Abstract
Tomography has made a revolutionary impact on the physical, biological and medical sciences. The mathematical foundation of tomography is to reconstruct a three-dimensional (3D) object from a set of two-dimensional (2D) projections. As the number of projections that can be measured from a sample is usually limited by the tolerable radiation dose and/or the geometric constraint on the tilt range, a main challenge in tomography is to achieve the best possible 3D reconstruction from a limited number of projections with noise. Over the years, a number of tomographic reconstruction methods have been developed including direct inversion, real-space, and Fourier-based iterative algorithms. Here, we report the development of a real-space iterative reconstruction (RESIRE) algorithm for accurate tomographic reconstruction. RESIRE iterates between the update of a reconstructed 3D object and the measured projections using a forward and back projection step. The forward projection step is implemented by the Fourier slice theorem or the Radon transform, and the back projection step by a linear transformation. Our numerical and experimental results demonstrate that RESIRE performs more accurate 3D reconstructions than other existing tomographic algorithms, when there are a limited number of projections with noise. Furthermore, RESIRE can be used to reconstruct the 3D structure of extended objects as demonstrated by the determination of the 3D atomic structure of an amorphous Ta thin film. We expect that RESIRE can be widely employed in the tomography applications in different fields. Finally, to make the method accessible to the general user community, the MATLAB source code of RESIRE and all the simulated and experimental data are available at https://zenodo.org/record/7273314.
Subject terms: Applied physics, Mathematics and computing, Nanoscale materials, Imaging techniques
Introduction
Tomography has had a radical impact on diverse fields ranging from the 3D determination of the individual atoms in matter to the diagnosis of disease in medicine1–4. In the physical sciences, atomic electron tomography (AET) has been developed to resolve the atomic structure of crystal defects and amorphous materials in unprecedented 3D detail1,3,5–10. In the biological sciences, cryo-electro nmicroscopy has become a powerful tool for 3D structural determination of macromolecules with identical or similar conformations at near-atomic resolution2,11–14. For pleomorphic biological structures, cryo-electron tomography is a method of choice with a resolution in the range of 2–5 nm15,16. Tomography has also been combined with coherent diffractive imaging methods17–19 to perform quantitative 3D imaging of thick samples with a resolution of tens of nanometers20–29. In medicine, computed tomography has been routinely used as a diagnostic imaging procedure4. Although the applications of tomography are wide and diverse, a central problem associated with its mathematical and experimental implementation is similar, that is, how to accurately reconstruct a 3D structure from noisy and incomplete projection data30. When there are a large number of projections with low noise and no missing data, a direct inversion technique, named filtered back-projection (FBP)2,4,30, is accurate and fast. However, for most applications, there are usually missing data and high noise due to the radiation damage to the samples and/or the geometric constraint on the tilt range. To deal with these issues, real-space iterative algorithms have been developed such as algebraic reconstruction technique (ART)30,31, simultaneous algebraic reconstruction technique (SART)32 and simultaneous iterative reconstruction technique (SIRT)33,34. These algorithms minimize the difference between measured and calculated projections using least-square optimization, which can be implemented in parallel computing with a fast running time. Although ART, SIRT and SART usually outperform FBP, the forward and back projection steps in the real-space iterative algorithms are based on mathematical approximations that are not accurate enough to produce high-quality reconstructions. Fourier-based iterative algorithms have been developed to overcome this accuracy limitation, such as equal slope tomography (EST), generalized Fourier iterative reconstruction (GENFIRE) and others8,35–39, which rely on the Fourier transform and the inversion as the accurate forward and back projection steps, respectively. Numerical simulation and experimental results have demonstrated that EST and GENFIRE perform better than FBP, ART, SIRT and SART when there are only a limited number of projections8,35,36,40–43. EST and GENFIRE have also played an essential role in the development of AET for the 3D determination of crystal defects at the single-atom level1,3,5–10,44. Albeit powerful, Fourier-based iterative algorithms require a large memory to assemble a 3D Fourier array in each reconstruction and are also computationally expensive due to the difficulty in implementing parallel computing9,35,36. Here, we report the development of RESIRE for accurate tomographic reconstruction. Compared with real-space iterative algorithms, RESIRE uses the Fourier slice theorem or the Radon transform as the forward projection and a linear transformation as the back projection, both of which are accurate. Compared with EST and GENFIRE, RESIRE is a real-space iterative algorithm and can be implemented in parallel computing with a fast running time. Our numerical simulation and experimental results indicate that RESIRE outperforms all existing algorithms for accurate tomographic reconstruction. Furthermore, RESIRE has been used by AET to determine the 3D atomic structure of amorphous materials for the first time45,46.
Methods
Mathematical foundation of RESIRE
The tomographic reconstruction can be formulated as a least-square optimization problem which minimizes the following sum of squared errors (SSE)
| 1 |
where and are the projections and their corresponding tilt angles measured from a 3D object O. We denote that is a linear projection operator with respect to the tilt angle . For simplicity, we represent Eq. (1) with a single tilt axis, but RESIRE works as well for multiple tilt axis case with three Euler angles. The error metric can be decomposed into a sum of which is the error metric for each projection .
| 2 |
The gradient of the error metric was computed with respect to the object O using the calculus of variation method47. The following derivation is a discrete version of the continuous problem. For a voxel with a 3D coordinate , the gradient is calculated by:
| 3 |
where is the rotation matrix. The first line of Eq. (3) is obtained by the vanilla chain rule and the second line is based on an assumption that each voxel of the object is independent from each other, that is, if and 0 otherwise. As the transpose of a rotation matrix is its inverse, we derive the following transformation
| 4 |
where is the (i, j)th element of the rotation matrix , and all the voxels are on a Cartesian grid (integer numbers) but the coordinates are not.
Using Eqs. (3)–(4), RESIRE iterates between an updated object and the measured projections with each iteration consisting of a forward and back projection step. In the forward projection step, the projections, , are calculated from the object O of the current iteration using one of the two approaches: the Fourier slice theorem (FST) or the Radon transform. The FST approach first pads zeros to the object and then computes its oversampled Fourier transform, where padding zeros in real space is equivalent to oversampling in reciprocal space and can improve the accuracy of the approach48. 2D projections can then be calculated by taking the inverse Fourier transform of the corresponding 2D slices through the origin of the oversampled 3D Fourier transform. Alternatively, the projections, , can be computed from object O via the Radon transform49,50. Each voxel of the object is divided into equally sub-voxels, each of which is independently projected along specific tilt directions to calculate 2D projections with sub-pixel precision. The finer the sub-pixel is, the better the precision is, but at the expense of more computational power. In the “back projection” step, the measured projections are subtracted from the forward projections to obtain the differences . The gradient of the error metric is computed by applying on this difference . Equation (4) shows how to “back-project” a 2D image to 3D domain. The “back-projection” is in fact a linear transformation which includes a rotation and a translation, that is, each (u, v) slice of the gradient is a linear transformation of the difference. In addition, the (u, v) slices where corresponds to the translation of the (u, v) slice where . The amount of the shift is the zero order term in Eq. (4).
Remarks
Regarding performance, the FST requires the oversampling ratio OR to be greater than 3; however, OR larger than 4 makes little or no further improvement. To balance the trade-off between performance and efficiency, we choose in all our experiments.
The “forward projection” using the FST and the Radon transform are slightly different. While the FST produces smoothed images, the Radon transform constructs sharper figures. Based on specific applications, users can choose either the FST or the Radon transform to perform the forward projection step.
Although the FST has been commonly used for 3D volume assembly in single particle cryo-EM38,39 via a software package named RELION, the limitation is the memory usage and the interpolation in the Fourier domain. RELION uses a statistical approach and works well in the case of numerous measurements. But when the measurements are limited, such as AET, which only has around 50–120 projections in a range of degrees, tomography reconstruction becomes an ill-posed problem. Real-space constraints and regularizers can help to stabilize the reconstruction. Thus, one can further improve the Radon transform-based RESIRE by iteratively applying sparse and Tikhonov regularizes similar to the optimization techniques, such as Alternating Direction Methods of Multiplier51 (ADMM), Douglas Rachford52 (DR), Primal-Dual Hybrid Gradient53 (PDHG), and Gradient Projection Method54 (GPM).
RESIRE convergence
To prove the convergence of RESIRE, we need to find the Lipchitz constant L of the gradient that satisfies the inequality
| 5 |
The Lipchitz property by Eq. (5) will guarantee that RESIRE converges with the step size 1/L. When applying the inequality to RESIRE, we first assume y is the single tilt axis and reduce the analysis to the 2D case where the reconstruction is a 2D array of size . For simplicity, let us assume that it is a square array with , and the object has a compact support with a circle shape of diameter and the center coinciding with the origin of the reconstruction. We then vectorize x by stacking its columns into a vector, i.e. . Next we decompose the projection operator into a product of two operators and , i.e. where is a rotation operator for angle , and S is projection operator along the z axis:
where is a vector of all ones. As all the elements of are in range [0, 1] and each row of is summed to 1, the rotation operator is non-expansive, that is, where is the induced matrix norm. Furthermore, the rotation of an object must be an orthogonal operator, i.e. . Since we approximate the rotation by linear interpolation, we have , implying that is semi-positive definite. Based on the above analysis, we derive the following inequalities
| 6 |
where all the inequalities are obtained by the triangle inequality. is a block-diagonal matrix, containing identical blocks which is a rank-one matrix with all elements equal to one. Since has exactly one non-zero eigenvalue , it results in . Since is non-expansive and , we approximate the Lipschitz constant
| 7 |
This inequality is an important result of the step size analysis, showing that is -Lipshitz. Since there are n projections contributing to the gradient, the Lipschitz constant increases by a factor of n, that is, the accumulated Lipchitz constant becomes . The gradient descent step size is determined by where is the normalized step size. Although our analysis assumes , in practice we usually have . Since Eq. (7) shows the upper bound of the Lipchitz constant, the same step size t is still applicable when is not too small relative to . Furthermore, we can choose larger t when the object is sparse. Our experimental results show that RESIRE converges well with . This t value has been chosen for all our experimental results reported in this paper. Finally, we derive the iterative equation of RESIRE:
| 8 |
where the superscript k represents the kth iteration. Having shown that is Lipchitz continuous with constant , we can conclude that RESIRE with a fixed step size after k iterations will yield a solution which satisfies:
| 9 |
where and are the optimal error value and solution, respectively. In addition, Eq. (9) shows that the sequences produced by the iteration in the Eq. (8) will converge to the optimal solution in the first order where the rate of convergence is . The proof can be found elsewhere55.
Although the above analysis focuses on the single axis rotation case, RESIRE is generalized to the multiple axis rotation reconstructions. For the general case, we use the three Euler angles to describe the orientation of a 3D object, corresponding to the rotations around the z, y, and x-axes, respectively. The composed rotation matrix is where , and are the rotation matrices around the z, y, and x-axes, respectively. By replacing with in Eq. (3), RESIRE can be used for the general rotation case.
RESIRE pseudocode
The pseudocode of the RESIRE algorithm is described below, which can be implemented with parallel computing with a fast running time.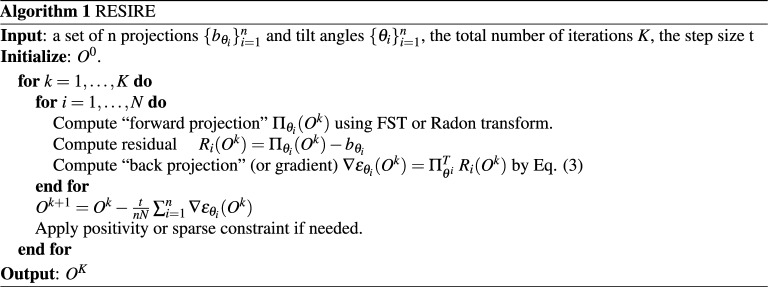
Figure 1 shows the schematic layout of the RESIRE algorithm. To reconstruct a 3D object from given experimental projections, RESIRE minimizes the L2-norm metric using gradient descent. Each iteration of the algorithm includes four steps:
RESIRE first computes the forward projections of an arbitrary initial object using FST or the Radon transform, where an empty object is often used.
The second step computes the differences between the calculated and experimental projections
The third step back-projects the difference to 3D real space, yielding the gradient of the object:
Lastly, the initial object is updated using the gradient: . Optional real-space constraints, such as positivity and support, if applicable, are enforced in RESIRE for better convergence and accuracy.
Repeat these four steps until the algorithm converges or the L2-norm error does not change. Typically, RESIRE reconstructs high-quality object with sufficiently optimized error metrics after several hundreds of iterations.
Figure 1.
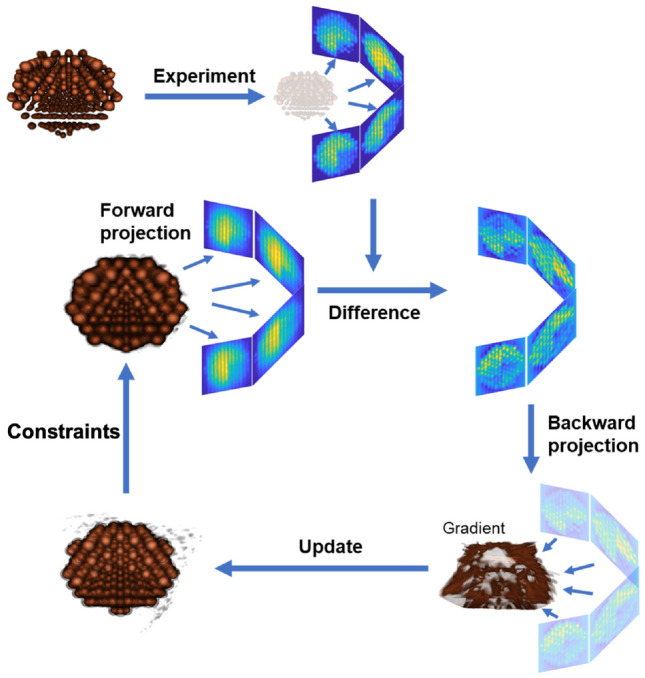
Schematic layout of the RESIRE algorithm. RESIRE first calculates the forward projections of an arbitrary initial object using FST or the Radon transform. The differences between the calculated and measured projections are back-projected to yield the gradient. The initial object is then updated using the gradient. Optional constraints such as positivity and support can be enforced for better convergence and accuracy. The updated object is used for the next iteration. Usually, after several hundreds of iterations, the algorithm converges.
Angular refinement
In many tomography experiments, the measured tilt angles are not always accurate due to instrument misalignment, motor slipping, beam-induced motion, vibration, thermal effects, and/or software error. RESIRE implements the following angular refinement procedure to improve the tilt angle accuracy. Using the current tilt angles, an initial 3D object is reconstructed by RESIRE. For each jth measured projection, a series of 2D projections are calculated from the 3D object by varying the three Euler angles , , and are the current tilt angles. Since our angular refinement uses brutal force, we suggest the searching range is within the initial angles for the sake of efficiency, i.e. . Each calculated 2D projection is then compared with the corresponding measured projection via an error metric. The three Euler angles with the smallest R-factor are recorded as the new angles for the jth measured projection. After repeating this step for all the measured projections, a new 3D object is reconstructed using the new angles. The refinement procedure is repeated until no further improvement can be made. This angular refinement procedure can improve the tilt angle accuracy and the quality of the 3D reconstruction as demonstrated with numerical simulation and experimental data36.
When working with other imaging sciences, such as single-particle, one can consider other refinement processes, such as the EMC algorithm56,57. There is a potential to combine RESIRE with such spatial refinements.
Results
RESIRE reconstruction of a simulated biological vesicle
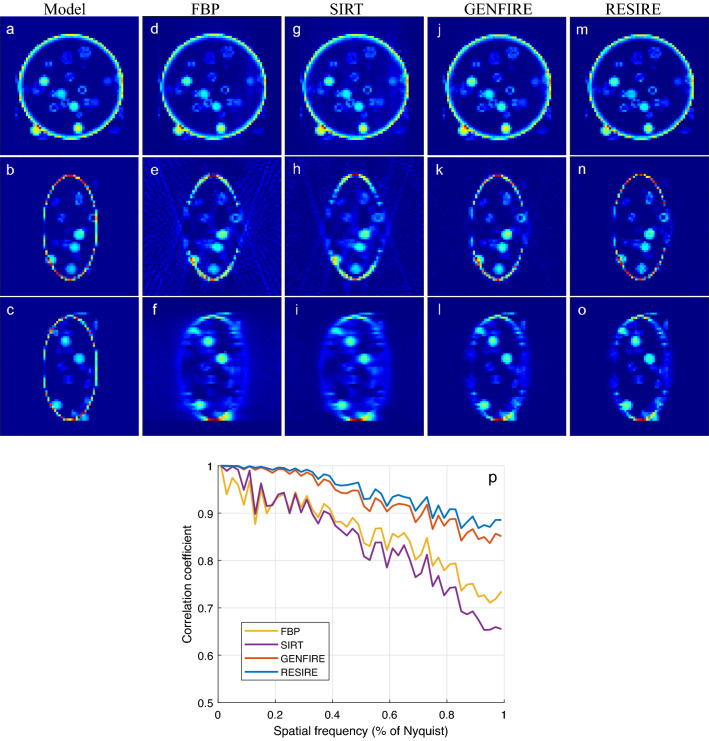
A voxel model of a biological vesicle (Fig. 2a–c) is used in this test to demonstrate the performance of RESIRE relative to the other tomographic reconstruction methods. In the simulation, the tilt axis is the y-axis and the other two Euler angles and are set to zero. When there is enough measurement and no missing wedge, the performance of all Tomography algorithms are all great; hence we omit this test. Instead, we focus on the missing wedge case and large angular increment. The missing wedge problem is believed to be more severe than the coarser tilt angles and causes trouble to Tomographic reconstructions. A tilt series of 41 projections are calculated from the 3D model with a tilt range of and an angular increment of . Poisson noise is added to each projection to make the simulation data more realistic. Reconstructions are performed using FBP, SIRT GENFIRE and RESIRE. To monitor the convergence, an R-factor is used as an error metric to quantify the difference between the calculated and measured projections.
| 10 |
All the iterative algorithms RESIRE, GENFIRE and SIRT are run with 150 iterations. ASTRA Toolbox58,59 is used to perform the SIRT reconstruction and IMOD60 is used for FBP. The R-factors are calculated to be 11.7, 23.9, 12.9 and 9.08% for FBP, SIRT GENFIRE, and RESIRE, respectively. By minimizing the least square through iteration, RESIRE and SIRT obtain lower than FBP and GENFIRE. FBP has the largest because it is a direct inversion method. Although its is slightly larger, GENFIRE produces a more accurate reconstruction than SIRT as demonstrated below.
Figure 2.
3D reconstruction of a simulated biological vesicle. (a–c) Three 10-voxel-thick central slices of the vesicle model in the XY, XZ and YZ planes, respectively. The corresponding three reconstructed slices with FBP (d–f), SIRT (g–i), GENFIRE (j–l), and RESIRE (m–o), where the missing wedge direction is along the z-axis. (p) The FSC between the model and the reconstructions showing that RESIRE produces a better reconstruction than FBP, SIRT, and GENFIRE at all spatial frequencies.
Figure 2 show 10-voxel-thick central slices of the 3D reconstructions in the XY (top row), XZ (middle row) and YZ (bottom row) planes by FBP, SIRT, GENFIRE, and RESIRE, respectively, where the z-axis is the missing wedge direction. The XY central slices from all four methods show a good agreement with the model. Although the missing wedge effects are present in all the four reconstructions of the XZ and YZ slices, RESIRE exhibits less peripheral noise, more easily detectable boundaries and reduces missing wedge effects than FBP, SIRT, and GENFIRE.
To quantify the results, we calculate the Fourier shell correlation (FSC) between the model and each reconstruction. Figure 2p confirms that that RESIRE outperforms the other three methods at all spatial frequencies.
Next, we perform the angular refinement test with the same simulation data, deviating the tilt angles by adding Gaussian noise with a normal distribution (standard deviation equals 1.0) to the ground truth angles. The angular refinement starts with these initial guesses and searches for optimal values in the range of within the initial ones. After a couple of rounds of refinement, the Root Mean Square Error (RMSE) between the ground truth angles and the refined ones is reduced from 1.00 to 0.16. The 3D image obtained by the angular refinements is almost indistinguishable from the regular RESIRE with the ground truth angles. At the same time, the two FSC curves are nearly identical (so the figures are omitted here).
RESIRE reconstruction of the experimental data of a biomineral
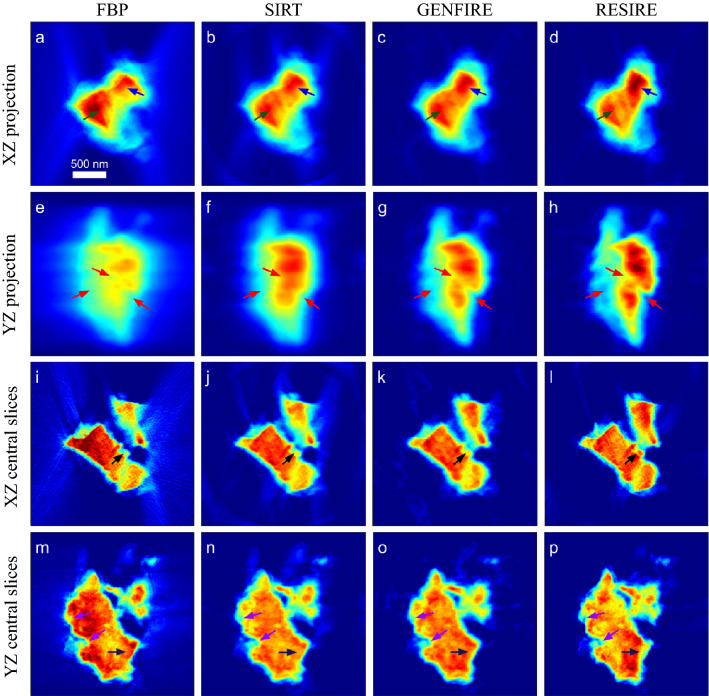
To test RESIRE with experimental data, we use a tomographic tilt series acquired from an aragonite coral sample with an annular dark-field (ADF) scanning transmission electron microscope (STEM). The tilt series consists of 69 projections with a tile range from to and an angular increment of . The projections were pre-processed with background subtraction, normalization, and alignment. Details of the experiment and pre-processing can be found elsewhere61.
Using the same experimental data set, we perform FBP, SIRT, GENFIRE, and RESIRE, reconstructions with of 25.4%, 13.5%, 7.29% and 5.30%, respectively. RESIRE produces the smallest , indicating that its 3D reconstruction is most consistent the 69 experimental projections. Figure 3a–h shows the XZ and YZ projections of the four 3D reconstructions, where the z-axis (horizontal) is the missing wedge direction. Compared with FBP, SIRT and GENFIRE, RESIRE reduces the reconstruction artifacts in the missing wedge direction as indicated by arrows. Figure 3i–p shows the 30-nm-thick central slices of the four reconstructions in the XZ, and YZ planes. RESIRE exhibits less peripheral noise, more reduced missing wedge effects, and sharper fine features than the other three methods (arrows).
Figure 3.
3D reconstruction of an aragonite coral sample. XZ and YZ projections of the FBP (a, e), SIRT (b, f), GENFIRE (c, g) and RESIRE (d, h) reconstructions, respectively. 30-nm-thick central slices of the FBP (i, m), SIRT (j, n), GENFIRE (k, o) and RESIRE(l, p) reconstructions in the XZ and YZ planes, respectively. The arrows show features that are better reconstructed by RESIRE than by the other three methods. The arrows show that RESIRE produces a reconstruction with less peripheral noise, more reduced missing wedge effects, and sharper fine features than FBP, SIRT and GENFIRE.
We also report the computational time of RESIRE in the case of GPU parallel computing. On an intel core i7-7800 computer equipped with a NVIDIA GeForce GTX1080Ti GPU, RESIRE needs 0.15 s per iteration to reconstruct the 3D volume with size pixels. Totally, the entire reconstruction accomplishes 200 iterations in 30 s.
RESIRE reconstruction of the 3D atomic structure of an amorphous film
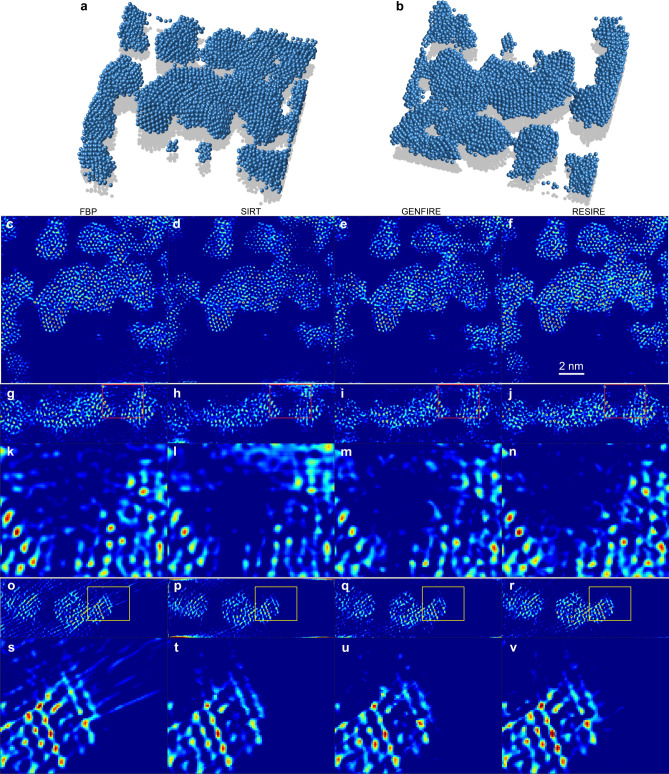
To perform the tomographic reconstruction of an extended object, we use an amorphous Ta thin film with an average thickness 5 nm, which was fabricated by physical vapor deposition. A tilt series of 46 ADF-STEM projections with a tilt range from to were acquired using the TEAM 1 microscope at NCEM and LBNL. The detailed experimental parameters and data pre-processing can be found elsewhere46.
The reconstruction of the amorphous thin film is performed by FBP, SIRT, GENFIRE, and RESIRE. Figure 4a–b shows the 3D atomic model of the amorphous thin film at two different orientations reconstructed by RESIRE, where the atomic tracing procedure is reported elsewhere46. Figure 4c–f, shows the 1.61-Å-thick central slices of the amorphous thin film in the XY plane, reconstructed by FBP, SIRT, GENFIRE and RESIRE, respectively, in which RESIRE produces better atomic features than the other methods. Figure 4g–j, (o–r) and their magnified regions (k–n) and (s–v) shows the 1.61-Å-thick central slices in the YZ and XZ planes by the four methods, where more missing wedge effects are clearly visible in the FBP and SIRT reconstructions. Due to the extended nature of the thin film, there are some the artifacts near the top and bottom edges of the slices in the YZ and XZ planes. Compared with FBP, SIRT and GENFIRE, RESIRE exhibits less peripheral noise, more easily detectable atomic features and reduced missing wedge effects.
Figure 4.
3D reconstruction of an amorphous Ta film at atomic resolution. (a–b), 3D atomic model of the amorphous Ta thin film at two different orientations, reconstructed by RESIRE. (c–f), 1.61-Å-thick central slices of the amorphous thin film in the XY plane, reconstructed by FBP, SIRT, GENFIRE and RESIRE, respectively. (g–j), 1.61-Å-thick central slices in the YZ plane of the reconstructions, where the missing wedge direction is along the z-axis. (k–n), magnified regions in (g–j) (red squares), showing that RESIRE produces a high quality reconstruction with least artifacts. (o–r), 1.61-Å-thick central slices in the XZ plane of the reconstructions. (s–v), magnified regions in (o–r) (yellow squares). Compared with the other methods, RESIRE shows less peripheral noise, more easily detectable atomic features and reduces missing wedge effects.
Remark
In the first test, we compared the 3D reconstructions of FBP, SIRT, GENFIRE, and RESIRE from 41 projections. In the 3D reconstruction from the experimental data of a biomineral in the second test, we used 69 projections. In the reconstruction of the 3D atomic structure of an amorphous film in the last test, 46 projections were used. The range of the tilt angles is approximately between and . In all these cases, RESIRE outperforms other tomographic algorithms. Thus, on the performance of the RESIRE on the projection numbers, we conclude that RESIRE consistently performs more accurate 3D reconstructions than other existing tomographic algorithms when there are a different number of 2D projections.
Conclusion
We have developed a real-space iterative algorithm, termed RESIRE, for accurate tomographic reconstruction, which uses gradient descent to solve the least-square problem . Compared with other real-space iterative algorithms such as ART, SIRT and SART, RESIRE implements a more accurate forward and back projection step in the iterative process. The forward projection step is calculated by either FST with oversampling or Radon transform with sub-voxel precision. The back projection step is computed by a linear transformation. Compared with Fourier-based iterative algorithms such as EST and GENFIRE, RESIRE can be used to reconstruction extended objects in parallel computing. Numerical simulations of a biological vesicle and experimental results of an aragonite coral sample and an amorphous thin film have shown that RESIRE outperforms FBP, SIRT, and GENFIRE with less peripheral noise, more easily detectable boundaries and features, and reduced missing wedge effects. As the power of RESIRE has been demonstrated with the successful reconstructions of amorphous solids and 3D vector ptychography of magnetic materials45,46, we expect that RESIRE can be applied to a wide range of imaging modalities in different disciplines, such as AET1,3,5–10, coherent diffractive imaging16–28,62,63, cryo-electron tomography14,15, x-ray absorption and phase contrast imaging2,42,64–68, and medical computed tomography30,34,41.
Acknowledgements
This work was supported by the US Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES), Division of Materials Sciences and Engineering under grant number DE-SC0010378 on the electron tomography experiments as well as supported by STROBE: A National Science Foundation Science and Technology Center under grant number DMR 1548924 on the development of the RESIRE algorithm. The ADF-STEM imaging with TEAM I was performed at the Molecular Foundry, Lawrence Berkeley National Laboratory, which is supported by the Office of Science, Office of Basic Energy Sciences of the US DOE under contract no. DE-AC02-05CH11231.
Author contributions
J.M. directed the research; M.P. developed the main algorithm with help from Y.Y. and A.R.; M.P and Y.Y. performed the numerical simulation and the reconstruction of the experimental data; M.P., J.M. and Y.Y wrote the manuscript with contributions from all authors. All authors reviewed the manuscript.
Data availability
The MATLAB source codes the simulated vesicle data, and the experimental data of the biomineral and the amorphous film are available at https://zenodo.org/record/7273314.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Minh Pham, Email: minhrose@ucla.edu.
Jianwei Miao, Email: miao@physics.ucla.edu.
References
- 1.Miao J, Ercius P, Billinge SJL. Atomic electron tomography: 3D structures without crystals. Science. 2016;aaf353:2157. doi: 10.1126/science.aaf2157. [DOI] [PubMed] [Google Scholar]
- 2.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State. Oxford: Oxford University Press; 2006. [Google Scholar]
- 3.Chen C-C, et al. Three-dimensional imaging of dislocations in a nanoparticle at atomic resolution. Nature. 2013;496:74–77. doi: 10.1038/nature12009. [DOI] [PubMed] [Google Scholar]
- 4.Momose A, Takeda T, Itai Y, Hirano K. Phase-contrast x-ray computed tomography for observing biological soft tissues. Nat. Med. 1996;2:473–475. doi: 10.1038/nm0496-473. [DOI] [PubMed] [Google Scholar]
- 5.Scott M, et al. Electron tomography at 2.4-ångström resolution. Nature. 2012;483:444–447. doi: 10.1038/nature10934. [DOI] [PubMed] [Google Scholar]
- 6.Xu R, et al. Three-dimensional coordinates of individual atoms in materials revealed by electron tomography. Nat. Mater. 2015;14:1099–1103. doi: 10.1038/nmat4426. [DOI] [PubMed] [Google Scholar]
- 7.Haberfehlner G, et al. Formation of bimetallic clusters in superfluid helium nanodroplets analysed by atomic resolution electron tomography. Nat. Commun. 2015;6:8779. doi: 10.1038/ncomms9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, et al. Deciphering chemical order/disorder and material properties at the single-atom level. Nature. 2017;542:75–79. doi: 10.1038/nature21042. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, et al. Observing crystal nucleation in four dimensions using atomic electron tomography. Nature. 2019;570:500–503. doi: 10.1038/s41586-019-1317-x. [DOI] [PubMed] [Google Scholar]
- 10.Tian X, et al. Correlating the three-dimensional atomic defects and electronic properties of two- dimensional transition metal dichalcogenides. Nat. Mater. 2020;19:867–873. doi: 10.1038/s41563-020-0636-5. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartesaghi A, et al. 2.2 Å resolution cryo-EM structure of -galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogales E. The development of cryo-EM into a mainstream structural biology technique. Nat. Methods. 2016;13:24–27. doi: 10.1038/nmeth.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Leiro R, Scheres SHW. Unravelling biological macromolecules with cryo-electron microscopy. Nature. 2016;537:339–346. doi: 10.1038/nature19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lučić V, Förster F, Baumeister W. Structural studies by electron tomography: From cells to molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 16.Oikonomou CM, Jensen GJ. A new view into prokaryotic cell biology from electron cryotomography. Nat. Rev. Microbiol. 2016;14:205–220. doi: 10.1038/nrmicro.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao J, Charalambous P, Kirz J, Sayre D. Extending the methodology of X-ray crystallography to allow imaging of micrometre-sized non-crystalline specimens. Nature. 1999;400:342–344. doi: 10.1038/22498. [DOI] [Google Scholar]
- 18.Rodenburg JM, et al. Hard-x-ray lensless imaging of extended objects. Phys. Rev. Lett. 2007;98:034801. doi: 10.1103/PhysRevLett.98.034801. [DOI] [PubMed] [Google Scholar]
- 19.Miao J, Ishikawa T, Robinson IK, Murnane MM. Beyond crystallography: Diffractive imaging using coherent X-ray light sources. Science. 2015;348:530–535. doi: 10.1126/science.aaa1394. [DOI] [PubMed] [Google Scholar]
- 20.Miao J, et al. Three-dimensional GaN– core shell structure revealed by X-ray diffraction microscopy. Phys. Rev. Lett. 2006;97:215503. doi: 10.1103/PhysRevLett.97.215503. [DOI] [PubMed] [Google Scholar]
- 21.Nishino Y, Takahashi Y, Imamoto N, Ishikawa T, Maeshima K. Three-dimensional visualization of a human chromosome using coherent X-ray diffraction. Phys. Rev. Lett. 2009;102:18101. doi: 10.1103/PhysRevLett.102.018101. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, et al. Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy. Proc. Natl. Acad. Sci. 2010;107:11234–11239. doi: 10.1073/pnas.1000156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dierolf M, et al. Ptychographic X-ray computed tomography at the nanoscale. Nature. 2010;467:436–439. doi: 10.1038/nature09419. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, et al. Three-dimensional coherent X-ray diffraction imaging of molten iron in mantle olivine at nanoscale resolution. Phys. Rev. Lett. 2013;110:205501. doi: 10.1103/PhysRevLett.110.205501. [DOI] [PubMed] [Google Scholar]
- 25.Miao J, Ishikawa T, Robinson IK, Murnane MM. Beyond crystallography: Diffractive imaging using coherent X-ray light sources. Science. 2015;348:530–535. doi: 10.1126/science.aaa1394. [DOI] [PubMed] [Google Scholar]
- 26.Holler M, et al. High-resolution non-destructive three-dimensional imaging of integrated circuits. Nature. 2017;543:402–406. doi: 10.1038/nature21698. [DOI] [PubMed] [Google Scholar]
- 27.Donnelly, C. Hard X-ray Tomography of Three Dimensional Magnetic Structures. Ph.D. Thesis, ETH Zurich (2017).
- 28.Deng J, et al. Correlative 3D X-ray fluorescence and ptychographic tomography of frozen-hydrated green algae. Sci. Adv. 2018;4:eaau4548. doi: 10.1126/sciadv.aau4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michelson A, et al. Three-dimensional visualization of nanoparticle lattices and multi-material frameworks. Science. 2022;376:203–207. doi: 10.1126/science.abk0463. [DOI] [PubMed] [Google Scholar]
- 30.Kak AC, Slaney M. Principles of Computerized Tomographic Imaging. Philadelphia: Society for Industrial and Applied Mathematics; 2001. [Google Scholar]
- 31.Gordon R, Bender R, Herman GT. Algebraic reconstruction techniques (ART) for three-dimensional electron microscopy and X-ray photography. J. Theor. Biol. 1970;29:471–481. doi: 10.1016/0022-5193(70)90109-8. [DOI] [PubMed] [Google Scholar]
- 32.Andersen AH, Kak AC. Simultaneous algebraic reconstruction technique (SART): A superior implementation of the ART algorithm. Ultrason. Imaging. 1984;6:81–94. doi: 10.1177/016173468400600107. [DOI] [PubMed] [Google Scholar]
- 33.Gilbert P. Iterative methods for the three-dimensional reconstruction of an object from projections. J. Theor. Biol. 1972;36:105–117. doi: 10.1016/0022-5193(72)90180-4. [DOI] [PubMed] [Google Scholar]
- 34.Herman GT. Fundamentals of Computerized Tomography: Image Reconstruction from Projections. Berlin: Springer Science & Business Media; 2009. [Google Scholar]
- 35.Miao J, Förster F, Levi O. Equally sloped tomography with oversampling reconstruction. Phys. Rev. B. 2005;72:52103. doi: 10.1103/PhysRevB.72.052103. [DOI] [Google Scholar]
- 36.Pryor A, et al. GENFIRE: A generalized Fourier iterative reconstruction algorithm for high-resolution 3D imaging. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-09847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connor YZ, Fessler JA. Fourier-based forward and back-projectors in iterative fan-beam tomographic image reconstruction. IEEE Trans. Med. Imaging. 2006;25:582–589. doi: 10.1109/TMI.2006.872139. [DOI] [PubMed] [Google Scholar]
- 38.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elmlund D, Elmlund H. Simple: Software for ab initio reconstruction of heterogeneous single-particles. J. Struct. Biol. 2012;180:420–427. doi: 10.1016/j.jsb.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Lee E, et al. Radiation dose reduction and image enhancement in biological imaging through equally-sloped tomography. J. Struct. Biol. 2008;164:221–227. doi: 10.1016/j.jsb.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahimian BP, Mao Y, Cloetens P, Miao J. Low-dose X-ray phase-contrast and absorption CT using equally sloped tomography. Phys. Med. Biol. 2010;55:5383. doi: 10.1088/0031-9155/55/18/008. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, et al. High-resolution, low-dose phase contrast X-ray tomography for 3D diagnosis of human breast cancers. Proc. Natl. Acad. Sci. 2012;109:18290–18294. doi: 10.1073/pnas.1204460109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fahimian BP, et al. Radiation dose reduction in medical X-ray CT via Fourier-based iterative reconstruction. Med. Phys. 2013;40:31914. doi: 10.1118/1.4791644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, Jeong C, Lee T, Ryu S, Yang Y. Direct observation of three-dimensional atomic structure of twinned metallic nanoparticles and their catalytic properties. Nano Lett. 2022;22:665–672. doi: 10.1021/acs.nanolett.1c03773. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, et al. Determining the three-dimensional atomic structure of an amorphous solid. Nature. 2021;592:60–64. doi: 10.1038/s41586-021-03354-0. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Y, et al. Three-dimensional atomic packing in amorphous solids with liquid-like structure. Nat. Mater. 2022;21:95–102. doi: 10.1038/s41563-021-01114-z. [DOI] [PubMed] [Google Scholar]
- 47.Aubert G, Kornprobst P. Mathematical Problems in Image Processing: Partial Differential Equations and the Calculus of Variations. Berlin: Springer Science & Business Media; 2006. [Google Scholar]
- 48.Miao J, Sayre D, Chapman HN. Phase retrieval from the magnitude of the Fourier transforms of nonperiodic objects. J. Opt. Soc. Am. A. 1998;15:1662–1669. doi: 10.1364/JOSAA.15.001662. [DOI] [Google Scholar]
- 49.Bracewell RN. Two-Dimensional Imaging. Hoboken: Prentice-Hall Inc.; 1995. pp. 505–537. [Google Scholar]
- 50.Lim JS. Two-Dimensional Signal and Image Processing. Hoboken: Prentice-Hall Inc; 1990. pp. 42–45. [Google Scholar]
- 51.Boyd S, et al. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends Mach. Learn. 2011;3:1–122. doi: 10.1561/2200000016. [DOI] [Google Scholar]
- 52.Eckstein J, Bertsekas DP. On the Douglas–Rachford splitting method and the proximal point algorithm for maximal monotone operators. Math. Program. 1992;55:293–318. doi: 10.1007/BF01581204. [DOI] [Google Scholar]
- 53.Zhu M, Chan T. An efficient primal–dual hybrid gradient algorithm for total variation image restoration. Ucla Cam Report. 2008;34:8–34. [Google Scholar]
- 54.Rosen JB. The gradient projection method for nonlinear programming. Part I Linear constraints. J. Soc. Ind. Appl. Math. 1960;8:181–217. doi: 10.1137/0108011. [DOI] [Google Scholar]
- 55.Gower, R. M. Convergence theorems for gradient descent. In Lecture Notes for Statistical Optimization (2018).
- 56.Cho D, et al. High-throughput 3D ensemble characterization of individual core-shell nanoparticles with X-ray free electron laser single-particle imaging. ACS Nano. 2021;15:4066–4076. doi: 10.1021/acsnano.0c07961. [DOI] [PubMed] [Google Scholar]
- 57.Loh N-TD, Elser V. Reconstruction algorithm for single-particle diffraction imaging experiments. Phys. Rev. E. 2009;80:026705. doi: 10.1103/PhysRevE.80.026705. [DOI] [PubMed] [Google Scholar]
- 58.Van Aarle W, et al. Fast and flexible X-ray tomography using the ASTRA toolbox. Opt. Express. 2016;24:25129–25147. doi: 10.1364/OE.24.025129. [DOI] [PubMed] [Google Scholar]
- 59.Van Aarle W, et al. The ASTRA toolbox: A platform for advanced algorithm development in electron tomography. Ultramicroscopy. 2015;157:35–47. doi: 10.1016/j.ultramic.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J. Struct. Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 61.Lo, Y. H. Developments in Mesoscale Correlative Multimodal X-ray Microscopy. Ph.D. thesis, University of California, Los Angeles (2019).
- 62.Rana A, et al. Three-dimensional topological magnetic monopoles and their interactions in a ferromagnetic meta-lattice. Nat. Nanotechnol. 2023;18:227–232. doi: 10.1038/s41565-022-01311-0. [DOI] [PubMed] [Google Scholar]
- 63.Sung D, et al. Stochastic chromatin packing of 3D mitotic chromosomes revealed by coherent X-rays. Proc. Natl. Acad. Sci. 2021;118:e2109921118. doi: 10.1073/pnas.2109921118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeiffer F, Weitkamp T, Bunk O, David C. Phase retrieval and differential phase-contrast imaging with low-brilliance X-ray sources. Nat. Phys. 2006;2:258–261. doi: 10.1038/nphys265. [DOI] [Google Scholar]
- 65.Larabell CA, Nugent KA. Imaging cellular architecture with X-rays. Curr. Opin. Struct. Biol. 2010;20:623–631. doi: 10.1016/j.sbi.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibbs JW, et al. The three-dimensional morphology of growing dendrites. Sci. Rep. 2015;5:11824. doi: 10.1038/srep11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meirer F, et al. Three-dimensional imaging of chemical phase transformations at the nanoscale with full-field transmission X-ray microscopy. J. Synchrotron Radiat. 2011;18:773–781. doi: 10.1107/S0909049511019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krenkel M, Markus A, Bartels M, et al. Phase-contrast zoom tomography reveals precise locations of macrophages in mouse lungs. Sci. Rep. 2015;5:9973. doi: 10.1038/srep09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MATLAB source codes the simulated vesicle data, and the experimental data of the biomineral and the amorphous film are available at https://zenodo.org/record/7273314.