Abstract
Tandem repeats (TRs) are one of the largest sources of polymorphism, and their length is associated with gene regulation. Although previous studies reported several tandem repeats regulating gene splicing in cis (spl-TRs), no large-scale study has been conducted. In this study, we established a genome-wide catalog of 9537 spl-TRs with a total of 58,290 significant TR–splicing associations across 49 tissues (false discovery rate 5%) by using Genotype-Tissue expression (GTex) Project data. Regression models explaining splicing variation by using spl-TRs and other flanking variants suggest that at least some of the spl-TRs directly modulate splicing. In our catalog, two spl-TRs are known loci for repeat expansion diseases, spinocerebellar ataxia 6 (SCA6) and 12 (SCA12). Splicing alterations by these spl-TRs were compatible with those observed in SCA6 and SCA12. Thus, our comprehensive spl-TR catalog may help elucidate the pathomechanism of genetic diseases.
Tandem repeats (TRs) are DNA sequences tandemly repeated twice or more in succession. TRs represent one of the largest sources of polymorphism and may have substantial effects on genome functions (de Koning et al. 2011). Indeed, TRs are well known to regulate gene expression in cis (Gymrek et al. 2016; Fotsing et al. 2019; Bakhtiari et al. 2021; Eslami Rasekh et al. 2021; Lu et al. 2021) and contribute to the pathogenesis of both rare (Lalioti et al. 1997; O'Hearn et al. 2015; LaCroix et al. 2019) and common diseases (Gymrek et al. 2016; Fotsing et al. 2019; Bakhtiari et al. 2021).
Several studies have suggested that TRs may also regulate gene splicing in cis, as summarized in Supplemental Figure S1 (Hui et al. 2003; Sathasivam et al. 2013; De Roeck et al. 2018; Pacheco et al. 2019). For example, a (TG)n repeat near the 3′ splice site of exon 9 of CFTR, the causative gene of cystic fibrosis, was shown to function as an intronic splicing silencer (Buratti and Baralle 2001). The repeat was bound by TARDBP (also known as TDP-43), an RNA-binding protein (RBP) silencing downstream exon inclusion (Tollervey et al. 2011) and, when expanded, enhanced exon 9 skipping. The lack of exon 9 leads to the production of nonfunctional proteins, thereby contributing as a pathogenic allele for cystic fibrosis. Another example is a 25-bp intronic TR in ABCA7. Its expansion was reported to be associated with skipping of the downstream exon putatively due to SRSF9 (also known as SRp30c) binding (Wang et al. 2005; De Roeck et al. 2018). The exon skipping led to ABCA7 dysfunction and posed a risk for Alzheimer's disease (De Roeck et al. 2018). Although these scattered studies have reported splicing-associated TRs (spl-TRs) and provided a glance at the potential mechanism and relevance to disease, no large-scale study on this issue has been conducted. Here, we map spl-TRs in a genome-wide manner across tissues throughout the whole body by using the Genotype-Tissue expression (GTEx) v8 data set, a population-scale resource of whole-genome sequencing (WGS) and RNA sequencing (RNA-seq).
Results
Genome-wide identification of spl-TRs across 49 tissues
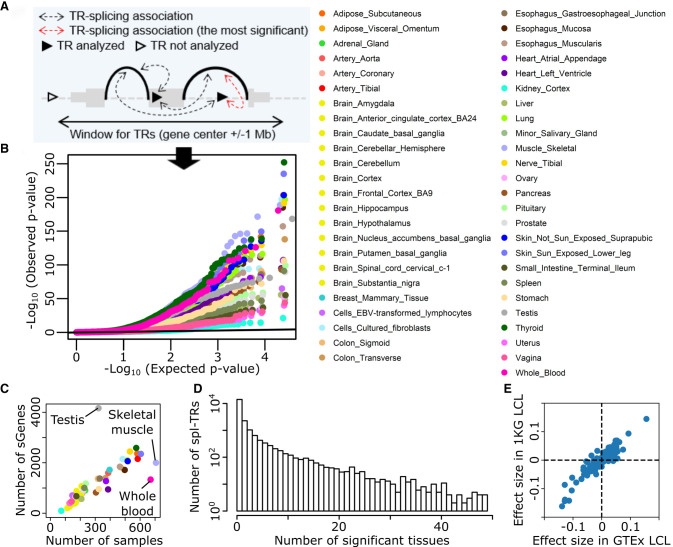
We discovered cis spl-TRs in GTEx v8 data, consisting of WGS of 838 donors and RNA-seq of 15,253 samples from 49 tissues, by using FastQTL (Fig. 1; Supplemental Table S1; Ongen et al. 2016; The GTEx Consortium 2020). TRs were sized using GangSTR, which handles TRs with motifs of up to 20 bp (n = 40,598 after quality control) (Mousavi et al. 2019). The sum of TR sizes at both alleles was defined as the TR dosage, and the association between TR dosage and splicing quantity was tested using linear regression whereas controlling for covariates including sex, population structure, and sequencing protocols. Among all TR–splicing pairs of each gene (Fig. 1A), only the top association in the linear regression was considered, and its P-value was adjusted for all TR–splicing pairs in each gene by employing a FastQTL permutation scheme. The adjusted P-values were further corrected for multiple gene testing by the Benjamini–Hochberg (BH) method. Consequently, there were a total of 58,290 significant TR–splicing associations across 49 tissues (gene-level false discovery rate [FDR] 5%) with 9537 unique spl-TRs and 21,832 unique TR–splicing associations (Fig. 1B). This spl-TR mapping was robust to various thresholds for GangSTR genotyping quality (gangstr-min-call-Q option); nominal P-values of the 1332 significant associations in whole blood (FDR 5%) were almost identical to those when using 0.8 or 0.95 quality threshold unless the TR site was filtered out (78 for 0.8 and 3 for 0.95 quality threshold) (Supplemental Fig. S2A,C). Most of the 1422 associations remained the most significant association of each gene (FDR 5%) when using those thresholds (1217 for 0.8 and 1186 for 0.95 quality threshold) (Supplemental Fig. S2B,D).
Figure 1.
Discovery of spl-TRs across 49 tissues. (A) Selection of the most significant TR–splicing association in each gene. (B) QQ plot of P-values obtained from spl-TR mapping. Dots represent the most significant TR–splicing association in each gene. The black y = x line corresponds to the null hypothesis. (C) Number of associations per tissue as a function of tissue sample size. Tissue color codes are shown in B. (D) Sharing of TR–splicing associations across tissues. The x-axis: the number of tissues sharing a given association; the y-axis: the number of associations shared across a given number of tissues. (E) Comparison of effect sizes of 563 TR–splicing pairs between GTEx and 1 KG LCL data sets. Dots denote TR–splicing pairs.
The number of significant TR–splicing associations varied across tissues from 95 in kidney cortex to 4164 in testis (Supplemental Table S1). The number grew along with the tissue sample size (Spearman's rank correlation test P = 1 × 10−21; ρ = 0.93). Testis, skeletal muscle, and whole blood escaped the trend as in previous works on GTEx data because the number of expressed genes were more in testis and less in skeletal muscle and whole blood than other tissues (Fig. 1C; The GTEx Consortium 2020; Garrido-Martín et al. 2021). A majority of TR–splicing associations were observed solely in either of the 49 tissues (68%), whereas some other associations were observed in five or more tissues (13%) (Fig. 1D). To compare the effect size of a TR–splicing association across tissues, we employed ψ, the proportion of split reads from an intron relative to all split reads sharing the same junction as the intron (Mertes et al. 2021), and defined the effect size of a TR–splicing association as the difference in ψ per repeat unit. When clustering pairwise correlation of effect sizes across tissues, tissue similarities were captured: brain subregions clustered together whereas testis, whole blood, and skeletal muscle were outliers (Supplemental Fig. S3). To examine the reproducibility of the associations, we utilized an independent data set of WGS and lymphoblastoid cell line transformed with Epstein-Barr virus (LCL) RNA-seq in 1000 genomes (1 KG). Among the 707 significant TR–splicing associations in GTEx LCL data, 563 could be assessed in 1 KG LCL data, whereas the rest could not be assessed due to differences in minor allele frequency and RNA-seq coverage. Effect sizes of the 563 associations correlated between GTEx and 1 KG data (Spearman's rank correlation test P = 8 × 10−190; ρ = 0.89), and the direction of effect sizes was concordant in the vast majority of the 563 associations (88%, one-sided sign test P = 5 × 10−83) (Fig. 1E). Altogether, these findings validate our TR–splicing associations.
Spl-TRs explain splicing variation independently of nearby variants
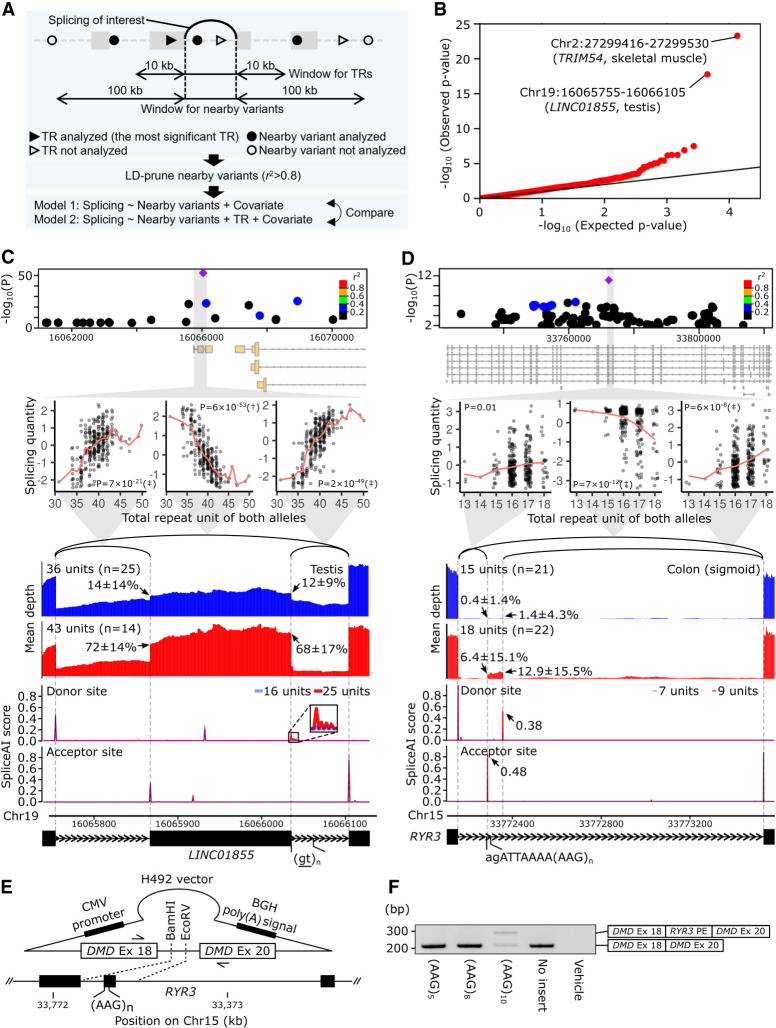
These 58,290 TR–splicing associations might be attributable to nearby true causal variants in linkage disequilibrium (LD) with the TRs. To address this issue, we determined whether TRs could explain splicing variation independently of nearby variants such as single-nucleotide polymorphisms (SNPs), small indels, and structural variations. Two linear regression models explaining the variation of each splicing event were compared using analysis of variance (ANOVA): one including nearby variants and the other additionally including a TR between the 10-kb upstream position from the donor site and the 10-kb downstream position from the acceptor site of the intron as an explanatory variable (Fig. 2A). Some splicing events have no qualified TR, and 11,435 TR–splicing associations remained for subsequent ANOVA tests. The resulting P-values indicated significant deviation from the uniform distribution (Fig. 2B, black line) (Kolmogorov–Smirnov test P < 2.2 × 10−16), suggesting that at least some of the TR–splicing associations cannot be explained by nearby variants.
Figure 2.
spl-TRs explain splicing variation independently of nearby variants. (A) Overview of ANOVA tests comparing two linear regression models explaining the variation of each splicing event. One model used TR dosage and nearby variants, whereas the other used nearby variants only. (B) QQ plot of P-values of ANOVA tests comparing the two models for 11,435 splicing events. The black y = x line corresponds to the null hypothesis. (C,D) Representative spl-TRs associated with splicing variation independently of nearby variants. Shown are associations of the spl-TR and neighboring variants with splicing quantity (top panel), correlations between TR dosages and splicing quantities (second panel from the top), mean RNA-seq depth of samples with a smaller or larger TR dosage (third panel from the top), SpliceAI scores of splice donor or acceptor site for sequences having a shorter or longer TR allele (fourth panel from the top), and exon–intron structures (bottom). (Top panel) Only variants whose nominal P-value was <0.01 are shown. Purple diamond: spl-TR; circle: other variants color-coded according to Pearson correlation coefficient between the variant and spl-TR dosages. (Second panel from the top) The splicing quantity indicates the normalized proportion in clustered splicing events (see Methods subsection “Mapping of spl-TRs”). The red line indicates the mean at each TR dosage. Nominal P-values of linear regression analysis in FastQTL are shown in each plot. (†) The top association in the gene (Q-value < 0.05); (‡) other significant associations (P-value < 8.7 × 10−5 for LINC01855 and 7.8 × 10−6 for RYR3; see Methods subsection “Identification of all significant TR–splicing pairs in each gene”). (Third panel from the top) Arrow: mean ± standard deviation of ψ5 or ψ3. (Fourth panel from the top) Because SpliceAI scores are almost the same between shorter (blue) and longer (red) TR alleles, the bars look purple in the most plotted region. The inset is a magnified image of junctions of splicing events whose alteration by TRs was supported by SpliceAI. (E,F) Experimental validation of the spl-TR at RYR3 using minigene assay. (E) Schematic representation of H492 minigene vector. The vector is constructed to have DMD exons 18 and 20, and between these exons the RYR3 pseudoexon and flanking sequences were inserted. For the synthesis of mRNA, the vector contains a cytomegalovirus (CMV) enhancer–promoter and a bovine growth hormone (BGH) polyadenylation signal. The vector was transfected into HeLa cells, and extracted RNA was amplified using the primers indicated by arrows in a reverse-transcription polymerase chain reaction (RT-PCR) assay. (F) RT-PCR products of minigene assay for RYR3 pseudoexon (PE). Here, 284- and 208-bp transcripts were generated from the vector carrying the (AAG)10 allele (A), whereas only a 208-bp transcript was generated from the vectors carrying the (AAG)3 or (AAG)8. Shown on the right is a schematic description of the RT-PCR products. The longer product consists of DMD exon 18, RYR3 PE, and DMD exon 20, whereas the smaller product does not contain the RYR3 PE.
This analysis simultaneously allowed us to discover TRs directly regulating splicing. We adjusted the P-values of all ANOVA tests of each gene using the Bonferroni method and selected the minimum P-value in each gene to ensure the inter-independence of inputs for the following FDR calculation. We then corrected the Bonferroni-adjusted P-values for the number of genes analyzed in the ANOVA tests and obtained FDR. There were 38 TR–splicing associations not attributable to nearby variants (FDR 20%) (e.g., associations at LINC01855, NARS2, and RYR3) (Supplemental Table S2). For several examples, RNA-seq BAM files were downloaded, and regions around the altered splicing events were visualized (Fig. 2C,D; Supplemental Fig. S4). In these examples, the spl-TR had a more significant association than neighboring variants, and TR dosages and splicing alterations were correlated clearly across a wide range of repeat lengths, which cannot be easily explained by tagged variants. We further investigated whether the causality of these spl-TRs for splicing alterations is supported by SpliceAI, a model predicting whether each position of a DNA sequence is a splice donor or acceptor site (Jaganathan et al. 2019). We compared two sequences with shorter or longer TR repeat for SpliceAI scores at each position, which correlate with the splicing amount of the position. The direction of difference in SpliceAI scores was compatible with the splicing alterations in most of these spl-TRs (Fig. 2C,D; Supplemental Fig. S4). We experimentally validated some of the spl-TRs using minigene assay in HeLa cells (Fig. 2E,F; Supplemental Fig. S5C,D), although for some genes splicing patterns could not be reproduced in this artificial system and the TR variation effects could not be evaluated (Supplemental Fig. S5A,B,E,F; Tran et al. 2006). In this system, an RYR3 pseudoexon harboring (AAG)10, which is a binding motif of serine/arginine (SR)-proteins including SRSF1, SRSF4, SRSF6, and SRSF9 and acts as an exonic splicing enhancer, was spliced into a proportion of transcripts (mean [SD]: 58% [1%]) whereas pseudoexons harboring (AAG)3 or 8 were not spliced (mean [SD]: 0% [0%]) (Welch's t-test P < 0.01) (Fig. 2E,F; Slišković et al. 2022). A NARS2 pseudoexon flanking a (GTTTTTT)4–containing intron was also spliced (mean [SD]: 14% [1%]) whereas ones flanking a (GTTTTTT)6 or 10–containing intron were not (mean [SD]: 0% [0%]) (Welch's t-test P < 0.01), as expected (Supplemental Fig. S5C,D). Taken together, these results suggest that some of our spl-TRs may directly regulate splicing.
Mechanisms of splicing regulation by spl-TRs
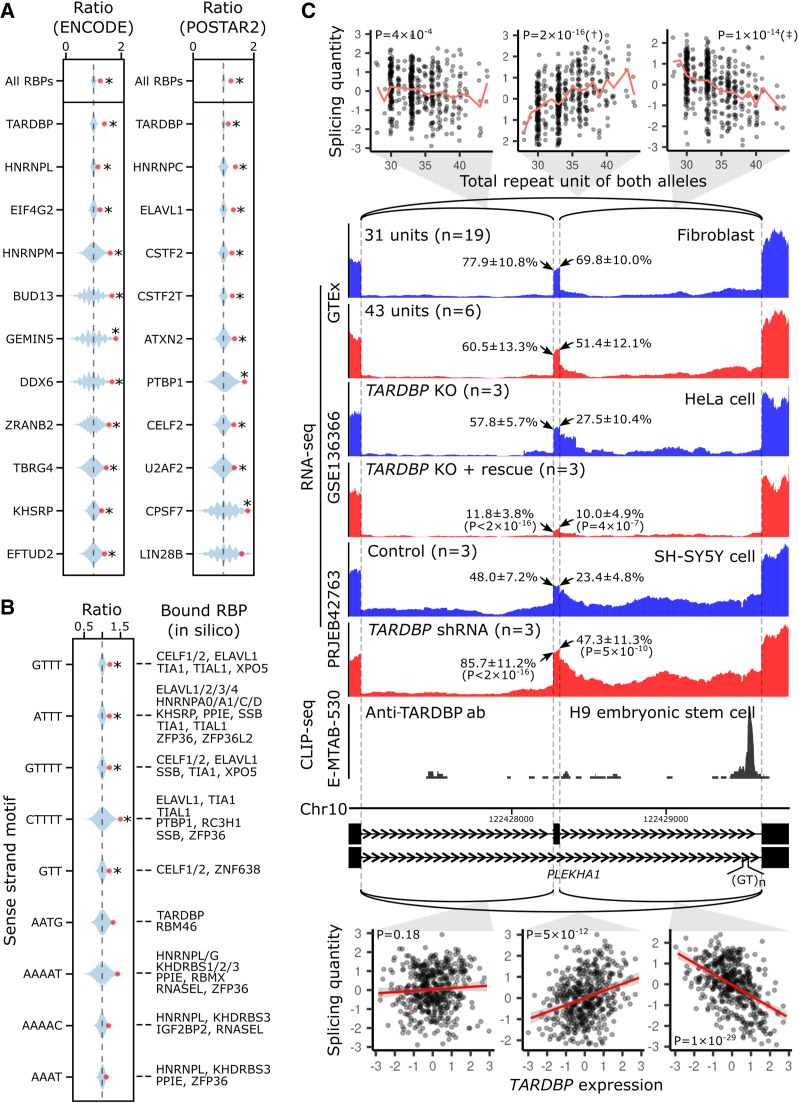
Previous studies have suggested that some TRs act as regulatory elements encompassing RBP binding motifs and that TR expansion might alter splicing in cis via RBP binding (Cuppens et al. 1998; Buratti and Baralle 2001; Buratti et al. 2001; Hui et al. 2003; Sathasivam et al. 2013; De Roeck et al. 2018). Indeed, our intronic or exonic spl-TRs were enriched for RBP binding in cross-linking immunoprecipitation sequencing (CLIP-seq) databases, The Encyclopedia of DNA Elements (ENCODE) and POSTAR2 (empirical P < 1 × 10−4). Specifically, intronic spl-TRs were enriched for 11 and 10 RBPs (Q-value < 0.1) in ENCODE and POSTAR2, respectively (Fig. 3A), whereas exonic spl-TRs were not due to the small sample size (Supplemental Tables S3, S4). The most significant RBP was TARDBP, in both the databases. The enriched RBPs except TDP-43 differed between ENCODE and POSTAR2 because they cover a different collection of RBPs (Supplemental Tables S3, S4). In parallel with this, intronic spl-TRs were enriched for five repeat motifs (Q-value < 0.1). Then, using SpliceAid and ATtRACT, we predicted which RBPs bind to the motifs (Fig. 3B; Supplemental Table S5; Piva et al. 2009; Giudice et al. 2016). These experimental and in silico analyses pointed out the several common RBPs. Next, we sought to identify examples of spl-TRs regulated by RBPs. Among the 58,290 spl-TRs, there were 33 unique TR–splicing pairs (127 across 49 tissues) (1) to which binding by the same RBPs was indicated by in silico tools (SpliceAid or ATtRACT) and CLIP-seq databases (ENCODE or POSTAR2), and (2) the causality of which was supported by SpliceAI (>0.01), with a total of 160 TR-RBP-splicing combinations (Supplemental Table S6). In 20 of the 160 combinations, bound RBP expression and splicing level were significantly correlated (P < 3 × 10−4 [Bonferroni-adjusted threshold for 160 tests]). As an example, a GT repeat of PLEKHA1 was associated with the increased skipping of a flanking exon (Fig. 3C). The repeat was predicted to bind TDP-43 as the binding motif has been reported as UG repeats (Tollervey et al. 2011; Humphrey et al. 2017) and indeed the binding was observed in CLIP-seq using an anti-TARDBP antibody (Fig. 3C; Tollervey et al. 2011). The endogenous TARDBP expression level was associated with the increased skipping of a PLEKHA1 exon in GTEx data, and concordantly, the exogenous expression and knockdown of TARDBP led to the increase and decrease in the skipping, respectively (Fig. 3C; Roczniak-Ferguson and Ferguson 2019; Brown et al. 2022). Thus, the PLEKHA1 GT repeat is a good candidate for TRs regulating splicing through RBP binding (Buratti et al. 2001; Hui et al. 2003; Sathasivam et al. 2013; De Roeck et al. 2018; Pacheco et al. 2019).
Figure 3.
Mechanisms underlying spl-TRs. (A,B) Shown from top to bottom are the most enriched RBPs (A) or repeat motifs (B) among intronic spl-TRs (n = 5660). The count of each RBP or repeat motif observed in the 5660 intronic sTRs is shown by the red point against blue violin plots representing the kernel density estimation of counts in sets (n = 10,000 and 100,000 for repeat motifs and RBPs, respectively) of 5660 randomly selected intronic TRs among all the intronic TRs (n = 18,139). The counts are represented as the ratio to the median of the negative control sets. For each repeat motif, RBPs predicted to bind it by SpliceAid or ATtRACT are shown on the right (B). (*) Q-value < 0.1. (C) The spl-TR at PLEKHA1. Shown are correlations between TR dosages and splicing quantities in GTEx data (top panel), RNA-seq depth of samples with a smaller or larger TR dosage in GTEx data (second panel from the top), TARDBP-knockout HeLa cell lines rescued by exogenously re-expressing TARDBP or not (third panel from the top), and SH-SY5Y cells stably expressing a shRNA for TARDBP or control SH-SY5Y cells (fourth panel from the top), CLIP-seq depth of H9 embryonic stem cells using an anti-TARDBP antibody (fifth panel from the top), exon–intron structures (sixth panel from the top), and correlations between TARDBP expression and splicing quantities in GTEx data (bottom). These panels are drawn as in Figure 2C and D. (Third and fourth panels from the top) The splice site usages were compared between the two cell types using binomial generalized linear model, and the resulting P-values are shown in brackets. (Bottom) The significance of linear regression is given in the plots (threshold: 3 × 10−4). The red line and gray shading represent the regression line and 95% confidence interval, respectively. The expression and splicing levels are normalized (see Methods subsection “Correlation between RBP expression level and splicing quantity”).
Another mechanism underlying spl-TRs may be the relative position change of regulatory elements, such as branch site, in pre-mRNA. We noted that the spl-TR at NARS2, experimentally validated above (Supplemental Fig. S5C,D), lies between the 3′ splice site and branch sites of the regulated pseudoexon (Supplemental Fig. S4). The spl-TR, when expanded to six units, increases the distance from 35 to 49 bp, which exceeds a typical distance (19 to 37 bp), and reduced the efficiency of splicing the pseudoexon (Supplemental Fig. S4; Mercer et al. 2015). In silico mutagenesis of the six-unit allele using SpliceAI showed that mutations creating a branch site sequence TNA, except ones concurrently creating an acceptor motif AG, recovered the splicing efficiency (Supplemental Fig. S6A; Leman et al. 2020). These trends were irrespective of the repeat motif, GTTTTTT, because even random sequences (NNNNNNN) reduced the splicing efficiency, whereas those containing TNA recovered it (Supplemental Fig. S6B). Thus, in the regulatory machinery that SpliceAI learned, the NARS2 GTTTTTT repeat regulates splicing through lengthening the distance from the 3′ splice site to branch sites.
Repeat expansion diseases relevant to spl-TRs
Leveraging our spl-TR catalog, we investigated whether TRs contribute to disease pathomechanisms via in-cis splicing alterations. We examined the overlap between our 9537 spl-TRs and 52 known loci responsible for repeat expansion diseases (Supplemental Table S7), and five of them overlapped (Supplemental Table S8). In three of these five, the spl-TR was located near the associated splicing, namely, within the spliced/removed intron or flanking exons: spinocerebellar ataxia (SCA) 6, SCA12, and myotonic dystrophy 1 (DM1).
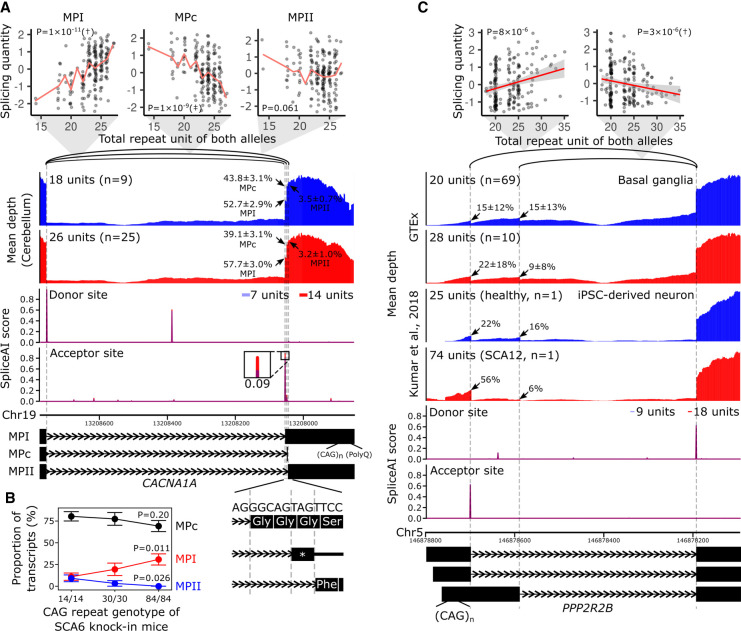
SCA6 is caused by heterozygous expansion of the CAG repeat in the last exon of CACNA1A, which produces toxic proteins with a polyglutamine tract (Paulson et al. 2017). The repeat correlated with an intron (Chr 19: 13,208,054–13,208,755) usage in the cerebellum in our catalog (Q-value = 5 × 10−7 and nominal P = 2 × 10−11, Supplemental Table S8), where Purkinje cells are lost in patients. Besides, the repeat length was also correlated, to a similar degree, with another intron (Chr 19: 13,208,049–13,208,755) usage (nominal P = 1 × 10−9; significance threshold for all TR–splicing pairs of CACNA1A: 5.1 × 10−6), whereas not with the other event (Chr 19: 13,208,046–13,208,755) among the cluster (nominal P = 0.061) (Fig. 4A; Paulson et al. 2017). These three splicing patterns correspond to the isoforms conventionally named as MPI, MPII, and MPc (Watase et al. 2008). The CAG repeat, when longer, preferred MPI to MPc (Fig. 4A), which is in good agreement with findings in knock-in mouse models of SCA6 carrying 14, 30, or 84 CAG repeats at the humanized last exon of Cacna1a; reverse-transcription polymerase chain reaction, followed by subcloning and sequencing, of Purkinje cells revealed that the proportions of MPI and MPc isoforms significantly increased and decreased as a function of the repeat length, respectively (Fig. 4B; Watase et al. 2008). In addition, Ishikawa et al. also qualitatively observed the MPI predominance in multiple brain regions, including cerebellar cortex, of SCA6 patients by using similar experiments (Ishikawa et al. 1999). Because MPI, but not MPII and MPc, encodes the polyglutamine tract, the splicing alterations may contribute to the pathogenesis by increasing the proportion of the toxic MPI isoform (Ishikawa et al. 1999; Watase et al. 2008).
Figure 4.
Splicing alterations by the spl-TR associated with SCA6 and SCA12. (A,C) Shown are correlations between TR dosages and splicing quantities (top), mean RNA-seq depth of samples with a smaller or larger TR dosage (second panel from the top), SpliceAI scores of splice donor or acceptor site for sequences having a shorter or longer TR allele (third panel from the top), and representative exon–intron structures (bottom). (Top panel) The splicing quantity indicates the normalized proportion in clustered splicing events (see Methods subsection “Mapping of spl-TRs”). The red line indicates the mean at each TR dosage (A); the red line and gray shading represent the regression line and 95% confidence interval, respectively (C). Nominal P-values of linear regression analysis in FastQTL are shown in each plot. (†) The top association in the gene (Q-value < 0.05); (‡) other significant associations (P-value threshold: CACNA1A, 5 × 10−6; PPP2R2B, 4 × 10−6; see Methods subsection “Identification of all significant TR–splicing pairs in each gene”). (Second panel from the top) iPSC-derived neurons of an SCA12 patient with 14/60 CAG repeats and a healthy individual with 9/16 units are shown (C) (Kumar et al. 2018); arrow: mean ± standard deviation of ψ5 or ψ3 of the junction. (Fourth panel from the top) The inset is a magnified image of junctions of splicing events whose alteration by TRs was supported by SpliceAI. The differences in SpliceAI scores between shorter and longer TR alleles are shown above or below the insets. Transcript models at the bottom are ENST00000360228 for MPI, ENST00000636473 for MPc, and ENST00000636389 for MPII (A) and ENST00000530902.5, ENST00000532154.5, and ENST00000394411.8 (C). (B) Proportions of MPI, MPII, and MPc isoforms in knock-in mouse models of SCA6 harboring 14, 30, or 84 CAG repeats at the humanized last exon of Cacna1a. The total number of sequenced clones was 55 from four Cacna1a14Q/14Q samples, 31 from two Cacna1a30Q/30Q samples, and 52 from three Cacna1a84Q/84Q samples. P-values of tests comparing proportions between Cacna1a14Q/14Q and Cacna1a84Q/84Q are shown.
SCA12 is an autosomal dominant disorder associated with CAG repeat expansion in PPP2R2B. The repeat size was negatively associated with an intron (Chr 5: 146,878,196–146,878,590) usage in basal ganglia, a brain subregion affected in SCA12 (Choudhury et al. 2018), in our spl-TR catalog (Q-value = 0.044 and nominal P = 3 × 10−6; Supplemental Table S8; Fig. 4C). Moreover, the repeat size positively correlated with Chr 5: 146,878,196–146,878,700 splicing to a similar degree (nominal P = 8 × 10−6), whereas all of the other 14 events among the splicing cluster did not correlate (nominal P > 0.1). The alterations were more remarkable in the neuron of an SCA12 patient, as evident from the RNA-seq coverage plot (Fig. 4C; Kumar et al. 2018). Longer CAG repeats preferred ENST00000530902.5 and ENST00000532154.5 isoforms, neither of which produces functional proteins as reported in Ensembl, to the ENST00000394411.8 isoform, which encodes the full-length protein. This switch may reduce the level of full-length protein, which seemed to be deleterious because PPP2R2B is loss-of-function-intolerant (pLI = 0.98 in gnomAD) (Karczewski et al. 2020).
DM1 is caused by expansion of a DMPK CTG repeat, whose dosage was significantly associated with the reduced usage of an intron (Chr 19: 45,770,641–45,770,970) in skeletal muscle in our catalog (Q-value = 0.012 and nominal P = 4 × 10−6, Supplemental Table S8; Supplemental Fig. S7). Because four other splicing events among the splicing cluster were not altered (nominal P > 0.05), we reasoned that the reduced intron usage might reflect the increased intron retention. We quantified the intron retention in skeletal muscle samples in GTEx (n = 706), and the TR dosage had a weak tendency for being associated with increased intron retention (nominal P = 4 × 10−3; significance threshold for all TR–splicing pairs in DMPK: 2 × 10−5) (Supplemental Fig. S7A,B). However, these alterations were not reproducible in publicly available RNA-seq data of biopsied skeletal muscle and cultured muscle cell samples of DM1 and healthy individuals (one-sided Wilcoxon rank-sum test P > 0.01, Bonferroni-corrected threshold for the number of tests [n = 5]) (Supplemental Fig. S7A,C). Thus, the alterations at DMPK observed in GTEx data might not be relevant to the disease mechanism.
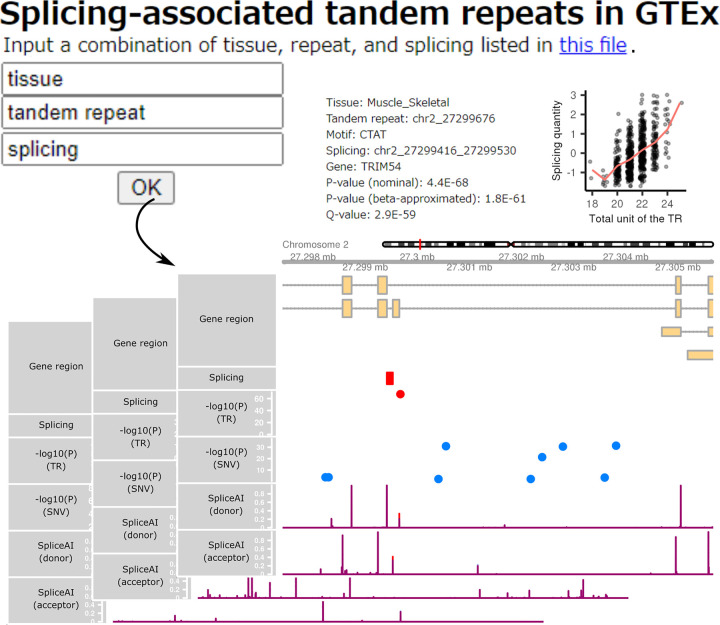
Lastly, we built a user-friendly platform, enabling public access to detailed information on the 58,290 significant TR–splicing associations across 49 tissues. By downloading the contents (https://doi.org/10.5281/zenodo.7086007) and giving combinations of TR, splicing, and tissue of interest, users can browse details on any of the associations (Fig. 5).
Figure 5.
Snapshots of our HTML pages presenting information on the 58,290 significant TR–splicing associations across 49 tissues. By providing combinations of TR, splicing, and tissue of interest on the portal page, users can browse the relationship between TR size and splicing quantity, the genomic coordinates of the TR, splicing, and genes, and the SpliceAI predictions of shorter and longer TR alleles for splice sites.
Discussion
Here, we present a comprehensive catalog of spl-TRs and their applications.
We provide insights into the mechanisms underlying a few spl-TRs (Fig. 3). However, the mechanisms of most spl-TRs are unknown, and their comprehensive exploration by experiments is laborious and unfeasible. Perturbation methods for deep learning models theoretically enable this, and indeed SpliceAI might disentangle the roles of DNA sequence in TR-mediated splicing (Supplemental Fig. S6), although SpliceAI is based solely on DNA sequences and does not give any clues to the roles of other factors such as RBPs. One solution to this is to construct another deep learning model using transcriptome data after the knockdown of each RBP or a model considering RBP binding sites as well as DNA sequences. Perturbations of these models should lead to comprehensive elucidation of TR-mediated splicing mechanisms.
Although the TR–splicing associations in our catalog are based on the range of polymorphic repeat lengths (>1% MAF), the associations in CACNA1A and PPP2R2B that we detected are in agreement with those observed in repeat expansion diseases (Fig. 4). Effects of TR dosage on splicing may be linear across broad ranges. Previous studies reported abnormal splicing events in a few repeat expansion diseases such as Huntington's disease, C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia, myotonic dystrophy type 2, and Fuchs endothelial corneal dystrophy (Neueder et al. 2017; Sznajder et al. 2018). However, we could not confirm these trends in the GTEx population because GangSTR could not confidently genotype these loci, thereby warranting QTL studies based on long-read technologies as discussed below. These TR-related splicing abnormalities could be alleviated by targeting the TRs with antisense oligonucleotides because the TRs may be bound by RBPs altering the splicing. Thus, our catalog may give insights into pathomechanisms and, at the same time, potential therapeutic targets for repeat expansion diseases.
We did not investigate the relevance of spl-TRs to common diseases because multiallelic sites such as TRs are not considered in conventional genome-wide association studies employing microarrays. However, WGS-based studies for common diseases will increasingly be conducted in the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) project, and the UK Biobank (Hu et al. 2021), which should eventually detect many susceptible TR loci. For the interpretation of their working mechanisms, our catalog will be an invaluable resource.
To maximize the benefit of these applications, our spl-TR catalog should be further expanded. Our catalog is based on GangSTR, which assesses TRs detected by the Tandem Repeats Finder (Mousavi et al. 2019). As many as 14,000 novel TR loci, not in the Tandem Repeats Finder list, were discovered in a previous study (Trost et al. 2020), and thus it is clear that our analysis in this study is unlikely to have covered all TR loci. Moreover, tools sizing TRs with more than 20 bp motifs, which GangSTR does not capture, have become increasingly available (Bakhtiari et al. 2021; Eslami Rasekh et al. 2021; Lu et al. 2021). More TRs are undoubtedly accessible by long-read WGS, although its high cost currently hampers large-scale QTL studies based on it. Thus, new informatics and sequencing technologies will enable the identification of more spl-TRs.
In conclusion, our spl-TR catalog may help elucidate functional mechanisms of TR-mediated splicing regulation and pathomechanisms of both rare and common genetic diseases, which could be targeted by employing the antisense oligonucleotide strategy. The catalog should be further expanded by using new technologies.
Methods
TR sizing and quality control
We genotyped TRs in 838 donors using GangSTR v2.4.3 (https://github.com/gymreklab/GangSTR) and a BED file for 832,380 TR regions (hg38_ver13.bed.gz, https://github.com/gymreklab/GangSTR) (Mousavi et al. 2019). We downloaded an hg38 reference file (resources_broad_hg38_v0_Homo_sapiens_assembly38.fasta, https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0/) and WGS CRAM files accessed from the NCBI database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under accession number phs000424.v8.p2. TR calls of each donor were merged and filtered by following the developer's recommendation (https://github.com/gymreklab/GangSTR/wiki/Filtering-GangSTR-output) and using TRTools v3.0.2 (https://github.com/gymreklab/trtools) (Mousavi et al. 2021). Specifically, the following options of the DumpSTR function were used: ‐‐gangstr-filter-spanbound-only: filtering all reads except spanning and bounding; ‐‐gangstr-filter-badCI: filtering regions where the maximum likelihood estimate is not in the confidence interval; ‐‐filter-regions: filtering sites in segmental duplication regions downloaded from UCSC Genome Browser (http://genome.ucsc.edu/); ‐‐gangstr-max-call-DP 1000 and ‐‐gangstr-min-call-DP 20: filtering calls with >1000× or <20× coverage; ‐‐min-locus-hwep 0.01: filtering sites with P-value < 0.01 from a two-sided binomial test comparing the observed and expected percentages of homozygous calls; ‐‐gangstr-min-call-Q 0.9: filtering calls with quality score <0.9; and ‐‐min-locus-callrate 0.8: filtering sites with call rate <0.8. In addition, alleles with allele counts of 16 or less, corresponding to <1% MAF, were removed, and sites with two or more alleles were kept. Repeat units on two alleles were summed at each site and regarded as TR dosage. When at least one allele was missing, TR dosage was not calculated. Consequently, 40,598 sites on autosomes remained. For 1 KG, we downloaded WGS CRAM files for 445 samples (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/) and performed processing as for GTEx data.
Mapping of spl-TRs
To map spl-TRs, we downloaded resources, based on the human reference genome build GRCh38, from GTEx portal (https://gtexportal.org/home/) and followed the sQTL mapping protocol developed by GTEx. Briefly, splicing level and covariates for 49 tissues were downloaded from GTEx portal (GTEx_Analysis_v8_sQTL_phenotype_matrices.tar and GTEx_Analysis_v8_sQTL_covariates.tar.gz). The splicing level was calculated in 49 tissues by The GTEx Consortium as follows: (1) Splicing was detected from RNA-seq BAM files using LeafCutter (Li et al. 2018); (2) splicing clusters, a group of splicing events sharing either of their junctions, were generated; (3) introns with few read counts or low diversity of counts across samples were filtered; (4) proportions of each splicing in splicing clusters were calculated and standardized across samples; and (5) the splicing proportions of all splicing events were quantile-normalized to the normal distribution in each sample. The covariate files include sex, library preparation protocol (PCR-based or PCR-free), sequencing platform (HiSeq 2000 or HiSeq X), five genotype principal components, and probabilistic estimation of expression residuals (PEER) factors. The PEER factors are calculated from the splicing quantities above by PEER, a data-driven approach detecting hidden confounders (Stegle et al. 2012).
Leveraging these downloaded resources and our TR dosages, we mapped spl-TRs using FastQTL as done in GTEx. Only TRs within 1 Mb from each gene center were considered (n = 36,020) because splicing-associated quantitative trait loci (sQTL) are enriched around genic regions (Battle et al. 2014; Takata et al. 2017; Walker et al. 2019). FastQTL performed linear regressions between TR dosages and splicing quantities and obtained a nominal P-value for each test. Only the top association among all TR–splicing pairs was selected for each gene using –grp option. The same regressions were performed in permutations of the sample labels. Permutation was performed using the adaptive permutation mode –permute 1000 10000, which adapts the number of permutations to the significance level to decrease the computational burden. The null distribution made of 1000–10,000 nominal P-values of the top association in each permutation was fitted to a β distribution for each gene. Using this fitted β distribution, the nominal P-value of the top association in the real data, not permuting sample labels, was extrapolated to obtain a β-approximated empirical P-value. From these empirical P-values, Q-values were calculated, and one spl-TR in each gene was identified (gene-level FDR 5%).
For 1 KG, we downloaded RNA-seq BAM files for 445 LCL samples (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/geuvadis/) and performed processing as for GTEx data by using script and docker files provided by The GTEx Consortium as follows. Splicing was quantified using the leafcutter_bam_to_junc.wdl and leafcutter_cluster.wdl script files and the broad-cga-francois-gtex/leafcutter (https://hub.docker.com/r/francois4/leafcutter) docker file; PEER factors were computed by using the run_PEER.R script in the broadinstitute/gtex_eqtl (https://hub.docker.com/r/broadinstitute/gtex_eqtl) docker file. Genotype principal components were computed from VCF files (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/) by using the ‐‐pca option of PLINK v1.90b (Chang et al. 2015) after selecting variants with >10% MAF and pruning them with the ‐‐indep 50 5 1.5 option.
Computation of ψ value
We downloaded LeafCutter junction read count files for GTEx RNA-seq data from AnVIL and merged the files of each tissue using the leafcutter_cluster.wdl script file and the broad-cga-francois-gtex/leafcutter (https://hub.docker.com/r/francois4/leafcutter) docker file mentioned above. From this merged file (perind_numers.counts.gz), ψ5 and ψ3 values, which are the proportions of split reads from an intron relative to all split reads sharing the same donor (ψ5) or acceptor (ψ3) as the intron, were computed. The mean of these two values, ψ5 and ψ3, of an intron was defined as ψ of the intron whereas ψ was not computed when either ψ5 or ψ3 was missing due to a lack of coverage.
Effect size correlation between data sets
To compare the effect size of TR–splicing associations, defined as change in ψ per repeat unit, among GTEx tissues and between GTEx and 1 KG LCL, we first regressed out the covariates used in the spl-TR mapping mentioned above from ψ values, followed by running the linear regression of the ψ residuals on TR size by using the Python stasmodels.api.OLS function. Based on the resulting regression coefficients of TR–splicing associations significant in at least one of the two data sets, Spearman's correlation coefficients were computed for every pair of GTEx tissues or GTEx and 1 KG LCL pair.
Analysis of independence of TR–splicing associations from nearby variants
To prove that the 58,290 TR–splicing associations were independent of nearby variants, we performed an analysis similar to that by Gymrek et al. (2016). For each of the 58,290 splicing events, we constructed two linear regression models explaining the splicing variation in the relevant tissue: first model: nearby variant dosages + covariates; second model: nearby variant dosages + spl-TR dosage + covariates. As the “nearby variants,” we considered SNVs and small indels (GTEx_Analysis_2017-06-05_v8_WholeGenomeSeq_838Indiv_Analysis_Freeze.SHAPEIT2_phased.vcf, downloaded from AnVIL) as well as SVs, discovered through LUMPY, Genome STRiP, and MELT (GTEx_v7.sv.low_pass.vcf.gz, downloaded from AnVIL and lifted over to hg38 with picard-tools-2.10. LiftOverIntervalList) (Scott et al. 2021). Because the information on SVs was available only for 613 individuals, 601 of whom overlapped with the 838 individuals analyzed for TRs, SNVs, and small indels, we restricted this analysis to these 601 individuals. In the two models, the nearby variants were defined as SNPs or small indels (1) with MAF > 1%, (2) within 100 kb up- and downstream of each splicing, and (3) with nominal P-value < 0.01 in linear regression as well as SVs (1) with MAF > 1% and (2) whose center position ([start position + end position]/2) was within 100 kb up- and downstream of each splicing; the spl-TR was defined as one of the 58,290 TRs within ±10 kb of each splicing; splicing events whose junction was within 900 kb (=1 Mb–100 kb) from the gene center position were selected. After this filtering, 11,435 remained among the 58,290 associations because some events had no qualified TR. For each association, the nearby variants, including SNVs, small indels, and SVs, in LD (>0.8 R2), were pruned using PLINK with ‐‐indep 50 5 0.8 for two reasons: (1) Too many explanatory variables, even when each of them was unrelated to splicing level, could explain it well and mask the contribution of TR. (2) Multicollinearity among explanatory variables can lead to inflated regression coefficients and test statistics, leading to error termination of the R lm function for linear regression. Then, whether the second model outperformed the first one was tested with ANOVA.
LD between TRs and other variants
For selected examples of TR–splicing associations independent of nearby variants, we calculated LD between TRs and nearby variants by employing an approach previously used by Fotsing et al. (2019). The LD was defined as a squared Pearson correlation coefficient between the TR dosage and the other variant dosage, obtained with Scipy.stats.pearsonr.
Identification of all significant TR–splicing pairs in each gene
In addition to top associations, we identified all significant TR–splicing pairs of each gene in a manner similar to that described in the GTEx original publication (The GTEx Consortium 2020). The significance threshold for a TR–splicing association was defined as the beta-approximated P-value closest to the 5% FDR threshold applied for top associations. The beta-approximated P-values were obtained after adjustment for all TR–splicing pairs in each gene with the permutation scheme; however, the permutation scheme can be applied only to the top association in each gene. Instead, associations whose nominal P-value exceeded the threshold after Bonferroni correction for the number of all TR–splicing pairs in each gene were considered significant.
RNA-seq coverage plot
We visualized the mean depth of multiple RNA-seq samples by ggsashimi (https://github.com/guigolab/ggsashimi) (Garrido-Martín et al. 2018). The BAM files of the RNA-seq data were downloaded from AnVIL (Analysis, Visualization, and Informatics Lab-Space, https://anvilproject.org/).
SpliceAI analysis
We predicted the effect of repeat length on splice site recognition by supplying two nucleotide sequences with a shorter or longer TR allele to SpliceAI (https://github.com/Illumina/SpliceAI) (Jaganathan et al. 2019). Because SpliceAI considers 10-kb sequences surrounding positions of interest, we gave sequences of the plotted region together with 5-kb upstream and downstream regions (Supplemental Fig. S4). For LINC01855, we filled the sequence of the upstream and downstream regions with “N” because otherwise SpliceAI did not recognize splice sites of LINC01855. This is possibly because SpliceAI may be less sensitive to noncoding genes or small genes such as LINC01855, which consists of three exons. To match the length of shorter and longer TR alleles to that of the reference allele when visualizing the scores, we filled in the middle part of the TR with zero scores in the shorter TR allele or deleted the middle part of the TR in the longer TR allele.
Minigene splicing assay
An H492 minigene vector containing two DMD exons and one intervening intron encompassing a multicloning site was kindly provided by Prof. Masafumi Matsuo (Tran et al. 2006). The target exons and flanking sequences (RYR3, Chr 15: 33,772,172–33,772,481; LINC01855, Chr 19: 16,065,776–16,066,086; CACNA1A, Chr 19: 13,207,811–13,208,253) were amplified from genomic DNA using primers containing recognition sites for BamHI or EcoRV restriction enzymes (New England Biolabs). The samples were selected from our genomic DNA and WGS data collection, based on repeat genotype determined with GangSTR. Amplified products were digested with the enzymes and ligated to the minigene using Mighty Mix DNA Ligation Kit (Takara Bio). The ligation mixture was cloned into Competent Quick DH5α cells (Toyobo), and minigene vectors were extracted with QIAprep Spin Miniprep Kit (Qiagen). Next, HeLa cells were grown in six-well plates in Dulbecco's Modified Eagle Medium containing 5% fetal bovine serum (Trace Biosciences). Minigenes (1 μg each) were transfected into the cells using Opti-MEM I Reduced Serum Media (Thermo Fisher Scientific) and polyethylenimine (PEI MAX; Polysciences, Inc.) in accordance with the manufacturer's protocol. Cells were harvested 24 h after transfection, and total RNA was extracted using RNeasy Mini Kit (Qiagen). Two micrograms of total RNA was subjected to reverse transcription (RT) using random hexamer primers (PrimeScript 1st strand cDNA Synthesis Kit, Takara Bio). PCR was carried out using forward and reverse primers corresponding to the upstream DMD exon (5′-GGTACCACAGCTGGATTACTCGCTC-3′) or the downstream DMD exon (5′-CTCGAGCAGCCAGTTAAGTCTCTCAC-3′). The cDNA was amplified using LA Taq Polymerase (Takara Bio) and PCR products were electrophoresed on a 2% agarose gel. The PCR products were cloned into Competent Quick DH5α cells with the TOPO TA Cloning Kit for Sequencing (Thermo Fisher Scientific), and colony PCR amplicons were subjected to Sanger sequencing. Band intensities were quantified with Image Lab software 5.2.1 (Bio-Rad Laboratories, Inc.) after local background intensities were subtracted using the Volume Tools function. These experiments were replicated independently twice.
For NARS2, pcDNA 3.1(+) vectors, similar to the H492 minigene vector, inserted with the two DMD exons and NARS2 exon-intron structure (Chr 11: 78,561,717–78,562,765) harboring (CAAAAAA)4, 6, or 10 at 78,562,032–78,562,066 were synthesized by using the GeneArt Gene Synthesis service (Thermo Fisher Scientific). The RT-PCR products of NARS2 were treated with T7 Endonuclease I (New England Biolabs) according to the manufacturer's protocol to digest heteroduplex products.
Enrichment analyses of repeat motifs and RBPs in spl-TRs
We tested the enrichment of repeat motifs among exonic or intronic spl-TRs significant in at least one tissue (n = 353 and 5660) compared with 100,000 negative data sets of 353 exonic or 5660 intronic TRs randomly sampled from all exonic or intronic TRs analyzed in spl-TR mapping (n = 721 and 18,139). TRs in each negative data set were matched to spl-TRs in terms of the distribution of 100 categories of distances to the nearest SJ. The distance category was made by binning all of the analyzed TRs (n = 36,020) into 100 categories of the same TR counts from the shortest to the longest distance. The enrichment was assessed only for 20 motifs observed more than five times in all of the exonic TRs (n = 721) or 53 motifs observed more than ten times in all of the intronic TRs (n = 18,139) (Supplemental Table S4). The empirical P-value of a motif was defined as the proportion of negative data sets where the motif count was more than that in the spl-TRs. The P-values were corrected for multiple testing using the BH method. Motif sequences on the sense strand were rearranged into all possible patterns (e.g., CAG to CAG, AGC, and GCA), and the lexicographically first sequence was defined as the motif of the TR. Exon and intron regions and strands were extracted from the gene model (gencode.v26.GRCh38.genes.gtf) above. We predicted RBPs binding to each repeat motif by SpliceAid and ATtRACT (Piva et al. 2009; Giudice et al. 2016). For each TR, the repeat sequence on the sense strand was tandemly repeated 100 times and, if the overall sequence contained an RBP binding motif existing in SpliceAid or ATtRACT, the repeat was considered to bind the RBP.
We also analyzed the enrichment of RBPs bound to spl-TRs. BED files for RBP binding sites detected through CLIP-seq were downloaded from the ENCODE portal (184 files) and POSTAR2 (human_RBP_binding_sites.txt) (Zhu et al. 2019; Luo et al. 2020). For ENCODE, BED files for the same RBP were merged, with a total of 122 files, and call cutoffs for each signal were set at fold-enrichment > 4 and P-value < 0.01. Because POSTAR2 data contained some of the ENCODE data, we removed them, and 85 RBPs remained. RBPs whose binding sites overlapped with at least one of the 36,020 TRs in spl-TR mapping were analyzed (n = 112 for ENCODE and 71 for POSTAR2). The enrichment was assessed only for RBPs observed more than ten times in all intronic TRs (n = 36 for ENCODE and 19 for POSTAR2).
Correlation between RBP expression level and splicing quantity
We assessed correlations between RBP expression level and splicing quantity using linear regression based on a Python module, statsmodels. The RBP expression levels for 49 tissues were downloaded from GTEx portal (GTEx_Analysis_v8_sQTL_expression_matrices.tar) and calculated by The GTEx Consortium as follows (The GTEx Consortium 2020): (1) Read counts were normalized between samples using the TMM (trimmed mean of M values) method (Robinson and Oshlack 2010); (2) genes were selected based on expression thresholds for TPM (transcripts per kilobase million) and read count; and (3) expression values for each gene were inverse normal transformed across samples. The splicing level was downloaded and processed as described above (see Methods subsection “Mapping of spl-TRs”).
CLIP-seq
We downloaded FASTQ files of CLIP-seq using anti-TDP43 antibody for H9 embryonic stem cells (E-MTAB-530) (Tollervey et al. 2011). NCBI Sequence Read Archives (SRA; https://www.ncbi.nlm.nih.gov/sra) files were converted to FASTQ files using the fasterq-dump-orig command of the SRA Toolkit (2.10.8-ubuntu64). FASTQ files were processed using the nf-core clip-seq workflow (https://nf-co.re/clipseq/1.0.0) of Amazon Genomics command-line interface briefly as follows (Ewels et al. 2020). Universal adapter sequences were trimmed, and reads shorter than 12 nucleotides were filtered out using cutadapt (Martin 2011). Remaining reads were premapped to rRNA and tRNA with Bowtie 2 (Langmead and Salzberg 2012). Resulting unmapped reads were aligned to the GRCh38 reference with STAR and deduplicated for PCR duplication using UMI-tools (Dobin et al. 2013; Smith et al. 2017).
Overlap of spl-TRs with known disease loci
We surveyed the literature and compiled a list of repeats whose expansion is related to rare diseases (n = 52, Supplemental Table S7; Tang et al. 2017; Yu et al. 2021). The overlap of these loci with the 9537 spl-TRs above was analyzed by using BEDTools intersect function (Quinlan and Hall 2010).
Processing of downloaded RNA-seq data
We downloaded SRA or FASTQ files of RNA-seq data from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) or the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) as follows: GSE136366: TARDBP-knockout HeLa cell lines rescued by exogenously re-expressing TARDBP or not (n = 3) (Roczniak-Ferguson and Ferguson 2019); PRJEB42763: SH-SY5Y cells stably expressing a shRNA for TARDBP or control SH-SY5Y cells (n = 3) (Brown et al. 2022); PRJNA391769: two technical replicates of SCA12 and control neuron derived from induced pluripotent stem cell (n = 1 for each); GSE86356: DM1 and control quadriceps femoris (n = 19 and 12), and DM1 and control tibialis anterior (n = 46 and 11); GSE128844: DM1 and control myoblast (n = 3 and 3); and GSE160916: DM1 and control myoblast (n = 3 and 3) and DM1 and control myotube (n = 3 and 3) (Kumar et al. 2018; Bargiela et al. 2019; Wang et al. 2019; Franck et al. 2022). SRA files were converted to FASTQ files using the fasterq-dump-orig command of the SRA Toolkit (2.10.8-ubuntu64). We determined the read length with FastQC-0.11.9 and noticed that read length was discordant between disease and control samples in GSE86356. Hence, we trimmed reads to 57 bp for tibialis anterior samples with Trimmomatic-0.40 (Bolger et al. 2014); for quadriceps femoris samples, we only analyzed samples of 60-bp read length (n = 12 for DM1 and n = 11 for control).
These downloaded files were processed following the GTEx pipeline (https://github.com/broadinstitute/gtex-pipeline/tree/master/rnaseq). Briefly, the FASTQ files were aligned to the human hg38 reference genome (Homo_sapiens_assembly38_noALT_noHLA_noDecoy.fasta, https://console.cloud.google.com/storage/browser/gtex-resources/references) with STAR-.2.5.3a (Dobin et al. 2013) and the gene annotation file (gencode.v26.GRCh38.genes.gtf, https://www.gtexportal.org/home/data sets) using a docker file (broadinstitute/gtex_rnaseq:V8, https://hub.docker.com/r/broadinstitute/gtex_rnaseq/tags). In the STAR mapping, we ensured that the “sjdbOverhang” parameter matched read length for each FASTQ file. Splicing levels were quantified using the provided scripts (leafcutter_bam_to_junc.wdl and leafcutter_cluster.wd) and the docker file (broad-cga-francois-gtex/leafcutter, https://hub.docker.com/r/francois4/leafcutter).
Quantification of the retention and splicing of Chr 19: 45,770,640–45,770,971 intron at DMPK
We quantified intron retention (Chr 19: 45,770,640–45,770,971) in DMPK in all available skeletal muscle samples in GTEx v8 (n = 706). We counted reads spanning either of the two exon–intron junctions (Chr 19: 45,770,640–45,770,641 and Chr 19: 45,770,970–45,770,971) using iREAD (Li et al. 2020) and averaged them in each sample. This intron retention event was added to the LeafCutter junction read count file of each sample downloaded from AnVIL. Then, the junction files were processed using the GTEx protocol described above (Methods subsection “Mapping of spl-TRs”). This processing treated the intron retention like a splicing event: (1) calculated the proportion of intron retention reads in a splicing and intron retention cluster and (2) normalized it across all samples and all events, as performed for splicing events. For DM1 and control samples (GEO; GSE86356, GSE128844, and GSE160916), no covariates were available, and we simply evaluated the difference in the proportion of Chr 19: 45,770,640–45,770,971 splicing and intron retention among the clustered splicing events by using one-sided Mann–Whitney U test (stats.mannwhitneyu). Aside from this regression analysis, we quantified the retention and splicing of the Chr 19: 45,770,640–45,770,971 intron with the theta (θ) value, which is almost identical to the ψ value but considers intron retention (Mertes et al. 2021).
Data access
The results of associations between all splicing events versus all TRs in 49 tissues and user-friendly web pages with detailed information are available in Zenodo (https://doi.org/10.5281/zenodo.7086007).
Supplementary Material
Acknowledgments
The authors thank Edanz (https://jp.edanz.com/ac) for editing the English text of a draft of this manuscript. This work was supported by Japan Agency for Medical Research and Development (AMED) (JP22ek0109486, JP22ek0109549, and JP22ek0109493 to N.M.); Japan Society for the Promotion of Science (JSPS) KAKENHI (JP20K16932 to K.H., JP20K17428 to N.T., JP21k15097 to Y.U., JP20K17936 to A.F., and JP20K07907 to S.M.); an intramural grant from Yokohama City University to K.H.; and the Takeda Science Foundation (to T.M. and N.M.).
Author contributions: K.H. conceptualized and designed the study, reviewed the literature, analyzed the data, and drafted the manuscript; D.Y., E.K., K.W., K.Mo., K.I., H.M., N.T., Y.U., A.F., K.Mi., and T.M. analyzed the data and revised the manuscript; S.M. and N.M. supervised all aspects of the study and revised the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277335.122.
Competing interest statement
The authors declare no competing interests.
References
- Bakhtiari M, Park J, Ding YC, Shleizer-Burko S, Neuhausen SL, Halldórsson BV, Stefánsson K, Gymrek M, Bafna V. 2021. Variable number tandem repeats mediate the expression of proximal genes. Nat Commun 12: 2075. 10.1038/s41467-021-22206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiela A, Sabater-Arcis M, Espinosa-Espinosa J, Zulaica M, Lopez de Munain A, Artero R. 2019. Increased Muscleblind levels by chloroquine treatment improve myotonic dystrophy type 1 phenotypes in in vitro and in vivo models. Proc Natl Acad Sci 116: 25203–25213. 10.1073/pnas.1820297116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, Haudenschild CD, Beckman KB, Shi J, Mei R, et al. 2014. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res 24: 14–24. 10.1101/gr.155192.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, Bampton A, Lee FCY, Masino L, Qi YA, et al. 2022. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603: 131–137. 10.1038/s41586-022-04436-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. 2001. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343. 10.1074/jbc.M104236200 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE. 2001. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20: 1774–1784. 10.1093/emboj/20.7.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4: 7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Chatterjee S, Chatterjee K, Banerjee R, Humby J, Mondal B, Anand SS, Shubham S, Kumar H. 2018. Clinical characterization of genetically diagnosed cases of spinocerebellar ataxia type 12 from India. Mov Disord Clin Pract 5: 39–46. 10.1002/mdc3.12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, Jorissen M, Droogmans G, Reynaert I, Goossens M, et al. 1998. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest 101: 487–496. 10.1172/JCI639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roeck A, Duchateau L, Van Dongen J, Cacace R, Bjerke M, Van den Bossche T, Cras P, Vandenberghe R, De Deyn P, Engelborghs S, et al. 2018. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease. Acta Neuropathol 135: 827–837. 10.1007/s00401-018-1841-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslami Rasekh M, Hernández Y, Drinan SD, Fuxman Bass JI, Benson G. 2021. Genome-wide characterization of human minisatellite VNTRs: population-specific alleles and gene expression differences. Nucleic Acids Res 49: 4308–4324. 10.1093/nar/gkab224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, Garcia MU, Di Tommaso P, Nahnsen S. 2020. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 38: 276–278. 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- Fotsing SF, Margoliash J, Wang C, Saini S, Yanicky R, Shleizer-Burko S, Goren A, Gymrek M. 2019. The impact of short tandem repeat variation on gene expression. Nat Genet 51: 1652–1659. 10.1038/s41588-019-0521-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck S, Couvreu De Deckersberg E, Bubenik JL, Markouli C, Barbé L, Allemeersch J, Hilven P, Duqué G, Swanson MS, Gheldof A, et al. 2022. Myotonic dystrophy type 1 embryonic stem cells show decreased myogenic potential, increased CpG methylation at the DMPK locus and RNA mis-splicing. Biol Open 11: bio058978. 10.1242/bio.058978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Martín D, Palumbo E, Guigó R, Breschi A. 2018. ggsashimi: sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput Biol 14: e1006360. 10.1371/journal.pcbi.1006360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Martín D, Borsari B, Calvo M, Reverter F, Guigó R. 2021. Identification and analysis of splicing quantitative trait loci across multiple tissues in the human genome. Nat Commun 12: 727. 10.1038/s41467-020-20578-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice G, Sánchez-Cabo F, Torroja C, Lara-Pezzi E. 2016. ATtRACT—a database of RNA-binding proteins and associated motifs. Database (Oxford) 2016: baw035. 10.1093/database/baw035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2020. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369: 1318–1330. 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, Daly MJ, Price AL, Pritchard JK, Sharp AJ, et al. 2016. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 48: 22–29. 10.1038/ng.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Stilp AM, McHugh CP, Rao S, Jain D, Zheng X, Lane J, Méric de Bellefon S, Raffield LM, Chen MH, et al. 2021. Whole-genome sequencing association analysis of quantitative red blood cell phenotypes: the NHLBI TOPMed program. Am J Hum Genet 108: 1165. 10.1016/j.ajhg.2021.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol 10: 33–37. 10.1038/nsb875 [DOI] [PubMed] [Google Scholar]
- Humphrey J, Emmett W, Fratta P, Isaacs AM, Plagnol V. 2017. Quantitative analysis of cryptic splicing associated with TDP-43 depletion. BMC Med Genomics 10: 38. 10.1186/s12920-017-0274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Fujigasaki H, Saegusa H, Ohwada K, Fujita T, Iwamoto H, Komatsuzaki Y, Toru S, Toriyama H, Watanabe M, et al. 1999. Abundant expression and cytoplasmic aggregations of α1A voltage-dependent calcium channel protein associated with neurodegeneration in spinocerebellar ataxia type 6. Hum Mol Genet 8: 1185–1193. 10.1093/hmg/8.7.1185 [DOI] [PubMed] [Google Scholar]
- Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, et al. 2019. Predicting splicing from primary sequence with deep learning. Cell 176: 535–548.e24. 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Dhapola P, Hussain A, Kutum R, Srivastava AK, Mukerji M, Mukherjee O, Faruq M. 2018. Transcriptomic dynamics of a noncoding trinucleotide repeat expansion disorder SCA12 in iPSC derived neuronal cells: signatures of interferon induced response. bioRxiv 10.1101/201137 [DOI]
- LaCroix AJ, Stabley D, Sahraoui R, Adam MP, Mehaffey M, Kernan K, Myers CT, Fagerstrom C, Anadiotis G, Akkari YM, et al. 2019. GGC repeat expansion and exon 1 methylation of XYLT1 is a common pathogenic variant in Baratela-Scott syndrome. Am J Hum Genet 104: 35–44. 10.1016/j.ajhg.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti MD, Scott HS, Buresi C, Rossier C, Bottani A, Morris MA, Malafosse A, Antonarakis SE. 1997. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 386: 847–851. 10.1038/386847a0 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman R, Tubeuf H, Raad S, Tournier I, Derambure C, Lanos R, Gaildrat P, Castelain G, Hauchard J, Killian A, et al. 2020. Assessment of branch point prediction tools to predict physiological branch points and their alteration by variants. BMC Genomics 21: 86. 10.1186/s12864-020-6484-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, Pritchard JK. 2018. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet 50: 151–158. 10.1038/s41588-017-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HD, Funk CC, Price ND. 2020. iREAD: a tool for intron retention detection from RNA-seq data. BMC Genomics 21: 128. 10.1186/s12864-020-6541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TY, Human Genome Structural Variation Consortium, Chaisson MJP. 2021. Profiling variable-number tandem repeat variation across populations using repeat-pangenome graphs. Nat Commun 12: 4250. 10.1038/s41467-021-24378-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. 2020. New developments on the encyclopedia of DNA elements (ENCODE) data portal. Nucleic Acids Res 48: D882–D889. 10.1093/nar/gkz1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. 2015. Genome-wide discovery of human splicing branchpoints. Genome Res 25: 290–303. 10.1101/gr.182899.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertes C, Scheller IF, Yépez VA, Çelik MH, Liang Y, Kremer LS, Gusic M, Prokisch H, Gagneur J. 2021. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat Commun 12: 529. 10.1038/s41467-020-20573-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi N, Shleizer-Burko S, Yanicky R, Gymrek M. 2019. Profiling the genome-wide landscape of tandem repeat expansions. Nucleic Acids Res 47: e90. 10.1093/nar/gkz501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi N, Margoliash J, Pusarla N, Saini S, Yanicky R, Gymrek M. 2021. TRTools: a toolkit for genome-wide analysis of tandem repeats. Bioinformatics 37: 731–733. 10.1093/bioinformatics/btaa736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neueder A, Landles C, Ghosh R, Howland D, Myers RH, Faull RLM, Tabrizi SJ, Bates GP. 2017. The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington's disease patients. Sci Rep 7: 1307. 10.1038/s41598-017-01510-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hearn EE, Hwang HS, Holmes SE, Rudnicki DD, Chung DW, Seixas AI, Cohen RL, Ross CA, Trojanowski JQ, Pletnikova O, et al. 2015. Neuropathology and cellular pathogenesis of spinocerebellar ataxia type 12. Mov Disord 30: 1813–1824. 10.1002/mds.26348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. 2016. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32: 1479–1485. 10.1093/bioinformatics/btv722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco A, Berger R, Freedman R, Law AJ. 2019. A VNTR regulates miR-137 expression through novel alternative splicing and contributes to risk for schizophrenia. Sci Rep 9: 11793. 10.1038/s41598-019-48141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Shakkottai VG, Clark HB, Orr HT. 2017. Polyglutamine spinocerebellar ataxias—from genes to potential treatments. Nat Rev Neurosci 18: 613–626. 10.1038/nrn.2017.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva F, Giulietti M, Nocchi L, Principato G. 2009. SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics 25: 1211–1213. 10.1093/bioinformatics/btp124 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Ferguson SM. 2019. Pleiotropic requirements for human TDP-43 in the regulation of cell and organelle homeostasis. Life Sci Alliance 2: e201900358. 10.26508/lsa.201900358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RL, Roos RA, Howland D, et al. 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci 110: 2366–2370. 10.1073/pnas.1221891110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AJ, Chiang C, Hall IM. 2021. Structural variants are a major source of gene expression differences in humans and often affect multiple nearby genes. Genome Res 31: 2249–2257. 10.1101/gr.275488.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slišković I, Eich H, Müller-McNicoll M. 2022. Exploring the multifunctionality of SR proteins. Biochem Soc Trans 50: 187–198. 10.1042/BST20210325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Heger A, Sudbery I. 2017. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res 27: 491–499. 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Parts L, Piipari M, Winn J, Durbin R. 2012. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7: 500–507. 10.1038/nprot.2011.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznajder ŁJ, Thomas JD, Carrell EM, Reid T, McFarland KN, Cleary JD, Oliveira R, Nutter CA, Bhatt K, Sobczak K, et al. 2018. Intron retention induced by microsatellite expansions as a disease biomarker. Proc Natl Acad Sci 115: 4234–4239. 10.1073/pnas.1716617115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata A, Matsumoto N, Kato T. 2017. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat Commun 8: 14519. 10.1038/ncomms14519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Kirkness EF, Lippert C, Biggs WH, Fabani M, Guzman E, Ramakrishnan S, Lavrenko V, Kakaradov B, Hou C, et al. 2017. Profiling of short-tandem-repeat disease alleles in 12,632 human whole genomes. Am J Hum Genet 101: 700–715. 10.1016/j.ajhg.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Župunski V, et al. 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458. 10.1038/nn.2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran VK, Takeshima Y, Zhang Z, Yagi M, Nishiyama A, Habara Y, Matsuo M. 2006. Splicing analysis disclosed a determinant single nucleotide for exon skipping caused by a novel intraexonic four-nucleotide deletion in the dystrophin gene. J Med Genet 43: 924–930. 10.1136/jmg.2006.042317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost B, Engchuan W, Nguyen CM, Thiruvahindrapuram B, Dolzhenko E, Backstrom I, Mirceta M, Mojarad BA, Yin Y, Dov A, et al. 2020. Genome-wide detection of tandem DNA repeats that are expanded in autism. Nature 586: 80–86. 10.1038/s41586-020-2579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RL, Ramaswami G, Hartl C, Mancuso N, Gandal MJ, de la Torre-Ubieta L, Pasaniuc B, Stein JL, Geschwind DH. 2019. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 179: 750–771.e22. 10.1016/j.cell.2019.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Gao L, Lafyatis R, Stamm S, Andreadis A. 2005. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c·SRp55 complex that either recruits or antagonizes htra2β1. J Biol Chem 280: 14230–14239. 10.1074/jbc.M413846200 [DOI] [PubMed] [Google Scholar]
- Wang ET, Treacy D, Eichinger K, Struck A, Estabrook J, Olafson H, Wang TT, Bhatt K, Westbrook T, Sedehizadeh S, et al. 2019. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum Mol Genet 28: 1312–1321. 10.1093/hmg/ddy432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watase K, Barrett CF, Miyazaki T, Ishiguro T, Ishikawa K, Hu Y, Unno T, Sun Y, Kasai S, Watanabe M, et al. 2008. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci 105: 11987–11992. 10.1073/pnas.0804350105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Deng J, Guo X, Shan J, Luan X, Cao L, Zhao J, Yu M, Zhang W, Lv H, et al. 2021. The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. Brain 144: 1819–1832. 10.1093/brain/awab077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Xu G, Yang YT, Xu Z, Chen X, Shi B, Xie D, Lu ZJ, Wang P. 2019. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res 47: D203–D211. 10.1093/nar/gky830 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.