ABSTRACT
Resistance to antibiotics is becoming a worldwide threat as infections caused by multidrug-resistant pathogenic microorganisms can overcome antibiotic treatments and spread quickly in the population. In the context of early life, newborns are at increased risk as their immune system is still under development, so infections and acquisition of resistance during childhood have short- and long-term consequences for the health. The moment of birth is the first exposure of infants to possible antibiotic-resistant microorganisms that may colonize their gut and other body sites. Different factors including mode of delivery, previous antibiotic exposure of the mother, gestational age and consumption of antibiotics in early-life have been described to modulate the neonate’s microbiota, and thus, the resistome. Other factors, such as lactation, also impact the establishment and development of gut microbiota, but little is known about the role of breastmilk in transferring Antibiotic Resistant Genes (ARG). A deeper understanding of vertical transmission of antibiotic resistance from mothers to their offspring is necessary to determine the most effective strategies for reducing antibiotic resistance in the early life. In this review, we aim to present the current perspective on antibiotic resistances in mother-infant dyads, as well as a new insight on the study of the human gut and breastmilk resistome, and current strategies to overcome this public health problem, toward highlighting the gaps of knowledge that still need to be closed.
KEYWORDS: Antibiotic, resistome, microbiota, gut, breastmilk, infant, mother, pregnancy
1. Antibiotics in mother-infant dyads
Antibiotic resistance occurs when microorganisms resist the effects of antibiotics, making them no longer effective in treating bacterial infections. This occurs as bacteria evolve and develop new strategies to counteract the impact of antibiotics, leading to the rise of Antibiotic-Resistant Microorganisms (ARMs)1. Resistance to antibiotics is a natural evolutionary process for microorganisms, but its progression is accelerated by the selective pressure exerted by the excessive use of antibacterial drugs2. Incorrect prescription, over-consumption and prolonged use of antibiotics in humans and animals have affected the microbiota equilibrium, resulting in an increased risk of encountering health issues in the future, and caused a global crisis of antibiotic resistance3. Indeed, the creation of a new antibacterial drug is soon followed by the emergence of a resistance to it. ARMs are widespread and can be easily transmitted from person to person or from animals to humans, including through consumption of animal-derived food4. Antibiotic-resistant bacteria carry a set of Antibiotic Resistance Genes (ARGs), thus allowing the bacteria to overcome the antibiotic effects. The set of ARGs present in a population of bacteria is known as the resistome.
The World Health Organization (WHO) has declared that the rise of antibiotic resistances around the world, including high- and low-income countries, is a major threat. To address this challenge, a comprehensive global action plan has been put in place to ensure that prevention and treatment of infectious diseases are carried out using safe and effective medicines5. Antibiotics are becoming increasingly ineffective due to the fast spread of drug resistances globally. As a result, hospital stays become extended and infected patients require more expensive and intensive care. The increasing prevalence of antibiotic-resistant microorganisms complicates medical procedures such as surgeries, chemotherapy, and organ transplants5. Accordingly, the WHO and the US Centers for Disease Control and Prevention (CDC) classify ARMs as a threat to human health worldwide6. Despite its importance, ARM infections are currently underreported, but it is estimated that there are more than 2.8 million AMR infections and up to 35,000 deaths per year in the United States alone, with an estimated derived cost of more than $4.7 billion7. In Europe, more than 33,000 deaths are attributed to acquired ARM infections each year, representing $1.5 billion in direct and indirect costs8. Furthermore, in developing countries, communicable diseases continue to be a leading cause of death, with a rise in newly emerging and reemerging infections9.
In 2017, the WHO published a list of 12 bacterial families for which new antimicrobial development is urgently needed10. Bacteria were classified into three categories based on the urgency to develop novel antibiotics to combat them: critical, high and medium priority11. The pathogens included in the most critical group are multidrug-resistant bacteria that are threat to hospitalized patients, causing nosocomial infections12,13. Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, describing the ESKAPE group pathogens, are recognized as a top priority for treatment14.
These pathogens are incredibly difficult to treat with antibiotics and can lead to severe, life-threatening illnesses such as blood infections and pneumonia15. Among these pathogens, extended-spectrum β-lactamase (ESBL) and carbapenem-resistant P. aeruginosa, A. baumannii, K. pneumoniae and Enterobacter spp., are considered critical priority pathogens. On the other hand, the high priority group includes vancomycin- and methicillin-resistant S. aureus (MRSA and VRSA) and vancomycin-resistant E. faecium (VRE)10.
Early human life (the perinatal and postnatal period) represents the first exposition to antibiotics, and at same time, it is when microbiota establishment and development take place in parallel with other biological processes (immune maturation, metabolic and neurocognitive processes). It is estimated that in high-income countries, 70% of all infants receive at least one course of antibiotics during the first year of life to treat skin, respiratory and gastrointestinal infections16. In low and middle-income nations, the use of antibiotics is higher, with children estimated to take an average of 11 rounds of antibiotics within their first 2 years of life17. Furthermore, the 80% of the drugs a women is subjected during pregnancy are antibiotics, and 35% of pregnant women in Western countries are exposed to antibiotics18. Pregnant mothers can receive antibiotics during the latter stages of pregnancy to prevent bacterial infections in their genital tract which could harm both the mother and the child19. Many women are exposed to antibiotics during delivery with the prophylaxis to prevent the vertical transmission of group B streptococci being the main reason for intrapartum antibiotics administration. For women conducting cesarean section procedures, the WHO recommends using a single dose of first-generation cephalosporin or penicillin20. For those with uncomplicated vaginal birth and without any associated risk factor routine antibiotic prophylaxis is not recommended21. Table 1 summarizes the most commonly used antibiotics during pregnancy and lactation and their indication of use.
Table 1.
Antibiotics commonly used during the perinatal period.
| Antibiotic | Class of antibiotic | Target infection |
|---|---|---|
| Penicillins22 | β-lactams | Group B streptococci22 infections, respiratory infections, urinary tract and skin infections, septicemia. |
| Cephalosporin23 | Respiratory tract, urinary tract and skin infections, gonorrhea. | |
| Carbapenems24 | Gram-negative bacterial infections of the urinary tract, kidney, gastrointestinal tract, respiratory tract, and urogenital and pelvic area. | |
| Azithromycin, erythromycin25 | Macrolides | Respiratory, intestine, gynecological, urogenital and skin infections, chlamydia infections and syphilis. |
| Ciprofloxacin, norfloxacin26,27 | Quinolones | Serious gram-negative bacterial infections. |
| Clindamycin28,29 | Lincomycin | Respiratory, skin, blood, female reproductive organ, and internal organ infections. |
| Gentamycin, streptomycin30,31 | Aminoglycosides | Pyelonephritis, urinary tract infection, gram-negative bacterial infections. |
| Metronidazole32 | Nitroimidazole | Infections affecting the reproductive system, gut, skin, heart, bones, joints, lungs, blood, and nervous system. |
| Vancomycin33,34 | Glycopeptide | Colitis, C. difficile–induced diarrhea, gram-positive bacterial infections. |
| Trimethoprim35 | Dyaminopyrimidines | Urinary tract infections, pneumonia, diarrhea. |
The risk of spreading antibiotic-resistant pathogens is particularly severe in newborns as their gut microbiota and immune system are not as mature and robust as those of adults. The worldwide problem of antibiotic resistance results in approximately 214,000 yearly newborn fatalities from septic infections, highlighting its devastating impact on infant health36.
Thus, the use of antibiotics in early stages of life can influence the microbiota-host crosstalk, resulting in a dysbiosis that can lead to short-term and long-term consequences37,38. Reported short-term consequences in premature infants include a correlation between decreased diversity of microbiota following an antibiotic treatment with and increased frequency of sepsis39. In addition, early antibiotic treatment in premature babies with extremely low body weight has been linked with an increased risk of necrotizing enterocolitis and death40–42.
Long-term consequences of early antibiotic exposure include pathologies related to the immune system maturation, and disruptions in the metabolic functions of the microbiota. Immune disorders, such as allergies, asthma, eczema or celiac disease, have been reported as a consequence of pre- and postnatal exposure to antibiotics43–47. Research has also established connections between early and repeated exposure to antibiotics and growth48, risk of obesity49–51 and inflammatory bowel disease52.
2. The maternal-infant resistome
The gut microbiome of adult humans is a source of ARGs that opportunistic pathogens can acquire. Bacterial pathogens carrying ARGs in pregnant mothers may impact ovum implantation, the pregnancy suspension, and delivery. This also increases the chance of early infections in the newborn and affects the establishment and development of the infant’s gut microbiome53.
The composition of the gut microbiota is a key factor in determining the presence of ARGs, as most of these genes are carried by a few specific taxa. Studies employing culture-based or metagenomics methods have shown that Gammaproteobacteria in the Pseudomonadota phylum are major carriers of ARGs, while Bacillota is also enriched in pregnant women and can carry ARGs53,54. Correlations between the phylum Proteobacteria and ARGs/mobile genetic elements (MGEs) have been found55,56. In addition, Proteobacteria are capable of DNA transfer through conjugation, which plays a critical role in the spread of ARGs57. The most abundant and unique ARGs in the infant gut belong to Escherichia, Klebsiella, Citrobacter, and Enterobacter species58.
On the contrary, Bifidobacterium spp. are negatively correlated with antibiotic resistance load15. Members of this genus are relevant commensals that dominate the breastfed infant gut microbiome, primarily due to their ability to use human milk oligosaccharides (HMOs) and other carbohydrates efficiently59,60. Bifidobacterium species are less likely to possess ARGs than other taxa such us Streptococcus, Staphylococcus, Enterococcus and Bacteroides spp.61,62.
Metagenomic shotgun sequencing has revealed the high levels of ARGs in the gut microbiome of infants compared to adults, even when not exposed to antimicrobials15,56,63–65. This is likely due to the main role of Pseudomonadota in the infant microbiota during the first months of life, being later replaced by other bacterial phyla. Further, infant resistome analyses at different time points have elucidated higher relative abundances of ARGs in younger infants in comparison to older children66,67. Different factors including delivery mode, type of breastfeeding and antibiotic exposure influence the ARG load in infants. Thus, the development of the infant’s resistome is linked to the evolution of the microbial composition, which contributes to the dynamics and stabilization of the infant resistome to adulthood67,68.
In this regard, the human resistome also includes the mobilome, which is defined as all Mobile Genetic Elements (MGEs) in a cell which are able to move around within or between genomes of different organisms68. They include plasmids, integrons, bacteriophages and transposable elements69.
There is evidence that an increased presence of MGEs in infant feces may facilitate the transmission of ARGs among and between species15. In the context of mother-infant transfer, it has been seen that the acquisition of the mobilome in early stages of life may be due to maternal influence and/or family-specific environment, with the latter being the most important influence at 6 months of life70.
The increasing use antibiotics during pregnancy and breastfeeding constitutes a risk factor for the development of ARM in the mother’s resistome, which can then be passed on to the offspring and impact their resistome71. Although load and variety of antibiotic resistance may vary between groups of infants according to different early-life factors, most predominant ARGs confer resistance to beta-lactams, tetracyclines, macrolides, aminoglycosides and quinolones72.
Beta-lactamases
Beta-lactamases have been prescribed for many decades, and are commonly used during pregnancy and labor, thus it is predictable that resistance appears in infant fecal samples58,72. The majority of beta-lactamases genes are extended-spectrum beta-lactamases (ESBLs), that can be grouped in: TEM beta-lactamases (class A), OXA beta-lactamases (class D), SHV beta-lactamases (class A) and CTX-M beta-lactamases (class A)73. ESBL-producing Enterobacteriaceae (ESBL-E), including Escherichia coli (E. coli) and Klebsiella pneumoniae, are a public health concern as they can potentially contribute to the neonatal mortality, and they are ranked as a priority among pathogens in need of new treatment by the WHO74. These multidrug-resistant bacteria cause a mortality rate of more than 70% in infants as they are widely spread in the gut microbiome75.
Tetracyclines
Genes conferring resistance to tetracycline (tet genes) have been reported to be the most abundant among infant fecal samples and are therefore the most representative resistant genes in the infant resistome76. The tet genes are the most frequently shared ARGs between mothers and infants, indicating that they are easily transmitted from the mother to the newborn56,58,71,77. The prevalence of tetracycline-resistant genes in various bacteria species, coupled with the common utilization of tetracycline to treat infections, may explain the acquisition of tet genes by women. Although this antibiotic is not recommended during pregnancy and early life due to potential dental staining effects, it is still extensively used in animals and, therefore, the environment and the diet may be sources of tet genes without direct exposure to the antibiotic77.
Macrolides
Macrolides are commonly used against infections in infants, such as trachoma, pertussis and Campylobacter enteritis49,78. Clindamycin and erythromycin are used to treat infections in skin and tissues produced by Staphylococci. Nevertheless, the expression of macrolide-lincosamide-streptogramin B resistance (MLSB) can reduce the efficacy of these antibiotics. The accumulation of macrolide resistance genes in the gut microbiome of newborns could present future consequences for their health. Klassert et. al 72 found that genes for macrolide resistance (mefA, ermB, and ermC) are the most widely distributed among mother-infant pairs and have the greatest likelihood of being transmitted vertically79.
Aminoglycosides
Aminoglycosides, usually combined with a beta-lactam, are widely used for neonatal sepsis. Antibiotics such as gentamicin, streptomycin, kanamycin, neomycin are examples of aminoglycosides. It has been identified in the infant gut microbiota the presence of genes for acetylation, phosphorylation and adenylation conferring aminoglycoside resistance80. The aac(6′)-aph(2′′), aadE and aph(3′)-I genes are the most commonly detected ARGs against aminoglycosides during the first year of life73.
Quinolones
Quinolone antibiotics are synthetic and widely used in medicine. Like other antibiotics, resistance to quinolones has emerged due to their frequent use, and it is now common in some bacterial pathogens. Although quinolones are rarely used in pediatrics, the presence of ciprofloxacin-resistant E. coli has been found in the feces of infants81. Additionally, when mothers are prescribed quinolones, there is an increased risk of their offspring acquiring community-acquired, quinolone-resistant E. coli 82.
3. Antibiotic resistance acquisition and transmission in early stages of life
It is important to distinguish between intrinsic, acquired and adaptative antibiotic resistance to determine how the resistance develops (Figure 1). Intrinsic resistance is naturally present in the microorganism; it is chromosomically encoded and is connected to the microorganism physiology83. The intrinsic resistance is conferred by several elements that contribute to antibiotic resistance, independent of earlier exposure to antibiotics, and can only be vertically transmitted84. These genes may be captured by MGEs during evolution and become mobile85. The acquired resistance is based on mutations in the bacterial chromosomic DNA and on the acquisition of mobile resistance genes, by either horizontal or vertical gene transfer. Adaptative resistance refers to a transient situation in which a susceptible bacterial population becomes resistant to antibiotics84.
Figure 1.
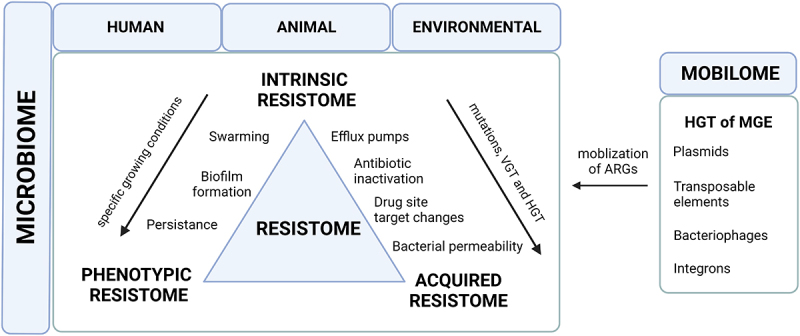
A schematic overview of the human resistome and mobilome. Intrinsic resistance is a property controlled by chromosomes and is related to the general physiology of the microorganism. The acquired resistance is encoded on plasmids and may be classified into four mechanisms: changes in the cell wall that make it less permeable to antibiotics, modifications of enzymes that inactivate antibiotics, changes in the target site of the drug, and efflux pumps that remove antibiotics from the cell. Phenotypic bacterial resistance appears in three categories: persistence, where a subpopulation of bacteria survive even though the majority is inhibited by the antibiotic; formation of biofilms, where bacteria form communities protected by a matrix; and swarming, where cells become hyper-flagellated, allowing them to colonize nutrient-rich environments and become less susceptible to antibiotics. MGE: mobile genetic elements; HGT: horizontal gene transfer; VGT: vertical gene transfer; ARG: antibiotic genetic elements. .
Despite their widespread presence in bacteria, not all bacteria harbor ARGs at comparable levels. In early stages of life, antibiotic resistance can be a result of horizontal transfer of genes between bacteria, vertical transmission from the mother during pregnancy, birth and lactation and/or antibiotic exposure of the infant71. The availability of new methods, especially culture-independent strategies, have enabled new insights on antibiotic resistance transmission (Box 1).
[Box 1. Methods to study the resistome].
- Culture-based approaches. This methodology continues to be essential to determine resistance phenotypes to multiple antimicrobials30. This approach stands alone or can also be used as a complement to metagenomic work by overcoming the depth bias inherent in metagenomic sequencing31. This methodology is only applicable in cultivable bacteria, not allowing the discovery of distant or unknown elements.
- Whole-genome sequencing. This methodology has high sensitivity, detecting species with low abundance and identifying potential ARGs. However, the full complexity of the resistome cannot be observed in vivo. Thus, it is necessary to validate using culture-based approaches to reduce the risk of false positives and determine that they are genuine resistance genes.
- Complete metagenomic sequencing. Methodology with which you can observe all the ARGs that would exist in a certain environment, taking into account that could be oversampled, because when these genes are overexpressed in a heterologous host may not be genuine resistance genes in their native contexts32.
- Long-read sequencing techniques. Previous research has demonstrated that utilizing long-read sequencing techniques can provide insight into the presence of ARGsin the resistome. Using a combination of both long-read (Oxford Nanopore) and short-read (Illumina) sequencing methods has been found to be an effective way to thoroughly examine the resistome in both human samples33 and in bacterial isolates34.
- Single-cellsequencing techniques. Refers to the process of analyzing the genomic, transcriptomic, and metabolomic functions of an individual cell through sequencing its genome or transcriptome35. Despite its high cost, this technique has been demonstrated as a powerful toolfor investigating non-cultivable organisms86.
- Databases. The detection and analysis of genetic elements require ongoing improvement, as it is crucial to track the development of resistance and its evolution over time. To achieve this, resistome databases must be frequently updated to incorporate newly discovered variations, such as variant sequences, insertions, and deletions, thereby enhancing our knowledge of resistance variations87. The most frequently cited specialized databases for ARGs that allow input of metagenomic data are ResFinder88, MEGARes89 and Resfams90.
Vertical transfer of antibiotic resistance from mother to offspring
The source of antibiotic-resistant bacteria in infants remains unclear, as they can be transmitted from other individuals, the mother or even the environment15. The resistance profiles of mothers and infants can differ in terms of prevalence and specific resistant strains, but there is evidence of vertical transmission of ARGs from mother to offspring. Many ARGs occur only in either maternal or in child samples56.
The resistome of infants, which is the collection of antibiotic resistance genes in the gut, was found to have resistance to several broad-spectrum beta-lactam antibiotics, such as aztreonam, piperacillin-tazobactam, and cefepime91, but ARGs to these antibiotics were not present in the mothers’ resistome. Despite the difference between the mothers’ and infants’ resistomes, there is evidence of vertical transmission of antibiotic resistance genes from mothers to infants15,92. In a cohort of 50 samples, 59 ARGs of a total of 249 had at least one significant family transmission56. Kozak et al.93 showed that highly similar isolates of antibiotic-resistant Lactobacillus fermentum, Lactobacillus gasseri, Bifidobacterium longum and Enterococcus faecalis were found in both mothers and infants, indicating that resistant strains were passed to infants through vertical transmission in early life.
Vertical transmission of ARGs at birth
ARGs have been found in neonates within hours of birth, indicating that colonization of the newborns with ARMs can occur at birth or soon after, likely through contact with the mother’s uterus and skin or from the hospital environment94. As reviewed by Patangia et al. 71, some studies have shown that ARMs can be present in a mother’s vagina and be transferred to her offspring. These include E. coli resistant to ampicillin and cefotaxime-resistant and multidrug-resistant E. coli 95–97, penicillin-resistant Streptococcus 98, and tetracycline-resistant bacteria including Prevotella, Lactobacillus, Ureaplasma, Gardnerella, Corynebacterium and Staphylococcus spp. carrying tetM, tetO and tetW genes99. However, there is still little evidence on the transmission of antibiotic resistance during pregnancy and birth and the associated health implications71.
Vertical transmission of ARGs during lactation
Breastfeeding provides numerous benefits for infants, including the transfer of maternal immune cells and antibodies, as well as vital nutrients and bacteria100. Human milk serves as a consistent source of commensal and potentially probiotic bacteria for the newborn’s gut, with Staphylococcus and Streptococcus spp. being the most dominant genera in a woman’s milk, followed by Lactobacillus and Bifidobacterium spp.101.
Human milk has been found to contain antibiotic-resistant bacteria. Studies have shown the presence of antibiotic-resistant strains of Staphylococcus, Streptococcus, Acinetobacter, Enterococcus, and Corynebacterium in human milk15,102. Multi-drug resistant profiles have also been detected in these species100.
Antibiotic resistant bacteria found in human milk have been linked to the administration of certain antibiotics to mothers during lactation, such as gentamicin, chloramphenicol, streptomycin, and quinupristin. Additionally, resistance to vancomycin, ampicillin, amoxicillin, oxacillin, and cephalothin has also been observed in human milk100. Multi-drug resistant strains of Streptococcus and Staphylococcus have been detected to be transferred to infants through breastfeeding103–105. This is concerning as Staphylococcus aureus is a major source of community and hospital infections106.
Metagenomic analyses have highlighted the high levels of ARGs and MGEs in breastmilk and the resemblance of the infant resistome to the mother’s milk resistome15. Antibiotic-resistant bacteria were observed in breastmilk as a potential source of ARM in infants [10]. Pärnänen et al.15 showed that infants shared gut resistomes and mobilomes with their own mother’s breastmilk microbiota with 70% of the ARGs detected in milk present in infant feces. Therefore, a mother can impact the infant’s gut resistome by transmitting resistant bacteria through her milk, although the resistance genes can also come from other parts of her body or external sources. The research found that most ARGs in the infant gut are carried by a limited number of microorganisms, and that antibiotics exposition has a major effect on the amount of resistance genes present during infancy.
Additional research is necessary to examine the transfer of antibiotic resistance from a mother to her infant, as it can affect the establishment of gut colonization. This will shed light into the source and dissemination of resistance genes and assist in the development of strategies to reduce this type of transmission.
4. Factors affecting antibiotic resistance in infants
Culture-based and metagenomic analyses have reported that the ARG load in the infant gut resistome can be affected and/or modulated by the type of feeding (maternal lactation or formula feeding), the duration of lactation, gestational and infant age (especially for preterm infants) and the antibiotic exposure in early stages of life (Figure 2).
Figure 2.
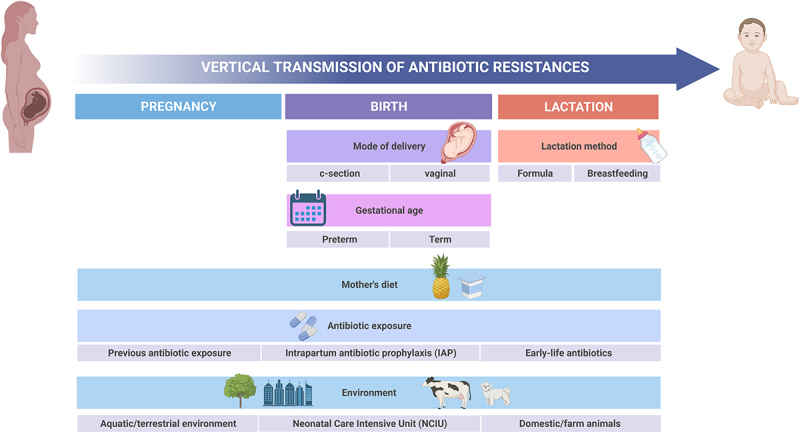
Factors influencing antibiotic resistance acquisition and vertical transmission.
Gestational Age
Premature infants are of particular interest as they often undergo prolonged and early antibiotic treatment soon after birth, which can significantly alter the gut microbiome107.
The amount of ARGs in a preterm infants is influenced by their gestational age, with a tendency to have a higher ARG load108.
Premature infants carry a gut microbiota characterized by multidrug-resistant strains of Klebsiella, Enterobacter, and Escherichia, which are related with hospital infections and in neonatal intensive care units (NICUs)63,109. A study conducted by Rose et al. 110 found several ARGs in fecal samples from preterm neonates. The average number of ARGs per infant was 13 and the identified bacteria included K. pneumoniae, C. perfringens, and members of the Enterobacter and Staphylococcus genera. The presence of these genes in healthy premature infants emphasizes the importance of understanding the potential impact it may have on future medical treatment plans.
Mode of Delivery
It is known that mode of delivery has an effect on neonatal microbiota development111. The gut microbiota of vaginal-delivered infants differs from those born via c-section over the first year of life, with higher levels of Bifidobacterium spp. and lower levels of Klebsiella and Enterococcus found in vaginally delivered infants112,113. Alterations in the composition of vaginal microbiota can have a substantial impact on the newborn’s gut microbiota15. Moreover, c-section delivery has been associated with reduced breastmilk microbiota diversity and richness114–116.
Recently, it has been shown that the way an infant is born and the amount of antibiotics they are exposed to during the perinatal period can affect the development of the gut microbiome establishment and have long-term impacts on their health, such as increasing the risk of asthma, inflammation, and obesity117,118.
Therefore, as delivery mode shapes maternal-infant and breastmilk microbiota, it can also influence the maternal, infant and milk resistome119. The WHO suggests that women who undergo a c-section should receive a single dose of penicillin or a first-generation cephalosporin as a preventive measure, instead of other types of antibiotics120. Some studies have elucidated the presence of ARGs in cesarean-section born babies121. Post-cesarean section periotonitis and surgical site infections have been related with higher rates of antibiotic resistance microorganisms in mothers122,123. However, there is no clear evidence on how mode of delivery may have an impact ARG load in breastmilk and on the mother-infant resistome development, and more research is needed to understand how mode of delivery, together with IAP, gestational age, maternal diet and other factors can shape the early-life resistance load.
Early-life feeding-type
Breastfeeding provides numerous benefits for infants, including the transfer of maternal immune cells and antibodies, and it serves as a consistent source of commensal and potentially probiotic bacteria for the newborn’s gut, being the genus Lactobacillus, Bifidobacterium, Staphylococcus and Streptococcus the most dominant genera in a woman’s milk101. Nevertheless, breastmilk plays a crucial role on the acquisition of antibiotic resistances in early-life. Human milk has been found to contain species of Staphylococcus, Streptococcus, Acinetobacter, Enterococcus, and Corynebacterium resistant to antibiotics and with multidrug-resistant profiles have been isolated in human milk15,100,102.
On the other hand, infants who are exclusively breastfed have a more varied gut microbiome and fewer antibiotic-resistant bacteria, compared to those who are formula-fed. Formula feeding can cause changes in the gut environment leading to a simpler microbiome dominated by bacteria carrying antibiotic resistance genes108. A study that analyzed the gut metagenomes of 46 neonates found that compared to breastfeeding, formula feeding was associated with a 70% increase in the abundance of ARGs. Infants only fed with human milk had a more diverse gut microbiome and are less likely to be exposed to antibiotic-resistant bacteria, compared to infants who are fed formula. The study showed that formula feeding is associated with an increase in the presence of antibiotic-resistant genes and a decrease in the presence of bacteria that are typically found in a healthy gut microbiome. The families of obligate anaerobic bacteria, including Veillonellaceae, Bifidobacteriaceae, Lachnospiraceae, Clostridiaceae, and Porphyromonadaceae, were reduced in formula-fed infants. On the other hand, the populations of facultative anaerobic and aerotolerant bacteria, such as Enterobacteriaceae, Enterococcaceae and Staphylococcaceae increased in these infants. Formula-fed neonates had clinically significant ARGs that could potentially lead to methicillin resistance in Staphylococcus aureus and ESBL phenotypes, compared to those who were exclusively fed with human milk108.
In addition, duration of breastfeeding has been reported to influence the load of ARGs. Breastfeeding for less than 6 months has been correlated with an enrichment of ARGs and MGE. Breastfeeding for at least 6 months decreases the abundance of Gammaproteobacteria, and as a result, the abundance of ARGs, and conversely increases the abundance of Bifidobacterium spp.15.
Antibiotic exposure during pregnancy, delivery and early life
Didier et al.124 reported that the use of antibiotics during the antenatal period can also increase the risk of antibiotic-resistant infections. In this regard, Li and collaborators reported that perinatal antibiotics impacted the transfer of bacteria from mothers to infants during the early stages of colonization. The administration of antibiotics led to an increase of horizontally transmitted bacteria from the environment, some of which were harmful strains commonly found in hospitals. In the group of mothers treated with antibiotics, the main source of ARGs in their offspring was not the microbiome of the maternal gut, while in the control group, most ARGs were passed on from the mother. This effect was particularly pronounced in the first 3–7 days after birth125.
Intrapartum Antimicrobial Prophylaxis (IAP) is a common clinical procedure during delivery that is used to prevent early-onset Group-B-Streptococcus (GBS) infections and in cases of pre-labor rupture of membranes or C-section deliveries. It is present in more than 30% of labors and is by far the most common exposure way to antibiotics in the perinatal and neonatal stages126,127. The effect of this treatment on the gut microbiome has gained attention the last decade and several articles have demonstrated an impact of IAP on the gut microbiota development. Several common findings highlighted the increment of Enterobacteriaceae and the reduction of strict anaerobes such as Bifidobacterium and Bacteroides genus 128. In addition, it has been observed a negative influence of IAP on the correct development of bifidobacterial community in newborns during the first months of life. Data accumulated during the last years suggest that IAP may also have lasting and significant effects on the infant’s resistome and mobilome. Two seminal studies demonstrated the impact of IAP on the prevalence of ARGs and mobile elements in vaginally-delivered full-term babies, showing also a beneficial effect of breastfeeding on the ARGs burden15,129. Most of the current studies concluded that the selective pressure exerted by IAP favors the presence of different bacteria carrying ARGs, most of them species belonging to Pseudomonadota phyla or Staphylococcus genus128. In addition, IAP has also been associated with increments of ARGs in other body microbiomes, such as the nasopharynx and oral microbiota128.
A recent study suggests that the use of broad-spectrum antibiotics for treating suspected early-onset neonatal sepsis (sEONS) has a significant impact on the microbial diversity, community composition, and selection of ARGs. The study found that the use of antibiotics resulted in a decrease in the abundance of Bifidobacterium species and an increase in the abundance of Klebsiella and Enterococcus species in the treated infants. The study also found that treatment with amoxicillin and cefotaxime had the strongest effect on both the microbial community composition and the ARG profile, while treatment with penicillin and gentamicin had the least effect130. In addition, B. longum and B. breve decreased in populations exposed to antibiotics, while Bacteroides increased131. Additionally, infants can inherit residual effects of past antibiotic use of the mother. Patangia et al. found that even before being exposed to antibiotics, newborns carry ARMs acquired from their mothers and the environment71. However, further research is required in this area.
Effects of other resistomes: One-Health perspective
In order to understand ARG transmission, the development of the human gut resistome in early life and the crossover between mother and offspring resistome, it is necessary to refer to the One-Health perspective (Human-Animal-Environment)132. There is increasing concern that the use of antibiotics in food-producing and pet animals may be a source of antimicrobial resistance to humans133. In this sense, animal and environmental resistome may have a direct effect on the establishment and development of the human resistome. Indeed, having pets or animals at home has been found to influence the growth and development of gut bacteria134,135 and, therefore, this may affect the ARG carriage through the impact on the microbiota and the potential transference of strains harboring ARGs. The transfer of pathogens or antibiotic resistance genes from pets to humans has been documented136–138. Although antibiotic usage in animals is more and more regulated, antibiotics are still given to animals to treat disease and to not only prevent infection but also the propagation of disease to other animals139. In addition, there is evidence of environmental transmission of ARGs among livestock and humans. Similar to humans, antibiotics in animals cause changes in gut bacterial composition: a decrease in the species of Lactobacillus, Bifidobacterium, and Ruminococcus and a rise in E. coli levels were observed, leading to alterations in ARG levels140. In addition, the animal resistome is not only developed in antibiotic-treated animals; wild animals also have ARGs141.
Humans can be exposed to ARGs from animals by direct contact with them but also by consuming vegetables that have been cultivated on soil fertilized with manure and swine farm wastewater, or by consuming animal meat. Both manure and wastewater from livestock farms have been observed to contain residues of antibiotics and resistant microorganisms, such as Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes 142. Downstream, ARGs and antibiotic residues have been detected in vegetables grown in manure-amended soils, which then may reach human microbiome promoting horizontal gene transfer143. Animal meat is also a source of antibiotic resistance in humans. Even though there are strict biosecurity measures established, meat may still contain traces of antibiotics if the proper time for withdrawal antibiotics from the treated animals is not efficient. Moreover, the process of dressing a carcass may result in contamination with ARMs, which can come from sources such as the animal’s skin and gut contents, the hands of the workers, and the slaughter environment141. Therefore, it is important to consider that all ecological niches harboring microbial communities will contain ARGs, including aquatic environments and terrestrial environments (agricultural sites, wastewater plants) and also hospital environments such as the NCIU144,145. Therefore, the One-Health initiative bas to be brought to the forefront of ARG research.
5. How can we reduce antibiotic resistance in mother-infant dyads?
Apart from strategies that involve antibiotic stewardship146, recent studies have focused on developing new approaches to ameliorate the antibiotic resistance load in the human microbiome, but further research into new mechanisms to reduce the resistance load, especially in infants, are needed.
Diet
Diet is significantly linked with the composition of microbiota, as it not only nourishes it but can also alter it, and, presumably, influence the resistome. Several studies have pointed the influence of maternal diet on their offspring gut microbiome, elucidating the role of the diet on the transfer of antibiotic resistances to the infant during delivery or lactation147–149. Recently, studies have started to explore the impact of diet on the dissemination of ARGs in the intestinal microbiota. For instance, it has been reported that a diverse, fiber-rich diet induces lower abundance of ARGs in human gut150. A diet high in sugar, fat, and protein can alter bacterial composition and diversity, increase the permeability of bacterial membranes, and activate the bacterial SOS response, resulting in gut inflammation and changes to gut bacteria and its by-products. These changes result in the amplification and transfer of exogenous ARGs in the gut151.
It has been established that different eating patterns, such as omnivore, ovolactovegetarian, and strict vegetarian diets, can have an impact on the resistome. It is believed that consuming fewer processed foods can help reduce the amount of ARGs in the gut microbiome152. However, other factors might also contribute to the resistome, as a study found that long-term dietary habits did not significantly affect resistome composition in the Dutch population153.
Therefore, diet seems to be a potential key factor on both acquisition and transmission of antibiotic resistances in all stages of life. For this reason, more knowledge is needed to develop dietary strategies that would help to ameliorate antibiotic resistances in the mother-infant environment.
Dietary supplements: - biotics family
It’s becoming more than obvious that - biotics family including prebiotics and probiotics might have an important role in the restoration of gut dysbiosis and resilience. In this context, the utilization of probiotics appears to be a highly hopeful strategy in this field, either through simultaneous use with antibiotics to prevent disruption or by taking advantage of their benefits to heal the altered microbiome after antibiotics have caused harm. Probiotic administration to pregnant women is recommended by the World Allergy Organization (WAO) for pregnant women at high risk for having an allergic child and women who breastfeed infants at high risk of developing allergies. On the other hand, infants at high risk of developing allergies are also recommended to receive probiotic supplementation154. There is no evidence that taking probiotics or prebiotics during pregnancy causes infant and maternal adverse pregnancy outcomes22, but significantly modulates the maternal gut microbiota and immune status23,24.
The presence of Bifidobacterium infantis EVC001 in the infant gut microbiome leads to a decrease in the amount of ARGs and the types of bacteria that carry and potentially spread them among individuals25. In premature babies, probiotic supplementation may increase protection against harmful bacteria and reduce the negative impact of antibiotics on the microbiome and resistome of the gut26. A different study demonstrated that administering probiotics to preterm infants during hospitalization after birth decreased the gut microbiome’s resistance gene diversity and prevented its persistence27.
Another metagenomic analysis reported that taking a probiotic supplement consisting of commonly used species, reduced the amount of antibiotic resistance genes in the gut, however, this positive outcome was limited to only certain individuals who were receptive to probiotic colonization28.
The significance of considering the ecological environment in which probiotics are given is emphasized by these contrasting effects. On the other hand, prebiotics, which are not made from live bacteria, are unlikely to lead to an increase in resistance genes. As a result, prebiotics might be a valuable resource in the battle against antibiotic resistance and infection, although their effectiveness has yet to be confirmed29.
Further studies are required to assess the potential reduction of horizontal transfer of resistance genes by probiotic administration. In addition, more information about the use of prebiotics during pregnancy and lactation to prevent antibiotic resistance transfer to offspring is needed.
6. Conclusions and Future Perspectives
It is widely accepted that the early infant gut microbiota has a crucial role in infant health, and that the close relationship between mothers and their offspring determines the formation and development of the microbiota of the neonate. Therefore, several maternal and external factors may contribute to the modulation of the infant’s resistome. Mode of delivery, gestational age, antibiotic use, or type of lactation may facilitate the colonization of ARMs, leading to a high risk of infections in early life. The term resistome appears to be a concept that has spread worldwide as antibiotic resistance transmission rises to dangerous levels. As next generation sequencing techniques have improved in recent years, the scientific community has gained more knowledge on the mother-infant resistome. However, there are still some to questions to answer. Do babies acquire the first ARGs during birth or are they born with them? Is lactation a crucial factor in the acquisition of antibiotic resistance? What impact do the mother’s dietary and lifestyle habits have on her infant’s resistome? Which is the best approach to study the resistome? There are few studies attempting to answer these gaps in our knowledge, but there is still a lot of work to do. Delving into antibiotic resistance acquisition and transmission in early stages of life may allow us to develop new strategies to overcome and reduce the resistance load. Diet appears to be one of the main tools to control antibiotic resistance transmission, as the benefits of a healthy diet on human gut microbiota are well known. Prebiotics and probiotics may also help beneficial bacteria colonize infant guts, displacing ARMs, but more studies are needed to know if biotic intervention should be applied to pregnant women prophylactically and/or via newborn feeding. All approaches for the reduction of antibiotic resistance load in infants may start at the beginning of the pregnancy, or even before, so the resistome of pregnant women is important to be taken into account. This review aims to present the latest evidence on mother-infant antibiotic resistance and reveal the challenges that lay ahead in the fight against multidrug-resistant bacteria.
Acknowledgments
We acknowledge Generalitat Valenciana-European Social Fund (ACIF/2021) for the predoctoral fellowship grant to Anna Samarra. Manuel Bernabeu wishes to thank the Post-PhD Program of University of Barcelona for the requalification of the Spanish University System from the Ministry of Universities of the Government of Spain, modality “Margarita Salas”, financed by the European Union, Next Generation EU. Maria Esteban-Torres thanks the support received from the MSCA-IF postdoctoral grant (MicroMi-898088). Raul Cabrera-Rubio wishes to thank Generalitat-Valenciana for the grant Plan GenT project (CDEIGENT 2020). Finally, authors also acknowledge support of Institute of Agrochemistry and Food Technology (IATA-CSIC). IATA-CSIC is a Centre of Excellence Severo Ochoa (CEX2021-001189-S MCIN/AEI/10.13039/501100011033).
Funding Statement
This work was supported by PROMETEO/GVA under grant “NeoHealth” project PROMETEO ref. 2020/012.
Abbreviations
- ARG
: antibiotic resistant genes
- ARM
: antibiotic resistant microorganisms
- HMO
: human milk oligosaccharides
- MGE
: mobile genetic elements
- WHO
: World Health Organization
- CDC
: Centers for Disease Control and Prevention
Disclosure statement
No potential conflict of interest was reported by the authors.
The figures were created using BioRender (biorender.com) under BioRender's Academic License Terms.
Contribution statement
All authors contributed to the conceptualization, critical review, revision, and approval of the final version of this manuscript.
Data sharing
Data sharing is not applicable to this article, as no new data were created or analyzed in this report.
References
- 1.Annunziato G. Strategies to overcome antimicrobial resistance (AMR) making use of non-essential target inhibitors: a review. Int J Mol Sci. 2019;20:20. doi: 10.3390/ijms20235844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ Health Prev Med. 2015;20:243. doi: 10.1007/s12199-015-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez J, Guarner F, Bustos Fernandez L, Maruy A, Sdepanian VL, Cohen H. Antibiotics as major disruptors of gut microbiota. Front Cell Infect Microbiol. 2020;10:731. doi: 10.3389/fcimb.2020.572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sir Alexander Fleming – Nobel Lecture - NobelPrize.org . [Internet]. cited 2022 Jul 25: Available from https://www.nobelprize.org/prizes/medicine/1945/fleming/lecture/
- 5.Antimicrobial resistance: global report on surveillance [Internet]. cited 2022 May 13: Available from https://apps.who.int/iris/handle/10665/112642
- 6.European Centre for Disease Prevention and Control & World Health Organization. Regional Office for Europe. Antimicrobial resistance surveillance in Europe. 2022–2020 data. 2022.
- 7.Centers for Disease Control U . 2019. Antibiotic resistance threats in the United States. Atlanta (GA): U.S. Department of Health and Human Services, CDC. [Google Scholar]
- 8.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19:56–20. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The state of the world’s antibiotics report in 2021 - Center for Disease Dynamics, Economics & Policy (CDDEP). [Internet]. cited 2022 Aug 12: Available from https://cddep.org/blog/posts/the-state-of-the-worlds-antibiotics-report-in-2021/
- 10.WHO publishes list of bacteria for which new antibiotics are urgently needed [Internet]. cited 2022 Aug 12: Available from https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- 11.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. Antimicrobial Resistance in ESKAPE Pathogens. Clin Microbiol Rev. 2020;33. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE Pathogens. Biomed Res Int. 2016;2016:1–8. cited 2022 Aug 12. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/27274985/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breijyeh Z, Jubeh B, Karaman R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25:1340. cited 2022 Aug 12. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/32187986/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ten Threats to global health in 2019. [cited 2022 Aug 12; ]; https://www.who.int/vietnam/news/feature-stories/detail/ten-threats-to-global-health-in-2019 [Internet] Available from. [Google Scholar]
- 15.Pärnänen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Available from: www.nature.com/naturecommunications [DOI] [PMC free article] [PubMed]
- 16.Slykerman RF, Thompson J, Waldie KE, Murphy R, Wall C, Mitchell EA. Antibiotics in the first year of life and subsequent neurocognitive outcomes. Acta Paediatr. 2017;106:87–94. doi: 10.1111/apa.13613. [DOI] [PubMed] [Google Scholar]
- 17.Fink G, D’acremont V, Leslie HH, Cohen J. Antibiotic exposure among children younger than 5 years in low-income and middle-income countries: a cross-sectional study of nationally representative facility-based and household-based surveys. Lancet Infect Dis. 2020;20:179–187. doi: 10.1016/S1473-3099(19)30572-9. [DOI] [PubMed] [Google Scholar]
- 18.Kuperman AA, Koren O. Antibiotic use during pregnancy: how bad is it? BMC Med. 2016;14. doi: 10.1186/s12916-016-0636-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thinkhamrop J, Hofmeyr GJ, Adetoro O, Lumbiganon P. Prophylactic antibiotic administration during second and third trimester in pregnancy for preventing infectious morbidity and mortality. Cochrane Database Syst Rev. 4. 2002. [DOI] [PubMed] [Google Scholar]
- 20.Survival Program, Guidelines Review Committee. WHO Recommendations for Prevention and Treatment of Maternal Peripartum Infections. 2015. [Google Scholar]
- 21.Intrapartum care for a positive childbirth experience WHO recommendations. 2018; [PubMed]
- 22.Jarde A, Lewis-Mikhael AM, Moayyedi P, Stearns JC, Collins SM, Beyene J, McDonald SD. Pregnancy outcomes in women taking probiotics or prebiotics: a systematic review and meta-analysis. BMC Pregnancy Childbirth. 2018; 18:1–14. cited 2023 Mar 6. [Internet]Available from: 10.1186/s12884-017-1629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Li Z, Tye KD, Luo H, Tang X, Liao Y, Wang D, Zhou J, Yang P, Li Y, et al. Probiotic supplementation during human pregnancy affects the gut microbiota and immune status. Front Cell Infect Microbiol. [Internet]Available from. 2019. cited 2023 Mar 6;9. https://pubmed.ncbi.nlm.nih.gov/31380297/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang T, Li Z, Tye KD, Chan SN, Tang X, Luo H, Wang D, Zhou J, Duan X, Xiao X. Probiotic supplementation during pregnancy alters gut microbial networks of pregnant women and infants. Front Microbiol [Internet]Available from. 2022;13. cited 2023 Mar 6. https://pubmed.ncbi.nlm.nih.gov/36532501/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, Smilowitz JT, Underwood MA. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2019;8. doi: 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esaiassen E, Hjerde E, Cavanagh JP, Pedersen T, Andresen JH, Rettedal SI, Støen R, Nakstad B, Willassen NP, Klingenberg C. Effects of probiotic supplementation on the gut microbiota and antibiotic resistome development in preterm infants. Front Pediatr. 2018;6:6. doi: 10.3389/fped.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guitor AK, Yousuf EI, Raphenya AR, Hutton EK, Morrison KM, McArthur AG, Wright GD, Stearns JC. Capturing the antibiotic resistome of preterm infants reveals new benefits of probiotic supplementation. Microbiome. 2022;10:1–16. doi: 10.1186/s40168-022-01327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montassier E, Valdamp R, Batard E, Zmora N, Dori-Bachash M, Suez J, Elinav E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. [DOI] [PMC free article] [PubMed]
- 29.Feehan A, Garcia-Diaz JB. Gut microbiome-modifying therapies to defend against multidrug resistant organisms. Microorganisms. 2020;8:166. doi: 10.3390/microorganisms8020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLain JE, Cytryn E, Durso LM, Young S. Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J Environ Qual. 2016;45:432–440. doi: 10.2134/jeq2015.06.0317. [DOI] [PubMed] [Google Scholar]
- 31.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 32.Escobar-Zepeda A, Godoy-Lozano EE, Raggi L, Segovia L, Merino E, Gutiérrez-Rios RM, Juarez K, Licea-Navarro AF, Pardo-Lopez L, Sanchez-Flores A. Analysis of sequencing strategies and tools for taxonomic annotation: defining standards for progressive metagenomics. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-30515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leggett RM, Alcon-Giner C, Heavens D, Caim S, Brook TC, Kujawska M, Martin S, Peel N, Acford-Palmer H, Hoyles L, et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat Microbiol. 2020;5:430–442. doi: 10.1038/s41564-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berbers B, Saltykova A, Garcia-Graells C, Philipp P, Arella F, Marchal K, Winand R, Vanneste K, Roosens NHC, De Keersmaecker SCJ. Combining short and long read sequencing to characterize antimicrobial resistance genes on plasmids applied to an unauthorized genetically modified Bacillus. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-61158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashima Y, Sakamoto Y, Kaneko K, Seki M, Suzuki Y, Suzuki A. Single-cell sequencing techniques from individual to multiomics analyses. Experimental & Molecular Medi. 2020;52(9):1419–1427. doi: 10.1038/s12276-020-00499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy SB, Bonnie M. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–9. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 37.Nogacka AM, Salazar N, Arboleya S, Suárez M, Fernández N, Solís G, de Los Reyes-Gavilán CG, Gueimonde M. Early microbiota, antibiotics and health. Cell Mol Life Sci. 2018;75:83–91. cited 2022 Jan 27. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/28988290/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkin K, Christophersen CT, Verhasselt V, Cooper MN, Martino D. Risk Factors for Gut Dysbiosis in Early Life. Microorganisms. 2021;9:2066. cited 2022 Feb 15. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/34683389/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aires J. First 1000 days of life: consequences of antibiotics on gut microbiota. Front Microbiol. 2021;12. doi: 10.3389/fmicb.2021.681427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michael Cotten C, Taylor S, Stoll B, Goldberg RN, Hansen NI, Sanchez PJ, Ambalavanan N, Benjamin DK Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. [cited 2022 Feb 7; ]; Pediatrics. 2009 123:58. doi: 10.1542/peds.2007-3423. [Internet] Available from :/pmc/articles/PMC2760222/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM, Tarr PI, Warner BB, Dantas G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbio. 2019. [cited 2022 Aug 13];4(12):2285–2297. [Internet]Available from https://www.nature.com/articles/s41564-019-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyman M, van Houten MA, Watson RL, Chu MLJN, Arp K, de Waal WJ, Schiering I, Plötz FB, Willems RJL, van Schaik W, et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: a randomized trial. Nat Commun. [Internet]Available from. 2022. cited 2022 Aug 13;13. https://pubmed.ncbi.nlm.nih.gov/35173154/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renz H, Skevaki C. Early life microbial exposures and allergy risks: opportunities for prevention. Nat Rev Immunol. 2021;21:177–191. cited 2022 Feb 15. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/32918062/. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z, Wang J, Wang H, Li Y, Jia Y, Yi M, Chen O. Association of infant antibiotic exposure and risk of childhood asthma: a meta-analysis. World Allergy Organ J [Internet]Available from. 2021;14. cited 2022 Feb 15. https://pubmed.ncbi.nlm.nih.gov/34934469/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhudasia MB, Spergel JM, Puopolo KM, Koebnick C, Bryan M, Grundmeier RW, Gerber JS, Lorch SA, Quarshie WO, Zaoutis T, et al. Intrapartum group B Streptococcal prophylaxis and childhood allergic disorders. Pediatrics. [Internet]Available from. 2021. cited 2022 Dec 19;147. https://pubmed.ncbi.nlm.nih.gov/33833072/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baron R, Taye M, der Vaart IB, van Ujčič-Voortman J, Szajewska H, Seidell JC, Verhoeff A, Seidell JC. The relationship of prenatal antibiotic exposure and infant antibiotic administration with childhood allergies: a systematic review. BMC Pediatr [Internet]Available from. 2020;20. cited 2022 Feb 15-20. https://pubmed.ncbi.nlm.nih.gov/32593308/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mårild K, Ludvigsson J, Sanz Y, Ludvigsson JF. Antibiotic exposure in pregnancy and risk of coeliac disease in offspring: a cohort study. BMC Gastroenterol [Internet]Available from. 2014;14. cited 2023 Mar 6. https://pubmed.ncbi.nlm.nih.gov/24731164/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uzan-Yulzari A, Turta O, Belogolovski A, Ziv O, Kunz C, Perschbacher S, Neuman H, Pasolli E, Oz A, Ben-Amram H, et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun. 2021;12. doi: 10.1038/s41467-020-20495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdellatif M, Ghozy S, Kamel MG, Elawady SS, Ghorab MME, Attia AW, le Huyen TT, Duy DTV, Hirayama K, Huy NT. Association between exposure to macrolides and the development of infantile hypertrophic pyloric stenosis: a systematic review and meta-analysis. Eur J Pediatr. 2019;178:301–314. cited 2022 May 14. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/30470884/. [DOI] [PubMed] [Google Scholar]
- 50.Aris IM, Lin PID, Rifas-Shiman SL, Bailey LC, Boone-Heinonen J, Eneli IU, Solomonides AE, Janicke DM, Toh S, Forrest CB, et al. Association of early antibiotic exposure with childhood body mass index trajectory milestones. JAMA Netw Open. 2021. cited 2022 Feb 15;4:e2116581. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/34251440/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng X, Zhu Y, Di H, Zhang M, Feng J, Xu M, Xia W, Tian Q, He Y, Gan Y, et al. Dose-response association of early-life antibiotic exposure and subsequent overweight or obesity in children: a meta-analysis of prospective studies. Obes Rev. [Internet]Available from. 2021. cited 2022 Feb 15;22. https://pubmed.ncbi.nlm.nih.gov/34328260/. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein CN. Review article: changes in the epidemiology of inflammatory bowel disease-clues for aetiology. Aliment Pharmacol Ther. 2017;46:911–919. doi: 10.1111/apt.14338. [DOI] [PubMed] [Google Scholar]
- 53. Khan I, Yasir M, Farman M, Kumosani T, Albasri SF, Bajouh OS, Azhar EI Evaluation of gut bacterial community composition and antimicrobial resistome in pregnant and non-pregnant women from Saudi population. [cited 2022 Feb 10; ]; Infect Drug Resist. 2019 12:1749. doi: 10.2147/IDR.S200213. [Internet] Available from :/pmc/articles/PMC6593780/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guernier-Cambert V, Chamings A, Collier F, Alexandersen S Diverse bacterial resistance genes detected in fecal samples from clinically healthy women and infants in Australia—A descriptive pilot study. [cited 2022 Feb 10; ]; Front Microbiol. 2021 12 doi: 10.3389/fmicb.2021.596984. [Internet] Available from :/pmc/articles/PMC8484959/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sosa-Moreno A, Comstock SS, Sugino KY, Ma TF, Paneth N, Davis Y, Olivero R, Schein R, Maurer J, Zhang L. Perinatal risk factors for fecal antibiotic resistance gene patterns in pregnant women and their infants. Plos One. 2020; 15:e0234751. cited 2022 May 13. [Internet]Available from: 10.1371/journal.pone.0234751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyöty H, Virtanen SM, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host & Microbe. [[Internet]Available from :/pmc/articles/PMC6091882/]. 2018. [cited 2022 Feb 10];24:146. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang X, Ellabaan MMH, Charusanti P, Munck C, Blin K, Tong Y, Weber T, Sommer MOA, Lee SY. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat Commun. 2017;8(1):1–7. doi: 10.1038/ncomms15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Stokholm J, Brejnrod A, Vestergaard GA, Russel J, Trivedi U, Thorsen J, Gupta S, Hjelmsø MH, Shah SA, et al. The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host & Microbe. 2021. cited 2022 Feb 10;29:975–987.e4. [Internet]Available from http://www.cell.com/article/S1931312821001451/fulltext. [DOI] [PubMed] [Google Scholar]
- 59.Lawson MAE, O’neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, Chalklen L, Hall LJ. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. Isme J. 2019. [cited 2022 Oct 18];14(2):635–648. [Internet]Available from https://www.nature.com/articles/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh C, Lane JA, van Sinderen D, Hickey RM. Human milk oligosaccharides: shaping the infant gut microbiota and supporting health. J Funct Foods. 2020;72:104074. doi: 10.1016/j.jff.2020.104074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere [Internet]Available from. 2018;3. cited 2022 Oct 21. https://pubmed.ncbi.nlm.nih.gov/30258040/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saturio S, Suárez M, Mancabelli L, Fernández N, Mantecón L, de Los Reyes-Gavilán CG, Ventura M, Gueimonde M, Arboleya S, Solís G. Effect of intrapartum antibiotics prophylaxis on the bifidobacterial establishment within the neonatal gut. Microorganisms. 2021;9:1867. cited 2022 Feb 8. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/34576761/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibson MK, Wang B, Ahmadi S, Burnham CAD, Tarr PI, Warner BB, Dantas G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbio. 2016. [cited 2022 Jul 21];1(4):1–10. [Internet]Available from https://www.nature.com/articles/nmicrobiol201624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore AM, Ahmadi S, Patel S, Gibson MK, Wang B, Ndao IM, Deych E, Shannon W, Tarr PI, Warner BB, et al. Gut resistome development in healthy twin pairs in the first year of life. Microbiome. 2015. cited 2022 Jul 21;3:1–10. [Internet]Available from https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-015-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. [Internet]Available from. 2016. cited 2022 May 13;8. https://pubmed.ncbi.nlm.nih.gov/27306663/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lebeaux RM, Coker MO, Dade EF, Palys TJ, Morrison HG, Ross BD, Baker ER, Karagas MR, Madan JC, Hoen AG. The infant gut resistome is associated with E. coli and early-life exposures. BMC Microbiol. 2021;21:1–18. cited 2022 Jul 21. [Internet]Available from https://bmcmicrobiol.biomedcentral.com/articles/10.1186/s12866-021-02129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe [Internet]. 2015. cited 2022 Jul 21;17:690–703. Available from http://www.cell.com/article/S1931312815001626/fulltext. [DOI] [PubMed] [Google Scholar]
- 68.Siefert JL. Defining the Mobilome. Methods Mol Biol. 2009; 532:13–27. cited 2022 Apr 13. [Internet]Available from: 10.1007/978-1-60327-853-9_2. [DOI] [PubMed] [Google Scholar]
- 69.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sosa-Moreno A, Comstock SS, Sugino KY, Ma TF, Paneth N, Davis Y, Olivero R, Schein R, Maurer J, Zhang L. Perinatal risk factors for fecal antibiotic resistance gene patterns in pregnant women and their infants. Plos One. 2020;15. doi: 10.1371/journal.pone.0234751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patangia DV, Ryan CA, Dempsey E, Stanton C, Ross RP. Vertical transfer of antibiotics and antibiotic resistant strains across the mother/baby axis. Trends Microbiol. 2022;30:47–56. doi: 10.1016/j.tim.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 72. Klassert TE, Zubiria-Barrera C, Kankel S, Stock M, Neubert R, Lorenzo-Diaz F, Doehring N, Driesch D, Fischer D, Slevogt H Early Bacterial Colonization and Antibiotic Resistance Gene Acquisition in Newborns. [cited 2022 May 14; ]; Front Cell Infect Microbiol. 2020 10:332. doi: 10.3389/fcimb.2020.00332. [Internet] Available from :/pmc/articles/PMC7366792/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loo EXL, Zain A, Yap GC, Purbojati RW, Drautz-Moses DI, Koh YQ, Chong YS, Tan KH, Gluckman PD, Yap F, et al. Longitudinal assessment of antibiotic resistance gene profiles in gut microbiomes of infants at risk of eczema. BMC Infect Dis. 2020;20:20. doi: 10.1186/s12879-020-05000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herindrainy P, Rabenandrasana AN, Andrianirina ZZ, Manitra F, Rakotoarimanana J, Padget M, De Lauzanne A, Ndir A, Kermorvant-Duchemin E, Garin B, et al. Acquisition of extended spectrum beta-lactamase-producing enterobacteriaceae in neonates: a community based cohort in Madagascar. Plos One. 2018;13:e0193325. doi: 10.1371/journal.pone.0193325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuwelker K, Langeland N, Löhr IH, Gidion J, Manyahi J, Moyo SJ, Blomberg B, Klingenberg C. Use of probiotics to reduce infections and death and prevent colonization with extended-spectrum beta-lactamase (ESBL)-producing bacteria among newborn infants in Tanzania (ProRIDE Trial): study protocol for a randomized controlled clinical trial. Trials. 2021;22. doi: 10.1186/s13063-021-05251-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Haddad EN, Comstock SS Archive for research in child health (ARCH) and baby gut: study protocol for a remote, prospective, longitudinal pregnancy and birth cohort to address microbiota development and child health. [cited 2022 Feb 10; ]; Methods Protoc. 2021 4:52. doi: 10.3390/mps4030052. [Internet] Available from :/pmc/articles/PMC8395764/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karami N, Nowrouzian F, Adlerberth I, Wold AE. Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob Agents Chemother [Internet]. 2006;50:156–161. cited 2022 Feb 10]. Available from https://pubmed.ncbi.nlm.nih.gov/16377681/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Same RG, Tamma PD. 2018Campylobacter Infections in Children Pediatr Rev [Internet]Available from:/pediatricsinreview/article/39/11/533/35101/Campylobacter-Infections-in-Childrencited 2022 May 14533–541 10.1542/pir.2017-028539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Severgnini M, Camboni T, Ceccarani C, Morselli S, Cantiani A, Zagonari S, Patuelli G, Pedna MF, Sambri V, Foschi C, et al. Distribution of ermb, ermf, tet(w), and tet(m) resistance genes in the vaginal ecosystem of women during pregnancy and puerperium. Pathogens. [[Internet]Available from:/pmc/articles/PMC8705968/]. 2021. [cited 2022 May 14];10:1546. doi: 10.3390/pathogens10121546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fouhy F, Ogilvie LA, Jones BV, Ross RP, Ryan AC, Dempsey EM, Fitzgerald GF, Stanton C, Cotter PD. Identification of aminoglycoside and β-lactam resistance genes from within an infant gut functional metagenomic library. Plos One. 2014;9:e108016. cited 2022 May 14. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/25247417/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gurnee EA, Ndao IM, Johnson JR, Johnston BD, Gonzalez MD, Burnham CAD, Hall-Moore CM, McGhee JE, Mellmann A, Warner BB, et al. Gut colonization of healthy children and their mothers with pathogenic ciprofloxacin-resistant Escherichia coli. J Infect Dis. 2015. cited 2022 Oct 18;212:1862–1868. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/25969564/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottesman BS, Low M, Almog R, Chowers M. Quinolone consumption by mothers increases their children’s risk of acquiring quinolone-resistant bacteriuria. Clinical Infectious Diseases. 2020;71:532–538. cited 2022 May 14. [Internet]Available from https://academic.oup.com/cid/article/71/3/532/5556473. [DOI] [PubMed] [Google Scholar]
- 83. Kim DW, Cha CJ Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission. [cited 2022 Jul 21; ]; Exp Mol Med. 2021 53:301. doi: 10.1038/s12276-021-00569-z. [Internet] Available from:/pmc/articles/PMC8080597/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olivares J, Bernardini A, Garcia-Leon G, Corona F, Sanchez MB, Martinez JL. The intrinsic resistome of bacterial pathogens. Front Microbiol. 2013;4:103. doi: 10.3389/fmicb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhang L, Kinkelaar D, Huang Y, Li Y, Li X, Wang HH Acquired antibiotic resistance: are we born with it? [cited 2022 May 13; ]; Appl Environ Microbiol. 2011 77:7134. doi: 10.1128/AEM.05087-11. [Internet] Available from:/pmc/articles/PMC3194877/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raghunathan A, Ferguson HR, Bornarth CJ, Song W, Driscoll M, Lasken RS. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dankó D, Blay JY, Garrison LP. Challenges in the value assessment, pricing and funding of targeted combination therapies in oncology. Health Policy (New York). 2019;123:1230–1236. doi: 10.1016/j.healthpol.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 88.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E, Rovira P, Abdo Z, Jones KL, Ruiz J, et al. Megares: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017;45:D574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. Isme J. 2015;9:207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moore AM, Ahmadi S, Patel S, Gibson MK, Wang B, Ndao MI, Deych E, Shannon W, Tarr PI, Warner BB, et al. Gut resistome development in healthy twin pairs in the first year of life. Microbiome. 2015;3. doi: 10.1186/s40168-015-0095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gosalbes MJ, Vallès Y, Jiménez-Hernández N, Balle C, Riva P, Miravet-Verde S, de Vries LE, Llop S, Agerso Y, Sørensen SJ, et al. High frequencies of antibiotic resistance genes in infants’ meconium and early fecal samples. J Dev Orig Health Dis. 2016. cited 2022 May 13;7:35–44. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/26353938/. [DOI] [PubMed] [Google Scholar]
- 93.Kozak K, Charbonneau D, Sanozky-Dawes R, Klaenhammer T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. [Internet] 2016. cited 2022 Oct 18; 6:341–351. Available from: https://www.tandfonline.com/action/journalInformation?journalCode=kgmi20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carvalho MJ, Sands K, Thomson K, Portal E, Mathias J, Milton R, Gillespie D, Dyer C, Akpulu C, Boostrom I, et al. 2022. Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Nat Microbio. 7(9):1337–1347. doi: 10.1038/s41564-022-01184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Devi U, Barman N, Barua P, Malik V, Das JK, Baruah P, Mahanta J. Vaginal carriage of antibiotic resistant Escherichia coli by pregnant women: a concern for the neonate. Clinical Microbiology: Open Access. 2014;03. doi: 10.4172/2327-5073.1000153. [DOI] [Google Scholar]
- 96.Sáez-López E, Guiral E, Fernández-Orth D, Villanueva S, Goncé A, López M, Teixidó I, Pericot A, Figueras F, Palacio M, et al. Vaginal versus obstetric infection Escherichia coli isolates among pregnant women: antimicrobial resistance and genetic virulence profile. Plos One. 2016;11:11. doi: 10.1371/journal.pone.0146531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dubois V, De Barbeyrac B, Rogues AM, Arpin C, Coulange L, Andre C, M’zali F, Megraud F, Quentin C. CTX-M-producing Escherichia coli in a maternity ward: a likely community importation and evidence of mother-to-neonate transmission. J Antimicrob Chemother. 2010;65:1368–1371. doi: 10.1093/jac/dkq153. [DOI] [PubMed] [Google Scholar]
- 98.McDonald LC, Bryant K, Snyder J. Peripartum transmission of penicillin-resistant Streptococcus pneumoniae. J Clin Microbiol. 2003;41:2258–2260. doi: 10.1128/JCM.41.5.2258-2260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alicea-Serrano AM, Contreras M, Magris M, Hidalgo G, Dominguez-Bello MG. Tetracycline resistance genes acquired at birth. Arch Microbiol. 2013;195:447–451. cited 2022 Oct 18. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/23483141/. [DOI] [PubMed] [Google Scholar]
- 100.Das L, Virmani R, Sharma V, Rawat D, Singh Y. Human milk microbiota: transferring the antibiotic resistome to infants. Indian J Microbiol. 2019;59:410–416. cited 2022 Jan 20]. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/31762502/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nadimpalli ML, Bourke CD, Robertson RC, Delarocque-Astagneau E, Manges AR, Pickering AJ. Can breastfeeding protect against antimicrobial resistance? BMC Med. 2020;18(1):1–11. doi: 10.1186/s12916-020-01862-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang MS, Cheng CC, Tseng SY, Lin YL, Min LH, Chen PW. Most commensally bacterial strains in human milk of healthy mothers display multiple antibiotic resistance. Microbiologyopen. 2019;8:e00618. cited 2022 Feb 14. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/29577668/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li X, Zhou Y, Zhan X, Huang W, Wang X. Breast milk is a potential reservoir for livestock-associated Staphylococcus aureus and community-associated Staphylococcus aureus in Shanghai, China. Front Microbiol. 2018;8:2639. doi: 10.3389/fmicb.2017.02639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen PW, Tseng SY, Huang MS. Antibiotic susceptibility of commensal bacteria from human milk. Curr Microbiol. 2016;72:113–119. doi: 10.1007/s00284-015-0925-4. [DOI] [PubMed] [Google Scholar]
- 105.Behari P, Englund J, Alcasid G, Garcia-Houchins S, Weber SG. Transmission of methicillin-resistant Staphylococcus aureus to preterm infants through breast milk. Infect Control Hosp Epidemiol. 2004;25:778–780. doi: 10.1086/502476. [DOI] [PubMed] [Google Scholar]
- 106.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol. 2012;15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 107.Gasparrini AJ, Crofts TS, Gibson MK, Tarr PI, Warner BB, Dantas G. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes. 2016;7:443–449. doi: 10.1080/19490976.2016.1218584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pärnänen KMM, Hultman J, Markkanen M, Satokari R, Rautava S, Lamendella R, Wright J, McLimans CJ, Kelleher SL, Virta MP. Early-life formula feeding is associated with infant gut microbiota alterations and an increased antibiotic resistance load. Am J Clin Nutr. 2021;115:407–421. doi: 10.1093/ajcn/nqab353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zou ZH, Liu D, Li HD, Zhu DP, He Y, Hou T, Yu JL. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Ann Clin Microbiol Antimicrob [Internet]Available from. 2018;17. cited 2022 Feb 10. https://pubmed.ncbi.nlm.nih.gov/29554907/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rose G, Shaw AG, Sim K, Wooldridge DJ, Li MS, Gharbia S, Misra R, Kroll JS. Antibiotic resistance potential of the healthy preterm infant gut microbiome. PeerJ. 2017;5:e2928. doi: 10.7717/peerj.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Selma-Royo M, García-Mantrana I, Calatayud M, Parra-Llorca A, Martínez-Costa C, Collado MC. Maternal microbiota, cortisol concentration, and post-partum weight recovery are dependent on mode of delivery. Nutrients. 2020;12:1779. doi: 10.3390/nu12061779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reyman M, van Houten MA, van Baarle D, Bosch AATM, Man WH, Chu MLJN, Arp K, Watson RL, Sanders EAM, Fuentes S, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun [Internet]Available from. 2019;10(11):1–12. cited 2022 Oct 5. https://www.nature.com/articles/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guo C, Zhou Q, Li M, Zhou L, Xu L, Zhang Y, Li D, Wang Y, Dai W, Li S, et al. Breastfeeding restored the gut microbiota in caesarean section infants and lowered the infection risk in early life. BMC Pediatr. 2020. cited 2022 Oct 5;20:1–6. [Internet]Available from https://bmcpediatr.biomedcentral.com/articles/10.1186/s12887-020-02433-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016;7:54–60. cited 2022 Oct 5. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/26286040/. [DOI] [PubMed] [Google Scholar]
- 115.Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr [Internet]Available from. 2019;6. cited 2022 Oct 5. https://pubmed.ncbi.nlm.nih.gov/30778389/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–551. cited 2022 Oct 5. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/22836031/. [DOI] [PubMed] [Google Scholar]
- 117.Liu Y, Qin S, Song Y, Feng Y, Lv N, Xue Y, Liu F, Wang S, Zhu B, Ma J, et al. The perturbation of infant gut microbiota caused by cesarean delivery is partially restored by exclusive breastfeeding. Front Microbiol. [Internet]Available from. 2019. cited 2022 Oct 5;10. https://pubmed.ncbi.nlm.nih.gov/30972048/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bossung V, Lupatsii M, Dashdorj L, Tassiello O, Jonassen S, Pagel J, Demmert M, Wolf EA, Rody A, Waschina S, et al.Timing of antimicrobial prophylaxis for cesarean section is critical for gut microbiome development in term born infants Gut Microbes [Internet]Available from2022cited 2022 Oct 514 10.1080/19490976.2022.2038855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arboleya S, Suárez M, Fernández N, Mantecón L, Solís G, Gueimonde M, De Los Reyes-Gavilán CG. C-section and the neonatal gut microbiome acquisition: consequences for future health. Ann Nutr Metab. 2018;73:17–23. doi: 10.1159/000490843. [DOI] [PubMed] [Google Scholar]
- 120.Prophylactic antibiotics for women undergoing caesarean section. [PubMed]
- 121.Dohou AM, Buda VO, Yemoa LA, Anagonou S, Van Bambeke F, Van Hees T, Dossou FM, Dalleur O. Antibiotic usage in patients having undergone caesarean section: a three-level study in benin. Antibiotics (Basel). 2022;11:11. doi: 10.3390/antibiotics11050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. [Internet]Available from 2019. [cited 2022 Oct 5];574(7776):117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Velin L, Umutesi G, Riviello R, Muwanguzi M, Bebell LM, Yankurije M, Faktor K, Nkurunziza T, Rukundo G, Gatete JDD, et al. Surgical site infections and antimicrobial resistance after cesarean section delivery in rural rwanda. Ann Glob Health. 2021. cited 2022 Oct 5;87:77. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/34430227/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Didier C, Streicher MP, Chognot D, Campagni R, Schnebelen A, Messer J, Donato L, Langer B, Meyer N, Astruc D, et al. Late-onset neonatal infections: incidences and pathogens in the era of antenatal antibiotics. Eur J Pediatr. 2012. cited 2022 Dec 19;171:681–687. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/22134805/. [DOI] [PubMed] [Google Scholar]
- 125.Li W, Tapiainen T, Brinkac L, Lorenzi HA, Moncera K, Tejesvi MV, Salo J, Nelson KE. Vertical transmission of gut microbiome and antimicrobial resistance genes in infants exposed to antibiotics at birth. J Infect Dis. 2021;224:1236–1246. doi: 10.1093/infdis/jiaa155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Dyke MK, Phares CR, Lynfield R, Thomas AR, Arnold KE, Craig AS, Mohle-Boetani J, Gershman K, Schaffner W, Petit S, et al. Evaluation of universal antenatal screening for group B streptococcus abstract. The New England journal of medicine. 2009;360(25): 2626–2636. [DOI] [PubMed] [Google Scholar]
- 127.Chai G, Governale L, McMahon AW, Trinidad JP, Staffa J, Murphy D. Trends of outpatient prescription drug utilization in US children, 2002-2010. Pediatrics. 2012;130:23–31. cited 2023 Feb 1. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/22711728/. [DOI] [PubMed] [Google Scholar]
- 128.Arboleya S, Saturio S, Gueimonde M, Linares R. Impact of intrapartum antibiotics on the developing microbiota: a review. Microbio Res Reports. 2022;1:22. cited 2023 Feb1. [Internet]Available from https://www.oaepublish.com/mrr/article/view/5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nogacka A, Salazar N, Suárez M, Milani C, Arboleya S, Solís G, Fernández N, Alaez L, Hernández-Barranco AM, de Los Reyes-Gavilán CG, et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome. [Internet]Available from. 2017. cited 2022 Oct 5;5. https://pubmed.ncbi.nlm.nih.gov/28789705/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reyman M, van Houten MA, Watson RL, Chu MLJN, Arp K, de Waal WJ, Schiering I, Plötz FB, Willems RJL, van Schaik W, et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: a randomized trial. Nat Commun. 2022;13. doi: 10.1038/s41467-022-28525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lebeaux RM, Madan JC, Nguyen QP, Coker MO, Dade EF, Moroishi Y, Palys TJ, Ross BD, Pettigrew MM, Morrison HG, et al. Impact of antibiotics on off-target infant gut microbiota and resistance genes in cohort studies. Pediatr Res . 2022;92:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Contributing to one world, one health* a strategic framework for reducing risks of infectious diseases at the animal-human-ecosystems interface. 2008;
- 133.Valles-Colomer M, Blanco-Míguez A, Manghi P, Asnicar F, Dubois L, Golzato D, Armanini F, Cumbo F, Huang KD, Manara S, et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature. 2023. cited 2023 Feb 3;614:7946–35 125–35. [Internet]Available from https://www.nature.com/articles/s41586-022-05620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, et al. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome. 2017. cited 2022 Apr 25;5:1–14. [Internet]Available from https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-017-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gómez-Gallego C, Forsgren M, Selma-Royo M, Nermes M, Carmen Collado M, Salminen S, Beasley S, Isolauri E. The composition and diversity of the gut microbiota in children is modifiable by the household dogs: impact of a canine-specific probiotic. Microorganisms. 2021;9:557. cited 2022 Apr 25. [Internet]Available from https://www.mdpi.com/2076-2607/9/3/557/htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pomba C, Belas A, Menezes J, Marques C. The public health risk of companion animal to human transmission of antimicrobial resistance during different types of animal infection Advs in Animal Health, Medi and Pro [Internet]Available from2020cited 2022 Apr 25265–278 10.1007/978-3-030-61981-7_14 [DOI] [Google Scholar]
- 137.Pomba C, Rantala M, Greko C, Baptiste KE, Catry B, van Duijkeren E, Mateus A, Moreno MA, Pyörälä S, Ružauskas M, et al. Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother. 2017;72:957–968. doi: 10.1093/jac/dkw481. [DOI] [PubMed] [Google Scholar]
- 138.Belas A, Menezes J, Gama LT, Pomba C. Sharing of clinically important antimicrobial resistance genes by companion animals and their human household members. [Internet] 2020. cited 2022 Apr 25; 26:1174–1185. Available from: 10.1089/mdr.2019.0380 [DOI] [PubMed] [Google Scholar]
- 139.Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 2012;127:4. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pi Y, Gao K, Peng Y, Mu CL, Zhu WY. Antibiotic-induced alterations of the gut microbiota and microbial fermentation in protein parallel the changes in host nitrogen metabolism of growing pigs. Animal. 2019;13:262–272. doi: 10.1017/S1751731118001416. [DOI] [PubMed] [Google Scholar]
- 141.Monger XC, Gilbert AA, Saucier L, Vincent AT. Antibiotic resistance: from pig to meat. Antibiotics (Basel). 2021;10:10. doi: 10.3390/antibiotics10101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang RM, Liu X, Wang SL, Fang LX, Sun J, Liu YH, Liao XP. Distribution patterns of antibiotic resistance genes and their bacterial hosts in pig farm wastewater treatment systems and soil fertilized with pig manure. Sci Total Environ. 2021;758:143654. doi: 10.1016/j.scitotenv.2020.143654. [DOI] [PubMed] [Google Scholar]
- 143.Zhang YJ, Hu HW, Chen QL, Singh BK, Yan H, Chen D, He JZ. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ Int. 2019;130:104912. doi: 10.1016/j.envint.2019.104912. [DOI] [PubMed] [Google Scholar]
- 144.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. The structure and diversity of human, animal and environmental resistomes. Microbiome [Internet]Available from. 2016;4. cited 2022 Jul 21. https://pubmed.ncbi.nlm.nih.gov/27717408/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hourigan SK, Subramanian P, Hasan NA, Ta A, Klein E, Chettout N, Huddleston K, Deopujari V, Levy S, Baveja R, et al.Comparison of infant gut and skin microbiota, resistome and virulome between neonatal intensive care unit (NICU) environments Front Microbiol [Internet]Available from:/pmc/articles/PMC6026636/2018cited 2022 Jul 219 10.3389/fmicb.2018.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Antimicrobial stewardship programmes in health-care facilities in low - and middle -income countries. [Internet]. cited 2022 Oct 18: Available from https://www.who.int/publications/i/item/9789241515481 [DOI] [PMC free article] [PubMed]
- 147.Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, Hoen AG. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018; 6:1–11. cited 2023 Mar 6. [Internet]Available from: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Savage JH, Lee-Sarwar KA, Sordillo JE, Lange NE, Zhou Y, O’connor GT, Sandel M, Bacharier LB, Zeiger R, Sodergren E, et al. Diet during pregnancy and infancy and the infant intestinal microbiome. J Pediatr. 2018. cited 2023 Mar 6;203:47–54.e4. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/30173873/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med [Internet]Available from. 2016;8. cited 2023 Mar 6. https://pubmed.ncbi.nlm.nih.gov/27503374/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oliver A, Xue Z, Villanueva YT, Durbin-Johnson B, Alkan Z, Taft DH, Liu J, Korf I, Laugero KD, Stephensen CB, et al. Association of diet and antimicrobial resistance in healthy U.S. Adults mBio. 2022;13. doi: 10.1128/mbio.00101-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tan R, Jin M, Shao Y, Yin J, Li H, Chen T, Shi D, Zhou S, Li J, Yang D. High-sugar, high-fat, and high-protein diets promote antibiotic resistance gene spreading in the mouse intestinal microbiota. Gut Microbes. 2022;14. doi: 10.1080/19490976.2021.2022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.da Silva SF, Reis IB, Monteiro MG, Dias VC, Machado ABF, da Silva VL, Diniz CG. Influence of human eating habits on antimicrobial resistance phenomenon: aspects of clinical resistome of gut microbiota in omnivores, ovolactovegetarians, and strict vegetarians. Antibiotics. 2021;10:1–12. doi: 10.3390/antibiotics10030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stege PB, Hordijk J, Shetty SA, Visser M, Viveen MC, Rogers MRC, Gijsbers E, Dierikx CM, van der Plaats RQJ, van Duijkeren E, et al. 2022. Impact of long-term dietary habits on the human gut resistome in the Dutch population. Sci Rep. 12(1):1–13. doi: 10.1038/s41598-022-05817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fiocchi A, Pawankar R, Cuello-Garcia C, Ahn K, Al-Hammadi S, Agarwal A, Beyer K, Burks W, Canonica GW, Ebisawa M, et al. World allergy organization-McMaster University guidelines for allergic disease prevention (GLAD-P): probiotics. World Allergy Organ J. 2015. cited 2023 Mar 6;8:4. [Internet]Available from https://pubmed.ncbi.nlm.nih.gov/25628773/. [DOI] [PMC free article] [PubMed] [Google Scholar]


