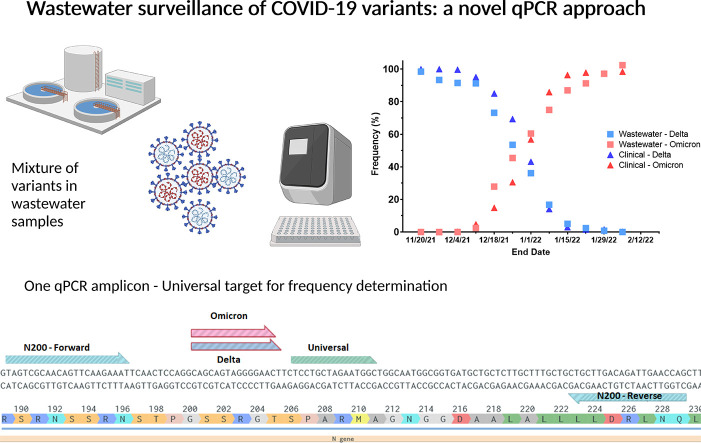
Abstract
Wastewater-based surveillance has become an effective tool around the globe for indirect monitoring of COVID-19 in communities. Variants of Concern (VOCs) have been detected in wastewater by use of reverse transcription polymerase chain reaction (RT-PCR) or whole genome sequencing (WGS). Rapid, reliable RT-PCR assays continue to be needed to determine the relative frequencies of VOCs and sub-lineages in wastewater-based surveillance programs. The presence of multiple mutations in a single region of the N-gene allowed for the design of a single amplicon, multiple probe assay, that can distinguish among several VOCs in wastewater RNA extracts. This approach which multiplexes probes designed to target mutations associated with specific VOC's along with an intra-amplicon universal probe (non-mutated region) was validated in singleplex and multiplex. The prevalence of each mutation (i.e. VOC) is estimated by comparing the abundance of the targeted mutation with a non-mutated and highly conserved region within the same amplicon. This is advantageous for the accurate and rapid estimation of variant frequencies in wastewater. The N200 assay was applied to monitor frequencies of VOCs in wastewater extracts from several communities in Ontario, Canada in near real time from November 28, 2021 to January 4, 2022. This includes the period of the rapid replacement of the Delta variant with the introduction of the Omicron variant in these Ontario communities in early December 2021. The frequency estimates using this assay were highly reflective of clinical WGS estimates for the same communities. This style of qPCR assay, which simultaneously measures signal from a non-mutated comparator probe and multiple mutation-specific probes contained within a single qPCR amplicon, can be applied to future assay development for rapid and accurate estimations of variant frequencies.
Keywords: SARS-CoV-2, Wastewater based epidemiology, VOC, RT-PCR, Omicron
Graphical abstract
1. Introduction
Communities around the world have quickly adopted wastewater monitoring of SARS-CoV-2 viral fragments to detect and infer prevalence of COVID-19 disease burden in communities (Ahmed et al., 2020; Cuevas-Ferrando et al., 2021; D'Aoust et al., 2021; Kumar et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Wu et al., 2020). Because wastewater-based surveillance (WBS) assays a single composite sample that includes all symptomatic or asymptomatic individuals who contribute to the sewershed, WBS has several advantages over more costly and invasive clinical testing. WBS for SARS-CoV-2 typically utilizes reverse transcription real-time polymerase chain reaction (RT-PCR) based techniques to target various regions of the SARS-CoV-2 genome, including the popular US-Centers for Disease Control (CDC) N1, N2 (nucleocapsid) and E (envelope), to test for the presence of SARS-CoV-2 RNA in wastewater (Hamouda et al., 2021; Kumblathan et al., 2021; review by: Kitajima et al., 2020).
Continuous emergence of variants of SARS-CoV-2 has presented additional challenges to the surveillance and has been an area where WBS can provide timely information. In addition to clinical tracking of abundance of SARS-CoV-2 infections, RT-PCR methods have been used to detect and monitor prevalence of emerging VOCs in wastewater (Carcereny et al., 2021; Graber et al., 2021; Heijnen et al., 2021; Lee et al., 2021; Peterson et al., 2022; Wolfe et al., 2022). This information provides an integrated estimation of the relative abundance of variants within populations. There have been many recent examples where RT-PCR assays for identification and quantification of VOCs in wastewater have provided useful information to communities (Tiwari et al., 2023). For example, an assay based on mutation D3L of the N gene, was applied for detection and quantification of relative proportion of the Alpha VOC in the wastewater of a community which was shown to be reflective of the reported Alpha clinical cases (Graber et al., 2021). The mutations N501Y and/or Δ69-70 del in the spike protein have been targeted by RT-qPCR assays to monitor for the presence of VOCs in wastewaters across Canada (Peterson et al., 2022; Xie et al., 2021) the Netherlands (Heijnen et al., 2021), Spain (Carcereny et al., 2021) and France (Wurtzer et al., 2022b). Using allele specific RT-qPCR primers with an artificial mismatch, three mutations, HV69/70del, Y144del and A570D, were targeted to discriminate and quantify the Alpha VOC relative to the wild type in wastewater of 19 communities in the USA (Lee et al., 2021). Others have recently targeted a variety of specific mutations, or deletions, to differentiate emerging VOCs (e.g. Alpha, Beta, Gamma, Delta) to be able to follow the transition of these variants in wastewater (Boogaerts et al., 2022; Farkas et al., 2023; Langan et al., 2022; Yaniv et al., 2021) as well as the later emergence of Omicron and its sublineages (Boehm et al., 2022; Lee et al., 2022; Mills et al., 2022; Wilhelm et al., 2022; Wolfe et al., 2022; Wurtzer et al., 2022a; Xu et al., 2022). These various studies demonstrate that WBS can rapidly provide information on incidence and prevalence of SARS-CoV-2 to authorities with sensitivity and lineage specificity.
RT-PCR assays can be specific and easily deployed rapidly across existing WBS networks. There are several challenges with using these assays, including ensuring the frequency estimates of mutations are accurate. To provide an estimate of the prevalence of a variant in a community, one must determine the amount of mutation present in a sample along with the total amount of SARS-CoV-2 in the sample. While the measurement of the mutation is relatively straight-forward, there are several different ways to determine the total amount of SARS-CoV-2 in a sample. One method is to use another amplicon (such as the N1 or E gene targets developed by the US CDC) to determine the prevalence of a variant. This method can potentially produce variable estimates of mutation/variant frequency since there can be variation in the abundance of two different amplicons of SARS-CoV-2 in a wastewater sample (Amman et al., 2022). Another method is to use the same amplicon and measure both the mutant and wild-type alleles. The sum of these alleles provides the total amount of SARS-CoV-2. This method can also be variable due to the need to quantitate the genes using two separate standard curves. In this study, we utilized a novel method of determining the total amount of SARS-CoV-2 in a variant RT-PCR assay, an intra-amplicon universal target, which eliminates both sources of variability mentioned above.
The objective of this study was to develop a multiplex RT-PCR assay that could be used rapidly and accurately estimate the prevalence of multiple VOCs. To accomplish this, a RT-PCR assay was designed that targets several mutations associated with VOCs within a single amplicon, along with a non-mutated region within this amplicon. The N gene region was selected for this assay for several reasons. This region is targeted by the N1 and N2 assays developed by the US CDC, which are commonly used for monitoring wastewater (Lu et al., 2020). There is evidence to suggest that assays developed for the N-gene of the SARS-CoV-2 genome have greater sensitivity for measuring various VOCs of SARS-CoV-2 in wastewater when compared to assays targeting the S-gene (Yaniv et al., 2021) or the envelope genes (Pérez-Cataluña et al., 2021). Additionally, the N-gene is rich in mutations unique to VOCs (Kiryanov et al., 2022). Specifically, the 121-basepair region in the N-gene open reading frame (ORF) (nucleotides 28,837 to 28,958) that includes single or multiple nucleotide mutations for each VOC (Table 1 ) was selected. According to Global Initiative on Sharing Avian Influenza Data (GISAID) data summarized by nextrain.org (Hadfield et al., 2018), the nucleotide 28,880–28,882 (AA N:203) has one of the highest entropy levels in the SARS-CoV-2 genome, at 0.972, with nucleotides 28,883–28,885 (AA N:204) also having high entropy level of 0.711 (nextstrain.org; accessed April 1, 2022). The non-synonymous functional polymorphisms present in this region include R203M, present in Delta, which increases the spread of the virus (Syed et al., 2021); R203K and G204R, (both present in Alpha, Gamma, Lambda, and Omicron), which are related to increased infectivity, fitness and virulence (Johnson et al., 2022; Lee et al., 2021; Wu et al., 2021); and T205I, present in Beta and Mu variants that were predicted to have reduced antigenicity and greater affinity for HLA-I (Pretti et al., 2022). The presence of multiple mutations in a single region allowed for the design of a single amplicon assay with multiple probes that can distinguish among several VOCs. Additionally, we introduce the concept of an intra-amplicon universal probe that targets an area of the amplicon observed to have a low number of observed mutations (<0.4 %, Table S1), which allows for the quantification of the total SARS-CoV-2 signal regardless of which variants are present in wastewater. The intra-amplicon universal probe allows for the measurement of the total SARS-CoV-2 signal as well as providing a basis for estimations of the relative abundance of individual or multiple VOCs. This intra-amplicon universal probe concept is novel and uniquely suited for monitoring wastewater samples as they often contain multiple VOCs present at different frequencies.
Table 1.
Nucleotide mutations targeted by probes in the N200 RT-PCR assay to differentiate variants of SARS-CoV-2.
| Wuhan sequence | 28,877 |
28,878 |
28,881 |
28,882 |
28,883 |
28,886 |
|---|---|---|---|---|---|---|
| A | G | G | G | G | C | |
| Alpha (B.1.1.7*) | – | – | A | A | C | – |
| Beta (B.1.351*) | – | – | – | – | – | T |
| Gamma (P.1*) | T | C | A | A | C | – |
| Delta (B.1.617.2*) | – | – | T | – | – | – |
| Omicron (BA.1.1.529*) | – | – | A | A | C | – |
Prior to being incorporated into a wastewater surveillance program, the N200 assay was validated with synthetic standards as well as wastewater samples. As an example of the utility of this assay, samples from six wastewater treatment facilities in Ontario, Canada, were examined with the N200 assay. Wastewater samples taken between November 2021 and January 2022 were tested for the presence and increased frequency of the R203K + G204R mutation (present in the Omicron VOC and subvariants), while comparing directly to the decrease in frequency of the R203M mutation (present in the Delta VOC), relative to the intra-amplicon universal marker.
2. Materials and methods
2.1. Target selection
To select an appropriate RT-PCR target for testing wastewater, mutations that were indicative of each VOC and encompassed amino acids N:202–205 were identified. Mutations selected for each VOC are presented in Table 1 (prevalence of these mutations can be found in Table S2). The template sequence used for designs were retrieved from accession numbers provided by TWIST Bioscience (South San Francisco, CA, USA) for synthetic controls 14 (Alpha), 16 (Beta), 17 (Gamma) and 23 (Delta) and sequences were aligned using MAFFT (Katoh et al., 2002).
2.2. Assay design
Candidate forward and reverse primers that encompassed an amplicon 121 base pairs long containing all the desired mutations were designed with a target predicted Tm around 61 °C and 40–60 % GC content. Candidate primers were screened for hairpins, homodimers and heterodimers in silico using OligoAnalyzer by Integrated DNA Technologies (IDT; RRID:SCR_001363) with the “qPCR” parameter selected (Table S3). To ensure that the primers used in the N200 assay did not have significant affinity for non-SARS-CoV-2 related targets, candidate primers with Δ G > -11 were queried individually via BLAST against NCBI's non-redundant nucleotide database. To maximize primer mismatches with as much non-target genetic material as possible, the top 100 hits were sorted by the number of mismatches, (human, microbes, food, etc., Table S4). Furthermore, to ensure that the primer used in the N200 assay did not have significant affinity to any human templates, Primer-BLAST to determine if any human (taxid = 9606) sequences were predicted to amplify with the N200 primers (Table S4).
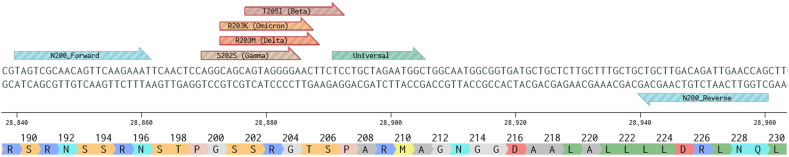
Four overlapping probes were designed to be specific for each of the VOC specific mutations flanked by the primers (Fig. 1 ). These probes were designed to target the N gene mutations S202S + R203K + G204R (Gamma), R203K + G204R (Alpha/Omicron), R203M (Delta), and T205I (Beta). Mutation-specific probes were designed with the mutated base(s) at their center using PrimerExpress v3.0.1 with a GC content of 40–60 %. BHQplus technology (LCG, Biosearch Technologies, Inc., California, USA) was used to increase the Tm of each probe, to have a predicted Tm of 69 °C, which enabled the design of short probes approximately 15 nucleotides long. A fifth probe was designed to target a highly conserved (bp 28,891–28,905, observed mutation rates <0.401 % as per Table S1) region of the amplicon to detect total SARS-CoV-2 (Table 2 , Fig. 1). Probes were then tested for hairpins, homodimers, and heterodimers as described above (Table S3). (Agarwala et al., 2016).
Fig. 1.
N200 assay amplicon with the location of the N200 forward and reverse primers and all probes are shown on a portion of the N gene of the SARS-CoV-2 genome. The sequence, nucleotide numbers, and amino acid displayed are based on the SARS-CoV-2 reference genome (Wuhan; NC_045512.2; figure generated using Benchling.com).
Table 2.
Forward (F) and Reverse (R) primer and probe designs for the N200 VOC assay. The reporter and quencher moieties, and final working concentrations are included for each oligonucleotide.
| Variant targeted | Oligo name | Sequence (5′ to 3′) | Reporter/quencher | Working concentration (nM) | |
|---|---|---|---|---|---|
| Primers | N200 forward | TAGTCGCAACAGTTCAAGAAAT | N/A | 500 | |
| N200 reverse | CTGGTTCAATCTGTCAAGCAG | N/A | 500 | ||
| Probes | All | Universal | TCCTGCTAGAATGGC | FAM/BHQ-1 plus | 100 |
| Omicron/Alpha | R203K + G204R | CAGCAGTAAACGAAC | Quasar670/BHQ-2 plus | 400 | |
| Beta | T205I | AGTAGGGGAATTTCTT | Cal Fluor Red 610/BHQ-2 plus | 100 | |
| Gamma | S202S | AGGCAGCTCTAAACGA | Quasar670/BHQ-2 plus | 100 | |
| Delta | R203M | CAGCAGTATGGGAACT | Cal Fluor Gold 540/BHQ-1 plus | 400 |
2.3. Assay validation
Initial tests of the N200 assay were performed on synthetic templates. Twist synthetic RNA controls 2, 14, 16, 17, 23, and 48 (TWIST Bioscience) were employed to generate standard curves (sequences representative in lieu of Wuhan-Hu-1 (Genbank MN908957.3), as well as the VOCs, Alpha, Beta, Gamma, Delta, and Omicron VOCs, respectively). The N200 standard curves were assessed for linearity and efficiency by use of each of the probes in singleplex. After validation with synthetic templates, assays were tested with wastewater RNA extracts to ensure successful amplification. Temperature gradients and primer/probe concentration gradients were performed to determine the optimal assay conditions in singleplex.
Cross-reactivity of probes was assessed in vitro by use of synthetic RNA controls. Each probe, in singleplex, was tested for reactivity with Twist RNA controls 2, 14, 16, 17 and 23 at a concentration of 2500 copies per reaction. Twist 48 was later tested at a concentration of 2048 copies per reaction with the Universal, Alpha and Delta probes once this control became available. Specificity was also determined in a triplex assay iteration (Universal, R203M, and R203K + G204R probes) by use of digital PCR (dPCR) at multiple concentrations of Twist RNA controls (see dPCR section for additional details).
The N200 assay was validated in multiplex by comparing the efficiency, R2 and y-intercept of standard curves. Sensitivity of the N200 assay with the Universal, R203K + G204R, and R203M probes were determined by use of a 12-point standard curve (Twist synthetic controls) with 15-replicates each in singleplex and duplex. The limit of detection (LOD, 95 % detection rate) and limit of quantification (LOQ; coefficient of variance threshold of 0.35) for singleplex and duplex, was calculated for Universal, R203M, and R203K + G204R probes as described by Klymus et al. (2020).
2.4. Wastewater sample collection, concentration, and extraction
Wastewater influent was collected from the Regions of Peel (wastewater treatment plant influents GE Booth and Clarkson), York (access points Humber Air Management Facility (AMF) and Warden) and Waterloo (wastewater treatment plant influents Kitchener and Waterloo) Ontario, Canada. At each site, wastewater was collected by plant operators using 24-hour cooled composite samplers (combined sample of three grabs at Warden) 3–5 times a week between November 28, 2021, and January 4, 2022. Samples were aliquoted into 250 mL HPDE bottles (Systems Plus, Baden, Canada) and transported on ice to the University of Waterloo. This work followed the Environmental Microbiology Minimum Information guidelines as indicated in the element checklist provided in supporting information (Borchardt et al., 2021).
To concentrate RNA, 250 mL samples were well-mixed by inversion, 40 mL poured into 50 mL conical tubes with 4 g PEG-8000 and 0.9 g NaCl, then mixed on an orbital shaker at low speed at 4 °C for 2 h before being stored at 4 °C overnight (Wu et al., 2020). Samples were then centrifuged at 12,000 ×g at 4 °C for 90 min with no brake. The supernatant was discarded, and the remaining sample was centrifuged again at 12,000 ×g at 4 °C for 5 min with no brake. The remaining supernatant was decanted and pipetted out and the wet weight of the resulting pellet recorded. Up to 250 mg of the pellet was used for extraction of RNA by use of the RNeasy PowerMicrobiome Kit (Qiagen, Germantown, MD) as per manufacturer's instructions and included the addition of 100 μL of TRIzol (Thermo Fisher, Mississauga, Canada) to the pellet before bead-beating. Total RNA was eluted in 100 μL of RNase-free water.
2.5. RT-qPCR
One-step RT-qPCR was performed using 4× One-Step Viral RT-PCR Master Mix (Thermo Fisher, Mississauga, Canada). The primer and probe sequences and final working concentrations are presented in Table 2. RT-qPCR reactions were run in triplicate using 5 μL RNA template and a final reaction volume of 20 μL. Cycling was performed on an OPUS or CFX 96 Touch qPCR thermocycler (BioRad, Hercules, CA) as follows: RT at 50 °C for 15 min, 95 °C for 2 min, 45 cycles of 95 °C for 3 s followed by 57 °C for 30 s. All qPCR plates were run with controls including no template controls (NTCs), non-reverse transcriptase (NRT) controls, a positive control (wastewater sample with previously determined amount of Delta variant) and a standard curve based on a homologous synthetic RNA (i.e., EDX, TWIST).
2.6. Digital PCR (dPCR)
For precise quantification, commercial standards were analyzed via singleplex one-step RT-dPCR (QIAcuity, Qiagen, Hilden, Germany). QIAquity One-Step Viral RT-PCR Kit (Qiagen) together with CDC_N1 probe and primers (Lu et al., 2020) were used to determine absolute copy number (cp) of RNA in the following SARS-CoV-2 genomic RNA standards: Twist Bioscience Controls 2, 14, 16, 17, 23, and 48. Testing for specificity of probes was also conducted using the N200 assay in triplex (Universal, R203M, and R203K + G204R probes) with two concentrations of Twist Bioscience Controls 23 and 48. Reactions consisted of 10 μL of RNA template, forward and reverse primers (working concentration of 500 nM each), probe (working concentration of 125 nM for N1, or 100, 400 and 400 nM for the Universal, R203M and R203K + G204R probes respectively), 10 μL of One-Step Viral RT-PCR Master Mix, 0.4 μL of reverse transcriptase, and 14.6 μL of PCR-grade water to make up the balance of the 40 μL reaction. All reactions, along with non-template controls, were performed in triplicate using 26 k 24-well QIAcuity Nanoplates. Cycling was performed on the QIAcuity as follows: RT at 50 °C for 30 min, 95 °C for 2 min, with 40 cycles at 95 °C for 3 s then annealing at 55 °C or 57 °C (for N1 and N200 assays respectively) for 30 s. Plate were read on all channels and data were analyzed with the QIAcuity Suite Software (version 2.0.20, Qiagen) using the automatically generated threshold and data expressed as gene copies/μL of template. The N200 assay was also tested on RT-dPCR in triplex (Universal, R203M, and R203K + G204R probes) using the TWIST Bioscience Controls 23 and 48 at multiple concentrations to determine the specificity of the assay.
2.7. Data analysis
Samples were quantified (copies/well) by use of a 7-point standard curve with the relevant TWIST standard. Samples were then corrected for elution volume and volume of wastewater subsampled and reported as copies/mL wastewater (Quantity). The proportion of total quantity associated with each VOC in a sample was determined by use of the equation mutation specific for the VOC (Eq. (1)).
| (1) |
2.8. Comparison with clinical sequencing
To compare the accuracy of the frequency of the Delta and Omicron variants of SARS-CoV-2 measure by this assay, qPCR data were compared with clinical whole genome sequencing (WGS) data measured in each of three communities being monitored. Clinical WGS data were obtained from the reports released by Public Health Ontario to the public between December 21st, 2021, and March 22nd, 2022 (Ontario Agency for Health Protection and Promotion (Public Health Ontario), 2021). In each of these reports, the number and percentage of cases by public health unit were given for a four-week period for the most prevalent Pango lineages (summarized in Table S5). To directly compare data, qPCR data for all treatment plants monitored in each region were averaged for the same four-week period, resulting in a single weekly data point.
3. Results
3.1. N200 assay conditions and performance
The N200 assay performed well with all probes in singleplex. When tested with a serial dilution of TWIST standards, the singleplex standard curves were linear, y-intercepts were near 40 Ct, and assays had slopes within the accepted range (Bivins et al., 2021) of −3.1 (110 %) to −3.58 (90 %) for each of the probe tested (Table 3 ). When probes were combined in, duplex and triplex, these parameters did not fall outside the accepted range (Table 3). All probes targeting mutations were found to be highly specific with little to no cross-reactivity for non-target templates using qPCR (in singleplex; Table 4 ). Using RT-qPCR, this was demonstrated by the lack of amplification on all VOC TWIST templates without the target mutation (Table 4). Some cross-reactivity was observed between the T205I probe and the Wuhan variant control (Twist Control 2; Table 4), but this accounted for <5 % of the signal observed on the homologous template. Positive amplification with similar sensitivity was seen on of all TWIST templates tested was observed with the Universal probe (Table 4). High target specificity of each probe was confirmed by RT-dPCR, where in the presence of a high concentration of non-target template, almost no amplification was detected (Table 5 ). Copy numbers reported by the mutation probes (R203M, R203K + G204R) were comparable to the copy numbers reported by the Universal probe in the RT-dPCR reactions at multiple concentrations, which further suggests that each probe has similar sensitivities and efficiencies (Table 5). The LOD (95 % detection) was lower when the assays were performed in singleplex than duplex and ranged from 2 to 4 copies per reaction (Table 6 ). The LOQ (coefficient of variance threshold of 0.35) was also lower in singleplex reaction compared to multiplex reactions and ranges from 6 to 17 copies per reaction (Table 6).
Table 3.
Performance metrics of the N200 VOC assay for each of the probes in singleplex, duplex or triplex.
| Assay | Slope | y-intercept | Efficiency | R2 | |
|---|---|---|---|---|---|
| Universal | Singleplex | −3.347 | 40.236 | 98.9 | 0.990 |
| Duplex (with R203M) | −3.366 | 39.50 | 98.2 | 0.991 | |
| Duplex (with R203K + G204R) | −3.299 | 38.793 | 101.0 | 0.990 | |
| Duplex (with S202S) | −3.311 | 40.207 | 100.5 | 0.990 | |
| Duplex (with T205I) | −3.461 | 39.773 | 94.5 | 0.990 | |
| Triplex (with R203M, R203K + G204R) | −3.443 | 39.661 | 95.2 | 0.995 | |
| R203K + G204R (Omicron/Alpha) |
Singleplex | −3.429 | 38.834 | 95.7 | 0.998 |
| Duplex (with Universal) | −3.334 | 39.585 | 99.5 | 0.991 | |
| Triplex (with Universal, R20K + G204R) | −3.464 | 39.902 | 94.4 | 0.995 | |
| T205I (Beta) |
Singleplex | −3.396 | 39.951 | 97.0 | 0.997 |
| Duplex (with Universal) | −3.533 | 39.952 | 91.9 | 0.994 | |
| S202S (Gamma) |
Singleplex | −3.249 | 40.363 | 103.1 | 0.998 |
| Duplex (with Universal) | −3.352 | 39.891 | 98.8 | 0.988 | |
| R203M (Delta) |
Singleplex | −3.541 | 40.365 | 91.6 | 0.998 |
| Duplex (with Universal) | −3.387 | 39.513 | 97.4 | 0.990 | |
| Triplex (with Universal, R203M) | −3.442 | 39.124 | 95.2 | 0.995 | |
Table 4.
Cycle Threshold (CT) value (mean) of N200 assay performed in singleplex with probes targeting a mutation (R203K + G204R; T205I; S202S; R203M) or a common region (Universal) of the SARS-CoV-2 genome. Individual probes were performed on each of five synthetic templates synthesized by TWIST Bioscience (San Francisco, USA), controls for the original Wuhan strain and for each VOC. ND = not detected, “-” = not tested.
| Universal (all) | R203K + G204R (Omicron/Alpha) |
T205I (Beta) |
S202S (Gamma) |
R203M (Delta) |
|
|---|---|---|---|---|---|
| Twist 2 (Wuhan) | 29.52 | ND | 35.90 | ND | ND |
| Twist 14 (Alpha) | 29.73 | 30.1 | ND | ND | ND |
| Twist 16 (Beta) | 29.49 | ND | 30.05 | ND | ND |
| Twist 17 (Gamma) | 29.40 | ND | ND | 27.69 | ND |
| Twist 23 (Delta) | 30.44 | ND | ND | ND | 29.16 |
| Twist 48 (Omicron) | 30.14 | 30.18 | – | – | ND |
Table 5.
Gene copies/μL of RNA (mean ± SD; n = 3) detected by the Universal, R203M, or R203K + G204R probes in the N200 assay (triplexed) when tested against TWIST controls for variants of SARS-CoV-2 using RT-dPCR.
| Dilution from stock (~1 ∗ 10^4 copies/μL) | Universal (all) | R203K + G204R (Omicron/Alpha) |
R203M (Delta) |
|
|---|---|---|---|---|
| Twist 23 (Delta) | 5× | 2769 ± 173 | 0 ± 0 | 2767 ± 173 |
| 500× | 25 ± 2 | 0 ± 0 | 25 ± 2 | |
| 5000× | 3 ± 1 | 0 ± 0 | 3 ± 1 | |
| Twist 48 (Omicron) | 5× | 824 ± 12 | 820 ± 22 | 2 ± 2 |
| 50× | 78 ± 3 | 77 ± 3 | 0 ± 0 | |
| 500× | 5 ± 0 | 5 ± 0 | 0 ± 0 |
Table 6.
Limit of detection (LOD) and limit of quantification (LOQ) in copies/reaction of the N200 assay when run in singleplex or duplex using qPCR with the Universal, R203K + G204R, or R203M probes.
| LOD (copies/reaction) |
LOQ (copies/reaction) |
|||||
|---|---|---|---|---|---|---|
| Universal | R203K + G204R | R203M | Universal | R203K + G204R | R203M | |
| Singleplex | 2.2 | 2.7 | 4.4 | 6.0 | 7.0 | 7.0 |
| Duplex | 4.0 | 4.1 | 4.0 | 11.0 | 17.0 | 10.0 |
3.2. Utilization of the N200 assay in wastewater samples
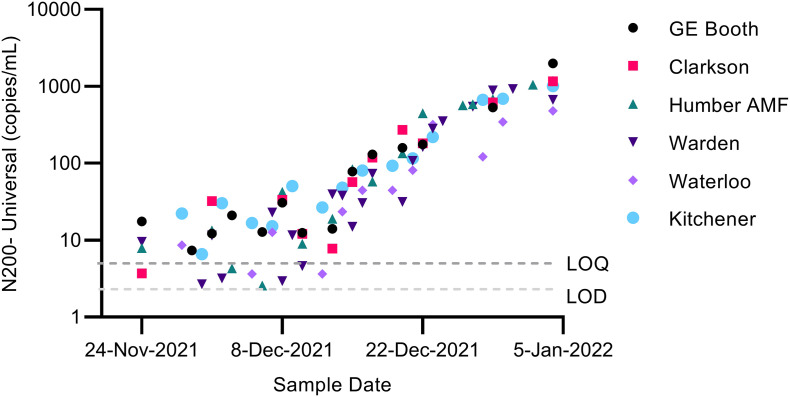
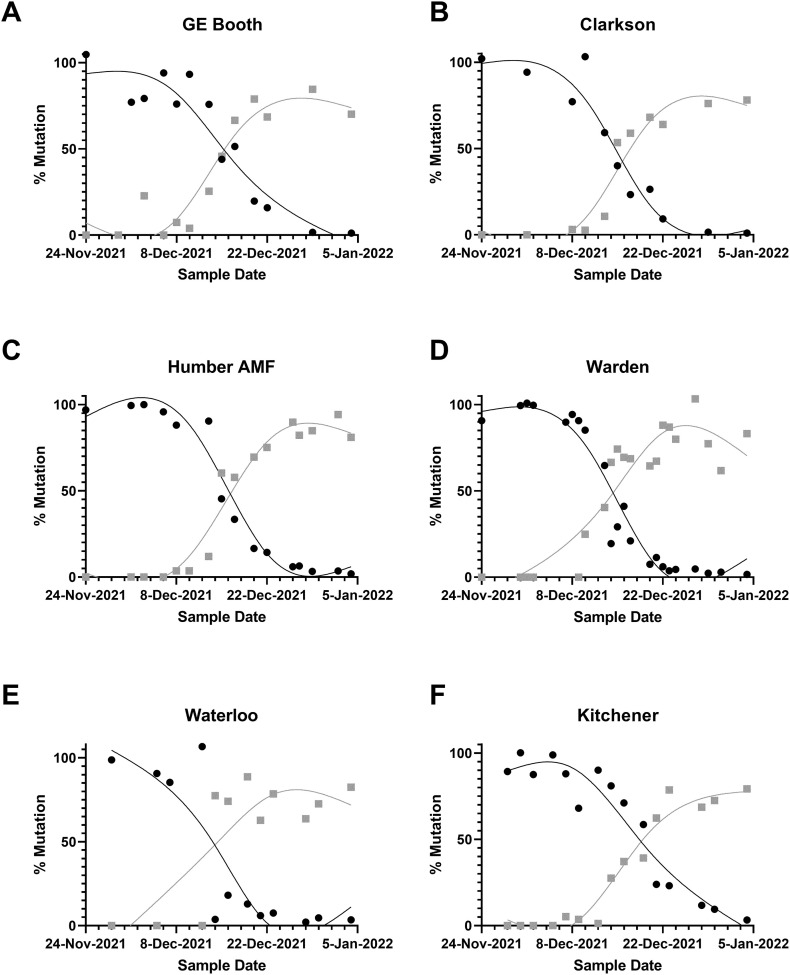
The triplex N200 assay using the Universal, R203M, or R203K + G204R probes was applied to wastewater samples throughout December 2021 and January 2022 to monitor for community transmission of the Omicron VOC. Detection of the Omicron VOC which would be signified by decrease in the proportion of the R203M (Delta) mutation and an increase in the R203K + G204R mutation (Omicron). The potential presence of Omicron was first detected in the Region of Peel in a sample collected on December 3rd, 2021, which tested positive for the R203K + G204R mutation. The latest introduction of this mutation occurred at the Waterloo Wastewater Treatment plant, with it's first detection of the R203K + G204R mutation occurring on December 14th, 2021. The total SARS-CoV-2 (Universal) signal in samples was initially near the LOD at the beginning of December 2021, but rapidly increased by over 100× to levels orders of magnitude greater than any previously detected by January 2022 (Fig. 2 ). During this time, we observed a rapid transition in the proportion of mutations from being dominated by R203M (Delta) to being dominated by R203K + G204R (Omicron) at all monitoring locations (Fig. 3 ). The transition from the Delta to the Omicron variant occurred simultaneously at all sites, with dominance of the Omicron variant R203K + G204M mutation (>50 % frequency) occurring between December 14 and 21, 2021 (Fig. 3).
Fig. 2.
SARS-CoV-2 signal measured in municipal wastewater using the N200 assay with the Universal probe in a multiplexed rection (Log scale, copies/mL). Some samples were included that were above the limit of detection (LOD) but below the limit of quantification (LOQ) as indicated in the figure. Samples from the Peel (GE Booth and Clarkson), York (Humber AMF and Warden) and Waterloo (Waterloo and Kitchener) Regions were collected during the period encompassing both the introduction of Omicron into these communities and the concomitant reduction of the endemic Delta VOC.
Fig. 3.
Application of the N200 assay to monitor the relative abundance (%) of the mutations R203M (black circles- presumed Delta) or R203K + G204R (grey squares – presumed Omicron) in municipal wastewater influent. Influent was collected from the Region of Peel (A, B), York Region (C, D) and Waterloo Region (E, F) using composite (A–B, D–F) and grab (C) sampling. Lines drawn spline smoothed curve calculated using GraphPad PRISM (v 9.5.0).
3.3. Comparison of N200 assay with clinical WGS
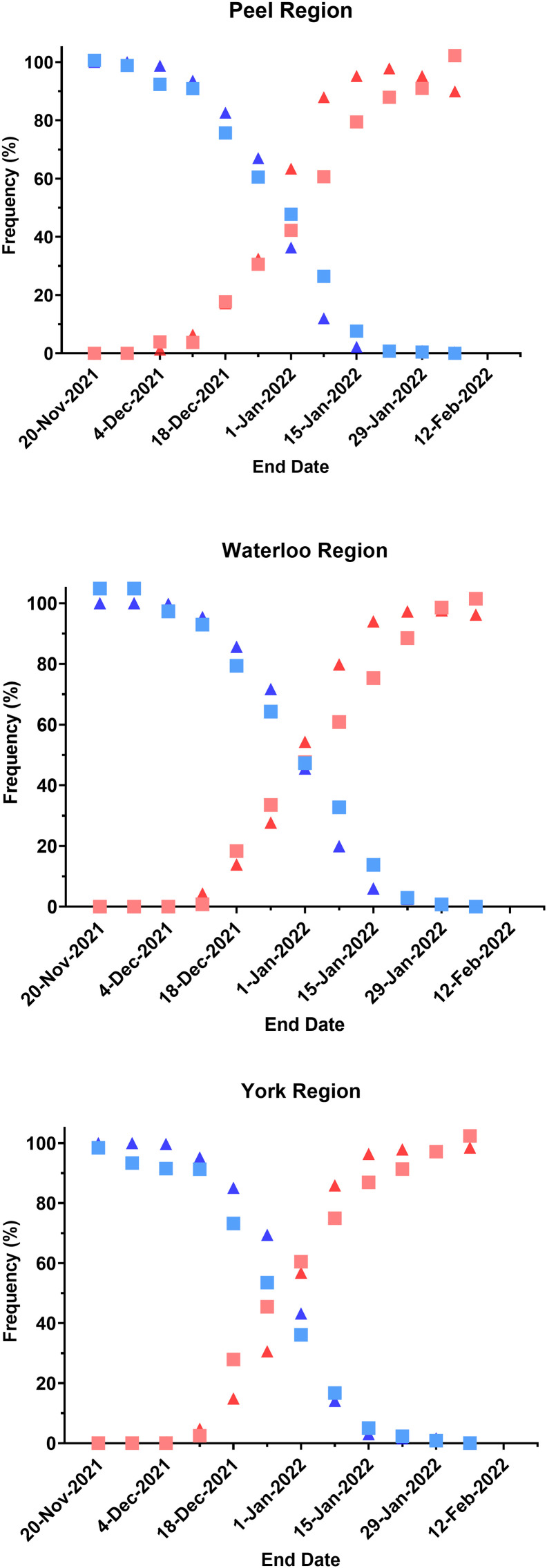
In all three regions examined, the frequency estimates of the Delta and Omicron variants by the N200 assay were very similar to those estimated by clinical WGS (Fig. 4 ). While not necessarily apparent in these figures, the date of first detection of Omicron in these communities with the N200 assay was either within the same Epi week (Peel, Waterloo) or in the week after (York) clinical WGS. While there was a slight delay in this date of detection by the N200 assay, the date of first detection was reported to the Public Health Units was earlier for the qPCR assay. For example, in the Peel Region, wastewater qPCR and Clinical sequencing both detected the presence of Omicron in the week of December 4th, but the wastewater qPCR data was reported to the Region on December 13th, whereas the presence of Omicron was first reported in the clinical WGS on December 21st, 2021.
Fig. 4.
Comparison of whole genome sequencing (WGS) data from clinical samples (triangles) and qPCR wastewater data (squares) during the transition from Delta (blue) to Omicron (red) in each of three regions; Peel Public Health (A), York Region Public Health (B), and Region of Waterloo Public Health (C). Clinical whole genome sequencing data points represent the frequency of each variant over a four-week period within a single public health unit. Wastewater qPCR data points represent the mean frequency of each variant at all sites assessed within a region within the same four-week period. The date on the x-axis represents the end of the four-week period.
4. Discussion
This study presents an RT-PCR approach for detection of SARS-CoV-2 and VOCs Omicron/Alpha, Beta, Gamma and Delta in a multiplexed assay with a single amplicon. The specificity and sensitivity of the N200 assay allow for simultaneous detection and quantification of mutations associated with VOCs. These individual assays were multiplexed and validated in duplex and triplex reactions, which allowed for rapid, cost-effective detection and quantitation of mutations associated with VOCs. Relative amounts of these mutations are then compared to a non-mutated region of the same amplicon to estimate the prevalence of current VOCs in a wastewater sample.
A major advantage of using an intra-amplicon universal probe, like the one described in the N200 assay, is reduced variability of mutation prevalence estimates. There are several methods that have been used to date to calculate variant frequency in a wastewater sample using qPCR. The differences occur in what gene(s) are used in the denominator of the equation. Some use another amplicon, such as the N1 or E genes as the denominator (Vogels et al., 2021; Wolfe et al., 2022; Wurtzer et al., 2022a). This can be problematic due to the variation in assay performance as well as the variation in the SARS-CoV-2 genome presence within a sample. For example, Amman et al. (2022) found that read coverage was not uniform across the SARS-CoV-2 genome, and that the areas of poor coverage were not consistent suggesting variation in sample composition as opposed to methodological issues. Thus, considerable variability may be experienced if frequency of a mutation is estimated using different regions of the genome. Comparing the frequencies of the mutant and “wild-type” (e.g., ancestral/endemic allele) at the same locus is another frequently used method to determine mutation/variant frequency (Chan et al., 2022; Graber et al., 2021; Lee et al., 2021; Peterson et al., 2022), where the abundance of the mutation and the wild-type alleles are summed to become the denominator. While these assays often produce accurate data, some variability can be expected due to the necessity of using multiple standards. Standard quantity can vary widely, as, observed in this study which is why standard quantification is imperative. Quantification of standards takes time, effort, and occasionally specialized equipment such as dPCR or digital droplet PCR. Additionally, when using this traditional means of determining variant frequency, all mutations of an allele must be measured (i.e. N:R203M, N:R204K). Otherwise, when both mutations are present in a sample, the total SARS-CoV-2 quantity (the denominator of a frequency estimation) will not be complete, and the frequency estimation would be inflated. Thus, a non-mutated intra-amplicon region (the universal probe used in this paper) can be used more readily and with a higher level of confidence in the accuracy of a mutation/variant frequency estimate than other methods. While we did not compare frequency estimation methods in this study, the estimations of Delta and Omicron prevalence calculated using the N200 intra-amplicon universal probe were comparable to frequencies of these variants in clinical WGS samples.
The N200 assay is a multiplex assay, which allows for the simultaneous measurement of one or more mutant alleles along with the intra-amplicon universal marker. This provides several advantages in addition to the accuracy of the denominator in the frequency estimate. Multiplexing ensures that the same sample aliquot (the RNA placed into each well of a RT-PCR reaction) is compared in the frequency estimate. Additionally, frequency of the mutation can be estimated in each well allowing for replicates. This demonstrates how multiplexing can further reduce variability and increase confidence of mutation/variant frequency estimates. The fact that this assay was multiplexed also reduced the volume of sample, cost, and time needed to determine the prevalence of multiple VOCs in wastewater samples.
Estimations of VOC prevalence are important during wastewater surveillance SARS-CoV-2 in that they can provide useful information to public health units in a timely manner. During analyses of wastewaters through late 2021 and early 2022, a rapid increase in the SARS-CoV-2 signal (Universal probe) as well as a quick change over of the dominant variant from Delta (R203M) to Omicron (R203K + G204R) was observed at all sites monitored. The assay first detected the R203K + G204R mutation in a sample collected from GE Booth (Peel Region) on December 3rd, 2021. While it is possible that this early detection of the R203K + G204R mutation could have been a variant other than Omicron, within a week of this detection, the R203K + G204R mutation was consistently detected in the wastewater at this site while R203M decreased in proportion. The use of two mutation targets provided confidence in the data as it was being collected and reported because both a decrease in the proportion of the R203M mutation (Delta) and an increase in the proportion of the R203K + G204R mutations (Omicron) occurred simultaneously. This data was reported to public health partners on December 13th and was immediately disseminated to the public by the Public Health Unit as part of a weekly Epi-Report (https://data.peelregion.ca/documents/profile-of-covid-19-cases/explore). In contrast, while clinical WGS detected Omicron in clinical samples in the Peel region in the week of December 4th, 2021, this was not reported to the public until December 21st. Rapid emergence of Omicron presented a major challenge for clinical testing and sequencing capacity. This was also confounded by the holiday season in Ontario. The wastewater data in general, quickly became important to Public Health Units as the clinical testing became less reliable because of changes to testing eligibility in Ontario. They were able to confirm the rapid replacement of Delta with the Omicron variant using wastewater that was independent of clinical testing (Arts et al., 2022).
In this assay and study, a single mutation was utilized to track the prevalence of variants of SARS-CoV-2 using qPCR. While this strategy was successful in this case study, there are limitations that one should keep in mind with this approach. Mutations are often associated with more than one lineage of SARS-CoV-2 and cannot always be utilized to differentiate between the diversity of variants circulating in a system. For example, the mutation used to track the introduction of Omicron (R203K + G204R) in this study is present in all sublineages of Omicron (i.e. BA.1, BA.2, BA.3, BA.4 and BA.5) and cannot differentiate between them. The R203K + G204R mutation used in this study is also present in the Alpha variant. While no Alpha was detected in clinical samples from the Regions sampled during this study (Ontario Agency for Health Protection and Promotion (Public Health Ontario), 2021), this mutation could be present and associated with a lineage other than Omicron or Alpha (cryptic variants). One could use multiple RT-PCR assays targeting two or more variant mutations to confirm the presence of a variant of SARS-Cov-2, however whole-gnome sequencing would be a more efficient way to monitor many mutations across the genome simultaneously. Although WGS and RT-PCR analysis of variants in wastewater are complementary and show similar trends (Galani et al., 2022), RT-PCR has several advantages and may be very useful for early detection when the targets are known (Lou et al., 2022). RT-PCR can cover a larger portion of the community at a lower cost and can usually provide data more rapidly than WGS. WGS is often necessary, however, to ensure confidence that the mutations being observed are associated with the assumed lineages of SARS-CoV-2. This is especially true during the introduction of a new lineage in a geographic area.
The N200 multiplex qPCR was developed to improve estimates of the proportion of various variants in wastewater, while also making the assay more efficient to use in terms of reduced use of extracts, reagents, and time. The assay was validated using synthetic templates and was successfully applied to monitor for the emergence of the Omicron variant in several communities in Ontario. Although, this assay is one among several now available for detection of VOCs in wastewater, the unique design concept, with a single amplicon, multiple target mutations, and intra-amplicon universal comparator will prove to be a useful in wastewater-based surveillance of SARS-CoV-2 as the virus continues to mutate.
CRediT authorship contribution statement
Meghan Fuzzen: Conceptualization, Methodology, Validation, Investigation, Data Curation, Visualization, Writing - original draft.
Nathanael B.J. Harper: Conceptualization, Methodology, Validation, Writing- original draft.
Hadi A. Dhiyebi: Conceptualization, Data Curation, Supervision, Writing-review.
Nivetha Srikanthan: Project Administration, Supervision.
Samina Hayat: Project Administration, Supervision, Investigation.
Leslie M. Bragg: Project Administration.
Shelley W. Peterson: Validation.
Ivy Yang: Validation.
J.X. Sun: Validation.
Elizabeth A. Edwards: Writing - review.
John P. Giesy: Writing - review.
Chand S. Mangat: Writing - review.
Tyson E. Graber: Writing - review.
Robert Delatolla: Writing - review.
Mark R. Servos: Supervision, Funding acquisition, Writing - review.
Funding
Funding for this project was provided by theOntario Ministry of the Environment, Conservation and Parks, NSERC Discovery Wastewater Surveillance Initiative(Grant No. TPA 2021-02-1-1564736554). In addition, the research pulished in this paper is part of the projects entitled, "Next generation solutions to ensure healthy water resources for future generations" and "Lake Futures" funded by theGlobal Water Futures program (Grant No.410295) funded in part byCanada First Research Excellence Fund. Additonal information is available at www.globalwaterfutures.ca. This research was also supported by an NSERC Discovery Grant (MS); the Canada Research Chairs program (MS, JG)
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Mark Servos reports financial support was provided by Ontario Ministry of the Environment Conservation and Parks. Mark Servos reports financial support was provided by Natural Sciences and Engineering Research Council of Canada. Mark Servos reports financial support was provided by Global Water Futures.
Acknowledgements
The Municipal and Public Health Unit staff contributed to sample collection and provided valuable advice. Codey Dueck (PHAC) for laboratory analysis supporting LOD determinations. Kirsten Nikel, Rhiannon Lagg, Heather Ikert, Yash Badlani, Carly Sing-Judge, Emily Dodsworth and Samantha Maerten assisted with sample preparation and analysis.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2023.163292.
Appendix A. Supplementary data
Supplementary figures
Data availability
Data will be made available on request.
References
- Agarwala R., Barrett T., Beck J., Benson D.A., Bollin C., Bolton E., Bourexis D., Brister J.R., Bryant S.H., Canese K., Charowhas C., Clark K., Dicuccio M., Dondoshansky I., Federhen S., Feolo M., Funk K., Geer L.Y., Gorelenkov V., Hoeppner M., Holmes B., Johnson M., Khotomlianski V., Kimchi A., Kimelman M., Kitts P., Klimke W., Krasnov S., Kuznetsov A., Landrum M.J., Landsman D., Lee J.M., Lipman D.J., Lu Z., Madden T.L., Madej T., Marchler-Bauer A., Karsch-Mizrachi I., Murphy T., Orris R., Ostell J., O’sullivan C., Panchenko A., Phan L., Preuss D., Pruitt K.D., Rodarmer K., Rubinstein W., Sayers E., Schneider V., Schuler G.D., Sherry S.T., Sirotkin K., Siyan K., Slotta D., Soboleva A., Soussov V., Starchenko G., Tatusova T.A., Todorov K., Trawick B.W., Vakatov D., Wang Y., Ward M., Wilbur W.J., Yaschenko E., Zbicz K. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7. doi: 10.1093/NAR/GKV1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman F., Markt R., Endler L., Hupfauf S., Agerer B., Schedl A., Richter L., Zechmeister M., Bicher M., Heiler G., Triska P., Thornton M., Penz T., Senekowitsch M., Laine J., Keszei Z., Klimek P., Nägele F., Mayr M., Daleiden B., Steinlechner M., Niederstätter H., Heidinger P., Rauch W., Scheffknecht C., Vogl G., Weichlinger G., Wagner A.O., Slipko K., Masseron A., Radu E., Allerberger F., Popper N., Bock C., Schmid D., Oberacher H., Kreuzinger N., Insam H., Bergthaler A. Viral variant-resolved wastewater surveillance of SARS-CoV-2 at national scale. Nat. Biotechnol. 2022;40:1814–1822. doi: 10.1038/s41587-022-01387-y. 2022 4012. [DOI] [PubMed] [Google Scholar]
- Arts E., Brown S., Bulir D., Charles T.C., Degroot C.T., Delatolla R., Desaulniers J.-P., Edwards E.A., Fuzzen M., Gilbride K., Gilchrist J., Goodridge L., Graber T.E., Habash M. Community surveillance of omicron in Ontario: wastewater-based epidemiology comes of age. Res. Sq. 2022:1–7. doi: 10.21203/RS.3.RS-1439969/V2. [DOI] [Google Scholar]
- Bivins A., Kaya D., Bibby K., Simpson S.L., Bustin S.A., Shanks O.C., Ahmed W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021;203 doi: 10.1016/J.WATRES.2021.117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A.B., Hughes B., Wolfe M.K., White B.J., Duong D., Chan-Herur V. Regional replacement of SARS-CoV-2 variant omicron BA.1 with BA.2 as observed through wastewater surveillance. Environ. Sci. Technol. Lett. 2022;9:575–580. doi: 10.1021/ACS.ESTLETT.2C00266/ASSET/IMAGES/LARGE/EZ2C00266_0003.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boogaerts T., Van den Bogaert S., Van Poelvoorde L.A.E., El Masri D., De Roeck N., Roosens N.H.C., Lesenfants M., Lahousse L., Van Hoorde K., van Nuijs A.L.N., Delputte P. Optimization and application of a multiplex digital PCR assay for the detection of SARS-CoV-2 variants of concern in Belgian influent wastewater. Viruses. 2022;14:610. doi: 10.3390/V14030610/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M.A., Boehm A.B., Salit M., Spencer S.K., Wigginton K.R., Noble R.T. The environmental microbiology minimum information (EMMI) guidelines: QPCR and dPCR quality and reporting for environmental microbiology. Environ. Sci. Technol. 2021;55:10210–10223. doi: 10.1021/ACS.EST.1C01767/ASSET/IMAGES/MEDIUM/ES1C01767_0006.GIF. [DOI] [PubMed] [Google Scholar]
- Carcereny A., Martínez-Velázquez A., Bosch A., Allende A., Truchado P., Cascales J., Romalde J.L., Lois M., Polo D., Sánchez G., Pérez-Cataluña A., Díaz-Reolid A., Antón A., Gregori J., Garcia-Cehic D., Quer J., Palau M., Ruano C.G., Pintó R.M., Guix S. Monitoring emergence of the SARS-CoV-2 B.1.1.7 variant through the Spanish national SARS-CoV-2 wastewater surveillance system (VATar COVID-19) Environ. Sci. Technol. 2021;55:11756–11766. doi: 10.1021/acs.est.1c03589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.T.-M., Leung J.S.-L., Lee L.-K., Lo H.W.-H., Wong E.Y.-K., Wong D.S.-H., Ng T.T.-L., Lao H.-Y., Lu K.K., Jim S.H.-C., Yau M.C.-Y., Lam J.Y.-W., Ho A.Y.-M., Luk K.S., Yip K.-T., Que T.-L., To K.K.-W., Siu G.K.-H. A low-cost TaqMan minor groove binder probe-based one-step RT-qPCR assay for rapid identification of N501Y variants of SARS-CoV-2. J. Virol. Methods. 2022;299 doi: 10.1016/j.jviromet.2021.114333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ferrando E., Pérez-Cataluña A., Allende A., Guix S., Randazzo W., Sánchez G. Recovering coronavirus from large volumes of water. Sci. Total Environ. 2021;762 doi: 10.1016/j.scitotenv.2020.143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Jüni P., MacKenzie A.E., Delatolla R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Pellett C., Williams R., Alex-Sanders N., Bassano I., Brown M.R., Denise H., Grimsley J.M.S., Kevill J.L., Khalifa M.S., Pântea I., Story R., Wade M.J., Woodhall N., Jones D.L. Rapid assessment of SARS-CoV-2 variant-associated mutations in wastewater using real-time RT-PCR. Microbiol. Spectr. 2023 doi: 10.1128/SPECTRUM.03177-22/SUPPL_FILE/SPECTRUM.03177-22-S0001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani A., Aalizadeh R., Kostakis M., Markou A., Alygizakis N., Lytras T., Adamopoulos P.G., Peccia J., Thompson D.C., Kontou A., Karagiannidis A., Lianidou E., Avgeris M., Paraskevis D., Tsiodras S., Scorilas A., Vasiliou V., Diopoulos M., Thomaidis N. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber T.E., Mercier É., Bhatnagar K., Fuzzen M., D’Aoust P.M., Hoang H.D., Tian X., Towhid S.T., Plaza-Diaz J., Eid W., Alain T., Butler A., Goodridge L., Servos M., Delatolla R. Near real-time determination of B.1.1.7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. NextStrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda M., Mustafa F., Maraqa M., Rizvi T., Aly Hassan A. Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 2021;759 doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Elsinga G., de Graaf M., Molenkamp R., Koopmans M.P.G., Medema G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/J.SCITOTENV.2021.149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Zhou Y., Lokugamage K.G., Vu M.N., Bopp N., Crocquet-Valdes P.A., Kalveram B., Schindewolf C., Liu Y., Scharton D., Plante J.A., Xie X., Aguilar P., Weaver S.C., Shi P.Y., Walker D.H., Routh A.L., Plante K.S., Menachery V.D. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 2022;18 doi: 10.1371/JOURNAL.PPAT.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K.I., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/NAR/GKF436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryanov S.A., Levina T.A., Konopleva M.V., Suslov A.P. Identification of hotspot mutations in the N gene of SARS-CoV-2 in Russian clinical samples that may affect the detection by reverse transcription-PCR. Diagnostics. 2022;12:147. doi: 10.3390/DIAGNOSTICS12010147. 2022, Vol. 12, Page 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymus K.E., Merkes C.M., Allison M.J., Goldberg C.S., Helbing C.C., Hunter M.E., Jackson C.A., Lance R.F., Mangan A.M., Monroe E.M., Piaggio A.J., Stokdyk J.P., Wilson C.C., Richter C.A. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA. 2020;2:271–282. doi: 10.1002/EDN3.29. [DOI] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumblathan T., Liu Y., Uppal G.K., Hrudey S.E., Li X.-F. Wastewater-based epidemiology for community monitoring of SARS-CoV-2: progress and challenges. ACS Environ. Au. 2021;1:18–31. doi: 10.1021/acsenvironau.1c00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan L.M., O'Brien M., Lovin L.M., Scarlett K.R., Davis H., Henke A.N., Seidel S.E., Archer N., Lawrence E., Norman R.S., Bojes H.K., Brooks B.W. Quantitative reverse transcription pcr surveillance of SARS-CoV-2 variants of concern in wastewater of two counties in Texas, United States. ACS ES&T Water. 2022 doi: 10.1021/ACSESTWATER.2C00103. [DOI] [PubMed] [Google Scholar]
- Lee W.L., Armas F., Guarneri F., Gu X., Formenti N., Wu F., Chandra F., Parisio G., Chen H., Xiao A., Romeo C., Scali F., Tonni M., Leifels M., Chua F.J.D., Kwok G.W., Tay J.Y., Pasquali P., Thompson J., Alborali G.L., Alm E.J. Rapid displacement of SARS-CoV-2 variant Delta by Omicron revealed by allele-specific PCR in wastewater. Water Res. 2022;221 doi: 10.1016/J.WATRES.2022.118809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Imakaev M., Armas F., McElroy K.A., Gu X., Duvallet C., Chandra F., Chen H., Leifels M., Mendola S., Floyd-O’Sullivan R., Powell M.M., Wilson S.T., Berge K.L.J., Lim C.Y.J., Wu F., Xiao A., Moniz K., Ghaeli N., Matus M., Thompson J., Alm E.J. Quantitative SARS-CoV-2 alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ. Sci. Technol. Lett. 2021;8:675–682. doi: 10.1021/acs.estlett.1c00375. [DOI] [Google Scholar]
- Lou E.G., Sapoval N., McCall C., Bauhs L., Carlson-Stadler R., Kalvapalle P., Lai Y., Palmer K., Penn R., Rich W., Wolken M., Brown P., Ensor K.B., Hopkins L., Treangen T.J., Stadler L.B. Direct comparison of RT-ddPCR and targeted amplicon sequencing for SARS-CoV-2 mutation monitoring in wastewater. Sci. Total Environ. 2022;833 doi: 10.1016/J.SCITOTENV.2022.155059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute tespiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mills M.G., Hajian P., Bakhash S.M., Xie H., Mantzke D., Zhu H., Perchetti G.A., Huang M.L., Pepper G., Jerome K.R., Roychoudhury P., Greninger A.L. Rapid and accurate identification of SARS-CoV-2 Omicron variants using droplet digital PCR (RT-ddPCR) J. Clin. Virol. 2022;154 doi: 10.1016/J.JCV.2022.105218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontario Agency for Health Protection and Promotion (Public Health Ontario) Epidemiologic Summary: SARS-CoV-2 Whole Genome Sequencing in Ontario, December 14, 2021 - March 22 2022. Queen's Print; Ontario: 2021. p. 27. [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/J.SCITOTENV.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Lidder R., Daigle J., Wonitowy Q., Dueck C., Nagasawa A., Mulvey M.R., Mangat C.S. RT-qPCR detection of SARS-CoV-2 mutations S 69–70 del, S N501Y and N D3L associated with variants of concern in Canadian wastewater samples. Sci. Total Environ. 2022;810 doi: 10.1016/J.SCITOTENV.2021.151283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretti M.A.M., Galvani R.G., Scherer N.M., Farias A.S., Boroni M. In silico analysis of mutant epitopes in new SARS-CoV-2 lineages suggest global enhanced CD8+ T cell reactivity and also signs of immune response escape. Infect. Genet. Evol. 2022;99 doi: 10.1016/J.MEEGID.2022.105236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A.M., Taha T.Y., Tabata T., Chen I.P., Ciling A., Khalid M.M., Sreekumar B., Chen P.-Y., Hayashi J.M., Soczek K.M., Ott M., Doudna J.A. Rapid assessment of SARS-CoV-2 evolved variants using virus-like particles. Science. 2021;eabl6184 doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A., Adhikari S., Zhang S., Solomon T.B., Lipponen A., Islam M.A., Thakali O., Sangkham S., Shaheen M.N.F., Jiang G., Haramoto E., Mazumder P., Malla B., Kumar M., Pitkänen T., Sherchan S.P. Tracing COVID-19 trails in wastewater: a systematic review of SARS-CoV-2 surveillance with viral variants. Water. 2023;15:1018. doi: 10.3390/W15061018. 2023, Vol. 15, Page 1018. [DOI] [Google Scholar]
- Vogels C.B.F., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., de Jesus J.G., Claro I.M., Ferreira G.M., Crispim M.A.E., Singh L., Tegally H., Anyaneji U.J., Hodcroft E.B., Mason C.E., Khullar G., Metti J., Dudley J.T., MacKay M.J., Nash M., Wang J., Liu C., Hui P., Murphy S., Neal C., Laszlo E., Landry M.L., Muyombwe A., Downing R., Razeq J., de Oliveira T., Faria N.R., Sabino E.C., Neher R.A., Fauver J.R., Grubaugh N.D., da Silva Sales F.C., Ramundo M.S., Candido D.S., Silva C.A.M., de Pinho M.C., Coletti T.de M., Andrade P.dos S., de Souza L.M., Rocha E.C., Gomes Jardim A.C., Manuli E., Gaburo N., Granato C., Levi J.E., Costa S., de Souza W.M., Salum M.A., Pereira R., de Souza A., Matkin L.E., Nogueria M.L., Levin A.S., Mayaud P., Alexander N., Souza R., Acosta A.L., Prete C., Quick J., Brady O., Messina J., Kraemer M., Gouveia N.da C., Oliva I., de Souza M., Lazari C., Alencar C.S., Thézé J., Buss L., Araujo L., Cunha M.S., Loman N.J., Pybus O.G., Aguiar R.S., Wilkinson E., Msomi N., Iranzadeh A., Fonseca V., Doolabh D., San E.J., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Kosakovsky Pond S.L., Weaver S., Giovanetti M., Alcantara L.C.J., Martin D., Bhiman J.N., Williamson C. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A., Agrawal S., Schoth J., Meinert-Berning C., Bastian D., Orschler L., Ciesek S., Teichgräber B., Wintgens T., Lackner S., Weber F.A., Widera M. Early detection of SARS-CoV-2 omicron BA.4 and BA.5 in German wastewater. Viruses. 2022;14:1876. doi: 10.3390/v14091876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M., Hughes B., Duong D., Chan-Herur V., Wigginton K.R., White B.J., Boehm A.B. Detection of SARS-CoV-2 variants mu, beta, gamma, lambda, delta, alpha, and omicron in wastewater settled solids using mutation-specific assays is associated with regional detection of variants in clinical samples. Appl. Environ. Microbiol. 2022;88 doi: 10.1128/AEM.00045-22/SUPPL_FILE/AEM.00045-22-S0001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020:5. doi: 10.1128/msystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Xing N., Meng K., Fu B., Xue W., Dong P., Tang W., Xiao Y., Liu G., Luo H., Zhu W., Lin X., Meng G., Zhu Z. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe. 2021;29:1788–1801.e6. doi: 10.1016/j.chom.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer Sebastien, Levert M., Dhenain E., Accrombessi H., Manco S., Fagour N., Goulet M., Boudaud N., Gaillard L., Bertrand I., Challant J., Masnada S., Azimi S., Gillon-Ritz M., Robin A., Mouchel J.M., Sig O., Moulin L. From Alpha to Omicron BA.2: new digital RT-PCR approach and challenges for SARS-CoV-2 VOC monitoring and normalization of variant dynamics in wastewater. Sci. Total Environ. 2022;848 doi: 10.1016/j.scitotenv.2022.157740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Levert M., Cluzel N., Almayrac J.L., Charpentier C., Masnada S., Gillon-Ritz M., Mouchel J.M., Maday Y., Boni M., Moulin L., Marechal V., Le Guyader S., Bertrand I., Gantzer C., Descamps D., Charpentier Charlotte, Houhou N., Visseaux B., Calvez V., Marcelin A.G., Marot S., Jary A., Chaix M.L., Delaugerre C., Salmona M., Legoff J., Gault E., Rameix-Welti M.A., L'honneur A.S., Mariaggi A.A., Rozenberg F., Roque A.M., Mouna L., Fenaux H., Leruez-Ville M., Morand-Joubert L., Lambert-Niclot S., Schnuriger A., Dubois C., Bourgeois-Nicolaos N., Brichler S., Delagrèverie H., Bouvier-Alias M., Fourati S., Rodriguez C., Pawlotsky J.M. SARS-CoV-2 genome quantification in wastewaters at regional and city scale allows precise monitoring of the whole outbreaks dynamics and variants spreading in the population. Sci. Total Environ. 2022;810 doi: 10.1016/J.SCITOTENV.2021.152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Challis J.K., Oloye F.F., Asadi M., Cantin J., Brinkmann M., McPhedran K.N., Hogan N., Sadowski M., Jones P.D., Landgraff C., Mangat C., Servos M.R., Giesy J.P. RNA in municipal wastewater reveals magnitudes of COVID-19 outbreaks across four waves driven by SARS-CoV-2 variants of concern. ACS Environ. Sci. Technol. Water. 2021 doi: 10.1021/ACSESTWATER.1C00349/ASSET/IMAGES/LARGE/EW1C00349_0004.JPEG. [DOI] [PubMed] [Google Scholar]
- Xu X., Deng Y., Ding J., Zheng X., Li S., Liu L., Chui H.kwong, Poon L.L.M., Zhang T. Real-time allelic assays of SARS-CoV-2 variants to enhance sewage surveillance. Water Res. 2022;220 doi: 10.1016/J.WATRES.2022.118686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Shagan M., Lakkakula S., Plotkin N., Bhandarkar N.S., Kushmaro A. Direct RT-qPCR assay for SARS-CoV-2 variants of concern (Alpha, B.1.1.7 and Beta, B.1.351) detection and quantification in wastewater. Environ. Res. 2021;201 doi: 10.1016/j.envres.2021.111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Data Availability Statement
Data will be made available on request.