Abstract
Neurodegeneration with brain iron accumulation (NBIA) represents a group of neurodegenerative disorders characterized by abnormal iron accumulation in the brain. In Parkinson’s Disease (PD), iron accumulation is a cardinal feature of degenerating regions in the brain and seems to be a key player in mechanisms that precipitate cell death. The aim of this study was to explore the genetic and genomic connection between NBIA and PD. We screened for known and rare pathogenic mutations in autosomal dominant and recessive genes linked to NBIA in a total of 4481 PD cases and 10,253 controls from the Accelerating Medicines Partnership Parkinsons’ Disease Program and the UKBiobank. We examined whether a genetic burden of NBIA variants contributes to PD risk through single-gene, gene-set, and single-variant association analyses. In addition, we assessed publicly available expression quantitative trait loci (eQTL) data through Summary-based Mendelian Randomization and conducted transcriptomic analyses in blood of 1886 PD cases and 1285 controls. Out of 29 previously reported NBIA screened coding variants, four were associated with PD risk at a nominal p value < 0.05. No enrichment of heterozygous variants in NBIA-related genes risk was identified in PD cases versus controls. Burden analyses did not reveal a cumulative effect of rare NBIA genetic variation on PD risk. Transcriptomic analyses suggested that DCAF17 is differentially expressed in blood from PD cases and controls. Due to low mutation occurrence in the datasets and lack of replication, our analyses suggest that NBIA and PD may be separate molecular entities.
Subject terms: Genetic association study, Genomics
Introduction
Neurodegeneration with brain iron accumulation (NBIA) represents a group of inherited heterogeneous neurodegenerative disorders characterized by iron accumulation and the presence of axonal spheroids in the basal ganglia and other brain areas1. The worldwide prevalence of NBIA in the general population is estimated between 1–3/1,000,000 individuals, adding these disorders to the group of ultra-rare orphan diseases2. In view of the rarity of these disorders and its diverse clinical presentation, NBIA can often go unrecognized, underdiagnosed, or misdiagnosed.
In the brain, the substantia nigra, putamen, globus pallidus and caudate nucleus have the highest iron concentration, and total iron content increases with age3. Brain iron is crucial for important processes such as the synthesis of myelin and neurotransmitters and oxygen transport3,4. Higher nigral iron concentrations have been shown in Parkinson’s Disease (PD) patients in postmortem studies repeatedly5,6. This excess of intracellular iron is associated with oxidative stress, lipid peroxidation, cellular dysfunction, and neuronal death in PD and thus potentially playing a role in PD pathogenesis7,8.
To date, mutations in 13 genes have been associated with autosomal dominant (FTL), autosomal recessive (ATP13A2, PANK2, PLA2G6, FA2H, CP, C19orf12, COASY, GTPBP2, DCAF17, VAC14) and X-linked forms of NBIA (WDR45, RAB39B), and are involved in a wide range of molecular processes affecting mitochondrial function, coenzyme A metabolism, lipid metabolism and autophagy9. Typically, NBIA disorders present with variable and complex phenotypes including parkinsonism, dystonia, intellectual disability, and cognitive decline1. The NBIA clinical spectrum is broad and there is increased awareness of clinical overlap between different NBIA disorders as well as with other diseases such as Parkinson’s disease (PD). In PD, it has been widely suggested that genetic components contributing to disease might include rare genetic variants of small or moderate effect, where functional and deleterious alleles might exist10. Furthermore, it has been previously predicted that variability in genes causing young-onset autosomal recessive neurological diseases contribute to late-onset neurological diseases. In fact, homozygous and compound heterozygous loss-of-function mutations in TREM2 have been previously associated with an autosomal recessive form of early-onset dementia, while when found in heterozygous state confer moderate risk for Alzheimer’s disease11,12. Similarly, homozygous and compound heterozygous mutations in GBA are responsible for Gaucher’s disease while heterozygous variants are a well-validated risk factor for PD13.
In PD, iron accumulation is a cardinal feature of degenerating regions in the brain and has been implicated in mechanisms that precipitate cell death14. Iron is a transition metal involved in several cellular functions such as neuronal metabolism, DNA synthesis, oxygen transport, mitochondrial respiration, and myelination in the brain15. Furthermore, it is implicated in production and turnover of some neurotransmitters such as dopamine, epinephrine or serotonin16. Iron tends to accumulate with age mainly in the cortex and the nuclei of the basal ganglia, including the globus pallidus, putamen, and caudate nucleus; and an excessive accumulation in these regions is associated with neurodegenerative disorders17. Evidence suggests abnormal iron levels in the brains of PD patients and a role for iron dysregulation in the disease18. Interestingly, NBIA-related genes are involved in mitochondrial function and autophagy, dysfunction of which is implicated in PD, suggesting there could be shared molecular pathways and gene networks between NBIA and PD19. Furthermore, autopsy examination of NBIA-genetically confirmed cases has demonstrated Lewy bodies in some subforms, linking the pathology of both diseases20.
The aim of the present study was to explore the relationship between genes known to contain mutations that cause autosomal dominant/recessive NBIA and PD etiology. Using the largest genetic and genomic datasets of PD cases and controls to date including the Accelerating Medicines Partnership Program for Parkinson’s Disease (AMP-PD) and the UKBiobank (UKB), we screened for the presence of known and rare pathogenic mutations linked to NBIA in PD. We further examined whether a genetic burden of variants in NBIA-linked genes could contribute to the risk of developing PD by performing single gene, gene-set, and single variant association analyses. Finally, to investigate the potential effect of changes in NBIA-linked genes expression in PD compared to healthy individuals, we assessed publicly available expression quantitative trait loci (eQTL) results from GTEx v8, BRAINEAC, and transcriptomics data from AMP-PD.
Results
Genetic screening of NBIA-related genes in whole-genome and whole-exome sequencing data of Parkinson’s disease cases and controls
ATP13A2
Genetic variants in the ATPase Cation Transporting 13A2 (ATP13A2) gene, located on chromosome 1, have been previously associated with Kufor-Rakeb syndrome, spastic paraplegia type 78, and parkinsonism21–23. A total of 36 protein-coding variants were present in the screened datasets, of which four missense variants had a higher frequency in cases versus controls. A summary of variants with a higher frequency in cases can be found in Table 1 while additional variant information is listed in the supplemental. The variants found in AMP-PD (p.I946F, p.A249V, p.T12M) have been previously associated with PD in individuals of European ancestry although their pathogenicity is still unclear24,25. The p.T402M variant, found in UKB, has been previously described in a homozygous individual with ataxia-myoclonus syndrome with an age at onset of 38 and lacking parkinsonism26,27. The report postulates that together, progressive ataxia with action myoclonus along with lack of parkinsonism may suggest a new phenotype linked to ATP13A2. In our UKB cohort, the p.T402M heterozygous mutation carrier was a male PD case with age at recruitment of 55 years. This mutation was absent in controls.
Table 1.
Summary of variants in NBIA related genes with higher frequency in PD cases versus controls, SNP = Single Nucleotide Polymorphism, p = P-value.
| SNP | Gene | p (AMP-PD) | p (UKB) | Cohort |
|---|---|---|---|---|
| chr1:16988161:T:A | ATP13A2 | 0.147 | – | AMP |
| chr1:16996487:G:A | ATP13A2 | – | 0.024 | UKB |
| chr1:17000494:G:A | ATP13A2 | 0.502 | – | AMP |
| chr1:17005754:G:A | ATP13A2 | 0.264 | – | AMP |
| chr3:149183513:C:T | CP | 0.067 | 0.887 | AMP |
| chr3:149198448:T:A | CP | – | 0.047 | UKB |
| chr3:149199783:G:A | CP | 0.648 | 0.260 | AMP |
| chr3:149206202:T:A | CP | – | 0.427 | UKB |
| chr16:74719154:G:A | FA2H | – | 0.024 | UKB |
| chr16:74774524:C:T | FA2H | 0.824 | 0.985 | AMP |
| chr16:74774662:G:C | FA2H | 0.875 | – | AMP |
| chr19:29702747:T:C | C19orf12 | – | 0.609 | UKB |
| chr19:29702977:C:A | C19orf12 | 0.875 | – | AMP |
| chr19:29702977:C:G | C19orf12 | 0.264 | – | AMP |
| chr19:29708268:G:A | C19orf12 | – | 0.024 | UKB |
| chr19:48966341:G:T | FTL | 0.875 | – | AMP |
| chr20:3889237:A:T | PANK2 | – | 0.694 | UKB |
| chr20:3907997:A:G | PANK2 | 0.114 | – | AMP |
| chr20:3910812:A:G | PANK2 | 0.439 | – | AMP |
| chr20:3912486:T:A | PANK2 | – | 0.024 | UKB |
| chr20:3912490:D:3 | PANK2 | – | 0.199 | UKB |
| chr20:3918695:G:A | PANK2 | 0.439 | 0.106 | AMP |
| chr22:38113560:C:T | PLA2G6 | – | 0.427 | UKB |
| chr22:38126390:T:C | PLA2G6 | 0.875 | 0.069 | AMP |
| chr22:38132881:C:T | PLA2G6 | 0.467 | 0.523 | AMP |
| chr22:38132917:C:T | PLA2G6 | 0.264 | – | AMP |
| chr22:38133007:G:A | PLA2G6 | 0.875 | – | AMP |
| chr22:38143275:C:T | PLA2G6 | 0.875 | – | AMP |
| chr22:38169326:G:A | PLA2G6 | 0.151 | 0.514 | AMP |
Bold entries indicate statistical significance at a nominal p value < 0.05.
CP
The Ceruloplasmin (CP) gene, located on chromosome 3, is closely related to iron metabolism in the brain28. Four out of 29 variants had a higher frequency in cases than controls. Notably, p.R793H and p.P477L, were seen in both AMP-PD and UKB. Reports have linked lower CP levels with PD development, and CP gene mutations have been associated with substantia nigra hyperechogenicity using transcranial sonography29. In addition, hyperechogenicity is a common finding in idiopathic PD patients and many studies suggest that it is an important risk marker of future PD30,31. One mutation originally associated with substantia nigra (SN) hyperechogenicity, although listed as likely benign, is p.R793H32. Our study identified the p.R793H variant in both homozygosity and heterozygosity with higher frequency in cases than controls both AMP-PD and UKB. However, it did not reach statistical significance for either dataset. Of note, we observed biallelic heterozygosity in one case and two controls from UKB carrying both the p.P477L and p.R793H mutations. The p.I392F variant was only present in UKB data. A three base pair deletion at this position has been associated with aceruloplasminemia, a disorder in which iron gradually accumulates in the brain and other organs33. In addition, this is a multi-allelic variant for which the risk allele identified in our analyses differs from those reported on HGMD.
FA2H
In the Fatty Acid 2-Hydroxylase (FA2H) gene, located on chromosome 16, we found 28 coding variants out of which two missense variants had a higher frequency in cases. The variant p.E78K appeared in both datasets and is associated with the fatty acid hydroxylase-associated neurodegeneration(FAHN)/hereditary spastic paraplegia (SPG35) phenotype although no conclusions were drawn about its significance34. The p.E78K mutation has also been reported in a patient with an age at onset of 10 with a positive family history of spastic paraplegia and was labeled as a putative pathogenic variant35. Our PD carriers with the p.E78K mutation had an age of inclusion of 70 and 52 for AMP-PD and 69 for UKB. The case/control frequency for p.E78K did not reach significance in our analysis. For the p.T207M mutation, all previously reported carriers were compound heterozygous with additional mutations in FA2H and had the spastic paraplegia phenotype22,34,36. In our study, we found one male UKB case with an age at inclusion of 65 carrying the p.T207M mutation. This mutation was absent in controls and was the only FA2H mutation that reached statistical significance.
C19orf12
Variation in the Chromosome 19 open reading frame 12 (C19orf12) gene, located on chromosome 19, has been associated with the mitochondrial membrane protein-associated neurodegeneration (MPAN) subtype of NBIA which presents with cognitive decline progressing to dementia, prominent neuropsychiatric abnormalities, motor neuronopathy, and parkinsonism movement abnormalities37,38. Out of 18 coding variants present, four missense variants in our datasets showed a higher frequency in cases versus controls, with only one proving significant and previously linked to the MPAN phenotype in the literature. In AMP-PD data, we found two multiallelic variants, p.G65V present in one case and one control and p.G65A present in just one case and no controls. In two patients reported in the literature, compound heterozygosity for p.G65V and p.G69RfsX10 variants led to the MPAN phenotype38. In addition, another two patients were reported to have this phenotype; one carrying the compound heterozygous mutations p.G65V and p.P60L and the other carrying a homozygous p.G65V variant39. The p.K142E variant, present it our data, has been seen in compound heterozygosity in patients with the MPAN NBIA phenotype40–42. Lastly, we found the p.P60L mutation at statistical significance present in one UKB case and absent in controls. The carrier under study was a female PD patient with an age at inclusion of 66 and no other notable C19orf12 mutations. The p.P60L variant has been previously detected in an MPAN patient who also carried the p.G65E C19orf12 mutation38.
FTL
Our study identified only one missense variant (p.E104) out of nine with a higher frequency in cases than controls in the Ferritin Light Chain (FTL) gene, located on chromosome 19. The p.E104 mutation was only of note in the AMP-PD dataset and was present in one case and one control (F_A = 2.02 × 10−4, F_U = 1.62 × 10−4). The p.E104 variant has been reported as causing an L-ferritin deficiency phenotype38,43.
PANK2
Variation in the Pantothenate Kinase 2 (PANK2) gene, located on chromosome 20, has been previously associated with pantothenate kinase-associated neurodegeneration (PKAN), the most common subtype of NBIA44. Clinical symptoms include dystonia, dysarthria, muscular rigidity, poor balance, and spasticity44,45. We found six out of 68 distinct coding variants in our dataset with a higher frequency in cases. The p.G521R mutation was the only one present in both AMP-PD and UKB. This variant results in an unstable and inactive PanK2 protein and has been reported as the most common variant in PANK246. It is often found in a compound heterozygous state in individuals presenting with the PKAN phenotype46,47. In AMP-PD, two PD cases, one male and one female with ages at inclusion of 54 and 67 respectively, carried the p.G521R mutation in heterozygous state compared to one control (F_A = 4.05 × 10−4, F_U = 4.05 × 10−4). In UKB, three cases, all males with an average age at inclusion of 62 years old, presented with the p.G521R mutation in heterozygous state compared to five controls (F_A = 1.36 × 10−3, F_U = 4.43 × 10−4). None of the PD p.G521R carriers from either cohort contained this mutation in compound heterozygous state.
PLA2G6
In the Phospholipase A2 Group VI (PLA2G6) gene, located on chromosome 22, we found 100 variants of which seven missense variants had a higher frequency in cases versus controls. These variants have been previously associated with both infantile neuroaxonal dystrophy (INAD) and PD48–52. INAD is a severe progressive psychomotor disorder, which presents before the third year of life, characterized by the presence of axonal spheroids throughout the central and peripheral nervous system52–54. The variant p.M470V is the only variant out of three associated with the INAD phenotype that was present in both the AMP-PD and UKB cohorts55. In addition, p.A343T has been associated with PD and we identified 77 cases and 86 controls carrying this mutation in AMP-PD (F_A = 1.58 × 10−2, F_U = 1.41 × 10−20) and 29 cases and 130 controls in UKB data (F_A = 1.31 × 10−2, F_U = 1.15 × 10−2)49,55. Of note, one of the p.A343T AMP-PD case carriers was a female homozygous for the mutation with an age at inclusion of 65. In addition, a heterozygous carrier of p.A343T in the UKB was a PD case and additionally carried the p.S34L mutation, also associated with PD49,55. This patient had an age at inclusion of 60. However, no PLA2G6 mutation with a higher frequency in cases reached statistical significance.
Transcriptomic analyses
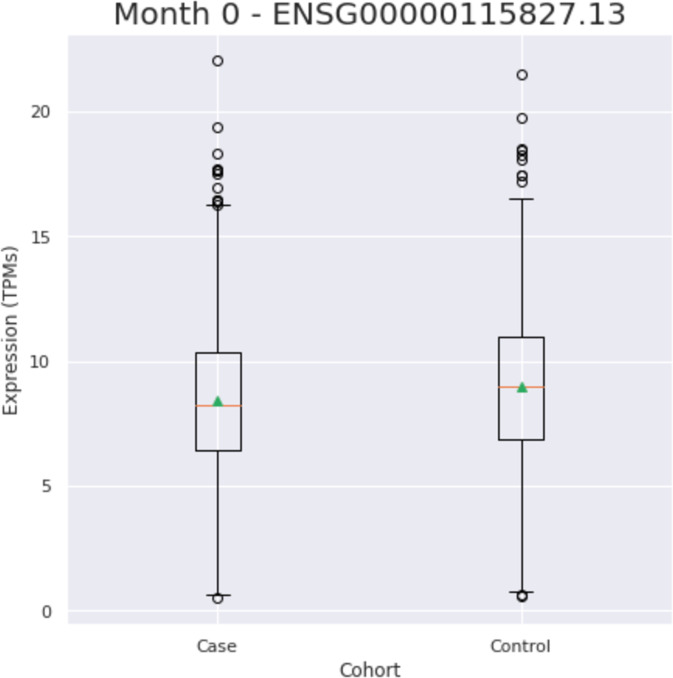
Two genes including FA2H (ENSG00000103089.8) and CP (ENSG00000047457.13) were removed for further analyses based on poor call rates (missingness rate > 0.33). Of the 10 NBIA genes left, DCAF17 expression (ENSG00000115827.13) was found to be significantly different in cases versus controls (Fig. 1)(Table 2). To further investigate these results we looked into SMR analysis for the DCAF17 gene. To prioritize SNV candidates, results were initially filtered by chromosome and base pair position for the DCAF17 gene in hg19. Afterwards, the resulting candidates were filtered by the SMR multi p-value at a threshold of p < 0.05. SMR analyses for the DCAF17 gene revealed four eQTL signals with a significant SMR p-value. Out of these four eQTL signals, only one was significant in both SMR analyses (p = 0.0024) and Nalls et al.67 GWAS (p = 0.0004). A summary of the DCAF17 SMR data can be found in the supplementary data.
Fig. 1. DCAF17 expression in blood of PD cases versus controls.
Center line indicates the median. The box bounds represent the first and third quartiles while the whiskers extend the box by 1.5× the interquartile range (IQR).
Table 2.
Results of transcriptomic analysis using blood PPMI data.
| Gene | Ensemble ID | T-statistic | P-value |
|---|---|---|---|
| ATP13A2 | ENSG00000159363.17 | −0.5126 | 0.6083 |
| DCAF17 | ENSG00000115827.13 | −4.036 | 5.54E−05 |
| GTPBP2 | ENSG00000172432.18 | −0.3596 | 0.7192 |
| VAC14 | ENSG00000103043.14 | −0.5266 | 0.5985 |
| COASY | ENSG00000068120.14 | 0.4632 | 0.6432 |
| C19orf12 | ENSG00000131943.17 | −1.446 | 0.1482 |
| FTL | ENSG00000087086.14 | 0.655 | 0.5125 |
| PANK2 | ENSG00000125779.22 | 0.8218 | 0.4113 |
DCAF17 is the only NBIA gene with significant differential expression in PD.
Bold entries indicate statistical significance at a nominal p value < 0.05.
Cumulative effect of genetic variation through single gene and gene-set burden analyses
When considering the cumulative effect of rare variation on PD risk, single gene and gene-set burden analyses did not reveal any statistically significant differences between cases and controls at MAF < 0.01 or MAF < 0.03 in any of the three cohorts and after meta-analysis (Table 3).
Table 3.
Results of burden analysis at two minor allele frequencies for NBIA genes in PD.
| Gene | Variant Group | MAF | P_AMP | P_UKB | P_Meta | WeightedZ |
|---|---|---|---|---|---|---|
| ATP13A2 | All | 0.01 | 0.0904 | 0.8813 | 0.2811 | 0.6518 |
| ATP13A2 | All | 0.03 | 0.6599 | 0.5547 | 0.7339 | −0.6615 |
| ATP13A2 | Coding | 0.01 | 0.9272 | 0.2949 | 0.6281 | −1.0192 |
| ATP13A2 | Coding | 0.03 | 0.6622 | 0.3033 | 0.5233 | −1.2470 |
| C19orf12 | All | 0.01 | 0.8789 | 0.5115 | 0.8089 | 1.6650 |
| C19orf12 | All | 0.03 | 0.7091 | 0.5115 | 0.7305 | 1.2089 |
| C19orf12 | Coding | 0.01 | 1.0000 | 0.4262 | 0.7897 | 1.6592 |
| C19orf12 | Coding | 0.03 | 1.0000 | 0.4262 | 0.7897 | 1.6592 |
| COASY | All | 0.01 | 0.5435 | 0.0832 | 0.1853 | −0.5970 |
| COASY | All | 0.03 | 0.5435 | 0.0832 | 0.1853 | −0.5970 |
| COASY | Coding | 0.01 | 1.0000 | 0.0936 | 0.3154 | 0.4017 |
| COASY | Coding | 0.03 | 1.0000 | 0.0936 | 0.3154 | 0.4017 |
| CP | All | 0.01 | 0.3392 | 0.4920 | 0.4657 | 1.0089 |
| CP | All | 0.03 | 0.6747 | 0.4339 | 0.6524 | 0.2643 |
| CP | Coding | 0.01 | 0.6615 | 0.3514 | 0.5717 | 0.4431 |
| CP | Coding | 0.03 | 0.6615 | 0.2169 | 0.4221 | 0.1493 |
| DCAF17 | All | 0.01 | 0.2382 | 0.2288 | 0.3721 | −0.9574 |
| DCAF17 | All | 0.03 | 0.2382 | 0.7715 | 0.8393 | 0.2144 |
| DCAF17 | Coding | 0.01 | 0.1146 | 0.4555 | 0.6522 | 0.8371 |
| DCAF17 | Coding | 0.03 | 0.1146 | 0.7161 | 0.8917 | 1.2486 |
| FA2H | All | 0.01 | 1.0000 | 0.3676 | 0.7355 | −0.4057 |
| FA2H | All | 0.03 | 0.8598 | 0.3676 | 0.6801 | −0.7388 |
| FA2H | Coding | 0.01 | 0.5816 | 0.5064 | 0.6546 | −0.9324 |
| FA2H | Coding | 0.03 | 0.5397 | 0.5064 | 0.6278 | −0.8240 |
| FTL | All | 0.01 | 0.4990 | 0.8460 | 0.7862 | −0.1314 |
| FTL | All | 0.03 | 0.4990 | 0.8460 | 0.7862 | −0.1314 |
| FTL | Coding | 0.01 | 0.5583 | NA | NA | 0.0555 |
| FTL | Coding | 0.03 | 0.5583 | NA | NA | 0.0555 |
| GTPBP2 | All | 0.01 | 0.5471 | 0.1980 | 0.3491 | −0.7209 |
| GTPBP2 | All | 0.03 | 0.2766 | 0.4241 | 0.3687 | −0.8013 |
| GTPBP2 | Coding | 0.01 | 0.0696 | 0.3808 | 0.1227 | −1.6865 |
| GTPBP2 | Coding | 0.03 | 0.6000 | 0.3712 | 0.5572 | 0.7064 |
| PANK2 | All | 0.01 | 0.3261 | 0.8209 | 0.6205 | 1.5979 |
| PANK2 | All | 0.03 | 0.4622 | 0.9000 | 0.7809 | 0.8418 |
| PANK2 | Coding | 0.01 | 0.7875 | 0.0507 | 0.1684 | 0.3729 |
| PANK2 | Coding | 0.03 | 0.7875 | 0.0507 | 0.1684 | 0.3729 |
| PLA2G6 | All | 0.01 | 0.7644 | 0.2124 | 0.4575 | 1.3405 |
| PLA2G6 | All | 0.03 | 0.7661 | 0.3815 | 0.6518 | 0.4891 |
| PLA2G6 | Coding | 0.01 | 0.9007 | 0.3588 | 0.6882 | −0.9407 |
| PLA2G6 | Coding | 0.03 | 0.8603 | 0.6690 | 0.8935 | 0.1710 |
| VAC14 | All | 0.01 | 0.2814 | 0.4390 | 0.3819 | −1.2446 |
| VAC14 | All | 0.03 | 0.3986 | 1.0000 | 0.7653 | −0.3362 |
| VAC14 | Coding | 0.01 | 0.3683 | 0.7708 | 0.6414 | 0.0051 |
| VAC14 | Coding | 0.03 | 0.3683 | 0.8155 | 0.6616 | −0.0728 |
MAF Minor Allele Frequency, P_AMP P value in AMP_PD cohort, P_UKB P value in UKB cohort, P_Meta P value in meta analysis, Weighted Z Weighted Z Statistic.
Discussion
Using the largest PD genetic and genomic datasets available to date, we conducted a comprehensive genetic assessment of NBIA related variants and genes for a potential role in PD etiology. We identified 29 previously reported NBIA coding mutations in a total of seven genes including ATP13A2, CP, FA2H, C19orf12, FTL, PANK2, and PLA2G6 with a higher frequency in PD cases than controls. All phenotypes previously linked to the reported variants in NBIA genes involve neurological disorders, adding to the conclusion of clinical overlap between neurological diseases and the strong link between brain iron accumulation and a number of neurodegenerative disorders. However, only four out of the 29 variants of interest we reported had a statistically significant difference in frequency between cases and controls (p < 0.05), all only significant in UKB data. Within these four variants, one was found in ATP13A2, one in FA2H, one in C19orf12, and one in PANK2, always in only one case and no controls. Notably, none of these four variants have been previously associated with PD in the literature. Noting the very low mutation frequency in the current data and the lack of replication, we cannot conclude that the mutation spectrum contributing to NBIA phenotype overlaps with PD etiology. As such, despite enrichment for the risk allele in PD cases versus controls in four variants, these data suggest that in their heterozygous state NBIA variants are not frequently associated with risk of PD.
We performed burden analyses at two different frequency levels, MAF < 0.01 and MAF < 0.03, on both individual genes and as a gene-set. Meta-analysis of SKAT-O results were not significant for any individual gene or as a gene-set at either frequency level, suggesting that a cumulative effect of rare variation on NBIA genes does not play a role on PD etiology (Fig. 2).
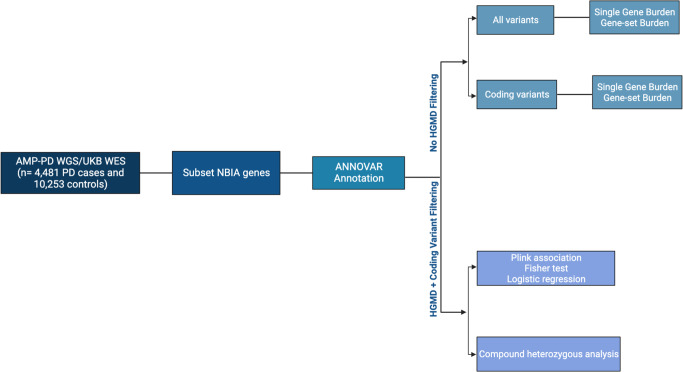
Fig. 2. Overview of methods for association and burden analyses.
Variants in NBIA genes were called in our PD and control cohorts and were then annotated and run through the analyses69.
Transcriptomic analyses revealed that DCAF17 is the only gene that is differentially expressed in blood from PD cases and controls. We further investigated this gene through Summary data-based Mendelian Randomization. One DCAF17 eQTL signal was significant for both GWAS and SMR. The DCAF17 gene is found on chromosome 2, encodes DDB1- and CUL4- associated factor 17, and is considered the cause of Woodhouse-Sakati syndrome, a rare neuroendocrine disorder that presents with neurological problems such as seizures and dystonia. While the exact role of the protein is still unclear, it is postulated that it may affect cellular activities such as growth, proliferation, and stress response specifically in the nucleolus of brain, liver, and skin cells56. It is possible that differing cellular stress responses between PD cases and controls may be leading to DCAF17 differential expression. However, it is still unclear what is driving the significant DCAF17 differential gene expression in our data and further assessments are needed.
This study has some limitations, mostly related to variant detection. Despite using the most well-powered PD datasets, it is nevertheless difficult to detect rare variants as evidenced by the number of variant counts of zero in both cases and controls. It is possible that other rare variants exist in NBIA genes that were not detected in our study. Therefore, we cannot fully elucidate the relationship between rare variants in NBIA genes and PD. In addition, due to a low number of non-European individuals found in the chosen datasets, we were underpowered to properly detect variation in other populations. Several the variants we identified have been reported in the literature in non-European populations, so a study encompassing a greater genetic diversity, with a larger number of non-European individuals, is warranted and would allow us to better characterize the relationship between NBIA and PD etiologies. In addition, as we have mainly focused on coding variants, it is also possible that non-detected variants, such as larger structural variant (SVs) events in NBIA genes, affect PD risk. There is increasing evidence that SVs are likely the main contribution to disease susceptibility and have a substantial effect in coding regions of the genome where they can result in deleterious alterations of the DNA sequence57. Therefore, further study into SVs would be an important step in identifying the relationship between NBIA and PD. Lastly, the cohorts currently available do not allow us to accurately perform a comprehensive clinical characterization of potential atypical symptoms nor are we able to perform clinicogenetic correlations exploring brain iron accumulation on MRI in PD cases with NBIA gene variants, therefore possibly drawing limited conclusions. In addition, as it has been previously suggested elevated nigral iron levels are unlikely to contribute to PD etiology but may vary with anti-parkinsonian drugs used for treatment58. Taking everything into account, we suggest that even though NBIA and PD share similar symptoms, they could be molecularly different entities supporting the notion that the mechanisms underpinning iron accumulation in PD are not shared with NBIA. Elevated nigral iron levels may not contribute to PD etiology and may vary with anti-parkinsonian drugs used for treatment or any other environmental factors.
Methods
Whole-genome sequencing data
Whole-genome sequencing (WGS) data was obtained from the Accelerating Medicines Partnership—Parkinson’s Disease Initiative release 2.5 (AMP-PD; www.amp-pd.org) which contained 3376 PD patients and 4610 healthy unrelated controls of European ancestry, with an average age at onset of 62 years in cases and age at collection of 72 years in controls. Ancestry was determined by principal component analysis versus the 1000Genomes populations and individuals deviating by more than six standard deviations from the European population mean were excluded from the analysis. Diagnosis criteria vary slightly between each subcohort within AMP-PD, but detailed cohort characteristics, as well as quality control procedures, are further described in https://amp-pd.org/whole-genome-data. Informed consent was obtained for all human participants in this cohort. Healthy unrelated controls include individuals with no neurodegenerative disease diagnosis nor any family history of such disease lacking any familial link to other individuals in our study. Data generation is described in detail by Iwaki and colleagues59. NBIA-related variants were extracted from the data by using PLINK1.9 and annotated with ANNOVAR60,61. Coding variants present in the Human Gene Mutation Database (HGMD) went through association analyses for risk of PD using Fisher’s exact test and logistic regression and adjusted by sex, age, and at least 5PCs to account for population substructure.
To assess the cumulative effect of multiple rare variants on the risk for PD, we performed single gene and gene-set burden analyses for all extracted variants under RVTESTS package v2.1.0 default parameters and adjusted by sex, age, and at least 5 PCs to account for population substructure62. Including all variants within the gene boundaries, a minimum allele count (MAC) threshold of 1 was applied. Sequence Kernel Association Test (SKAT-O) was performed considering two variant frequency levels: MAF < 1% and MAF < 3%. Age, sex, and at least 5 PCs, were accounted for as covariates.
Whole-exome sequencing data
Whole-exome sequencing (WES) data was obtained from the UK Biobank 2021 release (UK Biobank; https://www.ukbiobank.ac.uk/), which contained 1105 PD patients and 5643 healthy unrelated controls of European ancestry, with an average age at recruitment of 63 years in cases and 64 years in controls. Ancestry determination follows that of AMP-PD. Disease diagnosis of individuals is determined by self-reports and hospital codes and healthy unrelated controls include individuals with no neurodegenerative disease diagnosis nor any family history of such disease lacking any familial link to other individuals in our study. Detailed cohort characteristics, as well as quality control procedures, are further described in https://www.ukbiobank.ac.uk/enable-your-research/about-our-data/genetic-data. Informed consent was obtained for all human participants in this cohort. Variant extraction, association tests, and burden tests follow the AMP-PD WGS pipeline.
Transcriptomics data
Whole blood time progression gene expression data (gencode v29) from 0 to 24 months after first study visit was accessed from AMP-PD. We used PPMI (https://amp-pd.org/unified-cohorts/ppmi), PDBP (https://amp-pd.org/unified-cohorts/pdbp), and BioFIND (https://amp-pd.org/unified-cohorts/biofind) cohorts at the baseline of the study, 0 month time point (note BioFIND ‘Baseline’ is at ‘0.5’ month), including a total of 1886 cases and 1285 control samples. Expression data were quantified as transcripts per kilobase million (TPM) and quantile normalized before NBIA related genes values were extracted using Ensembl gene ids. Using the scikit-learn Python packages 19, residuals were calculated using linear regression, and the data were adjusted using age, sex, race, at least 5 PCs, cohort, and missingness rate as covariates. To test for significant differences in expression between the cases and controls, we performed a t-test with the residuals. We also used an additional dataset taken from the 24-month time point in the PDBP and PPMI cohorts of 841 cases and 386 controls to confirm our findings.
Summary based Mendelian Randomization
SMR is a mendelian randomization (MR) method that uses summary-level data to test if an exposure variable (i.e. gene expression) and outcome (i.e., trait) are associated because of a shared causal variant. In order to distinguish pleiotropy from linkage, the heterogeneity in dependent instruments (HEIDI) method was applied to each tested single nucleotide variant (SNV)63. SMR and HEIDI analysis were conducted using the SMR package64.
In order to conduct the analysis, we used PD GWAS summary statistics as well as methylation and eQTL meta-analysis summary statistics65,66. PD GWAS summary statistics from Nalls et al.67 contained 17 datasets which consisted of 37,688 PD cases, 18,618 proxy cases, and 1,417,791 controls67. The methylation data we used consists of brain and blood gene expression data from a meta-analysis conducted by Qi et al.65. All eQTL analyses were performed to test for potential in cis association between methylation site and SNV. In order to achieve this, SNVs within 1 Mb of the probes of interest were selected65. The eQTL summary statistic data used were generated from peripheral blood and came from the meta-analysis conducted by Wu et al.68. All of the SNPS from Wu et al. are located within 2 Mb of a probe68.
In order to prioritize SNP candidates, results were initially filtered by chromosome and base pair position for the DCAF17 gene in hg19. Afterwards, the resulting candidates were filtered by the SMR multi p-value at a threshold of p < 0.05.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Data used in the preparation of this article were obtained from Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Against Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). For a complete list of GP2 members see https://gp2.org. The work was carried out with the support and guidance of the ‘GP2 Trainee Network’. This work was supported by the Center for Alzheimer’s and Related Dementias, within the Intramural Research Program of the National Institute on Aging and the National Institute of Neurological Disorders and Stroke, National Institutes of Health, Department of Health and Human Services. Project number 1ZIA AG000534-04. AMP-PD: Data used in the preparation of this article were partly obtained from the Accelerating Medicine Partnership® (AMP®) Parkinson’s Disease (AMP PD) Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. The AMP® PD program is a public-private partnership managed by the Foundation for the National Institutes of Health and funded by the National Institute of Neurological Disorders and Stroke (NINDS) in partnership with the Aligning Science Across Parkinson’s (ASAP) initiative; Celgene Corporation, a subsidiary of Bristol-Myers Squibb Company; GlaxoSmithKline plc (GSK); The Michael J. Fox Foundation for Parkinson’s Research; Pfizer Inc.; Sanofi US Services Inc.; and Verily Life Sciences. ACCELERATING MEDICINES PARTNERSHIP and AMP are registered service marks of the U.S. Department of Health and Human Services. Clinical data and biosamples used in preparation of this article were obtained from the Michael J. Fox Foundation (MJFF) and National Institutes of Neurological Disorders and Stroke (NINDS) BioFIND study, Harvard Biomarkers Study (HBS), the NINDS Parkinson’s disease Biomarkers Program (PDBP) and MJFF Parkinson’s Progression Marker Initiative (PPMI). BioFIND is sponsored by The Michael J. Fox Foundation for Parkinson’s Research (MJFF) with support from the National Institute for Neurological Disorders and Stroke (NINDS). The BioFIND Investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit michaeljfox.org/news/biofind. The Harvard NeuroDiscovery Biomarker Study (HBS) is a collaboration of HBS investigators [full list of HBS investigator found at https://www.bwhparkinsoncenter.org/biobank/ and funded through philanthropy and NIH and Non-NIH funding sources. The HBS Investigators have not participated in reviewing the data analysis or content of the manuscript. PPM—a public-private partnership – is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, a full list of the PPMI funding partners can be found at www.ppmi-info.org/fundingpartners. The PPMI Investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit www.ppmi-info.org. Parkinson’s Disease Biomarker Program (PDBP) consortium is supported by the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. The PDBP Investigators have not participated in reviewing the data analysis or content of the manuscript. UKB: UK Biobank is a large-scale biomedical database and research resource containing genetic, lifestyle and health information from half a million UK participants. UK Biobank’s database, which includes blood samples, heart and brain scans and genetic data of the 500,000 volunteer participants, is globally accessible to approved researchers who are undertaking health-related research that’s in the public interest. UK Biobank recruited 500,000 people aged between 40–69 years in 2006–2010 from across the UK. With their consent, they provided detailed information about their lifestyle, physical measures and had blood, urine and saliva samples collected and stored for future analysis. UK Biobank’s research resource is a major contributor in the advancement of modern medicine and treatment, enabling better understanding of the prevention, diagnosis and treatment of a wide range of serious and life-threatening illnesses—including cancer, heart diseases and stroke. UK Biobank is generously supported by its founding funders the Wellcome Trust and UK Medical Research Council, as well as the Department of Health, Scottish Government, the Northwest Regional Development Agency, British Heart Foundation and Cancer Research UK. The organization has over 150 dedicated members of staff, based in multiple locations across the UK. 23andme: We thank the 23andMe research participants who consented to participate in research for enabling this study. We also thank the employees of 23andMe.
Author contributions
P.A.J., S.B.C., and R.D. designed the study and wrote the manuscript with inputs from all co-authors. P.A.J. analyzed the data and prepared the figures with supervision from S.B.C. C.X.A. performed the SMR analysis. A.M. performed the transcriptomic analysis. J.L.A. aided with initial study design and manuscript editing. M.B.M. provided meta-analysis scripts used and aided in manuscript editing. L.R.M., C.R., F.V.M., N.R.L., and P.S. aided in manuscript writing and editing. Z.G.O., K.J.B., C.B., A.J.N., and A.B.S. provided design supervision and aided in manuscript editing. All authors approved the final version of the manuscript. All authors take accountability for all aspects of the work.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Data availability
Data used for this publication is available through the AMP-PD and UKBiobank websites upon request. Data access is dependent on approval of a Data Usage Agreement. AMP-PD Main Website: www.amp-pd.org; AMP-PD Cohort Information: https://amp-pd.org/whole-genome-data; UKBiobank Main Website: https://www.ukbiobank.ac.uk/; UKBiobabank Cohort Information: https://www.ukbiobank.ac.uk/enable-your-research/about-our-data/genetic-data.
Code availability
Code used in this study can be found at our GitHub page: https://github.com/neurogenetics/NBIA_PD.
Competing interests
C.R. and A.B.S. declare no Competing Non-Financial Interests but the following Competing Financial Interests: C.R. held a predoctoral fellowship (FPU14/03473, MECD, Spain) from the Spanish Ministry of Education and Science. A.B.S. is an editor for npj Parkinson’s Disease. A.B.S. was not involved in the journal’s review of, or decisions related to, this manuscript. P.A.J., J.L.A., L.R.R., A.M., F.V.M., N.R.L., P.S., Z.G.O., C.X.A., M.B.M., K.J.B., C.B., A.J.N., A.B.S., R.D., and S.B.C. declare no competing interests.
Ethical approval
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this work is consistent with those guidelines.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-023-00496-y.
References
- 1.Schneider SA. Neurodegeneration with brain iron accumulation. Curr. Neurol. Neurosci. Rep. 2016;16:9. doi: 10.1007/s11910-015-0608-3. [DOI] [PubMed] [Google Scholar]
- 2.Hogarth P. Neurodegeneration with brain iron accumulation: diagnosis and management. J. Mov. Disord. 2015;8:1–13. doi: 10.14802/jmd.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014;13:1045–1060. doi: 10.1016/S1474-4422(14)70117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li KR, et al. Quantitative evaluation of brain iron accumulation in different stages of Parkinson’s disease. J. Neuroimaging. 2022;32:363–371. doi: 10.1111/jon.12957. [DOI] [PubMed] [Google Scholar]
- 5.Dexter DT, et al. Increased nigral iron content and alterations in other metal ions occurring in brain in Parkinson’s disease. J. Neurochem. 1989;52:1830–1836. doi: 10.1111/j.1471-4159.1989.tb07264.x. [DOI] [PubMed] [Google Scholar]
- 6.Sofic E, Paulus W, Jellinger K, Riederer P, Youdim MB. Selective increase of iron in substantia nigra zona compacta of Parkinsonian brains. J. Neurochem. 1991;56:978–982. doi: 10.1111/j.1471-4159.1991.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 7.Duran R, et al. Oxidative stress and aminopeptidases in Parkinson’s disease patients with and without treatment. Neurodegener. Dis. 2011;8:109–116. doi: 10.1159/000315404. [DOI] [PubMed] [Google Scholar]
- 8.Agil A, et al. Plasma lipid peroxidation in sporadic Parkinson’s disease. Role of the L-dopa. J. Neurol. Sci. 2006;240:31–36. doi: 10.1016/j.jns.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Arber CE, Li A, Houlden H, Wray S. Review: Insights into molecular mechanisms of disease in neurodegeneration with brain iron accumulation: unifying theories. Neuropathol. Appl. Neurobiol. 2016;42:220–241. doi: 10.1111/nan.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Hum. Mol. Genet. 2011;20:R158–R162. doi: 10.1093/hmg/ddr358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidransky E, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochizuki H, Yasuda T. Iron accumulation in Parkinson’s disease. J. Neural Transm. 2012;119:1511–1514. doi: 10.1007/s00702-012-0905-9. [DOI] [PubMed] [Google Scholar]
- 15.Masaldan S, Bush AI, Devos D, Rolland AS, Moreau C. Striking while the iron is hot: Iron metabolism and ferroptosis in neurodegeneration. Free Radic. Biol. Med. 2019;133:221–233. doi: 10.1016/j.freeradbiomed.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Carpenter KLH, et al. Magnetic susceptibility of brain iron is associated with childhood spatial IQ. Neuroimage. 2016;132:167–174. doi: 10.1016/j.neuroimage.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belaidi AA, Bush AI. Iron neurochemistry in Alzheimer’s disease and Parkinson’s disease: targets for therapeutics. J. Neurochem. 2016;139:179–197. doi: 10.1111/jnc.13425. [DOI] [PubMed] [Google Scholar]
- 18.Barbosa JHO, et al. Quantifying brain iron deposition in patients with Parkinson’s disease using quantitative susceptibility mapping, R2 and R2. Magn. Reson. Imaging. 2015;33:559–565. doi: 10.1016/j.mri.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Hinarejos I, Machuca-Arellano C, Sancho P, Espinós C. Mitochondrial dysfunction, oxidative stress and neuroinflammation in neurodegeneration with brain iron accumulation (NBIA) Antioxidants. 2020;9:1020. doi: 10.3390/antiox9101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z-B, et al. Neurodegeneration with brain iron accumulation: Insights into the mitochondria dysregulation. Biomed. Pharmacother. 2019;118:109068. doi: 10.1016/j.biopha.2019.109068. [DOI] [PubMed] [Google Scholar]
- 21.Ramirez A, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 22.Kara E, et al. Genetic and phenotypic characterization of complex hereditary spastic paraplegia. Brain. 2016;139:1904–1918. doi: 10.1093/brain/aww111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malakouti-Nejad M, et al. Identification of p.Gln858* in ATP13A2 in two EOPD patients and presentation of their clinical features. Neurosci. Lett. 2014;577:106–111. doi: 10.1016/j.neulet.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Djarmati A, et al. ATP13A2 variants in early-onset Parkinson’s disease patients and controls. Mov. Disord. 2009;24:2104–2111. doi: 10.1002/mds.22728. [DOI] [PubMed] [Google Scholar]
- 25.Di Fonzo A, et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology. 2007;68:1557–1562. doi: 10.1212/01.wnl.0000260963.08711.08. [DOI] [PubMed] [Google Scholar]
- 26.Podhajska A, et al. Common pathogenic effects of missense mutations in the P-type ATPase ATP13A2 (PARK9) associated with early-onset parkinsonism. PLoS ONE. 2012;7:e39942. doi: 10.1371/journal.pone.0039942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Michele G, et al. Ataxia-myoclonus syndrome due to a novel homozygous ATP13A2 mutation. Parkinsonism Relat. Disord. 2020;76:42–43. doi: 10.1016/j.parkreldis.2020.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Hellman NE, Gitlin JD. Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 2002;22:439–458. doi: 10.1146/annurev.nutr.22.012502.114457. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, et al. Ceruloplasmin in Parkinson’s disease and the nonmotor symptoms. Brain Behav. 2018;8:e00995. doi: 10.1002/brb3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasuhn J, et al. Neuroimaging correlates of substantia nigra hyperechogenicity in Parkinson’s disease. J. Parkinsons. Dis. 2022;12:1191–1200. doi: 10.3233/JPD-213000. [DOI] [PubMed] [Google Scholar]
- 31.Shafieesabet A, et al. Hyperechogenicity of substantia nigra for differential diagnosis of Parkinson’s disease: a meta-analysis. Parkinsonism Relat. Disord. 2017;42:1–11. doi: 10.1016/j.parkreldis.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Hochstrasser H, et al. Ceruloplasmin gene variations and substantia nigra hyperechogenicity in Parkinson disease. Neurology. 2004;63:1912–1917. doi: 10.1212/01.WNL.0000144276.29988.C3. [DOI] [PubMed] [Google Scholar]
- 33.Aydemir ST, Bulut O, Ceylaner S, Akbostancı MC. Aceruloplasminemia presenting with asymmetric chorea due to a novel frameshift mutation. Mov. Disord. Clin. Pr. 2020;7:S67–S70. doi: 10.1002/mdc3.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rattay TW, et al. FAHN/SPG35: a narrow phenotypic spectrum across disease classifications. Brain. 2019;142:1561–1572. doi: 10.1093/brain/awz102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shakya S, et al. Whole exome and targeted gene sequencing to detect pathogenic recessive variants in early onset cerebellar ataxia. Clin. Genet. 2019;96:566–574. doi: 10.1111/cge.13625. [DOI] [PubMed] [Google Scholar]
- 36.Magariello A, et al. Exome sequencing reveals two FA2H mutations in a family with a complicated form of Hereditary Spastic Paraplegia and psychiatric impairments. J. Neurol. Sci. 2017;372:347–349. doi: 10.1016/j.jns.2016.11.069. [DOI] [PubMed] [Google Scholar]
- 37.Hartig M, Prokisch H, Meitinger T, Klopstock T. Mitochondrial membrane protein-associated neurodegeneration (MPAN) Int. Rev. Neurobiol. 2013;110:73–84. doi: 10.1016/B978-0-12-410502-7.00004-1. [DOI] [PubMed] [Google Scholar]
- 38.Hogarth P, et al. New NBIA subtype: genetic, clinical, pathologic, and radiographic features of MPAN. Neurology. 2013;80:268–275. doi: 10.1212/WNL.0b013e31827e07be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory A, et al. Autosomal dominant mitochondrial membrane protein-associated neurodegeneration (MPAN) Mol. Genet. Genom. Med. 2019;7:e00736. doi: 10.1002/mgg3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartig MB, et al. Absence of an orphan mitochondrial protein, c19orf12, causes a distinct clinical subtype of neurodegeneration with brain iron accumulation. Am. J. Hum. Genet. 2011;89:543–550. doi: 10.1016/j.ajhg.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tschentscher A, et al. Analysis of the C19orf12 and WDR45 genes in patients with neurodegeneration with brain iron accumulation. J. Neurol. Sci. 2015;349:105–109. doi: 10.1016/j.jns.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Akçakaya NH, et al. Clinical and genetic spectrum of an orphan disease MPAN: a series with new variants and a novel phenotype. Neurol. Neurochir. Pol. 2019;53:476–483. doi: 10.5603/PJNNS.a2019.0062. [DOI] [PubMed] [Google Scholar]
- 43.Cozzi A, et al. Human L-ferritin deficiency is characterized by idiopathic generalized seizures and atypical restless leg syndrome. J. Exp. Med. 2013;210:1779–1791. doi: 10.1084/jem.20130315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svetel M, et al. NBIA syndromes: a step forward from the previous knowledge. Neurol. India. 2021;69:1380–1388. doi: 10.4103/0028-3886.329603. [DOI] [PubMed] [Google Scholar]
- 45.Hayflick SJ, Kurian MA, Hogarth P. Neurodegeneration with brain iron accumulation. Handb. Clin. Neurol. 2018;147:293–305. doi: 10.1016/B978-0-444-63233-3.00019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang T-W, et al. Partial deficit of pantothenate kinase 2 catalytic activity in a case of tremor-predominant neurodegeneration with brain iron accumulation. Mov. Disord. 2006;21:718–722. doi: 10.1002/mds.20797. [DOI] [PubMed] [Google Scholar]
- 47.DaRe JT, Vasta V, Penn J, Tran N-TB, Hahn SH. Targeted exome sequencing for mitochondrial disorders reveals high genetic heterogeneity. BMC Med. Genet. 2013;14:118. doi: 10.1186/1471-2350-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeksan M, Türk S, Polat M, Ciğli A, Erdoğan Y. Effects of 1,25 (OH)2D3 treatment on lipid levels in uremic hemodialysis patients. Int. J. Artif. Organs. 1992;15:704–707. doi: 10.1177/039139889201501204. [DOI] [PubMed] [Google Scholar]
- 49.Ghani M, et al. Mutation analysis of patients with neurodegenerative disorders using NeuroX array. Neurobiol. Aging. 2015;36:545.e9–14. doi: 10.1016/j.neurobiolaging.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daida K, et al. PLA2G6 variants associated with the number of affected alleles in Parkinson’s disease in Japan. Neurobiol. Aging. 2021;97:147.e1–147.e9. doi: 10.1016/j.neurobiolaging.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Morgan NV, et al. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat. Genet. 2006;38:752–754. doi: 10.1038/ng1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karaca E, et al. Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khateeb S, et al. PLA2G6 mutation underlies infantile neuroaxonal dystrophy. Am. J. Hum. Genet. 2006;79:942–948. doi: 10.1086/508572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iannello G, et al. A new PLA2G6 mutation in a family with infantile neuroaxonal dystrophy. J. Neurol. Sci. 2017;381:209–212. doi: 10.1016/j.jns.2017.08.3260. [DOI] [PubMed] [Google Scholar]
- 55.Arslan EA, Öncel İ, Ceylan AC, Topçu M, Topaloğlu H. Genetic and phenotypic features of patients with childhood ataxias diagnosed by next-generation sequencing gene panel. Brain Dev. 2020;42:6–18. doi: 10.1016/j.braindev.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Fozia F, et al. Novel splicing-site mutation in DCAF17 gene causing Woodhouse-Sakati syndrome in a large consanguineous family. J. Clin. Lab. Anal. 2022;36:e24127. doi: 10.1002/jcla.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pittman A, Hardy J. Genetic analysis in neurology: the next 10 years. JAMA Neurol. 2013;70:696–702. doi: 10.1001/jamaneurol.2013.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du G, et al. Dynamics of nigral iron accumulation in Parkinson’s disease: from diagnosis to late stage. Mov. Disord. 2022;37:1654–1662. doi: 10.1002/mds.29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwaki H, et al. Accelerating medicines partnership: Parkinson’s disease. genetic resource. Mov. Disord. 2021;36:1795–1804. doi: 10.1002/mds.28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience4 (2015). [DOI] [PMC free article] [PubMed]
- 61.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 64.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 65.Qi T, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 2018;9:2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McRae AF, et al. Identification of 55,000 replicated DNA methylation QTL. Sci. Rep. 2018;8:17605. doi: 10.1038/s41598-018-35871-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nalls MA, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 2018;9:918. doi: 10.1038/s41467-018-03371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adapted from “Flowchart 5”, by BioRender.com. Retrieved from https://app.biorender.com/biorender-templates (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for this publication is available through the AMP-PD and UKBiobank websites upon request. Data access is dependent on approval of a Data Usage Agreement. AMP-PD Main Website: www.amp-pd.org; AMP-PD Cohort Information: https://amp-pd.org/whole-genome-data; UKBiobank Main Website: https://www.ukbiobank.ac.uk/; UKBiobabank Cohort Information: https://www.ukbiobank.ac.uk/enable-your-research/about-our-data/genetic-data.
Code used in this study can be found at our GitHub page: https://github.com/neurogenetics/NBIA_PD.