Abstract
Human WASP and N-WASP are homologous proteins that require the binding of multiple regulators, including the acidic lipid PIP2 and the small GTPase Cdc42, to relieve autoinhibition before they can stimulate the initiation of actin polymerization. Autoinhibition involves intramolecular binding of the C-terminal acidic and central motifs to an upstream basic region and GTPase binding domain. Little is known about how a single intrinsically disordered protein, WASP or N-WASP, binds multiple regulators to achieve full activation. Here we used molecular dynamics simulations to characterize the binding of WASP and N-WASP with PIP2 and Cdc42. In the absence of Cdc42, both WASP and N-WASP strongly associate with PIP2-containing membranes, through their basic region and also possibly through a tail portion of the N-terminal WH1 domain. The basic region also participates in Cdc42 binding, especially for WASP; consequently Cdc42 binding significantly compromises the ability of the basic region in WASP, but not N-WASP, to bind PIP2. PIP2 binding to the WASP basic region is restored only when Cdc42 is prenylated at the C-terminus and tethered to the membrane. This distinction in the activation of WASP and N-WASP may contribute to their different functional roles.
Keywords: intrinsically disordered proteins, membrane binding, autoinhibition, PIP2, WASP, prenylation
Graphical Abstract
Introduction
Human Wiskott-Aldrich syndrome protein (WASP) and neural WASP (N-WASP) are homologous proteins that contain intrinsically disordered regions throughout their sequences [1]. Typically of intrinsically disordered proteins (IDPs), WASP and N-WASP engage in numerous intramolecular and intermolecular interactions. Intramolecular interactions result in autoinhibition [2–5] whereas intermolecular interactions result in activation and subsequent engagement in cellular activities. A major activity of WASP and N-WASP is to stimulate the initiation of branched actin polymerization [6]. WASP is expressed only in hematopoietic cells [7], whereas N-WASP, despite its prefix, is ubiquitously expressed [8]. Mutations in WASP are responsible for its namesake, an X-linked immunological disorder [7]. The fact that N-WASP in the hematopoietic cells of patients does not compensate for the defects caused by WASP mutations points to nonredundant roles of these two proteins. Indeed, different effects of WASP and N-WASP have been reported in filopodium formation [9], podosome-mediated extracellular matrix degradation [10], and T cell chemotaxis [11]. Phylogenetic analysis suggested a direct link between N-WASP and ancestral WASPs, whereas hematopoietic WASP branched off during the onset of vertebrates [12]. WASP and N-WASP are excellent models for studying how a single IDP binds multiple regulators to achieve full activation as well as for deciphering the differences between closely related IDPs in processes ranging from activation to cellular activities.
WASP and N-WASP share ~50% sequence homology [8] and have the same domain organization with one exception [1, 13] (Figure 1A). The functional element resides toward the C-terminus, where the verprolin-central-acidic (VCA) region binds G-actin (via the V motif) and Arp2/3 (via the CA motifs) to initiate actin polymerization [2, 3, 14, 15]. The exception to the shared domain organization is an additional V motif in N-WASP, which increases the activity for stimulating actin polymerization [16]. Upstream of the functional element are regulatory elements. At the N-terminus is a WASP homology 1 (WH1; also known as EVH1) domain that binds WASP-interacting proteins [17]. Following a linker of ~65 (or ~35 in N-WASP) residues, a basic region (BR) is present to bind the acidic lipid phosphatidylinositol 4,5-bisphosphate (PIP2) [3, 5, 18–20]. Higgs and Pollard [18] suggested that the WH1 domain of WASP may also participate in PIP2 binding. Immediate after the BR is a GTPase-binding domain (GBD), which binds Cdc42 and other small GTPases [9, 21]. In WASP, several Lys residues in the basic region, including K230KK232, are involved in electrostatic interactions with Cdc42 [22] and contribute to their binding rate [23–25]. Lastly, a proline-rich region (PRR) connects the GBD and the VCA region, and binds SH3-containing proteins such as Nck [26].
Figure 1.
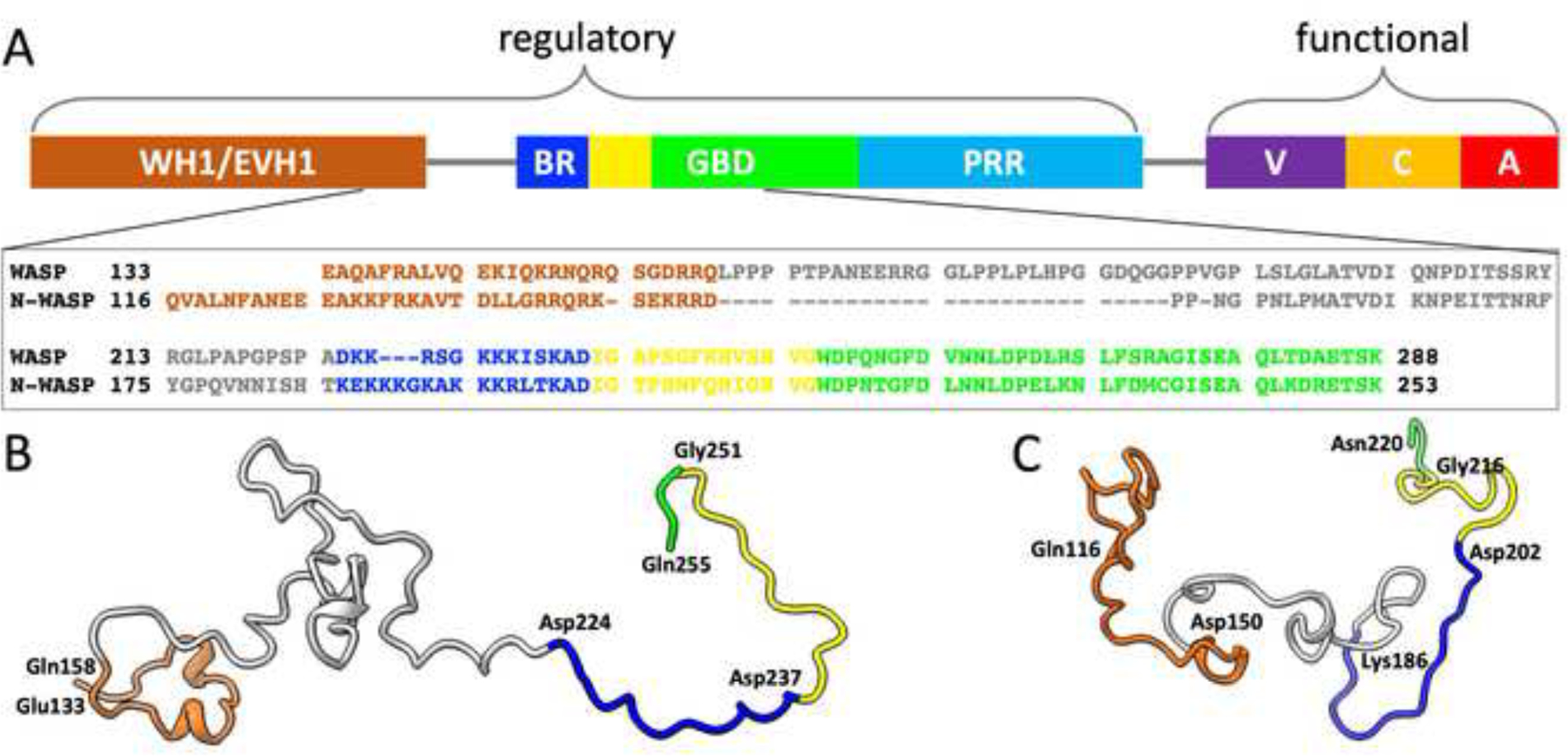
Sequences and conformations of WASP and N-WASP. (A) Domain organization of WASP and sequence alignment of WASP and N-WASP for residues implicated in PIP2 and Cdc42 binding. The domains and disordered regions are indicated by different colors. The initial portion of the GBD is a CRIB (Cdc42/Rac interactive binding) motif, colored yellow; the rest of the GBD is colored green. The domain organization is shared by N-WASP, with two exceptions: the linker between WH1 and BR is shorter by ~30 residues, and an extra V motif is inserted between PRR and VCA. The sequences are color-coordinated with the domain organization. Representative conformations of (B) WASP and (C) N-WASP while bound to PIP2-containing membranes. Colors of different regions are as in (A); the bordering residues between regions are labeled.
Under resting conditions, WASP and N-WASP are autoinhibited by intramolecular interactions, between the GBD and the C motif [3–5, 9] and between the BR and the A motif [1, 25, 27]. Binding to PIP2 and Cdc42 relieve N-WASP of autoinhibition, and the two regulators have synergistic effects on activation [3]. Endogenous Cdc42 is prenylated at the C-terminus and then tethered to membranes [28], but a follow-up study reported no difference between prenylated and soluble Cdc42 in their synergistic effects with PIP2 on N-WASP activation [19]. Prehoda et al. [5] also observed synergistic activation of NWASP by PIP2 and soluble Cdc42, though a follow-up study found higher activity when Cdc42 was prenylated and colocalized with PIP2 [20]. In contrast, synergistic activation of WASP by PIP2 and Cdc42 required Cdc42 prenylation [18]. Indeed, instead of doubling the effect of PIP2 when prenylated Cdc42 was added, a 30% decrease was observed when soluble Cdc42 was added. Both the synergistic effect of soluble Cdc42 on N-WASP activation and the negative effect of soluble Cdc42 on WASP activation were reproduced by Tomasevic et al. [29], but the reason for this contrast has remained a mystery.
The structures of the WASP GBD bound to the C motif (modeling the autoinhibited state) and to Cdc42 (modeling the activated state) have been determined by NMR spectroscopy [4, 22]. In the autoinhibited state, the C motif folds into an α-helix and docks to a three-helix platform formed by the GBD (Figure S1A). In the activated state, the platform falls apart as one of these helices, along with upstream segments, binds to Cdc42 (Figure S1B). Very little structural information is available for the binding of WASP or N-WASP with the other activator, i.e., PIP2. Binding studies [20] have shown that the N-WASP BR binds multiple PIP2 molecules and it is the number of basic residues, not the precise sequence, that determines the PIP2 binding affinity, typical of IDP binding to acidic membranes [30]. Structural characterization of IDP-membrane binding, due to its dynamic nature, presents challenges for experimental techniques, but this problem can now be addressed by molecular dynamics (MD) simulations [31–34].
Here we report MD simulation results on the binding of WASP and N-WASP with PIP2 and soluble and prenylated Cdc42. The simulations provided an opportunity to visualize how WASP and N-WASP simultaneously bind with two regulators, PIP2 and Cdc42, thereby fully releasing the VCA region for stimulating actin polymerization. For N-WASP, simultaneous binding can occur whether Cdc42 is soluble or prenylated. However, WASP can do so only when Cdc42 is prenylated. This result explains the previous puzzling observation that synergistic activation of WASP, but not N-WASP requires Cdc42 prenylation. The origin of this contrast lies in the differences between the BRs of WASP and N-WASP, in particular regarding the number of basic residues and their spacing from the GBD.
Results
We simulated WASP and N-WASP fragments binding to PIP2-containing membranes in the absence of Cdc42 and in the presence of soluble and prenylated Cdc42. The upper leaflet (facing the proteins) contained 10% PIP2 and 40% POPS; the remaining 50% was neutral lipids. In vitro studies often used PIP2 levels up to 10% and beyond [18–20]. In cellular membranes, PIP2 is locally enriched for signaling processes including those involving WASP and N-WASP [35]. The simulations for either WASP or N-WASP without Cdc42 were run in 16 replicates, each lasting 1360 ns. The simulations with either soluble or prenylated Cdc42 were run in seven replicates for WASP, each lasting 500–1000 ns, and in eight replicates for N-WASP, each lasting 500–700 ns. We quantify membrane binding by calculating the membrane contact probabilities of individual residues, defined as the fraction of MD frames in which a residue forms at least one contact with lipids. A contact is considered formed when two heavy atoms come within 3.5 Å. Structural and dynamic properties of the membranes demonstrate good lipid equilibration in the simulations (Figure S2). No systematic membrane curvature formation was detected due to compositional asymmetry between the leaflets.
The N-WASP BR shows stronger membrane association than the WASP counterpart.
For the simulations without Cdc42, the WASP and N-WASP fragments contained residues Glu133 to Gln255 and Gln116 to Asn220, respectively. They start from the tail portion of the WH1 domain and end just beyond the CRIB motif of the GBD. A snapshot of each fragment from the simulations is shown in Figure 1B, C. The WASP and N-WASP BRs consist of 14 and 17 residues (Asp224 to Asp237 and Lys186 to Asp202; Figure 1A), with 7 and 10 of them, respectively, being basic. As shown in Figure 2A, B, the BRs have high membrane contact probabilities, with basic residues making dominant contributions. All the basic residues have >10% membrane contact probabilities; the only other residue crossing this threshold is Ser228 in the WASP BR.
Figure 2.
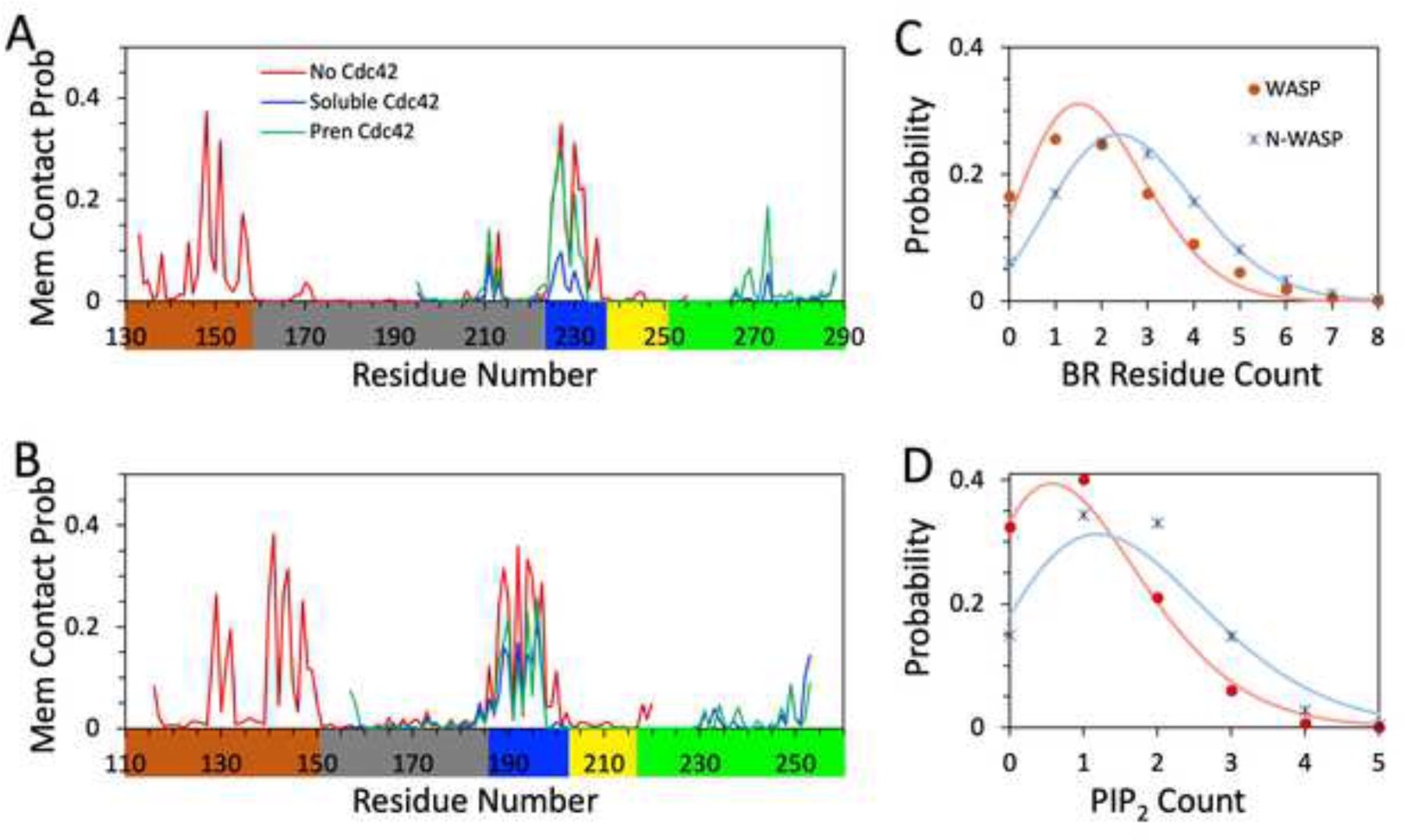
Membrane binding properties of WASP and N-WASP. Membrane contact probabilities of (A) WASP and (B) N-WASP residues. Traces in red, blue, and green display data from simulations without Cdc42, with soluble Cdc42, and with prenylated Cdc42, respectively. (C) Probabilities for having n BR residues simultaneously bound to membranes in simulations without Cdc42; n is BR residue count. (D) Probabilities for having m PIP2 lipids simultaneously bound to the BR in simulations without Cdc42; m is PIP2 count. In (C, D), data from the simulations of WASP and N-WASP fragments are shown as red and blue symbols, respectively; fits to a binomial distribution in (C) and Poisson distribution in (D) are displayed as curves in colors matching the corresponding symbols.
Because of its larger number of basic residues, the N-WASP BR shows stronger membrane association than the WASP counterpart. This difference can be measured in two ways. First, the fraction of MD frames where the BR is membrane-associated is significantly higher for N-WASP; it is (92 ± 4)%, compared to (83 ± 7)% for WASP (P value < 0.0001 in t-test; mean and standard deviation calculated using 16 replicate simulations for each protein). Second, in a given frame, more residues in the N-WASP BR are simultaneously membrane-bound than in the WASP BR. Figure 2C displays the probabilities of having exactly n BR residues bound to membranes at a given time, with n = 0, 1, 2, … Relative to WASP, the probability distribution of the N-WASP shifts toward higher n. Both of the probability distributions fit moderately well to a binomial distribution,
which assumes that the N BR residues (14 and 17 for WASP and N-WASP, respectively) each independently bind to the membrane with probability p. The predicted mean n, , represents the average number of simultaneously bound BR residues. The fits yield p = 0.135 for WASP and 0.159 for N-WASP. The resulting mean n is 1.9 for WASP and 2.7 for N-WASP. The higher value for N-WASP is due to both a longer BR and a higher membrane binding probability for each BR residue.
Most of the lipids interacting with the basic residues are PIP2; POPS lipids also participate in the interactions to some extent. Figure 2D displays the probabilities of having exactly m PIP2 lipids simultaneously bound to the BR. The probability distribution in m also shifts toward higher m values for N-WASP – just as more N-WASP BR residues participate in membrane interactions, we see more PIP2 lipids simultaneously bind to this BR. The probability distributions in m are close to a Poisson distribution,
which is the limit of the binomial distribution when N → ∞ while Np remaining at a constant value . The closeness to a Poisson distribution means that PIP2 molecules also largely work independently when binding to the BRs. This outcome is perhaps befitting to the relatively low level of PIP2 (10%) in the membranes. The mean is 1.6 for NWASP, compared to 1.0 for WASP.
Figure 3A displays a representative snapshot of the WASP fragment bound to the membrane. Several basic sidechains in the BR project downward and reach for lipid headgroups. The two BR sidechains with the highest membrane contact probabilities are Arg227 and Lys230 (Figure 2A). In the representative snapshot (Figure 3A, zoomed view), Arg227 forms a salt bridge with a PS headgroup while Lys230 hydrogen bonds with a PIP2. Two other sidechains, Lys232 and Lys235, are just outside the cutoff distance of 3.5 Å from lipids, and ready to form alternative or additional contacts with lipids.
Figure 3.
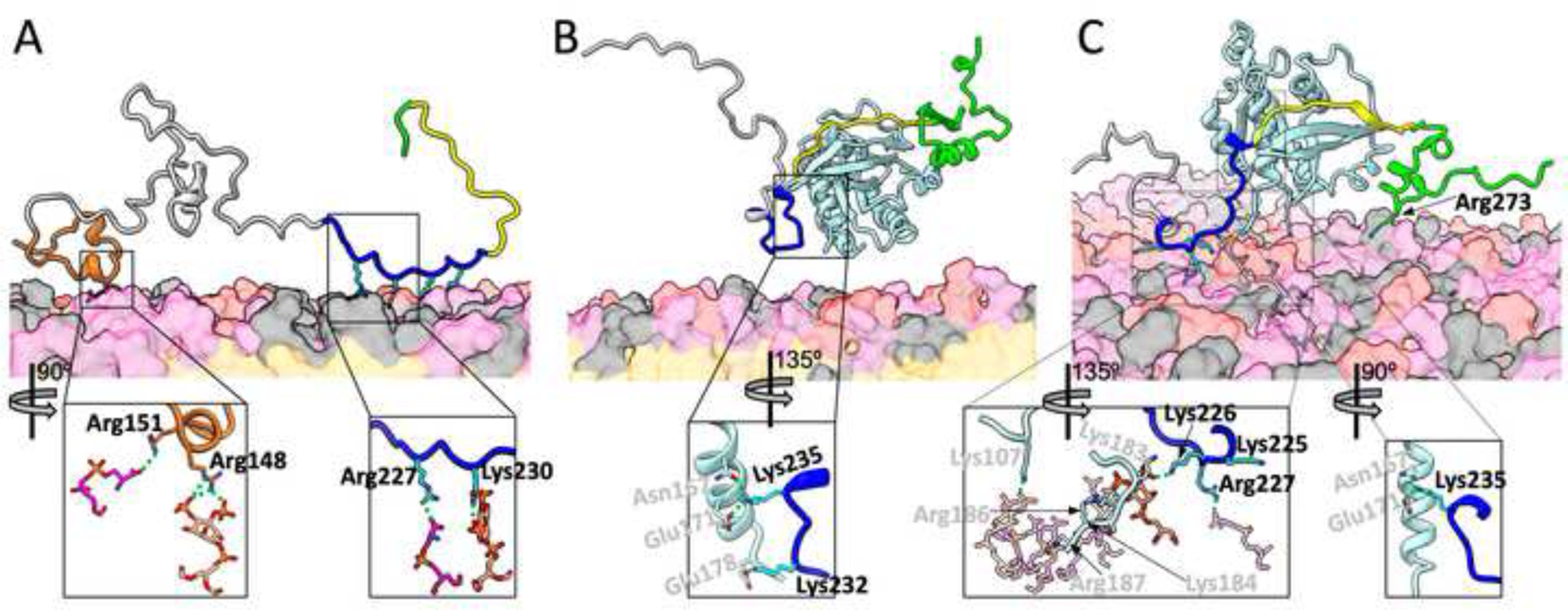
Representative snapshots from MD simulations of membrane binding of WASP. (A) Without Cdc42. (B) With soluble Cdc42. (C) With prenylated Cdc42. WASP residues are colored according to the scheme in Figure 1; Cdc42 is in light cyan. Lipids are shown as surface, with headgroups of PIP2, POPS, and neutral lipids (POPC, POPE, and cholesterol) in pink, magenta, and grey respectively; lipid tails are in orange. WASP sidechains that form or are about to form membrane contacts are shown as sticks. In the zoomed images, the lipid partners are also shown as sticks, as are BR-Cdc42 interactions. Residue labels are in black for WASP and gray for Cdc42, respectively. Carbon atoms have the same colors as the backbone representations, except that those in the BR are in cyan; O, N, and P atoms are in red, blue, and orange, respectively. In (C), the zoomed left image shows all but one lipid with dimmed intensity; the exception is a PIP2 that interacts with both WASP Lys226 and Cdc42 Lys183.
A representative snapshot of the N-WASP fragment is shown in Figure 4A. In this BR, the three sidechains with the highest membrane contact probabilities are Lys189, Lys192, and Lys194 (Figure 2B). In the representative snapshot (Figure 4A, zoomed view), Lys189 forms a salt bridge with a PIP2, Lys192 hydrogen bonds with a PIP2, and Lys194 forms a salt bridge with a PS. In this case, Lys195 is waiting to get closer to lipids.
Figure 4.
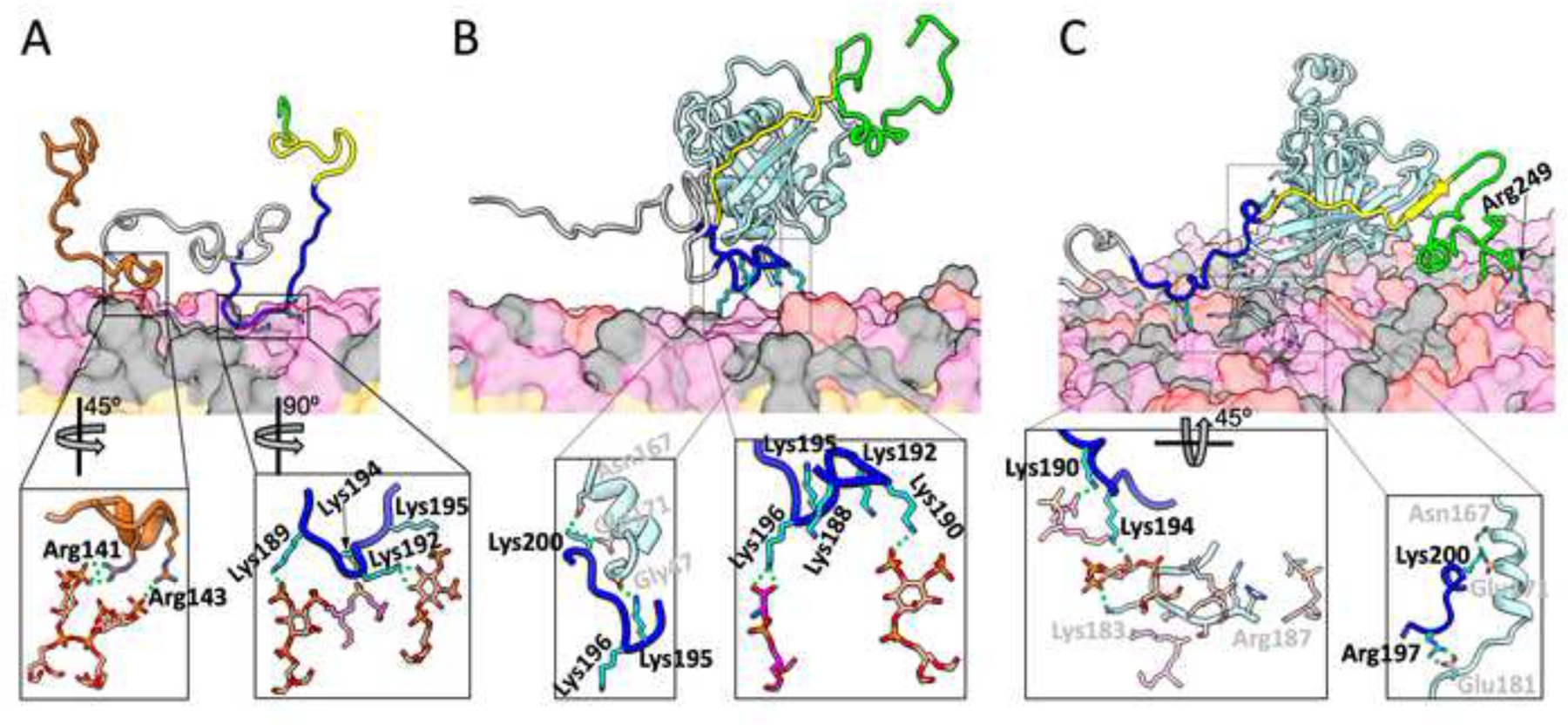
Representative snapshots from MD simulations of membrane binding of N-WASP. (A) Without Cdc42. (B) With soluble Cdc42. (C) With prenylated Cdc42. The representations and colors are the same as described in Figure 3, except that in (C), two salt bridges and a hydrogen bond between N-WASP and Cdc42 are shown in both the normal and zoomed views.
A tail portion of WH1 may assist with membrane association.
We included a portion of the WH1 domain in the WASP and N-WASP fragments for membrane association simulations in the absence of Cdc42, partly because Higgs and Pollard [18] suggested that the WASP WH1 may also participate in PIP2 binding. There was also an additional reason. We developed a sequence-based method called ReSMAP for predicting residue-specific membrane association propensities of IDPs, trained on MD simulation data including those from an initial portion of the WASP and N-WASP simulations [30]. We used an initial version of ReSMAP to select the WASP and N-WASP fragments for membrane association simulations. It already predicted significant membrane association propensities for the tail portions of the WASP and N-WASP WH1, due to a large number of basic residues there (7 and 11, respectively; Figure 1A). Hence the tail portions were included in the WASP and N-WASP fragments for membrane association simulations.
As shown in Figure 2A, B, the WH1 tail portions indeed show high membrane contact probabilities in the MD simulations. All but one basic residue (WASP Arg138) in the WH1 tails have >10% membrane contact probabilities, and all other residues in this region are below this threshold. Because N-WASP also has a larger number of basic residues in the WH1 tail than WASP, the same contrast between the two proteins described above for the BRs is repeated here. The probability that the WH1 tail is membrane-associated is 89% for N-WASP, higher than the 73% for WASP. Likewise, the average number of simultaneously bound WH1 residues is 2.9 for N-WASP, higher than a 2.0 for WASP, and the average number of PIP2 molecules bound to the WH1 is 1.4 for N-WASP, also higher than a 1.0 for WASP. These numbers are similar to the counterparts in BR, indicating nearly equal participation of these two regions in membrane association.
The representative snapshots in Figures 3A and 4A show that WASP and N-WASP are tethered to the membrane through both the WH1 tail and BR. The WH1 tails in both proteins have some tendency to form α-helices. In the zoomed images, WASP Arg148 and Arg151 form salt bridges with a PIP2 and a PS, respectively, whereas N-WASP Arg141 and Arg143 each form a salt bridge with a PIP2.
Based on the above observation that BR residues interact with the membrane largely independently of each other (Figure 2C), we expect independence between WH1 tail and BR in their association with the membrane. To test this idea, we counted the fractions of MD frames for four states: both regions are membrane-bound; only one of the two regions is bound; and neither region is bound. If the regions have membrane association probabilities of PW and PB on their own, then the independence model predicts the probabilities of the four states as PWPB, (1 – PW)PB, PW(1 – PB), and (1 – PW)(1 – PB), respectively. These predictions agree very well with the statistics from the MD simulations (Figure S3), confirming the independence of WH1 tail and BR in membrane association. The best fits yield PW = 73% and PB = 83% for WASP and PW = 89% and PB = 92% for N-WASP. These values once again confirm the stronger membrane association of N-WASP relative to WASP, as well as nearly equal contributions of WH1 and BR to membrane association in each protein.
N-WASP, but not WASP, can simultaneously bind PIP2 and soluble Cdc42.
We next simulated the membrane association of WASP and N-WASP when they are prebound to soluble Cdc42. Given the independence shown above between WH1 and BR in membrane association, in this set of simulations we left out the WH1 tails. The WASP and N-WASP fragments contained residues Leu195 to Lys288 and Leu157 to Lys253, respectively. They include a portion of the WH1-BR linker and extend to the end of the GBD residues found in the NMR structure of WASP bound with Cdc42 (Protein Data Bank entry 1CEE) [22]. For these simulations, the initial structure of WASP residues Lys230 to Lys288 in complex with Cdc42 (residues 1–179) was taken from 1CEE; the counterpart for N-WASP was built by homology modeling.
A striking difference between the WASP and N-WASP fragments prebound with soluble Cdc42 is that the former does not stably associate with the PIP2-containing membrane whereas the latter does. Cdc42-bound WASP only transiently comes into contact with the membrane, interrupted by long excursions away from the membrane surface. Not a single BR residue crosses the 10% threshold in membrane contact probability (Figure 2A). In contrast, Cdc42-bound N-WASP is always bound or close to the membrane; the membrane contact probabilities of seven BR residues are >10%: Lys189, Lys190, Lys192, Lys194, Lys195, Lys196, and Arg197 (Figure 2B). This contrast can also be seen by comparing the probability distributions in n, the number of BR residues bound to the membrane at a given time (Figure S4A, B). The probability distribution for Cdc42-bound WASP suffers a sharp shift toward n = 0, with the total probability for n ≥ 1 reduced to 21%, from 83% in the absence of Cdc42. Cdc42-bound N-WASP also experiences some loss in the total probability for n ≥ 1, but the loss is much more modest, down to 65% from 94%. The probability distributions in m, the number of BR-bound PIP2 molecules, show a similar difference between Cdc42-bound WASP and Cdc42-bound N-WASP (Figure S4C, D).
The reason that Cdc42 prebinding compromises WASP-membrane binding is that the BR is also involved in Cdc42 binding [22], and thus PIP2 must compete against Cdc42 for BR binding. In a representative snapshot of the Cdc42-bound WASP simulations (Figure 3B), the BR is away from the membrane. Instead, BR residue Lys232 forms a salt bridge with Cdc42 Glu178 and BR residue Lys235 interacts with two Cdc42 residues, Glu171 via a salt bridge and Asn167 via a hydrogen bond (Figure 3B, zoomed view). There are just too few BR basic residues that are free from Cdc42 interference and thus competent for PIP2 binding.
For N-WASP, despite the engagement of some BR basic residues with Cdc42, enough BR residues remain free to make membrane binding stable. A representative snapshot of the Cdc42-bound N-WASP simulations (Figure 4B) illustrates this scenario. BR residue Lys200, which aligns with WASP Lys235 (Figure 1A), forms the same interactions with Cdc42 Glu171 and Asn167 as found for WASP (Figure 4B zoomed view, left). In addition, BR residue Lys195 hydrogen bonds with the carbonyl of Gly47. However, the very next residue, Lys196 forms a salt bridge with a PS (Figure 4B zoomed view, right). In the upstream, Lys190 forms a salt bridge with a PIP2. In addition, Lys188 and Lys192 stand ready to contact lipids.
PIP2 binding of WASP is restored upon Cdc42 prenylation.
Endogenous Cdc42 is prenylated at the C-terminus and inserted into membranes. To model this situation, in our last set of simulations we appended nine residues, P180EPKKSRRC188, to the C-terminus of soluble Cdc42, added a geranylgeranyl group to the new terminal Cys residue, and inserted the geranylgeranyl group into the hydrophobic layer of the membrane.
Upon Cdc42 prenylation, membrane contact probabilities of WASP recover nearly to the levels without Cdc42 (Figure 2A). The number of BR residues with >10% membrane contact probabilities is now five; three residues, Lys231, Lys232, and Lys235 (see below), are missing from the corresponding list for WASP membrane binding in the absence of Cdc42. The restoration of WASP’s membrane association propensity is corroborated by the probability distributions in the number of membrane-bound BR residues and the number of BR-bound PIP2 molecules (Figure S4A, C), showing near matching without Cdc42 and when WASP is prebound to prenylated Cdc42.
The four basic residues, especially Arg186 and Arg187, in the C-terminal extension are known to contribute to the membrane association of prenylated Cdc42 [36, 37]. In our MD simulations, these residues form extensive salt bridges and hydrogen bonds with PIP2 and PS headgroups (Figures 3C and 4C). These interactions, along with the insertion of the geranylgeranyl group into the membrane hydrophobic region, firmly tether Cdc42 to the membrane (Figure S5). Occasionally, some other residues of Cdc42 also come into contact with the membrane, including Lys107, which in the snapshot in Figure 3C forms a salt bridge with a PIP2 molecule.
In the presence of prenylated Cdc42, the WASP BR residue Lys235 still forms a salt bridge with Cdc42 Glu171 and a hydrogen bond with Cdc42 Asn167 (Figure 3C zoomed view, right) while Lys232 also frequently forms a salt bridge with Cdc42 Glu178. However, moving just two residues upstream, Lys230 is already freed from Cdc42 and has a very high membrane contact probability (Figure 2A). The membrane-tethered Cdc42 apparently enhances the membrane contact probabilities of the upstream BR residues, making up for the loss at Lys231, Lys232, and Lys235. One way to achieve this enhancement is presented when basic residues in the Cdc42 C-terminal extension and in the WASP BR bind to the same PIP2 molecule. This scenario is seen in the representative snapshot in Figure 3C; in the zoomed view, left, Cdc42 Lys183 and WASP Lys226 bind to the same PIP2 molecule. In essence, the Cdc42 C-terminal extension recruits PIP2 molecules and raises their local concentration, therefore making it more likely for nearby BR residues to bind to these PIP2 molecules. In the same representative snapshot, WASP Arg227 forms a salt bridge with a PS and Lys225 stands ready for alternative or additional membrane contacts. A downstream residue, Arg273, also frequently binds the membrane (Figures 2A and 3C).
In contrast to the dramatic stabilization of the membrane association of the WASP BR, Cdc42 prenylation has little effect on the already strong membrane association of the N-WASP BR found when Cdc42 was in the soluble form (Figure 2B). The probability distributions in the number of membrane-bound BR residues and the number of BR-bound PIP2 molecules also look similar whether Cdc42 is in the soluble or prenylated form (Figure S4B, D).
Representative snapshots of N-WASP prebound with soluble and prenylated Cdc42 in Figure 4B, C illustrate their similarity in membrane association. With either soluble or prenylated Cdc42, a conserved Lys, Lys200, forms the same interactions with Cdc42 Glu171 and Asn167 (Figure 4B zoomed view, left; Figure 4C zoomed view, right) as found for WASP. In N-WASP prebound with prenylated Cdc42, the next upstream basic residue, Arg197, forms a salt bridge with Cdc42 Glu181 (Figure 4C zoomed view, right), instead of binding with the membrane, reminiscent of the hydrogen bonding of Lys195 with the Cdc42 Gly47 carbonyl when Cdc42 was in the soluble form. As found for WASP, the N-WASP BR can cooperate with the C-terminal extension of prenylated Cdc42 in membrane binding, as illustrated by the binding of Cdc42 Lys183 and N-WASP Lys194 to the same PIP2 molecule (Figure 4C zoomed view, left). However, this cooperation is not essential for the N-WASP BR, as it has more upstream basic residues for membrane binding. For example, in the same snapshot, Lys190 forms a salt bridge with a PS. A small difference between membrane association in the presence of soluble and prenylated Cdc42 is that, in the latter case, two downstream residues, Lys247 and Arg249 (Figures 2B and 4C) can sometimes contact the membrane.
In short, Cdc42 prenylation has a dramatic effect on WASP-membrane association, making it as strong, or perhaps even stronger, than that in the absence of Cdc42. However, N-WASP behaves similarly in membrane association when it is prebound to either soluble or prenylated Cdc42.
Discussion
Using MD simulations, we have visualized how WASP and N-WASP bind to both Cdc42 and PIP2 to achieve full activation. The simulations recapitulated the puzzling experimental observation of the contrasting effects of Cdc42 prenylation: it is absolutely required for WASP activation but makes no difference in N-WASP activation. In the presence of soluble Cdc42, the WASP BR loses the ability to bind PIP2; Cdc42 prenylation recovers this ability. In contrast, the N-WASP BR binds PIP2 equally well whether Cdc42 is soluble or prenylated.
The reason for the contrasting effects of Cdc42 prenylation can be attributed to the number of basic residues in BR and how close they are positioned relative to the GBD. GBD-proximal BR residues participate in Cdc42 binding; therefore there is potential for competition between Cdc42 binding and PIP2 binding. In the WASP BR, the number of basic residues is lower (7 compared to 10 in the N-WASP BR). In the presence of soluble Cdc42, there are just too few BR basic residues that are free from the interference of Cdc42 binding to sustain stable membrane binding. However, upon Cdc42 prenylation, basic residues in the Cdc42 C-terminal extension and the WASP BR cooperate in binding PIP2. Specifically, the basic residues in membrane-tethered C-terminal extension recruit PIP2 molecules and prime them for BR binding. In comparison, in the N-WASP BR, the basic residues are not only higher in number but also more spread out from the GBD (Figure 1A). Therefore, there are enough GBD-distal BR basic residues to sustain membrane binding even when N-WASP is prebound to Cdc42 (either soluble or prenylated); some of these residues could cooperate with the C-terminal extension of Cdc42 in PIP2 binding when Cdc42 is prenylated.
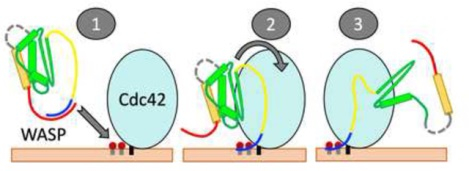
Given the foregoing understanding, we can speculate distinct mechanisms for the two homologous proteins when prenylated Cdc42 is presented in cellular membranes (Figure 5). Autoinhibited WASP would target its BR toward PIP2 molecules enriched around the C-terminal extension of Cdc42 (Figure 5A). Binding of the BR to PIP2 molecules would release the C-terminal A motif while binding of the GBD to Cdc42 would release the C-terminal C motif. Because this process requires coordination between PIP2 and Cdc42 in a single step, the free energy barrier would be relatively high (Figure 5B). In contrast, the BR of N-WASP would bind to PIP2 molecules anywhere in the cellular membrane, resulting in an intermediate where the A motif is released while the C motif is still latched onto the GBD (Figure 5C). Subsequent binding with Cdc42 would free the C motif. In this sequential binding process, the separate free energy barriers are relatively low (Figure 5D). The release of the A motif from the BR of N-WASP has been investigated recently by MD simulations, suggesting that the A motif is capable of engaging with Arp2/3 even before being fully released from the BR [38].
Figure 5.
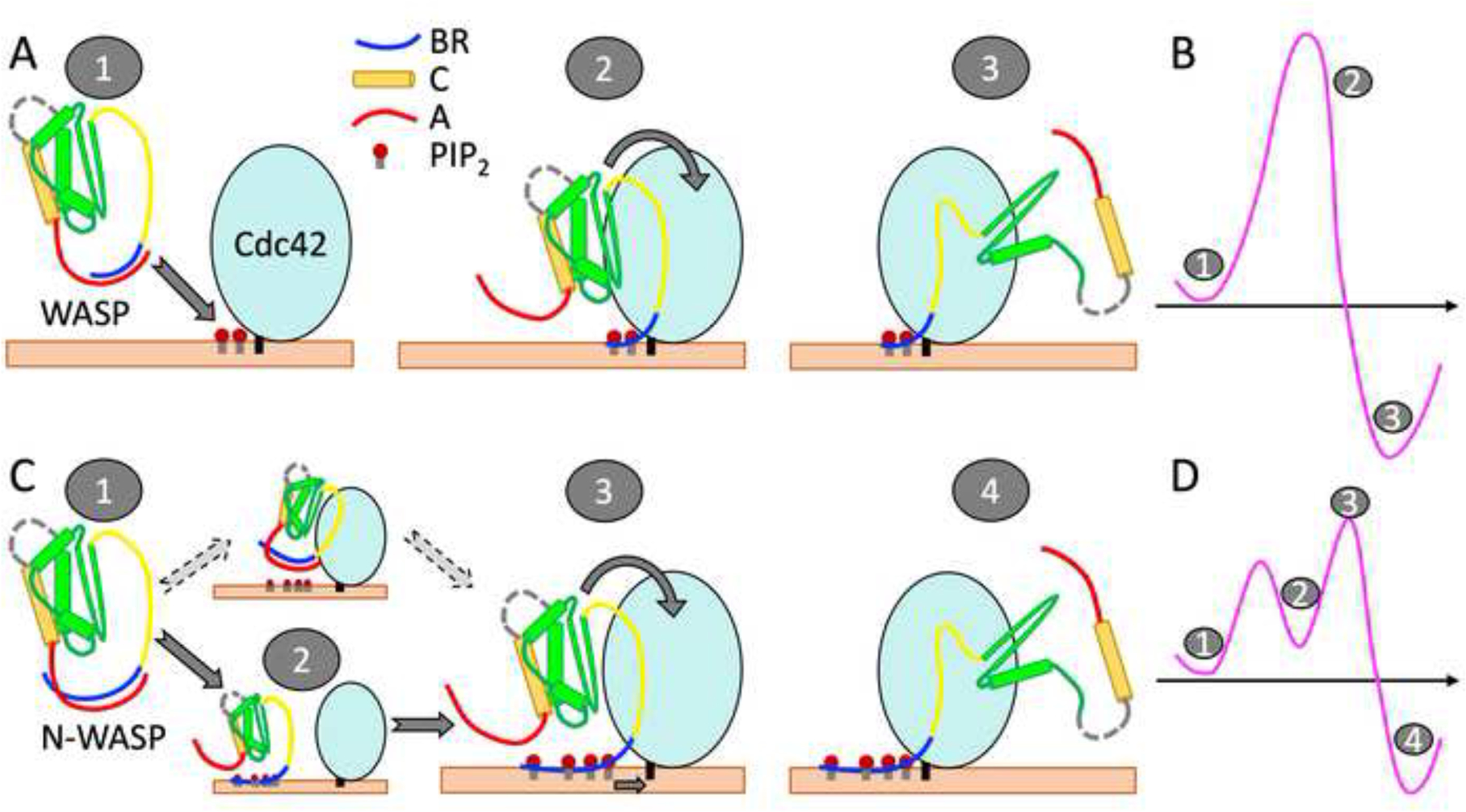
Distinct mechanisms of WASP and N-WASP activation. (A) Activation of autoinhibited WASP starts with targeting its A-bound BR to PIP2 molecules around the C-terminal extension of prenylated Cdc42 (state “1”). In a single concerted step, PIP2 molecules peel the BR away from the A motif (state “2”) and Cdc42 wrestles the GBD away from the C motif, leading to the activated state (“3”). (B) Free energy diagram of the WASP activation pathway. Note that state 2, in which PIP2 has replaced the A motif as the interaction partner of the BR, is just beyond the free energy barrier. (C) Activation of autoinhibited N-WASP (state “1”) starts with PIP2 displacing the A motif as the interaction partner of the BR, producing an intermediate state (“2”). (An alternative pathway, where N-WASP first targets Cdc42, should have a much higher free energy barrier and thus is much less likely.) Subsequently, PIP2-bound N-WASP engages with prenylated Cdc42 (state “3”). The complete binding of Cdc42 with the GBD and the proximal portion of the BR completes the activation (state “4”). Note that N-WASP has a longer BR than WASP and a correspondingly longer A motif. (D) Free energy diagram of the N-WASP activation pathway.
The simulation results for the simultaneous interactions of WASP and N-WASP with PIP2 and Cdc42 may prove insightful for the activation of other IDPs that require multiple regulators. One advantage of having multiple regulators is that they produce a sharp, or switch-like response. The sharpness increases with the degree of cooperativity between the regulators. Thus we expect that, other things being equal, the activation of WASP would be more switch-like than that of N-WASP. However, the sharper response comes at a cost: overcoming the corresponding higher free-energy barrier would require higher levels of Cdc42 or PIP2. This cost could be a contributing factor to why the expression of WASP is restricted to hematopoietic cells whereas the expression of N-WASP is ubiquitous.
Comparing the sequences of WASP and N-WASP, the former has a ~30-residue insertion in the linker between WH1 and BR whereas the latter has an additional V toward the C-terminus. Both of these insertions have direct functional consequences. The additional V motif in N-WASP increases the activity for stimulating actin polymerization [16], whereas the 30-residue insertion in WASP is responsible for the chemotaxis of T-cells toward a chemokine [11]. Here we show that even subtle sequence differences in a shared region, the BR, cause drastic disparity in the activation of the two homologous proteins, specifically in the effects of soluble Cdc42 and the effects of Cdc42 prenylation.
As noted above, one aspect of the differences in the BR sequences concerns the spacing of the basic residues from the GBD. Veltman and Insall [12] observed that the shift of basic residues toward the GBD is an evolutionary trend. This trend is evident when the 44 amino acids preceding the CRIB motifs of an ancestral (Dictyostelium discoideum, or Dd) WASP, invertebrate N-WASPs, vertebrate N-WASPs, and human WASP are lined up (Figure S6A). We can quantify this trend by using ReSMAP [30] to predict the membrane binding profiles of these sequences (Figure S6B). By fitting each profile to a Gaussian distribution, we identify the peak as the BR centroid and calculate its spacing from the start of the CRIB motif. The spacing is 20 for Dd WASP, then exhibits a sharp decline to 12 at the emergence of invertebrates, and finally tapers off to 9 for human WASP (Figure S6C). It will be interesting to use this wide set of WASP orthologs to validate the expected effects, based on the present study, of the BR centroid-CRIB spacing on WASP activity. For example, the wide spacing from GBD is expected to allow Dd WASP BR to bind PIP2 largely independent of GBD-Cdc42 interactions. Functional studies have indeed shown that this BR is crucial for Dd WASP to localize at sites rich in PIP2 (and PIP3) and stimulate actin polymerization during chemotaxis [39].
Computational Methods
System preparation.
To prepare for membrane association simulations, WASP and N-WASP fragments consisting of residues 133–255 and 116–220, respectively, were assigned initial conformations using TraDES [40]. These fragments were first simulated in water (with 100 mM NaCl) for 500 ns in quadruplicate with different random number seeds on GPUs using pmemd.cuda [41] in AMBER18 [42]. Following previous studies [33, 43, 44], the force fields were ff14SB [45] for proteins and TIP4P-D [46] for water. Four snapshots each for WASP and N-WASP were taken from these simulations to be used as starting structures for membrane association simulations without Cdc42. The ff14SB/TIP4P-D combination has been thoroughly validated for IDP simulations [33, 43, 44]. It also works well for structured proteins [47], and thus is well-suited for the simulations with or without Cdc42.
For membrane association simulations with Cdc42, the WASP and N-WASP fragments contained residues 195–288 and 157–253, respectively. The initial structure of WASP residues 195–229 was taken from a snapshot in a simulation without Cdc42; the remaining WASP residues and Cdc42 residues 1–179 were taken from 1CEE [22]. The initial structure of N-WASP prebound to Cdc42 was generated similarly, but with homology modeling using 1CEE as the template. Prenylated Cdc42 was generated by appending P180EPKKSRRC188 to the C-terminus of Cdc42 from 1CEE and then, in CHARMM-GUI [48], modifying the Cys188 sidechain with a geranylgeranyl group and adding a methyl group to the backbone terminal carboxyl group. Partial charge assignments for the modified Cys188 were made by the AM1-BCC charge model using the Antechamber module [49] of AmberTools18. The remaining force-field parameters were derived using the general AMBER force field (GAFF) [50], also using Antechamber. The proteins were placed atop a membrane (with the geranylgeranyl group, when present, aligned with lipid tails) and solvated using CHARMM-GUI. The upper leaflet contained 400 lipids, with the following composition: PIP2, 10%; POPS, 40%; POPC, 20%; POPE, 20%; cholesterol, 10%. In the lower leaflet, PIP2 and POPS were absent and replaced by POPC (at a slightly smaller number, because the surface area of POPC is ~10% larger than that of POPS). Na+ and Cl– ions were added to neutralize each system and make up 150 mM salt. The lipid force field was Lipid17 [51]. Each system contained ~430,000 atoms.
Molecular dynamics simulations.
Each system with the initial structure prepared above was first equilibrated in NAMD 2.12 [52] following a six-step protocol [48], in which, after 10,000 steps of conjugate-gradient minimization, constraints on protein and lipid atoms were gradually released. The timestep was 1 fs in the first three steps (two steps of 25 ps each at constant NVT and 1 step of 25 ps at constant NPT), and increased to 2 fs for the remaining steps of 100 ps each at constant NPT. All bonds involving hydrogens were restrained using the SHAKE algorithm [53]. The nonbonded cutoff was 12 Å (with force switching started at 10 Å for van der Waals interactions). Long-range electrostatic interactions were treated by the particle mesh Ewald method [54]. Temperature was maintained at 300 K using the Langevin thermostat [55] with a damping constant of 1.0 ps−1. Pressure was maintained at 1 atm by the Langevin piston [56] with an oscillation period of 50 ps and decay time of 25 ps.
The simulations were then continued in quadruplicate on GPUs using pmemd.cuda [41]. Pressure was now regulated by the Berendsen barostat [57]. Semi-isotropic scaling was applied in the x-y direction to maintain the overall shape of the system. In total, WASP and N-WASP simulations without Cdc42 were run in 16 replicates, each lasting 1360 ns; WASP simulations with soluble and prenylated Cdc42 were run in 4 copies, each lasting 1000 ns; and N-WASP simulations with soluble and prenylated Cdc42 were run in 4 copies, each lasting 700 ns. The first 360 ns of the simulations without Cdc42 and the first 100 ns of the simulations with Cdc42 were discarded. The remaining portions were saved at 20 ps intervals for analysis.
Data analysis:
Contacts between protein and lipid heavy atoms were calculated in cpptraj [58] and further analyzed using mdtraj [59]. A contact was defined between two heavy atoms within 3.5 Å.
Bilayer thickness and area per lipid were calculated using MEMPLUGIN [60]. Lateral diffusion constants were calculated using the diffusion coefficient tool [61], with the phosphorus atoms on PIP2 (internal position), POPS, POPC, and POPE and oxygen atom on cholesterol used for displacement. These properties were calculated on frames separated by 5 ns.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R35 GM118091.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- [1].Padrick SB, Rosen MK. Physical Mechanisms of Signal Integration by WASP Family Proteins. Annu Rev Biochem. 2010;79:707–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Miki H, Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem Bioph Res Co. 1998;243:73–8. [DOI] [PubMed] [Google Scholar]
- [3].Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. [DOI] [PubMed] [Google Scholar]
- [4].Kim AS, Kakalis LT, Abdul-Manan M, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–8. [DOI] [PubMed] [Google Scholar]
- [5].Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–6. [DOI] [PubMed] [Google Scholar]
- [6].Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Bioph Biom. 2007;36:451–77. [DOI] [PubMed] [Google Scholar]
- [7].Derry JMJ, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–44. [DOI] [PubMed] [Google Scholar]
- [8].Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–35. [PMC free article] [PubMed] [Google Scholar]
- [9].Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–6. [DOI] [PubMed] [Google Scholar]
- [10].Nusblat LM, Dovas A, Cox D. The non-redundant role of N-WASP in podosome-mediated matrix degradation in macrophages. Eur J Cell Biol. 2011;90:205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jain N, Thanabalu T. Molecular difference between WASP and N-WASP critical for chemotaxis of T-cells towards SDF-1α. Sci Rep. 2015;5:15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. [DOI] [PubMed] [Google Scholar]
- [14].Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–56. [DOI] [PubMed] [Google Scholar]
- [15].Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. [DOI] [PubMed] [Google Scholar]
- [16].Yamaguchi H, Miki H, Suetsugu S, Ma L, Kirschner MW, Takenawa T. Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proc Natl Acad Sci USA. 2000;97:12631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Higgs HN, Pollard TD. Activation by Cdc42 and Pip2 of Wiskott-Aldrich Syndrome Protein (Wasp) Stimulates Actin Nucleation by Arp2/3 Complex. J Cell Biol. 2000;150:1311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rohatgi R, Ho HYH, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J Cell Biol. 2000;150:1299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA. A polybasic motif allows N-WASP to act as a sensor of PIP2 density. Mol Cell. 2005;17:181–91. [DOI] [PubMed] [Google Scholar]
- [21].Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, et al. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–34. [DOI] [PubMed] [Google Scholar]
- [22].Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, et al. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–83. [DOI] [PubMed] [Google Scholar]
- [23].Hemsath L, Dvorsky R, Fiegen D, Carlier MF, Ahmadian MR. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol Cell. 2005;20:313–24. [DOI] [PubMed] [Google Scholar]
- [24].Pang XD, Zhou HX. Mechanism and rate constants of the Cdc42 GTPase binding with intrinsically disordered effectors. Proteins. 2016;84:674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wu D, Zhou HX. Designed Mutations Alter the Binding Pathways of an Intrinsically Disordered Protein. Sci Rep. 2019;9:6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rohatgi R, Nollau P, Ho HYH, Kirschner MW, Mayer BJ. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J Biol Chem. 2001;276:26448–52. [DOI] [PubMed] [Google Scholar]
- [27].Ou L, Matthews M, Pang X, Zhou H-X. The dock-and-coalesce mechanism for the association of a WASP disordered region with the Cdc42 GTPase. FEBS J 2017;284:3381–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tomasevic N, Jia Z, Russell A, Fujii T, Hartman JJ, Clancy S, et al. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry. 2007;46:3494–502. [DOI] [PubMed] [Google Scholar]
- [30].Qin SB, Hicks A, Dey S, Prasad R, Zhou HX. ReSMAP: Web Server for Predicting Residue-Specific Membrane-Association Propensities of Intrinsically Disordered Proteins. Membranes. 2022;12:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dey S, Zhou HX. Membrane Tethering of SepF, a Membrane Anchor for the Mycobacterium tuberculosis Z-ring. J Mol Biol. 2022;434:167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hicks A, Escobar CA, Cross TA, Zhou HX. Fuzzy Association of an Intrinsically Disordered Protein with Acidic Membranes. JACS Au. 2021;1:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Smrt ST, Escobar CA, Dey S, Cross TA, Zhou HX. An Arg / Ala-Rich Helix in the N-Terminal Region of M . tuberculosis FtsQ Anchors FtsZ to Membranes. biorxiv. 2022:DOI: 10.1101/2022.09.02.506397v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].MacAinsh M, Zhou HX. Partial Mimicry of the Microtubule Binding of Tau by Its Membrane Binding. Protein Sci. 2023:e4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gamper N, Shapiro MS. Target-specific PIP2 signalling: how might it work? The Journal of Physiology. 2007;582:967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Du Y, Bock BC, Schachter KA, Chao M, Gallo KA. Cdc42 induces activation loop phosphorylation and membrane targeting of mixed lineage kinase 3. J Biol Chem. 2005;280:42984–93. [DOI] [PubMed] [Google Scholar]
- [37].Johnson JL, Erickson JW, Cerione RA. C-terminal Di-arginine Motif of Cdc42 Protein Is Essential for Binding to Phosphatidylinositol 4,5-Bisphosphate-containing Membranes and Inducing Cellular Transformation. J Biol Chem. 2012;287:5764–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dey S, Zhou HX. N-WASP is competent for downstream signaling before full release from autoinhibition. J Chem Phys. 2023:DOI: 10.1063/5.0137908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Myers SA, Han JW, Lee Y, Firtel RA, Chung CY. A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol Biol Cell. 2005;16:2191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Feldman HJ, Hogue CWV. Probabilistic sampling of protein conformations: New hope for brute force? Proteins. 2002;46:8–23. [PubMed] [Google Scholar]
- [41].Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J Chem Theory Comput. 2013;9:3878–88. [DOI] [PubMed] [Google Scholar]
- [42].Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE, Cruzeiro VWD, et al. AMBER 2018, University of California, San Francisco. 2018. [Google Scholar]
- [43].Dey S, MacAinsh M, Zhou HX. Sequence-Dependent Backbone Dynamics of Intrinsically Disordered Proteins. J Chem Theory Comput. 2022;18:6310–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hicks A, Escobar CA, Cross TA, Zhou HX. Sequence-Dependent Correlated Segments in the Intrinsically Disordered Region of ChiZ. Biomolecules. 2020;10:946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Piana S, Donchev AG, Robustelli P, Shaw DE. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J Phys Chem B. 2015;119:5113–23. [DOI] [PubMed] [Google Scholar]
- [47].Hicks A, MacAinsh M, Zhou HX. Removing Thermostat Distortions of Protein Dynamics in Constant-Temperature Molecular Dynamics Simulations. J Chem Theory Comput. 2021;17:5920–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–65. [DOI] [PubMed] [Google Scholar]
- [49].Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–60. [DOI] [PubMed] [Google Scholar]
- [50].Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25:1157–74. [DOI] [PubMed] [Google Scholar]
- [51].Gould IR, Skjevik AA, Dickson CJ, Madej BD, Walker RC. Lipid17: A comprehensive AMBER force field for the simulation of zwitterionic and anionic lipids. In Prep. 2019. [Google Scholar]
- [52].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–41. [Google Scholar]
- [54].Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–93. [Google Scholar]
- [55].Pastor RW, Brooks BR, Szabo A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol Phys. 2006;65:1409–19. [Google Scholar]
- [56].Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J Chem Phys. 1995;103:4613–21. [Google Scholar]
- [57].Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. J Chem Phys. 1984;81:3684–90. [Google Scholar]
- [58].Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J Chem Theory Comput. 2013;9:3084–95. [DOI] [PubMed] [Google Scholar]
- [59].McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernandez CX, et al. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys J. 2015;109:1528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Guixà-González R, Rodriguez-Espigares I, Ramírez-Anguita JM, Carrió-Gaspar P, Martinez-Seara H, Giorgino T, et al. MEMBPLUGIN: studying membrane complexity in VMD. Bioinformatics. 2014;30:1478–80. [DOI] [PubMed] [Google Scholar]
- [61].Giorgino T Computing diffusion coefficients in macromolecular simulations: the Diffusion Coefficient Tool for VMD. J Open Source Softw. 2019;4:1698. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


