In a recent issue of HemaSphere, Pandzic and colleagues1 provide evidence that the use of next generation sequencing (NGS) in chronic lymphocytic leukemia (CLL) clinical practice is equally robust in detecting variants with allele frequency (VAF) between 5% and 10% respect to those with VAF ≥10%, and suggest to harmonize NGS methodologies across laboratories and to lower the canonical cutoff from ≥10% VAF to ≥5% VAF.2
This is an important issue to address, since it is now emerging from the scientific literature that the detection of TP53 mutations also at the subclonal level, that is, far below the 10% VAF cutoff, may have a clinical impact and should be therefore annotated for a more correct clinical management of CLL patients. In fact, since the first seminal observation,3 other more recent retrospective studies based on ultra-deep NGS have confirmed that TP53 mutations can be present in tumor cell populations at very low-VAF,4–7 and that the presence of these extremely small TP53 clones, even with VAF <1.0%, was associated with inferior survival in the context of chemoimmune therapy.5–7 Recently, it was reported that also in the setting of therapies with the novel BCR inhibitor ibrutinib, TP53 mutations/disruptions were independent negative prognostic factors,4,8 although the impact of low-VAF TP53 mutations upon treatment with targeted agents remains currently unexplored.
In general, all the above-mentioned studies,3–7 rather than identifying a threshold of clinical relevance, seem to point toward a “yes/no effect” of TP53 mutations on CLL outcome. Biologically speaking, this is in line with the basic principle of fitness where the fittest clones produce viable offspring capable of surviving and reproducing and, in the presence of selective pressures from the environment, for example, due to chemotherapy, eventually taking over the TP53 wild-type cell population.3
Nevertheless, the European Research Initiative on CLL (ERIC) recommend that only pathogenic/likely pathogenic TP53 variants with VAF >10% should be reported,2 a cautionary approach proposed to avoid not only the risk of possible technical artifacts in the identification of very low-VAF TP53 mutations, but also for the lack of solid evidence and/or prospective studies on the clinical impact of low-VAF TP53 variants. Recently, however, ERIC has launched an interlaboratory comparison of NGS methods to detect low-VAF variants; the multilaboratory initiative aimed to harmonize methods detecting TP53 variants below 10% VAF to be applied for the multicentric study analyzing their prognostic/predictive impact in CLL.9
Pandzic and colleagues1 thoroughly and brilliantly discussed all the limitations, drawbacks, and pitfalls of the use of the 10% VAF threshold, by also highlighting some paradoxical situations that may happen in the clinical practice for a different evaluation of the TP53 mutational burden due to the presence or not of a parallel deletion of the contralateral 17p allele. Yet, they provide extra-assay evidence (ie, by droplet digital polymerase chain reaction [PCR]) against a lowering of the threshold below the 5% VAF.1
Here we summarize the internal recommendations we are used to follow for NGS evaluation of TP53 mutational status in CLL, including wet laboratory and bioinformatics, as reported in a recent publication of ours,7 where we described the set-up of an ultra-deep NGS procedure capable to successfully detect mutations below the 1% VAF. A custom script of the bioinformatics pipeline, as described herein and in reference 7, can be retrieved at website address https://github.com/gamabunta313/TP_SNP-calling/. More details are reported in reference 7.
Briefly, analysis of TP53 mutations is performed with an amplicon-based strategy, covering exons 2-11, in keeping with ERIC recommendations.2 Amplicon libraries are generated starting from 40 ng of DNA (~6000 diploid genomes), a quantity capable to successfully detect mutations below the 1% VAF in the context of our procedures.7 Multiplex PCR products are generated using a high-fidelity DNA polymerase and tagged with specific indices and paired-end sequenced in an Illumina MiSeq instrument.
FASTQ files are aligned to the hg19 reference with Burrows-Wheeler Aligner-MEM algorithm, and allele variants are called by FreeBayes with nonstringent parameters. The overall strategy is designed to yield a minimum coverage of 2000X in 100% of the analyzed positions for each sequence, a prerequisite for ≤1% variant calling. To calculate random/systematic errors, a database of TP53 wild-type cases is utilized (362 cases in reference 7). A TP53 mutation is accepted as valid if (the following conditions have to be both fulfilled): (i) presented a VAF outdistancing for at least 2.75 standard deviations the mean of the transformed VAF at the specific TP53 position as retrieved in the wt cases; (ii) validated by Fisher exact test after Bonferroni correction (P < 0.01). The minimal allelic fraction found in reference 7 was 0.3%.
Even if robust bioinformatic workflows are employed to call true variants from background error noise, to exclude possible random polymerase errors despite the use of a high fidelity enzyme, it would be advisable to reanalyze mutation less than 2.0% of VAF. In addition, the detected variants should be checked using locus-specific database, that is, the TP53 Database (https://tp53.isb-cgc.org/), for interpretation of TP53 variants to discriminate between possible neutral and/or pathogenetic variants; programs like Mutalyzer (https://mutalyzer.nl/) and Sequence Variant Nomenclature (http://varnomen.hgvs.org/) are used to report the variant nomenclature according to the guidelines of the Human Genome Variation Society.
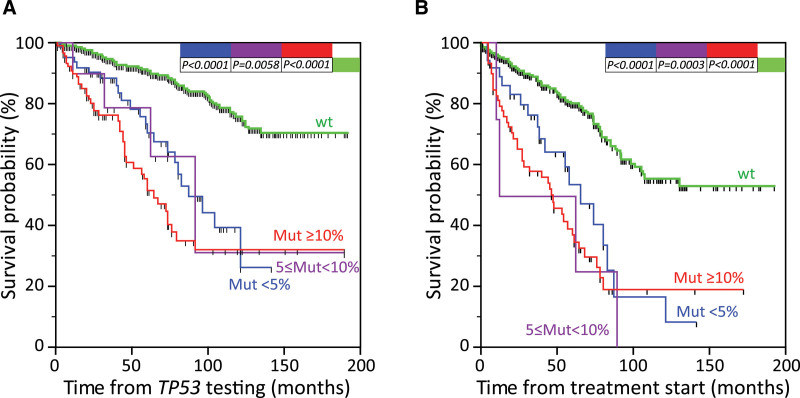
By such an approach, and re-classifying the retrospective cohort investigated in reference 7 (n = 1220) according to the cutoff suggested by 1, no difference was found in term of overall survival (OS) when comparing CLL cases with VAF lower than 5%, between 5% and 10% and upper than 10%, all the 3 categories experiencing significantly shorter OS intervals than TP53 wild-type CLL (Figure 1A). The same held true when the analysis was circumscribed to the 552 treated CLL cases, in which OS was computed from treatment initiation (Figure 1B).
Figure 1.
Overall survival of patients stratified according to the TP53 mutation load. CLL cases from a retrospective cohort originally published in7 were reclassified according to the standard cutoff for TP53 mutation evaluation (≥10% VAF), to the cutoff suggested by1 (≥5% VAF), and to the <5% VAF cutoff, as discussed in the present commentary. (A) Kaplan-Meier curves comparing OS computed from the date of TP53 mutation evaluation7 of CLL 1220 patients stratified into 4 classes according to the TP53 mutational load: TP53 wild-type cases (green line, n = 1052), cases with <5% VAF of TP53 mutation (blue line, n = 64), cases with VAF of TP53 mutations between 5.0% and 10.0% (purple line, n = 12), and cases with ≥10.0% VAF of TP53 mutation (red line, n = 92). (B) Kaplan-Meier curves comparing OS computed from the date of treatment7 of 552 treated CLL patients stratified into 4 classes according to the TP53 mutational load: TP53 wild-type cases (green line, n = 449), cases with <5% VAF of TP53 mutation (blue line, n = 38), cases with VAF of TP53 mutations between 5.0% and 10.0% (purple line, n = 4), and cases with ≥10.0% VAF of TP53 mutation (red line, n = 61). P values refer to the log-rank test. CLL = chronic lymphocytic leukemia; OS = overall survival; VAF = variants with allele frequency.
In conclusion, it is time to go beyond the 10% VAF cutoff for TP53 mutation evaluation in CLL by lowering the cutoff at least at the value of 5% VAF, as suggested by reference 1, or even below, when the technical quality is guaranteed by a solid laboratory and bioinformatics procedure; in this regard, some suggestions from reference 7 are summarized in the present commentary. However, it is to remind that any novel cutoff for TP53 mutation evaluation should be upfront validated by ad-hoc trials and cannot be introduced in the clinical practice in the absence of such a validation. Finally, TP53 variants analysis through a very sensitive NGS approaches should be centralized in few recognized/certified laboratories to fulfill all the criteria reported above and to guarantee the same standard of precision medicine for each patients.
AUTHOR CONTRIBUTIONS
All authors conceptualized, wrote, and finalized the manuscript.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
The authors declare no sources of funding.
REFERENCES
- 1.Pandzic T, Ladenvall C, Engvall M, et al. Five percent variant allele frequency is a reliable reporting threshold for TP53 variants detected by next generation sequencing in chronic lymphocytic leukemia in the clinical setting. HemaSphere. 2022;6:e761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malcikova J, Tausch E, Rossi D, et al. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia - update on methodological approaches and results interpretation. Leukemia. 2018;32:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brieghel C, Aarup K, Torp MH, et al. Clinical outcomes in patients with multi-hit TP53 chronic lymphocytic leukemia treated with ibrutinib. Clin Cancer Res. 2021;27:4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malcikova J, Pavlova S, Kunt Vonkova B, et al. Low-burden TP53 mutations in CLL: clinical impact and clonal evolution within the context of different treatment options. Blood. 2021;138:2670–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bomben R, Rossi FM, Vit F, et al. TP53 mutations with low variant allele frequency predict short survival in Chronic Lymphocytic Leukemia. Clin Cancer Res. 2021;27:5566–5575. [DOI] [PubMed] [Google Scholar]
- 8.Morabito F, Tripepi G, Del Poeta G, et al. Assessment of the 4-factor score: Retrospective analysis of 586 CLL patients receiving ibrutinib. A campus CLL study [Published online 2021]. Am J Hematol. 2021;96:E168–E171. [DOI] [PubMed] [Google Scholar]
- 9.Pavlova S, Malcikova J, Radova L, et al. Interlaboratory comparison of NGS methods for detection of TP53 Variants <10% VAF: the first phase of an ERIC multicenter study on the clinical impact of low-frequent TP53 variants in CLL. HemaSphere. 2020;4(S1):285–286. [Google Scholar]