Abstract
Ovarian cancers derived from endometrial cysts, also known as endometriosis in ovaries, are widespread histological types in Japan. Several studies suggest that zinc deficiency plays a role in endometriosis; however, the biological mechanism of zinc deficiency and endometrial cyst remains unknown. Thus, we investigated the association between zinc status and endometrial cysts. We measured the serum zinc levels in patients who had undergone surgery for endometrial cysts (n=19) and non-endometrial benign cysts (n=36). We analyzed cell proliferation, microarray data, and gene expression using N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine (TPEN), a zinc chelator, in human immortalized endometrial epithelial cells (EMosis). The endometrial cyst group had considerably lower serum zinc levels than the non-endometrial benign cyst group. After adjusting for age, body mass index, alcohol consumption, smoking, and supplement use, endometrial cysts were markedly associated with serum zinc levels. EMosis cells treated with 5 μM TPEN demonstrated extensively increased proliferation compared to untreated cells. In the microarray analysis of EMosis cells treated with 5 μM TPEN, the enriched cellular components contained nucleoplasm, nuclear parts, and nuclear lumen. The upregulated biological processes included responses to hypoxia and decreased oxygen levels. The upregulated Kyoto Encyclopedia of Genes and Genomes pathway included the hypoxia-inducible factor-1 signaling pathway. EMosis cells treated with 5 μM TPEN demonstrated increased activator 1 (SRA1) expression and decreased AT-rich interaction domain 1A (ARID1A) expression. Protein-protein interaction network analysis indicated that ARID1A and SRA1 were associated with SMARCD1 and ATF1 among the differentially expressed genes in the microarray. EMosis cells treated with 5 μM TPEN revealed increased SRA1 mRNA levels and decreased ARID1A mRNA expression, whereas EMosis cells treated with 5 μM TPEN together with 10 μM zinc did not reveal changes in the mRNA levels of SRA1 or ARID1A compared with those without TPEN. These results suggest that zinc deficiency contributes to endometrial cyst development. Accordingly, zinc supplementation may suppress endometrial cyst development.
Keywords: ARID1A, EMosis cell, ovarian endometrial cyst, endometriosis, SRA1, trace element, zinc
Introduction
Endometriosis is a common benign gynecological disease that occurs in 5-10% of women of reproductive age [1]. Endometriosis lesions extend primarily through the pelvic region. There are various types of lesions, some caused by endometriosis itself, while others due to inflammation. Ovarian endometriotic cysts are endometriotic lesions that develop in the ovaries [2]. Cyclic bleeding due to endometriosis lesions can cause inflammation, scarring, and adhesions, resulting in infertility, chronic pelvic pain, fatigue, dysmenorrhea, dyspareunia, dysuria, and dysmenorrhea [3]. Endometriosis is associated with autoimmune diseases, asthma/allergic diseases, and cardiovascular diseases [3,4]. Previous studies have shown that ovarian clear cell carcinoma and endometrioid carcinoma are derived from ovarian endometrial cysts [5,6]. Clear cell carcinoma is a widespread histological type in East Asia, accounting for more than 25% of ovarian cancers, and is the second most frequent histological subtype in Japan. Ovarian clear cell carcinoma is particularly chemotherapy-resistant [7]. Prevention of ovarian endometrial cyst will serve as a major advance in ovarian cancer treatment. Therefore, there is a critical need for studies on the development, mechanisms, and treatment of endometrial cysts, especially in Japan [8].
The current prevailing hypothesis for the development of endometriosis is Sampson’s theory. This theory proposes that regurgitated endometrium into the abdominal cavity during menstruation becomes attached to the pelvic cavity and grows in size [9]. Moreover, there is evidence that endometriosis involves disruption of female hormones, local inflammation, and immune processes [10]. Familial aggregation of endometriosis further suggests a genetic contribution to the disease [11]. Several genetic factors have been identified in genome-wide association studies [12]. Recent evidence suggests that environmental toxins (such as phthalates, bisphenol A, or organochlorine pollutants) may play a role in the development of endometriosis [13,14]. However, the only factors that, to date, have been robustly associated with endometriosis are reflected in increased exposure to menstruation (i.e., early menarche, short menstrual cycles, and nonpregnancy) and low body mass index [10,15]. No risk factors have yet been identified that would help in the primary prevention of endometriosis [4].
Zinc is an essential trace element involved in a wide variety of cellular processes, including cell proliferation, cell differentiation [16,17], DNA synthesis [18], cell membrane stabilization, structural maintenance [19,20], redox balance [21,22], and apoptosis [23,24]. A previous study revealed that low zinc intake was associated with endometriosis [25]. Patients with endometriosis have decreased zinc levels in their blood compared to patients without endometriosis [26,27], suggesting that zinc deficiency could contribute to endometriosis. However, previous studies have focused on all types of endometriosis patients, including patients with endometrial cysts [26,27]. Moreover, no studies have identified the mechanism by which low zinc leads to the development of endometriotic cysts. The biological mechanisms linking zinc deficiency and endometrial cysts have not yet been clarified.
In this study, we hypothesized that zinc deficiency causes changes in gene expression that promote endometrial cyst development. To verify this hypothesis, we conducted a study using clinical samples and immortalized epithelial cells derived from endometrial cysts [28].
Materials and methods
Measurement of zinc in serum
This study included 55 patients undergoing surgery for benign ovarian tumors at the Fukui University Hospital, Eiheiji-cho, Japan between 2018 and 2019. Two pathologists confirmed the ovarian tumor pathology in the specimens obtained after surgery. The exclusion criteria included malignancy, infectious bacterial peritonitis, renal failure, liver cirrhosis, corticosteroid treatment, immunosuppressive drug use, and chemotherapy. Written informed consent was obtained from all enrolled patients. Serum samples were collected approximately two weeks before surgery. Patients were instructed not to eat or drink anything other than water before blood sampling was carried out. We stored all samples in a freezer at -80°C until measurement. The colorimetric method was used to measure the serum zinc and copper levels. This study was approved by the Ethics Review Board of the Fukui University Hospital (approval no. 20160021).
Materials and cell culture
Human immortalized endometrial epithelial cells derived from endometrial cysts (EMosis) [28], provided by Prof. Satoru Kyo (Shimane University, Izumo, Japan) and Prof. Yoichi Kobayashi (Kyorin University, Tokyo, Japan), were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Biowest, Kansas, MO, USA) and 1.0% penicillin/streptomycin (Gibco, Billings, MT, USA) at 37°C and 5% CO2 in an incubator (Panasonic, Tokyo, Japan). The N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine (TPEN), a zinc chelator, and zinc sulfate were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell proliferation assay
We seeded EMosis cells (1×104 cells/well) in 96-well culture plates. After culturing for 24 h, the cells were exposed to 0, 5, and 10 μM TPEN for 4 or 24 h. A WST-1 reagent kit (Roche Japan, Tokyo, Japan) was used to determine cell proliferation. The experiments were performed six times.
Microarray
EMosis cells were plated in 6.0-cm dishes and incubated for 48 h. Thereafter, cells were incubated with fresh medium that contained either 0 or 5 μM TPEN for 4 h. Microarray analysis was performed according to a previously reported method [29]. The RNeasy Kit (Qiagen, Hilden, Germany) was used to collect total RNA from cells following the manufacturer’s instructions. RNA quality assessment was carried out using the Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). All samples had RNA integrity of 8.7 to 9.1, which is acceptable for a microarray. We performed the microarray using Clariom S assay (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. The Subio Platform (Subio Inc., Tokyo, Japan) was used to examine the data. The following criteria were used to designate differentially expressed genes (DEGs): (1) difference in expression >2-fold between cells treated with 0 and 5 μM TPEN, and (2) a significant difference (P<0.1, two-sided t-test) between cells treated with 0 and 5 μM TPEN. We used the shinyGO web tool (http://bioinformatics.sdstate.edu/go/) for Gene Ontology (GO) enrichment analysis to determine the GO cellular components (CC), GO biological processes (BP), and GO Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the DEGs. The false discovery rate (FDR) was calculated, and a p-value of less than 0.05 was defined as significant [30]. We performed protein-protein interaction (PPI) network analysis using the Network Analyst web tool (http://www.networkanalyst.ca) to identify DEGs that interacted with AT-rich interaction domain 1A (ARID1A) and steroid receptor RNA activator 1 (SRA1) [31].
Gene expression analysis
EMosis cells were plated in 6.0-cm dishes, grown for 48 h, and then incubated in a medium containing 5 μM TPEN and 5 μM TPEN with 10 μM zinc for 24 h. The RNeasy Kit and SuperScript IV VILO Master Mix kit (Invitrogen, Carlsbad, CA, USA) were used to collect total RNA from the cells and synthesize cDNA. Amplifying cDNA was done using the Power SYBR Green Master Mix kit (Applied Biosystems) with primers targeting gene sequences for 36B4, SRA1, and ARID1A. Gene expression was quantified and analyzed by real-time polymerase chain reaction (RT-PCR) using the StepOnePlus system (Applied Biosystems). The following gene-specific primer sequences were used: 36B4 forward, 5’-GCTGCAGCCCCAGCTAAGGT-3’; 36B4 reverse, 5’-TAAGTTGGTTGCTTTTTGGT-3’; SRA1 forward, 5’-CTCCCTTCTTACCACCACCA-3’; SRA1 reverse, 5’-TGCAGATACACAGGGAGCAG-3’ [32]; ARID1A forward, 5’-CAGTACCTGCCTCGCACATA-3’; and ARID1A reverse, 5’-GCCAGGAGACCAGACTTGAG-3’ [33]. Standardized SRA1 and ARID1A expression levels were calculated against 36B4 expression levels, and data are presented as the fold change in mRNA levels. All experiments were performed in triplicate.
Statistical analyses
Continuous and categorical variables were expressed as mean ± standard deviation and frequencies or proportions. Continuous variables were compared using the Student’s t-test, Welch’s test, Mann-Whitney U test, and one-way analysis of variance adjusted with the Dunnett’s method. The association between serum zinc levels and endometrial cysts or non-endometrial cysts was evaluated using multivariate linear regression analysis models. We used EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria), to conduct statistical analyses [34], and P<0.05 was defined as statistically significant.
Results
Patients with endometrial cysts had lower zinc concentrations in serum
Table 1 presents the patient data for analyzing the association of serum zinc levels with endometrial cysts. Nineteen patients had endometrial cysts, whereas 36 had non-endometrial cysts. The histology of non-endometrial cysts is presented in Table 2. Alcohol consumption, smoking, nutritional supplement use, albumin, aspartate aminotransferase, alanine aminotransferase, and serum copper levels were not significantly different between patients with endometrial cysts and those with non-endometrial cysts. Patients with endometrial cysts had significantly lower alkaline phosphatase levels (ALP) (P=0.011) and serum zinc levels (P=0.032) than those with non-endometrial cysts. After adjusting for age, BMI, alcohol consumption, smoking, and supplement use, a linear regression analysis revealed that endometrial cysts were significantly associated with serum zinc levels (regression coefficient 7.98, 95% confidence interval [CI] 0.36-15.59, P=0.040).
Table 1.
Serum zinc and patient characteristics
| Endometrial cyst | Non-endometrial cyst | p-value | |
|---|---|---|---|
| N | 19 | 36 | |
| *Age mean (SD), yrs. | 42.4 (7.7) | 47.0 (20.1) | 0.224 |
| ‡BMI median (range) | 21.6 (19-29.3) | 21.85 (17-40.8) | 0.832 |
| †Alcohol, % (n) | 63.2 (12) | 38.9 (14) | 0.099 |
| †Current or past smoking, % (n) | 36.8 (7) | 16.7 (6) | 0.109 |
| †Supplement | 21.1 (4) | 22.2 (8) | 1 |
| ¶Alb mean (SD) g/dL | 4.3 (0.3) | 4.4 (0.4) | 0.124 |
| ‡GOT median (range) IU/L | 19.3 (10.0-35.0) | 20.4 (13.0-44.0) | 0.729 |
| ‡GPT median (range) IU/L | 13.0 (7.0-38.0) | 15.0 (9.0-49.0) | 0.399 |
| ¶ALP mean (SD) IU/L | 176.3 (58.2) | 232.8 (83.3) | 0.011 |
| ¶Cu mean (SD) μg/dL | 101.4 (22.9) | 104.3 (20.9) | 0.64 |
| ¶Zn mean (SD) μg/dL | 77.00 (10.02) | 84.69 (13.30) | 0.032 |
Welch’s test for statistical analysis.
Fisher’s exact test for statistical analysis.
U test for statistical analysis.
T-test for statistical analysis.
Alb, albumin; ALP, alkaline phosphatase; BMI, body mass index; GOT, glutamate oxaloacetate transaminase; GPT, glutamate pyruvate transaminase; IU, international units.
Table 2.
Histology of non-endometrial cysts
| N | |
|---|---|
| Mature cystic teratoma | 12 |
| Serous adenoma | 15 |
| Mucinous adenoma | 8 |
| Fibroma | 1 |
Zinc deficiency enhanced the proliferation of immortalized endometrial epithelial cells
Figure 1 shows the results of the WST-1 assay in the EMosis cells. The proliferation of EMosis cells treated with 5 μM TPEN for 4 h was significantly higher than that of untreated cells (P<0.05). No significant difference was observed between the untreated cell and those treated with 10 μM TPEN for 4 h or 5 μM TPEN for 24 h. The proliferation of EMosis cells treated with 10 μM TPEN for 24 h was significantly lower than that of untreated cells (P<0.001).
Figure 1.
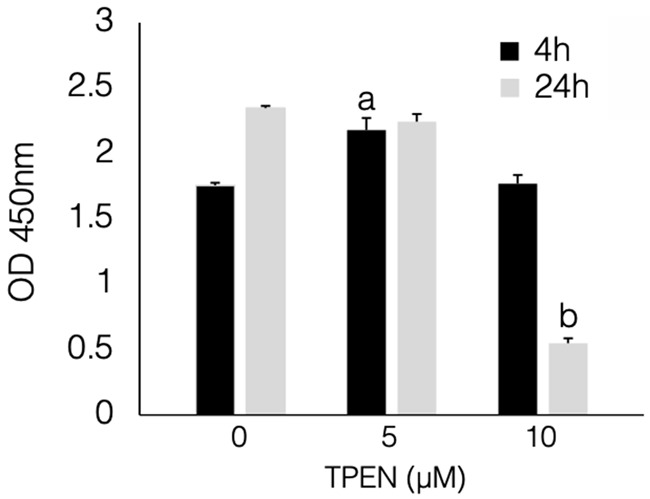
TPEN increased the proliferation of immortalized endometrial cells. Cells treated with 5 μM TPEN for 4 h demonstrated increased cell proliferation compared to cells without TPEN treatment (P<0.05). All data were analyzed using a one-way analysis of variance followed by Dunnett’s test. a; 4 h, 0 μM vs 5 μM, P<0.05. b; 24 h, 0 μM vs 10 μM, P<0.001. OD: optical density; TPEN: N,N,N’,N’-tetrakis 2-pyridylmethyl ethylenediamine.
Zinc deficiency is involved in BP and the KEGG pathway for hypoxia
Figure 2A-C shows the top 10 most enriched CC, BP, and KEGG pathways, ranked by -log10 (FDR p-value). All enriched CC, BP, and KEGG pathways, including the DEGs, are listed in Tables 3, 4 and 5. The enriched CC contained nucleoplasm, nuclear parts, and nuclear lumen. The enriched BPs included responses to hypoxia and decreased oxygen levels. The enriched KEGG pathways included the hypoxia-inducible factor-1 (HIF-1) signaling pathway.
Figure 2.
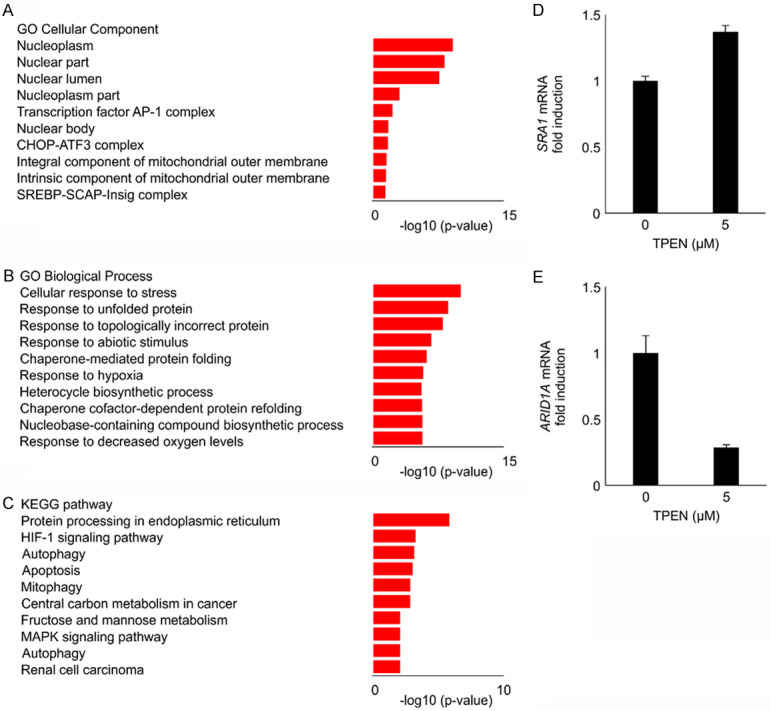
Zinc deficiency was associated with biological processes and the KEGG pathway for hypoxia. Microarray analysis was performed to determine the zinc deficiency-induced differentially expressed genes in EMosis cells. (A-C) The top 10 enriched cellular components (CC), biological processes (BP), and KEGG pathways in ascending order of -log 10 (FDR p-value). The enriched CC contained nucleoplasm, nuclear parts, and nuclear lumen. The enriched BPs included responses to hypoxia and decreased oxygen levels. The enriched KEGG pathways included the HIF-1 signaling pathway. The gene expression of SRA1 and ARID1A was validated by RT-PCR (D, E). The EMosis cells treated with 5 μM TPEN for 4 h demonstrated significantly increased SRA1 mRNA expression and decreased ARID1A mRNA expression compared to the cells without TPEN (all P<0.05). KEGG: Kyoto Encyclopedia of Genes and Genomes; EMosis cells: endometrial epithelial cells; FDR: false discovery rate; HIF-1: hypoxia-inducible factor-1; SRA1: steroid receptor RNA activator 1; ARID1A: AT-rich interaction domain 1A; RT-PCR: real-time polymerase chain reaction.
Table 3.
Activated Cellular Component in EMosis-CC/TERT1 cells treated with 5 μM TPEN
| Enrichment FDR | Genes in list | Functional Category | Genes |
|---|---|---|---|
| 6.44E-10 | 140 | Nucleoplasm | AFF4 EED ZBTB25 FBL KDM4C MYCT1 KDM6B MED10 SAP130 KANSL1L TRMT10A POLR2H SAP30 JUNB HDAC3 JUN HEXIM1 ZNF407 KDM7A BIRC3 ATP6V0A1 USP28 RRM2B MXD1 CTDP1 JMJD6 P4HA2 PIAS2 ALKBH5 SNAP23 BTAF1 RBFOX2 TRMU RNMT POLI NXT2 USP11 KLF5 HCFC1R1 IMPAD1 UBXN8 BNIP3L ZNF175 HBP1 PLEKHA8 SHB ACBD5 PTGES3 KDM3A ABCB6 CACYBP GADD45A HSPH1 ATF1 NQO2 OARD1 STK35 ZBTB1 THOC6 SLX1A USPL1 BHLHE40 HILPDA TXNL4B EIF4A3 CTSK PIP5K1A EAF1 FEM1C TNIP1 RBAK POMZP3 NPAT IPMK LSM11 FBXO32 EMSY RUNX1 CHTOP RYBP YEATS2 PCF11 RAB8B MAPK7 POLR3D ING2 UBE2V2 MAP3K2 PFKFB3 MLLT3 NMNAT1 CSTF3 SERTAD2 SIAH2 C11ORF54 SP1 STK40 POM121 LONP1 XRCC2 ADARB1 FANK1 HSPA1B HSPA1A PPP1R10 SRA1 ZNF432 ETV3 GZF1 THAP1 ASCC1 CIART PPID BNIP3 ARL6IP4 SNAPC1 NEDD4L RORA HSP90AA1 TRMT6 NR4A3 NR4A1 SNAI1 UPF3B IPPK EGLN3 DNAJB1 USP37 CENPO ANP32A ESCO1 SMYD2 METTL21A NOCT ATF3 DDIT3 IMP3 MAFF TRIM33 PPME1 |
| 4.06E-09 | 164 | Nuclear part | SNAPC1 SPAG4 AFF4 EED ZBTB25 NXT2 BNIP3L FBL KDM4C MYCT1 UPF3B PRKRIP1 KDM6B MED10 SAP130 CENPO NUP58 TXNL4B KANSL1L TRMT10A LSM11 POLR2H SAP30 POLR3D JUNB HDAC3 HSPA6 DDIT3 BNIP3 JUN IMP3 SPTY2D1 HEXIM1 POM121 ADARB1 SMG5 HSPA1B HSPA1A ZNF407 POM121C KDM7A BIRC3 ATP6V0A1 USP28 RRM2B MXD1 CTDP1 JMJD6 P4HA2 PIAS2 ALKBH5 SNAP23 BTAF1 SCD RBFOX2 TRMU RNMT POLI USP11 KLF5 HCFC1R1 IMPAD1 UBXN8 ZNF175 HBP1 PLEKHA8 SHB ACBD5 PTGES3 PARP11 KDM3A ABCB6 CACYBP GADD45A MXI1 HSPH1 ATF1 NR4A1 NQO2 OARD1 GZF1 STK35 ZBTB1 THOC6 THAP1 SLX1A WBP2 USPL1 BHLHE40 HILPDA DNAJB2 MFAP1 SLC16A3 EIF4A3 CTSK PIP5K1A EAF1 FEM1C TNIP1 RBAK POMZP3 NPAT IPMK PDK1 FBXO32 EMSY RUNX1 CHTOP ATF3 RYBP YEATS2 PCF11 RAB8B MAPK7 ING2 UBE2V2 ZEB2 MAP3K2 PFKFB3 PPID MLLT3 NMNAT1 CSTF3 SERTAD2 SIAH2 C11ORF54 DDX41 SP1 STK40 LONP1 XRCC2 FANK1 PPP1R10 SRA1 ZNF432 FAM156A GMCL1 RGS2 ETV3 HSPA2 DNAJB9 DNAJB1 ASCC1 AGPAT5 CIART ERN1 ARL6IP4 IPPK NEDD4L RORA HSP90AA1 TRMT6 NR4A3 SNAI1 EGLN3 USP37 ANP32A ESCO1 SMYD2 METTL21A NOCT MAFF TRIM33 PPME1 |
| 1.78E-08 | 151 | Nuclear lumen | AFF4 EED ZBTB25 FBL KDM4C MYCT1 UPF3B PRKRIP1 KDM6B MED10 SAP130 CENPO KANSL1L TRMT10A POLR2H SAP30 JUNB HDAC3 JUN IMP3 SPTY2D1 HEXIM1 ADARB1 ZNF407 KDM7A BIRC3 SNAPC1 ATP6V0A1 USP28 RRM2B MXD1 CTDP1 JMJD6 P4HA2 PIAS2 ALKBH5 SNAP23 BTAF1 SCD RBFOX2 TRMU RNMT POLI NXT2 USP11 KLF5 HCFC1R1 IMPAD1 UBXN8 BNIP3L ZNF175 HBP1 PLEKHA8 SHB ACBD5 PTGES3 KDM3A ABCB6 CACYBP GADD45A MXI1 HSPH1 ATF1 NQO2 OARD1 GZF1 STK35 ZBTB1 THOC6 THAP1 SLX1A WBP2 USPL1 BHLHE40 HILPDA TXNL4B EIF4A3 CTSK PIP5K1A EAF1 FEM1C TNIP1 RBAK POMZP3 NPAT IPMK PDK1 LSM11 FBXO32 EMSY RUNX1 CHTOP ATF3 RYBP YEATS2 PCF11 RAB8B MAPK7 POLR3D ING2 UBE2V2 ZEB2 MAP3K2 PFKFB3 PPID MLLT3 NMNAT1 CSTF3 SERTAD2 SIAH2 C11ORF54 SP1 STK40 POM121 LONP1 XRCC2 FANK1 HSPA1B HSPA1A PPP1R10 SRA1 ZNF432 GMCL1 RGS2 ETV3 HSPA2 DNAJB9 DNAJB1 ASCC1 CIART BNIP3 ARL6IP4 IPPK NEDD4L RORA HSP90AA1 TRMT6 NR4A3 NR4A1 SNAI1 EGLN3 USP37 ANP32A ESCO1 SMYD2 METTL21A NOCT DDIT3 MAFF TRIM33 PPME1 |
| 0.00090047 | 49 | Nucleoplasm part | AFF4 EED FBL KDM4C KDM6B MED10 SAP130 KANSL1L POLR2H SAP30 HDAC3 ZNF407 ATP6V0A1 USP28 PIAS2 ALKBH5 POLI IMPAD1 BNIP3L HBP1 GADD45A STK35 ZBTB1 THOC6 USPL1 BHLHE40 PIP5K1A EAF1 NPAT LSM11 CHTOP YEATS2 RAB8B MAPK7 POLR3D ING2 MLLT3 NMNAT1 C11ORF54 HSPA1B HSPA1A PPP1R10 ETV3 THAP1 ASCC1 EIF4A3 CIART ARL6IP4 PCF11 |
| 0.006030162 | 3 | Transcription factor AP-1 complex | JUNB JUN DDIT3 |
| 0.018260045 | 34 | Nuclear body | FBL ATP6V0A1 USP28 PIAS2 ALKBH5 POLI IMPAD1 BNIP3L HBP1 GADD45A STK35 ZBTB1 THOC6 USPL1 BHLHE40 SAP130 PIP5K1A EAF1 NPAT LSM11 CHTOP RAB8B MAPK7 POLR3D NMNAT1 C11ORF54 HSPA1B HSPA1A PPP1R10 THAP1 ASCC1 EIF4A3 CIART ARL6IP4 |
| 0.021715976 | 2 | CHOP-ATF3 complex | DDIT3 ATF3 |
| 0.029594035 | 4 | Integral component of mitochondrial outer membrane | MGARP FUNDC2 ABCB6 BNIP3 |
| 0.03197158 | 4 | Intrinsic component of mitochondrial outer membrane | MGARP FUNDC2 ABCB6 BNIP3 |
| 0.044981646 | 2 | SREBP-SCAP-Insig complex | INSIG2 INSIG1 |
Table 4.
Activated Biological Process in EMosis-CC/TERT1 celsl treated with 5 μM TPEN
| Enrichment FDR | Genes in list | Functional Category | Genes |
|---|---|---|---|
| 1.02E-10 | 100 | Cellular response to stress | USP28 ATG5 HSP90AA1 PPP1R15A HSPA2 EDA2R DNAJB2 GABARAPL1 RRAGA ANKZF1 MAP2K1 UBE2V2 MAP3K2 HSPA6 DDIT3 ERN1 HSPA1B HSPA1A LONP1 HERPUD1 UBA5 DNAJA1 TGFB2 GADD45B PGK1 VEGFA FAM162A IL1A GADD45A TNFRSF10B INSIG2 DNAJB1 WDR45B BAG3 ATF3 RELL2 HDAC3 EIF2AK3 ACER2 INSIG1 RRM2B PTGS2 PIK3C3 NFE2L1 ADNP2 POLI UBXN8 CCDC47 CHORDC1 STC2 ID2 ERRFI1 SLC2A1 NR4A3 ZBTB1 DNAJB9 EGLN3 SLX1A KDM6B EDEM1 ASCC1 PMAIP1 ADM KLF10 EMSY STC1 DDIT4 ING2 ZEB2 PRKCE FZD4 BNIP3 JUN XRCC2 ERO1A NDRG1 HILPDA BNIP3L PPP1R10 RORA SNAI1 OARD1 TMX1 SMYD2 PDK1 HK2 MAPK7 PJA2 MGARP FBXW11 ALKBH5 FKBP14 PTGES3 HSPH1 EGLN1 NUP58 CNOT8 POLR2H POM121 POM121C |
| 2.75E-09 | 24 | Response to unfolded protein | HSPA2 HSPA6 ERN1 HSPA1B HSPA1A EIF2AK3 DDIT3 HERPUD1 STC2 DNAJB9 EDEM1 ATF3 ERO1A DNAJB5 BAG3 HSPA4L HSP90AA1 DNAJA1 PPP1R15A FKBP14 HSPE1 HSPH1 DNAJB1 DNAJB2 |
| 9.63E-09 | 25 | Response to topologically incorrect protein | HSPA2 ANKZF1 HSPA6 ERN1 HSPA1B HSPA1A EIF2AK3 DDIT3 HERPUD1 STC2 DNAJB9 EDEM1 ATF3 ERO1A DNAJB5 BAG3 HSPA4L HSP90AA1 DNAJA1 PPP1R15A FKBP14 HSPE1 HSPH1 DNAJB1 DNAJB2 |
| 1.99E-07 | 63 | Response to abiotic stimulus | CLCN6 OPN3 HSP90AA1 VEGFA HSPA2 JUNB HSPA6 JUN HSPA1B HSPA1A USP28 ALKBH5 PGK1 FAM162A IL1A EGLN1 BAG3 DDIT4 PTGS2 NFE2L1 DDHD2 DNAJA1 CHORDC1 BIRC2 STC2 ID2 ERRFI1 SLC2A1 BHLHE40 DNAJA4 ADM STC1 HK2 LZIC ANGPTL4 PRKCE BNIP3 LONP1 XRCC2 ERO1A NDRG1 GADD45A TNFRSF10B EGLN3 HILPDA PMAIP1 BCL10 PLOD2 MAP3K2 BNIP3L RORA TGFB2 ZBTB1 PDK1 PPID EIF2AK3 MGARP PTGES3 HSPH1 DNAJB1 NUP58 POM121 POM121C |
| 7.84E-07 | 13 | Chaperone-mediated protein folding | HSPE1 HSPA2 DNAJB1 DNAJB2 DNAJB5 HSPA6 HSPA1B HSPA1A PTGES3 PPID ERO1A CHORDC1 HSPH1 |
| 2.16E-06 | 28 | Response to hypoxia | VEGFA ALKBH5 PGK1 FAM162A EGLN1 DDIT4 BIRC2 STC2 SLC2A1 ADM STC1 HK2 ANGPTL4 PRKCE BNIP3 LONP1 ERO1A PTGS2 NDRG1 EGLN3 HILPDA PMAIP1 PLOD2 BNIP3L RORA TGFB2 PDK1 MGARP |
| 2.37E-06 | 150 | Heterocycle biosynthetic process | SNAPC1 RRM2B CTDP1 GPBP1 PFKP EED NFE2L1 PGK1 KLF5 PTGES3 AMPD2 ETV3 KLF7 DNAJB1 MED10 SAP130 DNAJB5 ING1 KLF10 CIART HK2 SAP30 PCF11 ING2 JUNB HDAC3 DCAKD NMNAT1 JUN SPTY2D1 SP1 HEXIM1 SMG5 ID2 ZNF395 MXD1 RORA JMJD6 HSP90AA1 TGFB2 CREM RBFOX2 ZNF175 KDM4C VEGFA ABCB6 NR4A3 MXI1 BHLHE41 ATF1 NR4A1 SNAI1 INSIG2 GZF1 ZBTB1 EDA2R THAP1 BHLHE40 EGLN1 ASCC1 BCL10 NPAT RUNX1 AK4 ATF3 YEATS2 POLR2H ZEB2 MAP3K2 MLLT3 FZD4 DDIT3 MAFF INSIG1 FAM83G TRIM33 HSPA1A AK2 KDM7A ALDH18A1 AFF4 FBXW11 ZBTB25 UPRT ADNP2 POLI HBP1 ALDOC BIRC2 LOX IL1A KDM3A PANK2 PAICS PPAT ZNF331 WBP2 KDM6B MICAL2 FGF7 VGLL4 EAF1 RBAK ZNF41 VLDLR PDK1 AGPAT5 CNOT8 EMSY RYBP DDIT4 POLR3D PDE7B SERTAD2 SIAH2 DDX41 ZFP69B IL1RAP SERTAD1 SRA1 ZNF407 ZNF432 MAPK7 IPPK USPL1 SMYD2 BAG3 MAP2K1 PPID EIF2AK3 FANK1 HSPA1B RLF TNIP1 PIAS2 BTAF1 LDHA NEDD4L PTGS2 SCD RNMT PFKFB4 EGLN3 NUP58 NOCT LSM11 PFKFB3 CSTF3 POM121 POM121C |
| 2.37E-06 | 10 | Chaperone cofactor-dependent protein refolding | HSPE1 HSPA2 DNAJB1 DNAJB5 HSPA6 HSPA1B HSPA1A PTGES3 ERO1A HSPH1 |
| 2.44E-06 | 148 | Nucleobase-containing compound biosynthetic process | SNAPC1 RRM2B CTDP1 GPBP1 PFKP EED NFE2L1 PGK1 KLF5 PTGES3 AMPD2 ETV3 KLF7 DNAJB1 MED10 SAP130 DNAJB5 ING1 KLF10 CIART HK2 SAP30 PCF11 ING2 JUNB HDAC3 DCAKD NMNAT1 JUN SPTY2D1 SP1 HEXIM1 SMG5 ID2 ZNF395 MXD1 RORA JMJD6 HSP90AA1 TGFB2 CREM RBFOX2 ZNF175 KDM4C VEGFA NR4A3 MXI1 BHLHE41 ATF1 NR4A1 SNAI1 INSIG2 GZF1 ZBTB1 EDA2R THAP1 BHLHE40 EGLN1 ASCC1 BCL10 NPAT RUNX1 AK4 ATF3 YEATS2 POLR2H ZEB2 MAP3K2 MLLT3 FZD4 DDIT3 MAFF INSIG1 FAM83G TRIM33 HSPA1A AK2 KDM7A AFF4 FBXW11 ZBTB25 UPRT ADNP2 POLI HBP1 ALDOC BIRC2 LOX IL1A KDM3A PANK2 PAICS PPAT ZNF331 WBP2 KDM6B MICAL2 FGF7 VGLL4 EAF1 RBAK ZNF41 VLDLR PDK1 AGPAT5 CNOT8 EMSY RYBP DDIT4 POLR3D PDE7B SERTAD2 SIAH2 DDX41 ZFP69B IL1RAP SERTAD1 SRA1 ZNF407 ZNF432 MAPK7 IPPK USPL1 SMYD2 BAG3 MAP2K1 PPID EIF2AK3 FANK1 HSPA1B RLF TNIP1 PIAS2 BTAF1 LDHA NEDD4L PTGS2 SCD RNMT PFKFB4 EGLN3 NUP58 NOCT LSM11 PFKFB3 CSTF3 POM121 POM121C |
| 2.44E-06 | 28 | Response to decreased oxygen levels | VEGFA ALKBH5 PGK1 FAM162A EGLN1 DDIT4 BIRC2 STC2 SLC2A1 ADM STC1 HK2 ANGPTL4 PRKCE BNIP3 LONP1 ERO1A PTGS2 NDRG1 EGLN3 HILPDA PMAIP1 PLOD2 BNIP3L RORA TGFB2 PDK1 MGARP |
| 3.14E-06 | 147 | Macromolecule modification | UBE3C UBA6 USP28 NEDD4L ATG5 CTDP1 TRIB2 P4HA2 FBXW11 PIK3C3 UBA5 PPP1R15A ALKBH5 TGFB2 GADD45B TRMU ATG4A FBL VEGFA LOX KDM3A GADD45A P4HA1 PRKRIP1 EDA2R USP37 CCNG2 ESCO1 WDR45B RNF217 NGLY1 PLOD2 PIGA MAP2K1 UBE2V2 MAP3K2 B3GNT2 PRKCE HDAC3 HEXIM1 ADARB1 DZIP3 PJA2 PPME1 KDM7A BIRC3 JMJD6 PIAS2 HSP90AA1 DNAJA1 TRMT6 VCPKMT USP11 KDM4C BIRC2 PARP11 ERRFI1 PPP4R4 OARD1 EGLN3 USPL1 EGLN1 FGF7 BCL10 SMYD2 METTL21A TRMT10A TNIP1 FBXO32 KLHL21 RYBP YEATS2 RELL2 PPID EIF2AK3 RAB6A ERN1 TRIM33 HSPA1B HSPA1A SCYL3 HERPUD1 OPN3 PTGS2 EED NFE2L1 RNF24 METTL4 RNMT RIOK3 ZC3HAV1 FKBP14 WSB1 CHORDC1 PTP4A1 HBEGF CNPPD1 RGS2 RLF PPP1R3C NQO2 STK35 HSPA2 KDM6B MED10 EDEM1 HERC3 ARRDC4 KANSL1L FEM1C VLDLR PDK1 ATF3 SAP30 ABCA1 MAPK7 ING2 ZEB2 USP32 PPP1R3B NMNAT1 FZD4 JUN SIAH2 STK40 SLC39A10 FUT11 PPP1R10 KCTD11 WBP2 DNAJB2 CHTOP METTL2B ACER2 ARRDC3 METTL2A DDIT4 TNFRSF10B STC2 ASCC1 NUP58 RRAGA MCFD2 POM121 SERTAD1 ERO1A POM121C |
| 3.30E-06 | 149 | Aromatic compound biosynthetic process | SNAPC1 RRM2B CTDP1 GPBP1 PFKP EED NFE2L1 PGK1 KLF5 PTGES3 AMPD2 ETV3 KLF7 DNAJB1 MED10 SAP130 DNAJB5 ING1 KLF10 CIART HK2 SAP30 PCF11 ING2 JUNB HDAC3 DCAKD NMNAT1 JUN SPTY2D1 SP1 HEXIM1 SMG5 ID2 ZEB2 ZNF395 MXD1 RORA JMJD6 HSP90AA1 TGFB2 CREM RBFOX2 ZNF175 KDM4C VEGFA ABCB6 NR4A3 MXI1 BHLHE41 ATF1 NR4A1 SNAI1 INSIG2 GZF1 ZBTB1 EDA2R THAP1 BHLHE40 EGLN1 ASCC1 BCL10 NPAT RUNX1 AK4 ATF3 YEATS2 POLR2H MAP3K2 MLLT3 FZD4 DDIT3 MAFF INSIG1 FAM83G TRIM33 HSPA1A AK2 KDM7A AFF4 FBXW11 ZBTB25 UPRT ADNP2 POLI HBP1 ALDOC BIRC2 LOX IL1A KDM3A PANK2 PAICS PPAT ZNF331 WBP2 KDM6B MICAL2 FGF7 VGLL4 EAF1 RBAK ZNF41 VLDLR PDK1 AGPAT5 CNOT8 EMSY RYBP DDIT4 POLR3D PDE7B SERTAD2 SIAH2 DDX41 ZFP69B IL1RAP SERTAD1 SRA1 ZNF407 ZNF432 MAPK7 IPPK USPL1 SMYD2 BAG3 MAP2K1 PPID EIF2AK3 FANK1 HSPA1B RLF TNIP1 PIAS2 BTAF1 LDHA NEDD4L PTGS2 SCD RNMT PFKFB4 EGLN3 NUP58 NOCT LSM11 PFKFB3 CSTF3 POM121 POM121C |
| 4.39E-06 | 152 | Organic cyclic compound biosynthetic process | SNAPC1 RRM2B CTDP1 GPBP1 PFKP EED NFE2L1 PGK1 KLF5 PTGES3 AMPD2 ETV3 KLF7 DNAJB1 MED10 SAP130 DNAJB5 ING1 KLF10 CIART HK2 SAP30 PCF11 ING2 JUNB HDAC3 DCAKD NMNAT1 JUN SPTY2D1 SP1 HEXIM1 SMG5 ID2 ZEB2 ZNF395 MXD1 RORA JMJD6 HSP90AA1 TGFB2 CREM RBFOX2 ZNF175 KDM4C VEGFA ABCB6 NR4A3 MXI1 BHLHE41 ATF1 NR4A1 SNAI1 INSIG2 GZF1 ZBTB1 EDA2R THAP1 BHLHE40 EGLN1 ASCC1 BCL10 NPAT RUNX1 AK4 ATF3 YEATS2 POLR2H MAP3K2 MLLT3 FZD4 DDIT3 MAFF INSIG1 FAM83G TRIM33 HSPA1A AK2 KDM7A ALDH18A1 AFF4 FBXW11 ZBTB25 UPRT ADNP2 POLI HBP1 ALDOC BIRC2 LOX IL1A KDM3A PANK2 PAICS PPAT ZNF331 WBP2 KDM6B MICAL2 FGF7 VGLL4 EAF1 RBAK ZNF41 VLDLR PDK1 AGPAT5 CNOT8 EMSY RYBP DDIT4 POLR3D PDE7B SERTAD2 SIAH2 ACBD3 DDX41 ZFP69B IL1RAP SERTAD1 SRA1 ZNF407 ZNF432 MAPK7 IPPK USPL1 SMYD2 BAG3 MAP2K1 PPID EIF2AK3 FANK1 HSPA1B RLF TNIP1 PIAS2 BTAF1 LDHA NEDD4L PTGS2 SCD RNMT PFKFB4 EGLN3 NUP58 ADM NOCT LSM11 PFKFB3 CSTF3 POM121 POM121C |
| 5.36E-06 | 10 | De novo posttranslational protein folding | HSPE1 HSPA2 DNAJB1 DNAJB5 HSPA6 HSPA1B HSPA1A PTGES3 ERO1A HSPH1 |
| 6.00E-06 | 28 | Response to oxygen levels | VEGFA ALKBH5 PGK1 FAM162A EGLN1 DDIT4 BIRC2 STC2 SLC2A1 ADM STC1 HK2 ANGPTL4 PRKCE BNIP3 LONP1 ERO1A PTGS2 NDRG1 EGLN3 HILPDA PMAIP1 PLOD2 BNIP3L RORA TGFB2 PDK1 MGARP |
| 7.52E-06 | 21 | Protein folding | DNAJC25 PTGES3 HSPE1 HSPA2 DNAJB1 DNAJB2 DNAJB5 PPID HSPA6 ERO1A HSPA1B HSPA1A DNAJA4 HSP90AA1 DNAJA1 GRPEL1 CHORDC1 HSPH1 HSPA4L BAG3 NGLY1 |
| 7.91E-06 | 42 | Regulation of cellular response to stress | EDA2R DDIT3 HERPUD1 DNAJA1 PPP1R15A TGFB2 GADD45B VEGFA GADD45A RELL2 ERN1 INSIG1 HSPA1A PTGS2 NFE2L1 CHORDC1 NR4A3 EDEM1 ING2 MAP2K1 UBE2V2 ZEB2 FZD4 PPP1R10 SNAI1 PMAIP1 SMYD2 MAPK7 HDAC3 EIF2AK3 PJA2 DNAJB9 HSP90AA1 PTGES3 HSPH1 DNAJB1 NUP58 BAG3 MAP3K2 POM121 HSPA1B POM121C |
| 9.77E-06 | 10 | De novo protein folding | HSPE1 HSPA2 DNAJB1 DNAJB5 HSPA6 HSPA1B HSPA1A PTGES3 ERO1A HSPH1 |
| 1.40E-05 | 138 | Cellular protein modification process | UBE3C UBA6 USP28 NEDD4L ATG5 CTDP1 TRIB2 P4HA2 FBXW11 PIK3C3 UBA5 PPP1R15A TGFB2 GADD45B ATG4A FBL VEGFA LOX KDM3A GADD45A P4HA1 PRKRIP1 EDA2R USP37 CCNG2 ESCO1 WDR45B RNF217 NGLY1 PLOD2 PIGA MAP2K1 UBE2V2 MAP3K2 B3GNT2 PRKCE HDAC3 HEXIM1 DZIP3 PJA2 PPME1 KDM7A BIRC3 JMJD6 PIAS2 HSP90AA1 DNAJA1 VCPKMT USP11 KDM4C BIRC2 PARP11 ERRFI1 PPP4R4 OARD1 EGLN3 USPL1 EGLN1 FGF7 BCL10 SMYD2 METTL21A TNIP1 FBXO32 KLHL21 RYBP YEATS2 RELL2 PPID EIF2AK3 RAB6A ERN1 TRIM33 ADARB1 HSPA1B HSPA1A SCYL3 HERPUD1 OPN3 PTGS2 EED NFE2L1 RNF24 RIOK3 ZC3HAV1 FKBP14 WSB1 CHORDC1 PTP4A1 HBEGF CNPPD1 RGS2 RLF PPP1R3C NQO2 STK35 HSPA2 KDM6B MED10 EDEM1 HERC3 ARRDC4 KANSL1L FEM1C VLDLR PDK1 ATF3 SAP30 ABCA1 MAPK7 ING2 ZEB2 USP32 PPP1R3B NMNAT1 FZD4 JUN SIAH2 STK40 SLC39A10 FUT11 PPP1R10 KCTD11 WBP2 DNAJB2 CHTOP ACER2 ARRDC3 DDIT4 TNFRSF10B STC2 NUP58 RRAGA MCFD2 POM121 SERTAD1 ERO1A POM121C |
| 1.40E-05 | 138 | Protein modification process | UBE3C UBA6 USP28 NEDD4L ATG5 CTDP1 TRIB2 P4HA2 FBXW11 PIK3C3 UBA5 PPP1R15A TGFB2 GADD45B ATG4A FBL VEGFA LOX KDM3A GADD45A P4HA1 PRKRIP1 EDA2R USP37 CCNG2 ESCO1 WDR45B RNF217 NGLY1 PLOD2 PIGA MAP2K1 UBE2V2 MAP3K2 B3GNT2 PRKCE HDAC3 HEXIM1 DZIP3 PJA2 PPME1 KDM7A BIRC3 JMJD6 PIAS2 HSP90AA1 DNAJA1 VCPKMT USP11 KDM4C BIRC2 PARP11 ERRFI1 PPP4R4 OARD1 EGLN3 USPL1 EGLN1 FGF7 BCL10 SMYD2 METTL21A TNIP1 FBXO32 KLHL21 RYBP YEATS2 RELL2 PPID EIF2AK3 RAB6A ERN1 TRIM33 ADARB1 HSPA1B HSPA1A SCYL3 HERPUD1 OPN3 PTGS2 EED NFE2L1 RNF24 RIOK3 ZC3HAV1 FKBP14 WSB1 CHORDC1 PTP4A1 HBEGF CNPPD1 RGS2 RLF PPP1R3C NQO2 STK35 HSPA2 KDM6B MED10 EDEM1 HERC3 ARRDC4 KANSL1L FEM1C VLDLR PDK1 ATF3 SAP30 ABCA1 MAPK7 ING2 ZEB2 USP32 PPP1R3B NMNAT1 FZD4 JUN SIAH2 STK40 SLC39A10 FUT11 PPP1R10 KCTD11 WBP2 DNAJB2 CHTOP ACER2 ARRDC3 DDIT4 TNFRSF10B STC2 NUP58 RRAGA MCFD2 POM121 SERTAD1 ERO1A POM121C |
| 1.65E-05 | 122 | Positive regulation of metabolic process | NEDD4L TRIB2 NFE2L1 PPP1R15A TGFB2 GADD45B PGK1 VEGFA GADD45A UPF3B EDA2R MED10 DNAJB2 RNF217 ING1 ING2 MAP2K1 MAP3K2 JUNB HDAC3 JUN HSPA1B HSPA1A ID2 ZEB2 BIRC3 HERPUD1 RORA JMJD6 TFRC HSP90AA1 KLF5 ZNF175 ZC3HAV1 BIRC2 PTGES3 FAM162A IL1A KLF7 NR4A3 ATF1 NR4A1 INSIG2 FGF7 PMAIP1 BCL10 TNIP1 NPAT KLF10 RUNX1 ATF3 RELL2 MLLT3 SNX33 FZD4 DDIT3 ERN1 MAFF SP1 INSIG1 KDM7A GPBP1 FBXW11 PTGS2 EED CREM KDM4C HBEGF KDM3A CACYBP SNAI1 NQO2 HSPA2 WBP2 KDM6B MICAL2 EDEM1 EGLN1 VLDLR NOCT RYBP IL7R UBE2V2 PRKCE NMNAT1 BNIP3 ZFAND2A SERTAD2 DDX41 SECISBP2 SLC39A10 SERTAD1 SRA1 EGLN3 BNIP3L MAPK7 IPPK EIF4A3 BAG3 CNOT8 HK2 CHTOP EIF2AK3 ACER2 PJA2 FANK1 PPP1R10 ARRDC3 ARRDC4 SCD RLF SLX1A ACTC1 PIAS2 HSPE1 TNFRSF10B PFKFB4 HSPH1 ADM POLR2H PIGA PFKFB3 |
| 2.36E-05 | 16 | Cellular response to unfolded protein | HSPA2 HSPA6 ERN1 HSPA1B HSPA1A EIF2AK3 HERPUD1 STC2 ATF3 DDIT3 ERO1A BAG3 DNAJB9 PPP1R15A FKBP14 EDEM1 |
| 2.56E-05 | 21 | Response to temperature stimulus | HSP90AA1 HSPA2 HSPA6 HSPA1B HSPA1A IL1A BAG3 PTGS2 NFE2L1 DNAJA1 CHORDC1 DNAJA4 ADM EIF2AK3 PTGES3 HSPH1 DNAJB1 NUP58 POM121 ERO1A POM121C |
| 3.94E-05 | 22 | Response to endoplasmic reticulum stress | PPP1R15A DNAJB2 ANKZF1 DDIT3 ERN1 HERPUD1 UBA5 TNFRSF10B ATF3 EIF2AK3 HSPA1A NFE2L1 UBXN8 CCDC47 STC2 DNAJB9 EDEM1 ERO1A TMX1 JUN FKBP14 PMAIP1 |
| 5.14E-05 | 17 | Cellular response to topologically incorrect protein | HSPA2 ANKZF1 HSPA6 ERN1 HSPA1B HSPA1A EIF2AK3 HERPUD1 STC2 ATF3 DDIT3 ERO1A BAG3 DNAJB9 PPP1R15A FKBP14 EDEM1 |
| 5.68E-05 | 112 | Positive regulation of cellular metabolic process | TRIB2 NFE2L1 PPP1R15A TGFB2 GADD45B PGK1 VEGFA GADD45A UPF3B EDA2R MED10 DNAJB2 RNF217 ING1 ING2 MAP2K1 MAP3K2 JUNB HDAC3 JUN HSPA1B HSPA1A ID2 ZEB2 BIRC3 HERPUD1 RORA JMJD6 TFRC HSP90AA1 KLF5 ZNF175 ZC3HAV1 BIRC2 PTGES3 FAM162A KLF7 NR4A3 ATF1 NR4A1 INSIG2 FGF7 PMAIP1 BCL10 TNIP1 NPAT KLF10 RUNX1 ATF3 RELL2 MLLT3 SNX33 FZD4 DDIT3 ERN1 MAFF SP1 INSIG1 KDM7A GPBP1 FBXW11 PTGS2 EED CREM HBEGF IL1A KDM3A CACYBP SNAI1 NQO2 HSPA2 WBP2 KDM6B MICAL2 EDEM1 EGLN1 VLDLR RYBP UBE2V2 PRKCE NMNAT1 BNIP3 ZFAND2A SERTAD2 DDX41 SLC39A10 SERTAD1 SRA1 EGLN3 BNIP3L MAPK7 IPPK EIF4A3 BAG3 CNOT8 HK2 CHTOP EIF2AK3 ACER2 PJA2 FANK1 PPP1R10 ARRDC3 ARRDC4 RLF SLX1A PIAS2 HSPE1 TNFRSF10B PFKFB4 ADM PFKFB3 |
| 6.64E-05 | 17 | Ribonucleoside monophosphate biosynthetic process | PFKP PGK1 AMPD2 HK2 AK2 UPRT ALDOC PAICS PPAT AK4 DDIT4 LDHA PFKFB4 NUP58 PFKFB3 POM121 POM121C |
| 6.64E-05 | 112 | Positive regulation of macromolecule metabolic process | NEDD4L TRIB2 NFE2L1 PPP1R15A TGFB2 GADD45B VEGFA GADD45A UPF3B EDA2R MED10 DNAJB2 RNF217 ING1 ING2 MAP2K1 MAP3K2 JUNB HDAC3 JUN HSPA1B HSPA1A ID2 BIRC3 HERPUD1 RORA JMJD6 TFRC HSP90AA1 KLF5 ZNF175 ZC3HAV1 BIRC2 PTGES3 FAM162A IL1A KLF7 NR4A3 ATF1 NR4A1 INSIG2 FGF7 PMAIP1 BCL10 TNIP1 NPAT KLF10 RUNX1 ATF3 RELL2 MLLT3 SNX33 FZD4 DDIT3 ERN1 MAFF SP1 INSIG1 KDM7A GPBP1 FBXW11 PTGS2 EED CREM KDM4C HBEGF KDM3A CACYBP SNAI1 NQO2 HSPA2 WBP2 KDM6B MICAL2 EDEM1 EGLN1 VLDLR NOCT RYBP IL7R UBE2V2 ZEB2 PRKCE NMNAT1 ZFAND2A SERTAD2 DDX41 SECISBP2 SLC39A10 SERTAD1 SRA1 EGLN3 MAPK7 IPPK EIF4A3 CNOT8 CHTOP EIF2AK3 ACER2 PJA2 FANK1 PPP1R10 ARRDC3 ARRDC4 RLF SLX1A ACTC1 PIAS2 HSPE1 TNFRSF10B HSPH1 POLR2H |
| 6.97E-05 | 15 | Pyruvate metabolic process | PFKP PGK1 HK2 ALDOC NR4A3 PDK1 DDIT4 LDHA PFKFB4 NUP58 SLC16A3 SLC16A1 PFKFB3 POM121 POM121C |
| 6.97E-05 | 136 | Response to stress | USP28 ATG5 RORA HSP90AA1 PPP1R15A BNIP3L VEGFA HSPA2 EDA2R DNAJB2 GABARAPL1 IFIT5 RRAGA ANKZF1 MAP2K1 UBE2V2 MAP3K2 HSPA6 DDIT3 ERN1 HSPA1B HSPA1A LONP1 HERPUD1 UBA5 DNAJA1 ALKBH5 TGFB2 GADD45B PGK1 FAM162A IL1A GADD45A TNFRSF10B INSIG2 DNAJB1 EGLN1 WDR45B PMAIP1 BCL10 BAG3 ATF3 RELL2 DDIT4 POLR3D HDAC3 EIF2AK3 BNIP3 ACER2 INSIG1 HEXIM1 RRM2B PTGS2 PIK3C3 NFE2L1 ATRN ADNP2 POLI RIOK3 KLF5 BLOC1S6 UBXN8 ZNF175 ZC3HAV1 CCDC47 CHORDC1 PANX1 BIRC2 HBEGF LOX STC2 ID2 ERRFI1 SLC2A1 NR4A3 ZBTB1 DNAJB9 EGLN3 SLX1A KDM6B SYT11 EDEM1 ASCC1 RAB20 DNAJA4 TNIP1 ADM KLF10 EMSY STC1 HK2 ANGPTL4 ING2 ZEB2 PRKCE ETFDH NMNAT1 FZD4 JUN DDX41 IL1RAP XRCC2 ADARB1 ERO1A NDRG1 HILPDA DNAJB5 PLOD2 ABCA1 PPP1R10 SNAP23 KANK1 SNAI1 OARD1 TMX1 SMYD2 PDK1 MAPK7 PJA2 MGARP HSPA4L SP1 BIRC3 FBXW11 FKBP14 PTGES3 HSPE1 HSPH1 NUP58 FGF7 CTSK CNOT8 POLR2H MAFF POM121 POM121C |
Table 5.
Activated KEGG pathway in EMosis-CC/TERT1 cells treated with 5 μM TPEN
| Enrichment FDR | Genes in list | Functional Category | Genes |
|---|---|---|---|
| 1.33E-06 | 18 | Protein processing in endoplasmic reticulum | SEC24A HSPH1 DDIT3 ERN1 HSPA4L SEC61G PPP1R15A ERO1A DNAJB2 DNAJA1 HSPA2 HSPA6 HSP90AA1 DNAJB1 NGLY1 EIF2AK3 EDEM1 HERPUD1 |
| 0.000578518 | 11 | HIF-1 signaling pathway | EGLN3 HK2 LDHA PDK1 PFKFB3 PGK1 EGLN1 MAP2K1 SLC2A1 TFRC VEGFA |
| 0.000779509 | 12 | Autophagy | RRAGA ATG4A ERN1 GABARAPL1 PIK3C3 DDIT4 MAP2K1 BNIP3 ATG9A RAB33B EIF2AK3 ATG5 |
| 0.001001379 | 12 | Apoptosis | CTSK GADD45A DDIT3 ERN1 BIRC2 BIRC3 JUN GADD45B PMAIP1 MAP2K1 TUBA4A TNFRSF10B |
| 0.001557942 | 8 | Mitophagy | GABARAPL1 JUN BNIP3 BNIP3L SP1 ATG9A EIF2AK3 ATG5 |
| 0.001557942 | 8 | Central carbon metabolism in cancer | HK2 LDHA PDK1 PFKP MAP2K1 SLC2A1 SLC7A5 SLC16A3 |
| 0.009185506 | 5 | Fructose and mannose metabolism | ALDOC HK2 PFKFB3 PFKFB4 PFKP |
| 0.009185506 | 16 | MAPK signaling pathway | MAP3K2 GADD45A DDIT3 FGF7 FLNC NR4A1 HSPA2 HSPA6 IL1A IL1RAP JUN GADD45B MAPK7 MAP2K1 TGFB2 VEGFA |
| 0.009185506 | 5 | Autophagy | ATG4A GABARAPL1 PIK3C3 ATG9A ATG5 |
| 0.009185506 | 7 | Renal cell carcinoma | EGLN3 JUN EGLN1 MAP2K1 SLC2A1 TGFB2 VEGFA |
| 0.024622491 | 9 | FoxO signaling pathway | FBXO32 GADD45A GABARAPL1 IL7R GADD45B MAP2K1 BNIP3 TGFB2 CCNG2 |
| 0.028306059 | 7 | Colorectal cancer | GADD45A JUN GADD45B PMAIP1 MAP2K1 RALGDS TGFB2 |
| 0.036071076 | 7 | MRNA surveillance pathway | CSTF3 SMG5 PCF11 NXT2 UPF3B RNMT EIF4A3 |
| 0.039177861 | 6 | P53 signaling pathway | GADD45A GADD45B RRM2B PMAIP1 TNFRSF10B CCNG2 |
| 0.039177861 | 4 | Circadian rhythm | FBXW11 RORA BHLHE41 BHLHE40 |
| 0.039177861 | 21 | Pathways in cancer | EGLN3 GADD45A FGF7 GNG4 BIRC2 BIRC3 HSP90AA1 IL7R JUN GADD45B PMAIP1 EGLN1 MAP2K1 PTGS2 RALGDS SLC2A1 SP1 TGFB2 VEGFA FZD4 RUNX1 |
| 0.045146468 | 6 | Pancreatic cancer | GADD45A GADD45B MAP2K1 RALGDS TGFB2 VEGFA |
| 0.045513973 | 8 | Purine metabolism | PAICS AK2 AK4 AMPD2 PDE7B RRM2B PPAT ADPRM |
| 0.045513973 | 4 | SNARE interactions in vesicular transport | YKT6 BET1L VAMP1 SNAP23 |
| 0.045513973 | 10 | Transcriptional misregulation in cancer | GADD45A DDIT3 BIRC3 ID2 MLLT3 GADD45B ATF1 SP1 NR4A3 RUNX1 |
SRA1 expression was induced and ARID1A expression was suppressed by zinc deficiency
It is known that endometriosis is an estrogen-dependent disease, and nuclear receptors may contribute to its development [35]. Thus, we extracted DEGs using the GO molecular function of the nuclear receptor. Among the upregulated and downregulated DEGs, three upregulated and 11 downregulated genes were identified (Table 6). Previous studies have demonstrated that SRA1 and ARID1A are associated with estrogen [36,37]. SRA1 and ARID1A mRNA expression was validated (Figure 2D, 2E). The EMosis cells treated with 5 μM TPEN for 4 h revealed markedly increased SRA1 and decreased ARID1A mRNA expression compared to the untreated cells.
Table 6.
Upregulated and downregulated genes for the GO molecular function term “nuclear receptor”
| Genes | |
|---|---|
| Upregulated | NR4A3, NR4A1, SRA1 |
| Downregulated | NR2C1, ARID1A, NSD1, RBM14-RBM4, TRIP4, NCOA6, PNRC2, SMARCE1, BAZ2A, PPRC1, NCOA2 |
GO: Gene Ontology.
ARID1A and SRA1 were associated with SMARCD1 and ATF1
Figure 3A shows the results of the PPI analysis of SRA1 in EMosis cells. ATF1, ATF3, and DDIT3 were extracted from the upregulated DEGs. CREB1, EGR1, DDX5, UBE2Z, NCOA2, GTF2B, and GTF3C5 were extracted from downregulated DEGs. Figure 3B shows the PPI network analysis of ARID1A in the EMosis cells. ING1 was extracted from the upregulated DEGs. BAZ1B, SMARCD1, SMARCE1, and SMAD3 were extracted from the downregulated DEGs in PPI network analysis.
Figure 3.
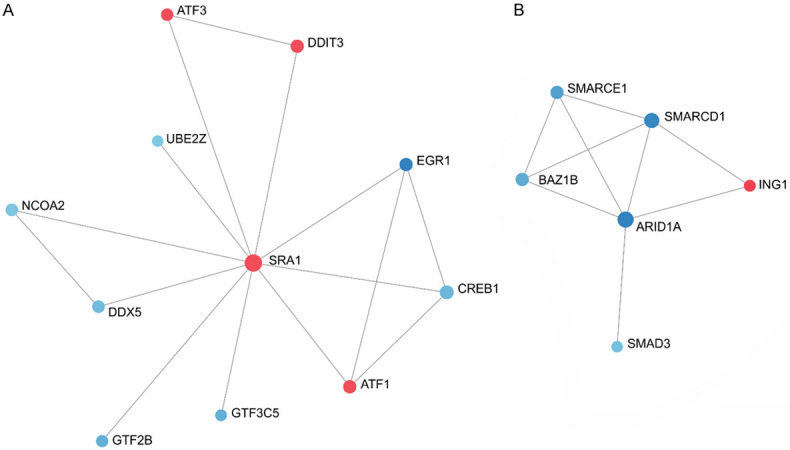
ARID1A and SRA1 were associated with SMARCD1 and ATF1. A, B. The results of the protein-protein network analysis for SRA1 and ARID1A in EMosis cells. ATF1, ATF3, and DDIT3 were extracted from the upregulated DEGs. CREB1, EGR1, DDX5, UBE2Z, NCOA2, GTF2B, and GTF3C5 were extracted from downregulated DEGs. ING1 was extracted from the upregulated DEGs. BAZ1B, SMARCD1, SMARCE1, and SMAD3 were extracted from downregulated DEGs. SRA1, steroid receptor RNA activator 1; ARID1A, AT-rich interaction domain 1A; DEGs, differentially expressed genes.
Zinc affected SRA1 gene expression and ARID1A expression in immortalized endometrial epithelial cells
SRA1 mRNA expression in EMosis cells is shown in Figure 4A. SRA1 mRNA levels increased in response to the 24-h incubation with 5 μM TPEN compared to cells without TPEN (P=0.039, Figure 4A). SRA1 mRNA levels were unchanged in response to 24-h incubation with 5 μM TPEN and 10 μM zinc compared with cells without TPEN (P=0.955, Figure 4A). Figure 4B shows the mRNA expression of ARID1A in the EMosis cells. ARID1A mRNA levels decreased in response to the 24-h incubation with 5 μM TPEN compared to the cells without TPEN (P<0.001, Figure 4B). ARID1A mRNA levels were unchanged in response to the 24-h incubation with 5 μM TPEN and 10 μM zinc compared to the cells without TPEN (P=0.927, Figure 4B).
Figure 4.
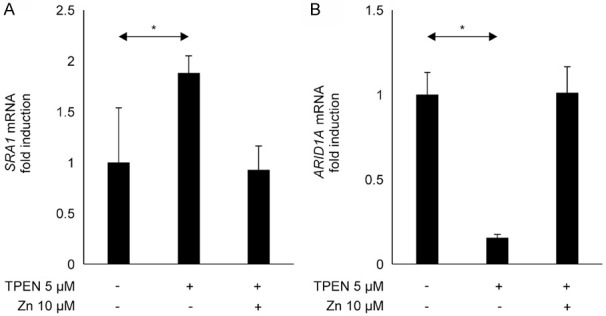
Zinc status affected SRA1 and ARID1A expression in immortalized endometrial cells. (A) SRA1 mRNA expression in the EMosis cells. SRA1 mRNA levels increased in response to 24-h incubation with 5 μM TPEN compared with cells without TPEN treatment (P=0.039, A) and did not change in cells treated with 5 μM TPEN and 10 μM zinc compared with cells without TPEN treatment (P=0.955, A). (B) ARID1A mRNA expression in the EMosis cells. ARID1A mRNA levels decreased in response to the 24-h incubation with 5 μM TPEN compared with the cells without TPEN (P<0.001, B) and did not change in cells treated with 5 μM TPEN and 10 μM zinc compared with the cells without TPEN treatment (P=0.927, B). *P<0.05, compared to the control. SRA1, steroid receptor RNA activator 1; ARID1A, AT-rich interaction domain 1A; EMosis cells, endometrial epithelial cells; TPEN, N,N,N’,N’-tetrakis 2-pyridylmethyl ethylenediamine.
Discussion
This study investigated whether zinc deficiency was associated with ovarian endometriosis (or endometrial cysts). We measured the serum zinc levels in patients with ovarian endometrial cysts and non-endometrial benign cysts and analyzed cell proliferation, microarray data, and gene expression in EMosis cells, which are immortalized human endometrial epithelial cells derived from endometrial cysts of the ovary [28]. Endometrial cyst patients had considerbaly lower zinc and ALP levels than non-endometrial benign cyst patients. Linear regression analysis adjusted for age, BMI, alcohol consumption, smoking, and supplement use revealed that endometrial cysts were significantly associated with serum zinc levels (regression coefficient 7.98, 95% CI 0.36-15.59, P=0.040). The proliferation of EMosis cells treated with TPEN 5 μM for 4 h was higher than that in untreated cells. Microarray analysis revealed that enriched CC was associated with the nucleus and enriched BP was associated with response to hypoxia. The enriched KEGG pathway included the HIF-1 signaling pathway. The EMosis cells treated with 5 μM TPEN demonstrated increased SRA1 mRNA levels and decreased ARID1A mRNA levels, and 10 μM zinc suppressed changes in SRA1 or ARID1A mRNA levels caused by 5 μM TPEN. These results indicate that zinc deficiency might be associated with ovarian endometrial cysts.
This is the first study to show that endometrial cysts are associated with lower serum zinc and ALP levels in Japan. Previous studies have shown that low zinc intake and low zinc levels in whole blood and serum are linked to endometriosis [25-27]. Our result was supported by those of previous studies. Patients with endometrial cysts had lower ALP levels than those with non-endometrial cysts, which seems plausible because zinc deficiency reduces ALP levels and activity [38,39]. In our study, 77.0 μg/dL of zinc was observed in patients with endometrial cysts, which is within the range of marginal zinc deficiency of 60-80 μg/dL [40], suggesting that marginal zinc deficiency might contribute to endometrial cysts.
Our study demonstrates that changes in gene expression caused by zinc deficiency are linked to endometrial cysts. Low TPEN concentrations treatment for 4 h caused cells to proliferate. TPEN increases oxidative stress by lowering zinc [41]. Oxidative stress has been reported to promote the proliferation of ovarian endometrial epithelial cells [42]. However, cell proliferation might be inhibited by the toxicity of long TPEN exposure [41]. For the first time, we revealed the changes in gene expression caused by zinc deficiency in EMosis cells. The enriched BPs included responses to hypoxia and decreased oxygen levels. In endometriosis, hyperoxia plays a crucial role [43] and enhances its progression [44,45]. The enriched KEGG pathways included HIF-1 signaling. This is plausible, as a previous study showed that zinc deficiency causes HIF-1α activation [46]. Another study showed that HIF-1α levels are markedly increased in patients with endometriosis [47]. Zinc deficiency might be responsible for the development of endometrial cysts.
SRA1 is a long noncoding RNA (lncRNA) that acts as a coactivator of estrogen receptor α (ERα), ERβ, and progesterone receptors. SRA1 plays a critical role in nuclear receptor-mediated hormone-dependent cancers such as estrogen-dependent breast cancer [48]. Oxidative stress and TGFβ expression could increase SRA1 expression in Emosis cells. TPEN increases oxidative stress by lowering zinc [41]. The previous study has shown that X-ray-induced oxidative stress increased SRA1 expression in TK6 cells [49]. An important factor in the development of endometriosis is oxidative stress [50]. Oxidative stress caused by low zinc might contribute to the development of endometriosis by promoting SRA1 expression.
SRA1 might play a role in TGF-β2 expression. A previous report has demonstrated SRA1 depletion reduced TGF-β2 expression in MCF-7cell [51]. Previous study has shown that endometriotic cell lines (12Z and 22B) secreted considerably higher levels of TGF-β2 compared to normal endometrial cell lines (T-HESC). TGF-β2 might enhance endometrial cell shedding by increasing Plasminogen activator inhibitor-1 (PAI-1) levels, which may contribute to endometriosis [52]. In our microarray analysis, TPEN significantly increased TGFβ2 gene expression in Emosis cells (P<0.001, fold change: 3.6254). Thus, TGFβ2 expression might be associated with SRA1 expression, promoting development of endometrial cyst.
Previous studies have revealed that SRA1 stimulates cellular proliferation, and SRA1 siRNA promotes apoptosis and decreases the proliferation of endometrial stromal cells [36,53]. The SRA1 gene has been reported to increase the transcriptional activity of ERα and ERβ [54]. ERα increases the expression of genes for cell survival/growth and cell cycle [55]. In a mouse model, ERα contributed to endometriosis development [56]. Furthermore, SRA1 is involved in the cell cycle; increased expression of SRA1 has been reported to downregulate CDKN1A and CDKN1B expression [57]. In our microarray analysis of Emosis cells, CDKN1A and CDKN1B expression were significantly decreased (CDKN1A; fold change: 0.6582 and P=0.0188, CDKN1B; fold change: 0.4565 and P<0.001), whereas that of SAR1 was increased. SRA1 might increase the transcriptional activity of ERα and decrease CDKN1A and CDKN1B expression. This might lead to endometrial cyst cell growth.
A previous study has shown that SRAP (SRA1 protein) expression is increased in ovarian endometriosis epithelial cells when ovarian mature cystic teratomas are used as controls [58]. SRAP has been found to interact with a variety of transcription factors, such as FOS [59], GATA1 [60], and ETS1 [61], which are essential transcription factors for tumorigenesis and development [62]. Among these, SRAP has been shown to interact with ATF1 [62]. There has been no study reporting how the interaction between ATF1 and SRAP regulates endometriosis. ATF1 plays a pivotal role in cell survival and proliferation. Previous studies have shown that increased expression of ATF1 in lymphoma and metastatic melanoma cells was associated with the proliferative potential of these tumor cells and cell survival [63,64]. Furthermore, ATF1 has been reported to be involved in ERK1/2 and p38 mitogen-activated protein kinase (MAPK) pathways in endometriosis [65]. Thus, increased SRAP and ATF1 interactions might be involved in endometrial cyst development. Therefore, SRA1 expression is associated with zinc deficiency in EMosis cells; increased SRA1 expression caused by zinc deficiency enhances the development of endometrial cysts.
In 20% of all human malignancies, genes encoding subunits of switch/sucrose non-fermenting (SWI/SNF) chromatin remodeling complexes are mutated. ARID1A is an SWI/SNF subunit gene, frequently mutated depending on the molecular and histological subtypes of cancer [66]. The suppression of ARID1A leads to increased proliferation, migration, and invasion of cells [67,68]. The previous study has shown oxidative stress using H2O2 suppressed ARID1A expression in primary cells from ovarian endometrial epithelial cells [42]. In the present study, the increased oxidative stress due to zinc deficiency might decrease ARID1A expression in EMosis cells. A previous report has shown that loss of ARID1A expression and activation of the PI3K/AKT pathway, such as via mutations in PIK3CA encoding the catalytic subunit p110α, of PI3K coexist in endometriosis and endometriosis-associated ovarian cancer tissues [69]. siRNA knockdown of ARID1A increases Akt phosphorylation in both endometrioid and nasopharyngeal cancer cell lines. This indicates that ARID1A regulates the PI3K/Akt pathway [68,70]. PIK3IP1 inhibits PI3K/Akt signaling. A previous study has shown that ARID1A binds to the PIK3IP1 promoter and promotes PIK3IP1 expression in ovarian clear cell carcinoma OVISE cells [71]. Based on these findings, ARID1A might be downregulated by oxidative stress due to low zinc content, which activates the PI3K/AKT pathway, thereby promoting the proliferation of endometrial epithelial cells. Uterine-specific Arid1a knock-out mice show increased epithelial proliferation as well as increased E2 signaling [72], suggesting that decreased ARID1A increased E2 signaling contributing to the development of endometriosis. Furthermore, transcriptome analysis for uterus from uterine-specific ARID1A knock-out mice has shown the role of ARID1A in repressing cell cycle-related genes and increasing progesterone receptors, especially PR-A and Kruppel-like factor 15 (KLF15), thereby inhibiting proliferation of epithelium [72]. Thus, reduction in ARID1A expression may contribute to the development of endometrial cysts.
A previous study has shown that ARID1A interacts with SMARCD1 [73]. SMARCD1, as well as ARID1A, is one of the components that form SWI/SNF complexes [74]. A decreased SMARCD1 level caused an increase in and invasion of endometriotic stromal cells [75]. SWI/SNF complexes normally have a role in tumor suppression [76]. SWI/SNF complexes coordinate with tissue-specific transcription factors to regulate the balance between lineage-specific gene activation and suppression of the proliferative program [76]. Loss of ARID1A destabilizes other SWI/SNF subunits, including SMRCD1, and reduces their association; degradation of ARID1A disrupts nucleosomes flanking pluripotent transcription factors [77]. Taken together, these reports suggest that a decrease in ARID1A causes a decreased interaction with SMARCD1, which reflects the SWI/SNF instability. That might lead to cancer pathway aggravation associated with the premalignant state. However, further studies are warranted.
Zinc supplements are effective in treating patients with dysmenorrhea, and a high-antioxidant diet containing zinc is effective in treating patients with endometriosis [25,78]. Endometriosis is mainly treated with hormonal drugs [79], and zinc supplementation may serve as a new treatment option for endometrial cysts. In addition, zinc supplementation may prevent ovarian cancer. Patients with ovarian cancer had lower serum zinc levels than patients with benign tumors and healthy controls. Lower zinc levels cause mutations owing to increased oxidative stress [80]. Mutations and loss of function of ARID1A are associated with the development of clear cell and endometrioid carcinoma derived from endometrial cysts [81]. Additionally, our study revealed that zinc deficiency directly impacted ARID1A expression. More research is needed to determine whether endometrial cysts in patients with low zinc levels might lead to clear cell carcinoma and ovarian endometrioid carcinoma.
This study has some limitations. The number of patients that participated in the study was small because the data were obtained from a single facility. Furthermore, zinc levels fluctuate according to the time of day and physical condition of the patient [82,83]. One strength of this study was that all serum samples were collected under the same conditions. Animal model analysis is needed to determine if zinc status or zinc supplementation affects the formation of endometrial cysts.
In conclusion, patients with endometrial cysts had lower serum zinc levels than those with benign non-endometrial cysts. In immortalized endometrial epithelial cells, zinc deficiency increased cell proliferation and enhanced cellular response to hypoxia and the HIF-1α pathway. Zinc deficiency increased SRA1 expression and decreased ARID1A expression, both of which are linked to the development of endometrial cyst. Zinc supplementation may represent a potential treatment for endometrial cyst.
Acknowledgements
This study was supported by the Japan Society for the Promotion of Science from 2019 to 2021 [grant number P19K09798].
The written informed consent of all participants was obtained in this study. Thus, this agreement was confirmed. Submitted manuscripts have been approved for publication by all authors. Patients signed a consent form to publish their data.
Disclosure of conflict of interest
None.
References
- 1.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 2.Brosens I, Gordts S, Puttemans P, Benagiano G. Pathophysiology proposed as the basis for modern management of the ovarian endometrioma. Reprod Biomed Online. 2014;28:232–238. doi: 10.1016/j.rbmo.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 3.Nnoaham KE, Hummelshoj L, Webster P, d’Hooghe T, de Cicco Nardone F, de Cicco Nardone C, Jenkinson C, Kennedy SH, Zondervan KT World Endometriosis Research Foundation Global Study of Women’s Health consortium. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil Steril. 2011;96:366–373. e8. doi: 10.1016/j.fertnstert.2011.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvaskoff M, Mu F, Terry KL, Harris HR, Poole EM, Farland L, Missmer SA. Endometriosis: a high-risk population for major chronic diseases? Hum Reprod Update. 2015;21:500–516. doi: 10.1093/humupd/dmv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidemann LN, Hartwell D, Heidemann CH, Jochumsen KM. The relation between endometriosis and ovarian cancer - a review. Acta Obstet Gynecol Scand. 2014;93:20–31. doi: 10.1111/aogs.12255. [DOI] [PubMed] [Google Scholar]
- 6.Guidozzi F. Endometriosis-associated cancer. Climacteric. 2021;24:587–592. doi: 10.1080/13697137.2021.1948994. [DOI] [PubMed] [Google Scholar]
- 7.Gadducci A, Multinu F, Cosio S, Carinelli S, Ghioni M, Aletti GD. Clear cell carcinoma of the ovary: epidemiology, pathological and biological features, treatment options and clinical outcomes. Gynecol Oncol. 2021;162:741–750. doi: 10.1016/j.ygyno.2021.06.033. [DOI] [PubMed] [Google Scholar]
- 8.Kajiyama H, Suzuki S, Yoshihara M, Tamauchi S, Yoshikawa N, Niimi K, Shibata K, Kikkawa F. Endometriosis and cancer. Free Radic Biol Med. 2019;133:186–192. doi: 10.1016/j.freeradbiomed.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Sampson JA. Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. Am J Pathol. 1927;3:93–110.43. [PMC free article] [PubMed] [Google Scholar]
- 10.Missmer SA, Cramer DW. The epidemiology of endometriosis. Obstet Gynecol Clin North Am. 2003;30:1–19. vii. doi: 10.1016/s0889-8545(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 11.Montgomery GW, Nyholt DR, Zhao ZZ, Treloar SA, Painter JN, Missmer SA, Kennedy SH, Zondervan KT. The search for genes contributing to endometriosis risk. Hum Reprod Update. 2008;14:447–457. doi: 10.1093/humupd/dmn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahmioglu N, Nyholt DR, Morris AP, Missmer SA, Montgomery GW, Zondervan KT. Genetic variants underlying risk of endometriosis: insights from meta-analysis of eight genome-wide association and replication datasets. Hum Reprod Update. 2014;20:702–716. doi: 10.1093/humupd/dmu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porpora MG, Resta S, Fuggetta E, Storelli P, Megiorni F, Manganaro L, De Felip E. Role of environmental organochlorinated pollutants in the development of endometriosis. Clin Exp Obstet Gynecol. 2013;40:565–567. [PubMed] [Google Scholar]
- 14.Buck Louis GM, Peterson CM, Chen Z, Croughan M, Sundaram R, Stanford J, Varner MW, Kennedy A, Giudice L, Fujimoto VY, Sun L, Wang L, Guo Y, Kannan K. Bisphenol A and phthalates and endometriosis: the endometriosis: natural history, diagnosis and outcomes study. Fertil Steril. 2013;100:162–9. e1–2. doi: 10.1016/j.fertnstert.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viganò P, Somigliana E, Panina P, Rabellotti E, Vercellini P, Candiani M. Principles of phenomics in endometriosis. Hum Reprod Update. 2012;18:248–259. doi: 10.1093/humupd/dms001. [DOI] [PubMed] [Google Scholar]
- 16.Pfaender S, Föhr K, Lutz AK, Putz S, Achberger K, Linta L, Liebau S, Boeckers TM, Grabrucker AM. Cellular zinc homeostasis contributes to neuronal differentiation in human induced pluripotent stem cells. Neural Plast. 2016;2016:3760702. doi: 10.1155/2016/3760702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haase H, Rink L. Multiple impacts of zinc on immune function. Metallomics. 2014;6:1175–1180. doi: 10.1039/c3mt00353a. [DOI] [PubMed] [Google Scholar]
- 18.Duke RC, Chervenak R, Cohen JJ. Endogenous endonuclease-induced DNA fragmentation: an early event in cell-mediated cytolysis. Proc Natl Acad Sci U S A. 1983;80:6361–6365. doi: 10.1073/pnas.80.20.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finamore A, Massimi M, Conti Devirgiliis L, Mengheri E. Zinc deficiency induces membrane barrier damage and increases neutrophil transmigration in Caco-2 cells. J Nutr. 2008;138:1664–1670. doi: 10.1093/jn/138.9.1664. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi Y, Tanabe S, Suzuki T. Cellular zinc is required for intestinal epithelial barrier maintenance via the regulation of claudin-3 and occludin expression. Am J Physiol Gastrointest Liver Physiol. 2016;311:G105–16. doi: 10.1152/ajpgi.00405.2015. [DOI] [PubMed] [Google Scholar]
- 21.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid Redox Signal. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 22.Bellomo E, Hogstrand C, Maret W. Redox and zinc signalling pathways converging on protein tyrosine phosphatases. Free Radic Biol Med. 2014;75(Suppl 1):S9. doi: 10.1016/j.freeradbiomed.2014.10.851. [DOI] [PubMed] [Google Scholar]
- 23.Truong-Tran AQ, Carter J, Ruffin RE, Zalewski PD. The role of zinc in caspase activation and apoptotic cell death. Biometals. 2001;14:315–330. doi: 10.1023/a:1012993017026. [DOI] [PubMed] [Google Scholar]
- 24.Sunderman FW Jr. The influence of zinc on apoptosis. Ann Clin Lab Sci. 1995;25:134–142. [PubMed] [Google Scholar]
- 25.Mier-Cabrera J, Aburto-Soto T, Burrola-Méndez S, Jiménez-Zamudio L, Tolentino MC, Casanueva E, Hernández-Guerrero C. Women with endometriosis improved their peripheral antioxidant markers after the application of a high antioxidant diet. Reprod Biol Endocrinol. 2009;7:54. doi: 10.1186/1477-7827-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai GL, Yeh CC, Yeh CY, Chen RY, Fu CL, Chen CH, Tzeng CR. Decreased zinc and increased lead blood levels are associated with endometriosis in Asian women. Reprod Toxicol. 2017;74:77–84. doi: 10.1016/j.reprotox.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Messalli EM, Schettino MT, Mainini G, Ercolano S, Fuschillo G, Falcone F, Esposito E, Di Donna MC, De Franciscis P, Torella M. The possible role of zinc in the etiopathogenesis of endometriosis. Clin Exp Obstet Gynecol. 2014;41:541–546. [PubMed] [Google Scholar]
- 28.Bono Y, Kyo S, Takakura M, Maida Y, Mizumoto Y, Nakamura M, Nomura K, Kiyono T, Inoue M. Creation of immortalised epithelial cells from ovarian endometrioma. Br J Cancer. 2012;106:1205–1213. doi: 10.1038/bjc.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onuma T, Mizutani T, Fujita Y, Yamada S, Yoshida Y. Copper content in ascitic fluid is associated with angiogenesis and progression in ovarian cancer. J Trace Elem Med Biol. 2021;68:126865. doi: 10.1016/j.jtemb.2021.126865. [DOI] [PubMed] [Google Scholar]
- 30.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia J, Benner MJ, Hancock RE. NetworkAnalyst--integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–74. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eoh KJ, Paek J, Kim SW, Kim HJ, Lee HY, Lee SK, Kim YT. Long non-coding RNA, steroid receptor RNA activator (SRA), induces tumor proliferation and invasion through the NOTCH pathway in cervical cancer cell lines. Oncol Rep. 2017;38:3481–3488. doi: 10.3892/or.2017.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erfani M, Hosseini SV, Mokhtari M, Zamani M, Tahmasebi K, Alizadeh Naini M, Taghavi A, Carethers JM, Koi M, Brim H, Mokarram P, Ashktorab H. Altered ARID1A expression in colorectal cancer. BMC Cancer. 2020;20:350. doi: 10.1186/s12885-020-6706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yilmaz BD, Bulun SE. Endometriosis and nuclear receptors. Hum Reprod Update. 2019;25:473–485. doi: 10.1093/humupd/dmz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin K, Zhan H, Ma J, Xu K, Wu R, Zhou C, Lin J. Silencing of SRA1 regulates ER expression and attenuates the growth of stromal cells in ovarian endometriosis. Reprod Sci. 2017;24:836–843. doi: 10.1177/1933719116670036. [DOI] [PubMed] [Google Scholar]
- 37.Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JE, Ferrando L, Selenica P, Ladewig E, Chan C, Da Cruz Paula A, Witkin M, Cheng Y, Park J, Serna-Tamayo C, Zhao H, Wu F, Sallaku M, Qu X, Zhao A, Collings CK, D’Avino AR, Jhaveri K, Koche R, Levine RL, Reis-Filho JS, Kadoch C, Scaltriti M, Leslie CS, Baselga J, Toska E. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet. 2020;52:198–207. doi: 10.1038/s41588-019-0554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weismann K, Høyer H. Serum alkaline phosphatase and serum zinc levels in the diagnosis and exclusion of zinc deficiency in man. Am J Clin Nutr. 1985;41:1214–1219. doi: 10.1093/ajcn/41.6.1214. [DOI] [PubMed] [Google Scholar]
- 39.Cho YE, Lomeda RA, Ryu SH, Sohn HY, Shin HI, Beattie JH, Kwun IS. Zinc deficiency negatively affects alkaline phosphatase and the concentration of Ca, Mg and P in rats. Nutr Res Pract. 2007;1:113–119. doi: 10.4162/nrp.2007.1.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kodama H, Tanaka M, Naito Y, Katayama K, Moriyama M. Japan’s practical guidelines for zinc deficiency with a particular focus on taste disorders, inflammatory bowel disease, and liver cirrhosis. Int J Mol Sci. 2020;21:2941. doi: 10.3390/ijms21082941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Z, Yu Z, Chen Z, Yang L, Ma M, Lu S, Wang C, Teng C, Nie Y. Zinc chelator TPEN induces pancreatic cancer cell death through causing oxidative stress and inhibiting cell autophagy. J Cell Physiol. 2019;234:20648–20661. doi: 10.1002/jcp.28670. [DOI] [PubMed] [Google Scholar]
- 42.Winarto H, Tan MI, Sadikin M, Wanandi SI. ARID1A expression is down-regulated by oxidative stress in endometriosis and endometriosis-associated ovarian cancer. Transl Oncogenomics. 2017;9:1177272716689818. doi: 10.1177/1177272716689818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaji M, Gotoh M, Takagi Y, Masuda H, Kimura Y, Uenoyama Y. Studies to determine the usefulness of the zinc clearance test to diagnose marginal zinc deficiency and the effects of oral zinc supplementation for short children. J Am Coll Nutr. 1998;17:388–391. doi: 10.1080/07315724.1998.10718781. [DOI] [PubMed] [Google Scholar]
- 44.Li WN, Wu MH, Tsai SJ. Hypoxia and reproductive health: the role of hypoxia in the development and progression of endometriosis. Reproduction. 2021;161:F19–F31. doi: 10.1530/REP-20-0267. [DOI] [PubMed] [Google Scholar]
- 45.Lee HC, Lin SC, Wu MH, Tsai SJ. Induction of pyruvate dehydrogenase kinase 1 by hypoxia alters cellular metabolism and inhibits apoptosis in endometriotic stromal cells. Reprod Sci. 2019;26:734–744. doi: 10.1177/1933719118789513. [DOI] [PubMed] [Google Scholar]
- 46.Morand J, Briançon-Marjollet A, Lemarie E, Gonthier B, Arnaud J, Korichneva I, Godin-Ribuot D. Zinc deficiency promotes endothelin secretion and endothelial cell migration through nuclear hypoxia-inducible factor-1 translocation. Am J Physiol Cell Physiol. 2019;317:C270–C276. doi: 10.1152/ajpcell.00460.2018. [DOI] [PubMed] [Google Scholar]
- 47.Zhan L, Wang W, Zhang Y, Song E, Fan Y, Wei B. Hypoxia-inducible factor-1alpha: a promising therapeutic target in endometriosis. Biochimie. 2016;123:130–137. doi: 10.1016/j.biochi.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Liu C, Wu HT, Zhu N, Shi YN, Liu Z, Ao BX, Liao DF, Zheng XL, Qin L. Steroid receptor RNA activator: biologic function and role in disease. Clin Chim Acta. 2016;459:137–146. doi: 10.1016/j.cca.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Chaudhry MA. Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells. Int J Mol Sci. 2013;14:9099–9110. doi: 10.3390/ijms14059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scutiero G, Iannone P, Bernardi G, Bonaccorsi G, Spadaro S, Volta CA, Greco P, Nappi L. Oxidative stress and endometriosis: a systematic review of the literature. Oxid Med Cell Longev. 2017;2017:7265238. doi: 10.1155/2017/7265238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foulds CE, Tsimelzon A, Long W, Le A, Tsai SY, Tsai MJ, O’Malley BW. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol Endocrinol. 2010;24:1090–1105. doi: 10.1210/me.2009-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sui C, Mecha E, Omwandho CO, Starzinski-Powitz A, Stammler A, Tinneberg HR, Konrad L. PAI-1 secretion of endometrial and endometriotic cells is Smad2/3- and ERK1/2-dependent and influences cell adhesion. Am J Transl Res. 2016;8:2394–2402. [PMC free article] [PubMed] [Google Scholar]
- 53.Lanz RB, Chua SS, Barron N, Söder BM, DeMayo F, O’Malley BW. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol. 2003;23:7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coleman KM, Lam V, Jaber BM, Lanz RB, Smith CL. SRA coactivation of estrogen receptor-alpha is phosphorylation-independent, and enhances 4-hydroxytamoxifen agonist activity. Biochem Biophys Res Commun. 2004;323:332–338. doi: 10.1016/j.bbrc.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 55.O’Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 56.Burns KA, Rodriguez KF, Hewitt SC, Janardhan KS, Young SL, Korach KS. Role of estrogen receptor signaling required for endometriosis-like lesion establishment in a mouse model. Endocrinology. 2012;153:3960–3971. doi: 10.1210/en.2012-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, Chen XW, Cawthorn WP, MacDougald OA, Koenig RJ. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin K, Ma J, Wu R, Zhou C, Lin J. Influence of ovarian endometrioma on expression of steroid receptor RNA activator, estrogen receptors, vascular endothelial growth factor, and thrombospondin 1 in the surrounding ovarian tissues. Reprod Sci. 2014;21:183–189. doi: 10.1177/1933719113492205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 60.Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chooniedass-Kothari S, Hamedani MK, Auge C, Wang X, Carascossa S, Yan Y, Cooper C, Vincett D, Myal Y, Jalaguier S, Cavailles V, Leygue E. The steroid receptor RNA activator protein is recruited to promoter regions and acts as a transcriptional repressor. FEBS Lett. 2010;584:2218–2224. doi: 10.1016/j.febslet.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 63.Hsueh YP, Lai MZ. Overexpression of activation transcriptional factor 1 in lymphomas and in activated lymphocytes. J Immunol. 1995;154:5675–5683. [PubMed] [Google Scholar]
- 64.Jean D, Tellez C, Huang S, Davis DW, Bruns CJ, McConkey DJ, Hinrichs SH, Bar-Eli M. Inhibition of tumor growth and metastasis of human melanoma by intracellular anti-ATF-1 single chain Fv fragment. Oncogene. 2000;19:2721–2730. doi: 10.1038/sj.onc.1203569. [DOI] [PubMed] [Google Scholar]
- 65.Uimari O, Rahmioglu N, Nyholt DR, Vincent K, Missmer SA, Becker C, Morris AP, Montgomery GW, Zondervan KT. Genome-wide genetic analyses highlight mitogen-activated protein kinase (MAPK) signaling in the pathogenesis of endometriosis. Hum Reprod. 2017;32:780–793. doi: 10.1093/humrep/dex024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathur R. ARID1A loss in cancer: towards a mechanistic understanding. Pharmacol Ther. 2018;190:15–23. doi: 10.1016/j.pharmthera.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 67.Cheng S, Wang L, Deng CH, Du SC, Han ZG. ARID1A represses hepatocellular carcinoma cell proliferation and migration through lncRNA MVIH. Biochem Biophys Res Commun. 2017;491:178–182. doi: 10.1016/j.bbrc.2017.07.072. [DOI] [PubMed] [Google Scholar]
- 68.Yang Y, Wang X, Yang J, Duan J, Wu Z, Yang F, Zhang X, Xiao S. Loss of ARID1A promotes proliferation, migration and invasion via the Akt signaling pathway in NPC. Cancer Manag Res. 2019;11:4931–4946. doi: 10.2147/CMAR.S207329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samartzis EP, Noske A, Dedes KJ, Fink D, Imesch P. ARID1A mutations and PI3K/AKT pathway alterations in endometriosis and endometriosis-associated ovarian carcinomas. Int J Mol Sci. 2013;14:18824–18849. doi: 10.3390/ijms140918824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, Guo W, Scherer SE, Carter H, Westin SN, Dyer MD, Verhaak RG, Zhang F, Karchin R, Liu CG, Lu KH, Broaddus RR, Scott KL, Hennessy BT, Mills GB. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih IM, Conejo-Garcia JR, Speicher DW, Zhang R. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim TH, Yoo JY, Wang Z, Lydon JP, Khatri S, Hawkins SM, Leach RE, Fazleabas AT, Young SL, Lessey BA, Ku BJ, Jeong JW. ARID1A is essential for endometrial function during early pregnancy. PLoS Genet. 2015;11:e1005537. doi: 10.1371/journal.pgen.1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caumanns JJ, Wisman GBA, Berns K, van der Zee AGJ, de Jong S. ARID1A mutant ovarian clear cell carcinoma: a clear target for synthetic lethal strategies. Biochim Biophys Acta Rev Cancer. 2018;1870:176–184. doi: 10.1016/j.bbcan.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 75.Takebayashi K, Nasu K, Okamoto M, Aoyagi Y, Hirakawa T, Narahara H. hsa-miR-100-5p, an overexpressed miRNA in human ovarian endometriotic stromal cells, promotes invasion through attenuation of SMARCD1 expression. Reprod Biol Endocrinol. 2020;18:31. doi: 10.1186/s12958-020-00590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 77.Blümli S, Wiechens N, Wu MY, Singh V, Gierlinski M, Schweikert G, Gilbert N, Naughton C, Sundaramoorthy R, Varghese J, Gourlay R, Soares R, Clark D, Owen-Hughes T. Acute depletion of the ARID1A subunit of SWI/SNF complexes reveals distinct pathways for activation and repression of transcription. Cell Rep. 2021;37:109943. doi: 10.1016/j.celrep.2021.109943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kashefi F, Khajehei M, Tabatabaeichehr M, Alavinia M, Asili J. Comparison of the effect of ginger and zinc sulfate on primary dysmenorrhea: a placebo-controlled randomized trial. Pain Manag Nurs. 2014;15:826–833. doi: 10.1016/j.pmn.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Rafique S, Decherney AH. Medical management of endometriosis. Clin Obstet Gynecol. 2017;60:485–496. doi: 10.1097/GRF.0000000000000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin S, Yang H. Ovarian cancer risk according to circulating zinc and copper concentrations: a meta-analysis and Mendelian randomization study. Clin Nutr. 2021;40:2464–2468. doi: 10.1016/j.clnu.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 81.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McMillan EM, Rowe DJ, Halberg F. Diurnal stage of circadian rhythm of plasma zinc in healthy and psoriatic volunteers. Prog Clin Biol Res. 1987;227B:295–303. [PubMed] [Google Scholar]
- 83.King JC, Hambidge KM, Westcott JL, Kern DL, Marshall G. Daily variation in plasma zinc concentrations in women fed meals at six-hour intervals. J Nutr. 1994;124:508–516. doi: 10.1093/jn/124.4.508. [DOI] [PubMed] [Google Scholar]


