Abstract
The structural complexity of glycans makes their characterization challenging, not only because of the presence of various isomeric forms of the precursor molecule but also because the fragments can themselves be isomeric. We have recently developed an IMS-CID-IMS approach using structures for lossless ion manipulations (SLIM) combined with cryogenic infrared (IR) spectroscopy for glycan analysis. It allows mobility separation and collision-induced dissociation of a precursor glycan followed by mobility separation and IR spectroscopy of the fragments. While this approach holds great promise for glycan analysis, we often encounter fragments for which we have no standards to identify their spectroscopic fingerprint. In this work, we perform proof-of-principle experiments employing a multistage SLIM-based IMS-CID technique to generate second-generation fragments, followed by their mobility separation and spectroscopic interrogation. This approach provides detailed structural information about the first-generation fragments, including their anomeric form, which in turn can be used to identify the precursor glycan.
Introduction
Glycans adorn the surfaces of virtually all cells and serve numerous biological functions, including cell-to-cell recognition, protein folding, and immune response.1,2 Despite their importance, their inherent isomeric complexity makes their characterization extremely challenging using mass spectrometry (MS) alone. Tandem mass spectrometry (MSn) can reveal information about linkage position and branching, but differentiation between all isomeric forms, especially the anomericity of the glycosidic linkages, remains challenging.3−7
Using MSn in conjunction with different separation techniques, such as liquid chromatography (LC) and capillary electrophoresis (CE), can be a powerful means to identify glycan isomers.8−14 Nevertheless, these methods often require derivatization steps and lengthy run times. Ion mobility spectrometry (IMS), which can separate gas-phase ions on a millisecond time scale based on their average collisional cross-section, has shown great promise for distinguishing glycan isomers,15,16 particularly when combined with MSn.16−28 While this combination has proven to be effective, isomeric glycans frequently produce isomeric fragments, which require additional IMS separation. There is a need for instruments that can perform IMSn experiments in which ions can be fragmented between consecutive IMS rounds. Toward this end, Li et al. developed a tandem IMS instrument to study a mixture of pentasaccharides and revealed isomeric heterogeneity of its fragments.29 Clemmer and co-workers developed an IMS-IMS analogue to MS-MS that allows mobility separation of ions prior to collisional activation inside a drift tube followed by mobility-separation of fragment ions in a second IMS region with the possibility of additional collisional activation before MS analysis.30 They applied it to distinguish a set of glycan isomers that differ by linkage anomericity, position, or both.31 Giles et al. have developed a high-resolution traveling wave-based cyclic IMS (cIMS) instrument in which mobility separated ions can undergo a series of selection, collision-induced dissociation (CID), and separation steps several times before MS detection,32 and this platform has been used for structural characterization of isomeric glycans.33−36
Over the past few years, it has become clear that gas-phase IR spectroscopy, particularly at cryogenic temperatures, can distinguish isomeric glycans with the subtlest of structural differences.37 We have recently demonstrated that a combination of high-resolution IMS based on structures for lossless ion manipulations (SLIM)38,39 with cryogenic IR spectroscopy can be used for unambiguous identification of glycan isomers via their IR fingerprint.40−45 We have also developed a SLIM-based IMS-CID-IMS-IR approach, which allows fragments of mobility-separated precursor ions to undergo further isomer separation before spectroscopic interrogation,46 and applied it to identify positional isomers and anomers of glycans.47−49 We illustrated how this approach can be used to expand an IR fingerprint database without heavy reliance on isomerically pure glycan standards. While this spectroscopic database approach holds great potential, we still encounter fragments for which the IR fingerprints are not contained in the database.
To solve this problem, we have developed a SLIM-based multistage IMS-CID technique combined with cryogenic IR spectroscopy. Using this approach, we can generate second-generation fragments from mobility-selected first-generation fragments, separate them once again by their mobility, and then measure their IR fingerprint. These mobility-selected second-generation fragments are then identified by comparing their IR fingerprints to reference spectra from our spectroscopic database. This allows us to identify the first-generation fragments that are not contained in the database, which in turn facilitates reconstruction of the precursor structure. After performing this procedure, our spectroscopic database can be expanded by adding the IR fingerprints of both the first-generation fragments and the precursor structures. This procedure only has to be done once per unknown species, since in subsequent analyses they can be detected directly by their IR fingerprint.
We demonstrate the proof-of-principle of this novel approach using human milk oligosaccharides (HMOs) as examples. While the structures of HMOs used in this work can be determined using tandem mass spectrometry, they serve as a good illustration of how this approach works.
Experimental Methods
Experiments were performed on a newly developed, home-built instrument, described in detail elsewhere,50 which couples traveling-wave ion mobility spectrometry (TW-IMS) using SLIM38,39 with CID and messenger-tagging spectroscopy. Briefly, ions generated by nanoelectrospray ionization (nESI) are guided by a dual-stage ion funnel assembly into an ion mobility module. In the SLIM-IMS device, the TW potentials are generated between electrodes on two parallel printed circuit boards to propel the ions through N2 drift gas maintained at a pressure of 2.2 mbar. Upon entering the SLIM module, the ions are accumulated in a 2 m long storage section51 and then released as packets into the separation section. Our SLIM device features a single-pass path length of 10 m, achieving a resolving power of ∼200, with the possibility to cycle the ions through the separation device multiple times to achieve higher resolution.50 The mobility-separated ions are then guided through differentially pumped stages into a cryogenic, planar trap maintained at a temperature of 45 K. Collisions with the cold buffer gas (He/N2, 80/20 mixture) lead to cooling and subsequent attachment of N2, which serves as a “messenger tag” to detect IR absorption.52,53 The vibrational spectrum is obtained by irradiating the N2-tagged ions with a continuous-wave mid-IR laser (IPG Photonics) operated at 0.5–1 W output power with a line width of ∼1 cm–1. When the laser wavenumber is in resonance with a vibrational band of the tagged ion, absorption of an infrared photon causes the N2 tag to boil off, which is detected by a commercial time-of-flight mass spectrometer (Tofwerk). We obtain the IR spectrum of the species by plotting the ratio of the tagged ion signal to the sum of the tagged and untagged ion signal as a function of laser wavenumber. All ions were studied in their singly sodiated charge state.
IMS3–IR Experiments
Our SLIM module features on-board traps that can be used for performing CID.46 We make use of two metallic grids placed at the entrance of the trap and separated by a distance of 0.8 mm to achieve a homogeneous electric field of up to 3300 V/cm for efficient CID.47,48,50 The (IMS)3 experiments consist of several steps. First, the precursor ions are separated by their mobility on the 10 m serpentine SLIM track. Then, ions of a certain mobility are diverted into the CID trap, which is held at a lower potential bias than the separation region. They are accelerated as they pass through the grids placed at the trap entrance and undergo fragmentation upon energetic collisions with the drift gas. The bias potential of the trap is then raised to the level of the separation region to allow both the precursor and first-generation fragment ions to exit the trap. The isomeric first-generation fragments are made to undergo additional separation cycle(s) on the SLIM device, during which time they are diverted once again into the on-board CID trap and fragmented further. The resulting second-generation fragments are then released from the trap and either exit the SLIM device directly or first undergo additional mobility-separation before being directed to the cryogenic ion trap, where we record their IR spectrum.
Materials
The glycans lacto-N-difucohexaose II (LNDFH II), lacto-N-tetraose (LNT), Lewis-a trisaccharide, and 3-fucosyllactose (3-FL) were purchased from Dextra Laboratories (UK). All glycans were used without further purification. The samples were prepared in a 50/50 solution of water/methanol to yield a concentration of 5–15 μM. All solvents used were HPLC grade. The experiments were performed using N2 as both a drift gas for ion mobility and a collision gas for CID.
Results and Discussion
Fragmentation of Mobility-Selected First-Generation Fragments
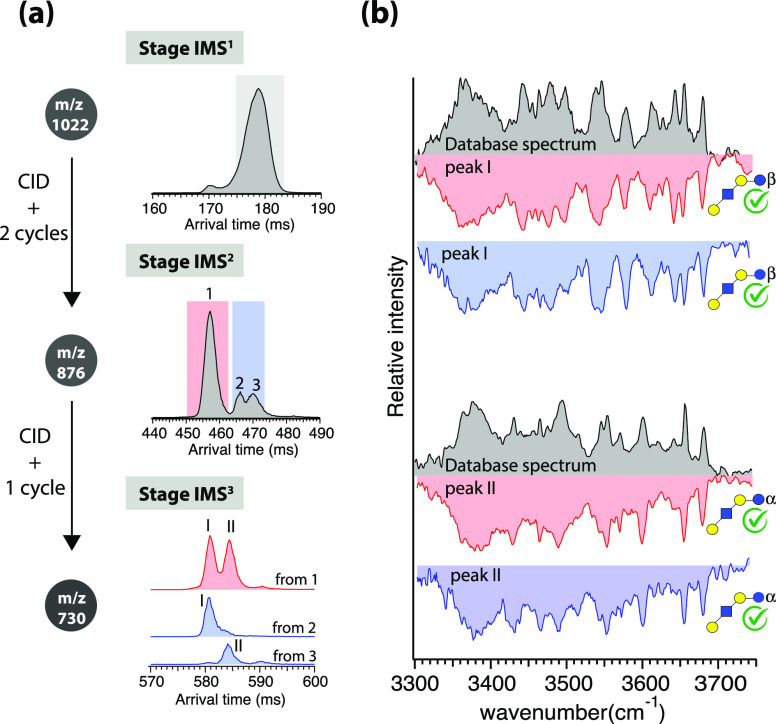
The first-generation fragment from LNDFH II at m/z 876 can be generated by the loss of fucose at the reducing end, producing Y1β, or the nonreducing end, yielding Y3α’’ (see Figure 1c for nomenclature). These fragments have the same composition but differ in the position of the remaining fucose. The ATD of the m/z 876 fragment after two separation cycles is displayed in Figure 1a (stage IMS2), which exhibits three drift peaks. To acquire further structural information, these first-generation fragments were mobility-selected, retrapped, and fragmented once again. The resulting mass spectra (Figure 1b) from the first ATD peak (red) and the second and third peaks together (blue) reveal a major difference in the appearance of the m/z 552 and 511 fragments. These differences indicate the presence of structural isomers of m/z 876, which could be characterized using these diagnostic fragments. The structure of the m/z 730 fragment, which results from the loss of two fucose residues from the precursor glycan, is common to all drift peaks and corresponds to the commercially available standard known as LNT.
Figure 1.
(a) ATD of singly sodiated LNDFH II after one separation cycle (stage IMS1) and that of the first-generation fragment (m/z 876) generated from the major peak of LNDFH II after two separation cycles (stage IMS2). (b) Mass spectrum generated by fragmentation of different drift peaks (red and blue) in the ATD of the m/z 876 fragment. (c) The nomenclature of glycan fragments, following that of Domon and Costello.54
These first results demonstrate the ability of our SLIM device to perform tandem IMS experiments. As described below, the IR fingerprints of the m/z 552, m/z 511, and mobility-selected m/z 730 s-generation fragments can provide detailed structural information about the first-generation m/z 876 fragment, including its anomericity.
Identification of Isomeric First-Generation Fragments Using IR Fingerprints of Second-Generation Fragments
While different methods have been used for identification of structural isomers of HMOs,26,55−57 we use IR fingerprints of the structurally diagnostic CID fragments. The second-generation m/z 511 fragment of LNDFH II has the same structure as 3-FL, which is commercially available. Since there are no other isomeric fragments with m/z 511 generated from LNDFHII, we recorded its IR spectrum without further mobility separation. Figure 2 compares the vibrational spectrum (blue) of the m/z 511 fragment with that of a 3-FL standard (gray). The good agreement in both the position and intensities of all vibrational bands confirms that the m/z 511 fragment is indeed 3-fucosyllactose. Given this, the m/z 876 first-generation fragment contained in peaks 2 and 3 of the arrival time distribution (blue shaded in left panel of Figure 2) must correspond to Y3α’’ produced from LNDFH II.
Figure 2.
Comparison of cryogenic IR spectra of second-generation fragments m/z 511 and m/z 552 generated with corresponding spectra from the database.
Assuming the dissociation of a single covalent bond under the CID conditions applied, the second-generation m/z 552 fragment, C2α, would have the same structure as that of a glycan standard known as the trisaccharide Lewis-a. Figure 2 shows the comparison of the IR fingerprint spectrum of the m/z 552 fragment (red) with that of a commercial Lewis-a standard (gray). While the spectra have strong similarities, the match is not perfect–the spectrum of Lewis-a displays additional bands at 3458, 3477, 3563, and 3645 cm–1. It is important to note that assuming the anomericity of the C-fragment is retained upon CID, as we and others have shown to be the case,46,58−60 the C2α fragment spectrum displayed in Figure 2 (red) would correspond to only the β anomer of the Lewis-a trisaccharide standard. Since the Lewis-a standard is a mixture of both α and β reducing-end anomers, the additional peaks in its IR spectrum compared to that of the C2α fragment are likely to come from the α reducing-end anomer. Data confirming this conclusion is presented in the Supporting Information.
Having spectroscopically identified the structure of the m/z 552 fragment, we can determine that the first drift peak in the ATD of the m/z 876 fragment to Y1β produced from LNDFH II, where the fucose at the reducing end is lost (see Figure 1c).
Identification of the Anomericity of First-Generation Fragments Using IR-Fingerprinting of Mobility-Selected Second-Generation Fragments
We now demonstrate how we can use the second-generation fragment with m/z 730 to determine the reducing-end anomericity of the m/z 876 fragments. Because we know the structure of LNDFH II, after the removal of two fucose units, we know that the structure of the m/z 730 fragment will be the same as that of a human milk oligosaccharide LNT, which is commercially available. The ATDs of the m/z 730 fragments generated from each drift peak of the m/z 876 fragment are displayed in Figure 3a. The ATD of m/z 730 fragments produced from the first peak of the m/z 876 fragment show two distinct drift peaks, suggesting the presence of two reducing-end anomers. Those generated from the second and third peaks yield a single mobility feature with slightly different arrival times, which suggests the presence of a single anomer.
Figure 3.
(a) ATDs of LNDFH II after one separation cycle (stage IMS1), m/z 876 first-generation fragment after two separation cycles (stage IMS2) generated from the major peak of LNDFH II, and m/z 730 s-generation fragment after one separation cycle (IMS3) generated from each drift peak of m/z 876 fragment. (b) Comparison of the cryogenic IR spectrum of mobility-separated second-generation m/z 730 fragment from peak 1 (red) and peaks 2 and 3 (blue) of m/z 876 fragment with those of α and β reducing-end anomers of LNT (gray) from the database.
While the arrival times suggest that the m/z 730 fragments generated from the second and third peaks of m/z 876 have structures identical to those generated from the first peak, this information alone is not sufficient to determine their reducing-end anomericity. We therefore recorded cryogenic IR fingerprints of each drift peak of the m/z 730 fragment separately. Figure 3b shows the comparison of these spectra (displayed in color) with previously recorded IR fingerprints for the α and β reducing-end anomers of LNT (displayed in gray)50 (see the Supporting Information). The IR fingerprints of the first and second drift peaks of the m/z 730 fragment generated from first mobility peak of m/z 876 provide a positive match for β and α reducing-end anomers, respectively. The IR fingerprints of those generated from the second and third mobility peaks of the m/z 876 fragment also correspond to the β and α reducing-end anomers of LNT, respectively.
Since the reducing-end anomericity of Y-fragments does not change upon CID,48,49 the first peak corresponds to a mixture of both α and β anomers of the Y1β fragment in the ATD of the m/z 876 fragment generated from LNDFH II. The second and third drift peaks belong to the β and α anomers of the Y3α’’ fragment from LNDFH II, respectively. The assignment of the anomericity of the first-generation fragments was only possible using the spectroscopic dimension.
Conclusions
In this work, we have demonstrated that (IMS)3-IR experiments allow us to completely elucidate the structure of first-generation fragment ions by fragmenting them further, followed by mobility-separation and spectroscopic interrogation of the second-generation fragments. We were able to not only distinguish between isomeric first-generation fragments but also identify their anomericity. Since our IR fingerprint database approach relies on identifying the fragments by comparing them to a previously recorded database, this method is particularly useful when we do not encounter a database match for the first-generation fragments. The (IMS)3-IR approach serves two purposes; it enables reconstruction of the unknown precursor glycan structure and the extension of our IR database with entries of newly identified structures.
These first results demonstrate application of SLIM-based (IMS)3 combined with cryogenic IR spectroscopy for glycan analysis. We envision these results as proof-of-principle for building a general (IMS)n approach, where mobility-separated ions can be retrapped, fragmented, and mobility-separated several times before spectroscopic interrogation until a positive database match is found. Such an approach should facilitate rapid structural identification of unknown glycans in their precise isomeric form.
Acknowledgments
The authors thank the Swiss National Science Foundation (Grants 200020_184838 and 206021_177004), the European Research Council (Grant 788697-GLYCANAL), and the EPFL for the financial support of this work.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.2c00361.
Identification of second-generation fragment with m/z 552 generated from LNDFHII and assignment of α and β reducing-end anomers of LNT (PDF)
Author Contributions
P.B. and T.R.R. designed the project; A.B.F. and S.W. designed and constructed the instrument. P.B. performed the experiments. P.B. wrote the original draft, and A.B.F., S.W., and T.R.R. were involved in editing it. T.R.R. supervised the project and acquired the funding.
The authors declare no competing financial interest.
Special Issue
Published as part of the Journal of the American Society for Mass Spectrometryvirtual special issue “Focus: Ion Chemistry and Electrospray Ionization”.
Supplementary Material
References
- Varki A. Biological roles of glycans. Glycobiology 2017, 27 (1), 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R. D.; Pierce J. M. The Challenge and Promise of Glycomics. Chem. Biol. 2014, 21 (1), 1–15. 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H. J.; Lebrilla C. B. Structure Elucidation of Native N- and O-Linked Glycans by Tandem Mass Spectrometry (Tutorial). Mass Spectrom. Rev. 2011, 30 (4), 560–578. 10.1002/mas.20283. [DOI] [PubMed] [Google Scholar]
- Chai W. G.; Piskarev V.; Lawson A. M. Branching pattern and sequence analysis of underivatized oligosaccharides by combined MS/MS of singly and doubly charged molecular ions in negative-ion electrospray mass spectrometry. J. Am. Soc. Mass. Spectrom. 2002, 13 (6), 670–679. 10.1016/S1044-0305(02)00363-X. [DOI] [PubMed] [Google Scholar]
- Harvey D. J. Collision-induced fragmentation of underivatized N-linked carbohydrates ionized by electrospray. J. Mass Spectrom. 2000, 35 (10), 1178–1190. . [DOI] [PubMed] [Google Scholar]
- Mechref Y.; Novotny M. V.; Krishnan C. Structural characterization of oligosaccharides using MALDI-TOF/TOF tandem mass spectrometry. Anal. Chem. 2003, 75 (18), 4895–4903. 10.1021/ac0341968. [DOI] [PubMed] [Google Scholar]
- Harvey D. J. Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass. Spectrom. 2005, 16 (5), 647–659. 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Ashwood C.; Lin C. H.; Thaysen-Andersen M.; Packer N. H. Discrimination of Isomers of Released N- and O-Glycans Using Diagnostic Product Ions in Negative Ion PGC-LC-ESI-MS/MS. J. Am. Soc. Mass. Spectrom. 2018, 29 (6), 1194–1209. 10.1007/s13361-018-1932-z. [DOI] [PubMed] [Google Scholar]
- Mechref Y.; Novotny M. V. Glycomic Analysis by Capillary Electrophoresis-Mass Spectrometry. Mass Spectrom. Rev. 2009, 28 (2), 207–222. 10.1002/mas.20196. [DOI] [PubMed] [Google Scholar]
- She Y. M.; Tam R. Y.; Li X. G.; Rosu-Myles M.; Sauve S. Resolving Isomeric Structures of Native Glycans by Nanoflow Porous Graphitized Carbon Chromatography-Mass Spectrometry. Anal. Chem. 2020, 92 (20), 14038–14046. 10.1021/acs.analchem.0c02951. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Wei J.; Costello C. E.; Lin C. Characterization of Isomeric Glycans by Reversed Phase Liquid Chromatography-Electronic Excitation Dissociation Tandem Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2018, 29 (6), 1295–1307. 10.1007/s13361-018-1943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.; Tang Y.; Bai Y.; Zaia J.; Costello C. E.; Hong P. Y.; Lin C. Toward Automatic and Comprehensive Glycan Characterization by Online PGC-LC-EED MS/MS. Anal. Chem. 2020, 92 (1), 782–791. 10.1021/acs.analchem.9b03183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Li S. Y.; Li C.; Wu S. L.; Xu W.; Chen Y. T.; Shameem M.; Richardson D.; Li H. J. Identification of Low Abundant Isomeric N-Glycan Structures in Biological Therapeutics by LC/MS. Anal. Chem. 2016, 88 (14), 7049–7059. 10.1021/acs.analchem.6b00636. [DOI] [PubMed] [Google Scholar]
- Zhou S. Y.; Huang Y. F.; Dong X.; Peng W. J.; Veillon L.; Kitagawa D. A. S.; Aquino A. J. A.; Mechref Y. Isomeric Separation of Permethylated Glycans by Porous Graphitic Carbon (PGC)-LC-MS/MS at High Temperatures. Anal. Chem. 2017, 89 (12), 6590–6597. 10.1021/acs.analchem.7b00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J.; Pagel K. Glycan Analysis by Ion Mobility-Mass Spectrometry. Angew. Chem., Int. Ed. 2017, 56 (29), 8342–8349. 10.1002/anie.201701309. [DOI] [PubMed] [Google Scholar]
- Gray C. J.; Thomas B.; Upton R.; Migas L. G.; Eyers C. E.; Barran P. E.; Flitsch S. L. Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Bba-Gen Subjects 2016, 1860 (8), 1688–1709. 10.1016/j.bbagen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Zhu F. F.; Lee S.; Valentine S. J.; Reilly J. P.; Clemmer D. E. Mannose7 Glycan Isomer Characterization by IMS-MS/MS Analysis. J. Am. Soc. Mass. Spectrom. 2012, 23 (12), 2158–2166. 10.1007/s13361-012-0491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J.; Stuckmann A.; Crispin M.; Harvey D. J.; Pagel K.; Struwe W. B. Identification of Lewis and Blood Group Carbohydrate Epitopes by Ion Mobility-Tandem-Mass Spectrometry Fingerprinting. Anal. Chem. 2017, 89 (4), 2318–2325. 10.1021/acs.analchem.6b03853. [DOI] [PubMed] [Google Scholar]
- Huang Y. T.; Dodds E. D. Discrimination of Isomeric Carbohydrates as the Electron Transfer Products of Group II Cation Adducts by Ion Mobility Spectrometry and Tandem Mass Spectrometry. Anal. Chem. 2015, 87 (11), 5664–5668. 10.1021/acs.analchem.5b00759. [DOI] [PubMed] [Google Scholar]
- Morrison K. A.; Clowers B. H. Differential Fragmentation of Mobility-Selected Glycans via Ultraviolet Photodissociation and Ion Mobility-Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2017, 28 (6), 1236–1241. 10.1007/s13361-017-1621-3. [DOI] [PubMed] [Google Scholar]
- Lee S.; Valentine S. J.; Reilly J. P.; Clemmer D. E. Analyzing a mixture of disaccharides by IMS-VUVPD-MS. Int. J. Mass spectrom. 2012, 309, 161–167. 10.1016/j.ijms.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both P.; Green A. P.; Gray C. J.; Sardzik R.; Voglmeir J.; Fontana C.; Austeri M.; Rejzek M.; Richardson D.; Field R. A.; Widmalm G.; Flitsch S. L.; Eyers C. E. Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat. Chem. 2014, 6 (1), 65–74. 10.1038/nchem.1817. [DOI] [PubMed] [Google Scholar]
- Harvey D. J.; Scarff C. A.; Edgeworth M.; Pagel K.; Thalassinos K.; Struwe W. B.; Crispin M.; Scrivens J. H. Travelling-wave ion mobility mass spectrometry and negative ion fragmentation of hybrid and complex N-glycans. J. Mass Spectrom. 2016, 51 (11), 1064–1079. 10.1002/jms.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey D. J.; Seabright G. E.; Vasiljevic S.; Crispin M.; Struwe W. B. Isomer Information from Ion Mobility Separation of High-Mannose Glycan Fragments. J. Am. Soc. Mass. Spectrom. 2018, 29 (5), 972–988. 10.1007/s13361-018-1890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y.; Ridgeway M. E.; Glaskin R. S.; Park M. A.; Costello C. E.; Lin C. Separation and Identification of Isomeric Glycans by Selected Accumulation-Trapped Ion Mobility Spectrometry-Electron Activated Dissociation Tandem Mass Spectrometry. Anal. Chem. 2016, 88 (7), 3440–3443. 10.1021/acs.analchem.6b00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.; Tang Y.; Ridgeway M. E.; Park M. A.; Costello C. E.; Lin C. Accurate Identification of Isomeric Glycans by Trapped Ion Mobility Spectrometry-Electronic Excitation Dissociation Tandem Mass Spectrometry. Anal. Chem. 2020, 92 (19), 13211–13220. 10.1021/acs.analchem.0c02374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Bendiak B.; Siems W. F.; Gang D. R.; Hill H. H. Jr. Determining the isomeric heterogeneity of neutral oligosaccharide-alditols of bovine submaxillary mucin using negative ion traveling wave ion mobility mass spectrometry. Anal. Chem. 2015, 87 (4), 2228–35. 10.1021/ac503754k. [DOI] [PubMed] [Google Scholar]
- Zhu M. L.; Bendiak B.; Clowers B.; Hill H. H. Ion mobility-mass spectrometry analysis of isomeric carbohydrate precursor ions. Anal. Bioanal. Chem. 2009, 394 (7), 1853–1867. 10.1007/s00216-009-2865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. L.; Bendiak B.; Siems W. F.; Gang D. R.; Hill H. H. Carbohydrate Structure Characterization by Tandem Ion Mobility Mass Spectrometry (IMMS)(2). Anal. Chem. 2013, 85 (5), 2760–2769. 10.1021/ac303273z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeniger S. L.; Merenbloom S. I.; Valentine S. J.; Jarrold M. F.; Udseth H. R.; Smith R. D.; Clemmer D. E. An IMS-IMS analogue of MS-MS. Anal. Chem. 2006, 78 (12), 4161–4174. 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- Gaye M. M.; Kurulugama R.; Clemmer D. E. Investigating carbohydrate isomers by IMS-CID-IMS-MS: precursor and fragment ion cross-sections. Analyst 2015, 140 (20), 6922–6932. 10.1039/C5AN00840A. [DOI] [PubMed] [Google Scholar]
- Giles K.; Ujma J.; Wildgoose J.; Pringle S.; Richardson K.; Langridge D.; Green M. A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 2019, 91 (13), 8564–8573. 10.1021/acs.analchem.9b01838. [DOI] [PubMed] [Google Scholar]
- Ujma J.; Ropartz D.; Giles K.; Richardson K.; Langridge D.; Wildgoose J.; Green M.; Pringle S. Cyclic Ion Mobility Mass Spectrometry Distinguishes Anomers and Open-Ring Forms of Pentasaccharides. J. Am. Soc. Mass. Spectrom. 2019, 30 (6), 1028–1037. 10.1007/s13361-019-02168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. L.; Nagy G. Toward Sequencing the Human Milk Glycome: High-Resolution Cyclic Ion Mobility Separations of Core Human Milk Oligosaccharide Building Blocks. Anal. Chem. 2021, 93 (27), 9397–9407. 10.1021/acs.analchem.1c00942. [DOI] [PubMed] [Google Scholar]
- Ollivier S.; Tarquis L.; Fanuel M.; Li A.; Durand J.; Laville E.; Potocki-Veronese G.; Ropartz D.; Rogniaux H. Anomeric Retention of Carbohydrates in Multistage Cyclic Ion Mobility (IMSn): De Novo Structural Elucidation of Enzymatically Produced Mannosides. Anal. Chem. 2021, 93 (15), 6254–6261. 10.1021/acs.analchem.1c00673. [DOI] [PubMed] [Google Scholar]
- Oganesyan I.; Hajduk J.; Harrison J. A.; Marchand A.; Czar M. F.; Zenobi R. Exploring Gas-Phase MS Methodologies for Structural Elucidation of Branched N-Glycan Isomers. Anal. Chem. 2022, 94 (29), 10531–10539. 10.1021/acs.analchem.2c02019. [DOI] [PubMed] [Google Scholar]
- Greis K.; Kirschbaum C.; von Helden G.; Pagel K. Gas-phase infrared spectroscopy of glycans and glycoconjugates. Curr. Opin. Struct. Biol. 2022, 72, 194–202. 10.1016/j.sbi.2021.11.006. [DOI] [PubMed] [Google Scholar]
- Deng L. L.; Webb I. K.; Garimella S. V. B.; Hamid A. M.; Zheng X. Y.; Norheim R. V.; Prost S. A.; Anderson G. A.; Sandoval J. A.; Baker E. S.; Ibrahim Y. M.; Smith R. D. Serpentine Ultralong Path with Extended Routing (SUPER) High Resolution Traveling Wave Ion Mobility-MS using Structures for Lossless Ion Manipulations. Anal. Chem. 2017, 89 (8), 4628–4634. 10.1021/acs.analchem.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid A. M.; Garimella S. V. B.; Ibrahim Y. M.; Deng L. L.; Zheng X. Y.; Webb I. K.; Anderson G. A.; Prost S. A.; Norheim R. V.; Tolmachev A. V.; Baker E. S.; Smith R. D. Achieving High Resolution Ion Mobility Separations Using Traveling Waves in Compact Multiturn Structures for Lossless Ion Manipulations. Anal. Chem. 2016, 88 (18), 8949–8956. 10.1021/acs.analchem.6b01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abikhodr A. H.; Yatsyna V.; Ben Faleh A.; Warnke S.; Rizzo T. R. Identifying Mixtures of Isomeric Human Milk Oligosaccharides by the Decomposition of IR Spectral Fingerprints. Anal. Chem. 2021, 93 (44), 14730–14736. 10.1021/acs.analchem.1c03190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Faleh A.; Warnke S.; Rizzo T. R. Combining Ultrahigh-Resolution Ion-Mobility Spectrometry with Cryogenic Infrared Spectroscopy for the Analysis of Glycan Mixtures. Anal. Chem. 2019, 91 (7), 4876–4882. 10.1021/acs.analchem.9b00659. [DOI] [PubMed] [Google Scholar]
- Dyukova I.; Ben Faleh A.; Warnke S.; Yalovenko N.; Yatsyna V.; Bansal P.; Rizzo T. R. A new approach for identifying positional isomers of glycans cleaved from monoclonal antibodies. Analyst 2021, 146 (15), 4789–4795. 10.1039/D1AN00780G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnke S.; Ben Faleh A.; Pellegrinelli R. P.; Yalovenko N.; Rizzo T. R. Combining ultra-high resolution ion mobility spectrometry with cryogenic IR spectroscopy for the study of biomolecular ions. Faraday Discuss. 2019, 217, 114–125. 10.1039/C8FD00180D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnke S.; Ben Faleh A.; Scutelnic V.; Rizzo T. R. Separation and Identification of Glycan Anomers Using Ultrahigh-Resolution Ion-Mobility Spectrometry and Cryogenic Ion Spectroscopy. J. Am. Soc. Mass. Spectrom. 2019, 30 (11), 2204–2211. 10.1007/s13361-019-02333-0. [DOI] [PubMed] [Google Scholar]
- Yalovenko N.; Yatsyna V.; Bansal P.; AbiKhodr A. H.; Rizzo T. R. Analyzing glycans cleaved from a biotherapeutic protein using ultrahigh-resolution ion mobility spectrometry together with cryogenic ion spectroscopy. Analyst 2020, 145 (20), 6493–6499. 10.1039/D0AN01206H. [DOI] [PubMed] [Google Scholar]
- Bansal P.; Yatsyna V.; AbiKhodr A. H.; Warnke S.; Ben Faleh A.; Yalovenko N.; Wysocki V. H.; Rizzo T. R. Using SLIM-Based IMS-IMS Together with Cryogenic Infrared Spectroscopy for Glycan Analysis. Anal. Chem. 2020, 92 (13), 9079–9085. 10.1021/acs.analchem.0c01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal P.; Ben Faleh A.; Warnke S.; Rizzo T. R. Identification of N-glycan positional isomers by combining IMS and vibrational fingerprinting of structurally determinant CID fragments. Analyst 2022, 147 (4), 704–711. 10.1039/D1AN01861B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Faleh A.; Warnke S.; Bansal P.; Pellegrinelli R. P.; Dyukova I.; Rizzo T. R. Identification of Mobility-Resolved N-Glycan Isomers. Anal. Chem. 2022, 94 (28), 10101–10108. 10.1021/acs.analchem.2c01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrinelli R. P.; Yue L.; Carrascosa E.; Ben Faleh A.; Warnke S.; Bansal P.; Rizzo T. R. A New Strategy Coupling Ion-Mobility-Selective CID and Cryogenic IR Spectroscopy to Identify Glycan Anomers. J. Am. Soc. Mass. Spectrom. 2022, 33 (5), 859–864. 10.1021/jasms.2c00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnke S.; Ben Faleh A.; Rizzo T. R. Toward High-Throughput Cryogenic IR Fingerprinting of Mobility-Separated Glycan Isomers. ACS Meas. Sci. Au 2021, 1 (3), 157–164. 10.1021/acsmeasuresciau.1c00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. L.; Nagy G.; Conant C. R.; Norheim R. V.; Lee J. Y.; Giberson C.; Hollerbach A. L.; Prabhakaran V.; Attah I. K.; Chouinard C. D.; Prabhakaran A.; Smith R. D.; Ibrahim Y. M.; Garimella S. V. B. Ion Mobility Spectrometry with High Ion Utilization Efficiency Using Traveling Wave-Based Structures for Lossless Ion Manipulations. Anal. Chem. 2020, 92 (22), 14930–14938. 10.1021/acs.analchem.0c02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamrath M. Z.; Garand E.; Jordan P. A.; Leavitt C. M.; Wolk A. B.; Van Stipdonk M. J.; Miller S. J.; Johnson M. A. Vibrational Characterization of Simple Peptides Using Cryogenic Infrared Photodissociation of H-2-Tagged, Mass-Selected Ions. J. Am. Chem. Soc. 2011, 133 (16), 6440–6448. 10.1021/ja200849g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk A. B.; Leavitt C. M.; Garand E.; Johnson M. A. Cryogenic Ion Chemistry and Spectroscopy. Acc. Chem. Res. 2014, 47 (1), 202–210. 10.1021/ar400125a. [DOI] [PubMed] [Google Scholar]
- Domon B.; Costello C. E. A Systematic Nomenclature for Carbohydrate Fragmentations in Fab-Ms Ms Spectra of Glycoconjugates. Glycoconjugate J. 1988, 5 (4), 397–409. 10.1007/BF01049915. [DOI] [Google Scholar]
- Mank M.; Welsch P.; Heck A. J. R.; Stahl B. Label-free targeted LC-ESI-MS2 analysis of human milk oligosaccharides (HMOS) and related human milk groups with enhanced structural selectivity. Anal. Bioanal. Chem. 2019, 411 (1), 231–250. 10.1007/s00216-018-1434-7. [DOI] [PubMed] [Google Scholar]
- Chai W. G.; Piskarev V.; Lawson A. M. Negative ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal. Chem. 2001, 73 (3), 651–657. 10.1021/ac0010126. [DOI] [PubMed] [Google Scholar]
- Delvaux A.; Rathahao-Paris E.; Guillon B.; Cholet S.; Adel-Patient K.; Fenaille F.; Junot C.; Alves S., Trapped ion mobility spectrometry time-of-flight mass spectrometry for high throughput and high resolution characterization of human milk oligosaccharide isomers. Anal. Chim. Acta 2021, 1180.338878. 10.1016/j.aca.2021.338878 [DOI] [PubMed] [Google Scholar]
- Gray C. J.; Schindler B.; Migas L. G.; Picmanova M.; Allouche A. R.; Green A. P.; Mandal S.; Motawia M. S.; Sanchez-Perez R.; Bjarnholt N.; Møller B. L.; Rijs A. M.; Barran P. E.; Compagnon I.; Eyers C. E.; Flitsch S. L. Bottom-Up Elucidation of Glycosidic Bond Stereochemistry. Anal. Chem. 2017, 89 (8), 4540–4549. 10.1021/acs.analchem.6b04998. [DOI] [PubMed] [Google Scholar]
- Pellegrinelli R. P.; Yue L.; Carrascosa E.; Warnke S.; Ben Faleh A.; Rizzo T. R. How General Is Anomeric Retention during Collision-Induced Dissociation of Glycans?. J. Am. Chem. Soc. 2020, 142 (13), 5948–5951. 10.1021/jacs.0c00264. [DOI] [PubMed] [Google Scholar]
- Schindler B.; Barnes L.; Renois G.; Gray C.; Chambert S.; Fort S.; Flitsch S.; Loison C.; Allouche A. R.; Compagnon I. Anomeric memory of the glycosidic bond upon fragmentation and its consequences for carbohydrate sequencing. Nat. Commun. 2017, 8, 973. 10.1038/s41467-017-01179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.