13
Infiltrating Myeloid Immune Cells Interact with Specific Mesenchymal Zone Clusters
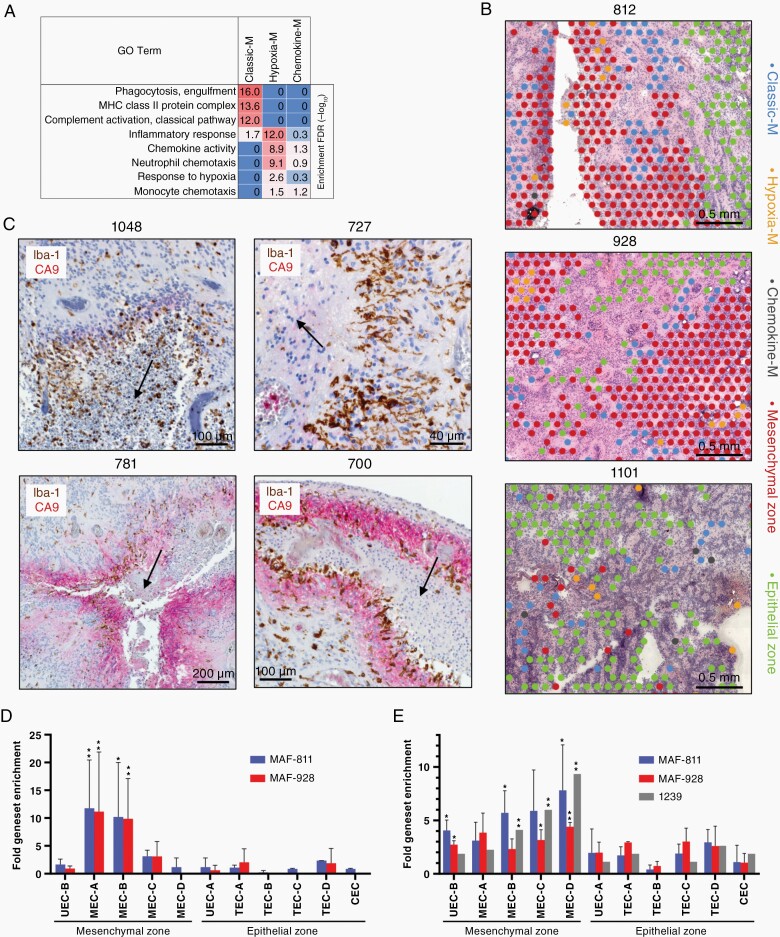
Molecular subgroup-specific immunophenotypes have previously been identified in EPN, implicating a prognostic tumor-modulatory role for tumor-infiltrating immune cells.8,14,15 In the present study ST further delineates the role of immune cells in EPN biology. Three major ST clusters were characterized by immune cell marker gene expression, and these clustered with other non-neoplastic cell types (Figure 1B). The most abundant immune ST cluster showed significant marker gene overlap with a previously identified myeloid scRNAseq subpopulation with gene expression characteristic of classic M1 polarization (Figure 1B). Gene ontologies included phagocytosis, MHC-class II and complement activation genes (Figure 5A and Supplementary Data 1, 2), and this ST cluster was therefore labeled classic-myeloid (classic-M). In contrast, the second most abundant immune cluster overlapped with a myeloid scRNAseq subpopulation with a hypoxia-response gene expression signature (Figure 1D). Gene ontologies associated with this ST cluster were primarily inflammatory response-related, consisting largely of genes involved with neutrophil chemotaxis, and was labeled hypoxia-myeloid (hypoxia-M). The smallest myeloid ST cluster, distinguished by particularly high expression of monocyte chemoattractants CCL3 and CCL4 (Supplementary Data 1), was labeled chemokine-myeloid (chemokine-M).
Fig. 5.
Myeloid lineage immune cells are spatially related to and interact with specific mesenchymal ST clusters in PFA. (A) Top GO term geneset enrichments for myeloid clusters [hypergeometric test FDR (−log10)]. (B) Representative spatial arrangement of myeloid clusters with respect to mesenchymal and epithelial zone clusters in three PFA samples. (C) Dual IHC for markers of myeloid cells (Iba-1; brown) and MEC zones (CA9; red) in representative PFA samples showing arrangement relative to areas of early necrosis (indicated by arrows). Top panels depict typical perinecrotic myeloid accumulation in the absence of CA9, lower left shows perinecrotic intermixing of myeloid with CA9+ mesenchymal cells, and lower right depicts sequential perinecrotic bands of myeloid accumulation and CA9+ mesenchymal cells. (D) Upregulation of specific ST cluster marker genesets in PFA cell lines MAF-811 (n = 2 experiments) and MAF-928 (n = 2) after exposure to CD14+ myeloid. After a 3 day coculture RNA was harvested and sequenced (RNAseq) to identify upregulated genes (FPKM fold change > 1.5) compared to untreated cell lines. Upregulated genes for each experiment were then compared to mesenchymal and epithelial zone ST spot cluster marker genesets (top 50) by hypergeometric analysis (*highest P-value within all cell line replicates < .005, **P < .0005). (E) Specific upregulation of ST cluster specific genes in PFA cell lines [MAF-811 (n = 3) and MAF-928 (n = 2)] and a short term PFA culture [1239 (n = 1)] after propagation in 2% hypoxia for 3 days was obtained from a previous study (Ref1) and analyzed as above. CEC, ciliated EPN cells; TEC, transportive EPN cells; UEC, undifferentiated EPN cells; MEC, mesenchymal EPN cells.
Spatial analysis revealed different patterns of classic-M and hypoxia-M spot localization in relation to mesenchymal zones. We observed instances where cohesive bands of classic-M ST cluster spots bordered large epithelial and mesenchymal zones (Figure 5B and Supplementary Figure 7). Proximity analysis of immune and neoplastic ST clusters revealed that classic-M was significantly closer to MEC-A than MEC-B (P = 1.3 × 10−5), MEC-C (P = 03.6 × 10−5) or MEC-D (P = 1.8 × 10−16) (Figure 4D). Dual IHC staining of myeloid marker Iba-1 and MEC marker CA9 was performed in an extended cohort of primary PFA samples including ST study samples (n = 38) to confirm this spatial relationship between myeloid cells, mesenchymal cells and necrotic zones. In those cases where early or late necrosis was present (n = 29) instances of perinecrotic myeloid bands were observed in the majority of cases. These occurred either discretely from areas of CA9+ mesenchymal cells (Figure 5C and Supplementary Figure 8A), intermixed with perinecrotic bands of CA9+ cells (Figure 5C and Supplementary Figure 8B) or forming parallel bands with CA9+ cells radiating from necrotic areas (Figure 5C and Supplementary Figure 8C). IHC analysis confirms the presence of perinecrotic myeloid cell accumulation and highlights the heterogeneous patterns of myeloid and mesenchymal cells in the complex PFA TME.
Hypoxia-M are also associated with mesenchymal zones, but in contrast to classic-M are embedded within mesenchymal areas rather than at their borders (Supplementary Figure 7), by proximity analysis being significantly closer to MEC-B than MEC-A (P = 5.5 × 10−8) or MEC-D (P = 3.0 × 10−14) (Figure 4D) suggesting a potential role in later stages of EMT, such as clearance of cellular debris.
The spatial colocalization of classic-M cells at the border of MEC-A suggests a potential immune cell role in initiation of EMT. We performed a functional test of this hypothesis by co-culturing CD14+ myeloid cells with PFA EPN cell lines MAF-811 and MAF-92816 in vitro for 3 days, followed by measurement of transcriptomic changes in cell lines by RNAseq. Cell lines co-cultured with CD14+ myeloid cells showed significant enrichment of mesenchymal zone cluster but not epithelial zone cluster signature genesets (Figure 5D). Specifically, we observed a predominant myeloid-mediated upregulation of MEC-A marker genes (including HLA-DRA, C1S, C1R, SERPINA3, CHI3L1) in both cell lines, whereas no significant enrichment of MEC-D signature genes was observed. In contrast, propagation of PFA cell lines (MAF-811 and MAF-928) and a short term culture (1239) in hypoxic conditions (3 days at 2% O2), previously shown to upregulate mesenchymal genes,1 specifically upregulated MEC-D marker genes (including CA9, LDHA) but not MEC-A markers (Figure 5E). These results further support the co-existence of two distinct EMT initiation pathways in PFA, where MEC-A is initiated by myeloid cell interaction, potentially triggered by immunogenic damage signals resulting from disruption of normal epithelial cell architecture, whereas MEC-D arises under hypoxic conditions that arise in the context of rapid tumor proliferation. MEC-A constituted on average 2.6% of tumor volume versus 1.1% for MEC-D (P = .016, paired two-tailed t-test, n = 11) revealing that myeloid cell-associated EMT is significantly more widespread than hypoxia-induced EMT which previously was considered to be the major cause of EMT in EPN.1
Discussion
Identification and characterization of mesenchymal and epithelial subpopulations using ST integrated with scRNAseq allows us to more accurately visualize and chart stages of tumor progression in PFA. This is illustrated by our discovery of co-existing progenitor-like clusters with distinct spatial characteristics restricting them to either epithelial or mesenchymal TME niches. A similar phenomenon of co-existing CSCs within the same primary tumor has been demonstrated in human breast cancer, identifying both proliferative epithelial-like CSCs and quiescent mesenchymal-like CSCs.13 These breast cancer CSCs show remarkable molecular and spatial similarity to the progenitor phenotypes in PFA in the present study. We contend that, as proposed in breast cancer, dual PFA progenitor populations are integrally involved in cellular plasticity via EMT. Importantly, identification of UEC-A as a highly proliferative progenitor that is associated with tumor progression provides a novel target for therapeutic development in PFA.
The increased resolution afforded by ST also reveals the existence of two independent EMT pathways—one associated with myeloid cell interaction, and a second induced by hypoxia. Our study demonstrates a novel application of ST, namely by providing an orthogonal delineation of EMT stages in situ in autochthonous tumors by incorporation of spatial context. Further work will be needed to determine whether the mesenchymal cells as a whole or distinct EMT trajectories contribute to the invasive phenotype of EPN. This is a critical clinically relevant question given the high recurrence rates in EPN attributed to microscopic disease that remains after gross total resection and radiation therapy, and that is potentially related to the invasive aspect of EMT. This hypothesized EMT-associated invasiveness may explain not only recurrence, but also the dire clinical outcomes experienced by children with recurrent EPN in whom standard treatment approaches usually fail.17
Myeloid-associated EMT in PFA, identified in the present study using ST, mirrors a recent study by Aubin et al.18 that identified pro-inflammatory factors as drivers of EMT in PFA. This inflammatory process may be triggered by homeostatic perturbations such as breakdown in epithelial architecture, hypoxia, or by an antitumor response driven by recognition of tumor associated antigens. An antitumor immune process is unlikely, given that PFA ependymoma has a mutational burden below the threshold considered necessary to trigger a robust tumor antigen-driven antitumor immune response. In support of a homeostatic role, the accumulation of myeloid cells around mesenchymal zones in PFA closely resembles myeloid corralling that has been identified in spinal cord injury,19 where myeloid cells form a barrier to protect surrounding “healthy” tissue from tissue that is undergoing inflammatory homeostatic remodeling. Thus, in PFA the host immune system may regard the tumor as self, initiating a potentially tumorigenic wound healing repair process. Subversion of the normal wound healing cycle by tumorigenic processes have been proposed.20 Consistent with this model, the collective spatial cellular and molecular insights revealed by ST analysis in the present study suggests that PFA EPN is a “wound that will not heal”. In normal skin, re-epithelialization involves the undamaged proximal epithelial layer, comprised of more differentiated epithelial cells, advancing en masse to repopulate the damaged area. This process is driven by a distal proliferative pool of epithelial cells repopulated by an epithelial stem cell. In PFA, we hypothesize that UEC-A mirrors this distal proliferative epithelial progenitor subpopulation, that underlies tumor recurrence and presents specific cellular target for therapeutic intervention.
Supplementary Material
Acknowledgements
The authors appreciate the contribution to immunohistochemical studies made by E. Erin Smith, HTL (ASCP) CMQIHC of the University of Colorado Denver Histology Shared Resource.
Contributor Information
Rui Fu, RNA Biosciences Initiative, University of Colorado Denver, Aurora, Colorado, USA.
Gregory A Norris, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Nicholas Willard, Department of Pathology, University of Colorado Denver, Aurora, Colorado, USA.
Andrea M Griesinger, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Kent A Riemondy, RNA Biosciences Initiative, University of Colorado Denver, Aurora, Colorado, USA.
Vladimir Amani, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Enrique Grimaldo, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Faith Harris, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Todd C Hankinson, Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA; Department of Neurosurgery, University of Colorado Denver, Aurora, Colorado, USA.
Siddhartha Mitra, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Timothy A Ritzmann, Children’s Brain Tumor Research Centre, School of Medicine, University of Nottingham, Nottingham, UK.
Richard R Grundy, Children’s Brain Tumor Research Centre, School of Medicine, University of Nottingham, Nottingham, UK.
Nicholas K Foreman, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Andrew M Donson, Department of Pediatrics, University of Colorado Denver, Aurora, Colorado, USA; Morgan Adams Foundation Pediatric Brain Tumor Research Program, Children’s Hospital Colorado, Aurora, Colorado, USA.
Funding
This study was supported by National Institutes of Health grant R01CA237608 (R.F., K.A.R., A.G., S.M., N.K.F., and A.M.D.). R.F and K.A.R. are supported as informatics fellows of the RNA Bioscience Initiative, University of Colorado School of Medicine. The University of Colorado Denver Genomics and Microarray, and Histology Shared Resources are supported by the National Institutes of Health/National Cancer Institute University of Colorado Cancer Center grant (P30CA046934). Additional support was provided by the Tanner Seebaum Foundation and the Morgan Adams Foundation.
Conflict of interests
The authors have no conflicts of interest to disclose
Authorship
A.M.D. performed experiments with assistance from A.M.G., V.A., E.G., F.H., S.M. and T.C.H. R.F performed bioinformatic analyses with assistance from K.A.R. N.W. assisted with neuropathology concerns. N.K.F., R.F. T.A.R, R.R.G and A.M.D. were responsible for the design of the study. All authors assisted with manuscript preparation.
References
- 1. Gillen AE, Riemondy KA, Amani V, et al. Single-cell rna sequencing of childhood ependymoma reveals neoplastic cell subpopulations that impact molecular classification and etiology. Cell Rep. 2020;32(6):108023:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gojo J, Englinger B, Jiang L, et al. Single-cell RNA-Seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. Cancer Cell. 2020;38(1):44–59.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods. 2019;16(12):1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zappia L, Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7(7):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Street K, Risso D, Fletcher RB, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19(1):477:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Riemondy KA, Venkataraman S, Willard N, et al. Neoplastic and immune single-cell transcriptomics define subgroup-specific intra-tumoral heterogeneity of childhood medulloblastoma. Neuro Oncol. 2022;24(2):273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donson AM, Birks DK, Barton VN, et al. Immune gene and cell enrichment is associated with a good prognosis in ependymoma. J Immunol. 2009;183(11):7428–7440. [DOI] [PubMed] [Google Scholar]
- 9. Wani K, Armstrong TS, Vera-Bolanos E, et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012;123(5):727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malgulwar PB, Nambirajan A, Pathak P, et al. Epithelial-to-mesenchymal transition-related transcription factors are up-regulated in ependymomas and correlate with a poor prognosis. Hum Pathol. 2018;82:149–157. [DOI] [PubMed] [Google Scholar]
- 11. Wolfsberger S, Fischer I, Hoftberger R, et al. Ki-67 immunolabeling index is an accurate predictor of outcome in patients with intracranial ependymoma. Am J Surg Pathol. 2004;28(7):914–920. [DOI] [PubMed] [Google Scholar]
- 12. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S, Cong Y, Wang D, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2(1):78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffman LM, Donson AM, Nakachi I, et al. Molecular sub-group-specific immunophenotypic changes are associated with outcome in recurrent posterior fossa ependymoma. Acta Neuropathol. 2014;127(5):731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Griesinger AM, Josephson RJ, Donson AM, et al. Interleukin-6/STAT3 pathway signaling drives an inflammatory phenotype in group A ependymoma. Cancer Immunol Res. 2015;3(10):1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amani V, Donson AM, Lummus SC, et al. Characterization of 2 novel ependymoma cell lines with chromosome 1q gain derived from posterior fossa tumors of childhood. J Neuropathol Exp Neurol. 2017;76(7):595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ritzmann TA, Rogers HA, Paine SML, et al. A retrospective analysis of recurrent pediatric ependymoma reveals extremely poor survival and ineffectiveness of current treatments across central nervous system locations and molecular subgroups. Pediatr Blood Cancer. 2020;67(9):e28426. [DOI] [PubMed] [Google Scholar]
- 18. Aubin RG, Troisi EC, Montelongo J, et al. Pro-inflammatory cytokines mediate the epithelial-to-mesenchymal-like transition of pediatric posterior fossa ependymoma. Nat Commun. 2022;13(1):3936: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou X, Wahane S, Friedl MS, et al. Microglia and macrophages promote corralling, wound compaction and recovery after spinal cord injury via Plexin-B2. Nat Neurosci. 2020;23(3):337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deyell M, Garris CS, Laughney AM. Cancer metastasis as a non-healing wound. Br J Cancer. 2021;124(9):1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ST data have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are publicly accessible through GEO accession number GSE195661. A browsable internet resource of the ST data is available at https://www.pneuroonccellatlas.org.