Abstract
The isolation chip method (iChip) provides a novel approach for culturing previously uncultivable microorganisms; this method is currently limited by the user being unable to ensure single-cell loading within individual wells. To address this limitation, we integrated flow cytometry-based fluorescence-activated cell sorting with a modified iChip (FACS-iChip) to effectively mine microbial dark matter in soils. This method was used for paddy soils with the aim of mining uncultivable microorganisms and making preliminary comparisons between the cultured microorganisms and the bulk soil via 16S rRNA gene sequencing. Results showed that the FACS-iChip achieved a culture recovery rate of almost 40% and a culture retrieval rate of 25%. Although nearly 500 strains were cultured from 19 genera with 8 FACS-iChip plates, only six genera could be identified via 16S rRNA gene amplification. This result suggests that the FACS-iChip is capable of detecting strains in the currently dead spaces of PCR-based sequencing technology. We, therefore, conclude that the FACS-iChip system provides a highly efficient and readily available approach for microbial ‘dark matter’ mining.
Electronic supplementary material
The online version of this article (10.1007/s42995-020-00067-7) contains supplementary material, which is available to authorized users.
Keywords: FACS-iChip, Microbial ‘dark matter’, In situ cultivation, Single cell sorting
Introduction
Microorganisms are an important genetic-diversity resource bank that is currently underused due to the limitations of detection and classification technologies. Microorganisms need to be cultivated to clearly determine their physiological characteristics, such as metabolism, environmental responses and growth characteristics. However, sequencing of environmental DNA reveals that most microbial lineages do not have pure cultures (Lloyd et al. 2018; Parks et al. 2017; Rinke et al. 2013). In addition, many studies of bacterial communities depend on a single gene, the 16S small subunit ribosomal RNA (rRNA) gene (Nemergut et al. 2011), even though this technique is limited by factors, such as sequencing errors (Quince et al. 2011), short read lengths (Poretsky et al. 2014), poor universal full-length primers (Klindworth et al. 2013) and operational taxonomic unit assessment difficulties (Huse et al. 2010). The microbial species that have not yet been cultivated are defined as microbial ‘dark matter’ (Dodsworth et al. 2013; Rinke et al. 2013) and it is imperative that new techniques are developed to mine this potential resource.
A significant limitation for current technologies is that traditional culture methods can only recover 1–5% of the microbial population (Pham and Kim 2012). To address this limitation, a high-throughput in situ cultivation method, described as an isolation chip (iChip), was developed (Nichols et al. 2010). The iChip consists of multiple micro-diffusion chambers and membranes with pore diameters large enough to allow growth factors to enter but small enough to prevent cells from moving between the diffusion chambers (Kaeberlein et al. 2002). In terms of in situ culturing of microorganisms, existing experimental data show a culture recovery rate of 40% in sea water and 50% in soil when using the iChip technique (Nichols et al. 2010). However, because the current iChip system relies on diluting the cell suspension to capture an average of one cell per diffusion chamber (Berdy et al. 2017), each chamber can potentially contain no cells, a single cell or multiple cells.
As devices, such as the iChip, rely on the delivery of single organism, single-cell-sorting techniques play an important role in cultivating environmental samples. In addition to dilution methods, single cells can also be separated by flow cytometer, laser capture microdissection, manual cell picking and microfluidics (Gross et al. 2015). The applicability of cell sorting techniques for culturing is affected by factors, such as sample preparation, cell properties and operating techniques; a fluorescent-activated cell sorter (FACS) was selected for single cell isolation in our study. This flow cytometer has the capacity to sort fluorescently labeled cells from a mixed cell population (Adan et al. 2017; Wilkerson 2012) and is widely used in the medical, biological and material science fields. This instrument is popular in high-throughput methods for separating environmental samples into individual cells because of its high speed, high throughput and sorting ability, which are based on cell characteristics, such as particle size and fluorescence. This method is, therefore, an ideal candidate for improving the iChip process.
For this special issue, we introduce the FACS-iChip system for mining previously unculturable microorganisms by optimizing the efficiency of iChip culturing by combining it with a single-cell-sorting flow cytometer. This method can be used to achieve efficient mining of microbial ‘dark matter’.
Results
FACS-iChip culturing
FACS-iChip culturing was achieved using microorganisms from paddy soils sorted by a flow cytometer and membrane-sealed iChips incubated in root boxes. For preliminary verification of the method, sterile Luria Broth (LB) medium and dyed Escherichia coli were loaded into each iChip well using a flow cytometer and incubated for two days. Visible colonies were observed in 99% of wells after incubation, indicating that the influences of fluorescence and dyes on microorganisms were negligible, and cell loading was successful.
For method validation, living cell sorting was performed with a flow cytometer (Fig. 1b) and a single living cell was loaded into each well of the iChip plate. An iChip is a sealed device consisting of two membranes and a 384-well culture plate with LB medium in every well (Fig. 1c). iChips were then incubated in root boxes with liquid media for 30 days or solid media for 30 and 50 days of culturing (Fig. 1d). Visible colonies were counted after incubation (Table 1). In the iChips that used solid medium and were cultured for 30 days, there were on average 250 wells (n = 2) with obvious colonies; this method achieved a 65% culture recovery rate (number of visible colony wells/total wells). For the same medium at 50 days, 85 wells (average; n = 3) had visible colonies after incubation; only a 22% culture recovery rate was achieved. Subsequent colony enrichment of solid medium wells with LB medium produced an average of 52% (30 days; n = 2) and 34% of iChip wells (50 days; n = 3) that could be subcultured. Liquid-medium iChips incubated for 50 days had a 35% culture recovery rate and 44% culture retrieval rate (average; n = 3; number of wells that can be subcultured/number of visible colony wells). In total, FACS-iChip achieved a culture recovery rate of almost 40% and a culture retrieval rate of 25%.
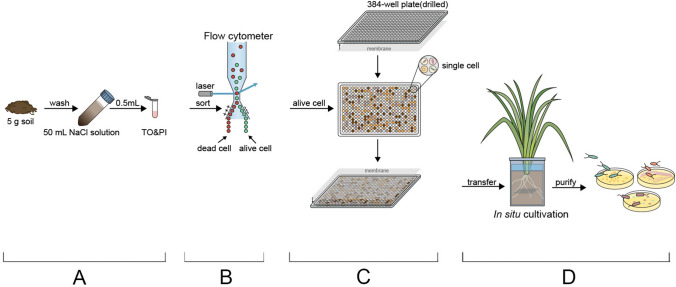
Fig. 1.
Schematic diagram of iChip operation. A Sample preparation. Cells were eluted from soil and dyed by TO (thiazole orange) and PI (propidium iodide). B Cell sorting. Dyed microbes were sorted (flow cytometer) into alive and dead cells. C Cell loading. Alive single cells were loaded into the drilled iChip and the whole device was sealed by membranes. D IChip culturing. The device was incubated in a root box for several days and was taken out to purify colonies
Table 1.
Recovery rate of FACS-iChip
| Number | Media type | Culture time | Wells with visible colonies | Culture recovery rate (%) | Colony count | Culture retrieval rate (%) | |
|---|---|---|---|---|---|---|---|
| 1 | Solid medium (LB) | 30 days | 257 | 66.93 | 139 | 54.09 | |
| 2 | Solid medium (LB) | 30 days | 242 | 63.02 | 123 | 50.83 | |
| 3 | Liquid medium (LB) | 50 days | 114 | 29.69 | 16 | 14.04 | |
| 4 | Liquid medium (LB) | 50 days | 152 | 39.58 | 57 | 37.50 | |
| 5 | Liquid medium (LB) | 50 days | 133 | 34.64 | 60 | 45.11 | |
| 6 | Solid medium (LB) | 50 days | 84 | 21.88 | 31 | 36.90 | |
| 7 | Solid medium (LB) | 50 days | 95 | 24.74 | 31 | 32.63 | |
| 8 | Solid medium (LB) | 50 days | 74 | 19.27 | 26 | 35.13 | |
Culture recovery rate means the ratio of number of visible colony wells to total wells. Culture retrieval rate means the ratio of number of wells that can be subcultured to number of visible colony wells. The LB medium was used in both preliminary culture and subculture. And the cloudy liquid was regarded as visible colonies in liquid medium
16S rRNA gene sequencing and taxonomic composition of FACS-iChip microbe mining
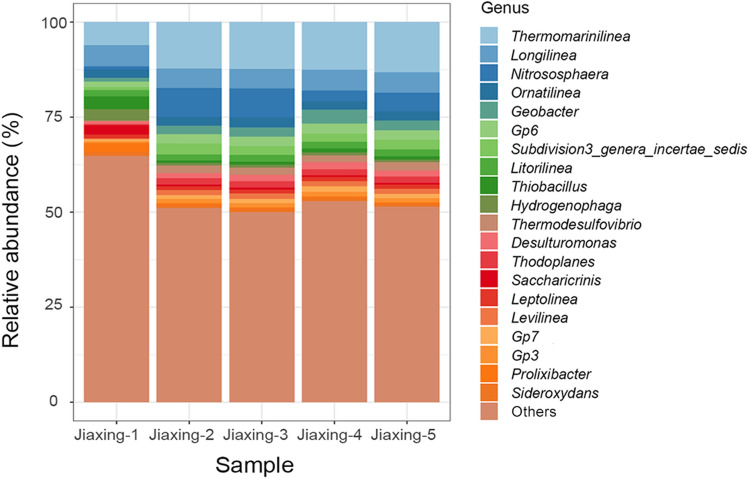
To compare the culture results against actual soil samples, five replicate DNA extraction samples from the test soil were analysed with 16S rRNA gene sequencing (Fig. 2). There was a large difference between culturing and soil at the genus level analysis with a total of 790 genera obtained in the test soils, dominated by Thermomarinilinea, Longilinea, Nitrososphaera, Ornatilinea and Geobacter.
Fig. 2.
Taxonomic composition (genus) for the five paddy soils displayed as relative abundance
In the culture results, a total of 19 genera (46 species) were observed from the cultivation of well samples (Table S1). Microorganisms were dominated by Bacillales, Aeromonas, Pseudomonas and Achromobacter; of these, only six genera were detected with 16S rRNA (Table 2). Interestingly, the ten most abundant genera detected by 16S rRNA gene sequencing were not observed in culture results.
Table 2.
Presence of cultured microorganisms in the 16S rRNA sequencing data (genus level)
| Number | Kingdom | Phylum | Class | Order | Family | Genus | Presence in sequencing resultsa |
|---|---|---|---|---|---|---|---|
| 1 | Bacteria | Proteobacteria | Gammaproteobacteria | Aeromonadales | Aeromonadaceae | Aeromonas | Yes |
| 2 | Bacteria | Actinobacteria | Actinomycetales | Micrococcineae | Microbacteriaceae | Microbacterium | No |
| 3 | Bacteria | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | Yes |
| 4 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Citrobacter | No |
| 5 | Bacteria | Firmicutes | Bacilli | Bacillales | Paenibacillaceae | Brevibacillus | Yes |
| 6 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Yersiniaceae | Serratia | No |
| 7 | Bacteria | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Delftia | No |
| 8 | Bacteria | Proteobacteria | Betaproteobacteria | Rhodocyclales | Rhodocyclaceae | Azospira | Yes |
| 9 | Bacteria | Firmicutes | Bacilli | Bacillales | Bacillaceae | Lysinibacillus | No |
| 10 | Bacteria | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas | No |
| 11 | Bacteria | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Comamonas | No |
| 12 | Bacteria | Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Achromobacter | Yes |
| 13 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | No |
| 14 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Enterobacter | Yes |
| 15 | Bacteria | Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | Sulfurospirillum | No |
| 16 | Bacteria | Firmicutes | Bacilli | Bacillales | Bacillaceae | Fictibacillus | No |
| 17 | Bacteria | Proteobacteria | Gamma Proteobacteria | Enterobacteriales | Erwiniaceae | Pantoea | No |
| 18 | Bacteria | Actinobacteria | Actinomycetales | Micrococcales | Micrococcaceae | Micrococcus | No |
| 19 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Atlantiacter | No |
The 19 genera listed in this table are the results obtained by cultivation
aThe genera that appeared in the 16S rRNA sequencing results were marked as “Yes”, otherwise “No”. Only six genera were appeared in sequencing results
Discussion
This study presents the results of method validation for a modified iChip culturing process, which we have called FACS-iChip. The original iChip from Nichols et al. (2010) included the central plate and the two symmetrical top and bottom plates, which were machined from blocks of hydrophobic plastic polyoxymethylene. In the method used here, 384-well plates were used, which are easy to obtain from the laboratory. Compared with the original iChip described by Nichols et al. (2010), which takes time to fix and disassemble, our iChip attaches the membranes to the sides of the plate to form a sealed device. After culturing is completed, the membranes can be directly opened for subsequent operation. The materials of the device are easy to obtain and it is easy to make and disassemble. It provides favorable conditions for batch operation.
When compared to the conventional iChip technique, the method proposed here used flow cytometer to more accurately load one cell per well. Two types of light scattering, forward scatter (FSC) and side scatter (SSC) occur when the laser in a FACS optical platform strikes the cells (Wilkerson 2012). FSC, which is suitable for sorting cell size and SSC, which is proportional to internal cell complexity, are used to sort fluorescent-labeled cells (Adan et al. 2017; Reggeti et al. 2011). Therefore, because of its optical platform combined with the principle of electrostatic deflection of charged droplets, FACS can achieve the separation of fluorescent-labeled cells in mixed cell populations. The flow cytometer works efficiently, and can load 384-well plates on average every 15 min, which can meet the needs of loading multiple times a day. Because the cell injected through the flow cytometer is single, the resultant growing colony is also single colony, which is equivalent to directly completing the purification of the colony. This method simplifies the step throughout the process of culturing cells. However, FACS also has its limitations. Different types of flow cytometers have different capabilities for sorting cell sizes and small cell sizes may not be separated from noise. Regular maintenance of the machine is also particularly important to prevent contamination during the sorting process. The choices of dye types or methods that can increase the fluorescence emission of dyes, such as an antireflective array (Xu et al. 2019), also have corresponding effects on cell sorting. In practice, if multiple colonies appear in a well, it means that contamination occured during the operation. At this time, the contaminated colonies can be repurified or discarded according to actual needs.
Solid media recovery rates were significantly higher than the liquid media (Table 1), possibly due to the slow growth of bacteria in an anaerobic environment (liquid media). It is also possible that, as this experiment used LB media, there was an effect of medium type. Therefore, changing the type of medium may lead to higher recovery rates and different microbial species, particularly as most microbial species cannot grow on conventional media (Rappe and Giovannoni 2003; Staley and Konopka 1985). Microorganisms with poor growth are, therefore, more likely to be new species that have not been cultivated in traditional media before. Incubation length also led to different culture results and, when compared with 30 day results, the recovery rate of 50 days was greatly reduced, possibly due to nutrient depletion of the medium restricting microorganism growth amplification. Cultivation time is related to nutrient richness in the environment with one to four weeks of incubation typically (Berdy et al. 2017). A culture recovery rate of 40% in sea water and 50% in soil has been reported by Nichols et al. (2010), with microbial recovery being calculated as the percentage of cells forming microcolonies. In our experiment, we also achieved culture recovery rate of almost 40% and the recovery rate of the solid medium, with an appropriate cultivation time, was even higher than this value. The colonies obtained by iChip were subcultured 1–2 times for domestication to obtain cultivable strain resources in the laboratory. During the domestication process, some colonies could not be cultured, and our results also reflected this problem. We suggest that as only visible colonies were selected for subsequent processing, a higher culture recovery might be achieved by culturing all well contents.
The FACS-iChip taxonomical results were significantly different to that obtained from bulk soil, and included many novel species that could not be identified (Table 2). This difference is possibly due to known biases and errors in 16S rRNA gene sequencing, such as amplification primers, being unable to fully cover the range of environmental microbial species (Klindworth et al. 2013); Clustering operational taxonomic units (OTUs) at a 97% similarity threshold (Westcott and Schloss 2017) may also be insufficient to approximate species (Edgar 2018); the threshold may need to be increased to at least 99%. Furthermore, incorrect predictions of 16S gene copy numbers cause noise in community profiles due to the widespread use of taxa estimations based on the relative read counts (Louca et al. 2018). Regardless of these limitations, FACS-iChip culturing allows the detection of novel species.
This proposed method is a prototype for the study of microorganism culturing, which optimizes the function of existing methods. The application of flow cytometer successfully achieved accurate single cell sorting. Further studies should explore the experimental conditions to achieve more representative or higher-purpose resource mining.
Materials and methods
Reagents and materials
Reagents for this method include: 0.9% physiological saline, TO (thiazole orange)/PI (propidium iodide) dyes, LB medium, agar powder, DNA extraction kit (MP FastDNA® Spin Kit for Soil, USA), PCR enzyme (Takara MightyAmp, Takara Tks Gflex, Japan), 384-hole plates, membranes (0.22 µm, poly tetra fluoroethylene), sterile silica gel, and root box.
The paddy soil used in this experiment was collected in Xiuzhou District, Jiaxing City, Zhejiang Province (30°50′8.74″ N, 120°43 3.68″ E), and the sampling depth was 0–20 cm. This study used paddy soil cells stained with TO and PI dyes (Fig. 1a). Cell sorting (live or dead) was performed using FACS flow cytometer after which an individual living cell was injected into individual iChip plate wells (Fig. 1b). An iChip is a sealed device consisting of two membranes and a 384-well culture plate (Fig. 1c). Membrane pore size is sufficiently large for the chemical diffusion while being small enough to prevent the cells moving from the wells.
IChip manufacture and single cell sorting
384-hole plates were sterilized with ultraviolet radiation and the bottom of the channel was drilled to form 384-hole culture chambers. An appropriate amount of silica gel was applied to the surface of the plate to act as adhesive, after which a sterile membrane (0.22 µm, poly tetra fluoroethylene) was attached to one side of each plate. The silica gel was allowed to completely solidify before further processing. 5 g paddy soil was diluted to 50 ml with 0.9% NaCl solution and vortexed for 5 min after which samples were centrifuged at 130 g for 5 min. The supernatant was filtered (300-mesh filter) to remove particulate matter (Bressan et al. 2015). This suspension was diluted to 106 cells per ml and 0.5 ml was added to a flow tube after which 5 μl of both TO and PI solutions was added (Fig. 1a). Samples were mixed well and the dye was allowed to develop 15 min in the dark. Cells were sorted (live or dead) using FACS flow cytometer and a single live cell was loaded into each iChip well (Fig. 1b). 30 μl LB medium for solid medium and 60 μl (the well was full) LB medium for liquid medium were added to each well after sorting after which the second membrane was attached with sterile silica gel to seal the plate (Fig. 1c). The silica gel was again allowed to completely solidify before further processing.
In situ culturing
IChips were incubated in root boxes. These boxes were half filled (volume) with the aforementioned paddy soil with an additional quarter of the root box filled with water to simulate a paddy soil under flood conditions. Soils were flooded for 3–5 days to activate soil microorganisms. After microbial activation, the assembled iChip was placed in the approximate middle of the root box and covered with paddy soil (Fig. 1d). Eight root boxes were prepared for this experiment; incubation conditions were maintained at 28 °C for 30–50 days.
IChip retrieval and enrichment purification
The iChip was gently removed from the soil and carefully washed with sterile water to remove environmental contaminants. Individual agar blocks (LB solid medium) were used to culture microorganisms from colonized wells. In the event of multiple strains growing on the same plate, purification was achieved using iterative culturing. Single colonies were purified to obtain a large number of monoclonal strains and assessed identified with 16S rRNA gene sequencing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (41991334), the Zhejiang Provincial Natural Science Foundation of China (LD19D060001, LQ20C030006) and the China Postdoctoral Science Foundation (2019M652097).
Author contributions
BM, RX and JX designed this study; HL and YW performed the experiments and HL wrote the manuscipt; ES revised the manuscript; SY drew the figure 1; all authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Animal and human rights statement
No animal or human materials were used in this study.
Footnotes
The original online version of this article was revised: The presentation of Table 2 was incorrect.
SPECIAL TOPIC: Cultivation of the uncultured microorganisms.
Haoze Liu, Ran Xue and Yiling Wang contributed equally.
Change history
11/11/2020
A Correction to this paper has been published: 10.1007/s42995-020-00079-3
References
- Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- Berdy B, Spoering AL, Ling LL, Epstein SS. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat Protoc. 2017;12:2232–2242. doi: 10.1038/nprot.2017.074. [DOI] [PubMed] [Google Scholar]
- Bressan M, Gattin IT, Desaire S, Castel L, Gangneux C, Laval K. A rapid flow cytometry method to assess bacterial abundance in agricultural soil. Appl Soil Ecol. 2015;88:60–68. doi: 10.1016/j.apsoil.2014.12.007. [DOI] [Google Scholar]
- Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, Chain PSG, Scholz MB, Lo CC, Raymond J, Quake SR, Hedlund BP. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Commun. 2013;4:1854. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 2018;34:2371–2375. doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P. Technologies for single-cell isolation. Int J Mol Sci. 2015;16:16897–16919. doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Steen AD, Ladau J, Yin JQ, Crosby L. Phylogenetically novel uncultured microbial cells dominate Earth microbiomes. Msystems. 2018;3:e00055–e118. doi: 10.1128/mSystems.00055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Doebeli M, Parfrey LW. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome. 2018;6:41. doi: 10.1186/s40168-018-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, Fierer N, Townsend AR, Cleveland CC, Stanish L, Knight R. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, Epstein SS. Use of Ichip for high-throughput in situ cultivation of “Uncultivable” microbial species. Appl Environ Microb. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- Pham VHT, Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30:475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Poretsky R, Rodriguez-R LM, Luo CW, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9:e93827. doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinform. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Reggeti F, Bienzle D. Flow Cytometry in veterinary oncology. Vet Pathol. 2011;48:223–235. doi: 10.1177/0300985810379435. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, SwanBK GEA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. Msphere. 2017;2:e00073–e117. doi: 10.1128/mSphereDirect.00073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MJ. Principles and applications of flow cytometry and cell sorting in companion animal medicine. Vet Clin North Am Small Anim Pract. 2012;42:53–71. doi: 10.1016/j.cvsm.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Xu H, Liu L, Teng F, Lu N. Emission enhancement of fluorescent molecules by antireflective arrays. Research. 2019;2019:3495841. doi: 10.34133/2019/3495841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.