Abstract
Fungi have evolved over millions of years and their species diversity is predicted to be the second largest on the earth. Fungi have cross-kingdom interactions with many organisms that have mutually shaped their evolutionary trajectories. Zygomycete fungi hold a pivotal position in the fungal tree of life and provide important perspectives on the early evolution of fungi from aquatic to terrestrial environments. Phylogenomic analyses have found that zygomycete fungi diversified into two separate clades, the Mucoromycota which are frequently associated with plants and Zoopagomycota that are commonly animal-associated fungi. Genetic elements that contributed to the fitness and divergence of these lineages may have been shaped by the varied interactions these fungi have had with plants, animals, bacteria, and other microbes. To investigate this, we performed comparative genomic analyses of the two clades of zygomycetes in the context of Kingdom Fungi, benefiting from our generation of a new collection of zygomycete genomes, including nine produced for this study. We identified lineage-specific genomic content that may contribute to the disparate biology observed in these zygomycetes. Our findings include the discovery of undescribed diversity in CotH, a Mucormycosis pathogenicity factor, which was found in a broad set of zygomycetes. Reconciliation analysis identified multiple duplication events and an expansion of CotH copies throughout the Mucoromycotina, Mortierellomycotina, Neocallimastigomycota, and Basidiobolus lineages. A kingdom-level phylogenomic analysis also identified new evolutionary relationships within the subphyla of Mucoromycota and Zoopagomycota, including supporting the sister-clade relationship between Glomeromycotina and Mortierellomycotina and the placement of Basidiobolus as sister to other Zoopagomycota lineages.
Keywords: comparative genomics, CotH, evolution, fungi, phylogenomics, zygomycetes
Significance.
Fungal phylogeny and the evolution of their early-diverging lineages have been conundrums. The study presents phylogenomic analyses across Kingdom Fungi using the largest collection of zygomycete genomes to date, which identified new phylogenetic relationships of the six subphyla. Phylum-specific genome content was also revealed to support the independent evolution of the two zygomycete phyla, including the evolution of the CotH, an important pathogenicity factor of Mucormycosis. Our work provides a large genomic resource for an understudied fungal group as well as a wide spectrum of fundamental views on the evolution of fungal pathogens with the global climate changes.
Introduction
Fungi play diverse ecological roles and interact with various organisms in both terrestrial and aquatic environments (James, Kauff, et al. 2006; Stajich et al. 2009; Spatafora et al. 2017; Fisher et al. 2020). Since their divergence from a common ancestor with animals over 1 billion years ago, fungi have evolved complex relationships with other organisms, including animals, bacteria, plants, protists, and other fungi (Currie et al. 2003; Frey-Klett et al. 2011; Parfrey et al. 2011; Gruninger et al. 2014; Uehling et al. 2017; Wang et al. 2018; Chambouvet et al. 2019; Malar et al. 2021). As a distinct eukaryotic kingdom, fungi are characterized by chitinous cell walls and osmotrophic feeding style, although neither of these characters is diagnostic for the kingdom (Richards et al. 2017; James et al. 2020). The versatile enzymes secreted by fungi facilitate their success in utilization of diverse polysaccharides and are key members of ecosystems supporting nutrient cycling processes (Hori et al. 2013; Chang et al. 2015; Solomon et al. 2016; Richards and Talbot 2018; Chang et al. 2022). Zygomycete fungi are a historically enigmatic group as their diversity and phylogenetic placement on the fungal tree of life remained somewhat cryptic based on morphological characters alone. The lineages emergence coincides with major transition of fungi from aquatic environment to terrestrial ecologies, which was characterized by the evolutionary loss of the flagellum (James, Kauff, et al. 2006; James, Letcher, et al. 2006; Chang et al. 2021). The zygomycete fungi are recognized by their gametangial conjugation, production of zygospore, and coenocytic aseptate or septate hyphae (White et al. 2006; Hibbett et al. 2007; Spatafora et al. 2017; Naranjo-Ortiz and Gabaldón 2020). Nevertheless, zygospore structures have not been observed for most members of zygomycete fungi due to their cryptic sexual stage or lack of appropriate culture approaches. Zygomycete fungi were found to be paraphyletic based on genome-scale evidence; as a result, two new phyla (Mucoromycota and Zoopagomycota) were established to accommodate the current members (Spatafora et al. 2016). However, incomplete sampling of zygomycete lineages has made resolution of the origin of terrestrial fungi difficult to resolve with standard phylogenetic approaches (Chang et al. 2021; Li et al. 2021).
Mycological and fungal cell biology research has been historically biased in favor of members of the Dikarya. Several established research model organisms have advanced fields of cell biology including the brewer's yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, the red bread mold Neurospora crassa, and the filamentous mold Aspergillus nidulans. These model organisms contributed to an expansion in the understanding of eukaryotes. Fungi were among the some of the first sequenced eukaryotic genomes (Goffeau et al. 1996; Wood et al. 2002; Galagan et al. 2003; Galagan et al. 2005). However, genomic research on zygomycete fungi had to wait for the first Mucoromycotina genome to be sequenced in 2009 (Ma et al. 2009). The majority of our existing knowledge of zygomycetes has come from studies of arbuscular mycorrhizae (Glomeromycotina) or saprophytes classified in Mucoromycota, such as the black bread mold Rhizopus stolonifer. Studies on the other zygomycete phylum, Zoopagomycota, are still rare, and the biodiversity of Zoopagomycota fungi is likely greatly underestimated and the research progress is largely hindered by the lack of axenic cultures. Culture independent studies have identified multiple zygomycetes as amplicon-based operational taxonomic units (OTUs) in unexplored ecological sites (Metcalf et al. 2016; Picard 2017; Pombubpa et al. 2020; Reynolds et al. 2021) and many “unknown” fungal OTUs will likely to be identified with the help of increasing fungal genomes, especially more representatives in the sparsely sequenced zygomycete lineages.
To fill this gap, our recent emphasis on sequencing zygomycete genomes through the ZyGoLife project (Spatafora et al. 2016; https://zygolife.org) has produced over a hundred genomes. The output has become the largest collection of genomic information for this fungal clade. Various techniques were also developed and employed to obtain genome sequences of the uncultured zygomycete species. The breakthroughs include the single-cell genomics as well as fungus-host co-culture techniques (Ahrendt et al. 2018) and sequencing of metagenomes of sporocarps (Chang et al. 2019). Progress on genomics and related multi-omics has greatly expanded our knowledge on zygomycetes. This includes the identification of a mosquito-like polyubiquitin gene in a zygomycete fungus inhabiting the gut of mosquitoes (Zancudomyces culisetae, Zoopagomycota) (Wang et al. 2016), the discovery of a photosynthetic mycelium using algal symbionts (Linnemannia elongata, Mortierellomycotina) (Du et al. 2019; Vandepol et al. 2020), the isolation of cicada behavior modifying alkaloids from Massospora (Entomophthoromycotina) (Boyce et al. 2019), and the expansion of secondary metabolite genes of amphibian gut fungi (Basidiobolus, Entomophthoromycotina) via Horizontal Gene Transfer from bacteria co-existing in the gastrointestinal tract (Tabima et al. 2020). However, a conundrum remains as to the evolutionary history of the zygomycete fungi. What evolutionary processes were associated with the divergence of the ancestors of Mucoromycota and Zoopagomycota into species which primarily associate with plants and plant material or animal and fungal hosts, respectively. We hypothesize that comparisons of gene content will enable identification of genetic elements that have contributed to their success in these ecologies and their reproductive strategies and may be reflected in lineage-specific genes, those with expanded copy number or enrichment in specific pathways or processes that underpin adaptations to these hosts and environments. In addition, the construction of a well-resolved phylogenetic tree incorporating the expanded collection of zygomycete genomes is an important framework to consider the complex natural history and relationships among these diverse fungi. Our work has contributed to the generation of 131 recent zygomycete genomes (supplementary table S1, Supplementary Material online), which were used to investigate the evolution and cryptic genetics behind the biology of these early diverging fungi.
The focus on these phyla is motivated by not only understanding their ecological roles and history, but also in the context of the increase in Mucormycosis, a deadly human-infectious disease, that has risen in prevalence and public attention due to high infection rates and co-morbidity during the COVID-19 pandemic (Garg et al. 2021; Revannavar et al. 2021). Mucormycosis is caused by members of Mucoromycotina, in particular many genera of the Mucorales fungi (Soare et al. 2020). We cataloged the prevalence of Mucormycosis pathogenicity factors across Mucorales genomes and profiled their evolutionary conservation among members of the Fungal Kingdom. We identified the genes for the Mucormycosis invasin factor in three Mortierellomycotina species as well (Dissophora ornate, Lobosporangium transversale, and Mortierella species), which all share a highly similar protein motif associated with the disease caused by Mucorales fungi indicating that these fungi may have additional potential for mammalian infection and the more ancient nature of this factor within these fungi. Our study highlights the importance of research on zygomycetes to characterize the unique and shared molecular components of their biology that can be examined as more genome sequences become available. The phylogeny with improved resolution will enhance the study of the evolutionary relationships for both organismal and molecular genetics of these important fungi.
Results
Phylogenetic Relationships and Genome Statistics of Zygomycete Fungi
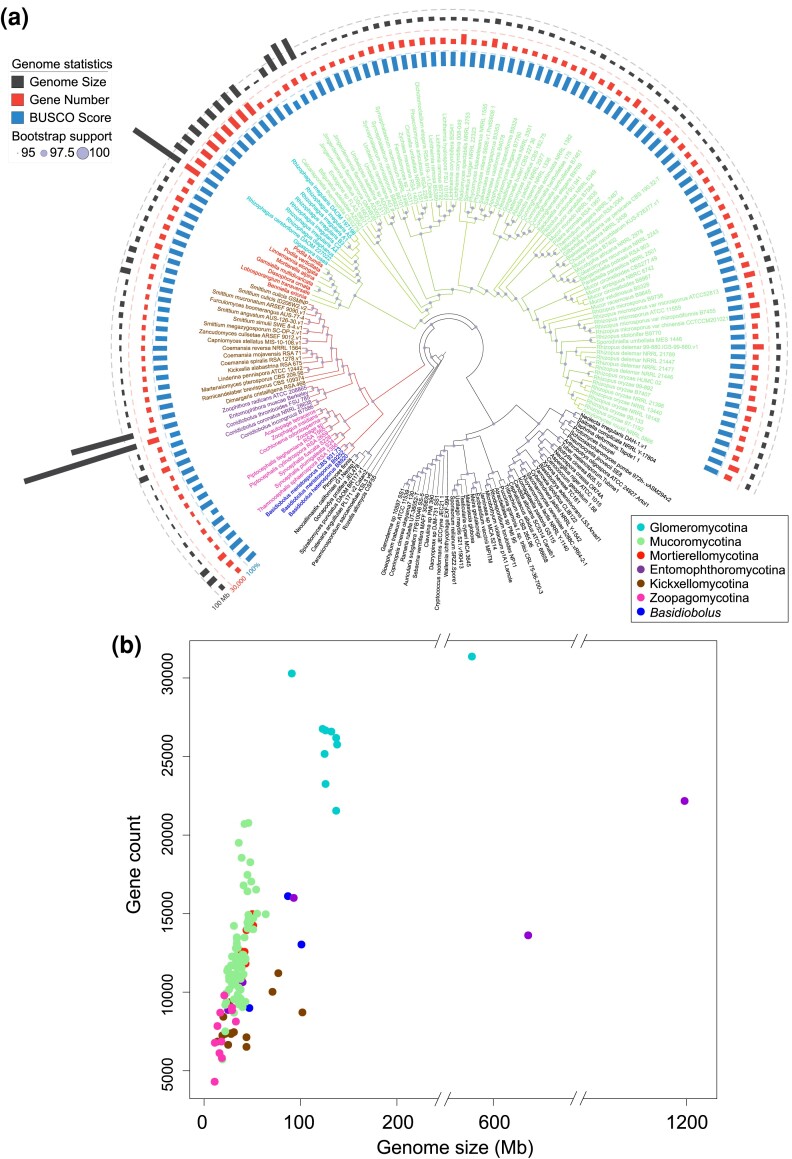
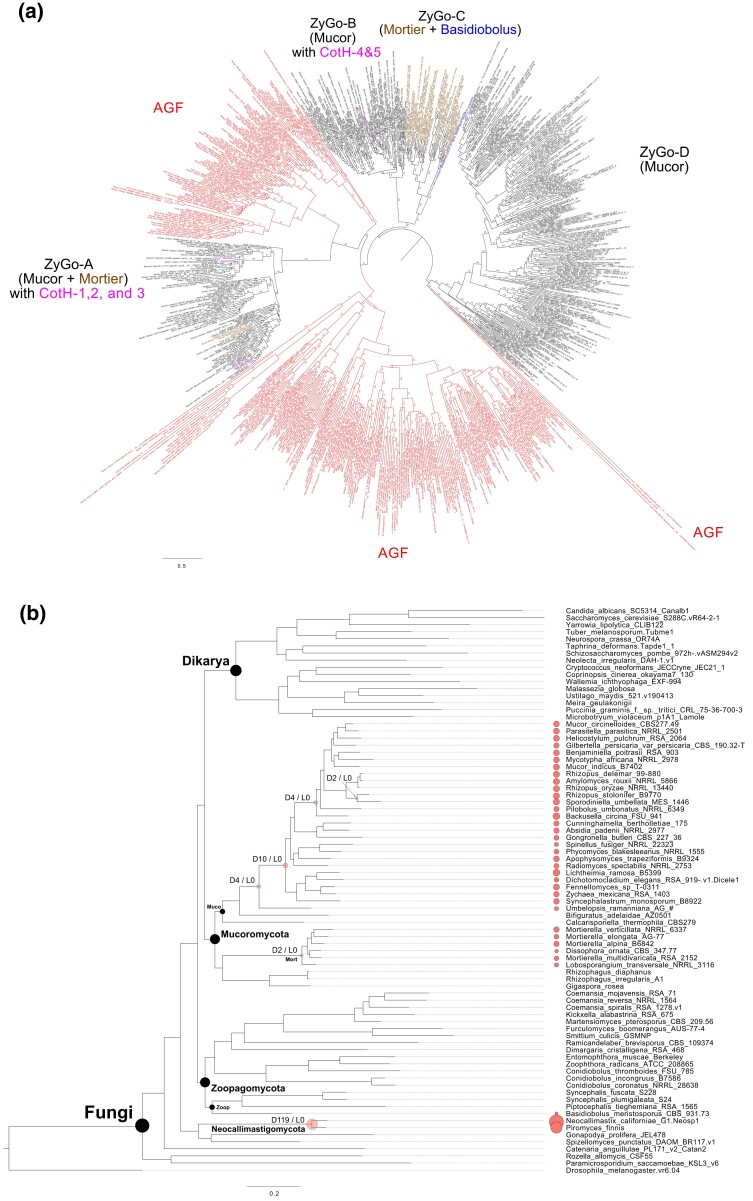
Collaborative efforts to sequence fungi have generated the 131 zygomycete genomes presented in this study and the relationships among these species have remained an open research question. Most of the assembled zygomycete genomes were assessed to have BUSCO scores higher than 80% (fig. 1a, supplementary table S1, Supplementary Material online). The phylogenetic analysis using all available zygomycete genomes and 50 additional representatives from other fungal clades (fig. 1a and supplementary fig. S1a, Supplementary Material online) provided an updated species tree representing the placement of these fungi in the kingdom. At the phylum level, the reconstructed phylogeny exhibits the same topology as presented in Spatafora et al. (2016). That is, Zoopagomycota forms a sister group to the clade comprising Mucoromycota and Dikarya, and the traditional zygomycete fungi (Mucoromycota and Zoopagomycota) remain paraphyletic. The increased sampling size and new set of phylogenetic markers provide additional confidence in these arrangements. This is in contrast to a kingdom-wide study that also uses protein-coding genes from BUSCO data sets suggests that zygomycetes could still be monophyletic with a different sampling strategy (Li et al. 2021). It should be noted that the marker sets used in this study (fungi_odb10 with 758 markers) and Li et al. (fungi_odb9 with 290 markers) differ, as well as the strategies to extract the hits—protein searches against genome annotations in this study and BUSCO predicted gene models in Li et al. Regardless of whether zygomycetes are paraphyletic or monophyletic, it is not controversial that Mucoromycota and Zoopagomycota are monophyletic phyla. At the subphylum level, however, new phylogenetic relationships were recovered with consistency in both the comprehensive tree (fig. 1a and supplementary fig. S1a, Supplementary Material online) and the backbone tree (fig. 2a and supplementary fig. S1b, Supplementary Material online). For example, Glomeromycotina grouped with Mortierellomycotina instead of being the earliest branch within Mucoromycota (Spatafora et al. 2016). Basidiobolus members were found grouped within Entomophthoromycotina (Spatafora et al. 2016); however, they were found as a sister lineage to the rest of the Zoopagomycota in this study (figs. 1a and 2a, and supplementary fig. S1, Supplementary Material online). The present subphylum-level classification received full bootstrap supports (100/100) in the comprehensive tree (fig. 1a), although gene/site concordance factors are relatively low (supplementary fig. S1a, Supplementary Material online). Tree topologies are identical in both the comprehensive (fig. 1a) and the backbone tree (fig. 2a). Two nodes within Zoopagomycota clade received relatively low support values in the backbone tree (82/100, fig. 2a); however, both were fully supported by bootstrap values in the comprehensive tree (supplementary fig. S1a, Supplementary Material online).
Fig. 1.
Phylogenetic relationships and genome statistics of zygomycete fungi. (a) The maximum-likelihood tree was inferred from a phylogenomic data set of 617 protein sequences identified in the included 181 genomes. Branches of Mucoromycota and Zoopagomycota were colored in green and red separately, while tip labels were in the color scheme according to the subphyla information. The bootstrap supports are indicated on each node relatively. Tracks from the inside to outside are mapped based on the BUSCO scores, protein-coding gene numbers, and genome size of included zygomycete fungi (detailed bootstrap values, concordance factors, and branch lengths are shown in supplementary fig. S1a, Supplementary Material online). (b) The density of protein-coding genes in each genome was plotted using genome sizes on the x-axis against the gene counts on the y-axis. Each dot was colored based on their phylogenetic placement shown in the legend.
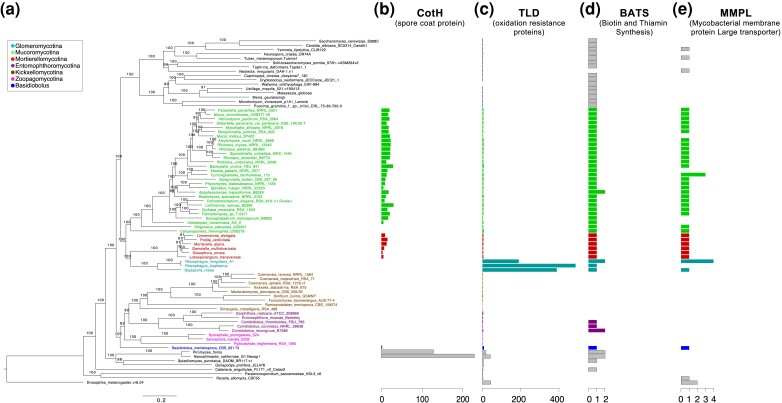
Fig. 2.
Phylogenetic backbone and highlighted genome content in zygomycete fungi. Zygomycete genomes that are well assembled (BUSCO score above 80%) and represent unique phylogenetic positions were selected to reconstruct the backbone phylogenomic tree. (a) The backbone phylogenomic tree of zygomycetes includes 80 taxa (rooted with Drosophila melanogaster). All bootstrap values (out of 100) were labeled on the branches (concordance factors are shown in supplementary fig. S1b, Supplementary Material online). (b–e) Protein family domains found with striking patterns in zygomycete fungi are plotted with the copy numbers individually.
Our results suggest that the saprobe, Calcarisporiella thermophila, is sister to the rest of the Mucoromycotina. Plant symbionts like Bifiguratus, Endogone, and Jimgerdemannia form a monophyletic clade which was placed between C. thermophila and Mucoromycotina (fig. 1a). Members of saprobes, pathogens, and mycoparasites were joined in more derived groups of Mucoromycotina.
In the Kickxellomycotina clade, the mycoparasite, Dimargaris cristalligena, is sister to the other members. Ramicandelaber brevisporus follows and leads to two separate monophyletic clades composed of insect symbionts (e.g., Furculomyces and Smittium) and soil saprobes (e.g., Coemansia, Kickxella, and Martnesiomyces). Both clades (insect symbionts and soil saprobes) are on relatively long branches implying early divergent evolution and underexplored biodiversity (fig. 2a and supplementary fig. S1, Supplementary Material online). Insect pathogens were grouped together on a separate lineage, Entomophthoromycotina, forming a sister clade to Kickxellomycotina (figs. 1a and 2a). The three included Conidiobolus species support a paraphyletic genus with the C. coronatus monophyletic with C. incongruus, while C. thromboides was more closely related to Zoophthora radicans and Entomophthora muscae. Zoopagomycotina is monophyletic and sister to the joined group of Entomophthoromycotina (excluding Basidiobolus) and Kickxellomycotina (figs. 1a and 2a).
The density of genes arranged in the genome of zygomycete fungi exhibited varying patterns among subphyla which was observed in plots of gene counts against genome sizes (fig. 1b). Most zygomycete fungi have genome sizes ranging from 20 to 100 Mb and gene counts range from 5 to 20 k. The Mucoromycotina fungi have relatively similar genome sizes with an average of 39 Mb, ranging from 19 to 75 Mb (excluding Endogone and Jimgerdemannia due to genome incompleteness), but gene counts vary from 6 to 21 k. The soil saprobes in Kickxellomycotina and the small animal associates in Zoopagomycotina have small genome sizes (10–20 Mb) and gene counts (4–8 k). On the other hand, Glomeromycotina fungi tend to have large genome sizes (>100 Mb) with the most abundant gene numbers (20–30 k) in all zygomycete fungi, which are among the largest fungal genome sizes sequenced to date. As an extreme case, the genome sizes of Entomophthoromycotina members exhibit the widest range and can be as large as 1.2 Gb according to the existing genome assemblies; however, their gene counts (9–23 k) are more modest. One recent genome announcement of Entomophthoromycotina members, Massospora cicadina, presents a large genome size (1.5 Gb) dominated by transposable elements and with fewer genes (7,532) (Stajich et al. 2022).
Orthologous Gene Families and Pfam Domains in Zygomycete Fungi
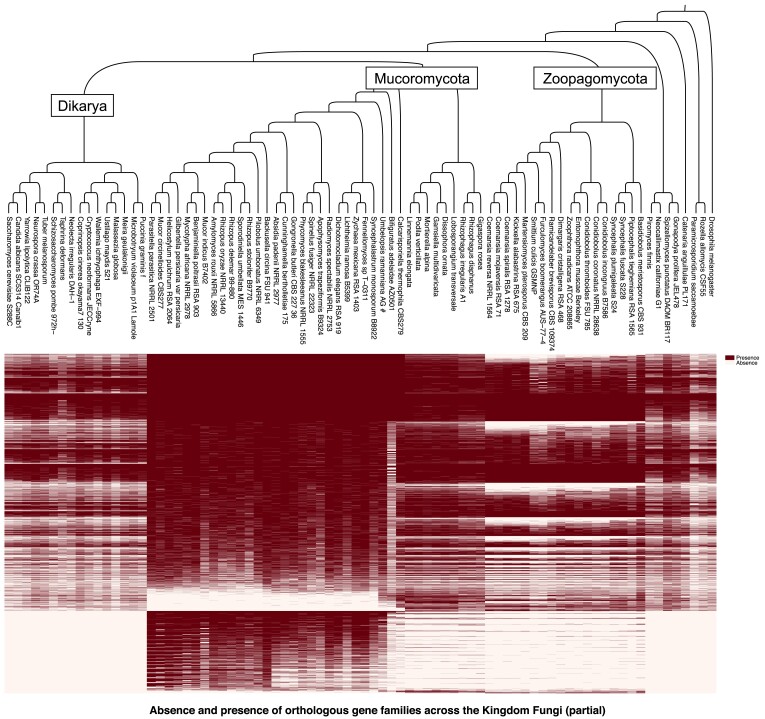
The 80 species used for the backbone tree were examined for orthologous gene families across the Kingdom Fungi. We identified 8,208 orthologous families which had genes from at least 11 of the 80 genomes. These gene families were subjected to more focused analyses to examine the presence/absence pattern of genome contents across the Kingdom Fungi, with a special attention on the divergent evolution between Mucoromycota and Zoopagomycota (fig. 3). The Mucoromycota members harbor 171 phylum-specific gene families that are present in at least two of the three Mucoromycota subphyla and absent in all other fungal lineages, while Zoopagomycota only have nine such gene families (table 1). At the subphylum level, there were considerably more lineage-specific gene families, ranging from 1,186 (in Zoopagomycotina) to 7,779 (in Mucoromycotina) (table 1).
Fig. 3.
Absence and presence of orthologous gene families across the Kingdom Fungi. Orthologous gene families were examined in the genomes included in the backbone tree. The 8,208 gene families were found present in at least 10 of the 80 taxa and thus included to examine the absence/presence pattern of genome content among different fungal lineages (a complete map showing the unfiltered 62,689 gene families was included in supplementary fig. S2, Supplementary Material online).
Table 1.
Summary of Phylum-Level and Subphylum-Level Lineage-Specific Genes and Pfam Domains in Zygomycete Fungi
| Phylum-level (>10 taxa) | Subphylum-level (>1 taxa) | |||||||
|---|---|---|---|---|---|---|---|---|
| Mucoromycota | Zoopagomycota | Mucoromycotina | Mortierellomycotina | Glomeromycotina | Kickxellomycotina | Entomorphthoromycotina | Zoopagomycotina | |
| Lineage-specific genes | 171 | 9 | 7,779 | 2,742 | 5,572 | 1,706 | 2,209 | 1,186 |
| Pfam domains | 2 | 0 | 32 | 11 | 24 | 0 | 5 | 1 |
We used protein domains cataloged in the Pfam database as an additional means to catalog unique and shared content. A total of 7,616 Pfam models had at least one similar sequence in the examined 80 genomes. Mucoromycota members possess two unique Pfam domains, with the CheR (PF01739) found in all three subphyla and the C9orf72-like (PF15019) in Mucoromycotina and Mortierellomycotina, while no phylum-specific Pfam domains were identified in the Zoopagomycota. At the subphylum level, a range of unique Pfam domains were observed, with 11 to 32 in the three subphyla of Mucoromycota and 0–5 in the ones in Zoopagomycota (table 1 and supplementary table S3, Supplementary Material online). Interestingly, the CotH domain (PF08757), a potential invasin factor of Mucormycosis, was found in Mortierellomycotina, Basidiobolus, and Neocallimastigomycota genomes (fig. 2b), but had previously only been described in the Mucoromycotina (Chibucos et al. 2016). In addition, the oxidation resistance protein domain (TLD, PF07534) has greatly expanded in copy number in the Glomeromycotina with up to 400 copies (fig. 2c). Kickxellomycotina and Zoopagomycotina members lacked Biotin and Thiamin synthesis associated domain (BATS, PF06968) and mycobacterial membrane protein large transporter domain (MMPL, PF03176) (fig. 2dand 2e). Interestingly, Basidiobolus meristosporus is the only Zoopagomycota member that maintains at least one copy of every examined domain (fig. 2b–e), including CotH and MMPL that are absent in all other Zoopagomycota members.
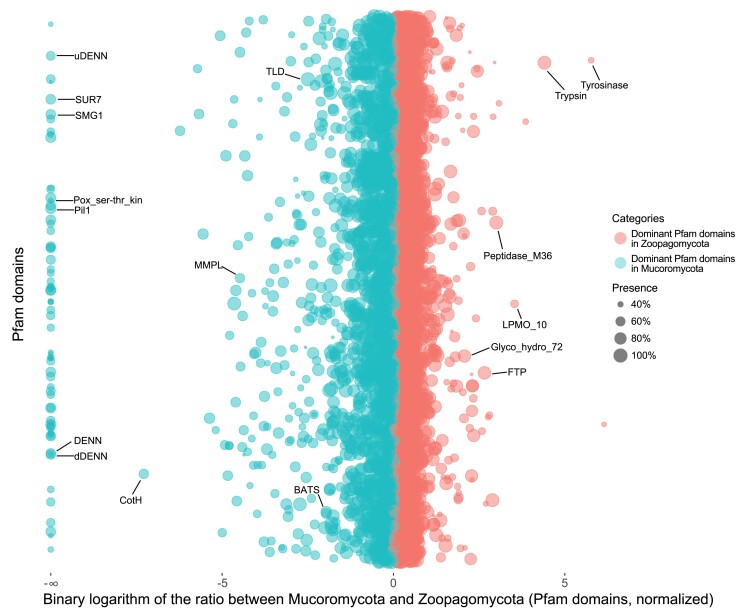
To identify Pfam domains that may contribute to the divergent evolution between Mucoromycota and Zoopagomycota, we calculated the relative abundance of each Pfam domain in their genomes. In total, 285 Pfam domains were present at least 4-fold differences (i.e., absolute value of the binary logarithm >2) between the two phyla with 243 of them in higher abundance in Mucoromycota while 42 in Zoopagomycota (fig. 4 and supplementary table S4, Supplementary Material online). Without consideration of nonzygomycete lineages, we found 70 Pfam domains in Mucoromycota that are completely missing in Zoopagomycota, whereas no such Pfam domains can be identified in Zoopagomycota. Zoopagomycota is a historically understudied fungal clade with few representative genomes until our recent studies. As a result, the lack of Zoopagomycota specific Pfam domains may be an artifact of insufficient sampling before domain curation in Pfam. To overcome this possibility, we examined the orthologous gene family data set to calculate the relative abundance of gene families to test for differences between the two phyla. This revealed 22 gene families in Zoopagomycota that were absent in all Mucoromycota members (supplementary fig. S3, Supplementary Material online and supplementary File 1, Supplementary Material online). Gene Ontology analysis shows that more than 50% of these genes are involved in binding, catalytic activity, cellular process, and metabolic process (supplementary fig. S4, Supplementary Material online). Finer scales of examination suggest they are closely related to nitrogen compound, organic substance, and primary metabolic process (supplementary fig. S5, Supplementary Material online).
Fig. 4.
Protein family (Pfam) domains with differentiated enrichment in Mucoromycota or Zoopagomycota. Each dot represents a Pfam domain found in zygomycete fungi. The x-axis is the binary logarithm of the Pfam copy ratios between Zoopagomycota and Mucoromycota, and the y-axis is used to rank the Pfam domains in alphabetical order. The Pfam domains enriched in Mucoromycota are shown on the left side in cyan color, and the Zoopagomycota-enriched ones are on the right side in red color. The bubbles (Pfam domains) with bigger sizes are shared by more zygomycetes members. The Pfam domains aligned on the left edge are domains only found in Mucoromycota and absent in Zoopagomycota. The domains discussed in the text were labeled with the Pfam name. A detailed chart including the names and ratios of all Pfam domains is also provided (supplementary table S4, Supplementary Material online).
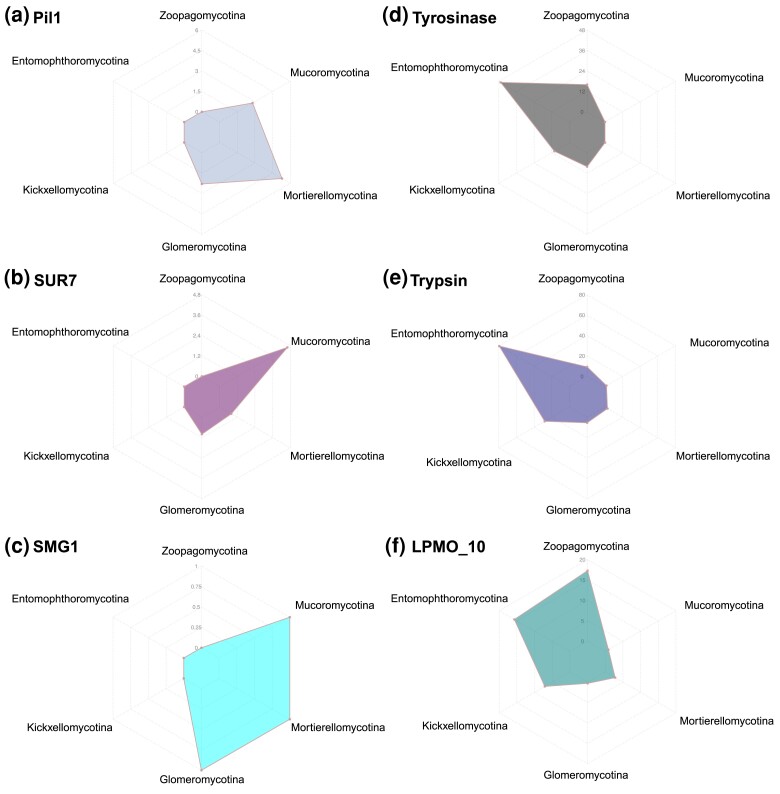
We found that many phylum-level distinct Pfam domains were favored unevenly in each subphylum group (fig. 5). For example, both Pil1 (PF13805) and SUR7 (PF06687) domains are eisosome components and are involved in the process of endocytosis. They are missing entirely from the Zoopagomycota but are encoded in the genomes of all (Pil1) or a majority (SUR7, except for Mortierella multidivaricata and Gigaspora rosea) of Mucoromycota members (figs. 4, 5a, and 5b). Interestingly, the Pil1 domain was enriched in copy number in the Mortierellomycotina (fig. 5a), and SUR7 domain has the largest copy number in Mucoromycotina (fig. 5b). The SMG1 domain (PF15785), a phosphatidylinositol kinase-related protein kinase, is a key regulator of growth. The Mucoromycota members maintain a single-copy SMG1 domain (except for Cunninghamella bertholletiae with 3 copies, and none in Mucor circinelloides, Phycomyces blakesleeanus, and Syncephalastrum monosporum), which is absent in Zoopagomycota species (fig. 5c). There are 67 additional Pfam domains including DENN (PF02141), uDENN (PF03456), dDENN (PF03455), and Pox_ser-thr_kin (PF05445) (supplementary table S4, Supplementary Material online) with a similar presence/absence pattern and may be important components to better understand and characterize the Mucoromycota fungi.
Fig. 5.
Subphylum-level distribution of six Pfam domains that may contribute to the divergent evolution of zygomycete fungi. The scales on each axis of the radar plots indicate the average copy number of the domain in each subphylum. (a–c) Pfam domains shared in all Mucoromycota subphyla and absent in the entire Zoopagomycota. (d–f) Distinct Pfam domains in Zoopagomycota subphyla and largely missing in Mucoromycota.
In contrast, while there are no Zoopagomycota-specific Pfam domains, there are some domains that exhibit copy number variance at the subphylum level. For example, the Tyrosinase domain (PF00264) is an important enzyme that controls the production of melanin and parasite encapsulation, especially in insects. It is also suggested that Tyrosinase may be involved in the host-microbe defensive mechanism. The Tyrosinase domains are found on average with 48 copies in the Entomophthoromycotina members but absent in nearly all Mucoromycotina (except for Calcarisporiella thermophila with 7 copies) and Mortierellomycotina (except for Mortierella verticillata with 1 copy) (fig. 5d). Similarly, Trypsin domain (PF00089), serine protease found in the digestive system of many vertebrates, was also enriched in copy number in the Entomophthoromycotina with 80 copies on average (fig. 5e). The domain LPMO_10 (PF03067) is found in lytic polysaccharide monooxygenases which can cleave glycosidic bonds in chitin and cellulose and is significantly enriched in Zoopagomycota (fig. 5f). All three examples (Trypsin, Tyrosinase, and LPMO_10) are related to animal-fungus interactions in the degradation of protein, chitin, and cellulose.
Discovery of CotH in Early-Diverging Fungi
The CotH domain as characterized in Mucorales fungi has positive correlations with the clinical pathogenesis of Mucormycosis (Chibucos et al. 2016). In our kingdom-wide study, we found additional copies of the CotH domain in a broader collection of fungi. Other than in Mucorales fungi, CotH was also found in Basidiobolus, Mortierellomycotina, and Neocallimastigomycota. The presence of this domain could indicate the potential of these fungi to support pathogenic interaction with animal hosts (fig. 2b). A total of 846 CotH copies were identified in 34 zygomycete genomes and two Neocallimastigomycota representatives (contributing 348 of the copies). Five CotH families (CotH 1–5) that were previously classified in Rhizopus oryzae were included in our phylogenetic analysis and helped us categorize the newly identified CotH copies (fig. 6a). Zygomycete CotH copies formed four distinct clades. ZyGo-A clade includes CotH families 1–3 that maintain true invasin motifs and are restricted to only Mucoromycotina and Mortierellomycotina members. ZyGo-B clade includes CotH families 4–5 with copies from Mucoromycotina. ZyGo-C clade is grouped with ZyGo-B with low support (34/100) and includes copies from Mortierellomycotina, and Basidiobolus. ZyGo-D clade has the largest number of members among the four but only includes copies from Mucoromycotina. Both ZyGo-C and ZyGo-D clades represent new families of CotH not previously described. Interestingly, the distantly related anaerobic gut fungi (AGF, Neocallimastigomycota) have homologs of the CotH domain and copies are found in several distinct clades. In total, 311 duplications, zero transfers, and 106 losses were identified along the evolution of CotH families in Kingdom Fungi. Six nodes were associated with more than one duplication event (fig. 6b). The absence of CotH in the most recent common ancestor of fungi was also inferred by Notung reconciliation analysis.
Fig. 6.
Phylogenetic analysis and evolution of CotH in Kingdom Fungi. (a) The 754 fungal CotH copies were identified from Mortierellomycotina (brown), Mucoromycotina (black), Basidiobolus (blue), and Neocallimastigomycota (red). The CotH phylogenetic tree was midpoint rooted and reconstructed using the maximum likelihood method with bootstrap supports (out of 100) labeled on each branch. The analysis included previously classified CotH families 1–5 (pink) to help categorize newly identified fungal CotH. (b) Reconstruction of CotH evolution in Kingdom Fungi with Notung. CotH copies identified in each genome were plotted at tree tips with proportional sizes. Nodes with more than one duplication event were highlighted with red bubbles and labeled with duplication (“D”) and loss (“L”) events. Node abbreviation: Muco, Mucoromycotina; Mort, Mortierellomycotina; Zoop, Zoopagomycotina.
Discussion
Genome Evolution of Zygomycete Fungi
Zygomycetes are important members of early diverging fungi and studying their evolutionary history can help us better understand the eukaryotic transition to terrestrial habitats. Zygomycete fungi are ubiquitous and can live as arbuscular mycorrhizae, ectomycorrhizae, saprobes, or symbionts of various organisms, including animals, bacteria, plants, and fungi. During the evolutionary adaptation and diversification of zygomycetes, many associated organisms (hosts, symbionts, etc.) may have mutually shaped the structure and content of their genomes. Mucoromycotina members have served as exemplars to investigate various evolutionary events at the genome-scale. For example, whole-genome duplications have been identified repeatedly in Mucoromycotina (Ma et al. 2009; Corrochano et al. 2016), which contributed to the large expansion of gene counts (5–20 k) among some Mucoromycotina members (fig. 1b). Phylogenomic analyses suggest that an early split of Mucoromycotina involved the evolution of thermophily (i.e., Calcarisporiella thermophila) (figs. 1a and 2a), which is followed by various lineages containing members of ectomycorrhizae, mycoparasites, plant, and animal pathogens. In addition, some genomes have been colonized to varying degrees by transposable elements (TEs) in some Mucoromycotina taxa, including Rhizopus oryzae (=R. delemar) (Ma et al. 2009) and Endogone sp. (Chang et al. 2019). The high proportion of TEs was also evident in other lineages of zygomycete fungi, like Gigaspora members (Morin et al. 2019) and Basidiobolus meristosporus (Muszewska et al. 2017). It has been suggested that TEs may have played a role in shaping transcriptional profiles, helped fungi adapt to different ecological niches, and contributed to the current fungal biodiversity (Castanera et al. 2016; Muszewska et al. 2017). It is still unclear what roles TE might have played in the evolution of Entomophthoromycotina members that exhibit the widest span of genome sizes (25–1,200 Mb) in Kingdom Fungi and what resulted in the gigantic size of Entomophthora muscae and Massospora cicadina. More samples from this and related lineages (e.g., Batkoa, Eryniopsis, Furia) may help us reconstruct the evolutionary history for the observed genome size modification in zygomycete fungi.
Phylogenomics of Zygomycetes and Basidiobolus
Zygomycete fungi hold important phylogenetic placement on the fungal tree of life. The former taxonomic unit, Zygomycota, has been recognized paraphyletic and thus been abandoned and replaced by Mucoromycota and Zoopagomycota to accommodate the six major lineages—Glomeromycotina, Mortierellomycotina, Mucoromycotina, Entomophthoromycotina, Kickxellomycotina, and Zoopagomycotina (James, Kauff, et al. 2006; White et al. 2006; Hibbett et al. 2007; Spatafora et al. 2016). Since the loss of flagella, the first evolutionary split of terrestrial fungi leads to Zoopagomycota and the clade of Mucoromycota and Dikarya (Chang et al. 2021). Mucoromycota is the sister clade of the subkingdom Dikarya clades (Ascomycota and Basidiomycota) (figs. 1a and 2a), and analysis of zygomycete fungi is essential to accurately reconstruct the evolutionary events that led to major lineages of terrestrial fungi. The arbuscular mycorrhizal fungi of Glomeromycotina with their distinct ecology formed a monophyletic clade with the soil saprobes and root endophytes of Mortierellomycotina (figs. 1a and 2a). Mucoromycota members are mostly associated with plants or more commonly as decomposers of plant carbohydrates. Zoopagomycota members are mostly animal associated (either as commensals or pathogens) or mycoparasites. The Entomophthoromycotina clade presents several interesting patterns. For example, our phylogenomic results confirm the nonmonophyly of Conidiobolus and encourage further work to reclassify this genus (Nie et al. 2020). Based on a four-gene phylogeny, three new genera (Capillidium, Microconidiobolus, and Neoconidiobolus) were proposed to delimitate the paraphyletic Conidiobolus. Conidiobolus thromboides has been renamed as a member of the Neoconidiobolus genus (Nie et al. 2020). In addition, our results suggest Basidiobolus, a traditional member of Entomophthoromycotina, as the earliest diverging lineage within Zoopagomycota (figs. 1a and 2a; supplementary fig. S1, Supplementary Material online).
Basidiobolus has been characterized as a “rogue” taxon and is often found with conflicting phylogenetic placements. Using nuclear rRNA genes (18S + 28S + 5.8S genes), Basidiobolus, Olpidium brassicae (a plant pathogen), and Schizangiella serpentis (a snake pathogen) were grouped together and placed at the earliest diverging branch within Zoopagomycota (White et al. 2006). In a separate study using four genes (nuclear 18S and 28S rDNA, mitochondrial 16S, and RPB2), Basidiobolus was interpreted as the earliest diverging member of Entomophthoromycotina (Gryganskyi et al. 2012). A genome-scale study based on 192 conserved orthologous proteins favored the Basidiobolus placement in Entomophthoromycotina as well (89/100 bootstrap support) (Spatafora et al. 2016). Interestingly, another genome-scale phylogenetic study examining the entire Kingdom Fungi found that Basidiobolus formed a sister clade to Mucoromycota instead of joining Zoopagomycota (Li et al. 2021) using the BUSCO fungi_odb9 marker set. In the present study, we included the largest collection of zygomycete genomes to date and employed the newly released 758 “fungi_odb10” markers. The results suggested that Basidiobolus is a distinct lineage within Zoopagomycota and is interpreted as the earliest diverging lineage (with 100/100 bootstrap, supplementary fig. S1, Supplementary Material online). The complex mixed history observed in the genomes of Basidiobolus is evidenced by their enriched secondary metabolite genes many of which are result of horizontal gene transfer from Bacteria, regionally duplicated genomes, and the broad range of animal hosts it can be found to inhabit including insects, amphibians, reptiles, and human beings (Henk and Fisher 2012; Tabima et al. 2020). This may explain the sources of phylogenetic conundrums that we have encountered in the last decades using different molecular markers. The phylogenetic and natural history of Basidiobolus may not be easily resolved until an appropriate approach can be carried out to parse their complex genome composed of redundant genes from various sources, such as large-scale gene duplications or horizontal gene transfers. In addition, the kingdom-wide comparison has helped discover many unique genome components in Basidiobolus, including the genes shared with the Mucoromycota clades (e.g., CotH and MMPL), which will be discussed in the following sections.
Divergent Evolution of Zygomycete Fungi
We identified gene content and Pfam domains favored by each of the zygomycete phyla, which can be interpreted to correspond to their disparate lifestyles (figs. 3 and 4). As suggested by the presence of both Pil1 and SUR7 domains, eisosome-mediated endocytosis and related active transportation are important facilitators to saprotrophic Mucoromycota fungi (Walther et al. 2006). Among the 70 Mucoromycota-featured domains (fig. 4 and supplementary table S4, Supplementary Material online), DENN, uDENN, and dDENN also serve as regulators during eukaryotic membrane trafficking events (Zhang et al. 2012). This implies that Mucoromycota fungi are able to transport particles via membrane trafficking domains, while Zoopagomycota fungi, as animal-associated microbes, may use different mechanisms. Noteworthy, the Pfam domain Pox_ser-thr_kin, a poxvirus serine/threonine protein kinase, specifically identified in Mucoromycota genomes (fig. 4 and supplementary table S4, Supplementary Material online) suggest that remnants of large DNA viruses are embedded in Mucoromycota genomes (Jacob et al. 2011). Mycoviruses have been extensively studied in Dikarya fungi, especially for plant pathogens (Ghabrial et al. 2015; Marzano et al. 2016). The existence of mycoviruses among early diverging fungi has not been examined until recently, which led to the discovery of Narnaviruses as members of fungal-bacterial–viral system in the plant pathogenic Rhizopus microsporus (Espino-Vázquez et al. 2020) and RNA mycoviruses in roughly one fifth laboratory cultures of early diverging fungal lineages (Myers et al. 2020). Our preliminary analyses suggest that Mucoromycota members contain genomic hallmarks that interact with both bacteria (MMPL domain, fig. 2e) and viruses (Pox_ser-thr_kin domain, supplementary table S4, Supplementary Material online). The “mycobacterial membrane protein large transporter” domain is well represented in all three subphyla of Mucoromycota as well as Basidiobolus (fig. 2e) consistent with the observations of fungal–bacterial interactions documented in these lineages (Uehling et al. 2017; Desirò et al. 2018; Chang et al. 2019; Bonfante and Venice 2020; Tabima et al. 2020). Although the TLD domain is universally present in almost all fungal lineages (except Wallemia ichthyophaga), the exceptionally large number of TLD domains identified in Glomeromycotina members is unusual (fig. 2c). It implies that TLD and related oxidation resistance proteins could provide protection of these arbuscular mycorrhizal fungi from reactive oxygen species (Blaise et al. 2012).
Zoopagomycota, on the other hand, lack exclusive Pfam domains, even though many domains are highly enriched suggesting important functions. One example is Tyrosinase which synthesize melanin via the amino acid L-tyrosine in melanosomes. Melanin is an important natural product and polymer that can protect organisms from diverse biotic and abiotic factors, including helping microbes counteract the attacks from host immune systems by neutralizing reactive oxygen species or other harmful molecules (Cordero and Casadevall 2020). As such, it is not surprising to find that Zoopagomycota fungi, especially the insect-associated ones, maintain a large number of melanin synthetic enzymes presumably helping them evade host immune responses. Trypsin is another Pfam domain featured in Zoopagomycota (fig. 4) which catalyzes the hydrolysis of peptide bonds to break proteins into smaller pieces and is extremely active in animal digestive systems. We discovered up to 59 copies (in Smittium culicis) of Trypsin domain in the insect gut-dwelling fungi (Harpellales, Kickxellomycotina). Interestingly, insect pathogenic species in Entomophthoromycotina were found heavily relying on hydrolases with 204 copies of Trypsin domains in Zoophthora radicans alone (43–138 copies in other Entomophthoromycotina members), while other zygomycete lineages maintain 0–18 copies variously (fig. 5e). Trypsin and Trypsin-like proteases have been studied in insects and entomopathogenic fungi for decades (Paterson et al. 1993; Dubovenko et al. 2010; Lazarević and Janković-Tomanić 2015). Results suggest that the Trypsin and Trypsin-like proteins are important for nutritional uptake and pathogenic processes of insect-associated fungi, which was also suggested with the potential to help develop new agents to control pest insects (Borges-Veloso et al. 2015; Lazarević and Janković-Tomanić 2015). The abundance of Trypsin domains identified in Zoopagomycota suggests that the expansion of Trypsin across fungal tree of life have occurred more than once (e.g., Ascomycota and Zoopagomycota) (Dubovenko et al. 2010). In addition, the emergence and detailed evolutionary patterns of Trypsin and Trypsin-like proteins in Ascomycota, Zoopagomycota, and insects deserve further examination. Many polysaccharides and protein degrading enzymes were also found expanded in Zoopagomycota, such as LPMO_10, Glyco_hydro_72 (PF03198), and Peptidase_M36 (PF02128) (fig. 4), suggesting their important functions during the interactions of Zoopagomycota fungi with small animals or other fungi. The fungalysin metallopeptidase (Peptidase_M36) and the associated fungalysin/thermolysin propeptide motif (FTP, PF07504) were both found expanded in the obligate mycoparasite Syncephalis (Lazarus et al. 2017). Both domains may help mycoparasites inhibit peptidases produced by the hosts, but their exact function has not been clearly known (Markaryan et al. 1996; Finn et al. 2016). Interestingly, the BATS domain involved in the biotin and thiamin synthesis is found absent in Kickxellomycotina and Zoopagomycotina members (fig. 2d). Both subphyla are short for available cultures, which is especially the case for the animal associated species. The inability to synthesize biotin and thiamin may be one of the culprits for the unsuccessful culture establishment in the lab. Supplementary biotin and thiamin could be suggested for future efforts on development of new cultures in these fungal lineages.
Human Infectious Diseases Caused by Zygomycete Fungi
Mucormycosis is a deadly human-infectious disease usually caused by Rhizopus, Mucor, and Lichtheimia. The current COVID-19 pandemic has triggered multiple cases of Mucormycosis in susceptible patients (Garg et al. 2021; Revannavar et al. 2021). The CotH was originally identified in bacteria as a spore-coat protein. It was later found in Mucorales fungi and identified as a potential invasin factor of the human-infectious Mucormycosis. The CotH was suggested to be directly involved in interactions between Mucorales pathogens and human endothelial cells (Chibucos et al. 2016). Our comparative genomic analyses provided a broader survey of CotH leading to discoveries of novel CotH families in Mucoromycotina strains and unexpected fungal lineages (Basidiobolus, Mortierellomycotina, and Neocallimastigomycota). CotH was maintained by almost every member of Mucoromycotina except the early diverging taxa—Calcarisporiella thermophila and Bifiguratus adelaidae. Unexpectedly, all members of Mortierellomycotina were also able to code CotH domains with the same or highly similar pathogenic motif “MGQTNDGAYRDPTDNN,” which was proposed as a key factor for Mucormycosis. This implies that the included Mortierellomycotina taxa (Dissophora ornate, L. transversale, and Mortierella species) may be facultative pathogens or have the potential to cause Mucormycosis or related human-infectious diseases if treated without caution. The results are informative to guide clinical practice as Mucormycosis may arise from many previously less documented situations, including the injuries during the natural disasters, unconscious contact, and triggered by other diseases like Novel Coronavirus Pneumonia (caused by COVID-19) (Neblett Fanfair et al. 2012; Revannavar et al. 2021). Basidiobolus is the only Zoopagomycota member that encodes CotH, albeit the copy number is low. On the other hand, Neocallimastigomycota members produce surprisingly high numbers of CotH domains with the largest duplication event (fig. 6b). It is not clear why anaerobic gut fungi maintain so many CotH copies since they serve as primary plant polysaccharide degraders and do not pose any identifiable harm to their mammal hosts. Phylogenetic analyses suggest that CotH domains in fungi can be classified into at least seven major groups (ZyGo-A, B, C, D, and three AGF groups; fig. 6a). The ZyGo-A is the only clade containing all known Mucormycosis invasin factors (i.e., CotH 2 and CotH 3) where Mortierellomycotina members are tightly clustered (Chibucos et al. 2016). The members in ZyGo-A, Mucoromycotina and Mortierellomycotina, should both have the potential to cause Mucormycosis.
There are additional emerging pathogens in Zoopagomycota. For example, members of the entomophthoralean fungi can cause infection in both insects and mammals, not only in immunocomprised patients, but also reported from immunocompetent individuals due to insect bites or other undetermined environmental contacts, especially in tropical and subtropical regions (Vilela and Mendoza 2018). Basidiobolus and Conidiobolus are two additional agents of human skin, subcutaneous, and gastrointestinal infections (Khan et al. 2001; Shaikh et al. 2016). Basidiobolus can be isolated from various types of environments, including soils or leaf litters, dung of frogs or lizards, and various insects (e.g., mosquitoes, mites, and springtails) (Lyon et al. 2001; Garros et al. 2008; Manning and Callaghan 2008; Werner et al. 2012). Recently, people also found that Basidiobolus can infect human eyes (Tananuvat et al. 2018; Vilela and Mendoza 2018). The two CotH copies identified in Basidiobolus genomes may be involved in the pathogenic processes. Conidiobolus, however, do not maintain CotH copies, suggesting that Conidiobolus may take different strategies to infect mammalian hosts. Our comparative genomic analyses provided a broader view regarding the molecular mechanism of human-infectious zygomycete fungi. As the quick accumulation of genomic resources for this fungal lineage, a detailed natural history and complete pathogenic pathways should be revealed in the near future.
Our combination of phylogenomic and comparative genomic study of zygomycete fungi provided a perspective on the phylogenetic relationships within the group. The identification of lineage-specific genome contents provides new understanding of their cryptic ecology and relationships with other organisms in the environment. The unexpected findings of the broad distribution of the CotH domain beyond the Mucorales fungi and in Mortierellomycotina, Basidiobolus, and Neocallimastigomycota give new clues to the evolution of this potentially important host-interaction factor. The application of comparative genomics in these zygomycete fungi helps further predict novel and unique biology of understudied fungi to aid study of their interactions with animals, plants, and ecosystems which appears to be altered in the era of global climate change. These presented results may further help mitigate damage and improve avenues of therapeutic research for the treatment and prevention of diseases caused by the human-infectious Mucormycosis.
Materials and Methods
Fungal Taxa and Genome Sampling
In total, 181 fungal genome sequences were analyzed in this study. Nine genomes were generated in this study and 172 were obtained from GenBank or the Joint Genome Institute MycoCosm portal (Grigoriev et al. 2014; https://mycocosm.jgi.doe.gov), with 136 produced by the ongoing 1,000 Fungal Genome Project (1KFG: http://1000.fungalgenomes.org/) and Zygomycetes Genealogy of Life Project (ZyGoLife: http://zygolife.org/). The data set includes 131 zygomycete genomes (supplementary table S1, Supplementary Material online), with 97 sampled from Mucoromycota clade and 34 from Zoopagomycota. In addition, we included 43 Dikarya genomes and seven representatives (supplementary table S2, Supplementary Material online) from other early diverging fungal lineages to enable kingdom-wide comparative analyses. The following nine genomes were produced for this study: Amylomyces rouxii NRRL 5866, Benjaminiella poitrasii RSA 903, Fennellomyces sp. ATCC 46495, Lichtheimia hyalospora FSU 10163, Mucor mucedo NRRL 3635, Parasitella parasitica NRRL 2501, Radiomyces spectabilis NRRL 2753, Spinellus fusiger NRRL 22323, and Piptocephalis tieghemiana RSA 1565.
Genome Sequencing and Assembly
The genome sequencing of S. fusiger NRRL 22323, Radiomyces spectabilis NRRL 2753, Mucor mucedo NRRL 3636, Benjaminiella poitrasii RSA 903 and Fennellomyces sp. ATCC 46495, was performed from 5 µg of genomic DNA, which was sheared to >10 kb using Covaris g-Tubes. The sheared DNA was treated with exonuclease to remove single-stranded ends and DNA damage repair mix was followed for end repair and ligation of blunt adapters using SMRTbell Template Prep Kit 1.0 (Pacific Biosciences). The library was purified with AMPure PB beads. PacBio Sequencing primer was then annealed to the SMRTbell template library and Version P6 sequencing polymerase was bound to them for S. fusiger, R. spectabilis, and Fennellomyces sp. ATCC 46495. The prepared SMRTbell template libraries were then sequenced on a Pacific Biosciences RSII sequencer using Version C4 chemistry and 1 × 240 sequencing movie run times. For B. poitrasii and M. mucedo, sequencing polymerase was bound to them using the Sequel Binding kit 2.1 and then the prepared SMRTbell template libraries were sequenced on a Pacific Biosystems’ Sequel sequencer using v3 sequencing primer, 1 M v2 SMRT cells, and Version 2.1 sequencing chemistry with 1 × 360 sequencing movie run times. Filtered subread data were then used to assemble all lineages using Falcon (version 0.4.2 for S. fusiger and R. spectabilis, version 1.8.8 for M. mucedo and B. poitrasii, and version 0.7.3 for Fennellomyces sp. ATCC 46495). Spinellus fusiger and R. spectabilis were then further improved using finisherSC version 2.0 (Lam et al. 2015). All assemblies were then polished using either Quiver version smrtanalysis_2.3.0.140936.p5 (S. fusiger, R. spectabilis and Fennellomyces sp. ATCC 46495) or Arrow version SMRTLink v5.1.0.26412 (M. mucedo and B. poitrasii).
Parasitella parasitica NRRL 2501, Piptocephalis tieghemiana, and Lichtheimia hyalospora were sequenced using the Illumina platform. For P. parasitica and P. tieghemiana, 100 ng of DNA was sheared to 300 bp using the Covaris LE220 and size selected using SPRI beads (Beckman Coulter). The fragments were treated with end-repair, A-tailing, and ligation of Illumina compatible adapters (IDT, Inc) using the KAPA-Illumina library creation kit (KAPA biosystems). Additionally, a 4-kb mate pair library was constructed for P. parasitica. For this, 5–10 µg of DNA was sheared using the Covaris g-TUBE(TM) and gel size selected for 4 kb. The sheared DNA was treated with end repair and ligated with biotinylated adapters containing loxP. The adapter ligated DNA fragments were circularized via recombination by a Cre excision reaction (NEB). The circularized DNA templates were then randomly sheared using the Covaris LE220 (Covaris). The sheared fragments were treated with end repair and A-tailing using the KAPA-Illumina library creation kit (KAPA biosystems) followed by immobilization of mate pair fragments on strepavidin beads (Invitrogen). Illumina compatible adapters (IDT, Inc) were ligated to the mate pair fragments and eight cycles of PCR were used to enrich for the final library (KAPA Biosystems). The prepared libraries were quantified using KAPA Biosystems’ next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit, v4. Sequencing of the flowcell was performed on the Illumina HiSeq2500 sequencer using HiSeq TruSeq SBS sequencing kits, v4, following a 2 × 150 indexed run recipe. Each fastq file was QC filtered for artifact/process contamination and subsequently assembled together with AllPathsLG version R49403 (Gnerre et al. 2011).
Piptocephalis tieghemiana is an obligate mycoparasite and was maintained as co-culture with Umbelopsis sp. nov. AD052. The P. tieghemiana contigs required further processing to separate these two assemblies. First, metagenomic scaffold sequences were binned into two groups using metabat (v2.12.1). The filtered reads were mapped to the sequences of the two bins and split into two separate data sets corresponding to each bin using bbsplit.sh in bbtools(ambiguous = all). The two data sets were then re-assembled separately. Scaffolds with length less than 2 kb were excluded. Then, four closely related genomes were used for reference genome to classify and filter re-assembled scaffolds based on BLASTN similarity (evalue < 1e−30). One included Piptocephalis related genome was Piptocephalis cylindrospora, and the others were Umbelopsis related genomes, Umbelopsis sp. AD052, Umbelopsis isabellina AD026, and Umbelopsis sp. PMI 123. If the scaffolds were covered more by Piptocephalis main genome than Umbelopsis main genomes, it would be classified to P. tieghemiana, and vice versa. The scaffolds without any similarity to the four genomes were discarded.
For L. hyalospora, 500 ng of DNA was sheared to 270 bp using the Covaris E210 (Covaris, Woburn, MA) and size selected using SPRI beads (Beckman Coulter, Brea, CA, USA). The fragments were treated with end-repair, A-tailing, and ligation of Illumina adapters using the TruSeq Sample Prep Kit (Illumina, San Diego, CA, USA), followed by quantification of libraries using KAPA Biosystem's next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were multiplexed and the pools were then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit, v3, and Illumina's cBot instrument to generate a clustered flowcell for sequencing. Sequencing of the flowcell was performed on the Illumina HiSeq2000 sequencer using a TruSeq SBS sequencing kit 200 cycles, v3, following a 2 × 150 indexed run recipe. Genomic reads were QC filtered for artifact/process contamination and subsequently assembled with Velvet. The resulting assembly was used to create a simulated 3-kbp insert long mate-pair library, which was then assembled together with the original Illumina library with AllPathsLG release version R42328.
Transcriptome Sequencing and Assembly
For all lineages except L. hyalospora, stranded cDNA libraries were generated using the Illumina Truseq Stranded RNA LT kit. mRNA was purified from 1 µg of total RNA using magnetic beads containing poly-T oligos. mRNA was fragmented and reversed transcribed using random hexamers and SSII (Invitrogen) followed by second strand synthesis. The fragmented cDNA was treated with end-pair, A-tailing, adapter ligation, and eight cycles of PCR. For L. hyalospora, plate-based RNA sample prep was performed on the PerkinElmer Sciclone NGS robotic liquid handling system using Illumina's TruSeq Stranded mRNA HT sample prep kit utilizing poly-A selection of mRNA following the protocol outlined by Illumina in their user guide, https://support.illumina.com/sequencing/sequencing_kits/truseq-stranded-mrna.html, and with the following conditions: total RNA starting material was 1 µg per sample and eight cycles of PCR were used for library amplification. The prepared libraries were then quantified using KAPA Biosystems’ next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit, v4. Sequencing of the flowcell was performed on the Illumina HiSeq2500 sequencer using HiSeq TruSeq SBS sequencing kits, v4, following a 2 × 150 indexed run recipe (2 × 100 for L. hyalospora).
Filtered fastq files were used as input for de novo assembly of RNA contigs. For all lineages except L. hyalospora and P. parasitica, reads were assembled into consensus sequences using Trinity version 2.1.1. Trinity was run with the −normalize_reads (in silico normalization routine) and –jaccard_clip (minimizing fusion transcripts derived from gene dense genomes) options. For L. hyalospora and P. parasitica, Rnnotator version 2.5.6 or later was used. Parasitella parasitica was further improved using eight runs of velveth (v. 1.2.07) performed in parallel, once for each hash length for the De Bruijn graph. Minimum contig length was set at 100. The read depth minimum was set to 3 reads. Redundant contigs were removed using Vmatch (v. 2.2.4) and contigs with significant overlap were further assembled using Minimus2 with a minimum overlap of 40. Contig postprocessing included splitting misassembled contigs, contig extension, and polishing using the strand information of the reads. Single base errors were corrected by aligning the reads back to each contig with BWA to generate a consensus nucleotide sequence. All nine new genomes in this study were annotated using the JGI Annotation pipeline (Grigoriev et al. 2014).
Phylogenomic Analyses
A set of 758 phylogenetic markers, “fungi_odb10”, from the Benchmarking Universal Single-Copy Orthologs (BUSCO) v4.0.5 was employed for the kingdom-wide phylogenomic analyses (Seppey et al. 2019). We used the PHYling pipeline (DOI: 10.5281/zenodo.1257002) to extract best hit copies using hmmsearch v3.3.2 (cutoff = 1E−10) from the genes predicted in each species against the marker set. A total of 617 (out of 758) well-conserved markers were identified as the best hit from the 181 fungal genomes. A backbone tree including 80 genomes, subsampled based on BUSCO scores and phylogenetic placement on the 181-taxon tree (except for the outgroup Drosophila melanogaster), recovered 604 orthologs. All orthologs were aligned separately using hmmalign v3.3.2 to the marker profile-HMM and then concatenated into a super-alignment with partitions defined by each marker. The best phylogenomic tree was searched and identified using the super-alignment file and partition scheme as the input with the best-fit model option for maximum likelihood analyses implemented in IQ-TREE v.1.5.5 (Nguyen et al. 2015; Kalyaanamoorthy et al. 2017). Branch supports were evaluated using 1,000 ultrafast bootstrap replicates (Hoang et al. 2017). Concordance factors were calculated as additional support for each branch using single gene alignments and concatenated tree file as instructed in the IQ-TREE package (v1.7-beta9).
Identification of Lineage-Specific Genes and Pfam Domains in Zygomycete Fungi
All orthologous groups of the 80 genomes included in the backbone tree were identified using a comparative genomic pipeline that utilized all-vs-all BLASTp search v2.6.0 (cutoff = 1E−5) (DOI: 10.5281/zenodo.1447224) (Altschul et al. 1990). Orthagogue v1.0.3 was used to infer putative orthologs and Markov-Clustering Algorithm v14-137 (MCL, inflation value of 1.5) was utilized to generate disjoint clusters (Van Dongen 2000; Ekseth et al. 2014). Shared genome components were counted using a permissive strategy that a gene family shared by at least 11 of the 80 included taxa was retained. Zygomycetes-specific genes are the ones that only exist in zygomycete fungi (Mucoromycota and Zoopagomycota) and are absent in all other lineages. The absence-presence pattern of gene families across the Kingdom Fungi was plotted using the “aheatmap” function in R package “NMF” (Gaujoux and Seoighe 2010). Protein domains coded by the 80 taxa were examined in a similar way. Each Protein Family (Pfam) entry in the Pfam database v31.0 was searched against the predicted proteomes of all included 80 taxa (using the threshold of 1E−3 with >50% overlap percentage). The Pfam domains dominated in either Mucoromycota or Zoopagomycota were inferred by the ratios of their copy numbers in Zoopagomycota and Mucoromycota. The disproportion was visualized by plotting the binary logarithm of the ratio for each Pfam entry so that dominated Pfam domains in each phylum will be isolated on the edge. The figure was plotted using R package “ggplot2” (Wickham 2016). Subphylum-level distribution of each discussed Pfam domain was plotted using the “radarchart” function implemented in R package “fmsb”. All lineage-specific genome content was summarized in table 1 (with detailed Pfam names listed in supplementary table S3, Supplementary Material online). Gene Ontology (GO) terms of Zoopagomycota “unique” genes were inferred and annotated using InterProScan v5.54 and WEGO v2.0 respectively (Jones et al. 2014; Ye et al. 2018).
Phylogenetic Analysis of the Spore Coating Protein (CotH) in Fungi
A total of 846 protein sequences that contain at least one CotH domain were identified in the 80 genomes included in the backbone tree. Absent in all Dikarya species, CotH genes were largely found in zygomycetes (all included 6 Mortierellomycotina members, 27 Mucoromycotina taxa, and 1 Basidiobolus) and in Neocallimastigomycota (including 2 taxa). Previously classified CotH families 1–5 (CotH 1–5) from Rhizopus oryzae were included in our phylogenetic analyses to categorize the newly identified CotH copies. Highly similar CotH sequences (>90%) were removed using CD-HIT v4.6.4 and poor-quality ones were manually excluded from the multiple sequence alignment using MUSCLE v3.8.31 (Edgar 2004; Fu et al. 2012). We employed IQ-TREE v1.5.5 to identify the most appropriate substitutional model and to reconstruct the phylogenetic tree of all fungal CotH copies with ultrafast bootstraps (1,000 replicates) (Nguyen et al. 2015; Hoang et al. 2017; Kalyaanamoorthy et al. 2017). The final input includes 754 sequences with 230 distinct patterns for CotH classification. Species-gene tree reconciliation analysis was conducted with Notung v3.0 BETA using the 80-taxa backbone tree as the species tree (Stolzer et al. 2012). We followed the phylogenomic workflows as recommended in the Notung v3.0 BETA manual to generate a summary report of gain, transfer, and loss events of CotH families in Kingdom Fungi. A threshold of 90% was applied to the rearrangement step to accommodate the ambiguities in the species tree and CotH gene tree.
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Science Foundation (DEB-1441604 to J.W.S. and DEB-1441715 to J.E.S.). The authors thank Drs. M. Catherine Aime, William J. Davis, Gunther Doehlemann, Toni Gabaldón, and Timothy Y. James for permission to use genomes ahead of publication. The work (proposals 10.46936/10.25585/60001019 and 10.46936/10.25585/60001062) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. J.E.S. is a paid consultant for Zymergen and Sincarne and CIFAR fellow in the program Fungal Kingdom: Threats and Opportunities.
Contributor Information
Yan Wang, Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, USA; Department of Biological Sciences, University of Toronto Scarborough, Canada; Department of Ecology and Evolutionary Biology, University of Toronto, Canada.
Ying Chang, Department of Botany and Plant Pathology, Oregon State University, Corvallis, USA; Division of Science, Yale-NUS College, Singapore, Singapore.
Jericho Ortañez, Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, USA.
Jesús F Peña, Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, USA.
Derreck Carter-House, Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, USA.
Nicole K Reynolds, Department of Plant Pathology, University of Florida, Gainesville, USA.
Matthew E Smith, Department of Plant Pathology, University of Florida, Gainesville, USA.
Gerald Benny, Department of Plant Pathology, University of Florida, Gainesville, USA.
Stephen J Mondo, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory; Department of Agricultural Biology, Colorado State University, Fort Collins.
Asaf Salamov, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Anna Lipzen, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Jasmyn Pangilinan, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Jie Guo, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Kurt LaButti, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
William Andreopolous, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Andrew Tritt, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Keykhosrow Keymanesh, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Mi Yan, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Kerrie Barry, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory.
Igor V Grigoriev, US Department of Energy (DOE) Joint Genome Institute (JGI), Lawrence Berkeley National Laboratory; Department of Plant and Microbial Biology, University of California, Berkeley.
Joseph W Spatafora, Department of Botany and Plant Pathology, Oregon State University, Corvallis, USA.
Jason E Stajich, Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, USA.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
Assembled genomes and annotation files are available at JGI MycoCosm website and are available in GenBank under genome accession numbers listed in supplementary table S1, Supplementary Material online. Alignment and tree files associated with this study are available at DOI: 10.5281/zenodo.7523466.
Literature Cited
- Ahrendt SR, et al. 2018. Leveraging single-cell genomics to expand the fungal tree of life. Nat Microbiol. 3:1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Blaise M, et al. 2012. Crystal structure of the TLDc domain of oxidation resistance protein 2 from zebrafish. Proteins 80:1694–1698. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Venice F. 2020. Mucoromycota: going to the roots of plant-interacting fungi. Fungal Biol Rev. 34:100–113. [Google Scholar]
- Borges-Veloso A, et al. 2015. In-depth characterization of trypsin-like serine peptidases in the midgut of the sugar fed Culex quinquefasciatus. Parasites Vectors 8:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce GR, et al. 2019. Psychoactive plant- and mushroom-associated alkaloids from two behavior modifying cicada pathogens. Fungal Ecol. 41:147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanera R, et al. 2016. Transposable elements versus the fungal genome: impact on whole-genome architecture and transcriptional profiles. PLoS Genet. 12:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambouvet A, et al. 2019. Intracellular infection of diverse diatoms by an evolutionary distinct relative of the fungi. Curr Biol. 29:4093–4101.e4. [DOI] [PubMed] [Google Scholar]
- Chang Y, et al. 2015. Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol Evol. 7:1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, et al. 2019. Phylogenomics of Endogonaceae and evolution of mycorrhizas within Mucoromycota. New Phytol. 222:511–525. [DOI] [PubMed] [Google Scholar]
- Chang Y, et al. 2021. Genome-scale phylogenetic analyses confirm Olpidium as the closest living zoosporic fungus to the non-flagellated, terrestrial fungi. Sci Rep. 11:3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, et al. 2022. Evolution of zygomycete secretomes and the origins of terrestrial fungal ecologies. iScience 25:104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibucos MC, et al. 2016. An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat Commun. 7:12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJB, Casadevall A. 2020. Melanin. Curr Biol. 30:R142–R143. [DOI] [PubMed] [Google Scholar]
- Corrochano LM, et al. 2016. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr Biol. 26:1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie CR, et al. 2003. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 299:386–388. [DOI] [PubMed] [Google Scholar]
- Desirò A, et al. 2018. Mycoplasma-related endobacteria within Mortierellomycotina fungi: diversity, distribution and functional insights into their lifestyle. ISME J. 12:1743–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZY, et al. 2019. Algal-fungal symbiosis leads to photosynthetic mycelium. Elife 8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovenko AG, et al. 2010. Trypsin-like proteins of the fungi as possible markers of pathogenicity. Fungal Biol. 114:151–159. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekseth OK, Kuiper M, Mironov V. 2014. Orthagogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 30:734–736. [DOI] [PubMed] [Google Scholar]
- Espino-Vázquez AN, et al. 2020. Narnaviruses: novel players in fungal–bacterial symbioses. ISME J. 14(7):1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, et al. 2020. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio 11:e00449-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Klett P, et al. 2011. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 75:583–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. [DOI] [PubMed] [Google Scholar]
- Garg D, et al. 2021. Coronavirus disease (Covid-19) associated mucormycosis (CAM): case report and systematic review of literature. Mycopathologia 186(2):289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garros C, Ngugi N, Githeko AE, Tuno N, Yan G. 2008. Gut content identification of larvae of the Anopheles gambiae complex in western Kenya using a barcoding approach. Mol Ecol Resour. 8:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. 2010. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N. 2015. 50-plus years of fungal viruses. Virology 479–480:356–368. [DOI] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau AA, et al. 1996. Life with 6000 genes. Science 274:546, 563–567. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, et al. 2014. Mycocosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42:D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruninger RJ, et al. 2014. Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol. 90:1–17. [DOI] [PubMed] [Google Scholar]
- Gryganskyi AP, et al. 2012. Molecular phylogeny of the Entomophthoromycota. Mol Phylogenet Evol. 65:682–694. [DOI] [PubMed] [Google Scholar]
- Henk DA, Fisher MC. 2012. The gut fungus basidiobolus ranarum has a large genome and different copy numbers of putatively functionally redundant elongation factor genes. PLoS One 7:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett DS, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol Res. 111:509–547. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Le SV. 2017. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori C, et al. 2013. Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 105:1412–1427. [DOI] [PubMed] [Google Scholar]
- Jacob T, Van den Broeke C, Favoreel HW. 2011. Viral serine/threonine protein kinases. J Virol. 85:1158–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, et al. 2006. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98:860–871. [DOI] [PubMed] [Google Scholar]
- James TY, et al. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822. [DOI] [PubMed] [Google Scholar]
- James TY, Stajich JE, Hittinger CT, Rokas A. 2020. Toward a fully resolved fungal tree of life. Annu Rev Microbiol. 74:291–313. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. 2014. Interproscan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan ZU, Khoursheed M, Makar R. 2001. Basidiobolus ranarum as an etiologic agent of gastrointestinal zygomycosis. J Clin Microbiol. 39:2360–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KK, Labutti K, Khalak A, Tse D. 2015. FinisherSC: a repeat-aware tool for upgrading de novo assembly using long reads. Bioinformatics 31:3207–3209. [DOI] [PubMed] [Google Scholar]
- Lazarević J, Janković-Tomanić M. 2015. Dietary and phylogenetic correlates of digestive trypsin activity in insect pests. Entomol Exp Appl. 157:123–151. [Google Scholar]
- Lazarus KL, Benny GL, Ho HM, Smith ME. 2017. Phylogenetic systematics of syncephalis (zoopagales, zoopagomycotina), a genus of ubiquitous mycoparasites. Mycologia 109:333–349. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. 2021. A genome-scale phylogeny of the kingdom Fungi. Curr Biol. 31:1653–1665.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GM, et al. 2001. Gastrointestinal basidiobolomycosis in Arizona: clinical and epidemiological characteristics and review of the literature. Clin Infect Dis. 32:1448–1455. [DOI] [PubMed] [Google Scholar]
- Ma L-J, et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5:e1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malar M, et al. 2021. The genome of Geosiphon pyriformis reveals ancestral traits linked to the emergence of the arbuscular mycorrhizal symbiosis. Curr Biol. 31:1–8. [DOI] [PubMed] [Google Scholar]
- Manning RJ, Callaghan AA. 2008. Pathogenicity of Conidiobolus spp. and Basidiobolus ranarum to arthropods co-occurring in leaf litter. Fungal Ecol. 1:33–39. [Google Scholar]
- Markaryan A, Lee JD, Sirakova TD, Kolattukudy PE. 1996. Specific inhibition of mature fungal serine proteinases and metalloproteinases by their propeptides. J Bacteriol. 178:2211–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzano SL, et al. 2016. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol. 90:6846–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf JL, et al. 2016. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 351:158–162. [DOI] [PubMed] [Google Scholar]
- Morin E, et al. 2019. Comparative genomics of Rhizophagus irregularis, R. cerebriforme, R. diaphanus and Gigaspora rosea highlights specific genetic features in Glomeromycotina. New Phytol. 222:1584–1598. [DOI] [PubMed] [Google Scholar]
- Muszewska A, Steczkiewicz K, Stepniewska-Dziubinska M, Ginalski K. 2017. Cut-and-paste transposons in fungi with diverse lifestyles. Genome Biol Evol. 9:3463–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JM, et al. 2020. Survey of early-diverging lineages of fungi reveals abundant and diverse mycoviruses. MBio 11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo-Ortiz MA, Gabaldón T. 2020. Fungal evolution: cellular, genomic and metabolic complexity. Biol Rev. 95(5):1198–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neblett Fanfair R, et al. 2012. Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. N Engl J Med. 367:2214–2225. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Yu DS, Wang CF, Liu XY, Huang B. 2020. A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. MycoKeys 66:55–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Knoll AH, Katz LA. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 108:13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson IC, Charnley AK, Cooper RM, Clarkson JM. 1993. Specific induction of a cuticle-degrading protease of the insect pathogenic fungus Metarhizium anisopliae. Microbiology 140:185–189. [DOI] [PubMed] [Google Scholar]
- Picard KT. 2017. Coastal marine habitats harbor novel early-diverging fungal diversity. Fungal Ecol. 25:1–13. [Google Scholar]
- Pombubpa N, Pietrasiak N, De Ley P, Stajich JE. 2020. Insights into dryland biocrust microbiome: geography, soil depth and crust type affect biocrust microbial communities and networks in Mojave Desert, USA. FEMS Microbiol Ecol. 96:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revannavar SM, Supriya PS, Samaga L, Vineeth VK. 2021. COVID-19 triggering mucormycosis in a susceptible patient: a new phenomenon in the developing world? BMJ Case Rep. 14(4):e241663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Jusino M, Stajich J, Smith M. 2021. Understudied, underrepresented, and unknown: methodological biases that limit detection of early diverging fungi from environmental samples. Mol Ecol Resour. 22(3):1065–1085. [DOI] [PubMed] [Google Scholar]
- Richards TA, Leonard GUY, Wideman JG. 2017. What defines the “Kingdom” Fungi? Microbiol Spectr. 5:1–21. [DOI] [PubMed] [Google Scholar]
- Richards TA, Talbot NJ. 2018. Osmotrophy. Curr Biol. 28:R1179–R1180. [DOI] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. In: Kollmar M, editor. Gene prediction: methods and protocols. New York (NY): Humana. p. 227–245. [DOI] [PubMed] [Google Scholar]
- Shaikh N, Hussain KA, Petraitiene R, Schuetz AN, Walsh TJ. 2016. Entomophthoramycosis: a neglected tropical mycosis. Clin Microbiol Infect. 22:688–694. [DOI] [PubMed] [Google Scholar]
- Soare AY, Watkins TN, Bruno VM. 2020. Understanding mucormycoses in the age of “omics.”. Front Genet. 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KV, et al. 2016. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351:1192–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, et al. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, et al. 2017. The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol Spectr. 5:3–34. [DOI] [PubMed] [Google Scholar]
- Stajich J, et al. 2009. The fungi. Curr Biol. 19:R840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, et al. 2022. An improved 1.5-gigabase draft assembly of Massospora cicadina (Zoopagomycota), an obligate fungal parasite of 13- and 17-year cicadas. Microbiol Resour Announc. 11:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzer M, et al. 2012. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabima JF, et al. 2020. Phylogenomic analyses of non-dikarya fungi supports horizontal gene transfer driving diversification of secondary metabolism in the amphibian gastrointestinal symbiont, basidiobolus. G3 Genes 10:3417–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tananuvat N, Supalaset S, Niparugs M, Chongkae S, Vanittanakom N. 2018. Ocular basidiobolomycosis: a case report. Case Rep Ophthalmol. 9:315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehling J, et al. 2017. Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ Microbiol. 19:2964–2983. [DOI] [PubMed] [Google Scholar]
- Vandepol N, et al. 2020. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 104:267–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S. 2000. Graph clustering by flow simulation [PhD thesis]. Utrecht, Netherlands: University of Utrecht. [Google Scholar]
- Vilela R, Mendoza L. 2018. Human pathogenic entomophthorales. Clin Microbiol Rev. 31:e00014-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, et al. 2006. Eisosomes mark static sites of endocytosis. Nature 439:998–1003. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. 2018. Comparative genomics reveals the core gene toolbox for the fungus-insect symbiosis. MBio 9:e00636-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, White MM, Kvist S, Moncalvo J-M. 2016. Genome-wide survey of gut fungi (Harpellales) reveals the first horizontally transferred ubiquitin gene from a mosquito host. Mol Biol Evol. 33:2544–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Peršoh D, Rambold G. 2012. Basidiobolus haptosporus is frequently associated with the gamasid mite Leptogamasus obesus. Fungal Biol. 116:90–97. [DOI] [PubMed] [Google Scholar]
- White MM, et al. 2006. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia 98:872–884. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. Ggplot2: elegant graphics for data analysis Hadley. New York: Springer-Verlag. [Google Scholar]
- Wood V, et al. 2002. The genome sequence of schizosaccharomyces pombe. Nature 415:871–880. [DOI] [PubMed] [Google Scholar]
- Ye J, et al. 2018. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46:W71–W75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. 2012. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Assembled genomes and annotation files are available at JGI MycoCosm website and are available in GenBank under genome accession numbers listed in supplementary table S1, Supplementary Material online. Alignment and tree files associated with this study are available at DOI: 10.5281/zenodo.7523466.