Abstract
The vertebrate liver is regarded as an organ essential to the regulation of immunity and inflammation as well as being central to the metabolism of nutrients. Here, we discuss the functions that the hepatic cecum of amphioxus plays in the regulation of immunity and inflammation, and the molecular basis of this. It is apparent that the hepatic cecum performs important roles in the immunity of amphioxus including immune surveillance, clearance of pathogens and acute phase response. Therefore, the hepatic cecum, like the vertebrate liver, is an organ functioning as a key integrator of immunity in amphioxus.
Keywords: Acute phase response, Amphioxus, Hepatic cecum, Immunity, Inflammation
Introduction
Amphioxus, a basal chordate, is the best available proxy for the proximate invertebrate ancestor of vertebrates. It has a vertebrate-like body plan including a circulatory system with an organization similar to that of vertebrates (Moller and Philpott 1973). However, amphioxus lacks lymphoid organs and has no free circulating blood cells (Moller and Philpott 1973; Yuan et al. 2015a). Instead, its digestive organs, including the gill, intestine and hepatic cecum, are regarded as sites of continuous immunological interaction and thus act as the major player of defense against infection (Gao and Zhang 2018; Huang et al. 2011b; Liao et al. 2017; Tao and Xu 2016). The hepatic cecum not only serves as a physical barrier but is also an integrator of the immune system. Numerous studies on morphology, embryology and molecular biology show that the hepatic cecum is homologous to the precursors of vertebrate liver (Guo et al. 2009; Müller 1844; Wang and Zhang 2011; Welsch 1975). The liver is the largest internal organ in vertebrates and is essential to the regulation of immunity and inflammation in addition to being central to the metabolism of nutrients and the clearance of toxins (Kubes and Jenne 2018). Similarly, the hepatic cecum of amphioxus plays a vital role in both metabolism and immunity. An array of liver-specific genes/proteins are predominantly expressed in the hepatic cecum, including the glutathione-S-transferase gene (Fan et al. 2007), alanine aminotransferase gene (Jing and Zhang 2011), fibrinogen-related protein gene (Fan et al. 2008), plasminogen-like gene (Liu and Zhang 2009), transferrin-like gene (Liu et al. 2009), C/EBPα/β gene (Wang et al. 2009), IGF-like gene (Guo et al. 2009), G6pase (Wang et al. 2015), complement system genes (Gao et al. 2014; He et al. 2008), and vitellogenin (Han et al. 2006). Most of these genes/proteins are linked to the immunity of amphioxus. Below, we discuss the roles the hepatic cecum plays in the immunity of amphioxus.
Gross anatomy and function
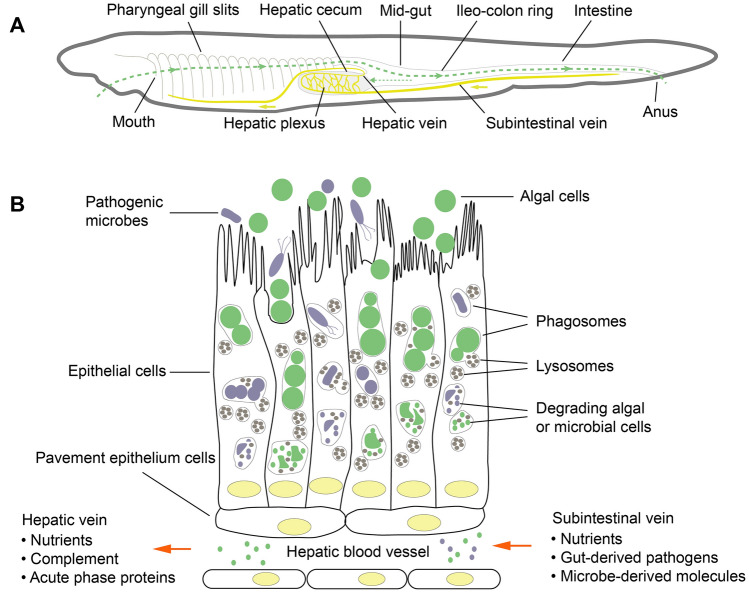
The hepatic cecum of amphioxus is a pouch that protrudes forward as an out-pocketing of the mid-gut and extends along the right side of the posterior part of the pharynx. The major component of the hepatic cecum is the pseudo-stratified columnar ciliated epithelium, in which the epithelial cells have functional features of both phagocytes and digestive cells (He et al. 2018). Amphioxus performs both intracellular and extracellular digestion. When food particles are transported into the mid-gut, the ileo-colon ring sorts some small particles containing algal and microbial cells (around 2 μm in diameter or less) into the lumen of the hepatic cecum. Some of the particles are swept back to the mid-gut again, while the detained particles in the cecum are phagocytized by the epithelial cells, sequestered in phagosomes and then degraded when the phagosomes fuse with lysosomes (Fig. 1). Almost all the epithelial cells in the hepatic cecum have a phagocytic function. The cells, similar to those of the vertebrate liver, generate large numbers of lysosomes to exert digestion and immune functions (He et al. 2018; Yuan et al. 2015a). The degraded pathogen molecules, as well as undegraded bacteria and viruses, can be detected by an array of pathogen recognition receptors, leading to rapid activation of immune responses.
Fig. 1.
Anatomical organization of the hepatic cecum as an immune organ of amphioxus. a The digestive system, including the pharyngeal gill slits, hepatic cecum and intestine, is regarded as the major line of defense against microbial infection. When food particles enter the pharynx, they are trapped by mucus and formed into a mucus cord, which is then transported posteriorly to the mid-gut. In the mid-gut, food particles within the mucus cord are mixed with digestive enzymes, and some small particles containing algal and microbial cells (around 2 μm in diameter or less) pass into the lumen of hepatic cecum, a structure arising from the mid-gut and expands anteriorly along the right side of the pharynx. b These algal and microbial cells are phagocytized by the epithelial cells of hepatic cecum and degraded by lysosomes. Then, the degrading products enter the hepatic blood vessel plexus, in which the blood is mainly delivered from intestine via the subintestinal vein which is ultimately joined to the hepatic vein. Gathering intestine-derived blood components and hepatic epithelial cells derived products, the blood in the hepatic plexus is rich in nutrients, pathogen-derived molecules, and even surviving pathogenic microbes. The cells of the hepatic cecum are responsible for the production of complement proteins and acute phase proteins
In amphioxus, the pharynx, hepatic cecum, mid-gut and intestine (also called hind-gut) are the major organs of the alimentary canal and are thus in continuous contact with pathogenic microbes (Fig. 1). However, their internal milieu and defense mechanisms differ. The pharynx, a structure responsible for filtering food particles suspended in seawater, contains numerous gill slits, that include lymphoid-like tissues and lymphocyte-like cells capable of executing an immune response to microbial challenge (Han et al. 2010; Huang et al. 2007), and an endostyle which secrets mucus to capture food particles (Nielsen et al. 2007). The food particles mix with mucus in the pharynx to form a mucus cord, which is then transported to the mid-gut and intestine. Importantly, the mucus cord ensures that the almost all algal and microbial cells are confined to the lumen, isolating them from the epithelial cells of the mid-gut and intestine (Nakashima et al. 2018). Any microbes that escape from the mucus cord can be phagocytized by the epithelial cells of intestine, or captured by the phagocytes in the intestinal blood vessels (Han et al. 2010). In contrast, the food particles released into the hepatic cecum are in a dispersed form and are phagocytized by the hepatic epithelial cells directly. Consistent with the difference in internal milieu between the hepatic cecum and intestine, the phagocytic ability of hepatic epithelial cells appears stronger than that of intestinal epithelial cells (He et al. 2018). However, whether phagocytes are present in hepatic blood vessels, such as the intestinal blood vessels, remains unclear. In the hepatic cecum, the blood vessel plexus consists of a network of spaces in the extracellular matrix, lined by the basement membrane of surrounding epithelial cells. Blood in the hepatic plexus is mainly delivered from the intestine via the sub-intestinal vein which ultimately joins to the hepatic vein (Monahan-Earley et al. 2013). Gathering intestine-derived blood components and hepatic epithelial cell-derived products renders the blood in the hepatic plexus rich in nutrients, pathogen-derived molecules [e.g., pathogen-associated molecular patterns (PAMPs)], and even surviving pathogenic microbes. In this complex microenvironment, the hepatic immune system must tolerate harmless molecules while at the same time remaining alert to possible infectious agents.
Immune recognition and signaling network
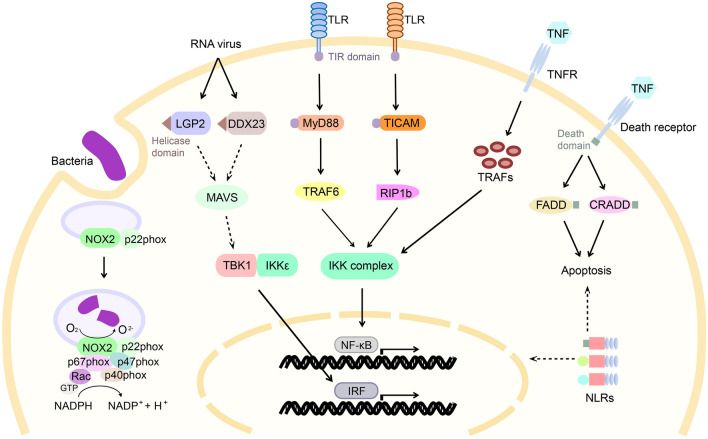
In the hepatic cells, pathogens and their PAMPs can be recognized by an array of pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), NOD-like receptors (NLRs), scavenger receptors (SRs) and RIG-I-like receptors (RLRs) (Liu et al. 2015b; Yuan et al. 2009a, 2016; Zhang et al. 2013). Upon pathogen/PAMPs recognition, the PRRs of hepatic cells trigger a series of signaling events that ultimately leads to activation of immune-related transcriptional factors such as nuclear factor-κB (NF-κB), interferon (IFN) regulatory factors (IRFs) and activator protein 1 (AP-1), that promote the expression of immune effectors and mediators (Fig. 2). Many immune-related gene families have been found to undergo lineage-specific expansion in amphioxus (Huang et al. 2008; Huang and Xu 2016). For example, the genome of Branchiostoma floridae has an expanded PRR repertoire including at least 22 TLRs, 73 NLRs and 144 SRs, and a huge intracellular intermediate signal-transducing network including 57 Toll/interleukin-1 receptor (TIR) domain-containing proteins, 24 tumor necrosis factor (TNF) receptor-associated factors (TRAFs), 45 caspases and hundreds of death-fold domain-containing proteins. In contrast, most of the kinases and transcription factors in this immune network, including IKK/TBK, MAPK/JNK, NF-κB, AP-1 and IRFs, have not undergone expansion, and their structure and number are similar to those in vertebrates (Huang et al. 2008).
Fig. 2.
Putative immune signaling network in amphioxus hepatic cells. The TLR signals can induce activation of the IKK complex and NF-κB through either the MyD88-TRAF6 or TICAM-RIP1b pathway. A group of TIR domain-containing adaptors are involved in the activation and regulation of the TLR pathway. The ligands of amphioxus TLRs remain to be determined. Putative antiviral mechanisms include viral RNA recognition by DExD/H-box helicase domain-containing receptors (e.g., LGP2 and DDX23) and activation of the TBK1-IKKɛ complex and IRFs. The transcription factor IRF family contains nine members which constitute a dynamic feedback regulatory framework among IRFs and NF-κB. In the scheme of the oxidative burst machinery, NOX2 and p22phox are transmembrane components, while p47phox, p67phox, p40phox and Rac are cytosolic subunits that are recruited to the NOX2-p22phox on activation. This phagocytic respiratory burst machinery, through releasing a large amount of ROS into the phagosome, participates in the bactericidal process of the hepatic epithelial cells. In the putative TNF system, TNF can either activate the IKK complex and NF-κB through the TNFR-TRAF pathway, or trigger caspase-dependent apoptosis through the Death receptor (TNFR with death domain)-FADD/CRADD pathway. Amphioxus has a large number of NLRs that share similar domain architectures containing a central NOD domain, a C-terminal LRR region and various N-terminal region. However, the NLR signaling pathway is unclear and needs further study. Solid lines indicate a pathway with experimental evidence; dashed lines represent no experimental support
Toll-like receptors
Vertebrate TLRs are expressed in immune cells (e.g., macrophage) as well as non-immune cells (e.g., epithelial cell), located on the cell surface or in endosomes, and responsible for recognition of a wide range of pathogen-derived molecules (Kawasaki and Kawai 2014). Totally, 30 TLRs have been identified in B. lanceolatum, 22 TLRs in B. floridae and 37 TLRs in B. belcheri. However, only one TLRs’ ligand recognition profiles have been determined: the TLR22 of B. lanceolatum was identified as a receptor for viral ligand poly I:C (Ji et al. 2018). Thus, it is still unclear if these TLRs serve as PAMP receptors, similar to the vertebrate TLRs that recognize PAMPs directly or indirectly with the aid of other proteins, or as cytokine receptors, similar to the Drosophila TLRs (Gangloff et al. 2003). In contrast, the downstream signaling of TLR system has been well studied in amphioxus (Yuan et al. 2015a). The amphioxus TLR signaling transduction system is composed of MyD88-dependent and TICAM-dependent pathways (Yang et al. 2011; Yuan et al. 2009a), both involving a group of TIR-containing adaptors and TRAFs, and ultimately activating NF-κB. The negatively regulatory molecules for the MyD88-dependent pathway (e.g., SARM, TRAF2a, TIRC, IRAK4 and Pelle) and the TICAM-dependent pathway (e.g., SARM, TRAF2a, TIRA and RIP1a) are essential in the regulation of TLR system in amphioxus (Li et al. 2011; Peng et al. 2015; Yan et al. 2020; Yuan et al. 2010).
NOD-like receptors
NLRs are a family of intracellular sensors that play important roles in apoptosis, innate immunity and inflammation. Amphioxus has 73 NLRs that share similar domain architectures containing a central NACHT domain (also called NOD domain) for oligomerization, a C-terminal LRR region for recognition of PAMP or endogenous harmful molecules, and various N-terminal regions [e.g., death domain, caspase activation and recruitment domain (CARD), or death effector domain (DED)] for signal transduction (Huang et al. 2011b). Compared to vertebrate NLRs, amphioxus NLRs have no pyrin domain (PYD, an important domain for innate immune signaling) in the N-terminal region (Ratsimandresy et al. 2013). In vertebrates, NLRs use CARD/PYD domains to participate in NF-κB signaling and in the assembly of inflammasomes, which are intracellular multiprotein complexes that sense danger signals and then trigger pro-inflammatory responses. These inflammasome formations generally require that the activated NLRs recruit the adaptor ASC (apoptosis-associated speck-like protein containing CARD) via PYD–PYD interaction, as well as caspase-1. Although these elements for the inflammasome-like complex have been determined in amphioxus, the mechanism should be different from those in vertebrates given their different structures (Xu et al. 2011).
RIG-I-like receptors
RLRs, including RIG-I, MDA5 and LGP2, are kinds of DExD/H-box RNA helicases that recognize intracellular PAMPs derived from viral genomes. Besides RLRs, several other DExD/H-box helicases, such as DHX9, DHX15, DHX36, DDX17, DDX23 and DDX41, also play important roles in detecting viral infection (Iwasaki 2012). Amphioxus possesses a large DExD/H-box helicase family, including LGP2, DHX9, DHX15 and DDX23, that have been found to be capable of binding poly I:C and are involved in antiviral response (Liu et al. 2015b; Ruan et al. 2019). The LGP2 (BjLGP2) gene of B. japonicum is predominantly expressed in the hepatic cecum, and up-regulated following challenge with poly I:C. In flounder gill cells, BjLGP2 has, upon poly I:C or viral challenge, the capacity to induce a RLRs signaling pathway via the interaction with MAVS which eventually leads to NF-κB- and IRF-3-dependent production of type I IFN and pro-inflammatory cytokines. In amphioxus, the key elements of the RLR signal transduction pathway, including MAVS, TBK1, IKK, STAT and IFN regulatory factors (IRFs), are present, but the canonical type I IFN is absent (Cao et al. 2020; Liu et al. 2015b; Yuan et al. 2015b). Nevertheless, one of the vertebrate IFN-stimulated genes, viperin, has been identified in B. japonicum, and it been shown capable of attenuating viral infectivity and propagation (Lei et al. 2015). Interestingly, neither lineage-specific virus nor virus-related diseases have thus far been reported in amphioxus. Moreover, no information is available to date regarding the antiviral cytokines and the cytokine-stimulated effectors in amphioxus. These deserve further study in the future.
Other PRRs
In the hepatic cecum of amphioxus, the signaling pathways of TLR, NLR and RLR act as the main defense line in antibacterial and antiviral immunity. Furthermore, several scavenger receptors (SRs) also serve as PRRs in the cecum. In vertebrates, liver cells express an array of SRs, and these SRs have a direct role in the detection and capture of pathogens (Jenne and Kubes 2013). For example, CD36 is a SR for sensing a wide range of ligands and is primarily located on the surface of macrophages and hepatocytes (Silverstein and Febbraio 2009). It has been shown that the CD36 gene of B. japonicum is mainly expressed in the hepatic cecum and up-regulated after feeding or E. coli stimulation, suggesting that it participates in both nutritional control and immune responses (Zhang et al. 2013). In addition, the leucine-rich repeat (LRR) domain-containing receptors and C-type lectin receptors are two vast repertoires in the amphioxus genome (Huang et al. 2008). In vertebrates, the LRR-containing receptors (e.g., variable lymphocyte receptor) and C-type lectin receptors (e.g., mannose receptor) often serve as PRRs (Boehm et al. 2012; Fraser et al. 1998), hinting that some of the proteins in amphioxus may also have a pathogen recognition function.
Although many genes encoding immune-associated receptors and adaptors have been found in the amphioxus genome, several questions remain unanswered. For example, how the PRRs distinguish distinct pathogens and how they direct downstream signaling when encountering such many adaptors. Due to the unique situation in the hepatic cecum with constant exposure to the commensal microflora, it is also necessary to distinguish between the harmless microflora and pathogenic microbes, triggering immune responses only in the pathogenic infection. Consequently, determining the specific ligands and adaptors of these PRRs will shed light on the immune surveillance function of the hepatic cecum.
Oxidative burst system
In vertebrates, oxidative burst occurs in the activated phagocytes, which rapidly release a large amount of reactive oxygen species (ROS) into the phagosome to kill ingested bacteria. In amphioxus, ROS production is also necessary for efficient antibacterial responses and oxidative burst, which mainly occurs in the epithelial cells of the hepatic cecum, intestine and gill upon bacterial stimulation, consistent with the recognition and phagocytosis functions of these epithelial cells (Yang et al. 2014). Like vertebrates, amphioxus also has a complete gene set for the classical oxidative burst system, in which NADPH-oxidase (NOX) 2 together with p22phox, p47phox, p67phox, p40phox and GTPase Rac, produces H2O2 which myeloperoxidase then converts into ROS. The genes encoding NOX2, p22phox, p47phox, p67phox, p40phox and Rac are mainly expressed in the hepatic cecum, intestine and gill, and can be rapidly up-regulated when amphioxus is challenged with bacteria. This phagocytic respiratory burst machinery participates in the initiation of the phagocytic process of the gut epithelial cells.
Cytokines
Cytokines are the signals that mediate intercellular communication in the immune system and are often responsible for the host inflammatory response which is an innate defense mechanism against infections and a variety of tissue injury. At the infection or injury site, local immune cells release cytokines which recruit and activate more immune cells to enhance the inflammatory response. The best-known cytokines in vertebrates are ILs, IFNs, TNFs, chemokines and TGF-β, and most members of these cytokine families have significant roles in pro- or anti-inflammatory processes. Among these cytokines, only the homologs of IL-17, TNFs and TGF-β genes have been identified in the amphioxus genome, although the function of the IL-17-like protein has not been determined. Amphioxus has a sophisticated TNF system containing 24 TNFs and 31 TNF-receptors, and downstream adaptors including 24 TRAFs and hundreds of death-fold domain-containing proteins (Huang et al. 2008; Yuan et al. 2009b). These adaptors can either activate NF-κB to enhance the transcription of immune response genes or initiate caspase cascade to induce apoptosis of the cells (Xu et al. 2011), suggesting that the TNF system plays important roles in the immune and inflammatory processes of amphioxus. The TGF-β molecule of B. japonicum has been characterized as a cytokine with bipolar properties, i.e., not only can it inhibit the production of TNF-α and IL-1β in LPS-activated mouse macrophages, suppressing the migration of LPS-activated mouse macrophages, but it can also enhance the migration of non-activated mouse macrophages (Wang et al. 2014). The bipolar properties of TGF-β proteins in immune regulation have also been reported in vertebrates (Cai et al. 2010).
An unregulated host response to infection is usually associated with the excessive activation and release of pro-inflammatory cytokines, termed a cytokine storm. LPS is a strong inducer of cytokine storms. In humans, LPS can activate the TLR signaling pathway that promotes the expression of inflammatory cytokines, and the outpouring of pro-inflammatory cytokines causes cytokine storms that contribute to tissue injury, sepsis and even septic shock (Opal 2007). In this situation, the liver has a central role in the regulation of excessive inflammation by producing anti-inflammatory cytokines or modulators (Strnad et al. 2017). Recently, a novel modulator of inflammatory networks, called BjIM1, has been identified in B. japonicum. BjIM1 is primarily released from cells of the hepatic cecum and is capable of inhibiting LPS-induced up-regulation of TLR pathway genes, such as MyD88, IKK, NF-κB1, Rel, p38, JNK and AP-1, as well as NOX2 (the core molecule of the oxidative burst system), indicating that BjIM1 may negatively regulate the TLR signaling pathway and oxidative burst system in the LPS-induced inflammatory response (Qu et al. 2020b). Interestingly, homologs of BjIM1 are only identified in invertebrates, but none in vertebrates, suggesting that they may be invertebrate-specific inflammatory modulators. BjIM1 shares little sequence similarity with any known functional proteins or domains, thus it is difficult to predict either its structure or receptor. Determining the mechanism of the BjIM1-mediated anti-inflammatory process will certainly shed light on the immune and inflammatory regulation function of the hepatic cecum.
Primitive adaptive immune system
Although not possessing classical adaptive immunity, amphioxus has obtained some raw and basic elements of the adaptive immune system, including lymphocyte-like cells, proto-MHC, protoRAG and variable immune receptors, implying the possible presence of alternative adaptive immunity (Feng et al. 2016; Huang et al. 2016). The appearance of variable immune receptors, e.g., immunoglobulin superfamily (IgSF), provides the possibility to recognize various antigens. In amphioxus, three types of IgSF protein with diversified Variable region (V region), i.e., V region-containing chitin-binding proteins (VCBPs), V and C domain-bearing protein (VCP), and an IgVJ-C-type protein (AmpIgVJ-C2), have been identified, all of which have a high expression pattern in the hepatic cecum (Cannon et al. 2002; Chen et al. 2018; Yu et al. 2005). Both VCBP and VCP can recognize a broad spectrum of bacteria (Dishaw et al. 2011; Yuan et al. 2015a). The details of their pathogen recognition and signal transduction mechanisms are, however, poorly characterized and deserve further study.
Humoral defensive molecule repertoire
In humans, a key and systemic function of the liver is to synthesize the majority of serum proteins, including complement proteins, fibrinogen, clotting factors and protease inhibitors. These proteins are involved in the regulation of systemic homeostasis and some also act as immune effectors. Amphioxus has extensive coelomic cavities and a closed circulatory system but lacks free-circulating blood cells. To date, many immune effectors have been identified in the humoral fluid of amphioxus, most of which originate from the hepatic cecum. This suggests that the hepatic cecum is essential for the humoral defensive system. Here we focus on the immune function of the humoral defensive molecules synthesized in the hepatic cecum.
Complement system
The human complement system is composed of more than 30 distinct plasma proteins and membrane-associated proteins, and ~ 90% of the plasma complement components are synthesized in the liver (Sarma and Ward 2011). Complement, which can be activated by pathogens either directly or indirectly with the aid of antibodies, initiates a cascade of cleavage and activation events leading to the formation of membrane attack complexes or the recruitment of immune cells (Merle et al. 2015a). The amphioxus genome contains multiple copies of a number of complement-related genes, such as 50 C1q-like, 41 ficolin-like, two MASP, two C3, three Bf/C2, five C6-like and 427 CCP (complement control protein)-containing genes. In this regard, the complement system of amphioxus seems more complex and diverse than that of vertebrates including mammals (Huang et al. 2008). The vertebrate complement system has three activation pathways, i.e., the classical, alternative and lectin pathways, as well as two terminal pathways, i.e., the cytolytic and opsonic pathways. In all the three activation pathways, C3-convertase cleaves and activates C3, triggering a cascade of further cleavage and activation events. Amphioxus complement can be activated through alternative (C3 autohydrolysis), lectin (ficolin-MASPs activating C3) as well as classical-like pathway (C1q-MASPs activating C3) (Gao et al. 2014, 2017; Huang et al. 2011a; Li et al. 2008). Several key molecules involved in the complement activation pathways have been characterized, such as C1q-like, ficolin, MASP1/3, C3, Bf/C2 and properdin (Endo et al. 2003; Gao et al. 2013, 2014, 2017; He et al. 2008; Huang et al. 2011a; Suzuki et al. 2002). They are all predominantly synthesized in the hepatic cecum except that the properdin gene is ubiquitously expressed, similar to the expression profile of vertebrate properdin (Cortes et al. 2012). In addition, a conserved microRNA (miR-92d) has been shown to be involved in the regulation of amphioxus complement pathway by targeting C3, the center molecule of complement system (Yang et al. 2013). The expression level of miR-92d in the hepatic cecum is higher than in any other tissues, and is down-regulated by challenge with bacteria (Yang et al. 2013). These findings indicate that the hepatic cecum plays a central role in the regulation of the complement system.
In mammals, the terminal cytolytic pathway of complement needs both C5 and four C6-like proteins (C6, C7, C8 and C9) to assemble the membrane attack complex that performs cytolytic effects against the targeted cells. Furthermore, the resulting fragmentary molecules of complement activation, such as C3a, C5a and C3b, are capable of killing bacteria directly or enhancing macrophage phagocytosis of pathogenic microbes (Merle et al. 2015b). Amphioxus has five C6-like proteins each of which has the membrane attack complex/perforin domain required for membrane perforation, but all lack the key domains responsible for interaction with C5 (Suzuki et al. 2002). Although complement-mediated cytolysis has been observed in amphioxus (Li et al. 2008; Zhang et al. 2003), the cytotoxic pore-forming mechanism is unclear and deserves further study. Amphioxus C3 can be cleaved into C3a and C3b (Huang et al. 2011a). The C3a fragment is directly bactericidal and capable of enhancing macrophage phagocytosis of bacteria (Gao et al. 2013). The presence of both the cytolytic and opsonic pathways indicates that amphioxus has a functional prototypic complement system (Fig. 3).
Fig. 3.
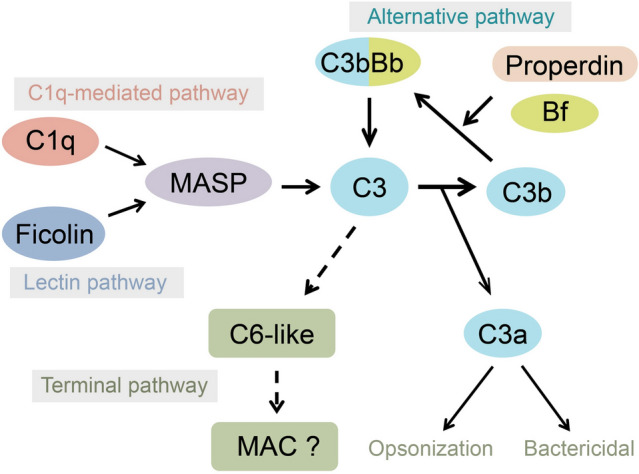
Amphioxus complement systems are activated and amplified by the formation of C3 convertases through the C1q-mediated, lectin and alternative pathways. C3 can be cleaved by C3 convertases to form C3a and C3b. The C3a fragment has bactericidal activity and is capable of enhancing phagocyte phagocytosis against bacteria. The C3b fragment can bind covalently to cell surface carbohydrates via its reactive thioester. Amphioxus has five C6-like proteins, but it is unclear whether they participate in the formation of the membrane attack complex (MAC). A solid arrow indicates that the pathway was supported by experimental data; a dashed arrow indicates that no experimental support is present
Phenoloxidase
Invertebrate phenoloxidase (PO), homologous to vertebrate tyrosinase, is a multi-function oxidase involved in sclerotization of the cuticle, defensive encapsulation, melanization of foreign microorganisms, and wound healing (Lemaitre and Hoffmann 2007). Intermediates produced in the melanization process can kill bacteria directly. PO normally exists as proPO zymogen. Upon recognition of the invasive microorganism by PRRs (e.g., PGRP and C-type lectins), a complex protease cascade is soon initiated to cleave proPO, resulting in the generation of active PO (Lu et al. 2014). In amphioxus, PO activity is detected in the humoral fluid, which is increased significantly after bacterial stimulation, suggesting that PO plays a crucial role in the humoral defensive system (Pang et al. 2009). Amphioxus PO is a tyrosinase-type enzyme (Pang et al. 2005), which is consistent with the fact that it has a close phylogenetic relationship with vertebrate tyrosinase. The tyrosinase-like gene is expressed in muscle, epidermis, hepatic cecum and some other tissues (Pang et al. 2013). However, the proteases and modulators of PO activation pathway remain to be identified in amphioxus.
Lectin and lectin-like protein
In the absence of free-circulating immune cells, amphioxus needs an efficient mechanism to prevent the dissemination of pathogens in the humoral fluid. This is seemingly achieved by lectin or lectin-like proteins. Amphioxus humoral fluid possesses bacterial-agglutinating activity towards both Gram-negative and Gram-positive bacteria (Pang et al. 2012). Over 1200 genes in the amphioxus genome, contain the C-type lectin domain (CTLD) (Huang et al. 2008). Hereafter, the CTLD refers to the carbohydrate recognition domain (CRD) of C-type lectins (a family of Ca2+-dependent lectins). CTLDs usually have sugar-binding motifs (mostly EPN/QPD + WND). In amphioxus, CTLDs have various sugar-binding motifs, such as EP(N/S/K/E/D) and QP(D/S/N), hinting that C-type lectins have diverse sugar-binding specificities in amphioxus. Half of these CTLD-containing proteins consist solely of a CRD domain. It has been demonstrated that in amphioxus, a C-type lectin consisting of a signal peptide and a single CRD, termed AmphiCTL1, can agglutinate Gram-positive bacteria and yeast cells, but has little binding activity toward Gram-negative bacteria. AmphiCTL1 can directly kill Staphylococcus aureus and Saccharomyces cerevisiae. Interestingly, the AmphiCTL1 gene is mainly expressed in the hepatic cecum and is dramatically up-regulated when challenged with S. aureus, S. cerevisiae and zymosan (Yu et al. 2007a). In addition, the C-type lectins with multiple domain combinations are in most cases associated with collagen, CCP and EGF domains. A C-type lectin containing EGF and low-density lipoprotein receptor domains has been identified in B. japonicum (BjCTL). This is a typical Ca2+-dependent carbohydrate-binding protein capable of binding to and agglutinating both Gram-negative and Gram-positive bacteria, though it is only slightly expressed in the hepatic cecum (Qu et al. 2016). Given the abundant presence of C-type lectins with diversified structures in the genome, amphioxus may have a pre-prepared defense network against almost all possible invading microorganisms.
Intelectin (a type of galactofuranose-binding lectin) and galectin (a type of galactoside-binding lectin) are two important groups of pattern recognition molecules capable of regulating immune and inflammatory responses (Boscher et al. 2011; Yan et al. 2013a). On the one hand, there are 22 intelectin homologs identified in amphioxus two of which, i.e., AmphiITLN71469 and AmphiITLN239631, have been characterized (Yan et al. 2012, 2013b). AmphiITLN71469 can strongly agglutinate Gram-positive bacteria in a Ca2+-dependent manner, but has lower agglutination activity towards Gram-negative bacteria, whereas AmphiITLN239631 can agglutinate both Gram-positive and Gram-negative bacteria in a Ca2+-independent manner, but its bacterial binding and agglutinating activity are both lower than that of AmphiITLN71469. On the other hand, amphioxus has two forms of galectins, i.e., dual-CRD tandem galectin (Gal-L) and its alternatively spliced mono-CRD isoform (Gal-S), both having β-galactoside binding activity (Yu et al. 2007b). The Gal-L gene is mainly expressed in the hepatic cecum, whereas the Gal-S gene is ubiquitously expressed in all the tissues. Bacterial or fungal (S. cerevisiae) stimulation can induce up-regulation of Gal-L expression. Like some mammalian galectins, amphioxus Gal-L and Gal-S are present both intracellularly and extracellularly, hinting that they are involved in the inflammatory response by cross-linking β-galactoside glycoconjugates or glycoprotein receptors on cell surfaces to mediate cell–cell or cell–matrix interactions.
Apextrins are a group of proteins capable of interacting with the major bacterial cell wall polymer, peptidoglycan (PGN). Amphioxus has nine apextrin-like genes, and two of them have been characterized in B. japonicum (BjALP1 and BjALP2) (Huang et al. 2014). Both BjALP1 and BjALP2 can interact with bacterial PGN and the minimal PGN motif muramyl dipeptide. BjALP1 is a secreted effector that agglutinates Gram-positive bacteria, but not Gram-negative bacteria. Neutralization of secreted BjALP1 by anti-BjALP1 monoclonal antibodies can cause serious damage to the gut epithelium and rapid death of the host animal after bacterial infection. The BjALP1 gene is mainly expressed in the hepatic cecum, intestine, gill and skin, and its expression in the hepatic cecum and intestine shows thousand-fold up-regulation after bacterial infection. BjALP2 is an intracellular sensor associated with the NF-κB signaling pathway and its expression shows hundred-fold up-regulation after bacterial infection.
Antimicrobial protein and peptide
Many well-known microbicidal proteins have been identified in amphioxus, e.g., PGN recognition protein (PGRP) (Yao et al. 2012), lysozymes (Liu et al. 2006; Xu et al. 2014), chitotriosidase (Xu and Zhang 2012), defensin (Teng et al. 2012), and apolipoprotein A-I (Wang et al. 2019), most of which are predominantly synthesized in the hepatic cecum (Table 1). Amphioxus has 17–18 PGRPs, all of which have Zn2+ binding and amidase active sites (Huang et al. 2011b). A short PGRP (PGRP-S) possessing a domain combination of CBD-PGRP has been identified in B. japonicum (Yao et al. 2012). The PGRP-S can bind to E. coli, S. aureus and Pichia pastoris, and displays enzymatic activity of amidase which is capable of hydrolyzing PGN. The bactericidal activity of PGRP-S against E. coli and S. aureus is mainly due to the PGRP domain, whereas the anti-P. pastoris activity relies on the CBD domain. Notably, this domain combination of CBD-PGRP has been found only in amphioxus, which might result from domain shuffling when a great expansion of amphioxus immune gene repertoire occurred, possibly broadening its recognition spectrum.
Table 1.
Antimicrobial proteins identified in amphioxus
| Proteins | Tissue expression | Activity (IC50) * | Reference |
|---|---|---|---|
| Alanine aminotransaminase | Mainly in hepatic cecum | G−: E. coli (< 1 μmol/L) | Jing and Zhang (2011) |
| AmphiCTL1 | Mainly in hepatic cecum |
G+: S. aureus (Not determined) F: S. cerevisiae (Not determined) |
Yu et al. (2007a) |
| Apolipoprotein A-I | Mainly in hepatic cecum | G−: Aeromonas hydrophila (1.47 μmol/L), Vibrio vulnificus (2.15 μmol/L), Pseudomonas aeruginosa (1.54 μmol/L) | Wang et al. (2019) |
| Big defensin | Hepatic cecum, muscle, gill and intestine |
G+: S. aureus (~ 5 μmol/L) G−: A. hydrophila (> 20 μmol/L) |
Teng et al. (2012) |
| BjAMP | Mainly in hepatic cecum, intestine, etc. |
G−: E. coli (3.2 μmol/L), Vibrio anguillarum (6.3 μmol/L) G+: S. aureus (6.3 μmol/L), Micrococcus luteus (0.8 μmol/L) |
Liu et al. (2015a) |
| C3a | Cleaved from C3 |
G−: E. coli (> 6.6 μmol/L), V. anguillarum (> 6.6 μmol/L) G+: S. aureus (6.6 μmol/L), Bacillus subtilis (1.6 μmol/L) |
Gao et al. (2013) |
| Chitotriosidase-like protein | Mainly in hepatic cecum | F: Candida albicans (Not determined) | Xu and Zhang (2012) |
| Creatine kinase | Not determined | G−: E. coli (~ 10 μmol/L) | An et al. (2009) |
| Fibrinogen-related protein | Mainly in hepatic cecum and intestine |
G−: E. coli (~ 3 μmol/L) G+ : S. aureus (< 0.5 μmol/L) |
Fan et al. (2008) |
| Lysozymes | g-type in hepatic cecum, i-type in gill, c-type in hepatic cecum, intestine and gill | Micrococcus lysodeikticus cell wall degradation activity of the three type lysozymes is in an order of i-type > c-type > g-type | Xu et al. (2014) |
| Miple | Mainly in ovary |
G−: A. hydrophila (0.25 μmol/L), E. coli (1 μmol/L) G+: S. aureus (0.25 μmol/L), B. subtilis (1 μmol/L) |
Gao et al. (2018) |
| PGRP-S | Mainly in hepatic cecum and muscle |
G −: E. coli (~ 7 μmol/L) G + : S. aureus (~ 7 μmol/L) F: Pichia pastoris (~ 7 μmol/L) |
Yao et al. (2012) |
| Ribosomal protein S15 | High expression in hepatic cecum |
G −: E. coli (0.5 μmol/L), A. hydrophila (0.5 μmol/L) G + : S. aureus (0.5 μmol/L), M. luteus (0.5 μmol/L) |
Qu et al. (2020a) |
| Ribosomal protein S23 | High expression in hepatic cecum and ovary |
G −: E. coli (4.2 μmol/L), A. hydrophila (4.2 μmol/L) G : S. aureus (3 μmol/L), M. luteus (3 μmol/L) |
Ma et al. (2020) |
| Tachylectin-related protein | Mainly in hepatic cecum and intestine | G −: E. coli (~ 7 μmol/L) | Ju et al. (2009) |
| Transferrin-like protein | Mainly in hepatic cecum, intestine and ovary | G −: E. coli (~ 8 μmol/L) | Liu et al. (2009) |
*The antimicrobial activities against different types of microbes are annotated as below: G− Gram-negative bacteria, G+ Gram-positive bacteria, F fungi. The IC50 is the concentration of protein needed to inhibit microbe growth by 50%
Lysozymes are a class of enzymes that are able to catalyze hydrolysis of the β-(1,4)-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine in PGN, and are produced in virtually all organisms, ranging from bacteriophages to humans. Most organisms are able to produce different types of lysozymes and/or multiple forms of the same lysozyme. It is presumed that different types/forms of lysozymes may have different or complementary functions. In amphioxus, lysozyme activity has been detected in the humoral fluid, and the lysozyme activity of humoral fluid increases significantly after exposure to bacteria (Pang et al. 2006, 2010, 2012). Peculiarly, amphioxus possesses three types of lysozymes, i.e., c-, g- and i-type lysozymes (Xu et al. 2014). The c- and g-type lysozymes are predominantly synthesized in the hepatic cecum, whereas the i-type lysozyme is primarily generated in the gill. All the three types of lysozyme have the capacity to degrade the Micrococcus lysodeikticus cell wall, but their levels of activity differ from each other in the order i-type > c-type > g-type.
In addition to these known bactericidal proteins, some well-known molecules are found to possess newly discovered antimicrobial functions. For example, alanine aminotransaminase (ALT) is an acute phase protein primarily synthesized in the liver and is regarded as an index for clinical diagnosis of liver function in humans. It has been revealed by Jing and Zhang (2011) that ALT has antibacterial activity. Amphioxus ALT can bind to LPS, and cause the Gram-negative bacterium E. coli to lyse. In contrast, it can neither bind nor kill Gram-positive bacterium S. aureus. LPS stimulation can induce a 40-fold increase in the expression of the amphioxus ALT gene, suggesting the important role of ALT in antibacterial defense. Recently, two ribosomal proteins of B. japonicum, BjRPS15 and BjRPS23, have been shown capable of killing both Gram-negative and Gram-positive bacteria (Ma et al. 2020; Qu et al. 2020a). The functional characteristics of BjRPS15 and BjRPS23 are similar in that both can interact with bacterial membranes via LPS and LTA, and cause membrane depolarization. They can also stimulate production of intracellular ROS in bacteria. Their expression in the hepatic cecum is significantly higher than in other tissues, and markedly up-regulated following challenge with bacteria, LPS or LTA. Moreover, both can be detected extracellularly in the humoral fluid, suggesting that they are “moonlighting” proteins, functioning not only as house-keeping proteins but also as antibacterial effectors.
Antimicrobial peptides (AMPs), usually composed of fewer than 100 amino acids, are endogenous antibiotics that are widely distributed in nature as ancient components of innate immunity. Most AMPs are cationic and amphipathic molecules that interact with microbial membranes and are capable of killing microbial cells, either by disrupting membrane integrity or by interacting with certain intracellular targets (Bahar and Ren 2013). More than 3000 AMPs have been recorded in the AMP database, only two of which are from amphioxus (Wang et al. 2016). This may be due to the fact that the study of AMPs in amphioxus is still in its infancy. The precursors of AMPs generally contain signal sequences and proregions, and these regions tend to be more conserved than mature AMPs. This advantage has been successfully employed to search for novel AMPs from databases within the same lineages of amphibians and fish (Juretic et al. 2011; Tessera et al. 2012). In the amphioxus B. japonicum, an AMP named BjAMP1 has been identified using the signal sequence of the jawless hagfish, HFIAP-1, a known AMP of the cathelicidin family. The mature peptide of BjAMP1, i.e., the C-terminal 21 residues, can directly kill a broad spectrum of microbes via a membrane active mechanism (Liu et al. 2015a). The other AMP known in amphioxus is big defensin, termed BjBD, identified by Teng et al. (2012). The BjBD gene is constitutively expressed in most of the tissues examined, and remarkably up-regulated following challenge with LPS, LTA, A. hydrophila and S. aureus. Moreover, recombinant BjBD is able to inhibit the growth of S. aureus, E. coli and A. hydrophila.
Other immune-relevant effectors
Besides the antibacterial molecules listed above, there are several other molecules, such as fibrinogen-related protein (Fan et al. 2008), transferrin (Liu et al. 2009), Gram-negative bacteria-binding proteins (Jin et al. 2012), alpha-2 macroglobulin (Pathirana et al. 2016), avidin (Guo et al. 2017), Miple (Gao et al. 2018), HSP5 and HSP90α (Yao et al. 2019), that play roles in the defense system of amphioxus. These molecules are primarily produced in the hepatic cecum. They either display bactericidal activity or bacteria-binding/agglutinating activity or both.
Acute phase response
Acute phase response (APR) is a physiological process occurring rapidly after the onset of infection, inflammation and trauma. One of the most prominent consequences of APR is the change in concentrations of a number of plasma proteins, collectively known as acute phase proteins (APPs). APRs function in a variety of defense-related activities, thereby protecting the body by preventing microbial growth, limiting tissue damage, and helping to restore metabolic homeostasis. In humans, the liver is the primary organ for APP production, which involves a range of processes to enhance inflammation and limit excessive inflammation (Strnad et al. 2017). The amphioxus hepatic cecum, like the vertebrate liver, plays a central role during APR (Wang and Zhang 2011). At least 58 vertebrate (zebrafish) liver-specific genes are expressed in a tissue-specific manner in the hepatic cecum of amphioxus, and 52 out of the 58 genes display similar expression profiles in both amphioxus hepatic cecum and zebrafish liver in response to LPS challenge, suggesting that these genes are commonly involved in APR in both animals. Liver-specific transcription factors such as HNF-1, HNF-4 and C/EBP are known to control the expression of APR-related genes (Armendariz and Krauss 2009; Babeu and Boudreau 2014; van der Krieken et al. 2015). Notably, in both amphioxus and zebrafish, the majority of the 52 APR-related genes possess binding sites for one or more of the above-mentioned transcription factors in the promoter sequences, suggesting that these APR-associated factors forms a similar network in both species for regulating the expression of APR-related genes (Wang and Zhang 2011).
The hepatic cecum secretes APPs into the humoral fluid and these usually have direct effector functions, including the clearance of pathogens and the regulation of inflammatory responses (Table 2). For example, the classical APPs, including alanine aminotransaminase (Jing and Zhang 2011), transferrin (Liu et al. 2009), alpha-2 macroglobulin (Liang et al. 2011), plasminogen (Liu and Zhang 2009), tachylectin-related protein (Ju et al. 2009), fibrinogen-related protein (Fan et al. 2008), and complement components C3 and Bf (Pan et al. 2011), all exhibit functional conservation in amphioxus and vertebrates. Most of the hepatic cecum-secreted APPs have antimicrobial activity or pro-inflammatory function, but BjIM1 has anti-inflammatory function. It is clear that the hepatic cecum-secreted APPs are important for maintaining systemic homeostasis of amphioxus. Consequently, the hepatic cecum can be regarded as a key integrator of immunity, similar to the vertebrate liver.
Table 2.
Humoral protein synthesis by the hepatic cecum during the acute phase response in amphioxus
| Proteins | Examples | Immune function |
|---|---|---|
| Complement proteins | C1q, Bf/C2 and C3 | Induce bacterial cell lysis, and enhance phagocytosis |
| Lectins | C-type lectins, ficolins, intelectins and apextrin 1 | Agglutinate bacteria, and active complement |
| Antimicrobial proteins | PGRP, lysozymes, chitotriosidase, defensin, apolipoprotein A-I, alanine aminotransaminase, fibrinogen-related protein, AMPs, etc. | Kill or inhibit the growth of microbes |
| Iron-binding proteins | Ferritin and transferrin | Act to reduce free iron in the humoral fluid, and antimicrobial functions |
| Inflammatory modulators | TGF-β and BjIM1 | Anti-inflammatory functions |
In summary, recent findings regarding the function of the hepatic cecum in immune surveillance, clearance of pathogens and acute phase response, all indicate that this organ is a crucial conductor of immune and inflammatory process in amphioxus. The similarities in liver/hepatic cecum-specific genes and immune functions between amphioxus and vertebrates supports the notion that the amphioxus hepatic cecum is the precursor of the vertebrate liver, acting as a key integrator of immunity.
Acknowledgements
This work was supported by the grants of National Natural Science Foundation of China (32070514; 31601862).
Author contributions
ZG and BQ wrote the manuscript and drew the figures; ZM analyzed the data; SZ improved and revised the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interests.
Animal and human rights statement
No animal and human rights are involved in this article.
References
- An Y, Fan N, Zhang S. Creatine kinase is a bacteriostatic factor with a lectin-like activity. Mol Immunol. 2009;46:2666–2670. doi: 10.1016/j.molimm.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Armendariz AD, Krauss RM. Hepatic nuclear factor 1-alpha: inflammation, genetics, and atherosclerosis. Curr Opin Lipidol. 2009;20:106–111. doi: 10.1097/MOL.0b013e3283295ee9. [DOI] [PubMed] [Google Scholar]
- Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol. 2014;20:22–30. doi: 10.3748/wjg.v20.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–220. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscher C, Dennis JW, Nabi IR. Glycosylation, galectins and cellular signaling. Curr Opin Cell Biol. 2011;23:383–392. doi: 10.1016/j.ceb.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Cai Z, Gao C, Li L, Xing K. Bipolar properties of red seabream (Pagrus major) transforming growth factor-β in induction of the leucocytes migration. Fish Shellfish Immunol. 2010;28:695–700. doi: 10.1016/j.fsi.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- Cao Y, Fang T, Fan M, Wang L, Lv C, Song X, Jin P, Ma F. Functional characterization of STATa/b genes encoding transcription factors from Branchiostoma belcheri. Dev Comp Immunol. 2020;114:103838. doi: 10.1016/j.dci.2020.103838. [DOI] [PubMed] [Google Scholar]
- Chen R, Zhang L, Qi J, Zhang N, Yao S, Wu Y, Jiang B, Wang Z, Yuan H, Zhang Q, Xia C. Discovery and analysis of invertebrate IgVJ-C2 structure from amphioxus provides insight into the evolution of the Ig superfamily. J Immunol. 2018;200:2869–2881. doi: 10.4049/jimmunol.1700906. [DOI] [PubMed] [Google Scholar]
- Cortes C, Ohtola JA, Saggu G, Ferreira VP. Local release of properdin in the cellular microenvironment: role in pattern recognition and amplification of the alternative pathway of complement. Front Immunol. 2012;3:412. doi: 10.3389/fimmu.2012.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LJ, Giacomelli S, Melillo D, Zucchetti I, Haire RN, Natale L, Russo NA, De Santis R, Litman GW, Pinto MR. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut-bacteria interactions. Proc Natl Acad Sci USA. 2011;108:16747–16752. doi: 10.1073/pnas.1109687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Nonaka M, Saiga H, Kakinuma Y, Matsushita A, Takahashi M, Matsushita M, Fujita T. Origin of mannose-binding lectin-associated serine protease (MASP)-1 and MASP-3 involved in the lectin complement pathway traced back to the invertebrate, amphioxus. J Immunol. 2003;170:4701–4707. doi: 10.4049/jimmunol.170.9.4701. [DOI] [PubMed] [Google Scholar]
- Fan C, Zhang S, Liu Z, Li L, Luan J, Saren G. Identification and expression of a novel class of glutathione-S-transferase from amphioxus Branchiostoma belcheri with implications to the origin of vertebrate liver. Int J Biochem Cell Biol. 2007;39:450–461. doi: 10.1016/j.biocel.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Fan C, Zhang S, Li L, Chao Y. Fibrinogen-related protein from amphioxus Branchiostoma belcheri is a multivalent pattern recognition receptor with a bacteriolytic activity. Mol Immunol. 2008;45:3338–3346. doi: 10.1016/j.molimm.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Feng Y, Huang S, Cai X, Xu A. Primitive adaptive immune system of amphioxus. In: Xu A, editor. Amphioxus immunity. Beijing: Academic Press; 2016. pp. 221–238. [Google Scholar]
- Fraser IP, Koziel H, Ezekowitz RA. The serum mannose-binding protein and the macrophage mannose receptor are pattern recognition molecules that link innate and adaptive immunity. Semin Immunol. 1998;10:363–372. doi: 10.1006/smim.1998.0141. [DOI] [PubMed] [Google Scholar]
- Gangloff M, Weber AN, Gibbard RJ, Gay NJ. Evolutionary relationships, but functional differences, between the Drosophila and human Toll-like receptor families. Biochem Soc Trans. 2003;31:659–663. doi: 10.1042/bst0310659. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang S. Cephalochordata: Branchiostoma. In: Cooper EL, editor. Advances in comparative immunology. Switzerland: Springer; 2018. pp. 593–636. [Google Scholar]
- Gao Z, Li M, Wu J, Zhang S. Interplay between invertebrate C3a with vertebrate macrophages: functional characterization of immune activities of amphioxus C3a. Fish Shellfish Immunol. 2013;35:1249–1259. doi: 10.1016/j.fsi.2013.07.049. [DOI] [PubMed] [Google Scholar]
- Gao Z, Li M, Ma J, Zhang S. An amphioxus gC1q protein binds human IgG and initiates the classical pathway: implications for a C1q-mediated complement system in the basal chordate. Eur J Immunol. 2014;44:3680–3695. doi: 10.1002/eji.201444734. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ma Z, Qu B, Jiao D, Zhang S. Identification and characterization of properdin in amphioxus: implications for a functional alternative complement pathway in the basal chordate. Fish Shellfish Immunol. 2017;65:1–8. doi: 10.1016/j.fsi.2017.03.052. [DOI] [PubMed] [Google Scholar]
- Gao Z, Qu B, Yao L, Ma Z, Cui P, Zhang S. Identification and functional characterization of amphioxus Miple, ancestral type of vertebrate midkine/pleiotrophin homologues. Dev Comp Immunol. 2018;89:31–43. doi: 10.1016/j.dci.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Guo B, Zhang S, Wang S, Liang Y. Expression, mitogenic activity and regulation by growth hormone of growth hormone/insulin-like growth factor in Branchiostoma belcheri. Cell Tissue Res. 2009;338:67–77. doi: 10.1007/s00441-009-0824-8. [DOI] [PubMed] [Google Scholar]
- Guo X, Xin J, Wang P, Du X, Ji G, Gao Z, Zhang S. Functional characterization of avidins in amphioxus Branchiostoma japonicum: evidence for a dual role in biotin-binding and immune response. Dev Comp Immunol. 2017;70:106–118. doi: 10.1016/j.dci.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Han L, Zhang S, Wang Y, Sun X. Immunohistochemical localization of vitellogenin in the hepatic diverticulum of the amphioxus Branchiostoma belcheri tsingtauense, with implications for the origin of the liver. Invert Biol. 2006;125:172–176. doi: 10.1111/j.1744-7410.2006.00050.x. [DOI] [Google Scholar]
- Han Y, Huang G, Zhang Q, Yuan S, Liu J, Zheng T, Fan L, Chen S, Xu A. The primitive immune system of amphioxus provides insights into the ancestral structure of the vertebrate immune system. Dev Comp Immunol. 2010;34:791–796. doi: 10.1016/j.dci.2010.03.009. [DOI] [PubMed] [Google Scholar]
- He Y, Tang B, Zhang S, Liu Z, Zhao B, Chen L. Molecular and immunochemical demonstration of a novel member of Bf/C2 homolog in amphioxus Branchiostoma belcheri: implications for involvement of hepatic cecum in acute phase response. Fish Shellfish Immunol. 2008;24:768–778. doi: 10.1016/j.fsi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- He C, Han T, Liao X, Zhou Y, Wang X, Guan R, Tian T, Li Y, Bi C, Lu N, He Z, Hu B, Zhou Q, Hu Y, Lu Z, Chen JY. Phagocytic intracellular digestion in amphioxus (Branchiostoma) Proc Biol Sci. 2018;285:20180438. doi: 10.1098/rspb.2018.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Xu A. Genomic and transcriptomic view of amphioxus immunity. In: Xu A, editor. Amphioxus immunity. Beijing: Academic Press; 2016. pp. 57–84. [Google Scholar]
- Huang G, Xie X, Han Y, Fan L, Chen J, Mou C, Guo L, Liu H, Zhang Q, Chen S, Dong M, Liu J, Xu A. The identification of lymphocyte-like cells and lymphoid-related genes in amphioxus indicates the twilight for the emergence of adaptive immune system. PLoS ONE. 2007;2:e206. doi: 10.1371/journal.pone.0000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Yuan S, Guo L, Yu Y, Li J, Wu T, Liu T, Yang M, Wu K, Liu H, Ge J, Huang H, Dong M, Yu C, Chen S, Xu A. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Huang S, Yu Y, Yuan S, Li R, Wang X, Zhao H, Li J, Yang M, Xu L, Chen S, Xu A. Functional characterization of a ficolin-mediated complement pathway in amphioxus. J Biol Chem. 2011;286:36739–36748. doi: 10.1074/jbc.M111.245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang X, Yan Q, Guo L, Yuan S, Huang G, Huang H, Li J, Dong M, Chen S, Xu A. The evolution and regulation of the mucosal immune complexity in the basal chordate amphioxus. J Immunol. 2011;186:2042–2055. doi: 10.4049/jimmunol.1001824. [DOI] [PubMed] [Google Scholar]
- Huang G, Huang S, Yan X, Yang P, Li J, Xu W, Zhang L, Wang R, Yu Y, Yuan S, Chen S, Luo G, Xu A. Two apextrin-like proteins mediate extracellular and intracellular bacterial recognition in amphioxus. Proc Natl Acad Sci USA. 2014;111:13469–13474. doi: 10.1073/pnas.1405414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Yu W, Pontarotti P, Escriva H, Le Petillon Y, Liu X, Chen S, Schatz DG, Xu A. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol. 2013;14:996–1006. doi: 10.1038/ni.2691. [DOI] [PubMed] [Google Scholar]
- Ji J, Ramos-Vicente D, Navas-Perez E, Herrera-Ubeda C, Lizcano JM, Garcia-Fernandez J, Escriva H, Bayes A, Roher N. Characterization of the TLR family in Branchiostoma lanceolatum and discovery of a novel TLR22-like involved in dsRNA recognition in amphioxus. Front Immunol. 2018;9:2525. doi: 10.3389/fimmu.2018.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zhou L, Song X, Qian J, Chen L, Ma F. Particularity and universality of a putative Gram-negative bacteria-binding protein (GNBP) gene from amphioxus (Branchiostoma belcheri): insights into the function and evolution of GNBP. Fish Shellfish Immunol. 2012;33:835–845. doi: 10.1016/j.fsi.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Jing X, Zhang S. An ancient molecule with novel function: Alanine aminotransferase as a lipopolysaccharide binding protein with bacteriocidal activity. Dev Comp Immunol. 2011;35:94–104. doi: 10.1016/j.dci.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Ju L, Zhang S, Liang Y, Sun X. Identification, expression and antibacterial activity of a tachylectin-related homolog in amphioxus Branchiostoma belcheri with implications for involvement of the digestive system in acute phase response. Fish Shellfish Immunol. 2009;26:235–242. doi: 10.1016/j.fsi.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Juretić D, Vukičević D, Petrov D, Novković M, Bojović V, Lučić B, Tossi A. Knowledge-based computational methods for identifying or designing novel, non-homologous antimicrobial peptides. Eur Biophys J. 2011;40:371–385. doi: 10.1007/s00249-011-0674-7. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubes P, Jenne C. Immune responses in the liver. Annu Rev Immunol. 2018;36:247–277. doi: 10.1146/annurev-immunol-051116-052415. [DOI] [PubMed] [Google Scholar]
- Lei M, Liu H, Liu S, Zhang Y, Zhang S. Identification and functional characterization of viperin of amphioxus Branchiostoma japonicum: implications for ancient origin of viperin-mediated antiviral response. Dev Comp Immunol. 2015;53:293–302. doi: 10.1016/j.dci.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang S, Wang C, Pang Q. Complement-mediated killing of Vibrio species by the humoral fluids of amphioxus Branchiostoma belcheri: implications for a dual role of O-antigens in the resistance to bactericidal activity. Fish Shellfish Immunol. 2008;24:215–222. doi: 10.1016/j.fsi.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Li J, Yuan S, Qi L, Huang S, Huang G, Yang M, Xu L, Li Y, Zhang R, Yu Y, Chen S, Xu A. Functional conservation and innovation of amphioxus RIP1-mediated signaling in cell fate determination. J Immunol. 2011;187:3962–3971. doi: 10.4049/jimmunol.1100816. [DOI] [PubMed] [Google Scholar]
- Liang Y, Pan A, Zhang S, Zhang Y, Liu M. Cloning, distribution and primary immune characteristics of amphioxus alpha-2 macroglobulin. Fish Shellfish Immunol. 2011;31:963–969. doi: 10.1016/j.fsi.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Liao X, Yang L, Chen X, Chen J. Identification of microRNA expression profiles in the gill, intestine and hepatic caecum of Branchiostoma belcheri. Protein Cell. 2017;8:302–307. doi: 10.1007/s13238-016-0365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang S. A kringle-containing protease with plasminogen-like activity in the basal chordate Branchiostoma belcheri. Biosci Rep. 2009;29:385–395. doi: 10.1042/BSR20080173. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang S, Liu Z, Li H, Xu A. Characterization, organization and expression of AmphiLysC, an acidic c-type lysozyme gene in amphioxus Branchiostoma belcheri tsingtauense. Gene. 2006;367:110–117. doi: 10.1016/j.gene.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang S, Li L. A transferrin-like homolog in amphioxus Branchiostoma belcheri: identification, expression and functional characterization. Mol Immunol. 2009;46:3117–3124. doi: 10.1016/j.molimm.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Liu H, Lei M, Du X, Cui P, Zhang S. Identification of a novel antimicrobial peptide from amphioxus Branchiostoma japonicum by in silico and functional analyses. Sci Rep. 2015;5:18355. doi: 10.1038/srep18355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang S, Huang Y, Qin Q, Zhang S. Evolutionary conservation of molecular structure and antiviral function of a viral receptor, LGP2, in amphioxus Branchiostoma japonicum. Eur J Immunol. 2015;45:3404–3416. doi: 10.1002/eji.201545860. [DOI] [PubMed] [Google Scholar]
- Lu A, Zhang Q, Zhang J, Yang B, Wu K, Xie W, Luan YX, Ling E. Insect prophenoloxidase: the view beyond immunity. Front Physiol. 2014;5:252. doi: 10.3389/fphys.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Qu B, Yao L, Gao Z, Zhang S. Identification and functional characterization of ribosomal protein S23 as a new member of antimicrobial protein. Dev Comp Immunol. 2020;110:103730. doi: 10.1016/j.dci.2020.103730. [DOI] [PubMed] [Google Scholar]
- Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system part I—molecular mechanisms of activation and regulation. Front Immunol. 2015;6:262. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle NS, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V, Roumenina LT. Complement system part II: role in immunity. Front Immunol. 2015;6:257. doi: 10.3389/fimmu.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller PC, Philpott CW. The circulatory system of amphioxus (Branchiostoma floridae). I. Morphology of the major vessels of the pharyngeal area. J Morphol. 1973;139:389–406. doi: 10.1002/jmor.1051390403. [DOI] [PubMed] [Google Scholar]
- Monahan-Earley R, Dvorak AM, Aird WC. Evolutionary origins of the blood vascular system and endothelium. J Thromb Haemost Suppl. 2013;1:46–66. doi: 10.1111/jth.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J. Über den Bau und die Lebenserscheinungen des Branchiostoma lubricum Costa, Amphioxus lanceolatus Yarrell. Berlin: Druckerei der Königliche Akademie der Wissenschaften; 1844. [Google Scholar]
- Nakashima K, Kimura S, Ogawa Y, Watanabe S, Soma S, Kaneko T, Yamada L, Sawada H, Tung CH, Lu TM, Yu JK, Villar-Briones A, Kikuchi S, Satoh N. Chitin-based barrier immunity and its loss predated mucus-colonization by indigenous gut microbiota. Nat Commun. 2018;9:3402. doi: 10.1038/s41467-018-05884-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SE, Bone Q, Bond P, Harper G. On particle filtration by amphioxus (Branchiostoma lanceolatum) J Mar Biol Ass UK. 2007;87:983–989. doi: 10.1017/S0025315407053519. [DOI] [Google Scholar]
- Opal SM. The host response to endotoxin, antilipopolysaccharide strategies, and the management of severe sepsis. Int J Med Microbiol. 2007;297:365–377. doi: 10.1016/j.ijmm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Pan J, Liu M, Zhang S. Interplay between amphioxus complement with fish macrophages: evidence for vertebrate-like alternative complement activation in the protochordate. J Ocean Univ China. 2011;10:357–361. doi: 10.1007/s11802-011-1842-1. [DOI] [Google Scholar]
- Pang Q, Zhang S, Shi X, Su F, Wu D. Purification and characterisation of phenoloxidase from amphioxus Branchiostoma belcheri tsingtauense. Fish Shellfish Immunol. 2005;19:139–148. doi: 10.1016/j.fsi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Pang Q, Zhang S, Liu X, Wu D. Humoral immune responses of amphioxus Branchiostoma belcheri to challenge with Escherichia coli. Fish Shellfish Immunol. 2006;21:139–145. doi: 10.1016/j.fsi.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Pang Q, Zhang S, Zhao B. Induction of phenoloxidases in the humoral fluids of amphioxus Branchiostoma belcheri by Vibrio alginolyticus and Escherichia coli. Fish Shellfish Immunol. 2009;26:669–671. doi: 10.1016/j.fsi.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Pang Q, Zhang S, Zhao B. Immune parameters in the humoral fluids of amphioxus Branchiostoma belcheri challenged with Vibrio alginolyticus. Fish Shellfish Immunol. 2010;28:232–234. doi: 10.1016/j.fsi.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Pang Q, Liu X, Zhao B, Sun H. Induction of phenoloxidase and other immunological activities in the humoral fluids of amphioxus Branchiostoma belcheri challenged with Lipopolysaccharide (LPS) Fish Physiol Biochem. 2012;38:1835–1842. doi: 10.1007/s10695-012-9680-7. [DOI] [PubMed] [Google Scholar]
- Pang Q, Liu X, Sun H, Zhang S, Song X, Zhang X, Zhang M, Bai Y, Gao L, Zhao B. Cloning, characterization and expression of tyrosinase-like gene in amphioxus Branchiostoma japonicum. Fish Shellfish Immunol. 2013;34:356–364. doi: 10.1016/j.fsi.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Pathirana A, Diao M, Huang S, Zuo L, Liang Y. Alpha 2 macroglobulin is a maternally-derived immune factor in amphioxus embryos: new evidence for defense roles of maternal immune components in invertebrate chordate. Fish Shellfish Immunol. 2016;50:21–26. doi: 10.1016/j.fsi.2015.10.040. [DOI] [PubMed] [Google Scholar]
- Peng J, Tao X, Li R, Hu J, Ruan J, Wang R, Yang M, Yang R, Dong X, Chen S, Xu A, Yuan S. Novel Toll/IL-1 receptor homologous region adaptors act as negative regulators in amphioxus TLR signaling. J Immunol. 2015;195:3110–3118. doi: 10.4049/jimmunol.1403003. [DOI] [PubMed] [Google Scholar]
- Qu B, Yang S, Ma Z, Gao Z, Zhang S. A new LDLa domain-containing C-type lectin with bacterial agglutinating and binding activity in amphioxus. Gene. 2016;594:220–228. doi: 10.1016/j.gene.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Qu B, Ma Z, Yao L, Gao Z, Zhang S. Preserved antibacterial activity of ribosomal protein S15 during evolution. Mol Immunol. 2020;127:57–66. doi: 10.1016/j.molimm.2020.08.024. [DOI] [PubMed] [Google Scholar]
- Qu B, Ma Z, Zhang Y, Gao Z, Zhang S. Characterization of a novel protein identified by proteomics analysis as a modulator of inflammatory networks in amphioxus. Fish Shellfish Immunol. 2020;96:97–106. doi: 10.1016/j.fsi.2019.12.003. [DOI] [PubMed] [Google Scholar]
- Ratsimandresy RA, Dorfleutner A, Stehlik C. An update on PYRIN domain-containing pattern recognition receptors: from immunity to pathology. Front Immunol. 2013;4:440. doi: 10.3389/fimmu.2013.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Cao Y, Ling T, Li P, Wu S, Peng D, Wang Y, Jia X, Chen S, Xu A, Yuan S. DDX23, an evolutionary conserved dsRNA sensor, participates in innate antiviral responses by pairing with TRIF or MAVS. Front Immunol. 2019;10:2202. doi: 10.3389/fimmu.2019.02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Tacke F, Koch A, Trautwein C. Liver—guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Satoh N, Nonaka M. C6-like and C3-like molecules from the cephalochordate, amphioxus, suggest a cytolytic complement system in invertebrates. J Mol Evol. 2002;54:671–679. doi: 10.1007/s00239-001-0068-z. [DOI] [PubMed] [Google Scholar]
- Tao X, Xu A. Basic knowledge of immunology. In: Xu A, editor. Amphioxus immunity. Beijing: Academic Press; 2016. pp. 15–42. [Google Scholar]
- Teng L, Gao B, Zhang S. The first chordate big defensin: identification, expression and bioactivity. Fish Shellfish Immunol. 2012;32:572–577. doi: 10.1016/j.fsi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Tessera V, Guida F, Juretic D, Tossi A. Identification of antimicrobial peptides from teleosts and anurans in expressed sequence tag databases using conserved signal sequences. FEBS J. 2012;279:724–736. doi: 10.1111/j.1742-4658.2011.08463.x. [DOI] [PubMed] [Google Scholar]
- Van der Krieken SE, Popeijus HE, Mensink RP, Plat J. CCAAT/enhancer binding protein β in relation to ER stress, inflammation, and metabolic disturbances. Biomed Res Int. 2015;2015:324815. doi: 10.1155/2015/324815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang S. Identification and expression of liver-specific genes after LPS challenge in amphioxus: the hepatic cecum as liver-like organ and “pre-hepatic” acute phase response. Funct Integr Genom. 2011;11:111–118. doi: 10.1007/s10142-010-0199-7. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang S, Zhao B, Lun L. Up-regulation of C/EBP by thyroid hormones: a case demonstrating the vertebrate-like thyroid hormone signaling pathway in amphioxus. Mol Cell Endocrinol. 2009;313:57–63. doi: 10.1016/j.mce.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Wang S, Li F, Hu L, Liu S, Li H, Zhang S. Structural and functional characterization of a TGFβ molecule from amphioxus reveals an ancient origin of both immune-enhancing and -inhibitory functions. Dev Comp Immunol. 2014;45:219–226. doi: 10.1016/j.dci.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang H, Li M, Gao Z, Zhang S. Identification, expression and regulation of amphioxus G6Pase gene with an emphasis on origin of liver. Gen Comp Endocrinol. 2015;214:9–16. doi: 10.1016/j.ygcen.2014.12.021. [DOI] [PubMed] [Google Scholar]
- Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Qu Q, Chen J. Identification, expression analysis, and antibacterial activity of Apolipoprotein A-I from amphioxus (Branchiostoma belcheri) Comp Biochem Phys B. 2019;238:110329. doi: 10.1016/j.cbpb.2019.110329. [DOI] [PubMed] [Google Scholar]
- Welsch U. The fine structure of the pharynx, cryptopodocytes and digestive caecum of amphioxus (Branchiostoma lanceolatum) Symp Zool Soc Lond. 1975;36:17–41. [Google Scholar]
- Xu N, Zhang S. Identification, expression and bioactivity of a chitotriosidase-like homolog in amphioxus: dependence of enzymatic and antifungal activities on the chitin-binding domain. Mol Immunol. 2012;51:57–65. doi: 10.1016/j.molimm.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Xu L, Yuan S, Li J, Ruan J, Huang S, Yang M, Huang H, Chen S, Ren Z, Xu A. The conservation and uniqueness of the caspase family in the basal chordate, amphioxus. BMC Biol. 2011;9:60. doi: 10.1186/1741-7007-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Pan J, Liu S, Xue Q, Zhang S. Three in one: identification, expression and enzymatic activity of lysozymes in amphioxus. Dev Comp Immunol. 2014;46:508–517. doi: 10.1016/j.dci.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Zhao Y, Zhang J, Bai C, Zhang C, Li K, Zhang H, Du X, Feng L. Identification of an amphioxus intelectin homolog that preferably agglutinates gram-positive over gram-negative bacteria likely due to different binding capacity to LPS and PGN. Fish Shellfish Immunol. 2012;33:11–20. doi: 10.1016/j.fsi.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Yan J, Xu L, Zhang Y, Zhang C, Zhao F, Feng L. Comparative genomic and phylogenetic analyses of the intelectin gene family: implications for their origin and evolution. Dev Comp Immunol. 2013;41:189–199. doi: 10.1016/j.dci.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Zhang Y, Li K, Xu L, Guo L, Kong Y, Feng L. Characterization and comparative analyses of two amphioxus intelectins involved in the innate immune response. Fish Shellfish Immunol. 2013;34:1139–1146. doi: 10.1016/j.fsi.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Yan X, Chen S, Huang H, Peng T, Lan M, Yang X, Dong M, Xu A, Huang S. Functional variation of IL-1R-associated kinases in the conserved MyD88-TRAF6 pathway during evolution. J Immunol. 2020;204:832–843. doi: 10.4049/jimmunol.1900222. [DOI] [PubMed] [Google Scholar]
- Yang M, Yuan S, Huang S, Li J, Xu L, Huang H, Tao X, Peng J, Xu A. Characterization of bbtTICAM from amphioxus suggests the emergence of a MyD88-independent pathway in basal chordates. Cell Res. 2011;21:1410–1423. doi: 10.1038/cr.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Zheng T, Cai X, Yu Y, Yu C, Guo L, Huang S, Zhu W, Zhu R, Yan Q, Ren Z, Chen S, Xu A. Genome-wide analyses of amphioxus microRNAs reveal an immune regulation via miR-92d targeting C3. J Immunol. 2013;190:1491–1500. doi: 10.4049/jimmunol.1200801. [DOI] [PubMed] [Google Scholar]
- Yang P, Huang S, Yan X, Huang G, Dong X, Zheng T, Yuan D, Wang R, Li R, Tan Y, Xu A. Origin of the phagocytic respiratory burst and its role in gut epithelial phagocytosis in a basal chordate. Free Radic Biol Med. 2014;70:54–67. doi: 10.1016/j.freeradbiomed.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Yao F, Li Z, Zhang Y, Zhang S. A novel short peptidoglycan recognition protein in amphioxus: identification, expression and bioactivity. Dev Comp Immunol. 2012;38:332–341. doi: 10.1016/j.dci.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Yao L, Qu B, Ma Z, Chen Y, Tan Y, Gao Z, Zhang S. Lectin-like and bacterial-agglutinating activities of heat shock proteins Hsp5 and Hsp90alpha from amphioxus Branchiostoma japonicum. Fish Shellfish Immunol. 2019;95:688–696. doi: 10.1016/j.fsi.2019.10.074. [DOI] [PubMed] [Google Scholar]
- Yu C, Dong M, Wu X, Li S, Huang S, Su J, Wei J, Shen Y, Mou C, Xie X, Lin J, Yuan S, Yu X, Yu Y, Du J, Zhang S, Peng X, Xiang M, Xu A. Genes “waiting” for recruitment by the adaptive immune system: the insights from amphioxus. J Immunol. 2005;174:3493–3500. doi: 10.4049/jimmunol.174.6.3493. [DOI] [PubMed] [Google Scholar]
- Yu Y, Huang H, Feng K, Pan M, Yuan S, Huang S, Wu T, Guo L, Dong M, Chen S, Xu A. A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J Immunol. 2007;179:8425–8434. doi: 10.4049/jimmunol.179.12.8425. [DOI] [PubMed] [Google Scholar]
- Yu Y, Yuan S, Huang H, Feng K, Pan M, Huang S, Dong M, Chen S, Xu A. Molecular and biochemical characterization of galectin from amphioxus: primitive galectin of chordates participated in the infection processes. Glycobiology. 2007;17:774–783. doi: 10.1093/glycob/cwm044. [DOI] [PubMed] [Google Scholar]
- Yuan S, Huang S, Zhang W, Wu T, Dong M, Yu Y, Liu T, Wu K, Liu H, Yang M, Zhang H, Xu A. An amphioxus TLR with dynamic embryonic expression pattern responses to pathogens and activates NF-κB pathway via MyD88. Mol Immunol. 2009;46:2348–2356. doi: 10.1016/j.molimm.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Yuan S, Liu T, Huang S, Wu T, Huang L, Liu H, Tao X, Yang M, Wu K, Yu Y, Dong M, Xu A. Genomic and functional uniqueness of the TNF receptor-associated factor gene family in amphioxus, the basal chordate. J Immunol. 2009;183:4560–4568. doi: 10.4049/jimmunol.0901537. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wu K, Yang M, Xu L, Huang L, Liu H, Tao X, Huang S, Xu A. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J Immunol. 2010;184:6874–6881. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
- Yuan S, Ruan J, Huang S, Chen S, Xu A. Amphioxus as a model for investigating evolution of the vertebrate immune system. Dev Comp Immunol. 2015;48:297–305. doi: 10.1016/j.dci.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Yuan S, Zheng T, Li P, Yang R, Ruan J, Huang S, Wu Z, Xu A. Characterization of amphioxus IFN regulatory factor family reveals an archaic signaling framework for innate immune response. J Immunol. 2015;195:5657–5666. doi: 10.4049/jimmunol.1501927. [DOI] [PubMed] [Google Scholar]
- Yuan S, Ruan J, Peng J, Xu A. Pattern recognition system in amphioxus. In: Xu A, editor. Amphioxus immunity. Beijing: Academic Press; 2016. pp. 85–119. [Google Scholar]
- Zhang S, Wang C, Wang Y, Wei R, Jiang G, Ju H. Presence and characterization of complement-like activity in the amphioxus Branchiostoma belcheri tsingtauense. Zool Sci. 2003;20:1207–1214. doi: 10.2108/zsj.20.1207. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xu Y, Li L, Wei S, Zhang S, Liu Z. Identification, evolution and expression of a CD36 homolog in the basal chordate amphioxus Branchiostoma japonicum. Fish Shellfish Immunol. 2013;34:546–555. doi: 10.1016/j.fsi.2012.11.043. [DOI] [PubMed] [Google Scholar]