Abstract
The morphology and molecular phylogeny of freshwater pleurostomatid ciliates are insufficiently explored. In the present study, we investigated three new Amphileptus species discovered in Lake Weishan and its vicinity, northern China, using standard alpha-taxonomic methods. Amphileptus paracarchesii sp. nov. is characterized by a lateral fossa (groove) in the posterior body portion, four macronuclear nodules, contractile vacuoles distributed along the dorsal margin, and 4–6 left and 44–50 right somatic kineties. Amphileptus pilosus sp. nov. differs from congeners by having 4–14 macronuclear nodules, numerous contractile vacuoles scattered throughout the cytoplasm, and 22–31 left and 35–42 right somatic kineties. Amphileptus orientalis sp. nov. is characterized by two ellipsoidal macronuclear nodules, three ventral contractile vacuoles, and about four left and 31–35 right somatic kineties. Phylogenetic analyses of nuclear small subunit ribosomal DNA (SSU rDNA) sequences indicate that the family Amphileptidae might be monophyletic while the genus Amphileptus is paraphyletic, as Pseudoamphileptus macrostoma robustly groups with Amphileptus sp. Although deep phylogenetic relationships of amphileptids are poorly resolved, multiple well-delimited species groups are recognizable within the genus Amphileptus.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42995-022-00143-0.
Keywords: Alpha-taxonomy, Morphology, New species, SSU rDNA
Introduction
The main goals of alpha-taxonomy are to describe new species and to re-describe insufficiently known species. Sound taxonomic research is the basis for all types of diversity and phylogenetic studies. Foissner et al. (2008) estimated that as much as 83‒89% of the ciliate diversity is still undescribed, which makes reporting of new species an important and timely objective. This holds also for pleurostomatids (order Pleurostomatida Schewiakoff, 1896), which are raptorial ciliates that are free-swimming or glide on substrates and are commonly found in a variety of aquatic environments. They are important constituents of aquatic microbial food webs due to their predation upon bacteria, algae, flagellates, and other ciliates, especially peritrichs (Foissner et al. 1995; Lynn 2008).
Amphileptidae Bütschli, 1889, are the second-most speciose family in the order Pleurostomatida. The name-bearing genus Amphileptus Ehrenberg, 1830 is the oldest genus of the family. It can be morphologically separated from other amphileptid genera by the following combination of features: (1) a narrowly rounded anterior body end that is not curved in a hook-like fashion; (2) right ciliary rows that form a suture in the anterior body half; (3) left somatic kineties that run meridionally and hence never encircle the cell; (4) a single perioral kinety that runs along the right and left side of the oral slit; and (5) perioral kineties that begin with dikinetids and continue posteriorly as monokinetids (Ehrenberg 1830; Foissner 1984; Foissner and Leipe 1995; Lynn 2008; Vd’ačný et al. 2015). However, some Amphileptus species deviate more or less significantly from this general pattern. Their anterior body end might be curved as in A. paracarchesii sp. nov., for example, the right ciliary rows could form an additional suture in the posterior body half (e.g., A. ensiformis, A. fusidens, A. fusiformis, A. litonotiformis, A. pleurosigma, and A. procerus) (Song and Wilbert 1989; Song 1991) and, rarely, the left somatic kineties form an additional, albeit inconspicuous, anterior suture (for example, in A. pilosus sp. nov.). Three species (A. meiianus, A. parafusidens, and A. yuianus) have three rather than two perioral kineties (Lin et al. 2005; Song and Wilbert 1989). Such an unusual variability in key taxonomic characters in amphileptids indicates their homoplastic nature and should be analyzed in the future with increased taxon and molecular marker sampling.
To date, about 60 nominal Amphileptus species have been reported (Foissner et al. 1995; Fryd-Versavel et al. 1975; Hu et al. 2019; Kahl 1931, 1933; Song and Wilbert 1989; Song et al. 2009; Stokes 1886; Vuxanovici 1960; Wu et al. 2021a). Like many other pleurostomatids, Amphileptus species often share a similar body shape, which makes the identification of living specimens very difficult. This is one of the reasons why the research history of Amphileptus is full of confusion and misidentifications. Therefore, it is necessary to circumscribe species of Amphileptus using a combination of molecular data and detailed observations of specimens both in vivo (including the shape and location of extrusomes, the number and position of contractile vacuoles, and the morphology and arrangement of cortical granules) and following protargol impregnation (including details of the nuclear apparatus and the somatic and oral ciliary pattern).
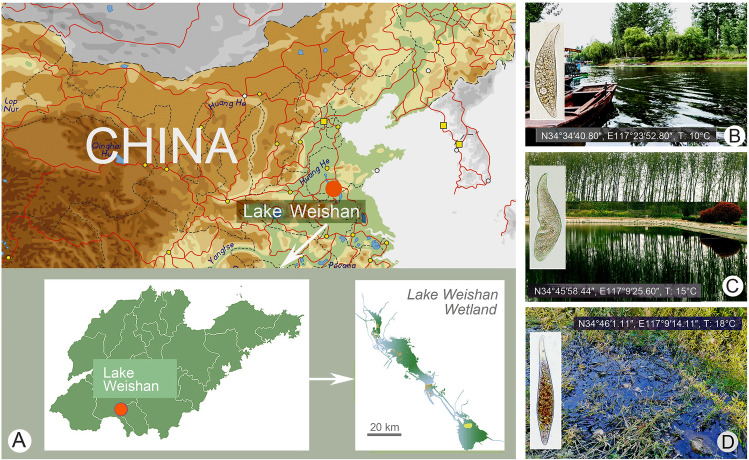
During the past two decades, our knowledge about the Amphileptidae has been significantly extended (e.g., Chen et al. 2011; Lin et al. 2005, 2007; Pan et al. 2014; Song et al. 2004; Sonntag and Foissner 2004; Wu et al. 2015, 2021a). Most of the recent studies have, however, focused mainly on marine and brackish species, while freshwater taxa remain comparatively understudied. Unbalanced taxon sampling affects the reliability of phylogenetic analyses. In the present study, we explored some freshwater habitats of northern China where we discovered three new Amphileptus species and determined their phylogenetic position using SSU rDNA sequences (Fig. 1).
Fig. 1.
Sampling locations and habitats. A Maps of China and Shandong Province, red circle shows the location of Lake Weishan. B Sampling site of A. paracarchesii sp. nov. C Sampling site of A. pilosus sp. nov. D Sampling site of A. orientalis sp. nov.
ZooBank registration number of this work: urn:lsid:zoobank.org:pub:FC380587-5D52-4991-BF7A-019EEB19012C.
Results
Family Amphileptidae Bütschli, 1889
Genus Amphileptus Ehrenberg, 1830
Amphileptus paracarchesii sp. nov. (Figs. 2, 3; Table 1)
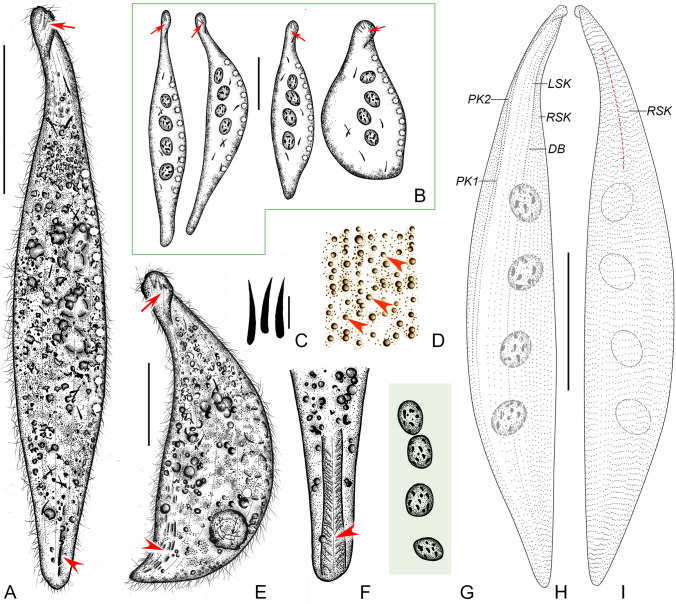
Fig. 2.
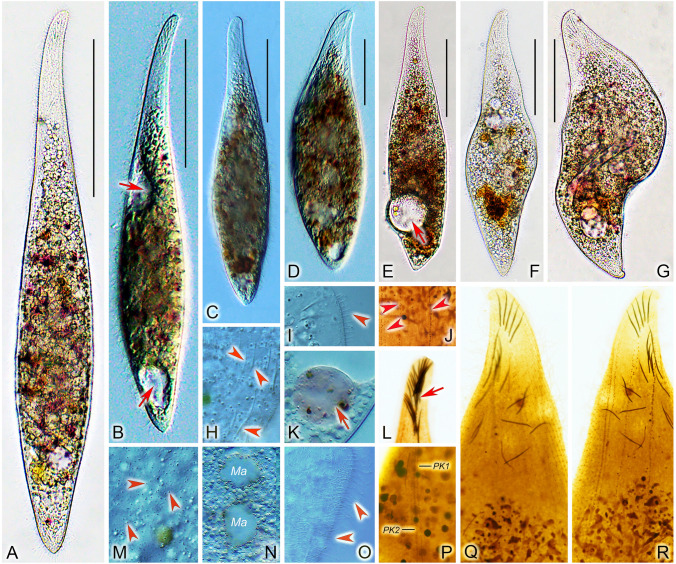
Amphileptus paracarchesii sp. nov. from life (A–G) and after protargol impregnation (H, I). A Left view of a representative individual, red arrow denotes the curved and twisted anterior body end, red arrowhead shows the lateral fossa (groove). B Shape variants, red arrows denote the anterior group of extrusomes. C Oral extrusomes. D Frontal view, showing cortical granules (arrowheads) of the left side. E A contracted individual, red arrow points to the apical group of extrusomes, red arrowhead marks the lateral groove. F Detail showing the lateral groove (arrowhead). G Nuclear apparatus. H Ciliary pattern of the left side of the holotype specimen. I Ciliary pattern of right the side of the holotype specimen, red dashed line shows the anterior suture. DB dorsal brush, LSK left somatic kineties, PK1 perioral kinety 1, PK2 perioral kinety 2, RSK right somatic kineties. Scale bars = 100 μm (A, B, E, H, I), 5 μm (C)
Fig. 3.
Amphileptus paracarchesii sp. nov. from life (A–H, J, L–M) and after protargol impregnation (I, K, N). A, B Right side view, arrows point to the groove, arrowhead shows the curved anterior body end. C Nuclear apparatus, arrowheads denote the four macronuclear nodules. D, G, J Shape variants, arrowheads mark the macronuclear nodules, arrow denotes the posterior groove. E Detail showing the apical group of extrusomes (arrow). F Detail showing the lateral groove situated in the posterior body region (arrow). H Contractile vacuoles (arrowheads). I Detail of the oral apparatus, showing a single perioral kinety right and left of the oral slit. K, N Detail of the anterior body portion, showing the ciliary pattern of the right and left sides of the holotype specimen. L A contracted individual, arrow shows the curved anterior body end. M Cytoplasmic extrusomes (arrowheads). Abbreviations: PK1, perioral kinety 1. PK2, perioral kinety 2. Scale bars = 100 μm
Table 1.
Morphometric characteristics of Amphileptus paracarchesii sp. nov. (upper line), Amphileptus pilosus sp. nov. (middle line) and Amphileptus orientalis sp. nov. (lower line). Data based on protargol-impregnated specimens
| Character | HT | Min | Max | Mean | Median | SD | CV | n |
|---|---|---|---|---|---|---|---|---|
| Body length (μm) | 233 | 184 | 334 | 239 | 233 | 52.91 | 22.3 | 22 |
| 298 | 215 | 359 | 278 | 283 | 40.80 | 13.4 | 20 | |
| 190 | 162 | 290 | 213 | 208 | 57.04 | 15.8 | 28 | |
| Body width (μm) | 55 | 47 | 74 | 59 | 60 | 6.44 | 11.0 | 22 |
| 84 | 59 | 91 | 72 | 69 | 9.63 | 13.4 | 20 | |
| 55 | 30 | 85 | 48 | 46 | 12.33 | 25.8 | 28 | |
| Number of right kinetiesa | 47 | 44 | 50 | 47 | 47 | 1.74 | 3.7 | 54 |
| 37 | 35 | 42 | 38 | 38 | 2.03 | 5.4 | 21 | |
| 33 | 31 | 35 | 33 | 33 | 1.29 | 3.9 | 31 | |
| Number of left kinetiesb | 6 | 4 | 6 | 5 | 5 | 0.50 | 9.5 | 55 |
| 27 | 22 | 31 | 27 | 28 | 2.23 | 8.3 | 20 | |
| 4 | 4 | 5 | 4 | 4 | 0.57 | 15.9 | 30 | |
| Number of dorsal brush dikinetids | 78 | 59 | 103 | 77 | 75 | 11.01 | 14.3 | 51 |
| 95 | 66 | 165 | 125 | 130 | 24.50 | 19.7 | 21 | |
| 56 | 47 | 74 | 63 | 65 | 7.85 | 12.5 | 22 | |
| Number of macronuclear nodules | 4 | 4 | 4 | 4 | 4 | 0 | 0 | 38 |
| 7 | 4 | 14 | 8 | 8 | 2.27 | 27.4 | 21 | |
| 2 | 2 | 2 | 2 | 2 | 0 | 0 | 30 | |
| Length of macronuclear nodule (μm) | - | 27 | 71 | 40 | 39 | 9.27 | 23.0 | 36 |
| 36 | 25 | 42 | 33 | 34 | 4.30 | 13.0 | 21 | |
| 50 | 32 | 67 | 43 | 42 | 8.88 | 20.8 | 29 | |
| Width of macronuclear nodule (μm) | - | 14 | 46 | 30 | 29 | 7.28 | 24.4 | 36 |
| 24 | 21 | 32 | 26 | 26 | 3.36 | 12.7 | 21 | |
| 3 | 20 | 50 | 30 | 27 | 7.57 | 25.4 | 29 |
CV coefficient of variation (%), HT holotype, Max maximum, Min minimum, n number of specimens investigated, SD standard deviation
aPerioral kinety 2 included
bPerioral kinety 1 and dorsal brush kinety included
Diagnosis. Body lanceolate, about 185–380 × 50–90 μm in vivo; a lateral fossa (groove) in posterior body portion; four macronuclear nodules; contractile vacuoles distributed along dorsal margin; extrusomes very narrowly ovate to clavate, arranged in an apical group and scattered throughout cytoplasm; cortical granules dot-like and colorless; 4–6 left and 44–50 right kineties; right anterior suture; perioral kinety 1 dikinetid in anterior one-third of body, monokinetid in posterior two-thirds; freshwater habitat.
Type material. A protargol slide with the holotype specimen circled by black ink, and two further slides with protargol-stained paratype specimens, have been deposited in Laboratory of Protozoology, Ocean University of China, with registration numbers ZGAT2020120701, ZGAT2020120702, and ZGAT2020120703, respectively.
Type locality. A touring boat port of Lake Weishan, China (N34°34′40.80″, E117°23′52.80″).
ZooBank registration number. Urn:lsid:zoobank.org:act:5324DEE9-57C2-4086-AE91-10314ABB2AE1.
Etymology. Composite of the Greek adjective “para-” (beside, near) and the species-group name carchesii, indicating the high morphological similarity of the new species to A. carchesii Stein, 1867.
SSU rDNA sequence. The SSU rDNA sequence of A. paracarchesii sp. nov. has been deposited in GenBank (accession no. OL828281). The sequence is 1563 nucleotides long and has a GC content of 42.48%.
Description. Body about 185–380 × 50–90 μm in vivo, typically lanceolate in lateral view, anterior end curved and twisted clockwise from right to left (Figs. 2A, B, E, 3A, L); highly contractile (Fig. 3A, L); neck region conspicuous occupying almost 1/4 of cell length, posterior region narrowed and tail-like occupying about 7% of cell length; fossa (groove) in posterior portion of left side, about 37–41 μm long (Fig. 3A, B, D, G, J). Nuclear apparatus in center of trunk region. Macronucleus invariably consists of four nodules; individual nodules ellipsoidal, about 15–25 × 8–12 μm in size in vivo; nucleoli globular to irregular, small to medium-sized, evenly distributed in macronuclear nodules (Figs. 2A, B, G, 3B–D). Micronuclei not observed. About 10 contractile vacuoles arranged in a row along dorsal body margin, 10–14 μm in diameter during diastole, pulsating every 30 s (Fig. 2A, B, H). Extrusomes very narrowly ovate, sometimes slightly curved, about 11.0–15.0 × 1.2‒1.5 μm in vivo; 2‒4 extrusomes attached to oral slit forming an apical group, numerous other extrusomes scattered throughout cytoplasm; impregnated deeply with protargol method used (Figs. 2A–C, E, 3E, M). Cortex very flexible; cortical granules dot-like, colorless, about 0.5 μm across, ordinarily spaced between adjacent left somatic kineties (Fig. 2D). Cytoplasm grayish, contains numerous granules (ca. 0.5–1.0 μm across) rendering cell opaque (Figs. 2A, E, 3A, L). Swims slowly while rotating about longitudinal body axis; feeds by attaching to stalk of sessile peritrichs using fossa as a sucker (Figs. 2F, 3A, B, F, G).
Somatic cilia about 10–13 μm long in vivo, very densely arranged on right side (Fig. 2A, E), sparsely distributed on left side and hence undetectable in vivo. Ciliary pattern as shown in Figs. 2H, I, 3I, K, N. About 44–50 right kineties including perioral kinety 2; intermediate kineties progressively shortened anteriorly forming a suture (Figs. 2H, I, 3K, N); 4‒6 left kineties including perioral kinety 1 and dorsal brush (Figs. 2H, 3N). Fossa lined by cilia that very likely have a thigmotactic function. Dorsal brush kinety composed of densely spaced dikinetids in anterior body third and of monokinetids in posterior two-thirds (Fig. 2H).
Oral slit extends over two-thirds down length of body, marked by dikinetids of perioral kineties. Perioral kinety 1 runs along left margin of oral slit, consists of densely spaced, oblique dikinetids in anterior body third and monokinetids in posterior two-thirds. Perioral kinety 2 extends along right margin of oral slit, consists of densely spaced, oblique dikinetids in anterior body half and monokinetids in posterior half (Figs. 2H, 3I). Nematodesmata not recognizable either in vivo or and after protargol impregnation.
Amphileptus pilosus sp. nov. (Figs. 4, 5; Table 1)
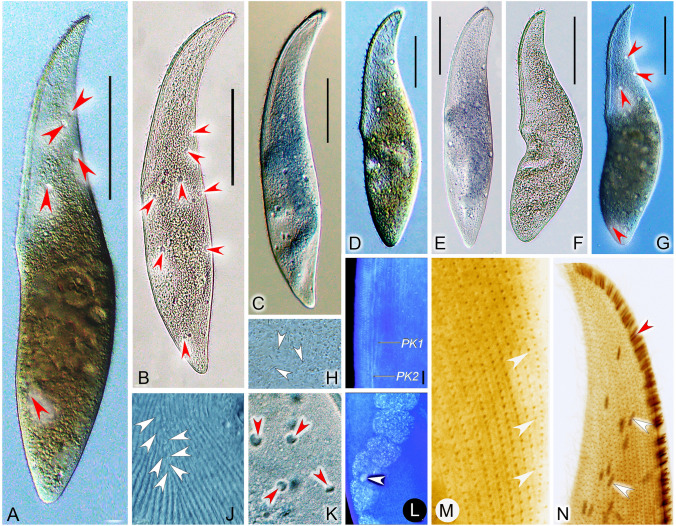
Fig. 4.
Amphileptus pilosus sp. nov. from life (A–E) and after protargol impregnation (F–H). A Left view of a representative individual, arrowheads point to the scattered contractile vacuoles. B Shape variants. C Oral extrusomes. D Nuclear apparatus. E Frontal view, showing cortical granules (arrowheads) of the left side. F Detail of the ventral margin of the posterior body region, showing the semi-suture made by progressively shortened ventral rightmost kineties and perioral kinety 2. G Ciliary pattern of the left side of the holotype specimen, asterisk marks the terminus of perioral kinety 1, dashed line denotes the rather indistinct anterior suture. H Ciliary pattern of the right side of the holotype specimen, dashed line marks the anterior suture, green-shaded area delimits the posterior suture. DB dorsal brush, PK1 perioral kinety 1, PK2 perioral kinety 2, RSK right somatic kineties. Scale bars = 100 μm in A, B, G, H; scale bars = 5 μm in C
Fig. 5.
Amphileptus pilosus sp. nov. from life (A–H, J–K) and after protargol impregnation (I, L–N). A–G Shape variants, red arrowheads denote the contractile vacuoles scattered throughout the body. H Cytoplasmic extrusomes (white arrowheads). I Detail of two perioral kineties, one right and one left of the oral slit. J Anterior suture made by abutting ciliary rows on the right side (white arrowheads). K Detail showing contractile vacuoles (red arrowheads). L Nuclear apparatus, white arrowhead marks the micronucleus closely associated with the moniliform macronuclear strand. M Detail of the ventral margin of the posterior body region of the holotype specimen, showing the suture made by progressively shortened right kineties (white arrowheads). N Detail of the anterior region of the right side of the holotype specimen, showing the oral extrusomes attached to the oral slit (red arrowhead) and the scattered cytoplasmic extrusomes (white arrowheads). PK1 perioral kinety 1, PK2 perioral kinety 2. Scale bars = 100 μm
Diagnosis. Body elongate-lanceolate, about 240–450 × 60–100 μm in vivo; macronucleus moniliform, composed of 4–14 nodules; numerous contractile vacuoles scattered throughout cell; extrusomes clavate, attached to anterior half of oral slit and scattered throughout cell; cortical granules dot-like, grayish; 22–31 left and 35–42 right kineties; right anterior suture, right posterior suture, right ventral semi-suture, and indistinct left anterior suture; perioral kinety 1 dikinetid, terminates above mid-portion of cell; freshwater habitat.
Type material. A protargol slide with the holotype specimen circled by black ink, and three further protargol slides with paratype specimens, have been deposited in Laboratory of Protozoology, Ocean University of China, with registration numbers ZGAT2020111601-1, ZGAT2020111601-2, ZGAT2020111601-3, and ZGAT2020111601-4, respectively.
Type locality. A fishpond located in the vicinity of Lake Weishan Wetland, China (N34°45′58.44″, E117°09′25.60″).
ZooBank registration number. urn:lsid:zoobank.org:act:976CCC1C-24E3-4576-A8D9-21F37A78A8FB.
Etymology. The Latin adjective pilosus (hairy) refers to the dense ciliation of the new species in comparison with congeners.
SSU rDNA sequence. The SSU rDNA sequence of A. pilosus sp. nov. has been deposited in GenBank (accession no. OL828282). The sequence is 1515 nucleotides long and has a GC content of 42.31%.
Description. Body about 240–450 × 60–100 μm in vivo, slightly contractile, elongate-lanceolate in lateral view, anterior end bluntly pointed to narrowly rounded, not twisted (Figs. 4A, B, 5A–G); neck region occupies about 15% of body length; posterior end gradually tapering, narrowly rounded, never tail-like (Figs. 4A, B, 5A–G). Nuclear apparatus extends through most of trunk. Macronucleus moniliform, consists of 4–14 ellipsoidal nodules about 23–30 × 13–15 μm in size in vivo; nucleoli globular to irregular, small to medium-sized, evenly distributed over macronuclear nodules (Fig. 4A, B, D). Single globular micronucleus, 8 μm in diameter after protargol impregnation, closely associated with one of the macronuclear nodules (Fig. 5L). Ten to 15 contractile vacuoles scattered throughout cell periphery, about 5–8 μm in diameter during diastole (Figs. 4A, B, 5A, B, G, K). Extrusomes clavate, almost straight or slightly curved, ca. 5.0–6.0 × 0.7–0.8 μm in vivo, some attached to anterior half of oral slit, others scattered throughout cell, impregnate strongly with the protargol method used (Figs. 4A–C, 5H, N). Cortex very flexible; cortical granules grayish, dot-like, ca. 0.5–1.0 μm in diameter, densely spaced between adjacent left somatic kineties (Fig. 4E). Cytoplasm grayish, studded with numerous granules and several 2.0–5.0 μm-sized food vacuoles rendering cell opaque (Figs. 4A, 5A–G). Locomotion by gliding slowly over substrate. When feeding, attaches to stalk of sessilid peritrich prey.
Somatic cilia about 7–9 μm long in vivo, very densely arranged on right side, not detected on left side in living specimens (Figs. 4A, 5J). Ciliary pattern as shown in Figs. 4F–H, 5I, M, N. About 35–42 right kineties including perioral kinety 2, intermediate kineties shortened anteriorly and posteriorly forming an anterior and a posterior suture (Figs. 4H, 5N), ventralmost kineties progressively shortened in posterior body region forming an inconspicuous semi-suture (Figs. 4F, 5M). About 22‒31 left kineties including perioral kinety 1 and dorsal brush; intermediate kineties shortened anteriorly forming an indistinct suture due to loosely spaced basal bodies (Fig. 4G). Dorsal brush kinety composed of closely spaced dikinetids in anterior body half and of monokinetids in posterior half (Fig. 4G).
Oral slit occupies about 40% of body length, marked by dikinetids of perioral kineties. Perioral kinety 1 runs along left margin of oral slit, terminates above cell equator, consists of densely spaced, obliquely oriented dikinetids (Figs. 4G, 5I). Perioral kinety 2 extends along right margin of oral slit, consists of narrowly spaced, obliquely oriented dikinetids in anterior body half and of monokinetids in posterior body half (Figs. 4G, 5I). Nematodesmata not recognizable in vivo or after protargol impregnation.
Amphileptus orientalis sp. nov. (Figs. 6, 7; Table 1)
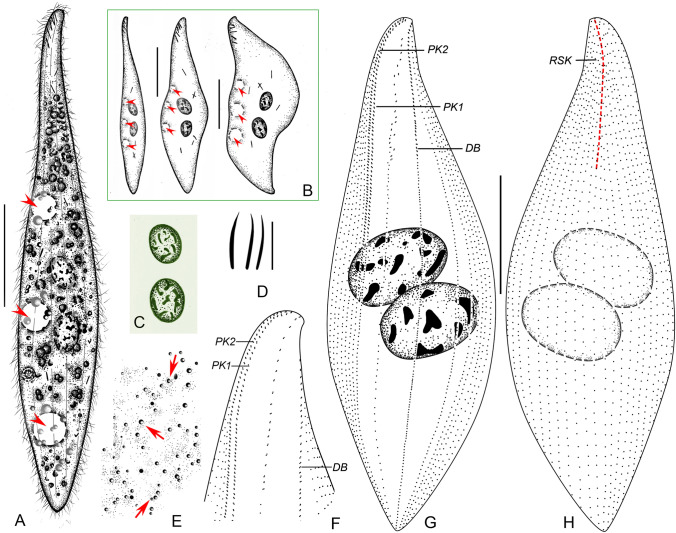
Fig. 6.
Amphileptus orientalis sp. nov. from life (A–E) and after protargol impregnation (F–H). A Left view of a representative individual, arrowheads point to the three ventral contractile vacuoles. B Shape variants, arrowheads denote the three contractile vacuoles. C Nuclear apparatus. D Oral extrusomes. E Cortical granules (arrows) of the left side. F Detail of the anterior region of the left side, showing the oral and somatic ciliary pattern. G Ciliary pattern of the left side of the holotype specimen. H Ciliary pattern of the right side of the holotype specimen, red dashed line denotes the anterior suture. DB dorsal brush, PK1 perioral kinety 1, PK2 perioral kinety 2, RSK right somatic kineties. Scale bars = 100 μm
Fig. 7.
Amphileptus orientalis sp. nov. from life (A–G, H, I, K, M–O) and after protargol impregnation (J, L, P–R). A–F Left side views of extended or only slightly contracted individuals, arrows mark the ventral contractile vacuoles. G Left side view of a contracted individual. H Cytoplasmic extrusomes (arrowheads). I Lateral view, showing the dorsal brush bristles (arrowhead). J Detail showing the very loosely arranged left somatic kineties (arrowheads). K Food vacuole (arrow). L Extrusomes attached to the oral slit (arrow). M Details of cell surface, showing cortical granules (arrowheads) of the left side. N Showing the two macronuclear nodules. O Somatic cilia (arrowheads) of lateral rightmost side kineties. P Detail of the oral apparatus, showing two perioal kineties, one right and one left of the oral slit. Q, R Details of the anterior region of the right (Q) and the left (R) side of the holotype specimen, showing the ciliary and extrusome patterns. Ma macronuclear nodules. PK1 perioral kinety 1, PK2 perioral kinety 2. Scale bars = 100 μm
Diagnosis. Body lanceolate, about 160–430 × 50–85 μm in vivo; two macronuclear nodules; three contractile vacuoles at ventral margin; extrusomes acicular, some attached to anterior 20–25% of oral slit, others mainly scattered in anterior body portion; cortical granules dot-like and colorless; 4–5 left and 31–35 right kineties; right anterior suture; perioral kinety 1 dikinetid in anterior portion, monokinetid in posterior portion; freshwater habitat.
Type material. A protargol slide with the holotype specimen circled by black ink, and one further protargol slide with paratype specimens, have been deposited in Laboratory of Protozoology, Ocean University of China, with registration numbers ZGAT20201023-1 and ZGAT2020102301-2, respectively.
Type locality. A wetland close to the mouth of the Xuehe River at Lake Weishan, China (N34°46′1.11″, E117°09′14.11″).
ZooBank registration number. urn:lsid:zoobank.org:act:13A8D2FC-710E-4DCE-8F0D-36415EC447A4.
Etymology. The Latin adjective orientalis (oriental) refers to the Chinese origin of the new species.
SSU rDNA sequence. The SSU rDNA sequence of A. orientalis sp. nov. has been deposited in GenBank (accession no. OL828283). The sequence is 1591 nucleotides long and has a GC content of 42.61%.
Description. Body about 160–430 × 50–85 μm in vivo; highly contractile; elongate-lanceolate in extended state, broadly lanceolate when contracted; anterior end bluntly pointed to narrowly rounded, not twisted; neck region occupies about 18% of body length, conspicuous in extended cells; posterior end gradually tapering and narrowly rounded, never tail-like (Figs. 6A, 7A–G). Nuclear apparatus in center of trunk. Invariably two macronuclear nodules; individual nodules separated from each other in vivo, while abutting in protargol-impregnated cells; nodules globular to ellipsoidal, about 30–50 × 20–40 μm in size in vivo; nucleoli usually irregular, medium to large-sized, evenly distributed over macronuclear nodules (Figs. 6A–C, 7N). Micronucleus not observed. Three contractile vacuoles along ventral margin, pulsating every 1 min; during diastole, anterior two vacuoles about 20 μm in diameter, subterminal vacuole up to 30 μm in diameter (Figs. 6A, B, 7B). Extrusomes acicular, usually almost straight, rarely curved; about 9.0–11.0 × 0.5–0.7 μm in vivo; some attached to anterior 20–25% of oral slit, others scattered mainly in anterior body portion; impregnate strongly with the protargol method used (Figs. 6A, B, D, 7H, L). Cortex very flexible; cortical granules colorless, dot-like, approximately 1.0 μm in diameter, ordinarily spaced between adjacent kineties on both right and left side of body (Figs. 6E, 7M). Cytoplasm grayish, studded with numerous granules and several 4–8 μm-sized food vacuoles rendering cell opaque (Figs. 6A, 7A–G, K). Locomotion by gliding on substrate or occasionally by swimming while rotating about long body axis.
Somatic cilia about 9–11 μm long in vivo, ordinarily arranged on right side, not detected on left side (Figs. 6A, 7I, O). Ciliary pattern as shown in Figs. 6F–H, 7J, P–R. About 31–35 right kineties including perioral kinety 2; intermediate kineties progressively shortened anteriorly forming a suture (Figs. 6H, 7Q). Four or five left kineties including perioral kinety 1 and dorsal brush (Figs. 6F, G, 7R). Left somatic kineties consisting of loosely spaced dikinetids in anterior body third, continues posteriorly as a row of loosely spaced monokinetids. Dorsal brush kinety composed of ordinarily spaced dikinetids in anterior body third, continues posteriorly as a row of ordinarily spaced monokinetids; brush bristles about 2 μm long in vivo (Figs. 6F, G, 7R).
Oral slit extends almost to mid-portion of cell, marked by dikinetids of perioral kineties. Perioral kinety 1 runs along left margin of oral slit, perioral kinety 2 runs along right margin; both perioral kineties extend to about mid-body with ordinarily spaced, oblique dikinetids and continue posteriorly with ordinarily to narrowly spaced monokinetids; dikinetidal portion of perioral kinety 2 slightly longer than that of kinety 1 (Figs. 6F, G, 7P, R). Nematodesmata not recognizable in vivo or after protargol impregnation.
Comparison of SSU rDNA sequences and phylogenetic analyses
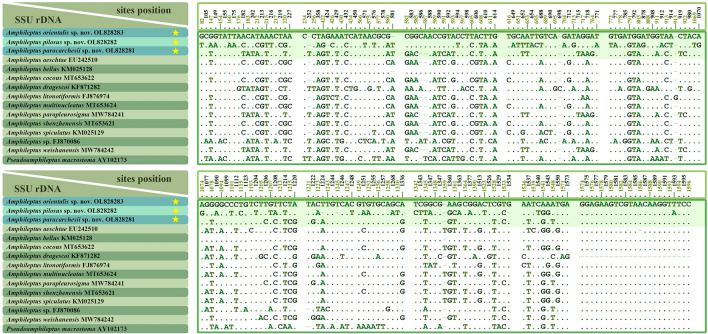
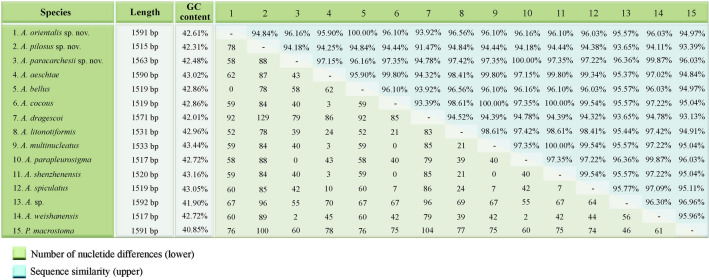
Sequence differences among amphiletid species range from none to 129 nucleotide positions. There are no nucleotide differences between A. paracarchesii sp. nov. and A. parapleurosigma Zhang et al., 2022, or between A. orientalis sp. nov. and A. bellus (Figs. 8, 9). On the other hand, A. paracarchesii sp. nov. and A. orientalis sp. nov. differ from other congeners by 2–79 and 52–92 nucleotide positions, respectively. Amphileptus pilosus sp. nov. differs from other amphileptids by 78–129 nucleotides, which corresponds to sequence similarities ranging from 91.47 to 94.84% (Figs. 8, 9).
Fig. 8.
Nucleotide differences in SSU rDNA sequences among the three new species (yellow asterisks) and other related taxa. Numbers represent nucleotide positions in the reference alignment. Dots represent matched sites, while dashes (‒) indicate deletions
Fig. 9.
Pairwise comparison of SSU rDNA sequences of 14 Amphileptus species and Pseudoamphileptus macrostoma. Numbers of different nucleotide positions are below the diagonal, while sequence similarities are above the diagonal
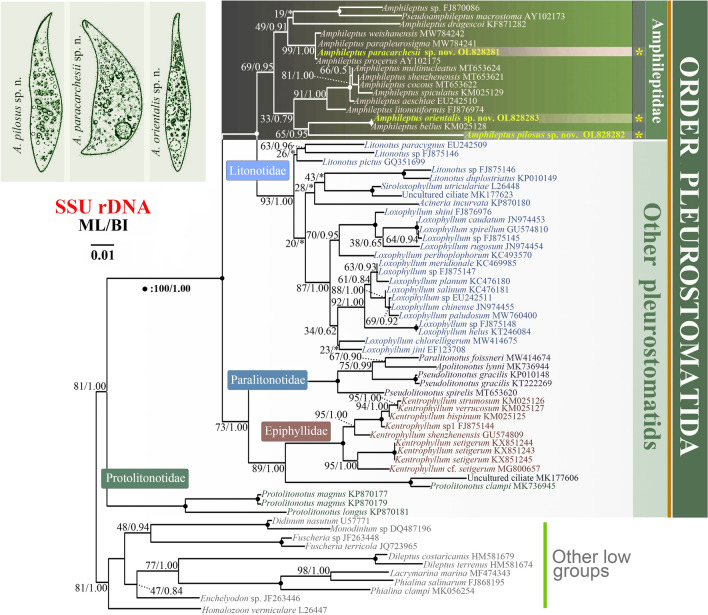
Topologies of phylogenetic trees generated by maximum likelihood (ML) and Bayesian inference (BI) are congruent, therefore only the ML tree is presented (Fig. 10). The family Amphileptidae (represented here by the genera Amphileptus and Pseudoamphiletus) is monophyletic although with low to moderate support (69% ML, 0.95 BI). The family Amphileptidae is divided into four more or less distinct subclades. The first subclade consists of Amphileptus sp. (FJ870086), Pseudoamphileptus macrostoma, and A. dragescoi (19% ML). The robust clustering of Amphileptus sp. with P. macrostoma causes paraphyly of the genus Amphileptus. The second subclade comprises A. paracarchesii sp. nov., A. weishanensis Zhang et al. 2022, A. parapleurosigma, and A. procerus (99% ML, 1.00 BI). The third subclade comprises six species (A. multinucleatus, A. shenzhenensis, A. cocous, A. spiculatus, A. aeschtae, and A. litonotiformis) (91% ML, 1.00 BI). Finally, Amphileptus orientalis sp. nov. and A. bellus form a fully supported clade that groups with A. pilosus sp. nov. though with variable support (65% ML, 0.95 BI).
Fig. 10.
Phylogenetic tree based on SSU rDNA sequences, showing the systematic positions of A. paracarchesii sp. nov., A. pilosus sp. nov., and A. orientalis sp. nov. Bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI) were mapped onto the best-scoring ML tree. The scale bar denotes one substitution per one hundred nucleotide positions
Discussion
Comparison of Amphileptus paracarchesii sp. nov. with similar species
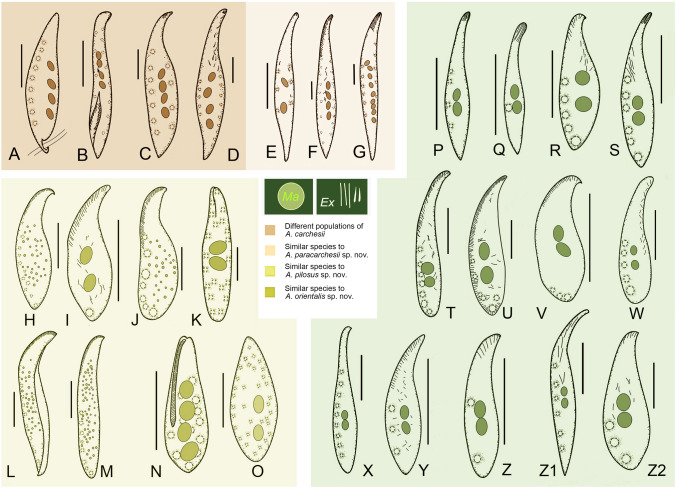
Amphiletpus paracarchesii sp. nov. resembles A. carchesii, A. parapleurosigma (Fig. 11E), A. quadrinucleatus (Dragesco and Njiné, 1971) Fryd-Versavel et al., 1975 (Fig. 11F), and A. weishanensis (Fig. 11G) in terms of its body size and numerous contractile vacuoles (Supplementary Table S1). The new species is most similar to A. carchesii. Besides having four macronuclear nodules, a dorsal row of contractile vacuoles, similar numbers of ciliary rows and a freshwater habitat, both species also share a peculiar fossa (groove) that is ciliated and situated in the posterior portion of the left body side. The fossa is used as a sucker, i.e., it serves for attaching to the stalk of sessile peritrichs, the preferred prey organisms of these species. Indeed, A. carchesii was first discovered on a colony of the sessilid peritrich ciliate Carchesium polypinum (Stein 1867). Subsequently, several populations were reported and studied using live observations (Canella 1960; Edmondson 1906; Gelei 1936; Kahl 1931, 1935; López-Ochoterena 1965; Schneider 1988). Foissner et al. (1995) reviewed previous studies, provided detailed diagnostic characters, and supplied also original in vivo and scanning electron microscope (SEM) photomicrographs, and hence their study might be considered as the authoritative redescription (Fig. 11A–D). Amphileptus paracarchesii sp. nov. can be clearly distinguished from A. carchesii by the morphology of the anterior body end and the absence/presence of a slime thread. The anterior body end is curved and twisted clockwise when viewed from the anterior aspect in A. paracarchesii sp. nov., while narrowly rounded and not twisted in A. carchesii. This distinguishing feature could be recognized in illustrations made by Edmondson (1906), Kahl (1931, 1935), and Canella (1960) as well as in the light and SEM micrographs provided by Foissner et al. (1995). Interestingly, some sort of twisting can be detected in Gelei’s (1936) drawings. Nevertheless, due to the lack of detailed morphological data and molecular information, the identity of Gelei’s specimens remains questionable. The other distinctive feature that separates these two species is the presence or absence of a slime thread. Amphileptus carchesii has a conspicuous slime thread that emerges from the posterior end of the lateral fossa. This thread is used as a lasso for attaching to the stalk of its peritrich prey. This peculiar structure and behavior were well documented by Edmondson (1906), Canella (1960), and Foissner et al. (1995). However, we have never observed this structure in A. paracarchesii sp. nov., hence we consider it a key species-specific character.
Fig. 11.
Species similar to Amphileptus paracarchesii sp. nov., A. pilosus sp. nov., and A. orientalis sp. nov. A Amphileptus carchesii, redrawn from Edmonson (1906). B Amphileptus carchesii, redrawn from Kahl (1935). C Amphileptus carchesii, redrawn from López-Ochoterena (1965). D Amphileptus carchesii, redrawn from Canella (1960). E Amphileptus parapleurosigma, redrawn from Zhang et al. (2022a). F Amphileptus quadrinucleatus, redrawn from Dragesco and Njiné (1971). G Amphileptus weishanensis, redrawn from Zhang et al. (2022a). H Amphileptus shenzhenensis, redrawn from Wu et al. (2021a). I Amphileptus litonotiformis, redrawn from Song (1991). J Amphileptus aeschtae, redrawn from Lin et al. (2007). K Apoamphileptus claparedii, redrawn from Foissner et al. (1995). L Amphileptus multinucleatus, redrawn from Wu et al. (2021a). M Amphileptus cocous, redrawn from Wu et al. (2021a). N Apoamphileptus robertsi, redrawn from Lin and Song (2004). O Amphileptus branchiarum, redrawn from Wenrich (1924). P Amphileptus ensiformis, redrawn from Song and Wilbert (1989). Q Amphileptus affinis, redrawn from Song and Wilbert (1989). R Amphileptus eigneri, redrawn from Lin et al. (2007). S Amphileptus gui, redrawn from Lin et al. (2005). T Amphileptus marinus, redrawn from Pan et al. (2014). U Amphileptus songi, redrawn from Song et al. (2004). V Amphileptus spiculatus, redrawn from Wu et al. (2015). W Hemiophrys rotundus, redrawn from Kahl (1931). X Hemiophrys pectinata, redrawn from Kahl (1931). Y Hemiophrys muscicola, redrawn from Kahl (1931). Z Hemiophrys bivacuolata, redrawn from Kahl (1931). Z1 Hemiophrys meleagris, redrawn from Kahl (1931). Z2 Amphileptus wilberti, redrawn from Pan et al. (2014). Scale bars = 80 μm
Amphileptus parapleurosigma and A. weishanensis can be separated from A. paracarchesii sp. nov. by the location of its contractile vacuoles (at ventral and dorsal margins vs. at dorsal margin only) and the number of right somatic kineties (19–24 in A. parapleurosigma and 56–61 in A. weishanenesis vs. 44–50 in A. paracarchesii sp. nov.) (Zhang et al. 2022a). Furthermore, A. parapleurosigma has two macronuclear nodules (vs. invariably four nodules in A. paracarchesii sp. nov.) and A. weishanensis possesses filiform extrusomes attached to the oral slit along its whole length (vs. narrowly ovate to clavate extrusomes attached only to the anterior portion of the oral slit in A. paracarchesii sp. nov.).
Amphileptus quadrinucleatus has four macronuclear nodules like A. paracarchesii sp. nov. (Dragesco and Njiné 1971). However, it can be distinguished from the latter by the location of its contractile vacuoles (at ventral and dorsal margins vs. dorsal margin only), by having fewer right somatic kineties (30–34 vs. 44–50), and by its habitat (marine vs. freshwater) (Dragesco and Njiné 1971).
Comparision of Amphileptus pilosus sp. nov. with similar species
Compared to its congeners, A. pilosus sp. nov. is unique in having the following combination of features: (1) an anterior and a posterior suture on the right side of the body; (2) an anterior suture on the left side of the body; (3) a semi-suture made by several posteriorly shortened kineties along the ventral margin of the right side of the body; and (4) perioral kinety 1 terminating above the mid-portion of the cell. Nevertheless, in terms of the body shape and the numerous scattered contractile vacuoles, eight species resemble the new species, namely, A. asechtae Lin et al., 2007 (Fig. 11J), A. branchiarum Wenrich, 1924 (Fig. 11O), A. cocous Wu et al., 2021 (Fig. 11M), A. litonotiformis Song, 1991 (Fig. 11I), A. multinucleatus Wang, 1934 (Fig. 11L), A. shenzhenensis Wu et al., 2021, (Fig. 11H), Apoamphileptus claparedii (Stein, 1867) Lin and Song, 2004 (Fig. 11K) and Apoamphileptus robertsi Lin et al., 2004 (Fig. 11N). Besides the unique ciliary pattern, A. pilosus sp. nov. can be distinguished from each of these by possessing more left somatic kineties (22–31) and clavate extrusomes. For further differences, see Supplementary Table S2.
Comparision of Amphileptus orientalis sp. nov. with its similar congeners
As concerns its nuclear apparatus and the location of its contractile vacuoles, A. orientalis sp. nov. resembles ten congeners and four Hemiophrys species (Supplementary Table S3), namely, A. affinis Song and Wilbert, 1989 (Fig. 11Q), A. bellus Wu et al., 2015, A. eigneri Lin et al., 2007 (Fig. 11R), A. ensiformis Song and Wilbert, 1989 (Fig. 11P), A. gui Lin et al., 2005 (Fig. 11S), A. marinus (Kahl 1931) Pan et al., 2014 (Fig. 11T), A. rotundus (Kahl, 1926) Foissner, 1988 (Fig. 11W), A. songi (Song, 2004) Pan et al., 2014 (Fig. 11U), A. spiculatus Wu et al., 2015 (Fig. 11V), A. wilberti Pan et al., 2014 (Fig. 11Z2), H. pectinata Kahl, 1926 (Fig. 11X), H. muscicola Kahl, 1931 (Fig. 11Y), H. bivacuolata Kahl, 1931 (Fig. 11Z), and H. meleagris (Ehrenberg, 1835) Kahl, 1931 (Fig. 11Z1). Although Foissner (1984) considered Hemiophrys Wrześniowski, 1866 (type species H. diaphanes Wrześniowski, 1866 by monotypy) to be a synonym of Amphileptus (type species A. cygnus Ehrenberg, 1830 by subsequent designation by Fromentel, 1875), the four aforementioned Hemiophrys species have not been formally transferred to Amphileptus. Because Kahl (1931) recognized both genera as valid, their type species are very insufficiently known and have no associated molecular information, we prefer to tentatively keep them in Hemiophrys and do not suggest any new combinations.
The new species is most similar to A. bellus, A. marinus, and A. wilberti. However, A. orientalis sp. nov. differs from A. bellus by having only one type of acicular extrusomes, (vs. two types, type I rod-shaped, type II spindle-like) and fewer left somatic kineties (4–5 vs. 6–7). Amphileptus orientalis sp. nov. can be distinguished from A. marinus and A. wilberti by having more right somatic kineties (31–35 vs. 13–21 in A. marinus and 15–19 in A. wilberti) and fewer left somatic kineties (4–5 vs. 5–8 in A. marinus and 7–8 in A. wilberti). Furthermore, the new species can be separated from A. marinus by having acicular (vs. fusiform) extrusomes (Pan et al. 2014; Song et al. 2004).
Amphileptus orientalis sp. nov. can be distinguished from A. ensiformis, A. affinis, A. eigneri, A. gui, and A. songi by the oral extrusome pattern. In A. orientalis sp. nov., the oral extrusomes are attached to the anterior 20–25% of the oral slit, whereas in A. songi they are distributed along the whole ventral margin and the posterior portion of the dorsal margin, and in the remaining species they form an apical group (Lin et al. 2005, 2007; Song and Wilbert 1989; Song et al. 2004). Amphileptus orientalis sp. nov. differs from A. spiculatus by having a longer body (160–430 μm vs. 85–150 μm), more right somatic kineties (31–35 vs. 11–14), fewer left somatic kineties (4–5 vs. 6–8), and acicular (vs. pyriform) extrusomes (Wu et al. 2015).
Kahl (1931) described five Hemiophrys species that resemble A. orientalis sp. nov. in terms of their morphology in vivo, namely, H. rotunda, H. pectinata, H. muscicola, H. bivacuolata, and H. meleagris. Hemiophrys rotunda was transferred to Amphileptus by Foissner (1988), while the remaining four species remain members of the genus Hemiophrys. Amphileptus orientalis sp. nov. is distinguished from all of these species except H. meleagris by its longer body (160–430 μm vs. 160–200 μm in A. rotundus, 200 μm in H. pectinata, 130 μm in H. muscicola, and 100–130 μm in H. bivacuolata) and in having more right somatic kineties (31–35 vs. 15–16 in A. rotundus, 10 in H. pectinata, 8 in H. bivacuolata). Hemiophrys meleagris differs from A. orientalis sp. nov. by having more contractile vacuoles (6 vs. 3), filiform (vs. acicular) extrusomes, and by the shape of the posterior end of the body (acutely tapered vs. rounded) (Kahl 1931).
Morphological and molecular evolution
The order Pleurostomatida is consistently recovered as a monophyletic group in morphological cladistic analyses and SSU rDNA phylogenies (Chi et al. 2021; Gao et al. 2008; Pan et al. 2020; Rajter and Vd’ačný 2017; Vd’ačný et al. 2011a, b, 2014, 2015; Wu et al. 2017, 2022; Zhang et al. 2012, 2022b; present study). Based on morphological and molecular data, pleurostomatids are currently divided into five families: Amphileptidae, Epiphyllidae, Litonotidae, Paralitonotidae, and Protolitonotidae. In the SSU rDNA tree, each family is monophyletic apart from Protolitonotidae, which is paraphyletic as Protolitonotus clampi clusters with Epiphyllidae rather than Protolitonotidae (Fig. 10).
As concerns the family Amphileptidae, the name-bearing genus Amphileptus is paraphyletic due to Pseudoamphileptus macrostoma (AY102173) nesting within it. The Amphileptidae consists of four subclades. The first subclade includes Amphileptus sp. (FJ870086), Pseudoamphileptus macrostoma and possibly A. dragescoi although the statistical support for the position of latter is low. The second subclade comprises A. paracarchesii sp. nov., A. weishanensis, A. parapleurosigma, and A. procerus. Interestingly, all members of the second subclade share an apical group of extrusomes. The third subclade comprises six species (A. multinucleatus, A. shenzhenensis, A. cocous, A. spiculatus, A. aeschtae, and A. litonotiformis) whose close kinship is morphologically supported by the possession of a row of contractile vacuoles along the ventral margin, an unusual feature in pleurostomatids. In the fourth subclade, which is made by A. pilosus sp. nov., A. orientalis sp. nov., and A. bellus, the former species has a comparatively long branch. Interestingly, A. pilosus sp. nov. has a peculiar ciliary pattern, i.e., the presence of the anterior and posterior sutures on the right side, an anterior suture on the left side, and a postoral semi-suture on the right side. Nonetheless, SSU rDNA phylogenies suggest that this deviating and complex ciliary pattern might be a species-level rather than a genus-level character (Fig. 10). This hypothesis is also corroborated by the rather high variability in the presence/absence of the right posterior suture within the genus Amphileptus (Table 2). Since there are some further peculiarities in the somatic ciliary pattern, the branch leading to A. pilosus sp. nov. is comparatively long and the statistical support for its position is variable, we cannot exclude the possibility that it represents a separate genus. However, we retain A. pilosus sp. nov. within the genus Amphileptus pending the availability of greater taxon sampling and sequences of more gene markers.
Table 2.
Comparison of Amphileptus species with respect to their somatic and oral ciliary patterns and habitat
| Species | RAS | RPS | LAS | RVS | Number of PK | Type of PK1 | Habitat | Reference |
|---|---|---|---|---|---|---|---|---|
| A. paracarchesii sp. nov | + | – | – | – | 2 | Type 1 | FW | Present work |
| A. orientalis sp. nov | + | – | – | – | 2 | Type 1 | FW | Present work |
| A. pilosus sp. nov | + | + | + | + | 2 | Type 2 | FW | Present work |
| A. aeschtae Lin et al., 2007 | + | – | – | – | 2 | Type 1 | MW | [1] |
| A. affinis Song and Wilbert, 1989a | + | + a | – | – | 2 | Type 1 | FW | [2] |
| A. agilis (Penard, 1922) Song and Wilbert, 1989a | + | – | – | – | 2 | Type 1 | FW | [2] |
| A. bellus Wu et al., 2015 | + | – | – | – | 2 | Type 1 | BW | [3] |
| A. cocous Wu et al., 2021 | + | + a | – | – | 2 | Type 1 | BW | [4] |
| A. eigneri Lin et al., 2007 | + | – | – | – | 2 | Type 1 | MW | [1] |
| A. ensiformis Song and Wilbert, 1989a | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. falcatus Song and Wilbert, 1989a | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. fusidens (Kahl 1926) Song and Wilbert 1989a | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. fusiformisa | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. gui Lin et al., 2005 | + | – | – | – | 2 | Type 1 | MW | [5] |
| A. litonotiformis Song, 1991a | + | + | – | – | 2 | Type 1 | MW | [6] |
| A. marinus (Kahl, 1931) Pan et al., 2014 | + | – | – | – | 2 | Type 1 | MW | [7] |
| A. meiianus Song and Wilbert, 1989a | + | – | – | – | 3 | Type 1 | FW | [2] |
| A. mutinucleatus Wang, 1934 | + | – | – | – | 2 | Type 1 | BW | [4] |
| A. parafusidens Song and Wilbert, 1989a | + | + | – | – | 3 | Type 1 | FW | [2] |
| A. parapleurosigma Zhang et al., 2022 | + | – | – | – | 2 | Type 1 | FW | [8] |
| A. pleurosigma (Stokes, 1884) Foissner, 1984a | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. proceroformis Song and Wilbert, 1989a | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. procerus (Penard, 1922) Song and Wilbert, 1989a | + | + | – | – | 2 | Type 1 | FW | [2] |
| A. puncatatus (Kahl, 1926) Foissner, 1984 | + | – | – | – | 2 | Type 1 | FW | [9] |
| A. shenzhenensis Wu et al., 2021 | + | – | – | – | 2 | Type 1 | FW | [4] |
| A. songi Pan et al., 2014 | + | – | – | – | 2 | Type 1 | MW | [10] |
| A. spiculatus Wu et al., 2015 | + | – | – | – | 2 | Type 1 | BW | [3] |
| A. yuianus Lin et al., 2005 | + | – | – | – | 3 | Type 1 | FW | [5] |
| A. weishanensis Zhang et al., 2022 | + | – | – | – | 2 | Type 1 | FW | [8] |
| A. wilberti Pan et al., 2014 | + | – | – | – | 2 | Type 1 | MW | [7] |
BW brackish water, FW freshwater, LAS left anterior suture, MW marine water, PK perioral kinety, RAS right anterior suture, RPS right posterior suture, RVS right ventral semi-suture
aData from illustrations
+, present; −, absent
Type 1: PK1 begins with dikinetids and continues posteriorly with monokinetids
Type 2: PK1 terminates above the mid-portion of cell with dikinetids
References: [1] Lin et al. (2007); [2] Song and Wilbert (1989); [3] Wu et al. (2015); [4] Wu et al. (2021a); [5] Lin et al. (2005); [6] Song (1991); [7] Pan et al. (2014); [8] Zhang et al. (2022a); [9] Foissner et al. (1995); [10] Song et al. (2004)
Hitherto, 30 Amphileptus species have been studied using protargol impregnation and, therefore, their ciliary pattern is known. These species consistently exhibit a right anterior suture, which was traditionally considered a generic character (Foissner and Leipe 1995; Vd’ačný et al. 2015). Only 12 species (including A. pilosus sp. nov.) have, in addition, a right posterior suture (Table 2). Interestingly, members of the family Epiphyllidae also possess both an anterior and a posterior suture on the right side of the body. Given their molecular phylogenies, the possession of two sutures on the right side is very likely a homoplastic character that evolved convergently in amphileptids and ephiphyllids. Another homoplastic feature of amphileptids might be the number of perioral kineties. According to Foissner and Leipe (1995), the family Amphileptidae was defined, inter alia, by having two perioral kineties. However, three Amphileptus species (A. meiianus, A. parafusidens and A. yuianus) have three perioral kineties, similar to members of the family Litonotidae (Lin et al. 2005; Song and Wilbert 1989). Due to the lack of molecular data, the generic affiliation of these three species could not be tested and remains questionable. It is noteworthy that A. pilosus sp. nov. also differs from its congeners by its oral ciliary pattern, i.e., its perioral kinety 1 terminates above the mid-portion of the cell and is entirely built from dikinetids, whereas it continues to the posterior end of the body as monokinetds in all other congeners (Table 2). Nevertheless, the molecular data support the classification of A. pilosus sp. nov. within the family Amphileptidae (Fig. 10).
It is well known that the SSU rDNA sequence does not necessarily carry a species-specific signal, i.e., distinct species could share an identical SSU rDNA sequence (e.g., Doerder 2019; Lynn and Strüder-Kypke 2006; Rataj and Vd’ačný 2021). This is the case both for A. paracarchesii sp. nov. and A. parapleurosigma, and for A. orientalis sp. nov. and A. bellus. However, A. paracarchesii sp. nov. distinctly differs from A. parapleurosigma by the nuclear apparatus (4 vs. 2 macronuclear nodules), the contractile vacuole pattern (dorsal row vs. dorsal and ventral rows of vacuoles), and the number of the right somatic kineties (44–50 vs. 19–24) (Zhang et al. 2022a). Amphileptus orientalis sp. nov. can be clearly distinguished from A. bellus by its extrusome pattern (extrusomes attached to the anterior 20%–25% of the oral slit vs. along the entire oral slit and tail), the number of left somatic kineties (4–5 vs. 6–7), and the habitat (freshwater vs. brackish). These findings support the ascertain that 100% identity of SSU rDNA sequences does not necessarily correlate with morphospecies conspecificity. Consequently, SSU rDNA does not appear to be an appropriate barcode for members of the genus Amphileptus and species identities need to be confirmed by morphological analyses and/or by faster evolving molecular markers such as ITS2 and COI gene sequences.
Materials and methods
Sample collection
Amphileptus paracarchesii sp. nov. was collected from a touring boat port (Fig. 1B) of Lake Weishan, China (N34°34′40.80″, E117°23′52.80″) on 7th December 2020. The water temperature was 10 °C, the pH was 8.28, and the DO was 11.75 mg/L.
Amphileptus pilosus sp. nov. was sampled from a fishpond (Fig. 1C) located in the vicinity of Lake Weishan, China (N34°45′58.44″, E117°09′25.60″) on 11th November 2020. The water temperature was 15 °C.
Amphileptus orientalis sp. nov. was isolated from a wetland close to the mouth of the Xuehe River (Fig. 1D) in Lake Weishan Wetland, China (N34°46′1.11″, E117°09′14.11″) on 23rd October 2020. The wetland was densely populated with aquatic plants and the water temperature was 18 °C.
Amphileptus paracarchesii sp. nov. and A. pilosus sp. nov. were collected using microscope slides that served as artificial substrates for the growth of biofilms when left immersed in the water for a sufficient period of time (Wu et al. 2021b). Samples were cultured in Petri dishes with habitat water at room temperature (about 20 °C). Some rice grains were added to stimulate the growth of bacteria that served as a food source for ciliates. Amphileptus paracarchesii sp. nov. was investigated immediately after collection as it was sufficiently abundant on setting up the culture. In contrast, A. pilosus sp. nov. only became sufficiently abundant after three days of cultivation in the laboratory. Amphileptus orientalis sp. nov. was sampled directly from the wetland using pipettes. After transportation to the laboratory, it was also cultivated in Petri dishes supplied with some rice grains.
Observation and identification
Live cells were observed using bright field and differential interference contrast microscopy (BX53, Olympus, Japan) at 100–1000 × magnification following the recommendations of Foissner (2014). The protargol impregnation of Wilbert (1975) was used to reveal the ciliary pattern and nuclear apparatus. The protargol reagent was synthesized according to the in-house protocol of Pan et al. (2013). Drawings of stained specimens were made with the help of a camera lucida and photomicrographs. Counts and measurements were conducted at a magnification of 1000 × . Terminology and systematics are mainly according to Foissner and Leipe (1995), Foissner and Xu (2007), and Wu et al. (2017).
DNA extraction, PCR amplification, and DNA sequencing
For each species, a single cell was isolated from raw cultures and washed five times with filtered habitat water to avoid contamination. Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. PCR amplification of the nuclear SSU rDNA was performed with the Q5 Hot Start High-Fidelity 2 × Master Mix DNA polymerase and the universal eukaryotic primers 82F (5’-GAA ACT GCG AAT GGC TC-3’) and 5.8S-R (5’-TAC TGA TAT GCT TAA GTT CAG CGG-3’) (Gao et al. 2012; Jerome et al. 1996) for A. pilosus sp. nov. and 18S-F (5’-AAC CTG GTT GAT CCT GCC AGT-3’) and 18S-R (5’-TGA TCC TTC TGC AGG TTC ACC TAC-3’) (Medlin et al. 1988) for A. paracarchesii sp. nov. and A. orientalis sp. nov. Cycling parameters followed the protocol of Chi et al. (2020). PCR products were sequenced in both directions using the Sanger method in Tsingke Biotechnology Co. Ltd., Qingdao, China, using the PCR primers and three internal primers: pro + B (5’-GGT TAA AAA GCT CGT AGT-3’), 900F (5’-CGA TCA GAT ACC GTC CTA GT-3’), and 900R (5’-ACT AGG ACG GTA TCT GAT CG-3’) (Wang et al. 2017). Sequencing fragments were assembled into contigs using SeqMan ver. 7.1 (DNAStar) and the final partial SSU rDNA sequences were edited in BioEdit ver 5.0.6 (Hall 1999).
Phylogenetic analyses
In addition to the three newly obtained Amphileptus sequences, SSU rDNA sequences of 57 pleurostomatids and 11 other free-living litostomateans (outgroup) were downloaded from the GenBank database (for accession numbers, see Fig. 8) for phylogenetic analyses. Sequences were aligned by the Muscle algorithm on the webserver Guidence (http://guidance.tau.ac.il/ver2/) with default settings (Sela et al. 2015). Sequences were trimmed to common length in BioEdit. The final alignment comprised 1652 characters, including 464 variable and 361 parsimony-informative sites. Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian inference (BI) analyses. ML analyses were carried out with RAxML-HPC2 (Stamatakis 2014) on XSEDE ver. 8.2.12 on the CIPRES Science Gateway (Miller et al. 2010) under the GTRGAMMA model and with 1000 rapid bootstrap pseudoreplicates. Bayesian inference analyses were conducted using MrBayes ver. 3.2.7 (Ronquist et al. 2012) on the CIPRES Science Gateway under the GTR + I + G model, which was selected as the best-fit model by MrModeltest ver. 2.2 via the Akaike Information Criterion (Nylander 2004). Bayesian analyses were run for ten million generations with a sampling frequency of 100. The first 10,000 trees were discarded as burn-in. MEGA ver. 10.2 was used to display the tree topologies (Kumar et al. 2018).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (project nos. 32030015, 32170533, 31961123002, 32111530116), the China Postdoctoral Science Foundation (project no. 2021M701276), the Slovak Research and Development Agency (project no. APVV-19-0076), and the King Saud University, Saudi Arabia (project no. RSP2022R7).
Author contributions
HP and PV conceived the study. GZ carried out the morphological experiments (live observation and protargol staining). GZ, YS and YL carried out DNA extraction, PCR and phylogenetic analyses. XC designed the field survey and provided institutional support. GZ contributed to the writing of the first draft of the manuscript. YS, YL, XC, SA, PV and HP contributed to the revision and all authors approved the final version.
Data availability
Sequence data are available in GenBank (Accession Numbers: OL828281–OL828283).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and human rights statements
We declare that all applicable international, national, and or institutional guidelines for sampling, care, and experimental use of organisms for the study have been followed and all necessary approvals have been obtained.
Footnotes
Special topic: Ciliatology.
Contributor Information
Peter Vďačný, Email: peter.vdacny@uniba.sk.
Hongbo Pan, Email: hbpan@shou.edu.cn.
References
- Canella MF. Contributo ad una revisione dei generi Amphileptus, Hemiophrys e Lionotus (Ciliata, Holotricha, Gymnostomata) Ann Univ Ferrara. 1960;2:47–95. [Google Scholar]
- Chen RM, Lin XF, Warren A. A new pleurostomatid ciliate, Amphileptus salignus n. sp. (Protozoa, Ciliophora), from mangrove wetlands in southern China. Zootaxa. 2011;3048:62–68. doi: 10.11646/zootaxa.3048.1.4. [DOI] [Google Scholar]
- Chi Y, Duan LL, Luo XT, Cheng T, Warren A, Huang J, Chen XR. A new contribution to the taxonomy and molecular phylogeny of three, well-known freshwater species of the ciliate genus Spirostomum (Protozoa: Ciliophora: Heterotrichea) Zool J Linn Soc. 2020;189:158–177. doi: 10.1093/zoolinnean/zlz115. [DOI] [Google Scholar]
- Chi Y, Wang Z, Lu BR, Ma HG, Mu CJ, Warren A, Zhao Y. Taxonomy and phylogeny of the dileptid ciliate genus Paradileptus (Protista: Ciliophora), with a brief review and redescriptions of two species isolated from a wetland in northern China. Front Microbiol. 2021;12:709566. doi: 10.3389/fmicb.2021.709566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fromentel E. Études sur les microzoaires ou infusoires proprement dits comprenant de nouvelles recherches sur leur organisation, leur classification et la description des espèces nouvelles ou peu connues. Paris: G Masson; 1875. [Google Scholar]
- Doerder FP. Barcodes reveal 48 new species of Tetrahymena, Dexiostoma, and Glaucoma: phylogeny, ecology, and biogeography of new and established species. J Eukaryot Microbiol. 2019;66:182–208. doi: 10.1111/jeu.12642. [DOI] [PubMed] [Google Scholar]
- Dragesco J, Njiné T. Compléments à la connaissance des ciliés libres du Cameroun. Ann Fac Sci Univ Féd Cameroun. 1971;7–8:97–140. [Google Scholar]
- Edmondson CH. The protozoa of Iowa. A study of species known to occur in the waters of this state. Proc Davenport Acad Sci. 1906;11:1–123. [Google Scholar]
- Ehrenberg CG. Beiträge zur Kenntnis der Organisation der Infusorien und ihrer geographischen Verbreitung, besonders in Sibirien. Abh Dt Akad Wiss Berl. 1830;1832:1–88. [Google Scholar]
- Foissner W. Taxonomie und Ökologie einiger Ciliaten (Protozoa, Ciliophora) des Saprobiensystems. I: Genera Litonotus, Amphileptus, Opisthodon. Hydrobiologia. 1984;119:193–208. doi: 10.1007/BF00015210. [DOI] [Google Scholar]
- Foissner W. Taxonomic and nomenclatural revision of Sládeček‘s list of ciliates (Protozoa: Ciliophora) as indicators of water quality. Hydrobiologia. 1988;166:1–64. doi: 10.1007/BF00017483. [DOI] [Google Scholar]
- Foissner W. An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int J Syst Evol Microbiol. 2014;27:271–292. doi: 10.1099/ijs.0.057893-0. [DOI] [PubMed] [Google Scholar]
- Foissner W, Leipe D. Morphology and ecology of Siroloxophyllum utriculariae (Penard, 1922) n. g., n. comb. (Ciliophora, Pleurostomatida) and an improved classification of Pleurostomatid ciliates. J Eukaryot Microbiol. 1995;42:476–490. doi: 10.1111/j.1550-7408.1995.tb05894.x. [DOI] [Google Scholar]
- Foissner W, Xu KD. Monograph of the Spathidiida (Ciliophora, Haptoria): Protospathidiidae, Arcuospathidiidae, Apertospathulidae. Dordrecht: Springer Verlag; 2007. [Google Scholar]
- Foissner W, Berger H, Blatterer H, Kohmann F. Taxonomische und ökologische revision der ciliaten des saprobiensystems—Band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte Des Bayer Landesamtes Für Wasserwirtschaft. 1995;1(95):1–540. [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodivers Conserv. 2008;17:345–363. doi: 10.1007/s10531-007-9254-7. [DOI] [Google Scholar]
- Fryd-Versavel G, Iftode F, Dragesco J. Contribution à la connaissance de quelques ciliés gymnostomes. II. Prostomiens, pleurostomiens: morphologie, stomatogenèse. Protistologica. 1975;4:509–530. [Google Scholar]
- Gao S, Song WB, Ma HW, Clamp JC, Yi ZZ, Al-Rasheid KAS, Chen ZG, Lin XF. Phylogeny of six genera of the subclass Haptoria (Ciliophora, Litostomatea) inferred from sequences of the gene coding for small subunit ribosomal RNA. J Eukaryot Microbiol. 2008;55:562–566. doi: 10.1111/j.1550-7408.2008.00360.x. [DOI] [PubMed] [Google Scholar]
- Gao F, Katz LA, Song WB. Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol Phylogenet Evol. 2012;64:308–317. doi: 10.1016/j.ympev.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Gelei JV. Beiträge zur Ciliatenfauna der Umgebung von Szeged. Zwei Gymnostomata-Arten: Amphileptus carchesii Stein und Bryophyllum hyalinum n. sp. Acta Biol. 1936;4:1–11. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hu XZ, Lin XF, Song WB. Ciliate atlas: species found in the South China Sea. Beijing: Science Press; 2019. [Google Scholar]
- Jerome CA, Lynn DH, Simon EM. Description of Tetrahymena empidokyrea n. sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can J Zool. 1996;74:1898–1906. doi: 10.1139/z96-214. [DOI] [Google Scholar]
- Kahl A. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria). 2. Holotricha außer den im 1. Teil Behandelten Prostomata Tierwelt Dtl. 1931;21:181–398. [Google Scholar]
- Kahl A. Ciliata libera et ectocommensalia. In: Grimpe G, Wagler E, editors. Die Tierwelt der Nord- und Ostsee 23. Leipzig: Akademische Verlagsgesellschaft Becker & Erler; 1933. pp. 147–183. [Google Scholar]
- Kahl A. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) 4. Peritricha Und Chonotricha Tierwelt Dtl. 1935;30:651–886. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Bio Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XF, Song WB. Establishment of a new amphileptid genus, Apoamphileptus nov. gen. (Ciliophora, Litostomatea, Pleurostomatida), with description of a new marine species, Apoamphileptus robertsi nov. spec. from Qingdao. China J Eukaryot Microbiol. 2004;51:618–625. doi: 10.1111/j.1550-7408.2004.tb00595.x. [DOI] [PubMed] [Google Scholar]
- Lin XF, Song WB, Warren A. Two new marine pleurostomatid ciliates from China, Amphileptus gui nov. spec. and Amphileptus yuianus nov. spec. (Ciliophora, Pleurostomatida) Eur J Protistol. 2005;41:163–173. doi: 10.1016/j.ejop.2005.01.002. [DOI] [Google Scholar]
- Lin XF, Song WB, Li JQ. Amphileptus aeschtae nov. spec. and Amphileptus eigneri nov. spec. (Ciliophora, Pleurostomatida), two new marine pleurostomatid ciliates from China. Eur J Protistol. 2007;43:77–86. doi: 10.1016/j.ejop.2006.10.002. [DOI] [PubMed] [Google Scholar]
- López-Ochoterena E. Ciliados mesosapróbicos de Chapultepec (sistematica, morfologia, ecologia) Rev Soc Mex Hist Nat. 1965;26:115–247. [Google Scholar]
- Lynn DH. The ciliated protozoa, characterization, classification, and guide to the literature. 3. Dordrecht: Springer; 2008. [Google Scholar]
- Lynn DH, Strüder-Kypke MC. Species of Tetrahymena identical by small subunit rRNA gene sequences are discriminated by mitochondrial cytochrome c oxidase I gene sequences. J Eukaryot Microbiol. 2006;53:385–387. doi: 10.1111/j.1550-7408.2006.00116.x. [DOI] [PubMed] [Google Scholar]
- Medlin L, Elwood H, Stickel S, Sogin M. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Work-shop (GCE). New Orleans, LA: pp 1–8
- Nylander JAA. MrModeltest v 2. Evolutionary Biology Centre: Uppsala University, Uppsala; 2004. [Google Scholar]
- Pan XM, Bourland WA, Song WB. Protargol synthesis: an in-house protocol. J Eukaryot Microbiol. 2013;60:609–614. doi: 10.1111/jeu.12067. [DOI] [PubMed] [Google Scholar]
- Pan HB, Li LF, Lin XF, Li JQ, Al-Rasheid KA. Morphology of three species of Amphileptus (Protozoa, Ciliophora, Pleurostomatida) from the South China Sea, with note on phylogeny of A. dragescoi sp. n. J Eukaryot Microbiol. 2014;61:644–654. doi: 10.1111/jeu.12146. [DOI] [PubMed] [Google Scholar]
- Pan HB, Zhang QQ, Dong JY, Jiang JM. Morphology and phylogeny of two novel pleurostomatids (Ciliophora, Litostomatea), establishing a new genus. J Eukaryot Microbiol. 2020;67:252–262. doi: 10.1111/jeu.12779. [DOI] [PubMed] [Google Scholar]
- Rajter Ľ, Vd’ačný P (2017) Constraints on phylogenetic interrelationships among four free-living litostomatean lineages inferred from 18S rRNA gene-ITS region sequences and secondary structure of the ITS2 molecule. Acta Protozool 56: 255–281
- Rataj M, Vd’ačný P (2021) Cryptic host-driven speciation of mobilid ciliates epibiotic on freshwater planarians. Mol Phylogenet Evol 161:107174 [DOI] [PubMed]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H. An der Nahrungsquelle angeseilt: der Glockentierfresser Amphileptus carchesii. Mikrokosmos. 1988;77:97–102. [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:7–14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WB. A new marine ciliate, Amphileptus litonotiformis nov. sp. (Protozoa, Ciliophora) Chin J Oceanol Limnol. 1991;9:300–305. doi: 10.1007/BF02850645. [DOI] [Google Scholar]
- Song WB, Wilbert N. Taxonomische Untersuchungen an Aufwuchsciliaten (Protozoa, Ciliophora) im Poppelsdorfer Weiher, Bonn. Lauterbornia. 1989;3:2–221. [Google Scholar]
- Song WB, Wilbert N, Hu XZ. Redescription and neotypification of Amphileptus marinus (Kahl, 1931) nov. comb. (Ciliophora, Pleurostomatida), and reactivation of A. parafusidens Song & Wilbert, 1989. Eur J Protistol. 2004;40:1–11. doi: 10.1016/j.ejop.2003.07.001. [DOI] [Google Scholar]
- Song WB, Warren A, Hu XZ. Free-living ciliates in the Bohai and Yellow Sea. Beijing: Science Press; 2009. [Google Scholar]
- Sonntag B, Foissner W. Urotricha psenneri n. sp. and Amphileptus piger (Vuxanovici, 1962) n. comb., two planktonic ciliates (Protozoa, Ciliophora) from an oligotrophic lake in Austria. J Eukaryot Microbiol. 2004;6:670–677. doi: 10.1111/j.1550-7408.2004.tb00607.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K (1867) Der Organismus der Infusionsthiere nach eigenen Forschungen in systematischer Reihenfolge bearbeitet. II. Abtheilung. 1) Darstellung der neuesten Forschungsergebnisse über Bau, Fortpflanzung und Entwickelung der Infusionsthiere. 2) Naturgeschichte der heterotrichen Infusorien. Verlag von Wilhelm Engelmann, Leipzig
- Stokes DAC. Some new infusoria from American fresh waters. Ann Mag Nat Hist. 1886;17:534–535. doi: 10.1080/00222938609460189. [DOI] [Google Scholar]
- Vd’ačný P, Bourland WA, Orsi W, Epstein SS, Foissner W (2011a) Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol Phylogenet Evol 59:510–522 [DOI] [PubMed]
- Vd’ačný P, Orsi W, Bourland WA, Shimano S, Epstein SS, Foissner W (2011b) Morphological and molecular phylogeny of dileptid and tracheliid ciliates: resolution at the base of the class Litostomatea (Ciliophora, Rhynchostomatia). Eur J Protistol 47:295–313 [DOI] [PMC free article] [PubMed]
- Vd’ačný P, Breiner HW, Yashchenko V, Dunthorn M, Stoeck T, Foissner W (2014) The chaos prevails: molecular phylogeny of the Haptoria (Ciliophora, Litostomatea). Protist 165:93–111 [DOI] [PubMed]
- Vd’ačný P, Rajter Ľ, Shazib SUA, Jang SW, Ji HK, Shin MK (2015) Reconstruction of evolutionary history of pleurostomatid ciliates (Ciliophora, Litostomatea, Haptoria): interplay of morphology and molecules. Acta Protozool 54:9–29
- Vuxanovici A. Contributii la studiul grupei subgenurilor Lionotus-Hemiophrys (Ciliata) Stud Cerc Biol (ser Biol Anim) 1960;12:125–139. [Google Scholar]
- Wang CD, Zhang TT, Wang YR, Katz LA, Gao F, Song WB. Disentangling sources of variation in SSU rDNA sequences from single cell analyses of ciliates: impact of copy number variation and experimental error. Proc R Soc B. 2017;284:20170425. doi: 10.1098/rspb.2017.0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenrich DH. A new protozoan parasite, Amphileptus branchiarum, n. sp., on the gilis of tadpoles. Trans Am Microsc Soc. 1924;43:191–199. doi: 10.2307/3221736. [DOI] [Google Scholar]
- Wilbert N. Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos. 1975;64:171–179. [Google Scholar]
- Wrześniowski A. Verzeichnis der Infusorien, welche in Warschau und seinen Umgebungen von 1861–1865 gesammelt wurden. Wykaz Szkoty Glownej Warszawskiej. 1866;5:15–28. [Google Scholar]
- Wu L, Yi ZZ, Li JQ, Warren A, Xu HL, Lin XF. Two new brackish ciliates, Amphileptus spiculatus sp. n. and A. bellus sp. n. from mangrove wetlands in Southern China, with notes on the molecular phylogeny of the family Amphileptidae (Protozoa, Ciliophora, Pleurostomatida) J Eukaryot Microbiol. 2015;62:662–669. doi: 10.1111/jeu.12225. [DOI] [PubMed] [Google Scholar]
- Wu L, Jiao XX, Shen Z, Yi ZZ, Li JQ, Warren A, Lin XF. New taxa refresh the phylogeny and classification of pleurostomatid ciliates (Ciliophora, Litostomatea) Zool Scr. 2017;46:245–253. doi: 10.1111/zsc.12193. [DOI] [Google Scholar]
- Wu L, Li JQ, Warren A, Lin XF. Species diversity of the pleurostomatid ciliate genus Amphileptus (Ciliophora, Haptoria), with notes on the taxonomy and molecular phylogeny of three species. Front Mar Sci. 2021;8:642767. doi: 10.3389/fmars.2021.642767. [DOI] [Google Scholar]
- Wu T, Li YQ, Zhang TT, Hou J, Mu CJ, Warren A, Lu BR (2021b) Morphology and molecular phylogeny of three Epistylis species found in freshwater habitats in China, including the description of E. foissneri n. sp. (Ciliophora, Peritrichia). Eur J Protistol 78: 125767 [DOI] [PubMed]
- Wu L, Li JQ, Warren A, Lin XF. Taxonomy and systematics of a new pleurostomatid ciliate, Pseudolitonotus spirelis gen. et sp. n. (Protozoa, Ciliophora, Haptoria) Mar Life Sci Technol. 2022;4:31–41. doi: 10.1007/s42995-021-00123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QQ, Simpson A, Song WB. Insights into the phylogeny of systematically controversial haptorian ciliates (Ciliophora, Litostomatea) based on multigene analyses. Proc R Soc B. 2012;279:2625–2635. doi: 10.1098/rspb.2011.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GAT, Chi Y, Wang Z, Wang Y, Liu R, Warren A, Zhao Y, Pan HB. Taxonomic and phylogenetic studies on two new freshwater Amphileptus species (Ciliophora, Pleurostomatida), from Lake Weishan, northern China. Eur J Protistol. 2022;82:125854. doi: 10.1016/j.ejop.2021.125854. [DOI] [PubMed] [Google Scholar]
- Zhang GAT, Zhao Y, Chi Y, Warren A, Pan HB, Song WB (2022b) Updating the phylogeny and taxonomy of pleurostomatid ciliates (Protista, Ciliophora) with establishment of a new family, a new genus and two new species. Zool J Linn Soc 196:105–123
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available in GenBank (Accession Numbers: OL828281–OL828283).