To the Editor:
Philadelphia-like (Ph-like) B-cell precursor acute lymphoblastic leaukemia (B-ALL) is a high-risk subtype of leukaemia [1]. It is characterized by a gene expression profile similar to Philadelphia chromosome positive (Ph+) ALL, but lacking the BCR::ABL1 rearrangement. Instead, Ph-like B-ALL represents a heterogeneous subtype driven by a diverse range of genetic aberrations and chromosomal rearrangements that converge on activated tyrosine kinase signalling [2, 3]. Integration of tyrosine kinase inhibitors into treatment regimens of Ph-like ALL patients has demonstrated early evidence of therapeutic efficacy, although results from larger clinical trials are pending [4].
Here we describe a novel PDGFRB lesion in a pediatric patient with Ph-like B-ALL. Unlike other PDGFRB fusions, this CD74::PDGFRB variant does not form a chimeric protein, yet is constitutively active and sufficient to drive oncogenic transformation. We provide a model for a unique mechanism of kinase activation in Ph-like ALL which has implications for diagnostic fusion identification and provides key insights into kinase biology.
The patient, a 22-month-old girl, presented with B-ALL and 86% blasts in the bone marrow (BM). Microarray analysis of the BM sample showed losses of chromosome regions 5q32 (partial PDGFRB and CD74), 7p12.2 (46 kb, intragenic IKZF1 including exons 4 to 7), and 9p13.3-p13.1 (5 Mb, including PAX5, sub-clonal ~70 %). The patient was stratified as high-risk and treated on the COG AALL1131 trial. At the end of induction, residual blasts of 46.3% were still detected. Given indications of a PDGFRB lesion, the patient received dasatinib in combination with consolidation chemotherapy, but despite a measurable response, maintained persistent minimal residual disease (0.396%) at the end of consolidation. The patient proceeded to receive a Kymriah CAR T infusion. Additional clinical details are provided in the Supplemental Materials section.
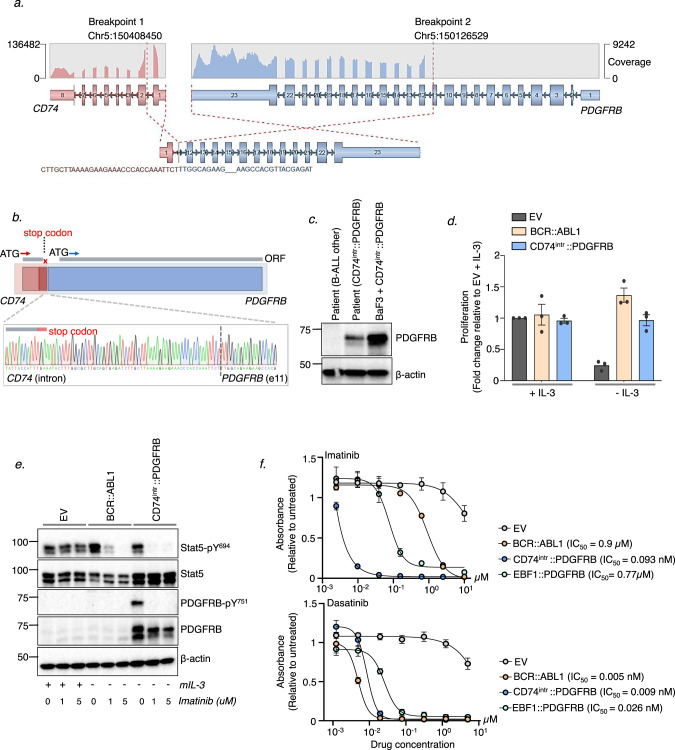
We performed RNA-sequencing on the diagnostic sample. Gene expression analysis identified a Ph-like signature [5], and highlighted aberrant expression of PDGFRB (Supplementary Fig. 1a). Sequence analysis confirmed a ~240 kb interstitial deletion in chromosome 5, resulting in a fusion between CD74 (exon 1) and PDGFRB (exon 11) (Fig. 1a). The fusion was validated by PCR amplification of the breakpoint (Supplementary Fig. 1b), and the full length CD74::PDGFRB cDNA was amplified and cloned into an MSCV-IRES-GFP (MIG) expression plasmid. Sanger sequencing revealed a retained 75 bp CD74 intronic sequence resulting in a stop codon before the fusion junction (Fig. 1b). We henceforth refer to this novel fusion as CD74intr::PDGFRB. A previous report described a CD74::PDGFRB variant resulting in an in-frame fusion linking CD74 exon 6 and PDGFRB exon 11 [6]. However, our patient’s fusion is distinguished by the absence of a clear open reading frame (ORF) to drive translation of an in-frame fusion protein. To determine whether a PDGFRB species is produced as a result of the CD74intr::PDGFRB fusion, we used western blotting to analyze leukemic mononuclear (MNC) cells from the patient (Fig. 1c). Strikingly, an immunoreactive PDGFRB species of ~60 kDa was detected in the patient MNCs corresponding to an equivalent product detected in murine Ba/F3 cells engineered to express the CD74intr::PDGFRB cDNA cloned directly from the patient.
Fig. 1. Identification of atypical PDGFRB fusion in Ph-like B-ALL patient.
a RNA-sequencing analysis identified a novel CD74::PDGFRB fusion between CD74 (exon 1) and PDGFRB (exon 11) on chromosome 5. Image shown is modified from a fusion visualization tool Arriba [12]. RNA read coverage is shown across the genes involved in the fusion. b Sanger sequencing of the breakpoint shows a retained CD74 intron sequence of 75 bp resulting in a stop codon before the fusion junction. A putative alternative open reading frame (ORF) starting at an ATG within PDGFRB exon 12 is also indicated. c Western blot analysis of PDGFRB in bone marrow MNCs from CD74intr::PDGFRB+ patient. Ba/F3 cells expressing CD74intr::PDGFRB and a second B-ALL patient with no known PDGFRB lesion were used as controls. d Proliferation of BCR::ABL1 and CD74intr::PDGFRB expressing Ba/F3 cells following 48-hour IL-3 withdrawal. Proliferation was measured by luminescence relative to the EV control in IL-3 (day 0), using the CellTiter-Glo® 2.0 reagent. Data shows Mean ± SEM (n = 3). e Western blot analysis of BCR::ABL1 and CD74intr::PDGFRB expressing Ba/F3 cells treated with 0, 1 or 5 μM imatinib for 6 hours. f Viability of BCR::ABL1, CD74intr::PDGFRB and EBF1::PDGFRB expressing Ba/F3 cells treated with a dose titration of imatinib (top panel) or dasatinib (lower panel) for 72 hours, measured using the CellTiter-Glo® 2.0 reagent. Data is normalized to untreated cells. IC50 values for each cell line is indicated. Data shows Mean ± SEM (n = 3).
To functionally characterize the novel CD74intr::PDGFRB fusion, we stably transduced IL-3-dependent mouse pro-B Ba/F3 cells with CD74intr::PDGFRB-MIG or an empty vector (EV) control. BCR::ABL1 was used as a positive control known to confer cytokine-independent growth of Ba/F3 cells. In the absence of IL-3, expression of CD74intr::PDGFRB was sufficient to drive cytokine independent proliferation similar to BCR::ABL1 (Fig. 1d). Consistent with constitutive activation of PDGFRB, transformed CD74intr::PDGFRB+ Ba/F3 cells grown in the absence of IL-3 showed phosphorylation of PDGFRB (Y751) and Stat5 (Y694) that was blocked by treatment with imatinib (Fig. 1e). These findings were recapitulated in primary murine IL-7 dependent pre-B-cells transduced with CD74intr::PDGFRB (Supplementary Fig. 1c). Importantly, CD74intr::PDGFRB-expressing Ba/F3 cells were highly sensitive to both imatinib and dasatinib in vitro (Fig. 1f).
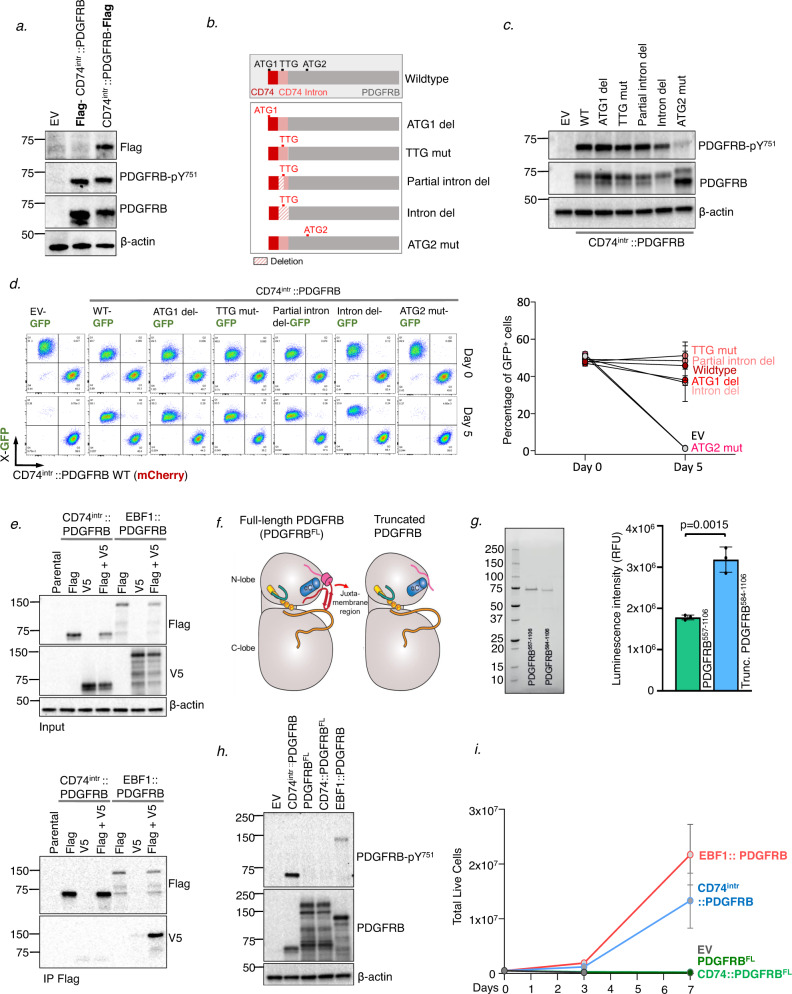
Given the unusual structure of the CD74intr::PDGFRB sequence, and the apparent lack of an ORF starting at the canonical ATG of CD74 exon 1, we investigated whether translation may be driven by an alternate start site. For this, N- and C-terminal FLAG-tagged versions of CD74intr::PDGFRB were expressed in Ba/F3 cells. While equivalent levels of p-PDGFRB (Y751) and total PDGFRB were detected in cells expressing the N- or C-terminal FLAG constructs (Fig. 2a), the FLAG epitope was only detected in cells expressing the C-terminal tag. This strongly suggests that translation of PDGFRB protein is not driven from the first ATG (ATG-1) within CD74. To validate this, and to more stringently dissect whether translation is driven from elsewhere within the CD74-portion, we generated a construct with deletion of the ATG-1 codon from the CD74intr::PDGFRB sequence. We also sequentially deleted or mutated putative translation start sites within the CD74intr::PDGFRB fusion including a non-canonical TTG-codon within the retained CD74-intronic region, and a putative ATG start site (ATG-2) in exon 12 of PDGFRB (Fig. 2b). These constructs were expressed in Ba/F3 cells, and levels of p-PDGFRB (Y751) and PDGFRB were measured by western blotting (Fig. 2c). This revealed that only mutation of ATG-2 disrupted expression of an active protein. Strikingly, growth competition analyses of Ba/F3 lines expressing the CD74intr::PDGFRB mutants (in GFP expression plasmids), competing with WT CD74intr::PDGFRB (in mCherry expression plasmid) confirmed that mutation of ATG-2 abolished the transforming potential of the CD74intr::PDGFRB fusion (Fig. 2d).
Fig. 2. CD74intr::PDGFRB is driven by a non-canonical translation start site.
a N- and C-terminal FLAG tagged CD74intr::PDGFRB constructs were transduced into Ba/F3 cells and analysed by Western blotting. b Schematic diagram of CD74intr::PDGFRB constructs with start codon mutations or deletions, and partial and full intron deletion. c Western blot analysis of CD74intr::PDGFRB mutants depicted in b, expressed in Ba/F3 cells. d Growth competition of Ba/F3 lines transduced with CD74intr::PDGFRB mutants. Ba/F3 lines expressing EV or CD74intr::PDGFRB constructs in GFP-expression plasmid, were mixed 1:1 with Ba/F3 cells expressing CD74intr::PDGFRB in mCherry-expression plasmid. GFP and mCherry populations were measured at day 0, and 5 days after mIL-3 withdrawal. Left panel shows representative FACS data, and right panel shows quantified GFP+ percentages (Mean ± SEM) from 3 independent experiments. e FLAG-co-immunoprecipitation in HEK293T cells expressing FLAG or V5-tagged CD74intr::PDGFRB constructs, alone, or in combination. EBF1::PDGFRB was used as a positive control. Shown is whole cell lysate input (top panel) and western blot analysis of FLAG-immunoprecipitate elutes probed for FLAG or V5 (bottom panel). f AlphaFold2 structural modelling of full length PDGFRB (PDGFRBFL) and CD74intr::PDGFRB highlighting loss of juxtamembrane region in CD74intr::PDGFRB. g Left Panel: Reducing SDS-PAGE analysis of purified PDGFRB visualized by stain-free imaging to illustrate the purity of each PDGFRB preparation. Right Panel: In vitro kinase activities of PDGFRB557–1106 and truncated PDGFRB584–1106 encoded by the CD74intr::PDGFRB fusion. Individual data points are plotted; the bar and error bars shown represent mean ± SD of three independent ADP-Glo assays. Presented data are representative of three technical replicates (n = 9). Statistical significance was calculated using an unpaired Student’s t test. h Western blot analysis of Ba/F3 lines expressing PDGFRB variants. i Proliferation of Ba/F3 cells following withdrawal of IL-3 measured by trypan blue exclusion. Data shows Mean ± SEM (n = 3). h Western blot analysis of Ba/F3 lines expressing PDGFRB variants. i Proliferation of Ba/F3 cells following withdrawal of IL-3 measured by trypan blue exclusion. Data shows mean ± SEM (n = 3).
Together, these results show that protein translation does not commence within CD74, and that the product from the CD74intr::PDGFRB fusion likely arises from an internal ATG within PDGFRB itself, encoding a truncated PDGFRB molecule. In an analogous approach, we assessed the capacity for the CD74-region to promote protein translation by generating an artificial hybrid construct of CD74 with the kinase domain of ABL1. This confirmed that the CD74-sequence from CD74intr::PDGFRB is unlikely to drive translation of a fusion protein (Supplementary Fig. 1d).
Our findings provide evidence for a remarkable scenario, in which aberrant expression of a ‘loose’ PDGFRB kinase domain in B-cell progenitors is sufficient to drive auto-activation and leukemic transformation. This is particularly unique in the context of Ph-like ALL, which is characterized by expression of chimeric fusion proteins in which tyrosine kinase domains are fused in-frame to distinct N-terminal partners (commonly lymphoid transcription factors). Constitutive kinase activation of oncogenic chimeric-fusions is driven by retained oligomerization domains of N-terminal partner proteins, which mediate fusion homodimerization and subsequent auto-phosphorylation of the kinase domains. Notably, oligomerization domains of the partner proteins are indispensable for constitutive activation of other PDGFRB fusions (e.g. ETV6::PDGFRB and EBF1::PDGFRB [7]). Interestingly, co-immunoprecipitation (coIP) of FLAG-tagged CD74intr::PDGFRB in HEK293T cells suggests that unlike EBF1::PDGFRB, CD74intr::PDGFRB does not strongly dimerize (Fig. 2e). However, this does not exclude the possibility of a weak or transient dimerization driven instead by the stoichiometry of the abundance of the fusion. We note, for instance, that recombinant truncated PDGFRB (PDGFRB584–1106) expressed and purified from insect cells elutes at a molecular weight consistent with a dimer (Supplementary Fig. 2a). Nonetheless, these findings suggest an alternative mechanism of activation for CD74intr::PDGFRB which may bypass the requirement for dimerization domains in the driver fusion.
To understand how a truncated PDGFRB kinase domain may be constitutively activated, we performed AlphaFold2 structural modelling comparing the intracellular portion of full-length (FL) human PDGFRB (PDGFRBFL) and CD74intr::PDGFRB (Fig. 2f, Supplementary Fig. 2b). This revealed that the juxtamembrane (JM) domain partially occupies the ATP binding site by running parallel with the αC helix, before forming a β-strand hairpin that abuts the C-lobe of the kinase domain, occluding the active site. This is consistent with the experimental structure of the related protein, PDGFRA, where the active site is analogously occluded by the JM region [8]. In contrast, the truncated form of PDGFRB, encoded by the CD74intr::PDGFRB fusion, lacks the JM region and models predict that the active site is not obstructed in this setting. To test this hypothesis, we generated recombinant proteins to measure the kinase activity of PDGFRB557–1106 relative to the truncated form that arises from CD74 fusion, PDGFRB584–1106, in an ADP-Glo assay with a Poly-(Glu,Tyr) peptide substrate. While the replete intracellular portion of PDGFRB (PDGFRB557–1106) exhibited some catalytic activity, the truncated PDGFRB584–1106 protein displayed approximately double in vitro kinase activity (Fig. 2g).
Hence, we hypothesized, that unlike the ‘truncated’ CD74intr::PDGFRB protein, expression of the full-length PDGFRB (PDGFRBFL) receptor may be ineffective in driving transformation as it is locked in an inactive state. To test this idea, we generated Ba/F3 lines expressing either the CD74intr::PDGFRB fusion or PDGFRBFL. We also included a hybrid between CD74 and PDGFRBFL (CD74::PDGFRBFL), fusing the CD74 region to PDGFRB exon 1. Immunofluorescence staining revealed that both CD74intr::PDGFRB and PDGFRBFL are dispersed through the cell cytoplasm (Supplementary Fig. 2c). Strikingly however, active PDGFRB protein (assessed by Y751 phosphorylation) was only observed in Ba/F3 cells expressing the CD74intr::PDGFRB construct, or EBF1::PDGFRB (Fig. 2h). Moreover, while Ba/F3 cells expressing CD74intr::PDGFRB proliferated in the absence of IL-3, cells expressing PDGFRBFL or the CD74::PDGFRBFL hybrid did not (Fig. 2i, Supplementary Fig. 2d).
It remains to be determined whether there are other examples of cancer driven by a non-chimeric truncated PDGFRB protein or whether this mechanism of oncogenic activation extends to other kinases. Nonetheless, our observations are broadly relevant to oncogenic signaling given the recognized autoinhibitory function of the juxtamembrane domain in other type III receptor tyrosine kinases including PDGFRB/A [8, 9], c-KIT [10] and CSF1R [11]. Interestingly, molecular dissection of the FIP1L1::PDGFRA fusion, which occurs in chronic eosinophilic leukemia, demonstrated that FIP1L1 is entirely dispensable for PDGFRA activation, and that instead it is the partial loss of the JM domain within PDGFRA that drives constitutive activation of the fusion [9]. In the setting of PDGFR fusions which retain the intact JM domains (e.g. ETV6::PDGFRA [9], EBF1::PDGFRB [7]), autoinhibition is instead overcome by enforced dimerization driven by the N-terminal fusion partner, representing two distinct mechanisms by which PDGFR-family kinases may become constitutively activated in malignancy.
We present a unique example of aberrant expression of a non-chimeric ‘loose kinase domain’, which is targetable by ABL-class inhibitors (e.g. imatinib), driving Ph-like B-ALL. Our findings have critical implications for current pipelines of sequencing-based fusion identification, in which putative cancer drivers may be disregarded following out-of-frame predictions. This work also highlights the merit of functional validation of structurally unusual lesions, which may ultimately provide an invaluable opportunity for administration of targeted compounds in treatment regimens.
Supplementary information
Supplementary Materials, Methods and Figures
Acknowledgements
We gratefully acknowledge the Research Genomics Facility of the Victorian Clinical Genetics Service. Tumour samples and coded data were supplied by the Children’s Cancer Centre Biobank at the Murdoch Children’s Research Institute and The Royal Children’s Hospital (mcri.edu.au/research/projects/childrens-cancer-centre-biobank). Establishment and running of the Children’s Cancer Centre is made possible through generous support by Cancer In Kids @ RCH (www.cika.org.au), The Royal Children’s Hospital Foundation and the Murdoch Children’s Research Institute and the Children’s Cancer Centre Tissue Bank (Murdoch Children’s Research Institute). This work is support by National Health and Medical Research Council Grants (1140626 to P.G.E.; 1172929 and 9000719 to JMM) and a SCOR Grant (7015-18) from the Lymphoma and Leukemia Society. We acknowledge the support of Perpetual Trustees and the Samuel Nissen Foundation. S.L.K. is supported by a fellowship from the Victorian Cancer Agency. This work was also supported by the Victorian Government’s Operational Infrastructure Support Program.
Author contributions
TS, FBJ, HJK, CRH, and LMB contributed to the design, conduct and analysis of experimental data presented in this manuscript. CRH and JMM designed performed and analyzed the experiments with recombinant proteins and generated the alpha-fold models. SE-K, MJC, NMD and AO analysed structural features of the fusion gene and contributed to the design of mutational experiments. LEAL, and SLK contribute to the identification of eligible samples, the collation and presentation of clinical data. TS, AO, JMM, CEdeB, APN, NMD, contributed to the concept and design of experiments and the analysis of experimental data. TS and PGE conceived and designed the project and wrote the manuscript. All authors contributed to the revision of manuscript drafts.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary information files. Any other data may be requested from the corresponding author
Competing interests
PGE, JMM, SLK and APN receive an annual payment related to the Walter and Eliza Hall Institute distribution of royalties scheme. PGE consults for Illumina.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Teresa Sadras, Fatimah B. Jalud.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-023-01843-x.
References
- 1.Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, et al. High Frequency and Poor Outcome of Philadelphia Chromosome-Like Acute Lymphoblastic Leukemia in Adults. J Clin Oncol. 2017;35:394–401. doi: 10.1200/JCO.2016.69.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran TH, Tasian SK. Has Ph-like ALL Superseded Ph+ ALL as the Least Favorable Subtype? Best Pr Res Clin Haematol. 2021;34:101331. doi: 10.1016/j.beha.2021.101331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt B, Brown LM, Ryland GL, Lonsdale A, Kosasih HJ, Ludlow LE, et al. ALLSorts: an RNA-Seq subtype classifier for B-cell acute lymphoblastic leukemia. Blood Adv. 2022;6:4093–7. doi: 10.1182/bloodadvances.2021005894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, et al. Genomic Profiling of Adult and Pediatric B-cell Acute Lymphoblastic Leukemia. EBioMedicine. 2016;8:173–83. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welsh SJ, Churchman ML, Togni M, Mullighan CG, Hagman J. Deregulation of kinase signaling and lymphoid development in EBF1-PDGFRB ALL leukemogenesis. Leukemia. 2018;32:38–48. doi: 10.1038/leu.2017.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang L, Yan XE, Yin Y, Yun CH. Structural and biochemical studies of the PDGFRA kinase domain. Biochem Biophys Res Commun. 2016;477:667–72. doi: 10.1016/j.bbrc.2016.06.117. [DOI] [PubMed] [Google Scholar]
- 9.Stover EH, Chen J, Folens C, Lee BH, Mentens N, Marynen P, et al. Activation of FIP1L1-PDGFRalpha requires disruption of the juxtamembrane domain of PDGFRalpha and is FIP1L1-independent. Proc Natl Acad Sci USA. 2006;103:8078–83. doi: 10.1073/pnas.0601192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan PM, Ilangumaran S, La Rose J, Chakrabartty A, Rottapel R. Autoinhibition of the kit receptor tyrosine kinase by the cytosolic juxtamembrane region. Mol Cell Biol. 2003;23:3067–78. doi: 10.1128/MCB.23.9.3067-3078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter M, Lucet IS, Patel O, Broughton SE, Bamert R, Williams NK, et al. The 2.7 A crystal structure of the autoinhibited human c-Fms kinase domain. J Mol Biol. 2007;367:839–47. doi: 10.1016/j.jmb.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Uhrig S, Ellermann J, Walther T, Burkhardt P, Frohlich M, Hutter B, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31:448–60. doi: 10.1101/gr.257246.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials, Methods and Figures
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary information files. Any other data may be requested from the corresponding author