Abstract
Scope
Circadian rhythm is an endogenous and self‐sustained timing system, responsible for the coordination of daily processes in 24‐h timescale. It is regulated by an endogenous molecular clock, which is sensitive to external cues as light and food. This study has previously shown that grape seed proanthocyanidins extract (GSPE) regulates the hepatic molecular clock. Moreover, GSPE is known to interact with some microRNAs (miRNAs). Therefore, the aim of this study is to evaluate if the activity of GSPE as modulator of hepatic clock genes can be mediated by miRNAs.
Methods and results
250 mg kg−1 of GSPE is administered to Wistar rats before a 6‐h jet lag and sacrificed at different time points. GSPE modulated both expression of Bmal1 and miR‐27b‐3p in the liver. Cosinor‐based analysis reveals that both Bmal1 and miR‐27b‐3p expression follow a circadian rhythm, a negative interaction between them, and the role of GSPE adjusting the hepatic peripheral clock via miRNA. Additionally, in vitro studies show that Bmal1 is sensitive to GSPE (25 mg L−1). However, this effect is independent of miR‐27b‐3p.
Conclusion
miRNA regulation of peripheral clocks via GSPE may be part of a complex mechanism that involves the crosstalk with the central system rather than a direct effect.
Keywords: circadian rhythm, Cosinor‐based rhythmometry, microRNA, peripheral molecular clock, polyphenols
Jet‐lagged rats and HepG2 cells were used to evaluate if the activity of grape seed proanthocyanidins extract (GSPE) as modulator of hepatic clock genes is mediated by miRNAs. GSPE modulates expression of hepatic Bmal1 and miR‐27b‐3p. Moreover, cosinor‐based analysis reveals that both Bmal1 and miR‐27b‐3p expression follow a circadian rhythm and that GSPE adjusted the hepatic peripheral clock via miRNA.

1. Introduction
Biological processes follow cycles of about 24 h during circadian oscillation dictated by light–dark cycles. These rhythms are regulated by the molecular clock, which is synchronized by external cues or zeitgebers (“time giver”, i.e., zeitgeber, synchronizes biological rhythm). The molecular clock has two differentiated parts: i) the master clock, which is hosted in the suprachiasmatic nucleus (SCN) of the hypothalamus and is mainly regulated by light[ 1 , 2 ] and ii) the peripheral clocks, which are present in almost all tissues and mainly reacts to nutritional cues.[ 3 ] An example is the liver, which has a pivotal role in the metabolism homeostasis and whose rhythmicity is regulated by nutritional signals.[ 4 ]
The molecular basis of circadian rhythms is the transcription‐translation feedback loop (TTFL), which regulates the clock genes. Essential clock genes are: aryl hydrocarbon receptor nuclear translocator‐like protein1 (Arntl1, normally called Bmal1) and circadian locomotor output cycles kaput (Clock). The heterodimer BMAL1:CLOCK works as a transcription factor of genes: period (Per), cryptochrome (Cry), nuclear receptor subfamily 1 group D (Rev‐Erba), and RAR‐related orphan receptor (Ror). PER and CRY proteins come back into the nucleus thus repressing their own transcription acting on BMAL1:CLOCK, giving rise to the negative feedback loop regulation. Protein products of clock genes set up complexes with epigenetic factors, which drive the expression of target genes (namely clock‐controlled genes).[ 5 ] Hence, these clock‐controlled genes, which are involved in metabolic processes, follow a rhythmic expression. An example is the nicotinamide phosphoribosyl transferase (Nampt) gene, which is a Bmal1 target gene. NAMPT protein is the rate limiting enzyme of NAD production, acting in the conversion of nicotinamide to nicotinamide mononucleotide in mammals that enables NAD+ biosynthesis.[ 6 ]
Significant changes in the light–dark cycles, such as sleep disruptions and jetlag, can produce misalignment of the clocks.[ 7 ] These rhythm disruptions can cause different negative metabolic consequences, which are related to the development of cardiometabolic, immune, neurologic, and psychiatric diseases.[ 8 , 9 ] Interestingly, it has been shown that some dietary compounds can act on molecular clock genes and consequently, they can exert beneficial effects on the disruption of the circadian rhythms.[ 10 , 11 ] In this regard, it has been reported that a grape seed proanthocyanidins, a class of dietary phenolic compounds, can alter the expression of the master clock genes including Bmal1 and Nampt [ 12 ] and of some components of the peripheral clock in liver, gut, and white adipose tissue, including Bmal1, Nampt, Sirt1, and NAD+.[ 13 , 14 , 15 ] These findings suggest that grape seed proanthocyanidins extract (GSPE) can modulate physiology and metabolism by adjusting both the peripheral and central molecular clock. Nevertheless, the mechanism involved in the GSPE modulation of the molecular clock machinery and clock‐controlled genes has not been investigated.
Modulation of microRNAs (miRNAs) has been identified as one of the molecular mechanisms involved in the effects of grape seed proanthocyanidins.[ 13 ] miRNAs are small single stranded non coding RNA (18–25 nucleotides) involved in RNA silencing and post‐transcriptional regulation of gene expression.[ 16 ] In this regard, it has been reported that GSPE regulates miR‐30b in HepG2 cells, whose target genes are involved in several pathways.[ 17 , 18 ] In addition, other studies evidenced that miRNAs can regulate both core molecular clock and clock‐controlled genes, pointing to a post‐transcriptional role in their modulation of endogenous clock.[ 19 ] For example, recent studies reported that miR‐27b‐3p functions as modulator at post‐transcriptional level of in peripheral clock like in liver.[ 19 ] Moreover, miR‐34a overexpression is associated to reduced levels of NAMPT, SIRT1, and NAD+.[ 20 ] Considering all the evidence, the aim of the present research was to evaluate whether the activity of grape seed proanthocyanidins, as a modulator of hepatic clock gene Bmal1 and clock‐controlled gene Nampt, could be mediated by miR‐27b‐3p and miR‐34a, respectively.
2. Experimental Section
2.1. Reactives
GSPE was kindly provided by Les Dérives Résiniques et Terpéniques (Dax, France). According to the manufacturer, the GSPE profile was composed of monomers or flavons‐3‐ols (21.3 %), dimers (17.4 %), trimers (16.3 %), tetramers (13.3 %), and oligomers 5–13 unit (31.7%). Gallic acid, protocatechuic acid, vanillic acid, proanthocyanidin B2, catechin, epicatechin, epigallocatechin gallate, p‐coumaric acid, quercetin, narigenin, and kaempferol were purchased from Fluka/Sigma‐Aldrich (Madrid, Spain). All reagents were analytical grade. High performance liquid chromatography (HPLC) grade acetic acid and acetonitrile were provided by Panreac (Barcelona, Spain).
HepG2 human hepatocarcinama cells were purchased by ECACC Sigma‐Aldrich. Minimum essential medium (DMEM), fetal bovine serum, l‐glutamine, penicillin, and streptomycin were bought in Lonza (Barcelona, Spain). N‐(2Hydroxyethyl) piperazine‐N′‐(2‐ethanesulfonic acid) (HEPES), non‐essential amino acids (NEAA), and horse serum were provided by Sigma‐Aldrich.
2.2. Characterization of the Phenolic Profile of the Grape Seed Proanthocyanidins Extract
Individual phenolic compounds from GSPE were identified and quantified by a HPLC coupled to a time of flight mass spectrometer (TOF 6210, Agilent), as previously described by Quiñones et al.,[ 21 ] using the same equipment and columns. Specific conditions in this study were: the flow rate and injection volume were 0.4 mL min−1 and 5 µL, respectively. Ionization in the mass spectrometer was performed by electrospray (ESI) in the negative mode, and the source parameters were as follows: capillary voltage, 3.5 kV; fragmentor, 120 V; source temperature, 150 °C; desolvation gas temperature, 350 °C, with a drying gas flow rate of 12 L min−1. Nitrogen was used as the cone gas. Individual phenols were quantified with a five‐point regression curve by using standard compounds obtained from commercial suppliers and analysis with Agilent MassHunter Qualitative Analysis software for extracted ion chromatogram (EIC) of each individual phenolic compound. Analysis was carried out in triplicate.
2.3. Experimental Procedure in Animals
Fifty four 8‐week‐old male Wistar rats (Crl: WI (Han)) were purchased from Charles River (Barcelona, Spain) and fed ad libitum with a standard chow diet (STD, Panlab 04, Barcelona, Spain) and tap water. Rats were divided into several groups (Figure 1 ):
Figure 1.
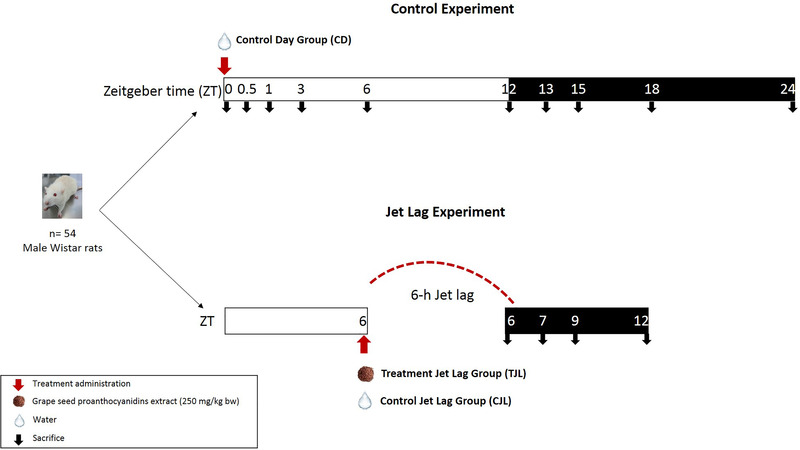
Experimental design used in the animal study.
Control day (CD) group: rats were singly caged in animal quarters at 22 °C with a 12 h light/dark cycle. After 3 weeks of adaptation, rats were administered tap water and were sacrificed by beheading at different zeitgeber times (ZT; unit of time based on the period of a zeitgeber such as light stimuli). Animals were sacrificed at ZT0 (light turned on), ZT0.5, ZT1, ZT3, ZT6, ZT12 (light off), ZT13, ZT15, ZT18, and ZT24 (n = 3 for groups). This experimental design was carried out to obtain a 24‐h curve to study the circadian rhythm of hepatic genes merging all the ZT data into one single curve.
6‐h jet lag groups: Rats were singly caged in animal quarters at 22 °C with a 12 h light/dark cycle. After 3 weeks of adaptation, rats were orally gavaged with tap water (Control jet lag group, CJL) or 250 mg of GSPE kg−1 body weight (bw) dissolved in tap water (Treatment jet lag group, TJL) at ZT6. Then, animals were immediately moved to a dusk room, thus giving rats a jet lag of 6 h (they lost 6 h of light). Rats were sacrificed by beheading at ZT6, ZT7, ZT9, and ZT12 (n = 3 for each group), all of them in the dark period. In all the groups, livers were excised and immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
All procedures involving the use and care of animals were approved by The Animal Ethics Committee of the Universitat Rovira i Virgili and by Generalitat de Catalunya (permit number 4249). All experiments were performed in accordance with relevant guidelines and regulations.
2.4. Dosage Information
A single acute dose of 250 mg kg−1 bw of GSPE, dissolved in tap water, or tap water were administered to rats by oral gavage. The dose of GSPE was chosen based on previous studies of the group where this dose showed effect on the regulation of clock genes.[ 12 ] This dose corresponded to 40 mg kg−1 bw in adult humans, when considering the body surface area according to Reagan‐Shaw et al.[ 22 ] People who consumed polyphenol‐rich diets, such as the Mediterranean diet, were most likely to consume flavonoids, with fruit, red wine, and tea. The daily intake of flavonoids was around 70 mg day−1[ 23 ] although in Spanish adults, the mean dietary flavonoid intake was approximately 300 mg day−1[ 24 ] and some reports had shown that the daily intake could be up to 1700 mg day−1.[ 25 ]
2.5. Cell Culture Procedure
HepG2 human hepatocarcinama cells were routinely propagated in a 5% CO2 humidified atmosphere at 37 °C in DMEM supplemented with 10% v/v fetal bovine serum, 2 mM l‐glutamine, 25 mM HEPES, 0.1 mM NEAA, and penicillin (100 U mL−1)/streptomycin (100 µg mL−1). The day before the experiment, cells at approximately 80% of confluence were plated at a density of 1 000 000 cells per well in a 6‐well plate (Cellstar, Tarragona, Spain). After 24 h, the cells were synchronized by serum shock, adding DMEM:horse serum: (1:1, v:v) to wells and leaving it for 2 h. Then, this medium was replaced with DMEM, supplemented as indicated before, and GSPE at 25 mg L−1 was added. Cells were lysed at 0, 3, 12, 15, 21, or 24 h after GSPE addition. Cells were lysed with the RNA‐Solv Reagent (miRNA kit, E.Z.N.A., Omega Bio‐tek) left 1 min at room temperature and stored at −80 °C until RNA extraction. For GSPE preparation, this extract was initially dissolved in 100% ethanol and then, diluted in supplemented DMEM until the desire concentration. Final concentration of ethanol in the well was 0.02% v/v. Non‐toxic effects in the cells were observed at this concentration. Control was carried out incubating cells with the same concentration of ethanol.
2.6. RNA Extraction and cDNA Synthesis
RNA extraction and cDNA synthesis were performed as described in Supplementary material, Supporting Information (Section 2.6.).
2.7. mRNA Quantification by RT‐qPCR
mRNA quantification by RT‐qPCR was performed as described in Supplementary material, Supporting Information (Section 2.7.).
2.8. microRNA Quantification by Real‐Time Quantitative Real‐Time Polymerase Chain Reaction (RT‐qPCR)
microRNA quantification by real‐time quantitative polymerase chain reaction (RT‐qPCR) was performed as described in Supplementary material, Supporting Information (Section 2.8.)
2.9. Circadian‐Rhythm Analysis
The analysis of the potential circadian rhythms of the different hepatic mRNAs and miRNAs expression was carried out using a Cosinor‐based rhythmometry method. For that, a script was developed using PyCharm software (v.2018.2.4, JetBrains s.r.o., Prague, Czech Republic) with Python version 3.7.4 and circadian‐rhythm estimated was plotted using CosinorPy package (v.1.1) (Ljubljana, Slovenia). The Cosinor method estimated the rhythmicity of the measured parameters by means of a mathematical regression model from non‐equidistant data.[ 26 ] The method allowed to obtain a model of the oscillation from the data, a p‐value that indicated whether this model fitted the data provided most closely and calculated the rhythmic parameters of this model with confidence intervals fitted to the data. Thus, the presence of circadian rhythm was considered when p < 0.05. The rhythmometric parameters analyzed were the following: the acrophase, the phase of the maximal value assumed by the curve; the amplitude, the difference between the peak and the mean value of a wave curve and the MESOR, an estimation of central tendency of the distribution of values of an oscillating variable. The rhythmicity of the both jet lag groups was calculated considering the data obtained from the common points of the CD group (ZT0, ZT0.5, ZT1, and ZT3) plus the data of the four ZT (ZT6, ZT7, ZT9, ZT12) of the different jet lag groups itself for each tested parameter.
2.10. Statistical Analysis
The results were presented as the mean value with the standard deviation (SD). The data were analyzed using Student's t‐test to determine the significant difference in relative microRNAs expression in each time point (ZT) and a two‐way ANOVA to determine if there was an interaction between time (t) and treatment (T), using SPSS Statistics 22 software (SPSS Inc., Chicago, IL, USA). The correlation coefficient was calculated with Pearson coefficient test. p values <0.05 were considered statistically significant.
3. Results and Discussion
3.1. Characterization of Phenolic Profile of GSPE
Phenolic compounds in vegetables and fruits exert a wide variety of health benefits.[ 27 ] Among the fruits, grapes are rich in phenolic compounds and many studies used grape‐derived products (skin, seed, stems) to obtain phenolic‐enriched extracts with different functional properties.[ 28 , 29 ] An example is the grape seed proanthocyanidins, which have been shown to improve of dyslipidemia, inflammation, insulin resistance, mitochondrial dysfunctionality, hypertension, and oxidative stress in animals with metabolic syndrome.[ 30 , 31 ] However, the phenolic composition of an extract can greatly vary depending on factors such as grape variety, growing grape condition, or methodology of extraction.[ 32 ] Thus, the phenolic composition of an extract is important, as it is known that phenolic health effects depend on dose administration and phenolic concentration and profile.[ 33 ] Therefore, the individual phenolic profile of the GSPE used in the present study was analyzed by HPLC–MS. Results indicated that GSPE had 250.77 mg g−1 of total phenolic content. In addition, 16 compounds were identified, being the most abundant phenolic compounds procyanidin dimer (87.51 mg g−1 of extract), catechin (58.36 mg g−1), epicatechin (50.35 mg g−1), gallic acid (35.16 mg g−1), and procyanidin trimer (9.77 mg g−1) (Table S1, Supporting Information). Individual concentration of the rest of phenolic compounds was lower than 5 mg g−1 of extract. The phenolic profile observed in this extract is similar to others previously reported in the literature.[ 21 ]
3.2. Acute Administration of GSPE Modulates Bmal1 via miR‐27b‐3p in the Liver of Jet‐Lagged Rats
Phenolic compounds and their effects on circadian rhythms is a recent frontier in chrononutrition.[ 34 , 35 , 36 , 37 ] It is known that GSPE acts as a non‐photic cue in the hypothalamus and on the peripheral clock, significantly modifying the expression of Bmal1 and Nampt in the liver of rats exposed to 6‐h jet lag.[ 38 ] GSPE counteracted the rhythm disruption produced by a 6‐h jet lag and thus, the clock genes recovered their normal rhythmicity.[ 12 ] This effect was only observed when animals suffered a disruption of circadian rhythm as GSPE administered to Wistar rats did not alter hepatic Bmal1 expression neither when the extract was administered during the day nor during the night.[ 12 ] Previous studies elucidated that GSPE acts on Bmal1 in the liver mimicking melatonin action, which has a membrane receptor on hepatocytes, MT1. However, MT1 was not involved in GSPE action.[ 4 ] Therefore, GSPE must act through other molecular mechanisms on Bmal1. Moreover, it is known that hepatic transcripts are modulated by the rhythmic food intake and specifically GSPE is involved in the modulation of miRNAs.[ 17 ] Thus, we hypothesized that the effect of grape seed proanthocyanidins on disrupted circadian rhythm of clock genes could be mediated by miRNAs. Considering this, the first objective of this study was to evaluate if miR‐27b‐3p could be target of GSPE, which in turn acting at post‐transcriptional level modulating the expression level of Bmal1.
To achieve this objective, animals were administered 250 mg kg−1 bw of GSPE and immediately were submitted to 6‐h jet lag to produce a rhythm disruption. The rats were administered with vehicle (control jet lag group, CJL) or GSPE (treatment jet lag, TJL). Then, animals were sacrificed at different time points (ZT) and the livers were used to determine gene expression. In addition, another group of animals (control day group, CD) consuming vehicle and housed in a standard photoperiod was used as control. Animals were sacrifice at different time points (ZT0, ZT0.5, ZT1, ZT3, ZT6, ZT12, ZT13, ZT15, ZT18, ZT24) and their hepatic expressions were used to know the rhythmicity of liver gene expression without a disruption.
Bmal1 and miR‐27b‐3p expression was determined in the liver and their potential circadian rhythm was analyzed by the Cosinor‐based method, assuming that the cycle of each curve was 24 h and considering the rhythmometric parameters as amplitude, acrophase, and MESOR. Results obtained from Bmal1 gene expression showed that the three experimental groups exhibited a circadian rhythm (Figure 2A), which were significantly different between each other's (p < 0.01) (Figure 2B and Table S2, Supporting Information). The differences were observed in the amplitude of the rhythms (Figure 2A). As it was aforementioned, we observed that GSPE was able to modulate Bmal1 expression in the liver of animals under 6‐h jet lag.[ 12 ] No significant differences were observed in the acrophase of Bmal1 expression rhythm of the different groups (Figure 2A). The acrophase was found between ZT0.5 and ZT2.4. Other studies carried out in mice observed that the acrophase of Bmal1 expression during the middle stage of the dark period. This difference could be explained by the different experimental design since in the model used by Zhang et al.[ 39 ] the animals were subjected to constant darkness.
Figure 2.
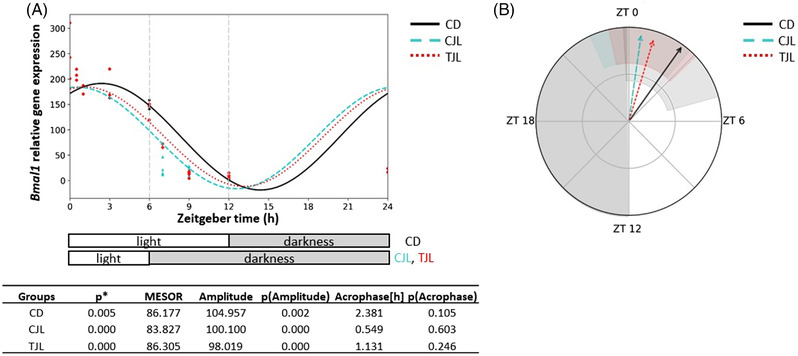
Estimated circadian rhythm of hepatic Bmal1 expression of the three animal groups analyzed by Cosinor method. A) Estimated circadian rhythm and rhythmometric parameters. B) Representation of the acrophases and the amplitude of the estimated rhythm. Control day group (CD group): animals housed at 12 h light/dark cycle, administered water (ZT0) and sacrificed at ZT0, 0.5, 1, 3, 6, 12, 13, 15, 18, and 24 h; Control jet lag group (CJL group): animals housed at 12 h light/dark cycle, administered water at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. Treatment jet lag group (TJL group): animals housed at 12 h light/dark cycle, administered GSPE (250 mg kg−1 bw) at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. *p ≤ 0.05 indicates circadian rhythm. Acrophase: time at which the peak of a rhythm occurs ([h] hours); Amplitude: difference between the peak and the mean value of a wave; MESOR: a circadian rhythm‐adjusted mean. p < 0.05 indicates significant differences.
Figure 3A shows the expression levels of hepatic miR‐27b‐3p in the dark period. It was observed that the CD group had the highest expression of miR‐27b‐3p 1 h after the lights were switched off. However, this effect was disrupted by jet lag, as a downregulation of this miRNA in the CJL group was observed at this time point. However, animals administered GSPE counteracted the jet lag effect. In this regard, it was observed a significant upregulation of the miR‐27b‐3p expression in the treated jet lag group (TJL) 3 h after the lights were switched off while a tendency to increase its value was observed 1 h after the lights were switched off (p‐value 0.07, Student's t‐test). Interestingly, a two‐way ANOVA test analysis indicated a significant interaction between time and treatment, namely GSPE administered before the jet‐lag induction significantly altered miR‐27b‐3p expression in the liver (t × T, p = 0.019). In this regard, previous studies showed that GSPE can regulate miR‐33 and miR‐122 expression, which are the main regulators of lipid hepatic metabolism. For example, GSPE modulated miRNA‐33 and miRNA‐122 in Wistar rats administered different doses of GSPE (5, 15, 25, and 50 mg kg−1 bw) and its effects were dose dependant.[ 40 ] In addition, miRNAs exert a post‐transcriptional regulation on clock gene expression.[ 19 ] For example, Choi et al.[ 20 ] observed that miR‐219 and miR‐132 can modulate the central circadian clock in the brain, which was tissue specific. They revealed that miRNAs can respond to photic cue, thus influencing and regulating the circadian clock in the SCN.[ 41 ] Moreover, it has been observed that miR‐27b‐3p regulated the rhythmicity of Bmal1 expression in mouse liver, opening a new point of view through which molecular clock could be regulated.[ 39 ] This miRNA binds Bmal1 at 3′ UTR underlying the pivotal role in circadian rhythm.[ 39 ]
Figure 3.
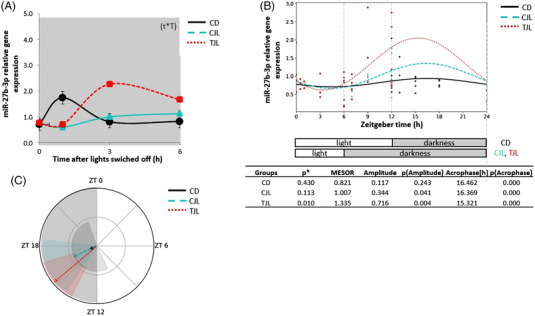
Relative gene expression of hepatic miR‐27b‐3 and estimated circadian rhythm of the hepatic miR‐27b‐3 expression of the three animal groups analyzed by Cosinor method. A) Relative gene expression of hepatic miR‐27b‐3 in the dark phase. B) Estimated circadian rhythm and rhythmometric parameters. C) Representation of the acrophases and the amplitude of the estimated rhythm. Control day group (CD group): animals housed at 12 h light/dark cycle, administered water (ZT0) and sacrificed at ZT0, 0.5, 1, 3, 6, 12, 13, 15, 18, and 24 h; Control jet lag group (CJL group): animals housed at 12 h light/dark cycle, administered water at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. Treatment jet lag group (TJL group): animals housed at 12 h light/dark cycle, administered GSPE (250 mg kg−1 bw) at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. *p ≤ 0.05 indicates circadian rhythm. Acrophase: time at which the peak of a rhythm occurs ([h]. hours); Amplitude: difference between the peak and the mean value of a wave; MESOR: a circadian rhythm‐adjusted mean. p < 0.05 shows significant differences. t × T indicates interaction between time and treatment (two‐way ANOVA).
In order to evaluate if GSPE could regulate miR‐27‐b‐3p rhythmicity a Cosinor based method analysis was performed. The rhythmicity of miR‐27b‐3p expression and rhythmometric parameters as amplitude, acrophase, and MESOR are shown in Figure 3B. Results showed that miR‐27b‐3p expression did not follow a circadian rhythm neither CD nor CJL groups, since the p value obtained for both groups was higher than 0.05. However, TJL group showed a circadian rhythm for this miRNA, behavior different to the observed by the other two groups (p < 0.05). The acrophase in this treated group was estimated at ZT15 in the dark phase (Figure 3C). In contrast to our findings, Zhang et al.[ 39 ] observed the lowest expression (nadir) of this miRNA at ZT12 in mice. Differences between studies could be due to differences by animal model and experimental design as mice in the mentioned study were exposed to a continuous darkness. In addition, it was also observed significant differences in the amplitude of TJL rhythm in comparison to the observed by CD group (Table S2, Supporting Information).
Furthermore, the Pearson correlation coefficient was determined to evaluate the degree of correlation between Bmal1 and miR‐27b‐3p expression (Figure 4 ). No correlations were observed for Bmal1 and miR‐27b‐3p expression in CD (Figure 4A) and CJL (Figure 4B). However, previous studies carried out in mouse model under continuous darkness observed that the highest levels of Bmal1 expression corresponded to the lowest values of miR‐27b‐3p expression levels.[ 39 ] Regarding TJL group, a negative correlation was observed (Pearson R‐0.389) (Figure 4C). This means that when Bmal1 decreases, the miRNA expression increases and vice versa. All the evidence indicates that the administration of GSPE to healthy rats exposed to 6 h jet lag induces circadian rhythm expression of miR‐27b‐3p. Moreover, when GSPE upregulated this miRNA expression, miR‐27b‐3p represses its target in the liver, corroborating the hypothesis that GSPE modulates hepatic peripheral clock at post‐transcriptional level, modulating cell functionality. Although previous studies have already suggested the implication of miRNA in the regulation of peripheral circadian clock, to the best of our knowledge, this is the first study in which is showed that the consumption of proanthocyanidins can drive the adaptation to a change of external condition by modulating a key peripheral clock gene, Bmal1, through miR‐27b‐3p.
Figure 4.
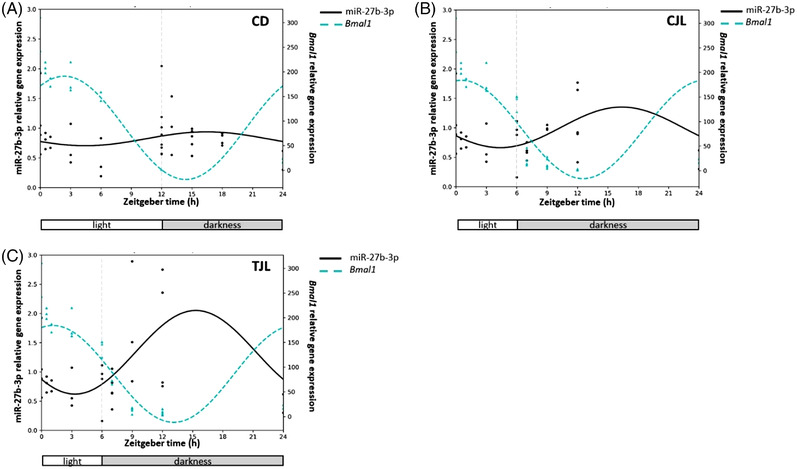
Correlation of the estimated circadian rhythm of the hepatic Bmal1 and miR‐27b‐3 expression of CD A), CJL B), and TJL C) analyzed by Cosinor method. Control day group (CD group): animals housed at 12 h light/dark cycle, administered water (ZT0) and sacrificed at ZT0, 0.5, 1, 3, 6, 12, 13, 15, 18, and 24 h; Control jet lag group (CJL group): animals housed at 12 h light/dark cycle, administered water at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. Treatment jet lag group (TJL group): animals housed at 12 h light/dark cycle, administered GSPE (250 mg kg−1 bw) at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12.
3.3. Acute Administration of GSPE Modulates Nampt Independent of miR‐34a
In addition, the second objective was to study if GSPE could modulate Nampt, a clock‐controlled gene, via miR‐34a. NAMPT is implicated in many aspects of the metabolism, being the rate limiting enzyme of NAD+.[ 42 ] Previous results showed that GSPE modulates Nampt expression in the liver of jet‐lagged rat.[ 12 ] Moreover, it is known that miR‐34a is able to target Nampt and it responds to nutritional cues.[ 20 ]
Thus, Nampt and miR‐34a expression was determined in the liver of the three experimental groups. Circadian rhythm of Nampt analyzed by the Cosinor method revealed three different circadian rhythms (p < 0.001) (Figure 5 ). Looking at the rhythmometric parameters, the amplitude showed a significant difference between CD versus CJL and CJL versus TJL groups (Table S2, Supporting Information). In addition, Figure 6A shows miR‐34a expression levels in liver of the three experimental groups once the lights were switched off. No significant differences were observed among the groups. However, other studies showed that GSPE can modified several miRNAs expression in HepG2 cells such as miR‐30b. Moreover, GSPE could act differently on miRNA expression depending on experimental model and phenolic profile of GSPE. In fact, it has been observed that the polymerization degree affects in the modulation of miRNA, while using just one phenolic compound such as epigallocatechin gallate (EGCG) fewer miRNA result affected due to EGCG low polymerization degree.[ 17 ]
Figure 5.
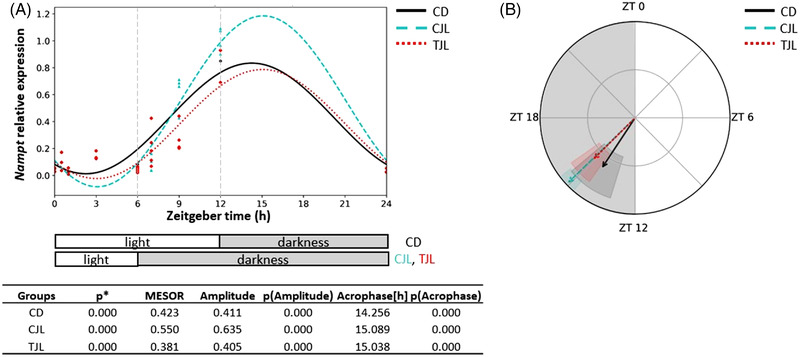
Estimated circadian rhythm of hepatic Nampt expression of the three animal groups analyzed by Cosinor method. A) Estimated circadian rhythm and rhythmometric parameters. B) Representation of the acrophases and the amplitude of the estimated rhythm. Control day group (CD group): animals housed at 12 h light/dark cycle, administered water (ZT0), and sacrificed at ZT0, 0.5, 1, 3, 6, 12, 13, 15, 18, and 24 h; Control jet lag group (CJL group): animals housed at 12 h light/dark cycle, administered water at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. Treatment jet lag group (TJL group): animals housed at 12 h light/dark cycle, administered GSPE (250 mg kg−1 bw) at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. *p ≤ 0.05 indicates circadian rhythm. Acrophase: time at which the peak of a rhythm occurs ([h]. hours); Amplitude: difference between the peak and the mean value of a wave; MESOR: a circadian rhythm‐adjusted mean. p < 0.05 indicates significant differences.
Figure 6.
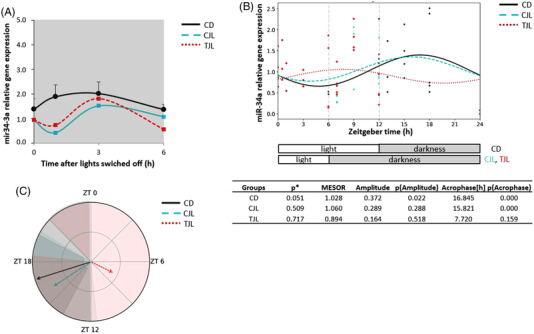
Relative gene expression of hepatic miR‐34a and estimated circadian rhythm of the hepatic miR‐34a expression of the three animal groups analyzed by Cosinor method. A) Relative gene expression of hepatic miR‐34a in the dark period. B) Estimated circadian rhythm and rhythmometric parameters. C) Representation of the acrophases and the amplitude of the estimated rhythm. Control day group (CD group): animals housed at 12 h light/dark cycle, administered water (ZT0) and sacrificed at ZT0, 0.5, 1, 3, 6, 12, 13, 15, 18, and 24 h; Control jet lag group (CJL group): animals housed at 12 h light/dark cycle, administered water at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. Treatment jet lag group (TJL group): animals housed at 12 h light/dark cycle, administered GSPE (250 mg kg−1 bw) at ZT6 and immediately moved to dark and sacrificed at ZT6, 7, 9, and 12. *p ≤ 0.05 indicates circadian rhythm. Acrophase: time at which the peak of a rhythm occurs ([h]. hours); Amplitude: difference between the peak and the mean value of a wave; MESOR: a circadian rhythm‐adjusted mean. p < 0.05 shows significant differences. t × T indicates interaction between time and treatment (two‐way ANOVA).
In addition, Cosinor analysis showed that miR‐34a expression followed a circadian rhythm in the CD group (Figure 6B). To the best of our knowledge, this is the first proof that miR‐34a exhibits circadian rhythmicity in hepatocytes. However, when animals were induced to 6‐h jet lag, the rhythmicity of miR‐34a expression was lost (Figure 6B). Similar results were found in the TJL, indicating that GSPE was not able to restore the miR‐34a rhythmicity. In addition, rhythmometric parameters did not differ between rhythm groups when compared between them (Table S2, Supporting Information). These results suggest that GSPE modulates Nampt expression levels independently from miR‐34a since the expression level of this miRNA is not altered by GSPE administration. Moreover, Pearson correlation carried out between Nampt and miR‐34a indicated that there is a positive correlation. However, other studies have evidenced that miR‐34a is able to target Nampt and it responds to nutritional cues[ 20 ] in contrast to our findings. Therefore, other molecular mechanisms are involved in Nampt regulation by GSPE, which in turn regulates energetic metabolic level in the liver. Thus, further studies are needed to identify the possible mechanisms of action of GSPE over Nampt.
3.4. 25 mg L−1 of GSPE is not Able to Modulate Bmal1 via miR‐27b‐3p in HepG2 Cells
In order to go deeper into the direct effects of nutritional signals on hepatic clock genes (Bmal1), it was carried out an in vitro experiment using human hepatocytes (HepG2 cells), which were synchronized by serum shock and then treated with 25 mg L−1 of GSPE. Cell lysis was done at different time points (0, 3, 12, 15, 21, 24 h) and Bmal1 expression was determined. The hepatic molecular clock is regulated by SCN through hormonal signals but it also responds to nutritional cues.[ 43 ] Thus, studying the circadian clock in vitro, it is a good way to circumvent hormonal and neuronal signals that come from the SCN and drive the peripheral clock. HepG2 cells were selected according to our previous studies as they respond to GSPE treatment, which was time dependant.[ 44 ] Previous studies showed that Bmal1 is undoubtedly the clock gene most sensitive to GSPE in hepatocytes and is the principal regulator of the molecular machinery architecture.[ 4 ] Furthermore, our previous study showed that a dose of 100 mg L−1 of GSPE modulated core clock genes and clock‐controlled genes in HepG2 cells. In this present work a dose of 25 mg L−1 was selected as it is a lower dose and it could be resembled to a physiological dose.
Results showed that the dose of 25 mg L−1 of GSPE, modulated Bmal1 expression in HepG2 cells at ZT12. Specifically, Bmal1 expression at this time point was significantly downregulated in treated cells compared to untreated cells (Figure 7A). Two‐way ANOVA test indicated that there was an interaction between time and treatment for Bmal1 expression levels. Similarly, it has been observed that GSPE at a dose of 100 mg L−1 also modulated Bmal1 expression in HepG2 cells at ZT12. However, this higher dose than the used in our present study also showed effect on Bmal1 expression at other time points.[ 4 ] In fact, prior research has shown that the efficacy of GSPE depends on its dose, bioactivity, and when it is consumed.[ 45 ] These findings clearly showed the importance of the GSPE concentration in the modulation of hepatic transcripts. Thus, in the present study it is demonstrated that GSPE at low dose (25 mg L−1) dose can modulate of peripheral molecular clocks (Bmal1), at a specific time point (ZT12) and independently from light signals.
Figure 7.
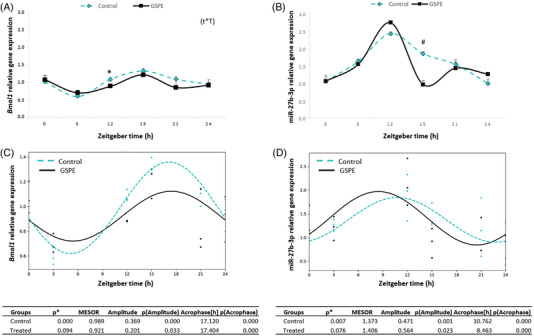
Relative gene expression of Bmal1 A) and miR‐34a B) obtained from HepG2 cells control and treated grape seed proanthocyanidins extract (GSPE; 25 mg L−1). Estimated circadian rhythm of the Bmal1 C) and miR‐34a D) expression of both cell groups and rhythmometric parameters. Acrophase: time at which the peak of a rhythm occurs ([h]. hours); Amplitude: difference between the peak and the mean value of a wave; MESOR: a circadian rhythm‐adjusted mean. p < 0.05 or * shows significant differences. # indicates tendency (p < 0.01). t × T indicates interaction between time and treatment (two‐way ANOVA).
Moreover, it was studied the miR‐27b‐3p expression of this cells in order to evaluate if GSPE could modulate Bmal1 via this miRNA, as seen in vivo study, but without the photic cue regulation. It would allow us to clarify if it only depends on nutritional response independently of light signals, which would involve the SCN. Figure 7B shows the expression levels of miR‐27b‐3p in control and treated HepG2 cells. The miR‐27b‐3p expression levels oscillated along the different collected time points and no significant differences were found between them. The acrophase of the miR‐27b‐3p expression was found at ZT12 in both cell groups. This is the first study in which is evaluated the role of GSPE in modulating miR‐27b‐3p. However, studies carried out in FAO and HepG2 cells evidenced that GSPE downregulated the expression of other miRNAs such as miR‐33a and miR‐122 at a dose of 25 mg L−1.[ 18 ] In addition, it was observed that effects on miRNAs depended of type of phenolic compounds and polymerization degree as resveratrol or EGCG modulated the expression of both miR‐33a and miR‐122 whereas dimer B2 and gallic acid only downregulated the expression of miR‐122 in HepG2 cells at a dose of 50 µM.[ 17 , 18 ] However, effects on miRNAs can be also different depending on the cell type as the same authors observed that gallic acid modulated the expression of miR‐33a in FAO cell but no effects were observed in HepG2 cells.
Furthermore, the rhythmicity of Bmal1 and miR‐27b‐3p expression was analyzed with the Cosinor method. Results showed that the expression of both followed a circadian rhythm in control cells (p < 0.001) but these circadian rhythms tended to be lost by GSPE treatment (p = 0.094 and p = 0.076 for Bmal1 and miR‐27b‐3p, respectively) (Figure 7C,D). Moreover, Pearson's correlation between Bmal1 and miR‐27b‐3p expression levels did not highlight any significant correlations among the genes studied in HepG2 cells (Figure 8A,B) (Pearson R = 0.204 and Pearson R = −0.223 for control and GSPE groups, respectively). Thus, although GSPE modulated Bmal1 expression, it seems that this modulation is not mediated via miR‐27b‐3p in absence of a light cue. However, it should be not discarded the role of GSPE at higher doses in the regulation of this miRNA, as a dose of 100 mg L−1 of GSPE produced different effects in Bmal1 [ 4 ] than the ones observed for our study using a dose of 25 mg L−1. In addition, it should be considered that GSPE, before reaching the hepatocytes is previously metabolized by microbiota and II phase enzymes in the intestine,[ 46 ] so the differences found between in vitro and in vivo studies could also be addressed to this fact. Additionally, it is worthy to mention that signals from the central system (SCN) could play an important role in the crosstalk and the action of GSPE via miRNA. In this sense it should be considered that this effect could be consequence of multiple factors rather than a direct nutritional system in the hepatocytes.
Figure 8.
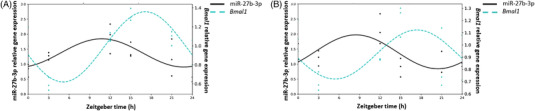
Correlation of the estimated circadian rhythm of Bmal1 and miR‐27b‐3 expression of HepG2 cells control A) and treated with a grape seed proanthocyanidins extract at a dose of 25 mg L−1 B).
3.5. Concluding Remarks
In conclusion, GSPE showed beneficial effects in the disruption of biological time, restoring the rhythmicity acting on the molecular peripheral clock via miRNA. Interestingly, our study suggests that one of the mechanisms of action of GSPE in peripheral molecular clock (Bmal1) of the liver occurs via miR‐27b‐3p. These findings open a new point of view in the understanding of how the circadian clock may be regulated and contribute to elucidate the molecular mechanism of the crosstalk between metabolism and circadian rhythm. Moreover, further research could be pursed to investigate the role of GSPE, acting as a non‐photic cue to restore a normal rhythmicity in central clock trough miRNAs.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
Conceptualization, A.A.‐A., M.S., and B.M.; formal analysis, F.M., J.R.S.‐R.; funding acquisition, F.I.B., A.A.‐A., M.S., and B.M.; investigation, F.M, J.R.S.‐R., A.R.‐L.; methodology, F.M., J.R.S.‐R., A.R.‐L.; supervision, F.I.B., A.A.‐A., M.S., and B.M.; writing—original draft, F.M, F.I.B; writing—review and editing, F.M, F.I.B, A.A.‐A., M.S., and B.M. All authors have read and agreed to the published version of the manuscript.
Supporting information
Supporting Information
Acknowledgements
The authors thank Niurka Dariela Llópiz and Rosa Maria Pastor, from the Universitat Rovira i Virgili, for the valuable support in the laboratory. F.M. is a recipient of a predoctoral fellowship from Universitat Rovira i Virgili (Grant number: 2019PMF‐PIPF‐19). JR.S.‐R. is a recipient of a predoctoral fellowship (Grant number: BES‐2017‐080919) from the Ministerio de Ciencia e Innovación MCIN/AEI/10.13039/501100011033 and FSE “El FSE invierte en tu futuro”. F.I.B and A.A‐A are Serra Húnter Fellows. This work has been supported by the Ministerio de Ciencia e Innovación MCIN/AEI/10.13039/501100011033/FEDER “Una manera de hacer Europa” (Grant number: AGL2016‐77105‐R).
Manocchio F., Soliz‐Rueda J. R., Ribas‐Latre A., Bravo F. I., Arola‐Arnal A., Suarez M., Muguerza B., Grape Seed Proanthocyanidins Modulate the Hepatic Molecular Clock via MicroRNAs. Mol. Nutr. Food Res. 2022, 66, 2200443. 10.1002/mnfr.202200443
Contributor Information
Francisca Isabel Bravo, Email: franciscaisabel.bravo@urv.cat.
Anna Arola‐Arnal, Email: anna.arola@urv.cat.
Manuel Suarez, Email: manuel.suarez@urv.cat.
Data Availability Statement
Research data are not shared.
References
- 1. Ribas‐Latre A., Eckel‐Mahan K., Mol. Metab. 2016, 5, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asher G., Sassone‐Corsi P., Cell 2015, 161, 84. [DOI] [PubMed] [Google Scholar]
- 3. Koronowski K. B., Kinouchi K., Welz P., Baldi P., Benitah S. A., Sassone‐corsi P., Koronowski K. B., Kinouchi K., Welz P., Smith J. G., Zinna V. M., Shi J., Cell 2019, 177, 1448.e14.31150621 [Google Scholar]
- 4. Ribas‐Latre A., Del Bas J. M., Baselga‐Escudero L., Casanova E., Arola‐Arnal A., Salvadó M. J., Bladé C., Arola L., J Funct Foods 2015, 13, 336. [DOI] [PubMed] [Google Scholar]
- 5. Sahar S., Sassone‐Corsi P., Trends Endocrinol. Metab. 2012, 23, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamezaki F., Sonoda S., Nakata S., Muraoka Y., Okazaki M., Tamura M., Abe H., Tekeuchi M., Otsuji Y., Hypertens. Res. 2013, 36, 398. [DOI] [PubMed] [Google Scholar]
- 7. Vosko A. M., Colwell C. S., Avidan A. Y., Nat. Sci. Sleep 2010, 2, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Potter G. D. M., Skene D. J., Arendt J., Cade J. E., Grant P. J., Hardie L. J., Endocr. Rev. 2016, 37, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fishbein A. B., Knutson K. L., Zee P. C., J. Clin. Invest. 2021, 131, e148286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soliz‐Rueda J. R., López‐Fernández‐Sobrino R., Bravo F. I., Aragonès G., Suarez M., Muguerza B., Nutrients 2022, 14, 1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng H., Liu Z., Wu G., Ho C. T., Li D., Xie Z., J. Funct. Foods 2021, 78, 104370. [Google Scholar]
- 12. Ribas‐Latre A., Baselga‐Escudero L., Casanova E., Arola‐Arnal A., Salvadó M. J., Bladé C., Arola L., Sci. Rep. 2015, 5, 10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ribas‐Latre A., Baselga‐Escudero L., Casanova E., Arola‐Arnal A., Salvadó M. J., Arola L., Bladé C., J. Nutr. Biochem. 2015, 26, 112. [DOI] [PubMed] [Google Scholar]
- 14. Aragonès G., Suárez M., Ardid‐Ruiz A., Vinaixa M., Rodríguez M. A., Correig X., Arola L., Bladé C., Sci. Rep. 2016, 6, 24977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casanova E., Baselga‐Escudero L., Ribas‐Latre A., Cedó L., Arola‐Arnal A., Pinent M., Bladé C., Arola L., Salvadó M. J., J. Nutr. Biochem. 2014, 25, 1003. [DOI] [PubMed] [Google Scholar]
- 16. Bartel D. P., Cell 2018, 173, 20.29570994 [Google Scholar]
- 17. Arola‐Arnal A., Bladé C., PLoS One 2011, 6, e25982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baselga‐Escudero L., Blade C., Ribas‐Latre A., Casanova E., Suárez M., Torres J. L., Salvadó M. J., Arola L., Arola‐Arnal A., Nucleic Acids Res. 2014, 42, 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torres M., Becquet D., Franc J. L., François‐Bellan A. M., Wiley Interdiscip. Rev. RNA 2018, 9, e1467. [DOI] [PubMed] [Google Scholar]
- 20. Choi S. E., Fu T., Seok S., Kim D. H., Yu E., Lee K. W., Kang Y., Li X., Kemper B., Kemper J. K., Aging Cell 2013, 12, 1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quiñones M., Guerrero L., Suarez M., Pons Z., Aleixandre A., Arola L., Muguerza B., Food Res. Int. 2013, 51, 587. [Google Scholar]
- 22. Reagan‐Shaw S., Nihal M., Ahmad N., FASEB J. 2008, 22, 659. [DOI] [PubMed] [Google Scholar]
- 23. Bawaked R. A., Schröder H., Barba L. R., Cárdenas G., Peña‐Quintana L., Rodrigo C. P., Fíto M., Majem L. S., Peer J. 2017, 5, e3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zamora‐Ros R., Knaze V., Rothwell J. A., Hémon B., Moskal A., Overvad K., Tjønneland A., Kyrø C., Fagherazzi G., Boutron‐Ruault M. C., Touillaud M., Katzke V., Kühn T., Boeing H., Förster J., Trichopoulou A., Valanou E., Peppa E., Palli D., Agnoli C., Ricceri F., Tumino R., de Magistris M. S., Peeters P. H. M., Bueno‐de‐Mesquita H. B., Engeset D., Skeie G., Hjartåker A., Menéndez V., Agudo A., et al., Eur. J. Nutr. 2015, 55, 1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grosso G., Stepaniak U., Topor‐Mądry R., Szafraniec K., Pająk A., Nutrition 2014, 30, 1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cornelissen G., Theor. Biol. Med. Model 2014, 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D'Archivio M., Filesi C., Di Benedetto R., Gargiulo R., Giovannini C., Masella R., Ann. Ist. Super. Sanita 2007, 43, 348. [PubMed] [Google Scholar]
- 28. López‐Fernández‐Sobrino R., Torres‐Fuentes C., Bravo F. I., Muguerza B., Crit. Rev. Food Sci. Nutr. 2022, 10.1080/10408398.2022.2049202. [DOI] [PubMed] [Google Scholar]
- 29. Chakka A. K., Babu A. S., Appl. Food Res. 2022, 2, 100058. [Google Scholar]
- 30. Bladé C., Aragonés G., Arola‐Arnal A., Muguerza B., Bravo F. I., Salvadó M. J., Arola L., Suárez M., BioFactors 2016, 42, 5. [DOI] [PubMed] [Google Scholar]
- 31. Mas‐Capdevila A., Iglesias‐Carres L., Arola‐Arnal A., Suárez M., Bravo F. I., Muguerza B., Food Funct. 2020, 11, 8735. [DOI] [PubMed] [Google Scholar]
- 32. Bustamante L., Saez V., Hinrichsen P., Castro M., Vergara Rosales C., von Carola D., Mardones C., J. Agric. Food Chem. 2017, 65, 2793. [DOI] [PubMed] [Google Scholar]
- 33. Kumar N., Goel N., Biotechnol. Rep. 2019, 24, e00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ávila‐Román J., Soliz‐Rueda J. R., Bravo F. I., Aragonès G., Suárez M., Arola‐Arnal A., Mulero M., Salvadó M. J., Arola L., Torres‐Fuentes C., Muguerza B., Trends Food Sci. Technol. 2021, 113, 77. [Google Scholar]
- 35. Arola‐Arnal A., Cruz‐Carrión Á., Torres‐Fuentes C., Ávila‐Román J., Aragonès G., Mulero M., Bravo F. I., Muguerza B., Arola L., Suárez M., Nutrients 2019, 11, 2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Torres‐Fuentes C., Suárez M., Aragonès G., Mulero M., Ávila‐Román J., Arola‐Arnal A., Salvadó M. J., Arola L., Bravo F. I., Muguerza B., Mol. Nutr. Food Res. 2022, 2100990, 10.1002/mnfr.202100990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ibarz‐Blanch N., Morales D., Calvo E., Ros‐Medina L., Muguerza B., Bravo F. I., Suárez M., Nutrients 2022, 14, 1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ribas‐Latre A., Del Bas J. M., Baselga‐Escudero L., Casanova E., Arola‐Arnal A., Salvadó M. J., Arola L., Bladé C., Mol. Nutr. Food Res. 2015, 59, 865. [DOI] [PubMed] [Google Scholar]
- 39. Zhang W., Wang P., Chen S., Zhang Z., Liang T., Liu C., FASEB J. 2016, 30, 2151. [DOI] [PubMed] [Google Scholar]
- 40. Baselga‐Escudero L., Blade C., Ribas‐Latre A., Casanova E., Salvadó M. J., Arola L., Arola‐Arnal A., J. Nutr. Biochem. 2014, 25, 151. [DOI] [PubMed] [Google Scholar]
- 41. Cheng H. Y. M., Papp J. W., Varlamova O., Dziema H., Russell B., Curfman J. P., Nakazawa T., Shimizu K., Okamura H., Impey S., Obrietan K., Neuron 2007, 54, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone‐Corsi P., Science 2009, 324, 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oike H., Nagai K., Fukushima T., Ishida N., Kobori M., PLoS One 2011, 6, e23709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guerrero L., Margalef M., Pons Z., Quiñones M., Arola L., Arola‐Arnal A., Muguerza B., J. Nutr. Biochem. 2013, 24, 2092. [DOI] [PubMed] [Google Scholar]
- 45. Rodríguez‐Pérez C., García‐Villanova B., Guerra‐Hernández E., Verardo V., Nutrients 2019, 11, 2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Margalef M., Pons Z., Bravo F. I., Muguerza B., Arola‐Arnal A., J. Nutr. Biochem. 2015, 26, 987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Research data are not shared.


