Abstract
Head and neck squamous carcinoma (HNSCC) affects more than half a million individuals and ranks the ninth leading cause of death globally each year. Many patients develop treatment resistance leading to poor clinical outcomes. The poor treatment responses are in part due to the heterogeneity of HNSCC tumor and tumor microenvironment (TME). The interaction of tumor cells with their TME has been studied vigorously in recent years because of their pivotal roles in tumorigenesis and determining the treatment response. Cancer-associated fibroblasts (CAFs) are one of the most abundant tumor-infiltrating cells, which have been shown to associate with the aggressive behavior of HNSCC. Hence, targeting and disrupting the tumor-CAFs interactions represents a rational therapeutic approach. To develop targeted therapeutic drugs against CAFs, the identification of CAF-associated gene signature is essential. Here, we analyzed multiple sequencing databases including microarrays and single-cell RNA-sequencing databases and identified SPARC/MMP9/CD44 as HNSCC targetable gene signatures encompassing cancer-associated fibroblasts (CAFs). We found SPARC/MMP9CD44 signature was highly expressed in HNSC tissues compared to adjacent normal tissues. Increased SPARC/MMP9/CD44 signature levels strongly correlated with tumor-infiltrating CAFs, suggesting the functional importance of this signature for HNSCC-CAFs interaction and progression. Subsequently, we utilized a genomics approach and identified midostaurin as the top-ranking drug candidate for targeting SPARC/MMP9/CD44 signature. For validation, we performed molecular docking of midostaurin in complex with SPARC/MMP9/CD44 and demonstrated midostaurin’s high binding affinities compared to their respective standard inhibitors. In summary, our study provided a rapid genomics approach for identifying targetable gene signature and drug candidate for HNSCC.
Keywords: Head and neck squamous cancers (HNSCC), cancer-associated fibroblasts (CAFs), drug resistance, midostaurin, molecular docking
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease and the 9th most common malignancy with high morbidity and mortality rates globally, constituting almost 90% of all head and neck cancers [1,2]. More than half a million new cases have been reported, and approximately 400,000 deaths are reported annually globally [3]. Current treatments include surgery, radiotherapy, chemotherapy, or chemo-radiotherapy, and they are decided based on the cancer stage and anatomic location [4-6]. However, despite these conventional treatment modalities, up to 40% of patients develop resistance to treatment due to local and distant metastasis post-surgery [7,8]. Thus, a better understanding of the molecular mechanism which promotes HNSCC development and progression is urgently needed for early diagnosis and development of personalized treatment for patients. The immune system plays a significant role in HNSC. Recent studies indicate that HNSCC is among the most highly immune infiltrated cancer types [9,10]. Moreover, current research has mainly focused on the HNSC tumor cells, with only a few studies on the interactions that occur with the tumor microenvironment (TME) [11-13]. A better understanding of the interactions between TME is vital as it will improve the treatment outcome for HNSCC. The TME consists of various cellular subsets, and these include cancer-associated fibroblasts (CAFs), macrophages, T cells, B cells, and natural killer (NK) cells, amongst others [14-17]. CAFs are heterogeneous stromal cells and the most abundant cells within the TME [18,19]. CAFs secret various immune-modulating and oncogenic cytokines and chemokines that contribute significantly toward tumor progression, therapy resistance, and metastasis [20]. In addition, overexpression CAFs markers and associated genes have been reported in HNSCC and participate in all stages of cancer progression [21,22]. Secreted protein acid and rich in cysteine (SPARC) is secreted by the fibroblasts, and its dysregulation has been reported to promote tumor growth, progression, and poor prognosis [23-26]. SPARC has been shown to induce the expression of MMP9 in the TME [27]. Matrix metalloproteinase-9 (MMP9) is a member of matrix metalloproteinases (MMPs) proteins, a family of highly homologous extracellular zinc- and calcium-dependent endopeptidases [28]. Overexpression of MMP9 has been reported to promote HNSCC progression, metastasis, and escape immune surveillance. Increased expression of SPARC and MMP9 have been shown to promote HNSCC progression [29], making them both targets for drug development.
Accumulating studies have shown that CAFs are crucial in promoting cell growth, metastasis, and resistance to therapy [30], also in facilitating the generation of cancer stem cells (CSC) [31]. One of the stemness markers, Cluster of Differentiation 44 (CD44), a cell surface receptor for hyaluronic acid, is overexpressed in HNSCC and correlated with poor prognosis [32,33]. Reportedly, CD44 provides cell surface receptor docking for MMP9 and promotes MMP9-mediated tumor invasion [34,35]. These findings strongly suggest crosstalk among SPARC/MMP9/CD44 oncogenes. In the present study, we explored bioinformatics analysis, to identify targetable oncogenic signatures of CAFs in HNSCC. Interestingly, from the Single cell RNA-seq analysis obtained from the gene expression omnibus (GEO) dataset; GSE103322 and GSE83519, we identified the overexpression of SPARC/MMP9/CD44 in HNSCC, and further used different web-tools to validate their upregulation. Moreover, we applied molecular docking to predict the interactions of midostaurin with SPARC/MMP9/CD44 oncogenic signatures. Midostaurin, previously known as PKC412, is a multi-kinase inhibitor that was originally developed for treatment of patients with solid malignancy [36], has already been approved by the FDA for treatment of acute myelocytic leukemia (AML) [36-41]. Here, we provide further mechanistic insights into midostaurin’s potential to inhibit SPARC/MMP9/CD44 in HNSCC.
Material and methods
Microarray data analysis
Differential expressed genes (DEGs) from HNSC patient samples were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) [42]. Two (2) datasets were obtained which included GSE103322 and GSE83519, and further analysed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2) [43]. which was used to identify DEGs between HNSC tumor samples and normal samples. The fold-change (FC) threshold was set to 2 and P < 0.05 was considered statistically significant. Futhermore, Bioinformatics & Evolutionary Genomics (BEG) online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/), was utilized to construct venn diagram.
Validation of SPARC/MMP9/CD44 expression levels in HNSCC
Expression levels of the SPARC/MMP9/CD44 oncogenes were analyzed with GEPIA 2 online web-tool (http://gepia2.cancer-pku.cn/). Expression levels of SPARC/MMP9/CD44 in HNSCC samples (red) were compared to adjacent normal samples (green), with P < 0.05 indicating statistical significance. Furthermore, we used UALCAN (http://ualcan.path.uab.edu/), an open-access public online tool for analysis of The Cancer Genome Atlas (TCGA) [44]. To analyze the tumor stages when SPARC/MMP9/CD44 genes are expressed in HNSCC. To Validate the overexpression and overall survival of these oncogenes, we used independent tool; online survival differentially expressed recurrence and metastasis (OSdream) and human protein atlas (https://www.proteinatlas.org/) [45]. P < 0.05 as statistically significant.
Protein-Protein Interaction (PPI) Network, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
Protein interactions were analyzed using the STRING tool (https://string-db.org/) to construct a PPI clustering network [46]. The interactions had an initial of 3 nodes, 3 edges, 2 average node degree and were increased to 18 nodes, 18 edges, and 4.5 average node degree. Moreover, the interactions also had an enrichment enrichment average local clustering coefficient of 0.85, PPI enrichment P-value of 6.71 × 10-4 and and the confidence cutoff value representing the interaction links was adjusted to 0.700 as the highest scoring link. Moreover, For the enrichment analysis, we explored Network Analyst tool (https://www.networkanalyst.ca/) using the SIGnaling Network Open Resource (SIGNOR 2.0) and selected the KEGGs database to analyze enriched co-expressed genes. The database for annotation, visualization, and integrated discovery (DAVID), (https://david.ncifcrf.gov/.jsp), was used to analyze enriched GO including biological processes and molecular functions involved, with the criterion set to P < 0.05.
Relations between abundance of tumor-infiltrating lymphocytes (TILs) and SPARC/MMP9/CD44 expression
The interaction between tumor and immune system plays a crucial role in both cancer development and treatment response. To investigate the correlation between HNSCC and the immune system, we used TISIDB (http://cis.hku.hk/TISIDB), an integrated repository portal for tumor-immune system interactions [47]. Herein, we used the GSEA method to characterize 28 subpopulations tumor-infiltrating lymphocytes (TILs), which displayed the prognostic cell type of HNSCC. Moreover, we analyzed the association of SPARC/MMP9/CD44 expression across the immune subtype which included; wound healing, IFN-gamma dominant, inflammatory, lymphocyte depleted, immunologically quiet and TGF-b dominant.
The correlation between SPARC/MMP9/CD44- and infiltrating immune cells in HNSCC patients
Correlations between SPARC/MMP9/CD44 expressions and tumor infiltration levels were analyzed with the Tumor Immune Estimation Resource (TIMER) (http://timer.cistrome.org/) an online computational tool used to analyze the nature of tumor immune interactions across a variety of cancers [48]. Herein, the gene DE sub-tool of TIMER to further investigate the expression of SPARC/MMP9/CD44 in HNSCCs compared to normal tissues. Distributions of gene expression levels are displayed using box plots. The statistical significance computed by the Wilcoxon test is annotated by the number of stars (*: P-value < 0.05; **: P-value < 0.01; ***: P-value < 0.001). To further analyze, we determined correlations of SPARC, MMP9, and CD44 with a set of gene markers of immune infiltration cells, particularly the cancer associated fibroblast (CAFs). The infiltration level was compared to the normal level using a two-sided Wilcoxon rank-sum test. In addition, we explored the online consensus survival analysis webserver for tumor microenvironment (OStme), a sub-tool from the biomedical informatics institute database (https://bioinfo.henu.edu.cn/), to identify abundance of macrophages, CAFs, NK cells and B cells in the TME as compared to other cells in HNSCC.
Drug sensitivity analysis of SPARC/MMP9/CD44 oncogenes
To determine the correlation between SPARC/MMP9/CD44 oncogenes and drug sensitivity of the genomics of drug sensitivity in cancer (GDSC) top 30 drugs in pancancer, we used the Gene Set Cancer Analysis (http://bioinfo.life.hust.edu.cn/web/%20GSCALite) [49], a web-based tool used to analyze differentially expressed genes (DEGs) and correlation to drug sensitivity. All the drugs approved by the Food and Drug Administration (FDA) were displayed. Moreover, we evaluated the evaluate the response of HNSCC cell lines, when treated with midostaurin, by using the GDSC dataset from the sanger institute (https://www.sanger.ac.uk/).
Molecular docking of protein-ligand interactions
In order to evaluate the strength of interactions of midostaurin with SPARC/MMP9/CD44 signature, we performed molecular docking analysis. Frirst, the crystal structures of SPARC (PDB:2V53), MMP9 (PDB:1I6J), and CD44 (1UUH), were downloaded from the Protein Data Bank (PDB) (https://www.rcsb.org/). The 3D structure of midostautin (CID:9829523) was retrieved from (https://pubchem.ncbi.nlm.nih.gov/compound/9829523). The 3D structures of established inhibitor for SPARC, mitomycin (CID:5746, molecular weight (MW: 334.53 g/mol), MMP9-illomastat (CID:132519, MW:388.5 g/mol) and CD44, sorafenib (CID:216239, MW:464.8 g/mol), were all downloaded from PubChem as SDF files. For further processing, we used PyMol software (https://pymol.org/2/) to visualize the ligands and convert them into PDB file format; these files were subsequently converted into PDBQT format using autodock (http://autodock.scripps.edu/resources/adt). For visualization and interpretation of the docking results, we used Discovery Studio software [50].
Statistical analysis
Pearson’s correlations were used to assess correlations of SPARC/MMP9/CD44 expressions in HNSCC database. The statistical significance of differentially expressed genes (DEGs) was evaluated using theWilcoxon test. *P < 0.05 was accepted as being statistically significant.
Results
Identification of DEGs in HNSC
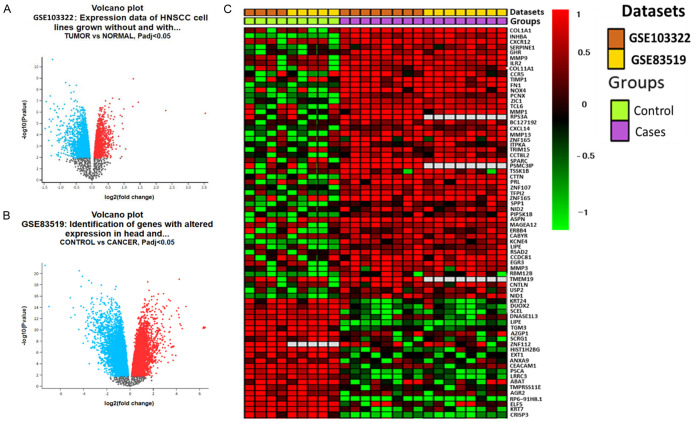
Gene expression genes from HNSC samples were extracted from the microarray dataset compared to normal samples from different studies. Results were analyzed from two (2) different datasets, i.e., GSE103322 and GSE83519. The volcano plots represent upregulated genes (red) as compared to downregulated (green) genes (Figure 1A, 1B). Moreover, the heatmap in (Figure 1C), shows overexpressed genes obtained from GSE103322 and GSE83519 datasets. P-value < 0.05 was considered statistically significant.
Figure 1.
Differentially-expressed genes (DEGs) in head and neck extracted from GSE103322 and GSE83519 microarray datasets. A, B. Volcano plots of DEGs from the two datasets with red and blue dots represent upregulated and downregulated genes (P < 0.05). C. The heatmap of overexpressed overlapping genes.
Identification of differential expression genes (DEGs) linked to cancer-associated fibroblasts in HNSCC
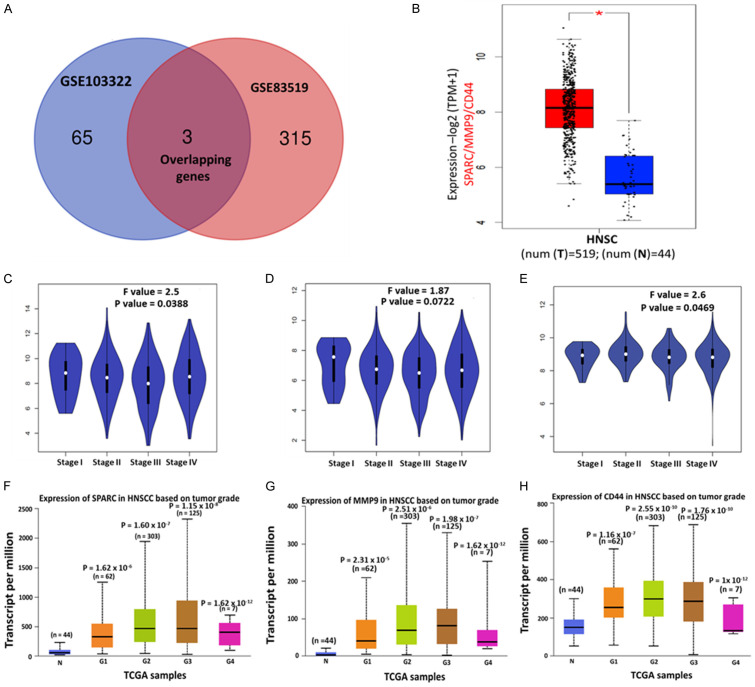
We utilized the Bioinformatics & Evolutionary Genomics (BEG) online tool to construct the Venn diagram, which displayed the overexpressed overlapping genes in HNSCC. Two overlapping genes were identified: SPARC and MMP9 (Figure 2A). Next, we incorporated CD44, an established marker for cancer stem cells, and epithelial-to-mesenchymal transition (EMT) [51] into the analysis of SPARC and MMP9 in the HNSCC database. Analyses performed using the GEPIA2 web tool revealed that the SPARC/MMP9/CD44 expressions were significantly higher in HNSCC tissues than their normal counterparts (Figure 2B). To further investigate whether these genes are associated with HNSCC progression, we analyzed their expression level through the tumor stages using the limma method from the GEPIA2 tool. The SPARC/MMP9/CD44 expression in stage 1 samples was lower than in stages 2, 3, and 4. These findings suggest that SPARC/MMP9/CD44 oncogenes may be involved in the progression of HNSCC (Figure 2C-E). The UALCAN algorithm using the TCGA samples also demonstrated that SPARC/MMP9/CD44 gene expression levels were significantly higher tumor grade (G). Interestingly, the expression of these oncogenes was lower in G1 and compared to G2 and G3 samples. Notably, G4 samples were also significantly lower, and this might have been due to insufficient number of samples (Figure 2F-H).
Figure 2.
SPARC/MMP9/CD44 signature is overexpressed in HNSCC and linked to progression. Expression levels of (A) Venn diagram of 3 selected overexpressed overlapping DEGs. (B) Overexpression of SPARC/MMP9/CD44 oncogenes in tumor tissues compared to normal tissues. (C-E) SPARC/MMP9/CD44 were more expressed in stage II to stage IV on HNSCC than in stage I. The transcript levels of (F) SPARC, (G) MMP9, and (H) CD44 were significantly overexpressed in tumors starting from grade 1 to grade 4 as compared to normal tissue. The numbers in parenthesis in (F-H) indicate sample numbers, and a P-value less than 0.05 is considered statistically significant SPARC (G1: P = 1.62 × 10-6, G2: P = 1.60 × 10-7, G3: P = 1.15 × 10-8 and G4: P = 1.62 × 10-12). MMP9: (G1: P = 2.31 × 10-5, G2: P = 2.51 × 10-6, G3: P = 1.98 × 10-7 and G4: P = 1.62 × 10-12) CD44: (G1: P = 1.16 × 10-7, G2: P = 2.55 × 10-10, G3: P = 1.76 × 10-10 and G4: P = 1 × 10-12).
Elevated SPARC/MMP9/CD44 expression is associated with distant metastasis in HNSCC
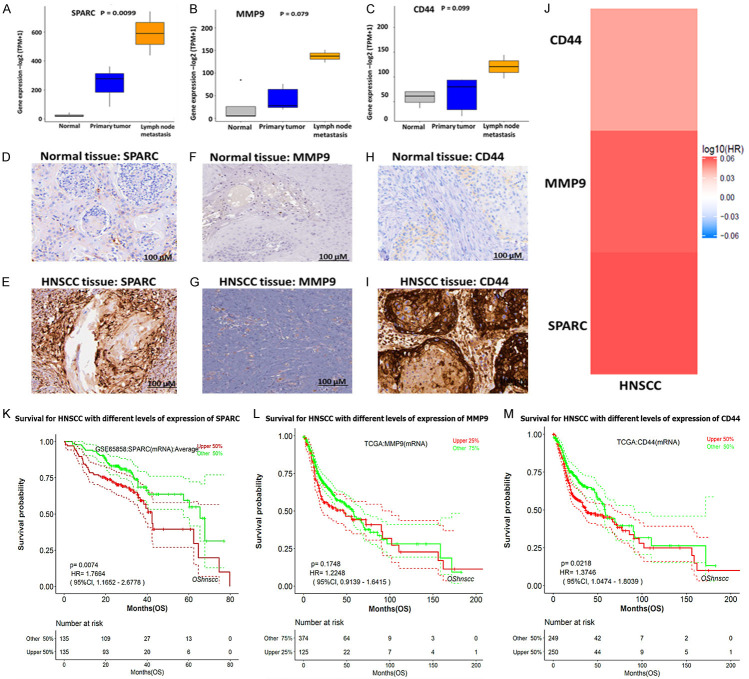
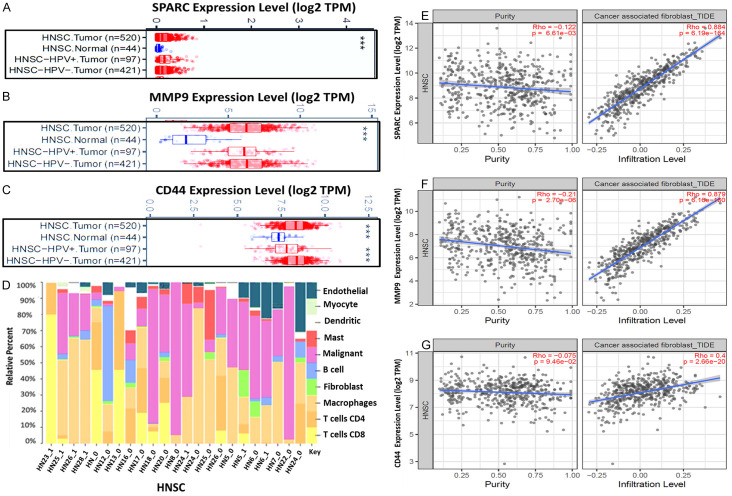
Tumor metastasis and recurrence are the major clinical challenges in cancer treatment; herein, we explored the online survival differentially expressed recurrence and metastasis (OSdream) database in determining the association of clinical characteristics between SPARC/MMP9/CD44 oncogenes in HNSCC patients. Notably, a significantly higher expression of SPARC/MMP9/CD44 was identified in patients with lymph node metastasis (Figure 3A-C). Subsequently, we queried the Human Protein Atlas (HPA) database and found that the protein expression levels of SPARC, MMP9, and CD44 were significantly higher in HNSCC tissues compared to those in normal tissues (Figure 3D-I), which was consistent with the mRNA and gene expression level results. Accordingly, SPARC showed moderate staining, moderate intensity, and high staining quantity > 75%. MMP9 exhibited low staining, with moderate intensity and a lower staining quantity of less than 25%. In contrast, CD44 expression exhibited high intensity and a higher staining quantity of > 75%. To verify the expression levels of SPARC/MMP9/CD44 in HNSCC in HNSCC, we used GEPIA2 software, which supported the above findings (Figure 3J). Based on the results, SPARC and MMP9 were more upregulated as compared to CD44, thus suggesting their association with cancer progression and metastasis. We then explored metastasis-free survival (MFS), a sub-tool of the OSdream, to analyze the survival probability of HNSCC patients harboring different risk factors. Patients with a high expression level of SPARC/MMP9/CD44 signature showed a shorter survival probability and disease-specific survival (DSS) time than those with lower expression, hence showing that SPARC/MMP9/CD44 signature might be associated with HNSCC recurrence and progression (Figure 3K-M).
Figure 3.
High expression levels of SPARC/MMP9/CD44 are associated with poor clinical outcomes in HNSCC. (A-C) Relative SPARC/MMP9/CD44 gene expression levels in normal tissue, primary tissue, and lymph node metastasis. Immunohistochemistry (IHC) analysis obtained from the HPA database revealed high protein expression levels of SPARC/MMP9/CD44 oncogenes in HNSCC tissues compared to those in normal tissues [52]. (D-I) Higher staining intensity of SPARC, MMP9, and CD44 in HNSCC tissues (lower panels) than the normal tissues (upper panels). (J) Bar graph showing significant upregulation of SPARC and MMP9, as compared to CD44 in HNSCC, suggesting their association with cancer metastasis. (K-M) Kaplan Meier survival curves revealed that a high expression level of SPARC/MMP9/CD44 signature was associated with shorter survival probability and disease-specific survival (DSS) time compared to those with lower expression.
The protein-protein interaction network and enrichment analysis SPARC/MMP9/CD44 signatures
SPARC/MMP9/CD44-proteins interaction networks were constructed STRING. After considering the gene neighborhood, gene co-occurrence, and coexpression, as anticipated, interactions were identified between SPARC with MMP9, SPARC with CD44, and MMP9 with CD44 within network clustering. The interactions had an initial of 3 nodes, 3 edges, 2 average node degrees and were increased to 18 nodes, 18 edges, and 4.5 average node degrees. Moreover, the interactions also had an average local clustering coefficient of 0.85, a PPI enrichment P-value of 6.71 × 10-4, and the confidence cutoff value representing the interaction links was adjusted to 0.700 as the highest scoring link (Figure 4A). For the enrichment analysis, we explored the Network Analyst tool using the SIGnaling Network Open Resource (SIGNOR 2.0) and selected the KEGGs database to analyze enriched co-expressed genes (Figure 4B). The signaling network analysis of KEGG pathway enrichment showed MMP9/CD44/STAT3/TIMP1 oncogenes coexpressions in the same network cluster. Subsequently, we utilized DAVID database to analyze the enriched biological processes and KEGG pathways, and further used the FunRich software to construct the sets (threshold set at P < 0.05). The enriched pathways included cMYC, integrins in angiogenesis, validated targets of c-MYC transcriptional activation mTOR signaling pathway, and Urokines e-type plasminogen activator mediated signaling (Figure 4C, 4D).
Figure 4.
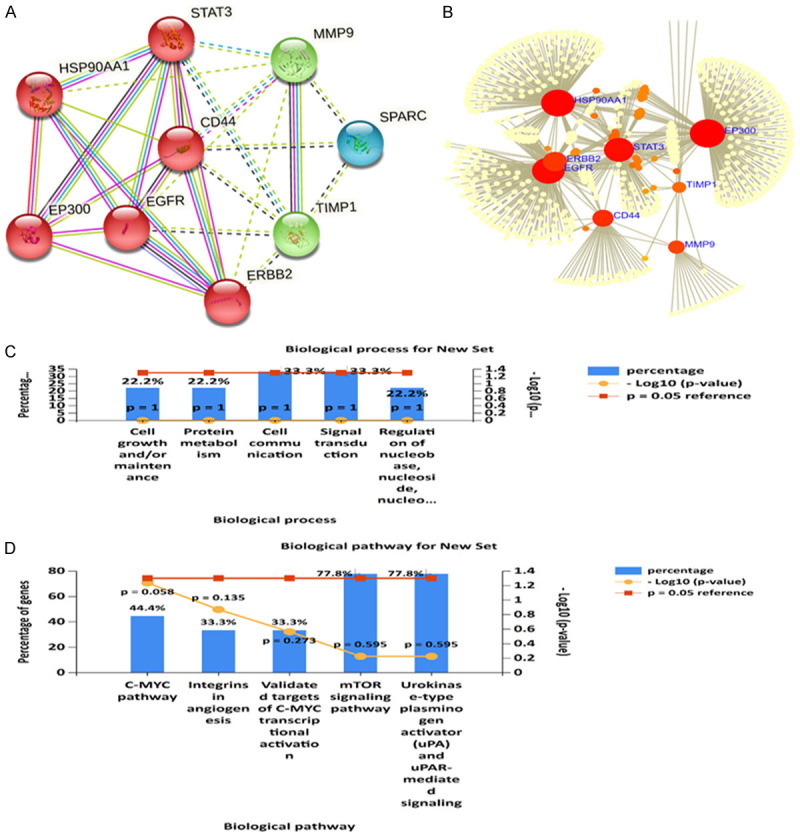
SPARC/MMP9/CD44-proteins interaction networks and enrichment analysis. A. PPI network in the STRING dataset with network clustering. B. Signaling network analysis of the KEGG pathway enrichment analysis showing coexpression of MMP9/CD44/STAT3/TIMP1 oncogenes in the same network cluster. C. Six major biological processes associated with SPARC/MMP9/CD44 signatures correlated gene clusters. D. KEGG pathway showing six affected pathways by SPARC/MMP9/CD44 oncogenic signature, with the criterion set to P < 0.05 in each panel.
The correlation between SPARC/MMP9/CD44 and immunomodulators in HNSCC patients
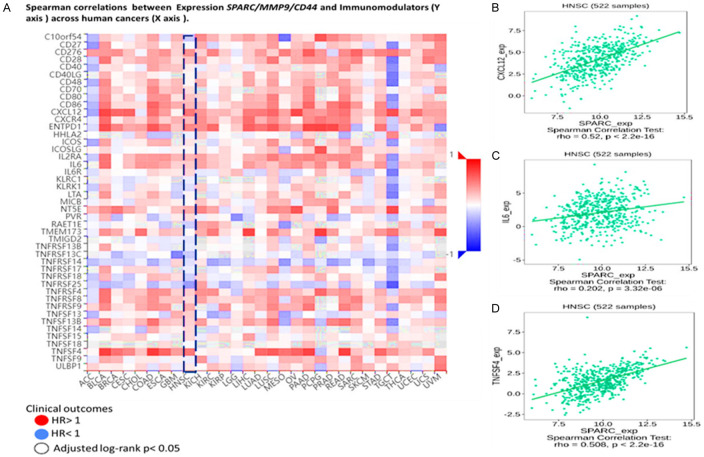
The interplay between tumor cells and their ability to infiltrate the immune system plays a vital role in tumorigenesis, progression, and the effectiveness of treatment. Herein we explored the TISIDB web portal to identify the correlation between SPARC/MMP9/CD44 across human cancers, particularly HNSCC. These immunomodulators were obtained from the study conducted previously [52]. Interestingly, among these immunomodulators were cancer-associated fibroblasts (CAF) derived cytokines and chemokines, including tumor necrosis factor (TNFs), Interleukin 6 (IL6), and C-X-C motif chemokine ligand 12 (CXCL12), which correlated with high SPARC/MMP9/CD44 oncogenic signature expression (Figure 5A).To further analyze, we explored spearman’s correlation analysis. Based on the results, expression of TNF, IL6, and CXCL12 positively correlated with CAF marker SPARC in HNSCC samples, with P < 0.05 considered statistically significant (Figure 5B).
Figure 5.
SPARC/MMP9/CD44 expression correlated with immunomodulators in HNSCC patients (A) Heatmap showing immunomodulators, including cytokines (TNF) and IL6, and chemokine (CXCL12), secreted by CAFs. HR > 1 shows poor prognosis, while HR < 1 blue shows better prognosis. (B-D) expression of TNF, IL6, and CXCL12 positively correlated with CAF marker SPARC in HNSCC samples, with p < 0.05 considered statistically significant.
Single-cell RNA sequencing revealed abundant infiltrating immune cells in HNSCC samples
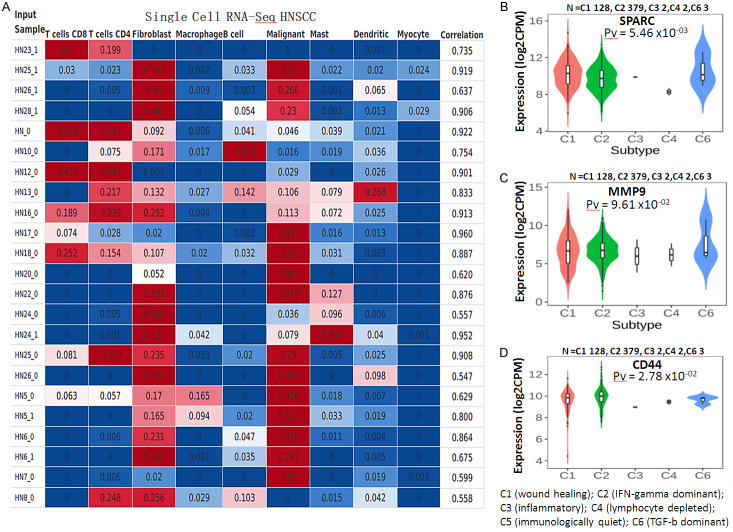
For further analysis, we explored the CIBERSORTx to profile the abundance of immune cell type profiling in HNSCC. Based on the analyzed results, NHSCC samples positively correlated with the abundance of T Cells CD8, T Cells CD4, malignant cells, and dendritic cells. Interestingly, the fibroblasts were more abundant than other HNSCC cells (Figure 6A). Moreover, we analyzed the association of SPARC/MMP9/CD44 expression across the immune subtype, which included; C1 (wound healing); C2 (IFN-gamma dominant); C3 (inflammatory); C4 (lymphocyte depleted); C5 (immunologically quiet); C6 (TGF-b dominant). Interestingly, SPARC/MMP9/CD44 oncogenes were expressed more in C1, C2 and C6 immune subtypes, however, the genes were not expressed in wound healing subtype, as shown in boxplot (Figure 6B-D).
Figure 6.
Single-cell RNA sequencing shows the abundance of infiltrating immune cells in HNSCC. (A) NHSCC samples positively correlated with the abundance of T Cells CD8, T Cells CD4, fibroblasts, malignant cells, and dendritic cells. Notably, fibroblasts were the most significantly correlated (B-D) SPARC/MMP9/CD44 were more expressed in C2 (IFN-gamma dominant); C3 (inflammatory); C4 (lymphocyte depleted); C5 (immunologically quiet); C6 (TGF-b dominant), as compared to the wound healing, thus suggesting that the overexpression of SPARC/MMP9/CD44 signature promotes tumor invasiveness, progression, and metastasis in HNSCC samples. P-value < 0.05 was considered statistically significant.
The correlation between SPARC/MMP9/CD44 and infiltrating immune cells in HNSCC patients
The tumor microenvironment plays a crucial role in cancer initiation and progression. However, the association between TME and tumor prognosis remains elusive. Herein, we utilized a web-based program (OStme) to explore our target genes’ correlation with the tumor microenvironment in HNSCC. Accordingly, we explored the TIMER database analysis to determine the correlation between SPARC, MMP9, and CD44 and infiltrating immune cells. Firstly, we applied the gene differential expression (DE) module within the TIMER database. Based on the results, we identified upregulation messenger (m)RNA levels of SPARC/MMP9/CD44 in HNSCC tumors compared to adjacent normal tissues. Distributions of gene expression levels are displayed using box plots (Figure 7A-C). HNSCC sample mixtures based on relative percentage fractions were compared using Pearson’s correlation coefficient (R). Based on the results, endothelial, myocyte, dendritic, mast, malignant, B cell, fibroblasts, and macrophages strongly correlated with HNSCC samples by displaying high relative percentage, as compared to T cell CD4 and T cell CD8, which showed lower relative percentage, indicating exhausted T cells (Figure 7D). To identify relations of SPARC/MMP9/CD44 expressions with selected immune cells, we applied a correlation analysis between the oncogenes mentioned above, and CAFs markers were adjusted by purity. As expected, the results showed correlations of SPARC/MMP9/CD44 with CAFs in HNSCC (Figure 7E-G).
Figure 7.
The correlation between SPARC/MMP9/CD44- and infiltrating immune cells in HNSCC patients (A-C) upregulated (m)RNA levels of SPARC/MMP9/CD44 in HNSCC tumor as compared to adjacent normal tissues. (D) HNSCC sample mixtures show a relatively high percentage of endothelial, myocyte, dendritic, mast, malignant, B cell, fibroblasts, and macrophages as compared to T cells. (E-G) SPARC/MMP9/CD44 expression level significantly correlates with CAF infiltration in HNSCC, where SPARC and MMP9 expression was the predominant determinant.
Single-cell RNA sequencing (scRNA-seq) reveals an immunosuppressive role of SPARC/MMP9/CD44 oncogenic signature within HNSCC TME
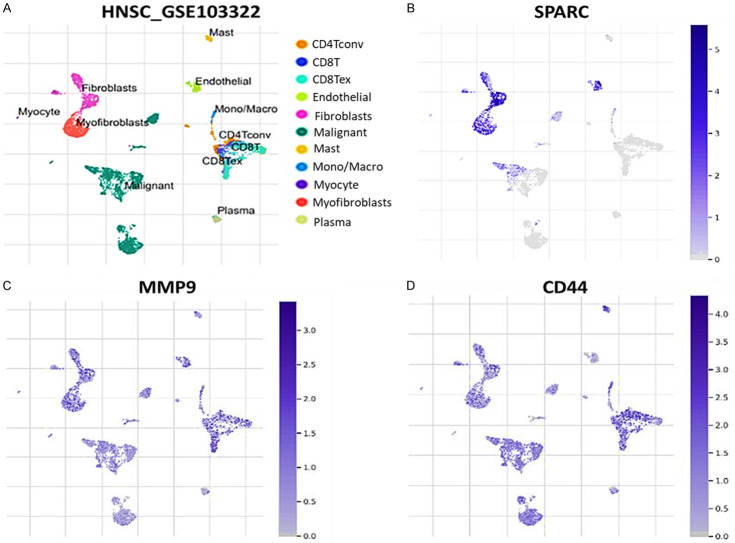
We explore the tumor immune single-cell hub (TISCH) datasets of HNSCC (GSE103322), which consists of scRNA-seq from primary HNSCC patients. The cell types obtained included CD4, CD8 T cells, endothelial, fibroblasts, malignant, mast, mono/macro, myocyte, myofibroblast and plasma (Figure 8A). Interestingly, we found that SPARC is overexpressed in endothelial cells, firoblasts, macrophages and malignant cells (Figure 8B). MMP9 is overexpressed in fibroblasts (Figure 8C). CD44 is highly expressed in endothelial cells, exhausted CD8 T cells, firoblasts, macrophages and malignant cells (Figure 8D). Thus suggesting that SPARC/MMP9/CD44 oncogenic signature is overexpressed in fibroblast with the HNSCC TME.
Figure 8.
Single-cell RNA sequencing (scRNA-seq) profiling shows that SPARC, MMP9 and CD44 oncogenic signature is associated with an immunosuppressive HNSCC TME. (A) UMAP projections of eleven major lineage cell types from single-cell RNA-sequencing data from GSE103322 datasets (B-D) UMAP plots showing expression distribution of SPARC/MMP9/CD44 oncogenic signature in single cell clusters.
Drug sensitivity analysis of SPARC/MMP9/CD44 oncogenes
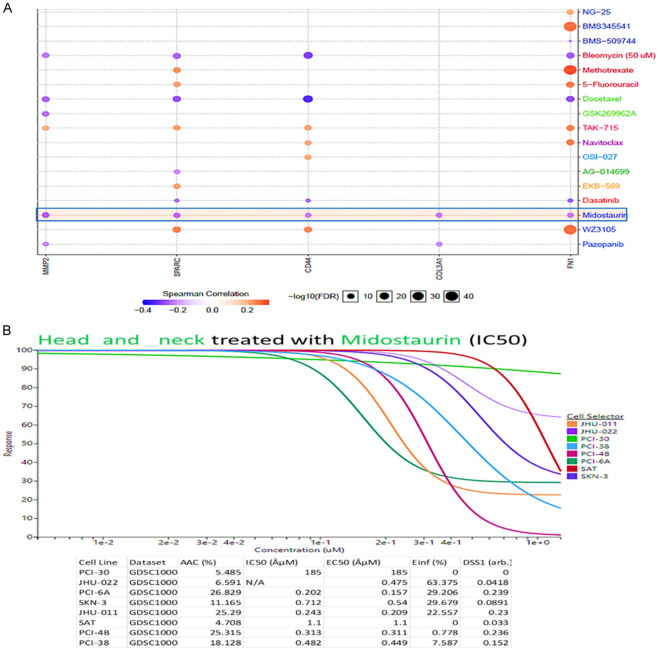
To determine the drug sensitivity of SPARC, MMP9 and CD44, we used the GSCA tool to analyze the drug response. The correlation coefficients analysis, shows that upregulated gene expression is associated with drug resistance. From our analysis of results, we identified increased mRNA expression levels of SPARC, MMP9, and CD44 (indicated in orange bubbles), to be less sensitive to the drugs. While blue bubbles indicated the sensitivity of SPARC, MMP9, and CD44 to the drugs, including bleomycin, docetaxel, dasatinib, and pazopanib, and interestingly, midostaurin was shown to be more effective as a potential drug target of SPARC, MMP9 and CD44 as compared to all the other FDA approved drugs mentioned above (Figure 9A). To further analyze, we explored the GDSC database to evaluate the response of different HNSCC cell lines obtained from the catalog of somatic mutations in cancer (COSMIC) project and identified with specific COSMIC ID. These included; PCI-30 (COSS1298529), JHU-022 (COSS1240162), PCI-6A (COSS1240206), SKN-3 (COSS1299059), JHU-001 (COSS1240161), SAT (COSS1299050), PCI-4B (COSS1298531), PCI-38 (COSS1240205) [53], Interestingly, most of the cell lines responded to midostaurin treatment with low concentration IC50 (Figure 9B).
Figure 9.
Drug response of SPARC/MMP9/CD44 Oncogenes to midostaurin in HNSCC cell lines.
In silico molecular docking analysis exhibited unique binding affinities between midostaurin and the SPARC/MMP9/CD44 signature
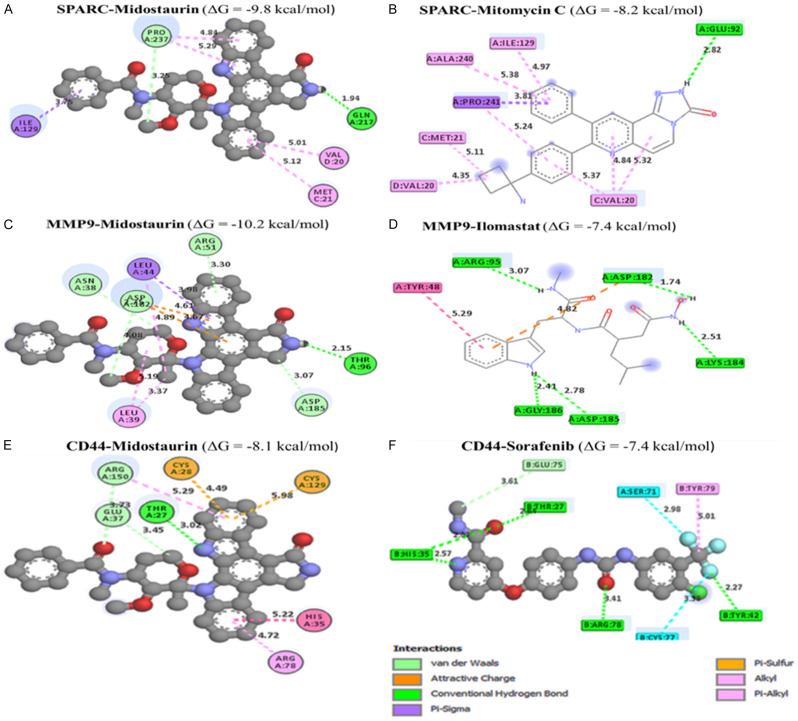
Docking results revealed potential inhibitory effects of midostaurin when bound to SPARC/MMP9/CD44 oncogenic signature. The binding affinity results of midostaurin in complex with SPARC, MMP9, and CD44 were much higher, with Gibbs free energies (ΔG) of (-9.8 kcal/mol, -10.2 kcal/mol, and -8.1 kcal/mol), respectively as compared with their standard inhibitors mitomycin C (SPARC), ilomastat (MMP9), and sorafenib (CD44) which exhibited lower binding energies (ΔG) of (-8.2 kcal/mol, -7.4 kcal/mol and 7.4 kcal/mol) respectively (Figure 10A-F). Subsequently, we used PyMol software and the discovery studio web tool for visualization to interpret the acquired results. Accordingly, several interactions were shown to stabilize the protein-ligand interactions, including a high number of conventional hydrogen bonds, van der Waals forces, carbon-hydrogen bonds, Pi anions, Pi-sigma, Pi-Pi stacked, and amide Pi-stacked, with their respective amino acids as shown in Table 1.
Figure 10.
Docking profiles of the SPARC/MMP9/CD44 oncogenes in complex midostaurin.
Table 1.
Analytical summary table showing interactions of midostaurin with the SPARC/MMP9/CD44 oncogenic signatures compared the standard inhibitors of these genes
| Midostaurin | Standard inhibitotrs | ||
|
|
|
||
| Midostaurin - SPARC Complex (ΔG = -9.8 Kcal/mol) | Mitomycin C - SPARC Complex (ΔG = -8.2 Kcal/mol) | ||
|
|
|
||
| Type of interactions and number of bonds | distance of interacting Amino acids | Type of interactions and number of bonds | distance of interacting Amino acid |
| Conventional Hydrogen bond (1) | GLN217 (1.94 Å) | Conventional Hydrogen bond (1) | GLU92 (2.82 Å) |
| Carbon Hydrogen bond | PRO237 | Pi-Sigma | PRO241 |
| Pi-Sigma | ILE 122 | Alkyl | ILE129, ALA248, MET121 VA20 |
| Pi-Alkyl | VAL28, MET21 | Pi-Alkyl | VAL20 |
|
|
|
||
| Midostaurin - MMP9 Complex (ΔG = -10.2 Kcal/mol) | Ilomastat - MMP9 Complex (ΔG = -7.4 Kcal/mol) | ||
|
|
|
||
| Type of interactions and number of bonds | distance of interacting Amino acids | Type of interactions and number of bonds | distance of interacting Amino acids |
| Conventional Hydrogen bond (1) | THR96 (2.15 Å) | Conventional Hydrogen Bonds (5) | ARG95 (2.64 Å), HIS35 (2.57 Å), LYS184 (2.51 Å), GLY186 (2.41 Å), ASP185 (2.78 Å) |
| Carbon Hydrogen Bond | ASP182, ASP185, ARG51 | Pi-Pi-T-shaped | TRY48 |
| Pi-sigma | LEU44 | ||
| Pi-alkyl | LEU39 | ||
|
|
|
||
| Midostaurin - CD44 Complex (ΔG = -8.2 Kcal/mol) | Sorafenib - CD44 Complex (ΔG = -7.5 Kcal/mol) | ||
|
|
|
||
| Type of interactions and number of bonds | distance of interacting Amino acids | Type of interactions and number of bonds | distance of interacting Amino acids |
| Conventional Hydrogen bond (1) | THR27 (3.02 Å) | Conventional Hydrogen bond (4) | THR27 (3.02 Å), THR27 (3.02 Å), ARG78 (3.41 Å), TRY42 (32.27 Å) |
| Carbon Hydrogen bond | ARG150, GLU37 | Carbon Hydrogen Bonds | GLU75 |
| Pi-sulfur | CYS28, CYS129 | Halogen | SER71, CYS77 |
| Pi-Pi-T-shaped | HIS35 | Pi-Alkyl | TYR79 |
| Pi-alkyl | ARG78 | ||
Discussion
The current multimodal strategies for the treatment of head and neck squamous (HNSCC) cell carcinoma, including surgical resection, radiotherapy and chemotherapy, have increased the overall survival of patients, however many patients still become resistant to therapy [54]. Most patients with recurrent or metastatic disease have poor clinical outcomes [12,55-57]. Moreover, HNSCCs are mostly heterogeneous, hence making it difficult to classify and develop personalized targeted therapy [9]. Thus, there is an urgent need for identification of novel biomarkers for the development of improved therapeutics. The tumor microenvironment (TME) of HNSCC consists of various cells that infiltrate the tumor cells [11,57-61]. Cancer-associated fibroblasts (CAFs) being the most abundant subsets of the TME, and have been associated with the aggressive nature of head and neck cancers (HNCs). In the present study, we explored bioinformatics analysis, to identify targetable oncogenic signatures of CAFs in HNSCC. Accordingly, we explored the NCBI-GEO database using two datasets; GSE103322 and GSE83519, and identify overexpression of CAFs targetable gene in HNSCC. These upregulated genes included SPARC, MMP9 and CD44.
To further analyze, we explored GEPIA and ULCAN online bioinformatics web tools, which verify the overexpression of SPARC/MMP9/CD44 messenger RNA (mRNA) expression in HNSCC. Interestingly, we also found that the expression of these oncogenes in HNSCC stage 1 and normal grade was lower than in stages 2-3 and grade 1-4. These findings suggested that SPARC/MMP9/CD44 oncogenes could be involved in cancer initiation and progression. To further analyze, we explored the metastasis-free survival (MFS) sub-tool of the OSdream, to analyze the survival probability of HNSCC patients harboring numerous risk factors. Based on the results, patients with high expression levels of SPARC/MMP9/CD44 oncogenes showed a shorter survival probability and disease-specific survival (DSS) time than those with lower expression, showing that SPARC/MMP9/CD44 signatures might be associated with HNSCC recurrence and progression. The protein-protein interactions (PPI) revealed coexpression of SPARC, MMP9, and CD44 with the same cluster, moreover the coexpression of these oncogenes enriched several biological pathways, including cMYC, integrins in angiogenesis, validated targets of c-MYC transcriptional activation mTOR signaling pathway, and Urokines e-type plasminogen activator mediated signaling, which was previously shown to associate with HNSCC metastasis and cancer stemness [62-64].
The interplay between tumors and their ability to evade the immune system plays a vital role in tumorigenesis, progression, and the effectiveness of treatment. We explored the single-cell RNA sequencing analysis obtained from the CIBERSORTx database, which revealed an abundance of immune cells, including T Cells, CD8, T Cells CD4, fibroblast cells, and dendritic cells. Interestingly, fibroblasts were the most significantly correlated to HNSCC samples, potentially promoting tumorigenesis [18,65,66]. The interaction between tumors and the immune system plays a crucial role in cancer development and treatment response [47]. We further profiled the tumor heterogeneity landscape and identified different types of cancer-associated fibroblasts (CAFs) involved, using the integrated repository portal for tumor-immune system interactions (TISI) database [67]. Our analysis revealed that the expression of SPARC/MMP9/CD44 oncogenic signatures was associated with myofibroblast-like cells (myoCAFs), including wound healing denoted as (C1) (Figure 5B-D). Moreover, we identified inflammatory - CAFs (iCAFs) related to immune inflammation. For instance, iCAFs included the expression of (C2)-IFN-γ (C3) inflammatory, and (C6) TGF-b dominant, which correlated to SPARC/MMP9/CD44 expression, suggesting the involvement of this signature in HNSCC tumor invasiveness, progression and metastasis [68-70]; this finding is in line with the study by Bello et al., which demonstrated that the infiltration of CAFs was associated with HNSCC aggressiveness and poor clinical outcome [71].
Furthermore, we identified the correlation between SPARC/MMP9/CD44 across human cancers, particularly HNSCC. Interestingly, among these immunomodulators were CAFs secreted cytokines and chemokines, including Tumor necrosis factor (TNFs), Interleukin 6 (IL6), and C-X-C motif chemokine ligand 12 (CXCL12), which correlated with high expressions SPARC/MMP9/CD44 oncogenic signatures. Moreover, we identified correlations of SPARC/MMP9/CD44 expressions with cancer-associated fibroblasts (CAFs) within the TME; this suggests its role in cancer initiation and progression. Thus, SPARC/MMP9/CD44 represents potential prognostic markers in HNSCC with high CAF infiltration. After exploring the Genomics of Drug Sensitivity in Cancer (GDSC), we identified midostaurin, a multi-kinase inhibitor initially developed for treating patients with other solid malignancies [36,38,72-74], as a sensitive drug for SPARC/MMP9/CD44 in HNSCC. We performed in silico molecular docking analysis to validate this finding by determining the protein-ligand interactions. Based on the docking results, we found midostaurin bound to SPARC, MMP9, and CD44, with high binding energies (ΔG, -9.8 kcal/mol, -10.2 kcal/mol, and -8.1 kcal/mol), respectively. Midostaurin showed higher binding affinities to SPARC/MMP9/CD44 than the standard inhibitors, mitomycin C (SPARC), ilomastat (MMP9), and sorafenib (CD44), ΔG: -8.2 kcal/mol, -7.4 kcal/mol and 7.4 kcal/mol, respectively. Our findings strongly suggested that midostaurin could function as an inhibitor of SPARC/MMP9/CD44 oncogenic signature in HNSCC. These results prompt us to access the therapeutic potential of midostaurin in HNSCC. The preclinical studies of midostaurin are currently undertaken in our laboratory.
Conclusions
In conclusion, we identified SPARC/MMP9/CD44 as a targetable signature correlated with infiltrating cancer-associated fibroblasts (CAF) in HNSCC. We found that the upregulation of these oncogenes promoted cancer initiation, invasion, progression, stemness, drug resistance, metastasis, and poor clinical outcome in HNSCC patients. Moreover, molecular docking of midostaurin in complex SPARC/MMP9/CD44 revealed putative binding affinities compared to their standard inhibitors. These findings strongly implicated midostaurin as a potential inhibitor of SPARC/MMP9/CD44 oncogenic signature in HNSCC and its potential for future investigation.
Acknowledgements
KC Lin is supported by the research grant from WanFang Hospital, Taipei Medical University (grant number: 111-wf-phd-06). This study is partially funded by the National Science and Technology Council, Taiwan (grant number: 110-2314-B-038 -032) to MC Liu.
Disclosure of conflict of interest
None.
References
- 1.Cohen EEW, Bell RB, Bifulco CB, Burtness B, Gillison ML, Harrington KJ, Le QT, Lee NY, Leidner R, Lewis RL, Licitra L, Mehanna H, Mell LK, Raben A, Sikora AG, Uppaluri R, Whitworth F, Zandberg DP, Ferris RL. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J Immunother Cancer. 2019;7:184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta B, Johnson NW, Kumar N. Global epidemiology of head and neck cancers: a continuing challenge. Oncology. 2016;91:13–23. doi: 10.1159/000446117. [DOI] [PubMed] [Google Scholar]
- 3.Du E, Mazul AL, Farquhar D, Brennan P, Anantharaman D, Abedi-Ardekani B, Weissler MC, Hayes DN, Olshan AF, Zevallos JP. Long-term survival in head and neck cancer: impact of site, stage, smoking, and human papillomavirus status. Laryngoscope. 2019;129:2506–2513. doi: 10.1002/lary.27807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfouzan AF. Radiation therapy in head and neck cancer. Saudi Med J. 2021;42:247–254. doi: 10.15537/smj.2021.42.3.20210660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel SR, Maiya AG, Fernandes DJ, Guddattu V, Saxena PUP, Kurian JR, Lin PJ, Mustian KM. Effectiveness of exercise-based rehabilitation on functional capacity and quality of life in head and neck cancer patients receiving chemo-radiotherapy. Support Care Cancer. 2019;27:3913–3920. doi: 10.1007/s00520-019-04750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra A, Meherotra R. Head and neck cancer: global burden and regional trends in India. Asian Pac J Cancer Prev. 2014;15:537–550. doi: 10.7314/apjcp.2014.15.2.537. [DOI] [PubMed] [Google Scholar]
- 7.Mei Z, Huang J, Qiao B, Lam AK. Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int J Oral Sci. 2020;12:16. doi: 10.1038/s41368-020-0084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 9.Chen SMY, Krinsky AL, Woolaver RA, Wang X, Chen Z, Wang JH. Tumor immune microenvironment in head and neck cancers. Mol Carcinog. 2020;59:766–774. doi: 10.1002/mc.23162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferris RL. Immunology and immunotherapy of head and neck cancer. J. Clin. Oncol. 2015;33:3293–3304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varilla V, Atienza J, Dasanu CA. Immune alterations and immunotherapy prospects in head and neck cancer. Expert Opin Biol Ther. 2013;13:1241–1256. doi: 10.1517/14712598.2013.810716. [DOI] [PubMed] [Google Scholar]
- 12.Mandal R, Şenbabaoğlu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, Lee KW, Ganly I, Hakimi AA, Chan TA, Morris LG. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829. doi: 10.1172/jci.insight.89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellone M, Hanley CJ, Thirdborough S, Mellows T, Garcia E, Woo J, Tod J, Frampton S, Jenei V, Moutasim KA, Kabir TD, Brennan PA, Venturi G, Ford K, Herranz N, Lim KP, Clarke J, Lambert DW, Prime SS, Underwood TJ, Vijayanand P, Eliceiri KW, Woelk C, King EV, Gil J, Ottensmeier CH, Thomas GJ. Induction of fibroblast senescence generates a non-fibrogenic myofibroblast phenotype that differentially impacts on cancer prognosis. Aging (Albany NY) 2016;9:114–132. doi: 10.18632/aging.101127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyauchi S, Kim SS, Pang J, Gold KA, Gutkind JS, Califano JA, Mell LK, Cohen EEW, Sharabi AB. Immune modulation of head and neck squamous cell carcinoma and the tumor microenvironment by conventional therapeutics. Clin Cancer Res. 2019;25:4211–4223. doi: 10.1158/1078-0432.CCR-18-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umansky V, Sevko A. Tumor microenvironment and myeloid-derived suppressor cells. Cancer Microenviron. 2013;6:169–177. doi: 10.1007/s12307-012-0126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Zhang M, Cheng M, Wang X, Li K, Chen J, Chen Z, Chen S, Chen J, Xiong G, Xu X, Wang C, Chen D. Tumor microenvironment in head and neck squamous cell carcinoma: Functions and regulatory mechanisms. Cancer Lett. 2021;507:55–69. doi: 10.1016/j.canlet.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Park YJ, Song B, Kim YS, Kim EK, Lee JM, Lee GE, Kim JO, Kim YJ, Chang WS, Kang CY. Tumor microenvironmental conversion of natural killer cells into myeloid-derived suppressor cells. Cancer Res. 2013;73:5669–5681. doi: 10.1158/0008-5472.CAN-13-0545. [DOI] [PubMed] [Google Scholar]
- 18.Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–1019. doi: 10.1101/gad.279737.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101:147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbazán J, Matic Vignjevic D. Cancer associated fibroblasts: is the force the path to the dark side? Curr Opin Cell Biol. 2019;56:71–79. doi: 10.1016/j.ceb.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Syed M, Flechsig P, Liermann J, Windisch P, Staudinger F, Akbaba S, Koerber SA, Freudlsperger C, Plinkert PK, Debus J, Giesel F, Haberkorn U, Adeberg S. Fibroblast activation protein inhibitor (FAPI) PET for diagnostics and advanced targeted radiotherapy in head and neck cancers. Eur J Nucl Med Mol Imaging. 2020;47:2836–2845. doi: 10.1007/s00259-020-04859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Tang Y, Tan Y, Wei Q, Yu W. Cancer-associated fibroblasts in radiotherapy: challenges and new opportunities. Cell Commun Signal. 2019;17:47. doi: 10.1186/s12964-019-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagliardi F, Narayanan A, Mortini P. SPARCL1 a novel player in cancer biology. Crit Rev Oncol Hematol. 2017;109:63–68. doi: 10.1016/j.critrevonc.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Podhajcer OL, Benedetti L, Girotti MR, Prada F, Salvatierra E, Llera AS. The role of the matricellular protein SPARC in the dynamic interaction between the tumor and the host. Cancer Metastasis Rev. 2008;27:523–537. doi: 10.1007/s10555-008-9135-x. [DOI] [PubMed] [Google Scholar]
- 25.Desai N, Trieu V, Damascelli B, Soon-Shiong P. SPARC expression correlates with tumor response to albumin-bound paclitaxel in head and neck cancer patients. Transl Oncol. 2009;2:59–64. doi: 10.1593/tlo.09109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida S, Asanoma K, Yagi H, Onoyama I, Hori E, Matsumura Y, Okugawa K, Yahata H, Kato K. Fibronectin mediates activation of stromal fibroblasts by SPARC in endometrial cancer cells. BMC Cancer. 2021;21:156. doi: 10.1186/s12885-021-07875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold S, Mira E, Muneer S, Korpanty G, Beck AW, Holloway SE, Mañes S, Brekken RA. Forced expression of MMP9 rescues the loss of angiogenesis and abrogates metastasis of pancreatic tumors triggered by the absence of host SPARC. Exp Biol Med (Maywood) 2008;233:860–873. doi: 10.3181/0801-RM-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren ZH, Wu K, Yang R, Liu ZQ, Cao W. Differential expression of matrix metalloproteinases and miRNAs in the metastasis of oral squamous cell carcinoma. BMC Oral Health. 2020;20:24. doi: 10.1186/s12903-020-1013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y, Zhu Y, Shi Y, He Y, Kuang Z, Sun Z, Wang J. Downregulation of SPARC expression inhibits the invasion of human trophoblast cells in vitro. PLoS One. 2013;8:e69079. doi: 10.1371/journal.pone.0069079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun LP, Xu K, Cui J, Yuan DY, Zou B, Li J, Liu JL, Li KY, Meng Z, Zhang B. Cancer‑associated fibroblast‑derived exosomal miR‑382‑5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol Rep. 2019;42:1319–1328. doi: 10.3892/or.2019.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez KE, Wu F, Keysar SB, Morton JJ, Miller B, Chimed TS, Le PN, Nieto C, Chowdhury FN, Tyagi A, Lyons TR, Young CD, Zhou H, Somerset HL, Wang XJ, Jimeno A. Cancer cell CD44 mediates macrophage/monocyte-driven regulation of head and neck cancer stem cells. Cancer Res. 2020;80:4185–4198. doi: 10.1158/0008-5472.CAN-20-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Jiang E, Shang Z. 3D co-culture of cancer-associated fibroblast with oral cancer organoids. J Dent Res. 2021;100:201–208. doi: 10.1177/0022034520956614. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Zhou J, Lu J, Xiong H, Shi X, Gong L. Significance of CD44 expression in head and neck cancer: a systemic review and meta-analysis. BMC Cancer. 2014;14:15. doi: 10.1186/1471-2407-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yüce I, Bayram A, Cağlı S, Canöz O, Bayram S, Güney E. The role of CD44 and matrix metalloproteinase-9 expression in predicting neck metastasis of supraglottic laryngeal carcinoma. Am J Otolaryngol. 2011;32:141–146. doi: 10.1016/j.amjoto.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Le Bras GF, Allison GL, Richards NF, Ansari SS, Washington MK, Andl CD. CD44 upregulation in E-cadherin-negative esophageal cancers results in cell invasion. PLoS One. 2011;6:e27063. doi: 10.1371/journal.pone.0027063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji N, Yang Y, Cai CY, Wang JQ, Lei ZN, Wu ZX, Cui Q, Yang DH, Chen ZS, Kong D. Midostaurin reverses ABCB1-mediated multidrug resistance, an in vitro study. Front Oncol. 2019;9:514. doi: 10.3389/fonc.2019.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stemler J, Koehler P, Maurer C, Müller C, Cornely OA. Antifungal prophylaxis and novel drugs in acute myeloid leukemia: the midostaurin and posaconazole dilemma. Ann Hematol. 2020;99:1429–1440. doi: 10.1007/s00277-020-04107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallogly MM, Lazarus HM. Midostaurin: an emerging treatment for acute myeloid leukemia patients. J Blood Med. 2016;7:73–83. doi: 10.2147/JBM.S100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peter B, Winter GE, Blatt K, Bennett KL, Stefanzl G, Rix U, Eisenwort G, Hadzijusufovic E, Gridling M, Dutreix C, Hoermann G, Schwaab J, Radia D, Roesel J, Manley PW, Reiter A, Superti-Furga G, Valent P. Target interaction profiling of midostaurin and its metabolites in neoplastic mast cells predicts distinct effects on activation and growth. Leukemia. 2016;30:464–472. doi: 10.1038/leu.2015.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ōmura S, Asami Y, Crump A. Staurosporine: new lease of life for parent compound of today’s novel and highly successful anti-cancer drugs. J Antibiot (Tokyo) 2018;71:688–701. doi: 10.1038/s41429-018-0029-z. [DOI] [PubMed] [Google Scholar]
- 41.Young DJ, Nguyen B, Li L, Higashimoto T, Levis MJ, Liu JO, Small D. A method for overcoming plasma protein inhibition of tyrosine kinase inhibitors. Blood Cancer Discov. 2021;2:532–547. doi: 10.1158/2643-3230.BCD-20-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pontén F, Schwenk JM, Asplund A, Edqvist PH. The Human protein atlas as a proteomic resource for biomarker discovery. J Intern Med. 2011;270:428–446. doi: 10.1111/j.1365-2796.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- 46.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 48.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 50.Temml V, Kaserer T, Kutil Z, Landa P, Vanek T, Schuster D. Pharmacophore modeling for COX-1 and -2 inhibitors with ligandscout in comparison to discovery studio. Future Med Chem. 2014;6:1869–1881. doi: 10.4155/fmc.14.114. [DOI] [PubMed] [Google Scholar]
- 51.May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 53.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10: Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokota T, Homma A, Kiyota N, Tahara M, Hanai N, Asakage T, Matsuura K, Ogawa T, Saito Y, Sano D, Kodaira T, Motegi A, Yasuda K, Takahashi S, Tanaka K, Onoe T, Okano S, Imamura Y, Ariizumi Y, Hayashi R. Immunotherapy for squamous cell carcinoma of the head and neck. Jpn J Clin Oncol. 2020;50:1089–1096. doi: 10.1093/jjco/hyaa139. [DOI] [PubMed] [Google Scholar]
- 55.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18:269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 56.Kaidar-Person O, Gil Z, Billan S. Precision medicine in head and neck cancer. Drug Resist Updat. 2018;40:13–16. doi: 10.1016/j.drup.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Gavrielatou N, Doumas S, Economopoulou P, Foukas PG, Psyrri A. Biomarkers for immunotherapy response in head and neck cancer. Cancer Treat Rev. 2020;84:101977. doi: 10.1016/j.ctrv.2020.101977. [DOI] [PubMed] [Google Scholar]
- 58.Elmusrati A, Wang J, Wang CY. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int J Oral Sci. 2021;13:24. doi: 10.1038/s41368-021-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young MR. Protective mechanisms of head and neck squamous cell carcinomas from immune assault. Head Neck. 2006;28:462–470. doi: 10.1002/hed.20331. [DOI] [PubMed] [Google Scholar]
- 60.Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, Cui Y. Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy. Front Cell Dev Biol. 2019;7:52. doi: 10.3389/fcell.2019.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Obradovic A, Graves D, Korrer M, Wang Y, Roy S, Naveed A, Xu Y, Luginbuhl A, Curry J, Gibson M, Idrees K, Hurley P, Jiang P, Liu XS, Uppaluri R, Drake CG, Califano A, Kim YJ. Immunostimulatory cancer-associated fibroblast subpopulations can predict immunotherapy response in head and neck cancer. Clin Cancer Res. 2022;28:2094–2109. doi: 10.1158/1078-0432.CCR-21-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waitzberg AF, Nonogaki S, Nishimoto IN, Kowalski LP, Miguel RE, Brentani RR, Brentani MM. Clinical significance of c-myc and p53 expression in head and neck squamous cell carcinomas. Cancer Detect Prev. 2004;28:178–186. doi: 10.1016/j.cdp.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 63.Murugan AK. mTOR: role in cancer, metastasis and drug resistance. Semin Cancer Biol. 2019;59:92–111. doi: 10.1016/j.semcancer.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 64.Wang XN, Su XX, Cheng SQ, Sun ZY, Huang ZS, Ou TM. MYC modulators in cancer: a patent review. Expert Opin Ther Pat. 2019;29:353–367. doi: 10.1080/13543776.2019.1612878. [DOI] [PubMed] [Google Scholar]
- 65.Dempsey LA. Antigen-presenting CAFs. Nat Immunol. 2022;23:645. doi: 10.1038/s41590-022-01207-0. [DOI] [PubMed] [Google Scholar]
- 66.Qin X, Yan M, Wang X, Xu Q, Wang X, Zhu X, Shi J, Li Z, Zhang J, Chen W. Cancer-associated fibroblast-derived IL-6 promotes head and neck cancer progression via the osteopontin-NF-kappa B signaling pathway. Theranostics. 2018;8:921–940. doi: 10.7150/thno.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng Z, Ye M, Ding H, Feng Z, Hu K. Spatial transcriptomics atlas reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment components in colorectal cancer. J Transl Med. 2022;20:302. doi: 10.1186/s12967-022-03510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan Q, Tan RJ, Liu Y. Myofibroblast in kidney fibrosis: origin, activation, and regulation. Adv Exp Med Biol. 2019;1165:253–283. doi: 10.1007/978-981-13-8871-2_12. [DOI] [PubMed] [Google Scholar]
- 69.Soliman H, Tung LW, Rossi FMV. Fibroblast and myofibroblast subtypes: single cell sequencing. Methods Mol Biol. 2021;2299:49–84. doi: 10.1007/978-1-0716-1382-5_4. [DOI] [PubMed] [Google Scholar]
- 70.Pakshir P, Noskovicova N, Lodyga M, Son DO, Schuster R, Goodwin A, Karvonen H, Hinz B. The myofibroblast at a glance. J Cell Sci. 2020;133:jcs227900. doi: 10.1242/jcs.227900. [DOI] [PubMed] [Google Scholar]
- 71.Bello IO, Vered M, Dayan D, Dobriyan A, Yahalom R, Alanen K, Nieminen P, Kantola S, Läärä E, Salo T. Cancer-associated fibroblasts, a parameter of the tumor microenvironment, overcomes carcinoma-associated parameters in the prognosis of patients with mobile tongue cancer. Oral Oncol. 2011;47:33–38. doi: 10.1016/j.oraloncology.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 72.Wang ES. Beyond midostaurin: which are the most promising FLT3 inhibitors in AML? Best Pract Res Clin Haematol. 2019;32:101103. doi: 10.1016/j.beha.2019.101103. [DOI] [PubMed] [Google Scholar]
- 73.Drugs and Lactation Database (LactMed) Bethesda (MD): National Library of Medicine (US); 2006. Midostaurin. [Google Scholar]
- 74.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Döhner K, Marcucci G, Lo-Coco F, Klisovic RB, Wei A, Sierra J, Sanz MA, Brandwein JM, de Witte T, Niederwieser D, Appelbaum FR, Medeiros BC, Tallman MS, Krauter J, Schlenk RF, Ganser A, Serve H, Ehninger G, Amadori S, Larson RA, Döhner H. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377:454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]