Abstract
Oxidoreductases catalyze oxidation–reduction reactions and comprise a very large and diverse group of enzymes, which can be subclassified depending on the catalytic mechanisms of the enzymes. One of the most prominent oxidative modifications in proteins is carbonylation, which involves the formation of aldehyde and keto groups in the side chain of lysines. This modification can alter the local macromolecular structure of proteins, thereby regulating their function, stability, and/or localization, as well as the nature of any protein–protein and/or protein–nucleic acid interactions. In this review, we focus on copper‐dependent amine oxidases, which catalyze oxidative deamination of amines to aldehydes. In particular, we discuss oxidation reactions that involve lysine residues and that are regulated by members of the lysyl oxidase (LOX) family of proteins. We summarize what is known about the newly identified substrates and how this posttranslational modification regulates protein function in different contexts.
Keywords: histones, LOX, LOXL, lysine, oxidation, posttranslational modifications, transcription factor
This review focuses on oxidation reactions regulated by members of the lysyl oxidase (LOX) family of proteins. We highlight newly identified substrates and how this posttranslational modification regulates protein function in different contexts. We also summarize several strategies currently used to identify protein oxidation and study its biological relevance.
Abbreviations
- ARP
aldehyde‐reactive probe
- ATR‐FTIR
attenuated total reflection‐Fourier transform infrared spectroscopy
- bFGF
fibroblast growth factor
- CAO
copper amine oxidase
- CBP
CREB‐binding protein
- CDH1
E‐cadherin gene
- CID
collision‐induced dissociation
- DDR
DNA damage repair pathway
- DNP
2,4‐dinitrophenol
- DNPH
2,4‐dinitrophenylhydrazine
- ECM
extracellular matrix
- EMT
epithelial‐to‐mesenchymal transition
- FADs
flavin adenine dinucleotides
- GRP
Girard’s P reagent
- GRT
Girard’s T reagent
- H3K4ox
oxidized histone H3
- LOX
lysine oxidase
- LOXL
lysine oxidase like
- LTQ
lysine tyrosylquinone
- MEF2
myocyte enhancer factor 2
- MS
mass spectrometry
- NAD+
nicotinamide adenine dinucleotide
- TAF
transcriptional enhancer factor
- TAF10
TBP‐associated factor 10
- TFIID
general transcription factor IID
- TNBC
triple‐negative breast cancer cells
- TPQ
2,4,5‐trihydroxyphenylalanine quinone
- VGLL3
vestigial-like 3
Introduction
Protein function is often regulated by selective posttranslational modifications, which are typically catalyzed by substrate‐specific enzymes. Many enzymes have common names that are not very informative; thus, an international classification system was established with six classes of enzymes: oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases. Oxidation was originally considered to be mainly an uncontrolled, nonenzymatic process mediated by free radicals and nonradical oxidants. However, many oxidation processes are enzymatically controlled and occur with specificity at the enzyme‐selected residues. The enzymes responsible for this modification are classified as oxidoreductases, which catalyze oxidation–reduction reactions by proton‐coupled electron transfer or hydride transfer. Oxidoreductases constitute a very large and diverse group, with different subcategories that reflect the catalytic mechanisms of the enzymes. Oxidases play a pivotal role in cell metabolism; they catalyze a large variety of oxidation reactions (including oxidation of amines and alcohols) and are responsible for oxidative ring closure and oxidative decarboxylation [1].
In this minireview, we will focus on copper‐dependent amine oxidases that catalyze oxidative deamination of amines to aldehydes, and in particular, on oxidation reactions that involve lysine residues and are regulated by members of the lysyl oxidase (LOX) family of proteins. (Note that there are excellent reviews that discuss in‐depth each separate topic, e.g., copper‐dependent amine oxidases or the LOX family of proteins) [2, 3, 4, 5]. Briefly, the copper amine oxidase (CAO) class of enzymes catalyzes the oxidative deamination of primary amines to aldehydes, generating hydrogen peroxide and ammonia as by‐products [2]. Many redox enzymes bind to cofactors that act as electron sinks (such as nicotinamide adenine dinucleotide [NAD+] or flavin adenine dinucleotides [FADs]), by accepting the electrons removed from the substrate. In contrast, CAOs deaminate primary amines using a different mechanism using a covalently linked quinone cofactor, which can be either 2,4,5‐trihydroxyphenylalanine quinone (TPQ) or lysine tyrosylquinone (LTQ). Precursor residues for these quinone cofactors are tyrosine for TPQ, and tyrosine and lysine for LTQ. The generation of both TPQ and LTQ is a self‐catalyzed process derived from posttranslational modification of a conserved tyrosine residue in the active side and also requires oxygen (O2) and a tightly bound copper ion (Cu2+). Based on the nature of these organic cofactors, two nonhomologous subgroups of copper amine oxidases can be distinguished: (a) TPQ‐dependent CAOs (herein, CAOs); and (b) LOXs, which are LTQ‐dependent lysyl oxidases.
The LOX family of proteins comprises five different amine oxidase enzymes, namely, LOX and four LOX‐like (LOXL) proteins (LOXL1–4), all of which are copper‐ and quinone‐dependent and catalyze the oxidation of the amino group in lysines. All members of the LOX family share a highly conserved carboxyl (C)‐terminal domain that contains the elements required for its catalytic activity: a His‐X‐His‐X‐His copper‐binding motif and residues for the formation of the lysine tyrosylquinone (LTQ) cofactor. A cytokine receptor‐like domain (CRL) is found in this part of the protein; however, its function and importance in the catalytic activity remains elusive. In contrast, the amino (N)‐terminal regions are highly variable among the members of the family [4, 5]. For instance, LOX and LOXL1 proteins contain prosequences and are initially secreted as inactive proenzymes; they are then activated extracellularly by cleavage of the prosequence by metalloproteinases, such as bone morphogenetic protein 1 (BMP‐1) [6, 7]. Furthermore, LOXL2, LOXL3, and LOXL4 contain four scavenger receptor cysteine‐rich (SRCR) domains, which appear to have a role in protein–protein interactions (as seen for other proteins) and have also been suggested to be catalytic domains that regulate protein deacetylation or deacetylimination [4, 8, 9].
LOX proteins and their involvement in biological functions have been widely studied for decades [10, 11, 12, 13, 14, 15]. Oxidation of proteins by the LOX family was first described to be involved in the covalent crosslinking of collagen and elastin fibers in the extracellular matrix (ECM), which is required for maintaining the tensile strength and structural integrity of many tissues [16, 17, 18]. Several studies in the recent years have revealed that lysine oxidation is more abundant than what we initially thought and regulates many proteins and not only the originally described extracellular substrates. Notably, LOX enzymes can posttranslationally modify a variety of cationic proteins, including intracellular and even nuclear proteins [8, 10, 11, 19, 20, 21].
In this review, we will highlight new substrates and functions of oxidation which are still unresolved or less studied, such as oxidation of histones and transcription factors.
Histone oxidation: A new histone posttranslational modification
Histones are basic proteins found in eukaryotic cell nuclei that pack DNA into the nucleosome structural units. Importantly, they contain an N‐terminal tail that protrudes away from the nucleosome core and contains many positively charged lysine and arginine residues, which can be posttranslationally modified [22]. Notably, lysine residues in histones can be oxidized by the LOX family of proteins. The presence of this peptidyl aldehyde in histones changes their macromolecular status and is particularly relevant for gene regulation, both by directly affecting chromatin structure and by recruiting effector proteins [8] (Fig. 1).
Fig. 1.
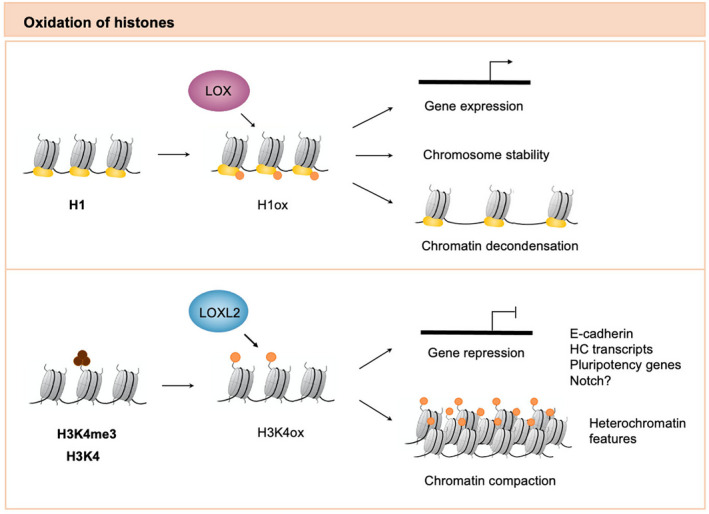
Oxidation of histone proteins by lysyl oxidases. Lysine residues in histones can be oxidized by the LOX family of proteins. Histone H1 interacts with the catalytic region of the LOX protein and can be oxidized by this enzyme. LOX activity on histone H1 has been described to regulate chromosome stability, chromatin decondensation, and gene expression. On the other hand, LOXL2 oxidizes H3K4me3 as well as (to a lesser extent) unmethylated H3. The resulting peptidyl aldehyde is referred to as oxidized histone H3 (H3K4ox), and this histone modification is important for regulating both gene repression and heterochromatin compaction. H1, histone H1; H1ox, oxidized histone H1; H3, histone H3; H3K4me4, trimethylated histone H3; H3K4ox, oxidized histone H3.
Oxidation of histone H1: A role in chromatin decondensation
The lysine‐rich histone H1 binds to the entry or exit sites of the DNA on the surface of the nucleosome core particle and facilitates three‐dimensional (3D) folding of chromatin fibers. Notably, histone H1 interacts with the catalytic region of LOX protein and can be oxidized by this enzyme. In fact, H1 shares some regions of homology with the enzymatically modified regions of the well‐known LOX substrate tropoelastin, which are lysine‐rich and contain the amino acid sequence AKAAAKAAAKA (or a part thereof) [23]. Importantly, deamination of lysine residues in histone H1 has been related to several effects in chromatin structure. First, this modification promotes the loss of positive charges in histone H1, thereby weakening/loosening its interaction with DNA [24]. Second, similar to other LOX substrates, it is likely to promote intra‐ and intermolecular crosslinks of histone H1 molecules. Third, H1 oxidation could be an alternative modification to other ones that have a similar effect on chromatin. It has been suggested that oxidized histone H1 hinders access to the nearby serine residues, preventing them from being phosphorylated; this histone modification would then cause H1 to detach from DNA and allows greater accessibility to DNA of enzymes and transcription factors [25].
In agreement with these effects, histone H1 oxidation by LOX protein has been found to regulate chromosome stability and chromosome condensation. For instance, LOX overexpression in COS‐7 cells leads to chromatin decondensation [24], which decreases cell proliferation and increases cell death preceded by multinucleation. Moreover, LOX has an important role in the regulation of the mammary tumor virus (MMTV) promoter, which depends on glucocorticoids and the phosphorylation status of histone H1 [25]. Overexpression of human recombinant LOX in MCF‐7 cells triggers MMTV activity, both in the presence and in the absence of glucocorticoids. Thus, it has been suggested that H1 oxidation leads to the removal of histone H1 from the promoter, leading to consequent changes in chromatin structure that allow access to the activating transcription factors. Finally, it is worth mentioning that histone H2 (H2b) can also interact with LOX, but through a different (but perhaps overlapping) region than H1. It has been suggested that histone H2 is important for orientating LOX toward its substrate H1, making the catalytic region available for the interaction with H1 [23]. Whether histone H2 can also be oxidized by LOX is still not known.
Oxidation of histone H3: A role in gene repression and chromatin compaction
Histone H3 has also been described as a substrate for oxidation. In this case, LOXL2 is the histone‐modifying enzyme, and the generated peptidyl aldehyde is referred as oxidized histone H3 (H3K4ox). In particular, LOXL2 has specificity for H3K4me3, but not for other lysine residues of the histone tail, such as H3K4me1/me2, H3K9me3, or H3K27me3, as demonstrated both in vitro and in vivo [26]. In addition, unpublished data from our laboratory show that recombinant LOXL2 can also oxidize unmethylated H3 in vitro, although to a lesser extent [8]. The reaction mechanism proposed by which LOXL2 is able to oxidize a tri‐methyl lysine involves the formation of an alcohol via deamination and subsequent oxidation of the alcohol. However, this mechanism is unprecedented for any LOX or CAO family members, and much work remains to be done to demonstrate its feasibility. Although in vitro reactions with recombinant LOXL2 and H3 peptide incubation have demonstrated by infrared spectrometry and mass spectrometry that an aldehyde group is formed, we cannot rule out that the in vivo reaction requires a demethylase to first remove the tri‐methyl group located in lysine 4 before LOXL2 reaction can occur.
Finally, it is highly likely that oxidized histone H3 is not a static modification, as the generation of an aldehyde group is highly reactive. Thus, maintenance of high levels of H3K4ox could be an active process that involves a constant exchange for an unmodified H3 and subsequent oxidation. The H3K4ox histone posttranslational modification has now been linked to gene repression in several contexts. First, it is required to downregulate the E‐cadherin gene (CDH1) at the onset of the epithelial‐to‐mesenchymal transition (EMT), when LOXL2 is recruited to the promoter of this gene together with the SNAIL1 transcription factor [26, 27]. Second, it is incorporated into the promoters of the pluripotency genes Nanog, Sox2, Klf4, and Oct4 and represses their expression during differentiation of embryonic stem cells into neural progenitors [28]. Third, H3K4ox has been suggested to repress NOTCH1 expression in the skin, thereby promoting squamous cell carcinoma progression (even though the levels of H3K4ox are still to be analyzed). Specifically, LOXL2 binds the NOTCH1 promoter and regulates demethylation of H3K4me3 in the proximal region, thereby reducing RNA polymerase II recruitment and the expression of this gene [29].
LOXL2 oxidation of histone H3 is not restricted to H3K4me3‐enriched regions: There is also strong evidence that H3K4ox plays crucial roles in heterochromatin regulation [30]. For instance, it is required for the downregulation of pericentromeric heterochromatin transcripts (major satellites) during EMT [31]. Further, it helps to maintain heterochromatin compaction in triple‐negative breast cancer cells (TNBC), which have particularly high levels of LOXL2 and H3K4ox. In these cells, H3K4ox is mainly located in heterochromatin; thus, while it is not directly involved in regulating gene expression in these regions, it is crucial for heterochromatin integrity and chromatin compaction. In addition, maintenance of these H3K4ox‐enriched heterochromatin domains is required for the oncogenic properties of TNBC cells as well as to protect them from an aberrant activation of the DNA damage repair pathway (DDR) [30].
Beyond histones: Oxidation of nonhistone proteins
In recent years, the LOX family of oxidases has been described to deaminate other intracellular substrates apart from histone proteins, affecting protein function, localization, stability, and/or interactions; thus, this modification is important for regulating different biological processes (Fig. 2). In Iturbide et al [28], a list of putative LOXL2 substrates was published, most of which await further validation. In this proteomic approach, 117 candidates were obtained, including previous identified LOX and LOXL2 substrates, such as H1 and H3. Surprisingly, 20 proteins (representing 17% of the total candidates) showed one or more putative SET7/9 histone methyltransferase recognition sequences [32], suggesting that the SET7/9 histone methyltransferase and LOX2 may share the same motif. As discussed above, H1 is a confirmed LOX substrate [23, 25, 33], with several methylation sites described for H1 [34], again reinforcing the idea the LOXL2 could recognize the same lysine methylation domain. It would be interesting to investigate how LOX and LOXL2, which have different sequences, could have the same substrate.
Fig. 2.

Oxidation of nonhistone proteins by lysyl oxidases. LOX oxidases can deaminate several intracellular substrates, allowing them to regulate protein function, localization, stability, and/or interactions. (i) TAF10, a member of the TFIID complex, can be oxidized by LOXL2, which alters its interaction with other members of the TFIID complex and leads to its release from pluripotency gene promoters in embryonic stem cells. (ii) LOXL3 deacetylates and deacetyliminates the transcription factor STAT3 on multiple acetyl‐lysine sites, disrupting its dimerization and inhibiting its transcriptional activity. (iii) Several studies suggest that the transcription factor SNAIL1 can be oxidized by LOXL2/3; this posttranslational modification could lead to an undefined conformational change that would protect SNAIL1 from nuclear export and/or proteasomal degradation. In addition, LOXL2 could also oxidize acetylated SNAIL1, thereby regulating its interactions with corepressors and expression of its target genes. (v) LOXL2/3 can be auto‐oxidized as a way to regulate its own enzymatic activity in response to treatment with trihydrophenolics. (v) The bFGF is oxidized by LOX, resulting in spontaneous and covalent crosslinking of bFGF monomers and the inhibition of its nuclear localization and mitogenic potential through ERK1/2MAP kinase activation. (vi) The transcriptional coactivator VGLL3 is oxidized by cytoplasmic LOX in myogenic progenitors; this modification promotes the translocation of VGLL3 into the nucleus, where it binds MEF2 and TEF and modulates large gene expression programs.
Here, we presented a list of confirmed LOX‐ and LOXL‐oxidized substrates:
TAF10
TBP‐associated factor 10 (TAF10) is a member of the general transcription factor IID (TFIID), a key complex in the regulation of gene transcription [35]. This subunit undergoes lysine methylation in vivo [36], and trimethylated TAF10 is a LOXL2 substrate. Mass spectrometry sequencing of TAF10 showed several oxidized lysine residues, but the specific lysine residues oxidized by LOXL2 remain to be determined [28]. Importantly, TAF10 methylation is restricted to the histone‐fold domain, and similarly to oxidation on H3, LOXL2 can oxidized unmodified TAF10 but to a lesser extent as compared to modified TAF10. TAF10 oxidation alters its interactions with other members of the TFIID complex and leads to its release from promoters, thereby blocking TFIID‐dependent gene transcription. This process is crucial for repressing pluripotency genes and is required for maintaining the pluripotent capacity of embryonic stem cells as well as the proper differentiation of these cells into neural progenitor cells [28].
STAT3
Another protein regulated by oxidation is the transcription factor STAT3. Ma et al. described that LOXL3 interacts with STAT3 in the nucleus, as a dual‐specificity enzyme that regulates both its deacetylation and deacetylimination on multiple acetyl‐lysine sites [9]. Interestingly, the authors showed that LOXL3 preferentially catalyzes acetyl‐STAT3 deacetylimination, rather than STAT3 deamination, suggesting that at least this LOX family member could prefer peptidyl acetyl‐lysine residues over peptidyl lysine residues as substrates.
LOXL3 activity on acetylated STAT3 alters modular protein interactions, disrupting STAT3 dimerization and inhibiting its transcriptional activity. Moreover, since LOXL3 has this dual role in regulating both deacetylation and deacetylimination, it might have a stronger or more efficient effect on STAT3 transcriptional activity than other protein deacetylases, such as HDAC or SIRT family members. Oxidation and deactivation of STAT3 by LOXL3 plays a critical role in regulating its functions, including on cell proliferation of both normal and cancer cells, cell differentiation of inflammatory Th17 and Treg upon stimulation, and production of inflammatory cytokines and chemokines such as interleukin (IL)‐6. It is also worth mentioning that, in this context, LOXL3 activity does not depend on the well‐described and highly conserved C‐terminal oxidase catalytic domain of LOX family members. Rather, the major deacetylase/deacetyliminase activities depend on the N‐terminal SRCR repeats, which are closely related in evolution to the C‐terminal domain but are functionally less well‐understood. This study therefore suggests that, similar to enzymes such as HDAC6 [37], LOX proteins could share two tandem catalytic activities that are important for regulating different modifications and/or cell substrates.
SNAIL1
The transcription factor SNAIL1 has a pivotal role in EMT: It interacts with and collaborates with corepressors, such as LOXL2, to regulate the downregulation of the E‐cadherin gene during this process. Notably, several studies have suggested that, apart from its role as a histone‐modifying enzyme, LOXL2 can also oxidize SNAIL1 and that this modification may be important for regulating the stability and function of SNAIL1. Peinado et al. demonstrated that SNAIL1 stability depends on Lys‐98 and/or Lys‐137 in its N‐terminal domain, residues that seem to be substrates for oxidation by LOXL2 [27]. Importantly, this posttranslational modification could lead to an undefined conformational change that would protect SNAIL1 from nuclear export and/or phosphorylation by GSK3β [38, 39], which leads to ubiquitinylation and proteasomal degradation. Along the same line, other authors also demonstrated that nuclear‐associated LOXL2 contributes to stabilizing SNAIL1 in an oxidase‐dependent reaction [40].
It has been also suggested that LOXL2 could oxidize acetylated SNAIL1 by removing acetylamine from acetyl‐lysine residues (deacetylimination) [9]. In fact, overexpression of LOXL2 in HEK293T cells leads to a decrease in the global levels of acetylated SNAIL1 [9]. Additionally, SNAIL1 deacetylation could be important for regulating its activity, although the specific residues that are deacetylated have not yet been determined. Notably, the function of SNAIL2 as a transcriptional activator is also regulated by acetylation of the CREB‐binding protein (CBP) at lysines 146 and 187 [41]. Thus, deacetylimination of SNAIL1 could be an important regulatory mechanism to avoid this function and allow its interaction with corepressors [42].
VGLL3
Yehezkely et al. found that the transcriptional coactivator vestigial‐like 3 (VGLL3) is oxidized by cytoplasmic LOX in myogenic progenitors [43]. This coactivator plays a crucial role in myogenic differentiation, regulating the activation/upregulation of the myocyte enhancer factor 2 (MEF2) and transcriptional enhancer factor (TAF) target genes [44]. Oxidation of VGLL3 promotes its translocation into the nucleus, where it binds to MEF2 and TEF and modulates large gene expression programs. This study demonstrates that LOX function in muscle regeneration is more extensive than its previously described role in modifying the ECM [43, 45]. The authors propose a model in which LOX plays a dual role both extracellularly and intracellularly, regulating ECM maturation and muscle progenitor cell differentiation, respectively. In this way, LOX activity is crucial to maintain homeostasis between distinct muscle components during muscle regeneration.
Moreover, in myogenic progenitors, this posttranslational modification is likely to be not only restricted to VGLL3. For instance, using a similar mechanism, LOX oxidation could also regulate proteins that conserve one lysine residue with VGLL3, such as VGLL2.
bFGF2
Another LOX substrate is the basic fibroblast growth factor (bFGF), a heparin‐binding polypeptide that regulates proliferation, differentiation, and migration of a variety of cell types. bFGF is a basic protein of 155 amino acids and contains 14 lysine residues that can be oxidized by LOX, as shown by fluorometric assays [46]. Certain substrates of this peptidyl lysine are accessible at the protein surface and are known to act as heparin‐binding sites, whereas others are located at the interface of two interacting bFGF molecules [46, 47, 48].
Oxidation of these lysine residues alters their cationic nature to nonionic aldehyde functions, resulting in spontaneous and covalent crosslinking of bFGF monomers to form dimers and higher‐order oligomers. In this way, this posttranslational modification blocks the roles of epsilon‐amino groups of bFGF and dramatically alters its biological properties. In Swiss 3T3 cells, LOX activity has an antiproliferative effect inhibiting both the nuclear localization of bFGF and its mitogenic potential through ERK1/2MAP kinase activation [46].
LOXL2
In an interesting twist, LOXL2/3 might also be auto‐oxidized as a way regulating its own function [49]. In particular, this auto‐oxidation can be induced by treatment with trihydrophenolics. These compounds promote LOXL2/3 auto‐oxidation at a specific lysine residue (K731), which irreversibly inhibits LOXL2 enzymatic activity. In this reaction, the trihydrophenolic is also converted to a previously undescribed metabolite that directly inhibits TβRI kinase. This combined inhibition of LOXL2 and TβRI activities by trihydrophenolics potently blocks TGF‐β1 responses, SNAIL1 expression, and collagen deposition in vivo in models of pulmonary fibrosis and collagen‐dependent lung cancer metastasis [46]. Although this oxidation is not due to its catalytic activity, it is important to highlight it, as it distinguishes this inhibition mechanism from previously described ones for other LOXL2 inhibitors and general LOX inhibitors [12].
Identification of oxidized proteins
As further studies are required to elucidate the biological relevance of lysine oxidation, one important point that we would like to highlight is how these oxidize proteins can be identified. Several strategies are currently used to study protein oxidation and the biological functions of this modification (Fig. 3).
Fig. 3.
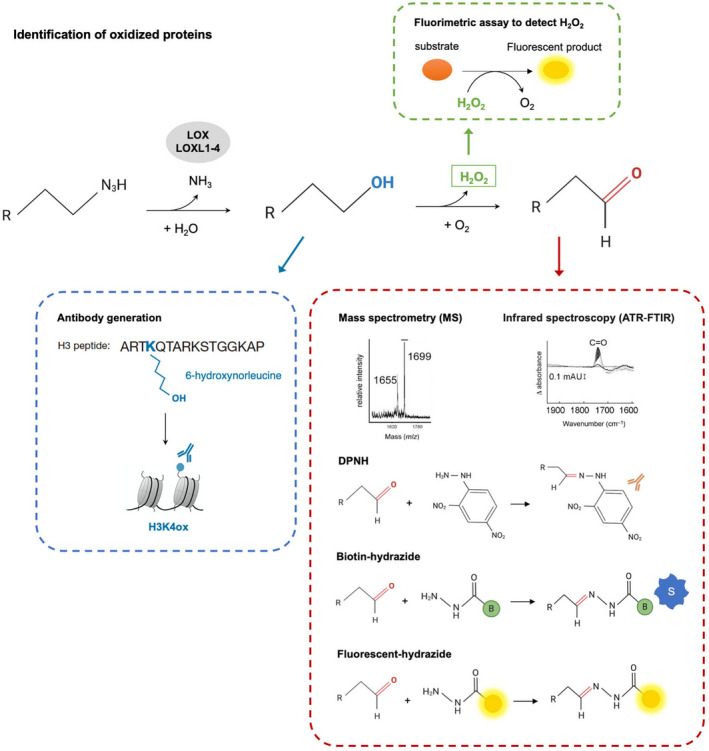
Identification of oxidized proteins. Aldehyde groups can be identified using enzymatic methods that rely on chemical derivatization of carbonyls with hydrazine, such as DNPH, biotin‐hydrazide, and hydrazides coupled to fluorescent molecules. Aldehyde groups can also be identified using amino acid analyzers, such as ATR‐FTIR and MS, with or without previous chemical derivatization. Oxidation can also be monitored by analyzing H202, one of the products of the reaction that is generated from the reoxidation of an internal cofactor; this compound can be analyzed with fluorometric assays. Finally, and considering that aldehyde groups are highly reactive, modification‐specific antibodies can be developed for the primary alcohol in the oxidation reaction and used as a readout of the final aldehyde.
First, some biochemical and analytical methods can specifically detect the formation/presence of aldehydes in proteins, such as by using enzymatic methods that rely on chemical derivatization of carbonyls with hydrazines and hydrazides [50]. These techniques are simple, easy to handle, and allow a large number of samples to be assayed in a short time. Many different carbonyl‐tagging reagents exist; the most widely used ones identify carbonylation in proteins and peptides [51, 52]. One of the first described methods was the use of 2,4‐dinitrophenylhydrazine (DNPH), a compound that reacts with aldehydes and ketones to generate dinitrophenylhydrazones [53, 54]. With this approach, the resulting hydrazone can be detected by spectrophotometry (as it has a strong absorbance at 375 nm) or via immunochemical techniques using an antibody specific for 2,4‐dinitrophenol (DNP), the derivative coupled to protein carbonyls. However, this technique often has a high level of nonspecific background noise and contaminating endogenous immunoglobulins, limiting its use when applied to tissue extracts from mammalian sources. To circumvent these problems, other chemically reactive compounds have been developed. For instance, biotin‐hydrazide is an activated biotin that selectively reacts with carbonyl groups; using it, labeled proteins can be detected using biotin–streptavidin/avidin affinity methods [55, 56]. As this approach is not affected by endogenous immunoglobulins, it is sensitive for detecting biotinylated proteins, with low background signals [26]. Alternatively, hydrazides can also be coupled to fluorescent molecules, such as fluorescein thiosemicarbazide, coumarin hydrazine, BODIPY FL‐hydrazide, and cyanine dyes (among others), allowing protein carbonyl groups to be detected by fluorometric assays [56, 57, 58]; these assays are widely used in gel‐based proteomics to directly detect protein carbonyls without immunoblotting.
Aldehyde groups can also be identified using amino acid analyzers, such as attenuated total reflection‐Fourier transform infrared spectroscopy (ATR‐FTIR) and mass spectrometry (MS) [26, 59]. These methods are very accurate but might be time‐consuming and usually require laborious treatment of samples. With ATR‐FTIR, aldehyde groups are detected based on the absorbance spectra of molecules, as each chemical group shows characteristic frequencies [59]. With MS, protein analysis is based on their conversion into gaseous ions and the characterization of mass‐to‐charge ratios (m/z) [60]. However, identification of allysine residues on peptides and proteins by MS can be challenging for various reasons. First, it requires high sensitivity and resolution equipment. Some oxidative modifications result in a very small, subtle mass shift (e.g., deamination of lysines is only 1 Da) that cannot be distinguished by fast‐scanning mass analyzers [60]. In addition, carbonylation is normally distributed at very low levels in complex biological samples, such as in cell or plasma proteomes [61, 62]. Second, the loss of positive charges from lysine residues after oxidation can reduce ionization efficiency and impair detection sensitivity for mass spectrometry analysis. Third, aldehyde groups may react with primary amines during peptide mapping and sample digestion process, complicating the subsequent analysis [63].
To address these challenges, some analytical methods use chemical derivatization prior to peptide mapping by MS to protect the carbonyl groups. This makes it possible to enrich the low‐abundance‐modified peptides or proteins, and in some cases, even to identify carbonylation sites, without the need for affinity enrichment or fractionation steps [60]. To date, different hydrazide reagents have been used in combination with MS to successfully identify carbonylated proteins, including biotin‐hydrazide, aldehyde‐reactive probe (ARP), Girard’s P reagent (GRP), and Girard’s T reagent (GRT) [51, 63, 64, 65, 66]. GRT is perhaps the one that is the most suitable for this approach: It is a small compound, has a relatively simple chemical structure (giving it good accessibility to carbonylation sites), and shows little self‐fragmentation of the tagged proteins/peptides during collision‐induced dissociation (CID) experiments. In addition, GRT adds a positive charge to the derived peptides, thereby increasing ionization efficiency, and it has high solubility in water, which allows higher reagent‐to‐protein molar ratio for derivatization and improves the solubility of the resulting peptides [63].
Beyond detection of aldehyde groups, oxidation can also be monitored by analyzing the presence of hydrogen peroxide (H202), one of the by‐products of the reaction. This compound can be detected using fluorometric assays in which samples are incubated with a substrate that reacts with H202 and generates a fluorescent product detectable by fluorometry or spectrophotometry [9, 26, 46, 67]. This method has high sensitivity, a high reproducibility level, is fast, and is easy to handle. In addition, H202 production in oxidation reactions can be studied either at a specific timepoint or continuously over time.
Finally, modification‐specific antibodies can also be generated to study oxidized proteins. However, due to the high reactivity of the aldehyde group, it is unfit for immunochemical studies and cannot be used as an immunogen. Alternatively, antibodies can be generated for the primary alcohol in the oxidation reaction; this intermediate provides a similar oxygen‐bearing functionality but is less reactive than the aldehyde group. Notably, such antibodies can be used for a readout of oxidation as well as for the final aldehyde. This strategy has already been used to develop an antibody for the histone modification H3K4ox. In this case, the antibody was obtained by immunizing with a synthetic H3 peptide that contains an artificial amino acid in position 4, giving it resemblance to the intermediate alcohol in LOXL2 reaction (termed 6‐hydroxy‐norleucine). Biochemical experiments with recombinant LOXL2 and nucleosomes have demonstrated that this intermediate alcohol is relatively stable (with a half‐life of about 2 h) and that it can be used as a readout of H3K4ox [30]. In addition, this antibody is highly specific for the H3K4ox peptide, with very low cross‐reactivity for unmodified H3 and no cross‐reactivity with other histone modifications, such as H3K9me3 or H3K4me3 [30].
Conclusions and future perspectives
Some structural and functional features of lysyl oxidase proteins are now clear, but much work remains to be done in order to fully understand the implications of these enzymes. We are now recognizing the role of oxidation as a posttranslational modification. Indeed, besides the classical ECM targets, these enzymes are also able to oxidize histones and transcription factors, implicating them in the regulation of crucial biological processes, including development, differentiation, proliferation, and cancer. Numerous novel and repurposed techniques will facilitate the detection of this highly reactive modification, allowing us to address the many questions about it that remain to be elucidated: to which extent are proteins oxidized in general? Why are the majority of the identified lysine substrates posttranslationally modified? Do specific domains for lysine recognition exist? Are allysines rapidly removed from the substrates? Do they serve as docking sites for other proteins? Are lysine oxidation levels altered in pathological situations, such as cancer? Addressing these open questions is likely to reveal new aspects about how posttranslational modification can regulate protein function.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
G Serra‐Bardenys and S Peiró prepared manuscript, wrote the article, and involved in discussion.
Acknowledgments
We would like to V.A. Raker for manuscript editing. This work was supported by grants from Instituto de Salud Carlos III (ISCIII) FIS/FEDER (PI12/01250; CP08/00223; PI16/00253 and CB16/12/00449), MINECO, and FPU14/04071 to G.S. We thank the FERO Foundation, La Caixa Foundation for financial support (LCF/PR/PR12/51070001), and Cellex Foundation for providing research facilities and equipment.
References
- 1. Lewis T & Stone W (2021) Biochemistry, Proteins Enzymes. [Updated 2021 May 4]. In StatPearls [Internet]. StatPearls Publishing, Treasure Island, FL. Available from: https://www.ncbi.nlm.nih.gov/books/NBK554481/ [PubMed] [Google Scholar]
- 2. Finney J, Moon HJ, Ronnebaum T, Lantz M & Mure M (2014) Human copper‐dependent amine oxidases. Arch Biochem Biophys 546, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lucero HA & Kagan HM (2006) Lysyl oxidase: an oxidative enzyme and effector of cell function. Cell Mol Life Sci 63, 2304–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Csiszar K (2001) Lysyl oxidases: a novel multifunctional amine oxidase family. Prog Nucleic Acid Res Mol Biol 70, 1–32. [DOI] [PubMed] [Google Scholar]
- 5. Molnar J, Fong KS, He QP, Hayashi K, Kim Y, Fong SF, Fogelgren B, Szauter KM, Mink M & Csiszar K (2003) Structural and functional diversity of lysyl oxidase and the LOX‐like proteins. Biochim Biophys Acta 1647, 220–224. [DOI] [PubMed] [Google Scholar]
- 6. Borel A, Eichenberger D, Farjanel J, Kessler E, Gleyzal C, Hulmes DJ, Sommer P & Font B (2001) Lysyl oxidase‐like protein from bovine aorta. Isolation and maturation to an active form by bone morphogenetic protein‐1. J Biol Chem 276, 48944–48949. [DOI] [PubMed] [Google Scholar]
- 7. Uzel MI, Scott IC, Babakhanlou‐Chase H, Palamakumbura AH, Pappano WN, Hong HH, Greenspan DS & Trackman PC (2001) Multiple bone morphogenetic protein 1‐related mammalian metalloproteinases process pro‐lysyl oxidase at the correct physiological site and control lysyl oxidase activation in mouse embryo fibroblast cultures. J Biol Chem 276, 22537–22543. [DOI] [PubMed] [Google Scholar]
- 8. Iturbide A, Garcia de Herreros A & Peiro S (2015) A new role for LOX and LOXL2 proteins in transcription regulation. FEBS J 282, 1768–1773. [DOI] [PubMed] [Google Scholar]
- 9. Ma L, Huang C, Wang XJ, Xin DE, Wang LS, Zou QC, Zhang YS, Tan MD, Wang YM, Zhao TC et al. (2017) Lysyl oxidase 3 is a dual‐specificity enzyme involved in STAT3 deacetylation and deacetylimination modulation. Mol Cell 65, 296–309. [DOI] [PubMed] [Google Scholar]
- 10. Barker HE, Cox TR & Erler JT (2012) The rationale for targeting the LOX family in cancer. Nat Rev Cancer 12, 540–552. [DOI] [PubMed] [Google Scholar]
- 11. Laczko R & Csiszar K (2020) Lysyl oxidase (LOX): functional contributions to signaling pathways. Biomolecules 10, 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kagan HM & Li W (2003) Lysyl oxidase: properties, specificity, and biological roles inside and outside of the cell. J Cell Biochem 88, 660–672. [DOI] [PubMed] [Google Scholar]
- 13. Lin HY, Li CJ, Yang YL, Huang YH, Hsiau YT & Chu PY (2020) Roles of lysyl oxidase family members in the tumor microenvironment and progression of liver cancer. Int J Mol Sci 21, 9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei S, Gao L, Wu C, Qin F & Yuan J (2020) Role of the lysyl oxidase family in organ development (Review). Exp Ther Med 20, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martínez‐González J, Varona S, Cañes L, Galán M, Briones AM, Cachofeiro V & Rodríguez C (2019) Emerging roles of lysyl oxidases in the cardiovascular system, New Concepts and Therapeutic Challenges. Biomolecules 9, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siegel RC & Martin GR (1970) Collagen cross‐linking. Enzymatic synthesis of lysine‐derived aldehydes and the production of cross‐linked components. J Biol Chem 245, 1653–1658. [PubMed] [Google Scholar]
- 17. Siegel RC (1974) Biosynthesis of collagen crosslinks: increased activity of purified lysyl oxidase with reconstituted collagen fibrils. Proc Natl Acad Sci USA 71, 4826–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vallet SD & Ricard‐Blum S (2019) Lysyl oxidases: from enzyme activity to extracellular matrix cross‐links. Essays Biochem 63, 349–364. [DOI] [PubMed] [Google Scholar]
- 19. Wen B, Xu LY & Li EM (2020) LOXL2 in cancer: regulation, downstream effectors and novel roles. Biochim Biophys Acta Rev Cancer 1874, 188435. [DOI] [PubMed] [Google Scholar]
- 20. Ye M, Song Y, Pan S, Chu M, Wang ZW & Zhu X (2020) Evolving roles of lysyl oxidase family in tumorigenesis and cancer therapy. Pharmacol Ther 215, 107633. [DOI] [PubMed] [Google Scholar]
- 21. Moon HJ, Finney J, Ronnebaum T & Mure M (2014) Human lysyl oxidase‐like 2. Bioorg Chem 57, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Campos EI & Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43, 559–599. [DOI] [PubMed] [Google Scholar]
- 23. Giampuzzi M, Oleggini R & Di Donato A (2003) Demonstration of in vitro interaction between tumor suppressor lysyl oxidase and histones H1 and H2: definition of the regions involved. Biochim Biophys Acta 1647, 245–251. [DOI] [PubMed] [Google Scholar]
- 24. Mello ML, Alvarenga EM, Vidal BEC & Di Donato A (2011) Chromatin supraorganization, mitotic abnormalities and proliferation in cells with increased or down‐regulated lox expression: Indirect evidence of a LOX‐histone H1 interaction in vivo. Micron 42, 8–16. [DOI] [PubMed] [Google Scholar]
- 25. Oleggini R & Di Donato A (2011) Lysyl oxidase regulates MMTV promoter: indirect evidence of histone H1 involvement. Biochem Cell Biol 89, 522–532. [DOI] [PubMed] [Google Scholar]
- 26. Herranz N, Dave N, Millanes‐Romero A, Pascual‐Reguant L, Morey L, Diaz VM, Lorenz‐Fonfria V, Gutierrez‐Gallego R, Jeronimo C, Iturbide A et al. (2016) Lysyl oxidase‐like 2 (LOXL2) oxidizes trimethylated lysine 4 in histone H3. FEBS J 283, 4263–4273. [DOI] [PubMed] [Google Scholar]
- 27. Peinado H, Iglesias‐de la Cruz MC, Olmeda D, Csiszar K, Fong KS, Vega S, Nieto MA, Cano A & Portillo F (2005) A molecular role for lysyl oxidase‐like 2 enzyme in snail regulation and tumor progression. EMBO J 24, 3446–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iturbide A, Pascual‐Reguant L, Fargas L, Cebria JP, Alsina B, Garcia de Herreros A & Peiro S (2015) LOXL2 Oxidizes Methylated TAF10 and Controls TFIID‐Dependent Genes during Neural Progenitor Differentiation. Mol Cell 58, 755–766. [DOI] [PubMed] [Google Scholar]
- 29. Martin A, Salvador F, Moreno‐Bueno G, Floristan A, Ruiz‐Herguido C, Cuevas EP, Morales S, Santos V, Csiszar K, Dubus P et al. (2015) Lysyl oxidase‐like 2 represses Notch1 expression in the skin to promote squamous cell carcinoma progression. EMBO J 34, 1090–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cebrià‐Costa JP, Pascual‐Reguant L, Gonzalez‐Perez A, Serra‐Bardenys G, Querol J, Cosín M, Verde G, Cigliano RA, Sanseverino W, Segura‐Bayona S et al. (2020) LOXL2‐mediated H3K4 oxidation reduces chromatin accessibility in triple‐negative breast cancer cells. Oncogene 39, 79–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Millanes‐Romero A, Herranz N, Perrera V, Iturbide A, Loubat‐Casanovas J, Gil J, Jenuwein T, Garcia de Herreros A & Peiro S (2013) Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial‐to‐mesenchymal transition. Mol Cell 52, 746–757. [DOI] [PubMed] [Google Scholar]
- 32. Nishioka K, Chuikov S, Sarma K, Erdjument‐Bromage H, Allis CD, Tempst P & Reinberg D (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kagan HM, Williams MA, Calaman SD & Berkowitz EM (1983) Histone H1 is a substrate for lysyl oxidase and contains endogenous sodium borotritide‐reducible residues. Biochem Biophys Res Commun 115, 186–192. [DOI] [PubMed] [Google Scholar]
- 34. Wisniewski JR, Zougman A, Krüger S & Mann M (2007) Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics 6, 72–87. [DOI] [PubMed] [Google Scholar]
- 35. Papai G, Weil PA & Schultz P (2011) New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev 21, 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kouskouti A, Scheer E, Staub A, Tora L & Talianidis I (2004) Gene‐specific modulation of TAF10 function by SET9‐mediated methylation. Mol Cell 14, 175–182. [DOI] [PubMed] [Google Scholar]
- 37. Zou H, Wu Y, Navre M & Sang BC (2006) Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun 341, 45–50. [DOI] [PubMed] [Google Scholar]
- 38. Dominguez D, Montserrat‐Sentis B, Virgos‐Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C & Garcia de Herreros A (2003) Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol 23, 5078–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M & Hung MC (2004) Dual regulation of Snail by GSK‐3beta‐mediated phosphorylation in control of epithelial‐mesenchymal transition. Nat Cell Biol 6, 931–940. [DOI] [PubMed] [Google Scholar]
- 40. Moon HJ, Finney J, Xu L, Moore D, Welch DR & Mure M (2013) MCF‐7 cells expressing nuclear associated lysyl oxidase‐like 2 (LOXL2) exhibit an epithelial‐to‐mesenchymal transition (EMT) phenotype and are highly invasive in vitro. J Biol Chem 288, 30000–30008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsu DS, Wang HJ, Tai SK, Chou CH, Hsieh CH, Chiu PH, Chen NJ & Yang MH (2014) Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell 26, 534–548. [DOI] [PubMed] [Google Scholar]
- 42. Baulida J, Díaz VM & Herreros AG (2019) Snail1: A transcriptional factor controlled at multiple levels. J Clin Med 8, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gabay Yehezkely R, Zaffryar‐Eilot S, Kaganovsky A, Fainshtain Malka N, Aviram R, Livneh I & Hasson P (2020) Intracellular role for the matrix‐modifying enzyme lox in regulating transcription factor subcellular localization and activity in muscle regeneration. Dev Cell 53, 406–417. [DOI] [PubMed] [Google Scholar]
- 44. Figeac N, Mohamed AD, Sun C, Schönfelder M, Matallanas D, Garcia‐Munoz A, Missiaglia E, Collie‐Duguid E, De Mello V, Pobbati AV et al. (2019) VGLL3 operates via TEAD1, TEAD3 and TEAD4 to influence myogenesis in skeletal muscle. J Cell Sci 132, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kutchuk L, Laitala A, Soueid‐Bomgarten S, Shentzer P, Rosendahl AH, Eilot S, Grossman M, Sagi I, Sormunen R, Myllyharju J et al. (2015) Muscle composition is regulated by a Lox‐TGFβ feedback loop. Development 142, 983–993. [DOI] [PubMed] [Google Scholar]
- 46. Li W, Nugent MA, Zhao Y, Chau AN, Li SJ, Chou IN, Liu G & Kagan HM (2003) Lysyl oxidase oxidizes basic fibroblast growth factor and inactivates its mitogenic potential. J Cell Biochem 88, 152–164. [DOI] [PubMed] [Google Scholar]
- 47. Eriksson AE, Cousens LS, Weaver LH & Matthews BW (1991) Three‐dimensional structure of human basic fibroblast growth factor. Proc Natl Acad Sci USA 88, 3441–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Faham S, Hileman RE, Fromm JR, Linhardt RJ & Rees DC (1996) Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120. [DOI] [PubMed] [Google Scholar]
- 49. Wei Y, Kim TJ, Peng DH, Duan D, Gibbons DL, Yamauchi M, Jackson JR, Le Saux CJ, Calhoun C, Peters J et al. (2017) Fibroblast‐specific inhibition of TGF‐β1 signaling attenuates lung and tumor fibrosis. J Clin Invest 127, 3675–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ros J (2017) Protein Carbonylation: Principles, Analysis, and Biological Implications. Wiley. [Google Scholar]
- 51. Madian AG & Regnier FE (2010) Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res 9, 3766–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yan LJ & Forster MJ (2011) Chemical probes for analysis of carbonylated proteins: a review. J Chromatogr B Analyt Technol Biomed Life Sci 879, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thompson CA & Burcham PC (2008) Protein alkylation, transcriptional responses and cytochrome c release during acrolein toxicity in A549 cells: influence of nucleophilic culture media constituents. Toxicol in Vitro 22, 844–853. [DOI] [PubMed] [Google Scholar]
- 54. Chen D, Fang L, Li H, Tang MS & Jin C (2013) Cigarette smoke component acrolein modulates chromatin assembly by inhibiting histone acetylation. J Biol Chem 288, 21678–21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hensley K (2015) Detection of protein carbonyls by means of biotin hydrazide‐streptavidin affinity methods. Methods Mol Biol 1314, 95–100. [DOI] [PubMed] [Google Scholar]
- 56. Eldridge GM & Weiss GA (2011) Hydrazide reactive peptide tags for site‐specific protein labeling. Bioconjug Chem 22, 2143–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anderson JM (1986) Fluorescent hydrazides for the high‐performance liquid chromatographic determination of biological carbonyls. Anal Biochem 152, 146–153. [DOI] [PubMed] [Google Scholar]
- 58. Georgiou CD, Zisimopoulos D, Argyropoulou V, Kalaitzopoulou E, Ioannou PV, Salachas G & Grune T (2018) Protein carbonyl determination by a rhodamine B hydrazide‐based fluorometric assay. Redox Biol 17, 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Socrates G (2001) Infrared characteristic group frequencies. Wiley‐Interscience Publication edn, Wiley & Sons.
- 60. Fedorova M, Bollineni RC & Hoffmann R (2014) Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev 33, 79–97. [DOI] [PubMed] [Google Scholar]
- 61. Rao RS & Møller IM (2011) Pattern of occurrence and occupancy of carbonylation sites in proteins. Proteomics 11, 4166–4173. [DOI] [PubMed] [Google Scholar]
- 62. Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E & Dukan S (2009) Rules governing selective protein carbonylation. PLoS One 4, e7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang Y, Stella C, Wang W, Schöneich C & Gennaro L (2014) Characterization of oxidative carbonylation on recombinant monoclonal antibodies. Anal Chem 86, 4799–4806. [DOI] [PubMed] [Google Scholar]
- 64. Mirzaei H & Regnier F (2005) Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Anal Chem 77, 2386–2392. [DOI] [PubMed] [Google Scholar]
- 65. Temple A, Yen TY & Gronert S (2006) Identification of specific protein carbonylation sites in model oxidations of human serum albumin. J Am Soc Mass Spectrom 17, 1172–1180. [DOI] [PubMed] [Google Scholar]
- 66. Chavez JD, Bisson WH & Maier CS (2010) A targeted mass spectrometry‐based approach for the identification and characterization of proteins containing α‐aminoadipic and γ‐glutamic semialdehyde residues. Anal Bioanal Chem 398, 2905–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trackman PC, Zoski CG & Kagan HM (1981) Development of a peroxidase‐coupled fluorometric assay for lysyl oxidase. Anal Biochem 113, 336–342. [DOI] [PubMed] [Google Scholar]


