Abstract
Objective
To evaluate the diagnostic accuracy of deep learning algorithms to identify age-related macular degeneration and to explore factors impacting the results for future model training.
Methods
Diagnostic accuracy studies published in PubMed, EMBASE, the Cochrane Library, and ClinicalTrails.gov before 11 August 2022 which employed deep learning for age-related macular degeneration detection were identified and extracted by two independent researchers. Sensitivity analysis, subgroup, and meta-regression were performed by Review Manager 5.4.1, Meta-disc 1.4, and Stata 16.0. The risk of bias was assessed using QUADAS-2. The review was registered (PROSPERO CRD42022352753).
Results
The pooled sensitivity and specificity in this meta-analysis were 94% (P = 0, 95% CI 0.94–0.94, I2 = 99.7%) and 97% (P = 0, 95% CI 0.97–0.97, I2 = 99.6%), respectively. The pooled positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and the area under the curve value were 21.77(95% CI 15.49–30.59), 0.06 (95% CI 0.04–0.09), 342.41 (95% CI 210.31–557.49), and 0.9925, respectively. Meta-regression indicated that types of AMD (P = 0.1882, RDOR = 36.03) and layers of the network (P = 0.4878, RDOR = 0.74) contributed to the heterogeneity.
Conclusions
Convolutional neural networks are mostly adopted deep learning algorithms in age-related macular degeneration detection. Convolutional neural networks, especially ResNets, are effective in detecting age-related macular degeneration with high diagnostic accuracy. Types of age-related macular degeneration and layers of the network are the two essential factors that impact the model training process. Proper layers of the network will make the model more reliable. More datasets established by new diagnostic methods will be used to train deep learning models in the future, which will benefit for fundus application screening, long-range medical treatment, and reducing the workload of physicians.
Introduction
Age-related macular degeneration (AMD) is one of the leading causes of severe irreversible vision impairment in developed countries [1, 2]. With the accelerated aging process of the global population, the number of AMD patients is expected to increase to 288 million by 2040 [3], and it has become one of the key topics in the research of ophthalmic blindness prevention.
Clinically, it is classified as dry AMD (dAMD) characterized by medium-sized drusen and retinal pigmentary changes, and wet AMD (wAMD) characterized by neovascular and atrophic [4]. Fundus photography (FP) and optical coherence tomography (OCT) are the most widely used auxiliary examinations in ophthalmology. FP is the cheapest and the most necessary fundus test in AMD, which can intuitively identify lesions and diagnose AMD. OCT uses low coherence light to scan biological tissues in cross-section and converts the acquired information into numbers. After computer processing, it displays the pathological changes of each layer of the retina clearly and provides quantitative diagnostic indicators. In addition, OPTOS ultra-widefield retinal images can clearly visualize peripheral retinal lesions, and when combined with angiography, it can clearly show peripheral choroidal neovascularization (CNV) [5], and produces better pseudocolor images than conventional 45° FP in diagnosis [6]. AMD first affects the retinal pigment epithelium, Bruch’s membrane, and choroidal capillaries in the macular area. AMD can be manifested as drusen, atrophy of the outer retinal structure, CNV, polypoid lesions, and pigment epithelial detachment in OCT images.
The rapid increase in the demand for screening and follow-up of AMD means that a large number of human and financial resources need to be provided by the healthcare systems of various countries. The use of deep learning (DL) model technology may be a long-term solution for screening and monitoring patients in primary eye care settings.
The DL model is a branch of machine learning, composed of neural networks that are good at computer vision, perception, and image recognition. DL model uses multilayer nonlinear information processing modules to extract supervised or unsupervised features from a set of training data and make the correct prediction. In recent years, DL models have been widely used in ophthalmology [7–9], dermatology [10], radiology [11, 12], pathology [13, 14], and many other image-centric specialties. In ophthalmology-related research, DL models are beginning to be widely used in the diagnosis and recognition of diseases including diabetic retinopathy [15–17], AMD [15, 18–20], glaucoma [21], refractive error [22], and prematurity retinopathy of prematurity [23–25].
To establish a DL system, technical network and the datasets are the most essential components. Although not all CNN (Convolutional Neural Network) belongs to deep learning, CNN is the most widely used technical network in AMD diagnostic research which can operate on the whole images without requiring radiologists or ophthalmologists to manually contour on images [26]. A CNN can be divided into input, hidden, and output layers. The hidden layers are usually composed of convolutional, pooling, full connection, and normalization layers. The core of the CNN is the convolutional layer, which transforms the input data by applying a set of filters (also known as kernels) that act as feature detectors. A CNN learns the values of these filters’ weights on its own during the training process [27]. Activations are used after convolution. The pooling layers can reduce the dimensionality and keep the most important information. The output of the convolutional and pooling layers represents high-level features of the input image. The purpose of the fully connected layer is to use these high-level features to classify the input image categories based on the training dataset. Afterwards, backpropagation is performed to calculate the network weights, and gradient descent is used to update all filters and parameter values to minimize the output error [27]. This process will be repeated many times.
The datasets for AMD detection are various. Most public databases were established using FP and OCT images. Peking University collected a structured FP database of 5,000 patients including normal, diabetes, glaucoma, cataract, AMD, hypertension (H), myopia, and other diseases/abnormalities in 2019. The database is named as Ocular Disease Intelligent Recognition (ODIR) [28]. iChallenge-AMD is composed of AMD and non-AMD (myopia, normal control, etc.) FPs [29]. Srinivasan et al [30] conducted an OCT database (Duke dataset) that was acquired from 45 patients: 15 normal patients, 15 patients with dry AMD, and 15 patients with DME in 2014. Established by Rasti et al [31] in 2017, the Noor dataset was acquired at Noor Eye Hospital in Tehran and is consisting of 50 normal, 48 dAMD, and 50 DME OCTs. Regarding the Kaggle dataset [32], OCT images were selected from retrospective cohorts of adult patients from the Shiley Eye Institute of the University of California San Diego, the California Retinal Research Foundation, Medical Center Ophthalmology Associates, the Shanghai First People’s Hospital, and Beijing Tongren Eye Center between July 1, 2013 and March 1, 2017. Kermany et al [33] established an OCT database (Mendeley dataset) that contains CNV, DME, Drusen and normal people in 2018. Gholami et al [34] established an AMD retinal OCT images database including 55 AMD images called OCTID (Optical Coherence Tomography Image Database) in 2019. Besides public datasets, plenty of studies choose self-built databases which obtained data from hospitals directly.
DL models, especially CNN, have flourished rapidly in AMD detection in recent years. Although most of the DL models show effective diagnostic accuracy, DL specialists are still trying to explore the best networks, diameters, and layers of the network for higher accuracy. This meta-analysis summarized the DL models for AMD diagnosis and aimed to evaluate the diagnostic accuracy of DL models and to explore the best settings for future AMD model training, which will benefit researchers interested in DL for the diagnosis of fundus disorder.
Methods
This systematic review and meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis of Diagnostic Test Accuracy Studies (PRISMA-DTA) [35], and the Cochrane handbook [36]. The PRISMA-DTA checklists are available in S1 and S2 Tables. This meta-analysis was registered on PROSPERO (ID: CRD42022352753).
Eligibility criteria
All peer-reviewed and preprint original articles that reported the sensitivity and specificity of DL models in detecting AMD were considered. The detailed inclusion criteria were as follows: (1) diagnosing AMD by DL model via various images; (2) true positive (TP), false positive (FP), true negative (TN), and false negative (FN) could be obtained or transferred from the study. Records without available data such as reviews, conference abstracts, letters, and replies were excluded. There was no restriction on the year of publication, language, country, or datasets.
Information sources, search strategy and study selection
The search engines used included PubMed, EMBASE, the Cochrane Library, Web of Science, Scopus, ScienceDirect ClinicalTrails.gov, and World Health Organization International Clinical Trial Registration Platform (WHO ICTRP), and Chinese Clinical Trail Registry (ChiCTR) by 11 August 11, 2022.
The search strategy using medical subject headings (MeSH and Emtree) combined with entry words for all search engines. Detailed search strategies in different search engines are detailed in S1 File.
Endnote 20 was adopted for the study selection process. Duplicate studies were excluded by automation tools. The titles and abstracts were independently identified for possible inclusion by two authors (Leng X. and Shi R.). Disagreements were resolved by a third researcher (Wu Y.). After full text selection, the reports assessed eligibility were included in this meta-analysis.
Data collection process and definitions for data extraction
The data from the included studies were extracted by an individual researcher (Cai X.) and were rechecked by another (Zhu S.). The data we extracted included the first author and published year, country, number of images, network layers, device, hardware, type of AMD datasets, total dataset size, type of images, TP, FP, FN, TN, AUC, sensitivity, and specificity.
AMD, including dAMD and wAMD, was considered as the target condition. The reference standard was clinically proven AMD, while the DL-based diagnosis was considered the index test.
Risk of bias and applicability
The study risk of bias assessment was conducted by two individual researchers using the QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies-2) tool (Leng X., Shi R.). Parameters included patient selection, index test, reference standard, flow and timing, and applicability concerns in terms of patient selection, index test, and reference standard. Disagreements were solved with consensus by the third researcher (Lu X.). Deeks’ funnel plot mapped by Stata 16.0 was applied to assess the potential publication bias. An asymmetrical funnel shape or a P < 0.05 means the presentation of publication bias [37].
Diagnostic accuracy measures and synthesis of results
To evaluate the diagnostic accuracy of deep learning in detecting AMD, the sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) along with a 95% Confidence Interval (CI) were calculated separately for each study. Random effects models were applied in the calculation of the pooled results.
Meta-analysis and additional analysis
Separate and summary results of sensitivity and specificity would be presented in a form of a forest plot. The heterogeneity of the meta-analysis was evaluated by the Cochran Q-test and I2 [38]. I2 exceeding 25%, 50% and 75% indicate the meta-analysis with low, medium, and high heterogeneity respectively [39]. Sensitivity analyses, subgroup analyses, and meta-regression were conducted to explore the sources of heterogeneity. All meta-analyses and additional analyses were performed using Metadisc 1.4 and Review manager 5.4.1.
Results
Study selection
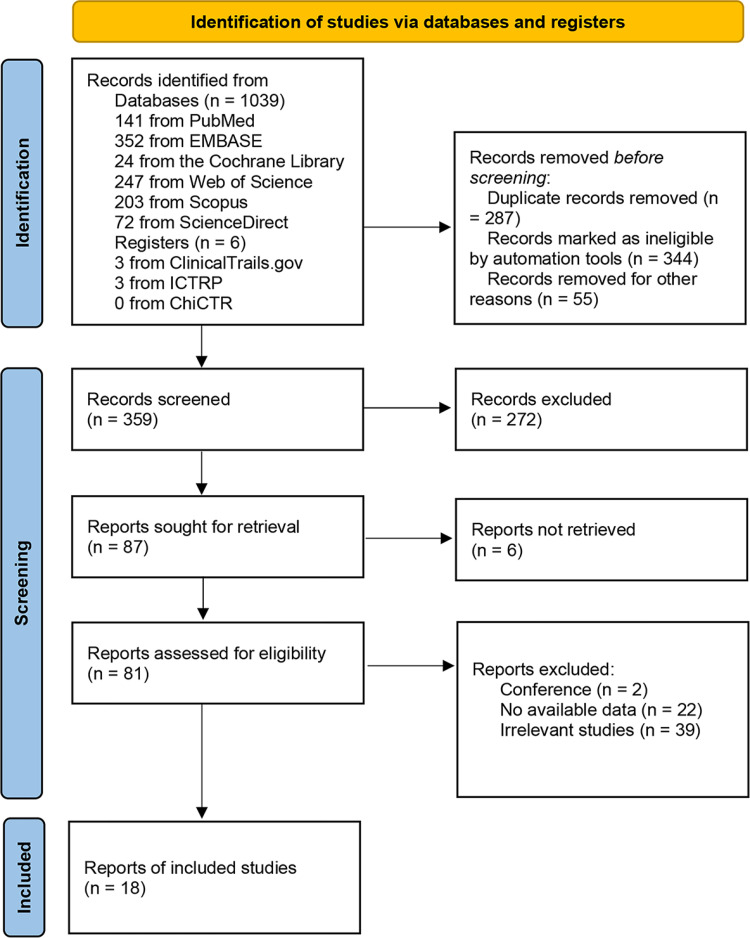
The detailed study selection process is described in Fig 1. 1045 records were searched using the present search strategy. 359 records remained after eliminating duplicate records and the ineligible records marked by automation tools. 272 records were excluded by screening titles and abstracts. 87 reports were sought for retrieval, of which 6 reports were not retrieved. 81 reports were assessed for eligibility through full text reading, and 2 conference abstracts, 39 irrelevant studies, and 22 no-available-data studies were excluded. Finally, 18 eligible studies were extracted from the remaining articles by full text review.
Fig 1. Study selection flow diagram.
Study characteristics
The detailed studies characteristics are summarized in Table 1. The 18 studies included were reported as full-text articles which consist of 56 models and summarized data from 778052 various images. OCT images, FPs, OPTOS ultra-widefield retinal images, and OCT images combined with FP images were included in 10, 5, 1, and 3 studies, respectively. All studies were published in 2017–2022, which were conducted in China, the USA, Japan, India, Jordan, the United Kingdom, Turkey, Russia, South Korea, Singapore, Norway and Spain. As for the variants, 5 studies used VGG, and 5 studies used ResNet. CapsNet, Darknet and other networks such as AlexNet, DenseNet, and self-created networks were adopted in one study, respectively. The layers of the network were divided into five classes including ≤10, 10–20, 20–50, 50–100, and >100, which were adopted in 2, 10, 6, 2 and 1 studies, respectively.
Table 1. Characteristics of included studies.
| First author & published year | Country | Variant of CNN | Layers | GPU | Datasets | Number of Images | Images | Other images in datasets | AMD |
|---|---|---|---|---|---|---|---|---|---|
| Alqudah 2019 [40] | Jordan | Self-created | 19 | Nvidia Tesla K40 (12GB) | Duke, Mendeley, Duke, Self-built | 135596 | OCT | CNV, DME, Normal | All |
| Bhatia 2019 [41] | UK | VGG | 16 | NA | Mendeley, Noor, Self-built | 5588 | OCT | CNV, DME, Normal | All |
| Celebi 2022 [42] | Turkey | CapsNet | 7 | Nvidia Tesla K40 (12GB) | Kaggle dataset, Self-built | 726 | OCT | Normal | All |
| Dong 2022 [43] | China | Darknet | 53 | NA | Multicenter Self-built | 208758 | FP | Normal | All |
| Gour 2020 [44] | India | VGG | 16 | NA | ODIR | 331 | FP | Cataract, Diabetes, Glaucoma, Hyperattention, Myopia, other abnormalities, Normal | All |
| He 2022 [45] | China | ResNet | 50 | NA | Duke, Mendeley | 795 | OCT | DME, Normal | All |
| Kadry 2021 [46] | Norway | VGG | 16 | NA | iChallenge-AMD database, OCTID | 6400 | OCT, FP | Non-AMD | All |
| 19 | |||||||||
| AlexNet | 11 | ||||||||
| ResNet | 50 | ||||||||
| Lee 2017 [47] | USA | VGG | 16 | NVIDIA Pascal Titan X (12GB) | Self-built | 101002 | OCT | Normal | All |
| Ma 2022 [48] | USA | ResNet | 34 | Nvidia V100 (32 GB) | Self-built | 73 | OCT | PCV | Wet |
| Mathews 2022 [49] | India | Self-created | 11 | NA | Duke, Mendeley | 75 | OCT | DME, Normal | Dry |
| Matsuba 2019 [50] | Japan | DCNN | 7 | NA | Self-built | 364 | OPTOS2 | Normal | Wet |
| Motozawa 2019 [51] | Japan | Unclear | 18 | GTX 1080 TI (11GB) | Self-built | 169 | OCT | Normal | All |
| Takhchidi 2021 [52] | Russia | ResNet | 50 | Nvidia RTX 2070 Max-Q (8GB) | Self-built | 1200 | FP | Normal | All |
| Tan 2018 [53] | Singapore | Unclear | 14 | NA | Self-built | 1110 | FP | Normal | All |
| Thomas 2021 [54] | India | Unclear | 19 | Nvidia RTX2080 (8GB) | Duke, Mendeley, Noor, OCTID | 1139 | OCT | Normal | All |
| Wang 2019 [55] | China | DenseNet | 121 | NVIDIA RTX 2080 TI (11G) | Duke, Noor | 8315 | OCT | DME, Normal | All |
| ResNet | 50 | ||||||||
| ResNext | 101 | ||||||||
| DPN | 92 | ||||||||
| CliqueNet | 10 | ||||||||
| Yoo 2018 [56] | Korea | VGG | 19 | NVIDIA GTX1060 (3GB); GTX980 (6GB) | Project Macula | 83 | OCT, FP | Normal | All |
| Zapata 2020 [57] | Spain | Self-created | 24 | NA | Optretina’s tagged dataset | 306302 | OCT, FP | GON3 | All |
NA, not applicable; OPTOS, OPTOS ultra-widefield retinal images; GON, glaucomatous optic neuropathy
Risk of bias and bias of publication
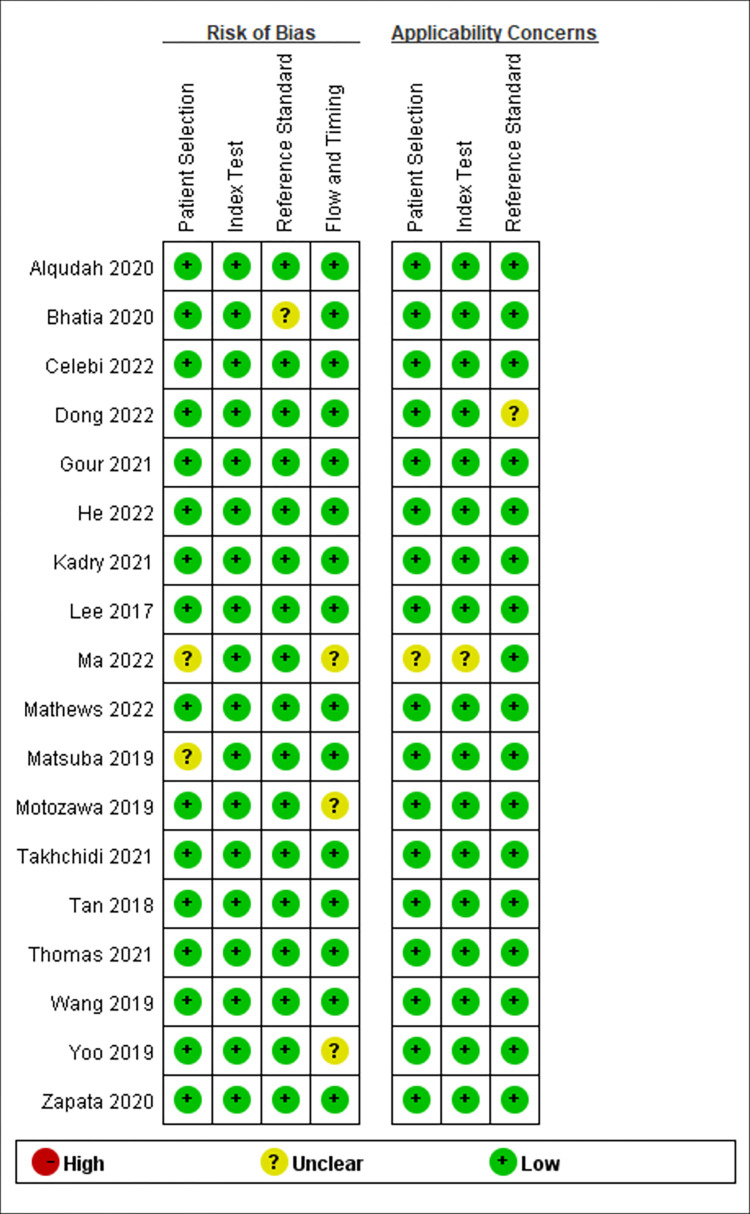
The results of the QUADAS-2 analysis are summarized in Fig 2. Generally, the risk of bias is low for this meta-analysis. The risk of patient selection was considered “low risk” in 16 studies and “unclear risk” in 2 studies. The risk of bias for the index test and reference standard was “low risk” in all studies. The risk of bias for reference standard was rated “low” in 17 studies and 1 were rated “unclear risk”. The risk of bias for flow and timing was rated “low” in 15 studies and 3 were rated “unclear risk”. Applicability concerns including patient selection, index test, and the reference standard only existed in one “unclear risk” study and the other 17 studies were rated “low risk”.
Fig 2. QUADAS-2 results in each study.
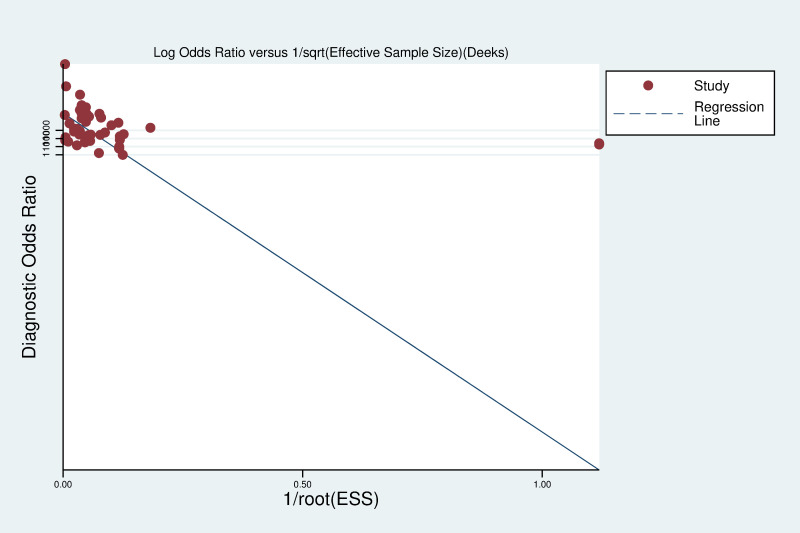
Deeks’ funnel plot (Fig 3) was adopted to investigate the potential bias of publication by Stata 16.0 (P = 0.375, 95%CI -292.9264 to 112.091), which indicated no obvious publication bias existed in this meta-analysis.
Fig 3. Deek’s funnel plot.
Results of individual studies
In this research, various DL models were mentioned in the included studies for AMD identification, including VGG, CapsNet, ResNet, AlexNet, DenseNet, ResNext, DPN, CliqueNet etc. The results show that these models have high sensitivity and specificity in AMD identification, which can meet the needs of practical clinical applications. The detailed results of individual studies are summarized in Table 2. Alqudah et al [40] used a 15-layer CNN to classify 136,187 OCT images from Mendeley, Duke, and Self-built datasets (4 classes including AMD, CNV, DME, and normal) for AMD identifying with a sensitivity of 100% and a specificity of 100%. Bhatia et al [41] used VGG-16 to classify 5588 OCT images from Mendeley, Duke, Noor, and Self-built datasets (4 classes including AMD, CNV, DME, normal) for AMD identifying with a sensitivity of 94% and a specificity of 90%. Celebi et al [42] used CapsNet with 7 layers to classify 726 OCT images form Kaggle and self-built datasets (2 classes including AMD and normal) for AMD identifying with a sensitivity of 100% and a specificity of 99%. Dong et al [43] used a joint CNN detector using Yolov3 to classify 208758 FP images from self-built multicenter real-world data (11 classes including AMD, DR, glaucoma, pathological myopia, retinal vein occlusion, macula hole, epiretinal macular membrane, hypertensive retinopathy, myelinated fibers, retinitis pigmentosa and normal) for AMD identifying with a sensitivity of 88% and a specificity of 98%. Gour et al [44] used VGG-16 to classify 331 FP images from ODIR dataset (8 classes including AMD, cataract, diabetes, glaucoma, hyperattention, myopia, and other abnormalities) for AMD identifying with a sensitivity of 6% and a specificity of 94%. He et al [45] used ResNet-50 to classify 795 OCT images from Mendeley and Duke datasets (3 classes including AMD, DME, and normal) for AMD identifying with a sensitivity of 96% and a specificity of 99%. Kadry et al [46] used VGG-16, VGG-19, AlexNet, and ResNet-50 to classify 3200 FP images and 3200 OCT images from iChallenge AMD database, OCTID (2 classes including AMD and Non-AMD) resulting in sensitivity of 88%, 84%, 88%, 88% and specificity of 85%, 87%, 85%, 84%, respectively. Lee et al [47] used VGG-16 to classify 101002 OCT images from self-built dataset (2 classes including AMD and normal) for AMD identifying with a sensitivity of 90% and a specificity of 91%. Ma et al [48] used ResNet-34 to classify 73 OCT images from self-built dataset (2 classes including AMD and polypoidal choroidal vasculopathy) for AMD identifying with a sensitivity of 92% and a specificity of 90%. Mathews et al [49] used a 11-layer lightweight CNN to classify 75 OCT images from Duke and Mendeley datasets (3 classes including AMD, DME, and normal) for AMD identifying with a sensitivity of 100% and a specificity of 100%. Matsuba et al [50] used a 7-layer CNN to classify 364 OPTOS ultra-widefield retinal images from self-built dataset (2 classes including AMD and normal) for AMD identifying with a sensitivity of 100% and a specificity of 97%. Motozawa et al [51] used an 18-layer CNN to classify 169 OCT images from self-built database (2 classes including AMD and normal) for AMD identifying with a sensitivity of 99% and a specificity of 100%. Takhchidi et al [52] used ResNet-50 to classify 1200 FP images from self-built dataset (2 classes including AMD and normal) for AMD identifying with a sensitivity of 90% and a specificity of 86%. Tan et al [53] used a 14-layer CNN to classify 1110 FP images from self-built dataset (2 classes including AMD and normal) for AMD identifying with a sensitivity of 96% and a specificity of 94%. Thomas et al [54] used a 14-layer CNN to classify 1139 OCT images from Mendeley, Duke, Noor, and OCTID datasets (2 classes including AMD and normal) for AMD identifying with a sensitivity of 99% and a specificity of 100%. Wang et al [55] used DenseNet, ResNet, ResNext, DPN, and CliqueNet to classify 8315 OCT images from Duke and Noor datasets (3 classes including AMD, DME and normal) resulting in sensitivity of 96%, 97%, 100%, 97%, 99% and specificity of 95%, 100%, 100%, 97%, 99% in dataset 1, and sensitivity of 95%, 100%, 99%, 100%, 93% and specificity of 95%, 99%, 95%, 99%, 98% in dataset 2. Yoo et al [56] used VGG-19 to classify three types of images (OCT, FP, and OCT combined with FP) from Project Macula (2 classes including AMD and normal) for AMD identifying with a pooled sensitivity of 84% and a pooled sensitivity of 59%. Zapata et al [57] used a 24-layer CNN to classify 306302 FP images and OCT images from Optretina’s tagged dataset (2 classes including AMD and glaucomatous optic neuropathy) for AMD identifying with a sensitivity of 83% and a specificity of 89%.
Table 2. Summary of each included studies.
| Study | Methodology | Datasets | Number of images | Classes | Other diseases | Sensitivity | Specificity | Limitations |
|---|---|---|---|---|---|---|---|---|
| Alqudah 2019 [40] | 15-layer CNN | Duke, Mendeley, Self-built | 136187 OCT images | 4 | CNV, DME, Normal | 100% | 100% | |
| Bhatia 2019 [41] | VGG-16 | Duke, Mendeley, Noor, Self-built | 5588 OCT images | 4 | CNV, DME, Normal | 94% | 90% | 1) Ignored bad quality pictures. |
| Celebi 2022 [42] | CapsNet | Kaggle dataset, | 726 OCT images | 2 | Normal | 100% | 99% | 1)Did not study other retinal diseases; 2)Ignored bad quality pictures and patients who had other retinal diseases. |
| Dong 2022 [43] | A joint CNN detector using Yolov3 | Multicenter Self-built | 208758 FP images | 11 | DR, Glaucoma, Pathological myopia, Retinal vein occlusion, Macula hole, Epiretinal macular membrane, Hypertensive retinopathy, Myelinated fibers, Retinitis pigmentosa, Normal | 88% | 98% | 1)Only small number of retinitis pigmentosa. |
| Gour 2020 [44] | VGG-16 | ODIR | 331 FP images | 8 | Cataract, Diabetes, Glaucoma, Hyperattention, Myopia, other abnormalities, Normal | 6% | 94% | 1)The dataset contained 8 types of diseases, but with a small dataset. |
| He 2022 [45] | ResNet-50 | Duke, Mendeley | 795 OCT images | 3 | DME, Normal | 96% | 99% | 1)Only contained one other diseases. |
| Kadry 2021 [46] | VGG-16 | iChallenge-AMD database, OCTID | 3200 FP and 3200 OCT images | 2 | Non-AMD | 88% | 85% | 1)The definition of non-AMD is not clear. |
| VGG-19 | 84% | 87% | ||||||
| AlexNet, | 88% | 85% | ||||||
| ResNet-50 | 88% | 84% | ||||||
| Lee 2017 [47] | VGG-16 | Self-built | 101002 OCT images | 2 | Normal | 90% | 91% | 1)Included only images from patients who met the study criteria, and the neural network was only trained on these images; 2) This model was trained using images from a single academic center, and the external generalizability is unknown |
| Ma 2022 [48] | ResNet-34 | Self-built | 73 OCT images | 2 | Polypoidal choroidal vasculopathy | 92% | 90% | 1) Small dataset |
| Mathews 2022 [49] | A 11-layer lightweight CNN | Duke, Mendeley | 10907 OCT images | 3 | DME, Normal | 100% | 100% | 1) This study used drusen macular degeneration for AMD diagnosis; 2)Only contain one other diseases. |
| Matsuba 2019 [50] | A 7-layer CNN | Self-built | 364 OPTOS images | 2 | Normal | 100% | 97% | 1) It is difficult to acquire precise images using OPTOS when the transmission of light into the eye is impaired by an intermediate translucent zone; 2) Most AMD patients accept treatment which may cause diagnostic error 3) Did not study other retinal diseases. |
| Motozawa 2019 [51] | An 18-layer CNN | Self-built | 169 OCT images | 2 | Normal | 99% | 100% | 1) Excluded low quality images and patients who had other concomitant diseases; 2) Did not study other retinal diseases. |
| Takhchidi 2021 [52] | ResNet-50 | Self-built | 1200 FP images | 2 | Normal | 90% | 86% | 1) Did not study other retinal diseases. |
| Tan 2018 [53] | A 14-layer CNN | Self-built | 1110 FP images | 2 | Normal | 96% | 94% | 1) Did not study other retinal diseases. |
| Thomas 2021 [54] | A 19-layer CNN | Mendeley, Duke, Noor, OCTID | 1139 OCT images | 2 | Normal | 99% | 100% | 1) Did not study other retinal diseases. |
| Wang 2019 [55] | DenseNet-121 | Duke, Noor | 8315 OCT images | 3 | DME, Normal | 96% in Duke, 95% in Noor | 95% in Duke, 95% in Noor | 1) Only contained one other diseases. |
| ResNet-50 | 97% in Duke, 100% in Noor | 100% in Duke, 99% in Noor | ||||||
| ResNext-101 | 100% in Duke, 99% in Noor | 100% in Duke, 95% in Noor | ||||||
| DPN-92 | 97% in Duke, 100% in Noor | 97% in Duke, 99% in Noor | ||||||
| CliqueNet-10 | 99% in Duke, 93% in Noor | 99% in Duke, 98% in Noor | ||||||
| Yoo 2018 [56] | VGG-19 | Project Macula | 83 FP and 83 OCT images | 2 | Normal | 84% | 59% | 1) Did not study other retinal diseases; 2) Small datasets; |
| Zapata 2020 [57] | A 24-layer CNN | Optretina’s tagged dataset | 306302 FP images and OCT images | 2 | Glaucomatous optic neuropathy | 83% | 89% | 1.No clear number of OCT or FP images. |
Results of synthesis
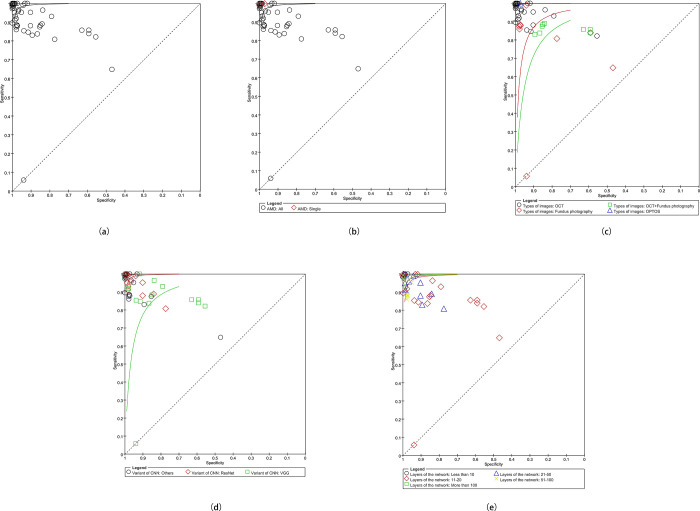
The pooled sensitivity and specificity in this meta-analysis were 94% (P = 0, 95% CI 0.94–0.94, I2 = 99.7%) and 97% (P = 0, 95% CI 0.97–0.97, I2 = 99.6%) (Fig 4, S1 Fig), respectively. The PLR, NLR, DOR, and AUC values were 21.77(95% CI 15.49–30.59), 0.06 (95% CI 0.04–0.09), 342.41 (95% CI 210.31–557.49) and 0.9925. The SROC (Summary Receiver Operating Characteristic) curves are showed in Fig 5(A).
Fig 4. The forest plot of the pooled sensitivity and specificity.
Fig 5.
The SROC (a) the pooled SROC; (b) the SROC of types of AMD; (c) the SROC of types of images; (d) the SROC of variants of CNNs; (e) the SROC of Networks.
Additional analysis
For the high heterogeneity, the additional analyses were conducted based on the results of sensitivity and specificity. Sensitivity analyses were conducted to investigate the sources of heterogeneity, however, neither the I2 of sensitivity nor specificity significantly decreased after excluding studies one by one. Therefore, subgroup analyses which included the type of AMD, type of images, variant of CNN, and variants were conducted (Fig 5B–5E). Meta regression indicated that the sources of heterogeneity were types of AMD (P = 0.1882, RDOR = 36.03) and layers of the network (P = 0.4878, RDOR = 0.74). All additional analyses results were summarized in Table 3. Original forest plots were available in S1 Fig.
Table 3. Subgroup analyses and meta regression results.
| Number of models | Sen1 | Spe2 | PLR | NLR | DOR | AUC | Meta regression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pooled (95%CI) | I 2 | Pooled (95%CI) | I 2 | Pooled (95%CI) | I 2 | Pooled (95%CI) | I 2 | Pooled (95%CI) | I 2 | P | RDOR | |||
| Layers | 0.4878 | 0.74 | ||||||||||||
| ≤10 | 5 | 1.00 (1.00–1.00) | 97.80% | 0.99 (0.99–0.99) | 60.20% | 104.30 (70.94–153.36) | 65.70% | 0 (0.00–0.03) | 98.60% | 35609.58(4947.73–256287.76) | 93.80% | 0.9992 | ||
| 10–20 | 28 | 0.91(0.92–0.91) | 99.70% | 0.97 (0.97–0.96) | 99.80% | 7.40 (5.04–10.86) | 99.20% | 0.10 (0.07–0.16) | 99.60% | 52.47 (33.55–82.06) | 98.50% | 0.9861 | ||
| 20–50 | 11 | 0.94(0.93–0.95) | 96.70% | 0.95 (0.95–0.96) | 97.80% | 28.68 (10.64–77.28) | 98.30% | 0.04 (0.02–0.09) | 96.40% | 770.17 (163.74–3622.58) | 97.90% | 0.9927 | ||
| 50–100 | 7 | 0.93(0.91–0.94) | 85.40% | 0.98 (0.97–0.98) | 72.10% | 36.07 (30.09–43.24) | 50.80% | 0.10 (0.06–0.16) | 70.70% | 366.71 (219.09–613.82) | 59.70% | 0.9935 | ||
| >100 | 4 | 0.95 (0.94–0.97) | 64.80% | 0.98 (0.97–0.99) | 93.70% | 76.90 (21.55–274.39) | 90.70% | 0.04 (0.03–0.08) | 57.10% | 1967.84 (464.67–8333.64) | 84.40% | 0.9914 | ||
| Type of AMD | 0.1882 | 36.03 | ||||||||||||
| Single | 2 | 1.00 (0.99–1.00) | 0% | 0.99 (0.98–1.00) | 89.0% | 122.44 (6.08–2463.89) | 87.70% | 0.00 (0.00–0.02) | 0% | 107120.15 (8471.87–1354450.49) | 0% | NA3 | ||
| All | 54 | 0.94 (0.94–0.94) | 99.70% | 0.97 (0.97–0.97) | 99.70% | 20.61 (14.59–29.12) | 99.30% | 0.06 (0.04–0.09) | 99.70% | 306.14 (187.38–500.18) | 99.20% | 0.9915 | ||
| Architectures | 0.0004 | 0.19 | ||||||||||||
| ResNet | 13 | 0.94 (0.93–0.95) | 96.10% | 0.96 (0.95–0.96) | 97.60% | 34.66 (13.66–87.94) | 98.20% | 0.04 (0.02–0.08) | 96.00% | 951.18 (232.03–3899.24) | 97.70% | 0.9941 | ||
| VGG | 16 | 0.90 (0.90–0.89) | 99.60% | 0.91 (0.91–0.90) | 98.50% | 4.29 (3.25–5.67) | 98.30% | 0.19 (0.13–0.27) | 99.50% | 30.62 (21.91–42.79) | 96.90% | 0.8972 | ||
| Others | 26 | 0.99 (0.99–0.99) | 99.50% | 0.99 (0.99–0.99) | 99.40% | 86.04 (33.97–217.95) | 99.50% | 0.01 (0.00–0.08) | 99.70% | 7354.18 (1231.62–43913.09) | 99.40% | 0.9987 | ||
| Types of Images | 0.0002 | 0.12 | ||||||||||||
| OCT | 36 | 0.94 (0.94–0.94) | 99.80% | 0.97 (0.97–0.97) | 99.70% | 52.33 (32.57–84.10) | 99.50% | 0.03 (0.02–0.04) | 99.70% | 2209.74 (1113.02–4387.10) | 99.40% | 0.9982 | ||
| FP | 11 | 0.83 (0.81–0.84) | 96.90% | 0.96 (0.96–0.97) | 98.20% | 9.26 (3.74–22.92) | 99.10% | 0.18 (0.06–0.53) | 99.10% | 51.02 (13.37–194.69) | 97.70% | 0.9592 | ||
| OCT & FP | 8 | 0.84 (0.83–0.85) | 45.70% | 0.88 (0.87–0.89) | 87.00% | 4.38 (3.11–6.17) | 91.30% | 0.18 (0.15–0.20) | 30.00% | 28.80 (20.38–40.71) | 70.70% | 0.9176 | ||
| OPTOS | 1 | 1.00 (0.97–1.00) | 0% | 0.97 (0.94–0.99) | 0 | 34.95 (20.45–59.73) | 0% | 0 | 0 | 9371.15 (523.79–167661.33) | 0% | NA | ||
Sen: Sensitivity; Spe: Specificity; NA: not applicable.
Discussion
This meta-analysis included 18 studies and 56 models aimed to investigate the performance of deep learning in detecting AMD. The results of the present study indicate a high accuracy in detecting AMD through CNN, but with high heterogeneity. The sources of heterogeneity were the types of AMD and layers of the network according to the meta-regression.
DL has been widely adopted in image recognition, speech recognition, and natural language processing, but is only beginning to impact healthcare, especially in ophthalmology [6]. DL is a subset of machine learning which has become possible with increasing computing power. Compared to traditional machine learning algorithms and shallow networks, current DL algorithms are characterized by large amounts of processable data, high computational power, and large network size [58, 59].
Fluorescein angiography, optical coherence tomography (OCT), optical coherence tomography angiography (OCTA), FP, fundus autofluorescence, and indocyanine green angiography are useful diagnostic tests in clinical practice to detect AMD [1], of which OCT and FP are the most commonly used. Plenty of public ophthalmic datasets are based on the above two types of images, which have facilitated the rapid development of artificial intelligence in ophthalmology, and will make telemedicine more convenient in the future. This meta-analysis reveals that DL detection through OCT, FP, and OPTOS ultra-widefield retinal images has a high accuracy in AMD diagnosing.
18 included studies were summarized in Table 2. All studies adopted CNN to conduct DL models. The non-saturating ReLU activation function was introduced in AlexNet to increase the training speed and the dropout method was used to minimize overfitting in the fully connected layers [60]. VGG has a deeper architecture, but cannot overcome the limitation of the vanishing gradient problem [61]. In the ResNet architecture, identity mapping is introduced to solve the vanishing gradient problem. ResNet can therefore be used to train deeper models [62]. DenseNets can alleviate the vanishing-gradient problem, strengthen feature propagation, encourage feature reuse, and substantially reduce the number of parameters [63]. Other studies mostly used a self-created CNN architectures with 7–20 layers. Duke, Mendeley, and Noor are the most used OCT databases. Most FP image datasets were built clinically. 10 studies [40, 41, 43, 46, 47, 52–55, 57] included more than 1000 images in their research. 4 studies [40, 43, 47, 57] included more than 100 thousand images. Only Matsuba et al [50] used OPTOS as the dataset, which is unique and pioneering among the 18 studies. Dong et al [43] and Gour et al [44] included 11 and 8 classes respectively, while other studies only contained 2–4 classes.
In this research, the type of AMD and the layers of the network were found to be the two essential factors that impact the accuracy of the diagnosis. However, the layers of the network are not positively correlated with diagnostic accuracy. Even though DOR and AUC are higher when the layers are less than 10, as layers of the network are more than 10, the diagnostic accuracy gradually grows as the number of layers increases. Cautiously, when the number of layers becomes too deep, overfitting may occur. Overfitting is a serious issue when training DL models, which may cause the trained models cannot be generalized in other data or datasets [64]. Predictably, deeper and more accurate networks will be placed in service soon. Meanwhile, different types of AMD may make the computation more difficult, but the prevailing datasets contain different types of AMD, which will make the trained models more generalized.
Although the meta-regression results did not show that the networks and types of images connected to the diagnostic accuracy, they are still significant. The DOR of ResNet showed superior than VGG, other variants cannot be assessed because they were only included in one study. That might be because ResNet with more layers was developed after VGG. ResNet belongs to deep residual networks with a higher amount of processable data [65]. ResNets can be trained easily without increasing the training error percentage, and are helpful in tackling the vanishing gradient problem using identity mapping [66]. Therefore, it is believed ResNet is an ideal architecture among the present variants of CNN. However, the influence due to the layers of the network impacts the results. This may be the reason the RDOR of networks in meta-regression is very low. As for the types of images, OCT images showed superior in detecting AMD. OCT images can reveal every layer of macular structures with more anatomical information than fundus images. Combined OCT images with fundus images had worse sensitivity, specificity, DOR, and AUC. We think it is because two images have more information. More information means more computation and the potential to be more accurate, which may require considering the layers of the network and the architecture of CNNs. Additionally, although only one study [50] reported the OPTOS ultra-widefield retinal images as self-dataset, the pooled sensitivity, specificity, and DOR were all highest in the four subgroups.
The detailed limitations for each study were summarized in Table 3. Generally, 8 studies [42, 47, 50–54, 56] did not study other retinal diseases. 3 studies [45, 49, 55] only contained one other diseases. 2 studies [48, 56] had small datasets with no more than 100 images. Bhatia et al [41] ignored bad quality pictures that may cause a generalization issue. Celebi et al [42] ignored bad quality pictures and patients who had other retinal diseases. Although Dong et al [43] established a database with 11 classes, the number of retinitis pigmentosa images is small. Gour et al [44] contained 8 types of diseases, but with a small dataset of 331 FP images. Kadry et al [46] used 4 CNN variants for classification, but the definition of non-AMD is not clear. Lee et al [47] included only images from patients who met the study criteria, and the neural network was only trained on these images. Meanwhile, the model was trained using images from a single academic center, and the external generalizability is unknown. Mathews et al [49] used drusen macular degeneration for AMD diagnosis. Matsuba et al [50] used OPTOS images, but it is difficult to acquire precise images using OPTOS when the transmission of light into the eye is impaired by an intermediate translucent zone. At the same time, most AMD patients accept treatment which may cause diagnostic errors. Motozawa et al [51] excluded low quality images and patients who had other concomitant diseases. Zapata et al did not report a clear number of OCT or FP images.
This meta-analysis and the included studies have several limitations. First, some variants of CNN including CapsNet, AlexNet, and DenseNet only existed once, and some studies used self-created CNN architectures. Therefore, the subgroup analysis of networks is not accurate. Second, we tried to establish more subgroups or to find more possible covariates such as hardware, network, and hyperparameters. However, these potential factors were not mentioned in many studies. Third, we concentrated more on diagnostic accuracy, but as DL develops, AMD diagnostics has become more diverse, more plentiful, and more useful in lesion segmentation and efficacy prediction, which will be highly considered for further research. Forth, the Duke and some self-built datasets have a small number of images for training.
Future challenges and direction
DL is still in the early stages of development in AMD diagnosis, but in the foreseeable future, widespread use could play a significant role in fundus applications, screening, telemedicine, reducing the workload of physicians, etc.
The purpose of DL algorithms for diagnosing AMD is to achieve an automated diagnosis of many kinds of fundus diseases. However, no matter in public databases or self-built databases, only several diseases were chosen for classification which is difficult for widespread use clinically. Establishing a database which covers heterogeneous and large image sets is still a serious challenge. Meanwhile, the DL algorithms concentrate more on images only, but the images are not the only data obtained clinically. Ideally, multimodal data containing clinical data, FP, and OCT, etc. may increase the diagnostic accuracy. At the same time, traditional fundus datasets mostly consist of FP and OCT images. However, with diagnostic tests developing, more new methods and technologies such as OPTOS ultra-widefield retinal images, OCTA, FFA, ICGA, etc. will be added as public or self-built datasets in future AMD detection. Finally, as the equipment evolves, the image quality of FP, OCT, OCTA etc. improves. More high definition images will increase the diagnostic accuracy.
Conclusions
CNNs are mostly adopted deep learning algorithms in AMD detection. All included DL algorithms adopted CNNs. CNNs, especially ResNets, are effective in detecting AMD with high diagnostic accuracy. The types of AMD and the layers of the network are the two essential factors that impact the model training process. Proper layers of the network will make the model more reliable. More datasets established by new diagnostic methods such as ultra-widefield retinal images, FFA, and ICGA will be used to train DL models in the future, which will be helpful in fundus application screening, long-range medical treatment, and reducing the workload of physicians.
Supporting information
(PDF)
(DOC)
(DOC)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
National Natural Science Foundation of China, grant number 82174444.
References
- 1.Flaxel CJ, Adelman RA, Bailey ST, Fawzi A, Lim JI, Vemulakonda GA, et al. Age-Related Macular Degeneration Preferred Practice Pattern(R). Ophthalmology. 2020;127(1):P1–P65. Epub 2019/11/24. doi: 10.1016/j.ophtha.2019.09.024 . [DOI] [PubMed] [Google Scholar]
- 2.Congdon N, O’Colmain B, Klaver CC, Klein R, Muñoz B, Friedman DS, et al. Causes and prevalence of visual impairment among adults in the United States. Archives of ophthalmology (Chicago, Ill: 1960). 2004;122(4):477–85. Epub 2004/04/14. doi: 10.1001/archopht.122.4.477 . [DOI] [PubMed] [Google Scholar]
- 3.Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng C-Y, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. The Lancet Global Health. 2014;2(2):e106–e16. doi: 10.1016/S2214-109X(13)70145-1 [DOI] [PubMed] [Google Scholar]
- 4.Mitchell P, Liew G, Gopinath B, Wong TY. Age-related macular degeneration. Lancet (London, England). 2018;392(10153):1147–59. Epub 2018/10/12. doi: 10.1016/S0140-6736(18)31550-2 . [DOI] [PubMed] [Google Scholar]
- 5.Tsui I, Jain A, Shah S, Schwartz SD, McCannel TA, editors. Ultra widefield imaging of peripheral exudative hemorrhagic chorioretinopathy. Seminars in ophthalmology; 2009: Taylor & Francis. [DOI] [PubMed] [Google Scholar]
- 6.Nagiel A, Lalane RA, Sadda SR, Schwartz SDJR. Ultra-widefield fundus imaging: a review of clinical applications and future trends. 2016;36(4):660–78. doi: 10.1097/IAE.0000000000000937 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunovic H. Artificial intelligence in retina. Prog Retin Eye Res. 2018;67:1–29. Epub 2018/08/05. doi: 10.1016/j.preteyeres.2018.07.004 . [DOI] [PubMed] [Google Scholar]
- 8.Ting DSW, Pasquale LR, Peng L, Campbell JP, Lee AY, Raman R, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103(2):167–75. Epub 2018/10/27. doi: 10.1136/bjophthalmol-2018-313173 ; PubMed Central PMCID: PMC6362807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018;172(5):1122–31 e9. Epub 2018/02/24. doi: 10.1016/j.cell.2018.02.010 . [DOI] [PubMed] [Google Scholar]
- 10.Chan S, Reddy V, Myers B, Thibodeaux Q, Brownstone N, Liao W. Machine Learning in Dermatology: Current Applications, Opportunities, and Limitations. Dermatol Ther (Heidelb). 2020;10(3):365–86. Epub 2020/04/08. doi: 10.1007/s13555-020-00372-0 ; PubMed Central PMCID: PMC7211783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saba L, Biswas M, Kuppili V, Cuadrado Godia E, Suri HS, Edla DR, et al. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14–24. Epub 2019/04/22. doi: 10.1016/j.ejrad.2019.02.038 . [DOI] [PubMed] [Google Scholar]
- 12.McBee MP, Awan OA, Colucci AT, Ghobadi CW, Kadom N, Kansagra AP, et al. Deep Learning in Radiology. Acad Radiol. 2018;25(11):1472–80. Epub 2018/04/03. doi: 10.1016/j.acra.2018.02.018 . [DOI] [PubMed] [Google Scholar]
- 13.Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun (Lond). 2020;40(4):154–66. Epub 2020/04/12. doi: 10.1002/cac2.12012 ; PubMed Central PMCID: PMC7170661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Yang DM, Rong R, Zhan X, Xiao G. Pathology Image Analysis Using Segmentation Deep Learning Algorithms. Am J Pathol. 2019;189(9):1686–98. Epub 2019/06/15. doi: 10.1016/j.ajpath.2019.05.007 ; PubMed Central PMCID: PMC6723214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318(22):2211–23. Epub 2017/12/14. doi: 10.1001/jama.2017.18152 ; PubMed Central PMCID: PMC5820739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abramoff MD, Lou Y, Erginay A, Clarida W, Amelon R, Folk JC, et al. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep Learning. Invest Ophthalmol Vis Sci. 2016;57(13):5200–6. Epub 2016/10/05. doi: 10.1167/iovs.16-19964 . [DOI] [PubMed] [Google Scholar]
- 17.Sahlsten J, Jaskari J, Kivinen J, Turunen L, Jaanio E, Hietala K, et al. Deep Learning Fundus Image Analysis for Diabetic Retinopathy and Macular Edema Grading. Sci Rep. 2019;9(1):10750. Epub 2019/07/26. doi: 10.1038/s41598-019-47181-w ; PubMed Central PMCID: PMC6656880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassmann F, Mengelkamp J, Brandl C, Harsch S, Zimmermann ME, Linkohr B, et al. A Deep Learning Algorithm for Prediction of Age-Related Eye Disease Study Severity Scale for Age-Related Macular Degeneration from Color Fundus Photography. Ophthalmology. 2018;125(9):1410–20. Epub 2018/04/15. doi: 10.1016/j.ophtha.2018.02.037 . [DOI] [PubMed] [Google Scholar]
- 19.Rim TH, Lee AY, Ting DS, Teo K, Betzler BK, Teo ZL, et al. Detection of features associated with neovascular age-related macular degeneration in ethnically distinct data sets by an optical coherence tomography: trained deep learning algorithm. Br J Ophthalmol. 2021;105(8):1133–9. Epub 2020/09/11. doi: 10.1136/bjophthalmol-2020-316984 ; PubMed Central PMCID: PMC8185637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldstein SM, Vogl WD, Bogunovic H, Sadeghipour A, Riedl S, Schmidt-Erfurth U. Characterization of Drusen and Hyperreflective Foci as Biomarkers for Disease Progression in Age-Related Macular Degeneration Using Artificial Intelligence in Optical Coherence Tomography. JAMA Ophthalmol. 2020;138(7):740–7. Epub 2020/05/08. doi: 10.1001/jamaophthalmol.2020.1376 ; PubMed Central PMCID: PMC7206537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhuiyan A, Govindaiah A, Smith RT. An Artificial-Intelligence- and Telemedicine-Based Screening Tool to Identify Glaucoma Suspects from Color Fundus Imaging. J Ophthalmol. 2021;2021:6694784. Epub 2021/06/18. doi: 10.1155/2021/6694784 ; PubMed Central PMCID: PMC8179760 conflicts of interest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varadarajan AV, Poplin R, Blumer K, Angermueller C, Ledsam J, Chopra R, et al. Deep Learning for Predicting Refractive Error From Retinal Fundus Images. Invest Ophthalmol Vis Sci. 2018;59(7):2861–8. Epub 2018/07/20. doi: 10.1167/iovs.18-23887 . [DOI] [PubMed] [Google Scholar]
- 23.Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Chan RVP, et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018;136(7):803–10. Epub 2018/05/26. doi: 10.1001/jamaophthalmol.2018.1934 ; PubMed Central PMCID: PMC6136045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell JP, Kim SJ, Brown JM, Ostmo S, Chan RVP, Kalpathy-Cramer J, et al. Evaluation of a Deep Learning-Derived Quantitative Retinopathy of Prematurity Severity Scale. Ophthalmology. 2021;128(7):1070–6. Epub 2020/10/31. doi: 10.1016/j.ophtha.2020.10.025 ; PubMed Central PMCID: PMC8076329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gensure RH, Chiang MF, Campbell JP. Artificial intelligence for retinopathy of prematurity. Curr Opin Ophthalmol. 2020;31(5):312–7. Epub 2020/07/23. doi: 10.1097/ICU.0000000000000680 ; PubMed Central PMCID: PMC7891849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan N, Adam R, Huang P, Maldjian T, Duong TQ. Deep Learning Prediction of Pathologic Complete Response in Breast Cancer Using MRI and Other Clinical Data: A Systematic Review. Tomography. 2022;8(6):2784–95. Epub 20221121. doi: 10.3390/tomography8060232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ting DSW, Peng L, Varadarajan AV, Keane PA, Burlina PM, Chiang MF, et al. Deep learning in ophthalmology: The technical and clinical considerations. Prog Retin Eye Res. 2019;72:100759. Epub 2019/05/03. doi: 10.1016/j.preteyeres.2019.04.003 . [DOI] [PubMed] [Google Scholar]
- 28.Peking university international competition on ocular disease intelligent recognition (ODIR-2019) 2020 [cited 2022 December 28, 2022]. Available from: https://odir2019.grand-challenge.org/dataset/.
- 29.grand-challenge.org. iChallenge-AMD 2021 [December 28, 2022]. Available from: https://refuge.grand-challenge.org/iChallenge-AMD/.
- 30.Srinivasan PP, Kim LA, Mettu PS, Cousins SW, Comer GM, Izatt JA, et al. Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images. Biomedical optics express. 2014;5(10):3568–77. doi: 10.1364/BOE.5.003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasti R, Rabbani H, Mehridehnavi A, Hajizadeh F. Macular OCT classification using a multi-scale convolutional neural network ensemble. IEEE transactions on medical imaging. 2017;37(4):1024–34. [DOI] [PubMed] [Google Scholar]
- 32.Mooney P. Retinal OCT Images (optical coherence tomography) 2017. [December 28, 2022]. Available from: https://www.kaggle.com/datasets/paultimothymooney/kermany2018. [Google Scholar]
- 33.Kermany D, Zhang K, Goldbaum M. Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley data. 2018;2(2). [Google Scholar]
- 34.Gholami P, Roy P, Parthasarathy MK, Lakshminarayanan V. OCTID: Optical coherence tomography image database. Computers & Electrical Engineering. 2020;81:106532. [Google Scholar]
- 35.McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM, and the P-DTAG, et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA. 2018;319(4):388–96. Epub 2018/01/25. doi: 10.1001/jama.2017.19163 . [DOI] [PubMed] [Google Scholar]
- 36.Leeflang MM, Deeks JJ, Takwoingi Y, Macaskill PJSr. Cochrane diagnostic test accuracy reviews. 2013;2(1):1–6. doi: 10.1186/2046-4053-2-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang S, Chang LJBim. Serum matrix metalloproteinase-9 level as a biomarker for colorectal cancer: a diagnostic meta-analysis. 2018;12(4):393–402. doi: 10.2217/bmm-2017-0206 [DOI] [PubMed] [Google Scholar]
- 38.Higgins JP, Thompson SG, Deeks JJ, Altman DGJB. Measuring inconsistency in meta-analyses. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins JP, Thompson SGJSim. Quantifying heterogeneity in a meta‐analysis. 2002;21(11):1539–58. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 40.Alqudah AM. AOCT-NET: a convolutional network automated classification of multiclass retinal diseases using spectral-domain optical coherence tomography images. Med Biol Eng Comput. 2020;58(1):41–53. Epub 2019/11/16. doi: 10.1007/s11517-019-02066-y . [DOI] [PubMed] [Google Scholar]
- 41.Bhatia KK, Graham MS, Terry L, Wood A, Tranos P, Trikha S, et al. DISEASE CLASSIFICATION OF MACULAR OPTICAL COHERENCE TOMOGRAPHY SCANS USING DEEP LEARNING SOFTWARE: Validation on Independent, Multicenter Data. Retina (Philadelphia, Pa). 2020;40(8):1549–57. Epub 2019/10/05. doi: 10.1097/IAE.0000000000002640 . [DOI] [PubMed] [Google Scholar]
- 42.Celebi ARC, Bulut E, Sezer A. Artificial intelligence based detection of age-related macular degeneration using optical coherence tomography with unique image preprocessing. Eur J Ophthalmol. 2022:11206721221096294. Epub 2022/04/27. doi: 10.1177/11206721221096294 . [DOI] [PubMed] [Google Scholar]
- 43.Dong L, He W, Zhang R, Ge Z, Wang YX, Zhou J, et al. Artificial Intelligence for Screening of Multiple Retinal and Optic Nerve Diseases. JAMA Netw Open. 2022;5(5):e229960. Epub 2022/05/04. doi: 10.1001/jamanetworkopen.2022.9960 ; PubMed Central PMCID: PMC9066285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gour N, Khanna P. Multi-class multi-label ophthalmological disease detection using transfer learning based convolutional neural network. Biomedical Signal Processing and Control. 2021;66. doi: 10.1016/j.bspc.2020.102329 [DOI] [Google Scholar]
- 45.He T, Zhou Q, Zou Y. Automatic Detection of Age-Related Macular Degeneration Based on Deep Learning and Local Outlier Factor Algorithm. Diagnostics (Basel). 2022;12(2). Epub 2022/02/26. doi: 10.3390/diagnostics12020532 ; PubMed Central PMCID: PMC8871377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadry S, Rajinikanth V, Crespo RG, Verdu E. Automated detection of age-related macular degeneration using a pre-trained deep-learning scheme. JOURNAL OF SUPERCOMPUTING. 2022;78(5):7321–40. doi: 10.1007/s11227-021-04181-w WOS:000716301900001. [DOI] [Google Scholar]
- 47.Lee CS, Baughman DM, Lee AY. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol Retina. 2017;1(4):322–7. Epub 2017/07/01. doi: 10.1016/j.oret.2016.12.009 ; PubMed Central PMCID: PMC6347658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma D, Kumar M, Khetan V, Sen P, Bhende M, Chen S, et al. Clinical explainable differential diagnosis of polypoidal choroidal vasculopathy and age-related macular degeneration using deep learning. Comput Biol Med. 2022;143:105319. Epub 2022/02/28. doi: 10.1016/j.compbiomed.2022.105319 . [DOI] [PubMed] [Google Scholar]
- 49.Mathews MR, Anzar STM. A lightweight deep learning model for retinal optical coherence tomography image classification. INTERNATIONAL JOURNAL OF IMAGING SYSTEMS AND TECHNOLOGY. doi: 10.1002/ima.22808 WOS:000856143200001. [DOI] [Google Scholar]
- 50.Matsuba S, Tabuchi H, Ohsugi H, Enno H, Ishitobi N, Masumoto H, et al. Accuracy of ultra-wide-field fundus ophthalmoscopy-assisted deep learning, a machine-learning technology, for detecting age-related macular degeneration. Int Ophthalmol. 2019;39(6):1269–75. Epub 2018/05/11. doi: 10.1007/s10792-018-0940-0 . [DOI] [PubMed] [Google Scholar]
- 51.Motozawa N, An G, Takagi S, Kitahata S, Mandai M, Hirami Y, et al. Optical Coherence Tomography-Based Deep-Learning Models for Classifying Normal and Age-Related Macular Degeneration and Exudative and Non-Exudative Age-Related Macular Degeneration Changes. Ophthalmol Ther. 2019;8(4):527–39. Epub 2019/08/14. doi: 10.1007/s40123-019-00207-y ; PubMed Central PMCID: PMC6858411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takhchidi K, Gliznitsa PV, Svetozarskiy SN, Bursov AI, Shusterzon KA. Labelling of data on fundus color pictures used to train a deep learning model enhances its macular pathology recognition capabilities. Bulletin of Russian State Medical University. 2021;(2021(4)). doi: 10.24075/brsmu.2021.040 [DOI] [Google Scholar]
- 53.Tan JH, Bhandary SV, Sivaprasad S, Hagiwara Y, Bagchi A, Raghavendra U, et al. Age-related Macular Degeneration detection using deep convolutional neural network. Future Generation Computer Systems. 2018;87:127–35. 10.1016/j.future.2018.05.001. [DOI] [Google Scholar]
- 54.Thomas A, Harikrishnan PM, Gopi VP, Palanisamy P. An Automated Method to Detect Age-Related Macular Degeneration from Optical Coherence Tomographic Images. Biomedical Engineering: Applications, Basis and Communications. 2021;33(05). doi: 10.4015/s1016237221500368 [DOI] [Google Scholar]
- 55.Wang DP, Wang LJ. On OCT Image Classification via Deep Learning. IEEE PHOTONICS JOURNAL. 2019;11(5). doi: 10.1109/JPHOT.2019.2934484 WOS:000484194500001. [DOI] [Google Scholar]
- 56.Yoo TK, Choi JY, Seo JG, Ramasubramanian B, Selvaperumal S, Kim DW. The possibility of the combination of OCT and fundus images for improving the diagnostic accuracy of deep learning for age-related macular degeneration: a preliminary experiment. Med Biol Eng Comput. 2019;57(3):677–87. Epub 2018/10/24. doi: 10.1007/s11517-018-1915-z . [DOI] [PubMed] [Google Scholar]
- 57.Zapata MA, Royo-Fibla D, Font O, Vela JI, Marcantonio I, Moya-Sanchez EU, et al. Artificial Intelligence to Identify Retinal Fundus Images, Quality Validation, Laterality Evaluation, Macular Degeneration, and Suspected Glaucoma. Clin Ophthalmol. 2020;14:419–29. Epub 2020/02/28. doi: 10.2147/OPTH.S235751 ; PubMed Central PMCID: PMC7025650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Survarachakan S, Prasad PJR, Naseem R, Perez de Frutos J, Kumar RP, Lango T, et al. Deep learning for image-based liver analysis—A comprehensive review focusing on malignant lesions. Artif Intell Med. 2022;130:102331. Epub 2022/07/10. doi: 10.1016/j.artmed.2022.102331 . [DOI] [PubMed] [Google Scholar]
- 59.Romond K, Alam M, Kravets S, Sisternes L, Leng T, Lim JI, et al. Imaging and artificial intelligence for progression of age-related macular degeneration. Exp Biol Med (Maywood). 2021;246(20):2159–69. Epub 2021/08/19. doi: 10.1177/15353702211031547 ; PubMed Central PMCID: PMC8718252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Communications of the ACM. 2017;60(6):84–90. [Google Scholar]
- 61.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014. [Google Scholar]
- 62.He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. [Google Scholar]
- 63.Zhu Y, Newsam S, editors. Densenet for dense flow. 2017 IEEE international conference on image processing (ICIP); 2017: IEEE. [Google Scholar]
- 64.Xie Z, He F, Fu S, Sato I, Tao D, Sugiyama M. Artificial Neural Variability for Deep Learning: On Overfitting, Noise Memorization, and Catastrophic Forgetting. Neural computation. 2021;33(8):2163–92. Epub 2021/07/27. doi: 10.1162/neco_a_01403 . [DOI] [PubMed] [Google Scholar]
- 65.Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, et al. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. 2021;8(1):1–74. doi: 10.1186/s40537-021-00444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, Xie Z, Zhang W, Xu X, editors. ResNet and Model Fusion for Automatic Spoofing Detection. Interspeech; 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(DOC)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.