Abstract
Submerged plants in wetlands play important roles as ecosystem engineers to improve self-purification and promote elemental cycling. However, their effects on the functional capacity of microbial communities in wetland sediments remain poorly understood. Here, we provide detailed metagenomic insights into the biogeochemical potential of microbial communities in wetland sediments with and without submerged plants (i.e., Vallisneria natans). A large number of functional genes involved in carbon (C), nitrogen (N) and sulfur (S) cycling were detected in the wetland sediments. However, most functional genes showed higher abundance in sediments with submerged plants than in those without plants. Based on the comparison of annotated functional genes in the N and S cycling databases (i.e., NCycDB and SCycDB), we found that genes involved in nitrogen fixation (e.g., nifD/H/K/W), assimilatory nitrate reduction (e.g., nasA and nirA), denitrification (e.g., nirK/S and nosZ), assimilatory sulfate reduction (e.g., cysD/H/J/N/Q and sir), and sulfur oxidation (e.g., glpE, soeA, sqr and sseA) were significantly higher (corrected p < 0.05) in vegetated vs. unvegetated sediments. This could be mainly driven by environmental factors including total phosphorus, total nitrogen, and C:N ratio. The binning of metagenomes further revealed that some archaeal taxa could have the potential of methane metabolism including hydrogenotrophic, acetoclastic, and methylotrophic methanogenesis, which are crucial to the wetland methane budget and carbon cycling. This study opens a new avenue for linking submerged plants with microbial functions, and has further implications for understanding global carbon, nitrogen and sulfur cycling in wetland ecosystems.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42995-021-00100-3.
Keywords: Submerged plants, Metagenome, Elemental cycling, Methanogenesis, Verstraetearchaeota
Introduction
Wetlands play important roles in global carbon (C), nitrogen (N) and sulfur (S) cycling, and contribute significantly to the processes of methane production (Borrel et al. 2011; Emilson et al. 2018), nitrogen transformation and removal (Harrison et al. 2009; Liu et al. 2018), and sulfur reduction and oxidation (Holmer and Storkholm 2001; Purcell et al. 2014). The resultant ‘hot spots’ of biogeochemical cycling involving plants and microorganisms may create an eco-buffer zone for environmental protection, regulation and management (Chen et al. 2014; Sims et al. 2013). As plant-microbe interactions could considerably influence element cycling in wetland sediments (Kofoed et al. 2012; Vilacosta et al. 2016), studying the effects of plants on the functional potential of microbial communities is very important to understand wetland ecosystem function.
Aquatic macrophytes (e.g., Vallisneria natans, Hydrilla verticillata and Typha latifolia) are commonly used as biological tools for reducing nitrogen and sulfur elements, and raising dissolved oxygen (DO) concentration in natural or constructed wetlands (Chen et al. 2014; Qiu et al. 2001; Zhu et al. 2016). They almost invariably show positive effects on improving water clarity and increasing the stability of aquatic ecosystems by a variety of different mechanisms (Scheffer et al. 1993; Zhu et al. 2016). Specifically, leaf and root exudates supply nutrients and create micro-niches for microorganisms (Michael et al. 2008; Wiessner et al. 2010) which stimulate ecosystem element cycling (Kaokniffin et al. 2010; Ullah et al. 2014). For example, the oxidation of sedimental NH4+-N to NO3−-N could be enhanced by submerged macrophyte rhizosphere microorganisms (Ottosen et al. 1999), which in turn benefit the plants by enhancing the supply of available N (Bodelier et al. 2006). Also, macrophytes could affect sedimental biogeochemical cycling by increasing the activity of microbes, such as methanogens, which would lead to high methane production (Emilson et al. 2018). In turn, the sediment microbial communities play key roles in the health of plants, for example, facilitating the oxidation of phytotoxic sulfide into non-toxic sulfate or protecting plants from pathogens (Frigaard and Dahl 2008; Ugarelli et al. 2019). Therefore, plants may regulate rhizosphere microorganisms to improve their own fitness (Zhalnina et al. 2018).
Although the effects of macrophytes on microbial community structure have been sufficient reports (Liu et al. 2018), further studies are needed to provide more direct insights into how vegetation impacts microbial processes and functions (Grossart et al. 2020). For example, vegetated sediments show a higher abundance of nirS and nirK genes than unvegetated sediments in oligotrophic shallow lakes, and some nitrogen cycling processes are promoted by vegetation (Vilacosta et al. 2016). Additionally, wetlands are considered to be a significant source of greenhouse gases (e.g., CH4 and N2O), which also could be regulated by aquatic macrophytes (Heilman and Carlton 2001; Veraart et al. 2011). In contrast, other studies found that submerged macrophytes had little effect on N2O production (Liu et al. 2018; Yao et al. 2018). However, the effects of submerged plants on microbial functions in C, N and S cycling have not yet been fully explored. Therefore, it is important to identify molecular markers (e.g., key functional genes involved in biogeochemical cycling) in microorganisms to predict the effects of submerged plants on the functional status and stability of wetland ecosystems.
The submerged macrophyte Vallisneria natans is a widely distributed perennial species with well-developed roots that provides valuable ecosystem services to improve water quality and filter nutrients (Qiu et al. 2001). A recent study indicated that V. natans can even resist the adverse impacts of algal blooms (Jiang et al. 2019). Although the feedback of microbial community composition to V. natans has previously been studied (Wang et al. 2017), it remains elusive how V. natans affects the functional potential of microbial groups involved in elemental cycling in wetland sediments. The main aim of this study was to determine the influence of V. natans on functional diversity of microbial communities. We hypothesized that submerged plants provide an important impetus (direct or indirect) for accelerating biogeochemical cycling through plant-microbe interactions in wetland sediments. To test this hypothesis, metagenomic analysis was performed to study microbial functions acclimating to V. natans in the Xixi National Wetland Park. We also reconstructed and characterized draft genomes to identify the main and potential microbial taxa involved in sedimental element cycling. This study thus provides insights to link submerged macrophytes with the functional capacity of sedimental microbial communities in urban wetlands.
Results
Metabolic genes involved in nitrogen cycling
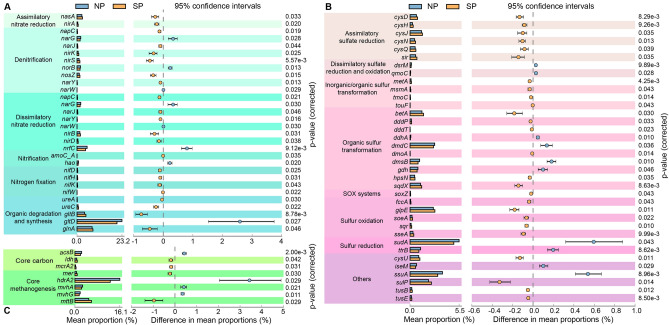
Key genes for microbial nitrogen metabolism were searched for in the metagenomes of two types of sedimental samples (i.e., with submerged plants, SP; with no plants, NP), and differences between SP and NP were clear regarding gene inventories. In total, more than 70 functional genes involved in nitrification, denitrification, assimilatory nitrate reduction, dissimilatory nitrate reduction, nitrogen fixation, anammox, organic degradation and synthesis were annotated in this study (Supplementary file 1). More than two-thirds of the functional genes involved in the pathways of nitrogen fixation, assimilatory nitrate reduction and denitrification were more abundant in the SP sediments than in the NP sediments (Fig. 1a). Specifically, genes encoding the glutamate synthase (NADPH/NADH) small chain (gltD) accounted for more than 20% of the total abundance of functional genes involved in the nitrogen cycle and had significantly higher abundance in SP than in NP (corrected p < 0.05). Similarly, the abundance of genes encoding enzymes of the nitrate reduction process, including assimilatory nitrate reductase catalytic subunit (nasA), ferredoxin-nitrite reductase (nirA), and nitrite reductase (NADH) (nirB/D), was significantly higher in SP than NP. In contrast, the abundance of genes encoding nitric oxide reductase (norB) and hydroxylamine dehydrogenase (hao) had significantly higher abundances (corrected p < 0.05) in NP than SP. For the nitrogen fixation pathway, genes encoding the nitrogenase molybdenum-iron protein (nifD/K), nitrogenase iron protein (nifH), and nitrogenase-stabilizing/protective protein (nifW) were more abundant in SP than NP. In addition, the abundance of genes encoding urease subunit alpha/gamma (ureC/A), the key enzyme in the conversion of organic nitrogen to ammonium nitrogen, was higher in the SP than in the NP sediments (Fig. S1 in Supplementary file 2). The correlation variations based on redundancy analysis (RDA) indicated that total phosphorus (TP), total nitrogen (TN) and C:N ratio (C/N) were the major factors shaping microbial communities (Fig. S2a in Supplementary file 2), and explain 59.66% of the community variances by the first two axes. The relationships between environmental factors and microbial functional genes involved in nitrogen metabolic pathways were further investigated by partial Mantel tests. We found the genes involved in assimilatory nitrate reduction (r = 0.512), denitrification (r = 0.425) and dissimilatory nitrate reduction (r = 0.372) were significantly (p < 0.05) correlated with C/N (Table 1).
Fig. 1.
Abundance comparison of key genes involved in nitrogen cycling (a), sulfur cycling (b), and methanogenesis (c) between sediments with (SP) and without (NP) submerged plants. The p values are based on Welch’s exact test and corrected by Benjamini-Hochberg false discovery rate (FDR) method
Table 1.
Summary statistics for partial Mantel tests of correlation between functional genes and environmental factors
| Gene category | pH | Conductivity (μs/cm) | TP (g/kg) | TN (g/kg) | TC (g/kg) | C/N |
|---|---|---|---|---|---|---|
| Nitrogen-cycling pathway | ||||||
| Anammox | − 0.124 (0.792) | − 0.311 (0.954) | 0.104 (0.263) | − 0.035 (0.578) | − 0.320 (0.956) | 0.110 (0.313) |
| Assimilatory nitrate reduction | 0.254 (0.055) | 0.347 (0.017) | − 0.220 (0.919) | − 0.438 (0.990) | 0.050 (0.412) | 0.512 (0.006) |
| Denitrification | 0.184 (0.094) | 0.380 (0.014) | 0.009 (0.428) | − 0.027 (0.560) | 0.299 (0.019) | 0.425 (0.008) |
| Dissimilatory nitrate reduction | 0.421 (0.020) | 0.446 (0.007) | − 0.198 (0.932) | − 0.086 (0.723) | 0.322 (0.016) | 0.372 (0.019) |
| Nitrification | 0.040 (0.384) | 0.206 (0.109) | 0.266 (0.100) | − 0.076 (0.642) | 0.284 (0.077) | 0.085 (0.274) |
| Nitrogen fixation | 0.453 (0.007) | 0.476 (0.003) | − 0.270 (0.968) | 0.207 (0.151) | 0.321 (0.024) | 0.049 (0.304) |
| Organic degradation and synthesis | − 0.081 (0.681) | 0.093 (0.294) | 0.127 (0.248) | − 0.256 (0.915) | − 0.004 (0.505) | 0.331 (0.049) |
| Sulfur-cycling pathway | ||||||
| Assimilatory sulfate reduction | 0.122 (0.222) | 0.331 (0.028) | − 0.208 (0.922) | − 0.361 (0.987) | 0.007 (0.511) | 0.603 (0.005) |
| Dissimilatory sulfate reduction and oxidation | 0.180 (0.120) | 0.257 (0.069) | − 0.191 (0.838) | − 0.273 (0.961) | − 0.050 (0.603) | 0.346 (0.013) |
| Link between inorganic and organic sulfur transformation | − 0.142 (0.812) | 0.066 (0.389) | − 0.056 (0.536) | − 0.285 (0.906) | − 0.104 (0.703) | 0.426 (0.050) |
| Organic sulfur transformation | 0.026 (0.475) | 0.147 (0.238) | − 0.212 (0.878) | − 0.301 (0.946) | − 0.091 (0.703) | 0.463 (0.021) |
| SOX systems | − 0.090 (0.717) | 0.059 (0.413) | 0.038 (0.364) | − 0.349 (0.968) | − 0.012 (0.535) | 0.456 (0.016) |
| Sulfur disproportionation | − 0.016 (0.559) | 0.069 (0.342) | 0.088 (0.268) | − 0.074 (0.676) | 0.064 (0.331) | − 0.061 (0.594) |
| Sulfur oxidation | 0.132 (0.167) | 0.286 (0.024) | − 0.110 (0.751) | − − 0.060 (0.650) | 0.097 (0.306) | 0.379 (0.026) |
| Sulfur reduction | 0.077 (0.354) | 0.137 (0.227) | 0.122 (0.209) | -0.226 (0.849) | 0.128 (0.292) | 0.222 (0.139) |
| Methanogenesis | ||||||
| Core methanogenesis | − 0.295 (0.977) | − 0.084 (0.692) | 0.164 (0.214) | − 0.221 (0.871) | − 0.122 (0.71) | 0.313 (0.101) |
Values are correlation coefficients with p values in brackets (p < 0.05 in bold)
TP: Total phosphorus; TC: Total carbon; TN: Total nitrogen; C/N: Total carbon/Total nitrogen
Metabolic genes involved in sulfur cycling
More than 200 functional genes involved in assimilatory sulfate reduction, sulfate reduction, dissimilatory sulfate reduction and oxidation, sulfur reduction, sulfur oxidization (SOX) systems, sulfur oxidation, sulfur disproportionation, organic sulfur transformation, and the link between inorganic and organic sulfur transformation, were identified in both NP and SP samples (Supplementary file 3). Among them, 26 genes encoding the key enzyme of sulfur cycling showed significantly higher (corrected p < 0.05) abundances in the SP than in the NP sediment samples (Fig. 1b). For the assimilatory sulfate reduction pathway, most relevant genes involved in encoding for sulfate adenylyltransferase subunit (cysD/N), phosphoadenosine phosphosulfate reductase (cysH), sulfite reductase (NADPH) flavoprotein alpha-component (cysJ) and sulfite reductase (ferredoxin) (sir) were significantly more abundant in SP than NP sediments. Also, SP had significantly higher abundances of genes encoding thiosulfate sulfurtransferase (glpE), sulfite dehydrogenase (quinone) subunit (soeA), sulfide-quinone oxidoreductase (sqr), and the key enzyme of sulfide oxidation (Fig. 1b). For the SOX systems, similar gene abundances were found in NP and SP, except for soxZ. In contrast, the NP microbial communities showed significantly higher (corrected p < 0.05) abundances of genes encoding 3-(methylthio)propanoyl-CoA dehydrogenase (dmdC; catalyzing organic sulfur oxidation), anaerobic dimethyl sulfoxide reductase subunit B (dmsB) and sulfide dehydrogenase subunit alpha (sudA). According to the RDA plots, the environmental factors TN, TP and C/N had significant influences on the relative abundances of functional genes involved in sulfur cycling, and explained 58.07% of the total variation by the first two axes (Fig. S2b in Supplementary file 2). Furthermore, C/N showed significant positive correlations with the functional genes involved in assimilatory sulfate reduction (r = 0.603, p < 0.05), dissimilatory sulfate reduction and oxidation (r = 0.346, p < 0.05), organic sulfur transformation (r = 0.463, p < 0.05), SOX systems (r = 0.456, p < 0.05), and the sulfur oxidation pathway (r = 0.379, p < 0.05) (Table 1).
Metabolic genes and potential pathways involved in methanogenesis
Genes involved in methanogenesis are summarized in Supplementary file 4. The detected methanogenic processes included methanol, methylamine, acetate, carbon dioxide, core carbon and core methanogenesis. Gene encoding for the methyl coenzyme M reductase system, component A2 (mcrA2/K00400) was more abundant in the SP than NP sediments. Similarly, abundances of genes encoding the trimethylamine-corrinoid protein Co-methyltransferase (mttB) and 5,10-methylenetetra-hydro-methanopterin reductase (mer) were significantly higher (corrected p < 0.05) in SP sediments than in NP sediments. In contrast, the abundance of gene associated with heterodisulfide reductase subunit A2 (hdrA2) was significantly higher in the NP than the SP sediments (corrected p < 0.05) (Fig. 1c).
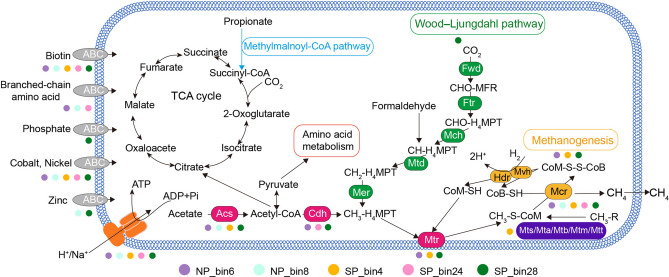
To reveal the diversity of metabolic potential for methanogenesis, we chose relatively high-quality genome bins (completeness > 80%) and performed bin annotation to identify the metabolic pathways of hydrogenotrophic, acetoclastic, and methylotrophic methanogenesis in urban wetland sediments (Fig. 2). Specifically, SP_bin28 had genes encoding conserved core enzymes of hydrogenotrophic methanogenesis, including Fwd, Ftr, Mch, Mtd, Mer, Mtr and Mcr. NP_bin6 and SP_bin28 contained genes encoding Acs, Cdh, Mtr, and Mcr enzymes for the utilization of acetate, which showed potential for acetoclastic methanogenesis. SP_bin4 contained genes encoding methyl-compound methyltransferase, such as Mts, Mta, Mtb, Mtm and Mtt. These genes showed a potential for methane production through the methylotrophic pathway. However, no complete methanogenesis pathways were identified in NP_bin8 and SP_bin24 due to the incompleteness of their genomes.
Fig. 2.
Predicted methanogenic pathways in the archaeal lineages based on the gene analysis of five MAGs with completeness > 80%. Genes involved in methanogenesis and various transporters are shown in different colors
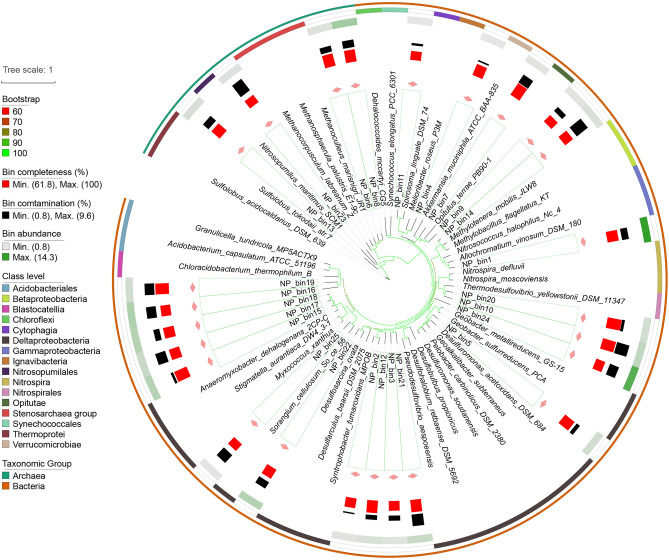
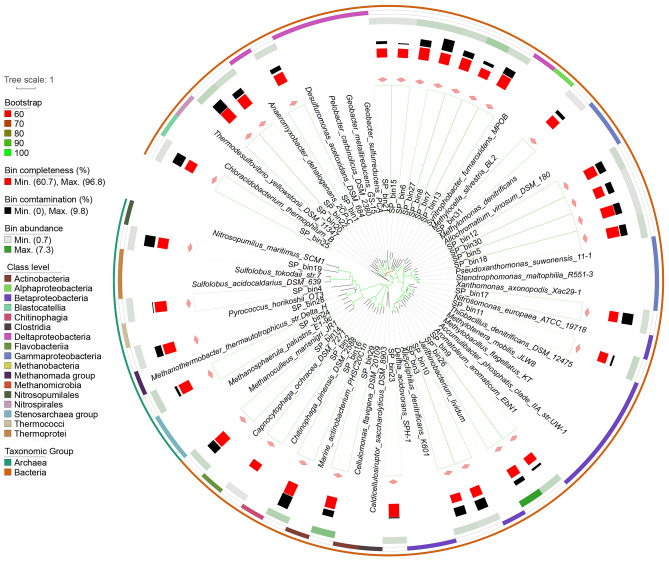
Phylogeny of metagenome-assembled genomes (MAGs)
The 56 retrieved MAGs (25 from NP and 31 from SP) could be assigned to more than Ten phyla, including one archaeal phylum that lacks cultivated representatives (Table S1 in Supplementary file 2). For NP metagenomes, 11 bins were assigned to the class Deltaproteobacteria, which was identified as the dominant taxon with a total bin abundance of 27.8 (the number of genome copies per million sequences). NP_bin1 was affiliated to Gammaproteobacteria with a bin abundance of 14.3. Two archaeal bins (NP_bin6 and NP_8) were assigned to the class Methanomicrobia with a total bin abundance of 7.4. NP_bin13 was affiliated to the order Methanomethylicales, with bin abundance of 1.9 (Fig. 3). In the SP metagenomes, the total abundances of bins affiliated to Betaproteobacteria (six bins), Deltaproteobacteria (eight bins) and Gammaproteobacteria (five bins) were 15.5, 13.6 and 5.9, respectively. SP_bin26 assigned to the order Burkholderiales was the most abundant bin (7.3). For archaea, four bins were affilitaed to the phyla Euryarchaeota, Thaumarchaeota and Candidatus Verstraetearchaeota. Phylogenetic analysis indicated that the dominant archaeal bin (SP_bin24, bin abundance 1.8) was affiliated to the genus Methanoculleus (Fig. 4).
Fig. 3.
Phylogenetic characterization of 25 moderate/high-quality MAGs recovered from sediments without submerged plants. The phylogeny was generated from orthology proteins using Orthofinder software. 63 microbial genomes using IQ-TREE with a best fit LG + F + R9 model using the Bayesian Information Criterion (BIC). Bootstraps were based on 1000 replicated trees. The alignment of 63 genomes was generated using MAFFT
Fig. 4.
Phylogenetic characterization of 31 moderate/high-quality MAGs recovered from sediments with submerged plants. The phylogeny was generated from orthology proteins using Orthofinder software. 66 microbial genomes using IQ-TREE with a best fit LG + F + R8 model using the Bayesian Information Criterion (BIC). Bootstraps were based on 1000 replicated trees. The alignment of 66 genomes was generated using MAFFT
To improve understanding of the uncultured archaeal phylum Verstraetearchaeota, a phylogenetic analysis of a potentially divergent cluster of mcrA genes from these Verstraetearchaeota genomes was conducted (Fig. S3 in Supplementary file 2). Results showed that sequences obtained from this study clustered with natural environmental clones. For example, mcrA from MAG NP_bin13 was close to sequences from hot spring sediments, and mcrA from MAG SP_bin4 clustered with sequences from freshwater spring sediments.
Discussion
Due to the prevalence of macrophytes in wetland ecosystems, understanding the influence of aquatic plants on the functional potential of microbial communities is becoming a hot topic in ecology and environmental science. In this study, detailed metagenomic analyses of microbial genes involved in the nitrogen cycle, sulfur cycle and methanogenesis affected by V. natans in urban wetlands of the Xixi National Wetland Park were performed. Our results showed that V. natans increased the relative abundances of most functional genes involved in the transformation of carbon, nitrogen and sulfur in the wetland sediments. Specifically, the relative abundances of functional genes involved in nitrogen fixation (nifD/H/K/W), assimilatory nitrate reduction (nasA and nirA), denitrification (nirK/S and nosZ), assimilatory sulfate reduction (cysD/H/J/N/Q and sir), and sulfur oxidation (glpE, soeA, sqr and sseA) in sediments with V. natans were all significantly higher than in unvegetated wetland sediments. Additionally, the recovered and annotated methanogen MAGs, including uncultured Verstraetearchaeota, indicated that hydrogenotrophic, acetoclastic, and methylotrophic methanogenesis may simultaneously contribute to methane production and therefore play important roles in carbon cycling in urban wetlands.
Submerged plants in wetland ecosystems can create aerobic microhabitats and provide rich nutrients in the rhizosphere (Srivastava et al. 2017), which facilitates the growth of specialized microorganisms and significantly affects the microbial mediation of biogeochemical cycling (Kaokniffin et al. 2010; Liu et al. 2018). We were able to annotate almost all nitrogen cycling genes in our wetland sediments, suggesting the presence of an active nitrogen cycling process in the Xixi National Wetland Park, which is consistent with a recent study (Liu et al. 2018). Moreover, many genes involved in nitrogen fixation were enriched in SP, suggesting that the potential nitrogen sink capacity of sediments was enhanced by V. natans. The gene abundances of ureA/C that catalyzes the conversion of organic nitrogen to ammonia nitrogen were significantly higher in SP than in NP sediments. The enhancement of the ammonification process is important for organic nitrogen removal and nitrogen cycling. The higher abundances of the genes nasA and nirA in SP sediments increased the transformation of NO3− to NH4+. Also, the abundances of nirS/K genes were significantly higher in SP than in NP sediment, which is consistent with a previous study on oligotrophic shallow lakes (Vilacosta et al. 2016). Furthermore, the denitrification process was not limited by carbon in vegetated sediments due to plant root exudates or the decay of plant tissue, both of which provide sufficient carbon sources for microorganisms (Vilacosta et al. 2016). The abundance of the nosZ gene was significantly higher in the vegetated than the unvegetated sediments, indicating that submerged plants may reduce N2O production from wetlands. However, García-Lledó et al. (2011) showed that Phragmites and Typha have the potential for high N2O emissions in vegetated sediments. Therefore, it is necessary to assess the effect of vegetation type on the production of N2O in wetlands, and more studies are needed to understand the relationship between wetlands and greenhouse gas emissions.
Although the concentration of sulfur compounds (e.g., sulfate and sulphide) in freshwater wetlands (e.g., lakes and ponds) is generally lower than in oceans, the sulfur cycle is still active in such habitats (Holmer and Storkholm 2001). Our studies detected almost all functional genes involved in the sulfur cycle in freshwater wetland sediments, suggesting that sulfur oxidizers and sulfate reducers are widespread in freshwater habitats. Sulfide oxidation is one of the important processes in the sulfur cycle and its metabolic pathway, which involves the genes dsrA/B/E/F/H and dsrC, is considered to be the reverse of the sulfite reduction pathway (Bell et al. 2020). The dsrD gene detected herein can facilitate sulfite reduction (Rabus et al. 2015). The lower abundance of the dsrM gene (which as a similar function to dsrD), detected in SP sediments indicates that submerged plants may weaken the sulfite reduction function. Additionally, the genes encoding sulfite dehydrogenase (quinone) subunit (soeA) and sulfide-quinone oxidoreductase (sqr) had higher abundances in SP than NP sediments, indicating that the conversion of sulfite to sulfate and oxidation of sulfide to elemental sulfur may be enhanced in the SP sediments. Such changes in biochemical processes could be attributed to two phenomena: (i) the aerobic environment around the roots of submerged plants could inhibit the reduction of sulfite to sulfide (Allen et al. 2002); (ii) inorganic sulfides are more cytotoxic than other sulfur compounds, so submerged plants may stimulate microbes to detoxify sulfide (Marcia et al. 2009; Xia et al. 2017). Sulfur is an essential element to maintain cell viability so sulfur compounds are synthesized through assimilation (Takahashi et al. 2011). Microorganisms play key roles in promoting sulfur uptake in plants by transforming organosulfur (Kertesz and Mirleau 2004). Cordovez et al. (2018) found that the dimethyl trisulfide released by root-associated Microbacterium promoted Arabidopsis growth via modulation of sulfur metabolism. These factors may partly explain why the abundances of many genes (such as cysD/H/J/N/Q and sir) involved in assimilatory sulfate reduction were significantly higher in SP than NP sediments.
Methanogenesis is one of the important parts of the carbon cycle in wetlands, and the annual methane release from wetlands accounts for more than 20% of global methane emissions (Laanbroek 2010). We detected hydrogenotrophic, acetoclastic and methylotrophic methanogenesis in the sediments, suggesting these metabolic pathways contributed to methane release in urban wetlands. However, when methyl substances are limiting, acetoclastic methanogens usually dominate methane production in freshwater wetlands (Lyu et al. 2018). The MAGs of NP_bin6 and SP_bin28 (Methanomicrobiales) have all the genes necessary for the utilization of acetate, however a previous study suggested that acetoclastic methanogens were only affiliated to the Methanosarcinales group (Borrel et al. 2011). A possible explanation for this inconsistency is that genes regulating the acetoclastic methanogenesis from non-Methanosarcinales taxa are not expressed.
Methanomicrobiales and Methanobacteriaceae were the dominant methanogenic archaean lineages in the wetland sediments, which is consistent with previous sequence-based studies of amplicons in freshwater wetlands (Lin et al. 2015; Liu et al. 2012). Furthermore, SP_bin28 (Methanobacteriaceae) harbored a large number of electron transporters (e.g., ABC-type transporters) encoded in their genomes, which could help microbial adaptation to low substrate (H2) environments (Browne et al. 2017). Notably, SP_bin4 (Methanomethylicales) with obligate H2-dependent methylotrophic methanogenesis belongs to the archaeal phylum Candidatus Verstraetearchaeota (Vanwonterghem et al. 2016). The near-complete genomes of Methanomethylicales from the freshwater wetland sediments were analyzed in current study. These genomes harbor the genes for heterodisulfide reductase (Hdr)/[Ni-Fe] hydrogenase complex for heterodisulfide coenzyme B coenzyme M (CoB-S-SCoM) reduction and H2 oxidation (Zhang et al. 2020).
The abundance of nirS/K and mcrA2 genes was higher in the SP than the NP sediments, suggesting a promotion of nitrite-dependent anaerobic methane oxidation (Hu et al. 2014). At the same time, the accumulation of plant-derived carbon sources may have given rise to a relatively high C:S ratio (Chen et al. 2014), which decreased the electron transfer between anaerobic methanotrophic archaea and sulfate-reducing bacteria (Wegener et al. 2015), finally resulting in poor methane oxidation. In addition, the presence of plants had different effects on the production of different greenhouse gases due to microbial processes. For example, the abundance of the nosZ gene, which encodes nitrous oxide reductase, increased in sediments with V. natans and may promote the conversion of N2O to N2. However, the abundance of mcrA2 in SP was slightly higher than in NP, which may favor methane production. Therefore, the impacts of vegetation on greenhouse gas production should also be considered when designing wetlands for ecological restoration. From the perspective of environmental protection, molecular markers, such as microbial functional genes, could provide useful information for improving the ecological evaluation of wetland ecosystems.
In summary, sedimental microorganisms from the Xixi National Wetland Park wetlands have strong biogeochemical potential, including microbially mediated methane, nitrogen and sulfur cycling. Vegetation significantly increases the abundance of functional genes involved in sedimental nitrogen fixation, assimilatory nitrate reduction, denitrification, assimilatory sulfate reduction, and sulfur oxidation. Moreover, we found a suitable C:N ratio was important for maintaining stable microbial community structure and function in wetland ecosystems. Additionally, hydrogenotrophic, acetoclastic, and methylotrophic methanogenesis may contribute significantly to the production of methane in wetland sediments. This study expands our understanding of functional diversity of urban wetland microbial communities and the effects of aquatic plants on them, which provides a basis for further mining of wetland microbial "dark matter".
Materials and methods
Study area and sampling procedures
All samples involved in this study were collected from the Xixi National Wetland Park (30°15′ ~ 30°17′ N and 120°1′ ~ 120°6′ E) in Hangzhou, China, which is the oldest urban wetland park in China and has a total area of 11.5 km2. The vegetation not only enhances the scenic amenity but also provides water purification functions in the wetlands. So, this is an excellent natural laboratory to study changes and adaptations of microbial functions in biogeochemical cycling. To reveal the effects of submerged plants on microbial functional genes and the corresponding changes in wetland ecosystem functioning, two types of sediment were investigated: five sites with submerged plants, primarily V. natans (SP) and five sites with no plants (NP). The upper layer of sediments (0–10 cm) was collected using a Peterson grab sampler in May 2018. All the collected samples were transported in a cooler box to the laboratory for further analysis.
Chemical measurements
After drying the sediments at 30 °C until they reached a constant weight, chemical characteristics including total nitrogen (TN) and total carbon (TC) were determined with an elemental analyzer (EA3000, EuroVector, German). Total phosphorus (TP) was measured by an inductively coupled plasma spectrometry (ICP) instrument (Prodigy7, Leeman Labs, USA). Conductivity and pH were determined as previously described (Wang et al. 2020). All samples were analyzed in three replicates (Table S2 in Supplementary file 2).
DNA extraction and purification
After removing impurities (e.g., plant roots and grit) from the sediment samples, microbial community DNA was extracted using a freeze-grinding plus sodium dodecyl sulfate (SDS) lysis method as described previously (Zhou et al. 1996), followed by purification using the PowerSoil DNA Extraction kit (MoBio Laboratories, CA, USA) according to manufacturer’s protocol. The extracted DNA was dissolved in 50 μl elution buffer. DNA concentrations were determined using a NanoDrop-2000 fluorospectrometer (Thermo Fisher Scientific, MA, USA). All DNA samples were stored at − 80 °C until further processing.
Shot-gun metagenome sequencing and analysis
For each sample, 1 μg of genomic DNA was used for library construction using VAHTSTM Universal DNA library Prep Kit for Illimina® V3 (ND607) and then sequenced on an Illumina NextSeq 550 platform (paired-end 150 bp). A total of 265.4 Gb raw reads generated for ten samples were qualified using the Read_qc module in metaWRAP, which is a flexible pipeline for genome-resolved metagenomic data analysis (Uritskiy et al. 2018). Only good quality reads (197.9 Gb) were used for downstream analysis (Supplementary file 3). These were annotated with biological functions (i.e., methane, nitrogen and sulfur cycling) using the Diamond tool (Buchfink et al. 2015) in Kyoto Encyclopedia of Genes and Genomes (KEGG), NCycDB (Tu et al. 2019) and ScycDB (Yu et al. 2021). Reads targeted by databases were extracted and used to build a relative abundance functional profile. A function for random subsampling each sample at the lowest sequencing depth was included (Tu et al. 2019).
Assembly, binning and annotation
The resulting sequences were put into metaWRAP for metagenomic assembly and binning. Briefly, the qualified reads were assembled using MEGAHIT with default parameters (Li et al. 2016). The resulting contigs are listed in Table S4 in Supplementary file 2. Binning of the assembled metagenome was computed and refined using MaxBin2 (Wu et al. 2016) and metaBAT2 (Kang et al. 2015). Levels of completion and contamination of all the bins were evaluated with CheckM (Parks et al. 2015). Bins with completion of > 60% and contamination of < 10% were subjected to annotation of taxonomy using Taxator-tk (Chaumeil et al. 2019). The abundance of bins in each sample was estimated, and the Salmon software was used to quantify individual contigs (Patro et al. 2017). Gene functions of those bins assigned to archaea were annotated with methanogenesis using the KEGG server (BlastKOALA) (Kanehisa et al. 2016) and eggNOG-mapper (Huertacepas et al. 2017). The annotated information is listed in Supplementary file 5.
Phylogenetic analyses
The phylogenetic tree of MAGs was constructed using OrthoFinder algorithm software (Emms and Kelly 2015) and IQ-TREE (Nguyen et al. 2015). Briefly, reference genomes were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov) database. Each gene at the amino acid sequence level was aligned using the MAFFT software integrated into OrthoFinder (Katoh and Standley 2014) and the phylogenomic tree was generated using IQ-TREE with a best-fit model.
The methyl coenzyme M reductase alpha subunit encoded by mcrA catalyzes methanogenesis, so the mcrA gene was used as a marker gene for detecting methanogens (Evans et al. 2015). The phylogeny of mcrA was used to identify the placement of the Verstraetearchaeota mcrA. The retrieved mcrA sequences were aligned using ClustalW (Larkin et al. 2007) with all Verstraetearchaeota mcrA sequences downloaded from the NCBI and IMG-M (https://img.jgi.doe.gov/cgi-bin/m/main.cgi) databases on August 3, 2020. A phylogenetic tree for the mcrA gene was generated using IQ-tree with parameters: iqtree -m TIM2 + F + R5 -bb 1000 -redo -alrt 1000. The phylogenomic trees were constructed using iTOL (Letunic and Bork 2016).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Yinan Zhang and Dandan Hu at the Hangzhou Normal University for field sampling and laboratory assistance. This work was supported by the National Natural Science Foundation of China (92051120), the Science & Technology Basic Resources Investigation Program of China (2017FY100300), the Fundamental Research Funds for the Central Universities (19lgzd28), and the Sun Yat-sen University (project no. 18821107).
Author contributions
BW: conceptualization, methodology, investigation, software, writing—original draft. XZ: methodology, writing—review and editing. HZ: resources. XY: data curation, methodology. YL: methodology. XY: methodology. HY: methodology. RH: methodology. ZH: writing—review and editing. FX: data curation, writing—review and editing, funding acquisition. QY: conceptualization, writing—review and editing, funding acquisition.
Data availability
All the raw DNA sequences (metagenomic reads) are available in the Genome Sequence Archive (GSA) under project PRJCA003534, publicly accessible at https://bigd.big.ac.cn/gsa/.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
Animal and human rights statement
No human or animal materials were used in this study.
Contributor Information
Fanshu Xiao, Email: xiaofansh@mail.sysu.edu.cn.
Qingyun Yan, Email: yanqy6@mail.sysu.edu.cn.
References
- Allen WC, Hook PB, Biederman JA, Stein OR. Temperature and wetland plant species effects on wastewater treatment and root zone oxidation. J Environ Qual. 2002;31:1010–1016. doi: 10.2134/jeq2002.1010. [DOI] [PubMed] [Google Scholar]
- Bell E, Lamminmaki T, Alneberg J, Andersson AF, Qian C, Xiong W, Hettich RL, Frutschi M, Bernierlatmani R (2020) Active sulfur cycling in the terrestrial deep subsurface. ISME J:1260–1272 [DOI] [PMC free article] [PubMed]
- Bodelier PL, Stomp M, Santamaria L, Klaassen M, Laanbroek HJ. Animal–plant–microbe interactions: direct and indirect effects of swan foraging behaviour modulate methane cycling in temperate shallow wetlands. Oecologia. 2006;149:233–244. doi: 10.1007/s00442-006-0445-9. [DOI] [PubMed] [Google Scholar]
- Borrel G, Jézéquel D, Biderre-Petit C, Morel-Desrosiers N, Morel J-P, Peyret P, Fonty G, Lehours A-C. Production and consumption of methane in freshwater lake ecosystems. Res Microbiol. 2011;162:832–847. doi: 10.1016/j.resmic.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Browne P, Tamaki H, Kyrpides NC, Woyke T, Goodwin L, Imachi H, Brauer SL, Yavitt JB, Liu W, Zinder SH. Genomic composition and dynamics among Methanomicrobiales predict adaptation to contrasting environments. ISME J. 2017;11:87–99. doi: 10.1038/ismej.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Chaumeil P, Mussig AJ, Hugenholtz P, Parks DH (2019) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927 [DOI] [PMC free article] [PubMed]
- Chen Y, Wen Y, Zhou J, Tang Z, Li L, Zhou Q, Vymazal J. Effects of cattail biomass on sulfate removal and carbon sources competition in subsurface-flow constructed wetlands treating secondary effluent. Water Res. 2014;59:1–10. doi: 10.1016/j.watres.2014.03.077. [DOI] [PubMed] [Google Scholar]
- Cordovez V, Schop S, Hordijk K, de Boulois HD, Coppens F, Hanssen I, Raaijmakers JM, Carrion VJ. Priming of plant growth promotion by volatiles of root-associated Microbacterium spp. Appl Environ Microbiol. 2018;84:e01865. doi: 10.1128/AEM.01865-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilson EJ, Carson MA, Yakimovich KM, Osterholz H, Dittmar T, Gunn J, Mykytczuk N, Basiliko N, Tanentzap A. Climate-driven shifts in sediment chemistry enhance methane production in northern lakes. Nat Commun. 2018;9:1801. doi: 10.1038/s41467-018-04236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D, Kelly SL. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- Frigaard N-U, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol. 2008;54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- García-Lledó A, Vilar-Sanz A, Trias R, Hallin S, Bañeras L. Genetic potential for N2O emissions from the sediment of a free water surface constructed wetland. Water Res. 2011;45:5621–5632. doi: 10.1016/j.watres.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Grossart HP, Massana R, McMahon KD, Walsh DA. Linking metagenomics to aquatic microbial ecology and biogeochemical cycles. Limnol Oceanogr. 2020;65:S2–S20. doi: 10.1002/lno.11382. [DOI] [Google Scholar]
- Harrison JA, Maranger R, Alexander RB, Giblin AE, Jacinthe P, Mayorga E, Seitzinger SP, Sobota DJ, Wollheim WM. The regional and global significance of nitrogen removal in lakes and reservoirs. Biogeochemistry. 2009;93:143–157. doi: 10.1007/s10533-008-9272-x. [DOI] [Google Scholar]
- Heilman MA, Carlton RG. Methane oxidation associated with submersed vascular macrophytes and its impact on plant diffusive methane flux. Biogeochemistry. 2001;52:207–224. doi: 10.1023/A:1006427712846. [DOI] [Google Scholar]
- Holmer M, Storkholm P. Sulphate reduction and sulphur cycling in lake sediments: a review. Freshw Biol. 2001;46:431–451. doi: 10.1046/j.1365-2427.2001.00687.x. [DOI] [Google Scholar]
- Hu BL, Shen LD, Lian X, Zhu Q, Liu S, Huang Q, He ZF, Geng S, Cheng DQ, Lou LP, Xu XY, Zheng P, He YF. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci USA. 2014;111:4495–4500. doi: 10.1073/pnas.1318393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertacepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Zhou Y, Ji X, Li H, Zheng Z, Zhang J. Responses of leaf-associated biofilms on the submerged macrophyte Vallisneria natans during harmful algal blooms. Environ Pollut. 2019;246:819–826. doi: 10.1016/j.envpol.2018.12.081. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaokniffin J, Freyre DS, Balser TC. Methane dynamics across wetland plant species. Aquat Bot. 2010;93:107–113. doi: 10.1016/j.aquabot.2010.03.009. [DOI] [Google Scholar]
- Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- Kertesz MA, Mirleau P. The role of soil microbes in plant sulphur nutrition. J Exp Bot. 2004;55:1939–1945. doi: 10.1093/jxb/erh176. [DOI] [PubMed] [Google Scholar]
- Kofoed MVW, Stief P, Hauzmayer S, Schramm A, Herrmann M. Higher nitrate-reducer diversity in macrophyte-colonized compared to unvegetated freshwater sediment. Syst Appl Microbiol. 2012;35:465–472. doi: 10.1016/j.syapm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Laanbroek HJ. Methane emission from natural wetlands: interplay between emergent macrophytes and soil microbial processes. A mini-review Ann Bot. 2010;105:141–153. doi: 10.1093/aob/mcp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Luo R, Liu C, Leung C, Ting H, Sadakane K, Yamashita H, Lam T. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Lin Y, Liu D, Ding W, Kang H, Freeman C, Yuan J, Xiang J. Substrate sources regulate spatial variation of metabolically active methanogens from two contrasting freshwater wetlands. Appl Microbiol Biotechnol. 2015;99:10779–10791. doi: 10.1007/s00253-015-6912-7. [DOI] [PubMed] [Google Scholar]
- Liu D, Ding W, Jia Z, Cai Z. The impact of dissolved organic carbon on the spatial variability of methanogenic archaea communities in natural wetland ecosystems across China. Appl Microbiol Biotechnol. 2012;96:253–263. doi: 10.1007/s00253-011-3842-x. [DOI] [PubMed] [Google Scholar]
- Liu W, Jiang X, Zhang Q, Li F, Liu G. Has submerged vegetation loss altered sediment denitrification, N2O production, and denitrifying microbial communities in subtropical lakes? Glob Biogeochem Cycle. 2018;32:1195–1207. [Google Scholar]
- Lyu Z, Shao N, Akinyemi T, Whitman WB. Methanogenesis. Curr Biol. 2018;28:R727–R732. doi: 10.1016/j.cub.2018.05.021. [DOI] [PubMed] [Google Scholar]
- Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide: quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TS, Shin HW, Hanna R, Spafford DC. A review of epiphyte community development: surface interactions and settlement on seagrass. J Environ Biol. 2008;29:629–638. [PubMed] [Google Scholar]
- Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottosen LDM, Risgaard-Petersen N, Nielsen LP. Direct and indirect measurements of nitrification and denitrification in the rhizosphere of aquatic macrophytes. Aquat Microb Ecol. 1999;19:81–91. doi: 10.3354/ame019081. [DOI] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell A, Mikucki JA, Achberger AM, Alekhina IA, Barbante C, Christner BC, Ghosh D, Michaud AB, Mitchell AC, Priscu JC. Microbial sulfur transformations in sediments from Subglacial Lake Whillans. Front Microbiol. 2014;5:594. doi: 10.3389/fmicb.2014.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Wu Z, Liu B, Deng J, Fu G, He F. The restoration of aquatic macrophytes for improving water quality in a hypertrophic shallow lake in Hubei Province, China. Ecol Eng. 2001;18:147–156. doi: 10.1016/S0925-8574(01)00074-X. [DOI] [Google Scholar]
- Rabus R, Venceslau SS, Wohlbrand L, Voordouw G, Wall JD, Pereira IAC. A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv Microb Physiol. 2015;66:55–321. doi: 10.1016/bs.ampbs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Scheffer M, Hosper SH, Meijer ML, Moss B, Jeppesen E. Alternative equilibria in shallow lakes. Trends Ecol Evol. 1993;8:275–279. doi: 10.1016/0169-5347(93)90254-M. [DOI] [PubMed] [Google Scholar]
- Sims A, Zhang Y, Gajaraj S, Brown PB, Hu Z. Toward the development of microbial indicators for wetland assessment. Water Res. 2013;47:1711–1725. doi: 10.1016/j.watres.2013.01.023. [DOI] [PubMed] [Google Scholar]
- Srivastava J, Chandra H, Kalra SJS, Mishra P, Khan H, Yadav P. Plant-microbe interaction in aquatic system and their role in the management of water quality: a review. Appl Water Sci. 2017;7:1079–1090. doi: 10.1007/s13201-016-0415-2. [DOI] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- Tu Q, Lin L, Cheng L, Deng Y, He Z. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. BMC Bioinf. 2019;35:1040–1048. doi: 10.1093/bioinformatics/bty741. [DOI] [PubMed] [Google Scholar]
- Ugarelli K, Laas P, Stingl U. The microbial communities of leaves and roots associated with turtle grass (Thalassia testudinum) and manatee grass (Syringodium filliforme) are distinct from seawater and sediment communities, but are similar between species and sampling sites. Microorganisms. 2019;7:4. doi: 10.3390/microorganisms7010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah S, Zhang H, Heathwaite AL, Heppell C, Lansdown K, Binley A, Trimmer M. Influence of emergent vegetation on nitrate cycling in sediments of a groundwater-fed river. Biogeochemistry. 2014;118:121–134. doi: 10.1007/s10533-013-9909-2. [DOI] [Google Scholar]
- Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:1–13. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, Tyson GW. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol. 2016;1:16170. doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- Veraart AJ, De Bruijne WJJ, De Klein JJM, Peeters ETHM, Scheffer M. Effects of aquatic vegetation type on denitrification. Biogeochemistry. 2011;104:267–274. doi: 10.1007/s10533-010-9500-z. [DOI] [Google Scholar]
- Vilacosta M, Pulido C, Chappuis E, Calvino A, Casamayor EO, Gacia E. Macrophyte landscape modulates lake ecosystem-level nitrogen losses through tightly coupled plant-microbe interactions. Limnol Oceanogr. 2016;61:78–88. doi: 10.1002/lno.10209. [DOI] [Google Scholar]
- Wang C, Liu S, Jahan TE, Liu B, He F, Zhou Q, Wu Z. Short term succession of artificially restored submerged macrophytes and their impact on the sediment microbial community. Ecol Eng. 2017;103:50–58. doi: 10.1016/j.ecoleng.2017.02.030. [DOI] [Google Scholar]
- Wang B, Zheng X, Zhang H, Xiao F, Gu H, Zhang K, He Z, Liu X, Yan Q. Bacterial community responses to tourism development in the Xixi National Wetland Park, China. Sci Total Environ. 2020;720:137570–137580. doi: 10.1016/j.scitotenv.2020.137570. [DOI] [PubMed] [Google Scholar]
- Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature. 2015;526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- Wiessner A, Rahman K, Kuschk P, Kästner M, Jechorek M. Dynamics of sulphur compounds in horizontal sub-surface flow laboratory-scale constructed wetlands treating artificial sewage. Water Res. 2010;44:6175–6185. doi: 10.1016/j.watres.2010.07.044. [DOI] [PubMed] [Google Scholar]
- Wu YW, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. BMC Bioinf. 2016;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- Xia Y, Lü C, Hou N, Xin Y, Liu J, Liu H, Xun L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017;11:2754–2766. doi: 10.1038/ismej.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Chen C, Liu G, Liu W. Sediment nitrogen cycling rates and microbial abundance along a submerged vegetation gradient in a eutrophic lake. Sci Total Environ. 2018;616:899–907. doi: 10.1016/j.scitotenv.2017.10.230. [DOI] [PubMed] [Google Scholar]
- Yu X, Zhou J, Song W, Xu M, He Q, Peng Y, Tian Y, Wang C, Shu L, Wang S, Yan Q, Liu J, Tu Q, He Z. SCycDB: a curated functional gene database for metagenomic profiling of sulfur cycling pathways. Mol Ecol Resour. 2021;21:924–940. doi: 10.1111/1755-0998.13306. [DOI] [Google Scholar]
- Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- Zhang C-J, Pan J, Liu Y, Duan C-H, Li M. Genomic and transcriptomic insights into methanogenesis potential of novel methanogens from mangrove sediments. Microbiome. 2020;8:1–12. doi: 10.1186/s40168-020-00876-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Cao T, Ni L, He L, Yi C, Yuan C, Xie P. Improvement of water quality by sediment capping and re-vegetation with Vallisneria natans L.: a short-term investigation using an in situ enclosure experiment in Lake Erhai. China Ecol Eng. 2016;86:113–119. doi: 10.1016/j.ecoleng.2015.10.031. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw DNA sequences (metagenomic reads) are available in the Genome Sequence Archive (GSA) under project PRJCA003534, publicly accessible at https://bigd.big.ac.cn/gsa/.