Abstract
Nanopore sequencing is being rapidly adopted in genomics. We recently developed SLOW5, a new file format with advantages for storage and analysis of raw signal data from nanopore experiments. Here we introduce slow5tools, an intuitive toolkit for handling nanopore data in SLOW5 format. Slow5tools enables lossless data conversion and a range of tools for interacting with SLOW5 files. Slow5tools uses multi-threading, multi-processing, and other engineering strategies to achieve fast data conversion and manipulation, including live FAST5-to-SLOW5 conversion during sequencing. We provide examples and benchmarking experiments to illustrate slow5tools usage, and describe the engineering principles underpinning its performance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-023-02910-3.
Keywords: Nanopore sequencing, Nanopore signal data, SLOW5, Toolkit
Background
Nanopore sequencing analyzes native DNA or RNA molecules with no upper limit on read length [1]. Popular devices from Oxford Nanopore Technologies (ONT) measure the displacement of ionic current as a DNA/RNA strand passes through a protein pore, recording time-series signal data in a format known as “FAST5.” This signal data is typically “base-called” into sequence reads and is further used during a wide range of downstream analyses [2–8].
ONT’s native FAST5 data format suffers from several inherent limitations, which we have articulated previously [9]. FAST5 file sizes are inflated by inefficient space allocation and metadata redundancy. Its dependence on Hierarchical Data Format (HDF) makes FAST5 unsuitable for efficient parallel access by multiple CPU threads. FAST5 files can only be read/written by the hdf5lib software library, which reduces usability and can make even relatively simple operations difficult and expensive.
We recently developed a new file format named SLOW5, which is designed to overcome the above limitations in FAST5 [9]. In its compressed binary form (BLOW5), the new format is ~20–80% smaller than FAST5 and permits efficient parallel access by multiple CPU threads. This enables order-of-magnitude improvements in the speed and cost of signal data analysis on common high-performance computing (HPC) and cloud systems [9]. SLOW5 is well-documented and fully open source. Data reading/writing is managed by an intuitive software API that is compatible with a wide range of system architectures. Overall, SLOW5 offers many benefits to the ONT user community.
To accompany the SLOW5 data format, we have developed a suite of tools for creating, handling, and interacting with SLOW5/BLOW5 files. Slow5tools includes all utilities required for novice and advanced users to integrate SLOW5 with their workflows and solves existing challenges in the management and analysis of ONT raw signal data. Slow5tools is implemented in C/C++ with minimal dependencies and utilizes multi-threading, multi-processing, and other software engineering strategies to achieve a fast and efficient performance. This article provides a series of examples that serve to articulate the design, usage, and performance of slow5tools. Slow5tools is freely available as an open-source program under an MIT license: https://github.com/hasindu2008/slow5tools.
Results
Overview of slow5tools
Slow5tools is a modular toolkit for working with signal data from ONT experiments. Slow5tools is written in C/C++ and uses two core libraries, slow5lib and hdf5lib, for reading/writing SLOW5/BLOW5 and FAST5 files, respectively (see the “Methods” section; Additional File 1: Fig. S1a). The software has the following basic usage, where “command” specifies the operation to be performed:
# generic usage for slow5tools
slow5tools command input.slow5 -o output.slow5
The various commands currently available in slowt5tools are summarized in Fig. 1 and Additional File 1: Table S1. Different commands within slow5tools employ various engineering strategies to achieve optimum performance. These strategies are articulated below, along with relevant benchmarking results.
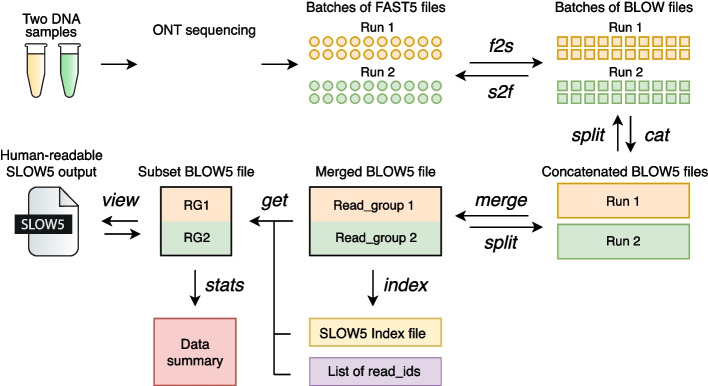
Fig. 1.
Schematic overview of nanopore raw signal data management with slow5tools. The software permits lossless data conversion from FAST5-to-SLOW5 (f2s command) or SLOW5-to-FAST5 (s2f command). Slow5tools executes a range of common operations for data management, including combining multiple SLOW5/BLOW5 datasets (merge or cat commands) or splitting a dataset into multiple parts (split command). The index command creates an index file for a SLOW5/BLOW5 file, which then enables a specific record(s) to be quickly fetched from the file using the get command. The view command is used to view the contents of a binary BLOW5 file/s in human-readable SLOW5 format and convert datasets between SLOW5 and BLOW5, or between different BLOW5 compression types. Finally, slow5tools can check the integrity of a SLOW5/BLOW5 file (quickcheck) or generate simple summary statistics describing the dataset (stats)
Fast, lossless data conversion
Because ONT devices currently write data in FAST5 format, data conversion to SLOW5/BLOW5 format will be the first step in most workflows. FAST5-to-SLOW5 conversion can be performed with slow5tools f2s, as follows:
# convert a single FAST5 file into SLOW5 ASCII format
slow5tools f2s file.fast5 -o file.slow5
# convert a directory of FAST5 files into binary BLOW5 files
slow5tools f2s fast5_dir -d blow5_dir
Data conversion is lossless by default, and the user may convert their data back to the original FAST5 format using slow5tools s2f, as follows:
# convert a directory of BLOW5 files to FAST5
slow5tools s2f blow5_dir -d fast5_dir
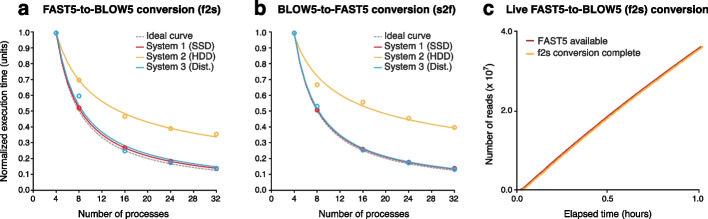
It is not possible to read/write FAST5 files using parallel CPU threads, due to their dependence on the hdf5lib library, which serializes any thread-based input/output (I/O) operations [9]. Therefore, f2s/s2f data conversion is instead parallelized through a multi-processing strategy illustrated in Additional File 1: Fig. S1b (see the “Methods” section). To evaluate this solution, we measured runtimes for data conversion on a typical ONT sequencing dataset (~9M reads; Additional File 1: Table S2), as executed with various numbers of processes on three machines with different disk systems (SDD, HDD, Distributed; Additional File 1: Table S3). Data conversion rates scaled with the number of processes for both f2s and s2f, and the observed trends were similar to the ideal curves, indicating highly efficient utilization of parallel compute resources, with the exception of System 2 (Fig. 2a, b). Data conversion on System 2 showed sub-optimal parallelism for two main reasons: (i) System 2 has a traditional HDD disk system that throttles when the number of I/O requests is high and (ii) using processes instead of threads makes these I/O requests completely separated and thus the associated overhead is high. This experiment demonstrates that multi-processing is a viable solution for the parallelization of FAST5/SLOW5 data conversion, in which multi-threading is not permitted, although this approach has limitations that will impact performance on some systems.
Fig. 2.
Efficient data conversion with slow5tools f2s and s2f commands. a, b Normalized execution times for conversion of a typical ONT dataset (~9M reads) from FAST5-to-SLOW5 format (a) or SLOW5-to-FAST5 format (b) when using increasing numbers of parallel processes. Data conversion was evaluated on three separate machines with different disk systems (SSD, HDD, Distributed). For each system, execution times are normalized relative to the execution time for the smallest number of processes (four) run on that system. Ideal curves (dashed lines) model the expected performance under optimal parallelism. c The rate of FAST5 data production (red) and FAST5-to-SLOW5 data conversion (orange) with realf2s.sh on a PromethION P48 device running at maximum sequencing load
With the speeds achieved by f2s above, live FAST5-to-SLOW5 data conversion during a sequencing experiment is also possible. To manage this process, we developed an accompanying script (realf2s.sh) that detects freshly created FAST5 files during sequencing, converts them to BLOW5, and checks the integrity of the data before (optionally) deleting the FAST5s (see the “Methods” section). We tested this feature on ONT’s PromethION P48 device, using the onboard computer for live data conversion (Additional File 1: Table S3), and found that f2s can easily match the pace of data production at maximum sequencing load (i.e., 48 flow cells running in parallel; Fig. 2c). Indeed, the average delay between the completion of an individual FAST5 file containing 4000 reads and its conversion to BLOW5 format was just ~20 s. Given the smaller size of BLOW5 vs FAST5 files (typically ~50% smaller), live f2s conversion increases the number of sequencing runs that can be performed in parallel before reaching the maximum storage capacity on an ONT sequencing device. We also anticipate that the use of live-converted BLOW5 format will also improve the capacity of live base-calling that is achievable on ONT sequencing devices in the future.
Merging and splitting datasets
The slow5tools merge command can also be used to combine multiple batches of reads or data from different sequencing runs into a single BLOW5 file, for example when aggregating data across replicate experiments. If data is merged from different runs/flow-cells, each will be assigned a separate read_group, with which all its constituent reads are tagged in the merged file. Merging can be performed as follows:
# merge all BLOW5 files in one or more directories into a single BLOW5 file
slow5tools merge blow5_dir1 blow5_dir2 -o file.blow5
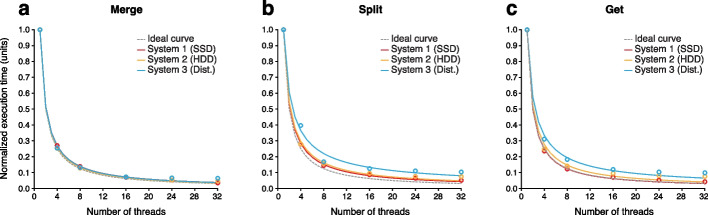
Data merging is accelerated via an interleaved multi-threading strategy illustrated in Additional File 1: Fig. S1c (see the “Methods” section), which is more resource-efficient than the multi-processing strategy described above. Evaluating this solution on the same machines as above, we observed near-optimal parallelism, with data merging times being reduced in proportion to the number of CPU threads used (Fig. 3a, Additional File 1: Fig. S2a).
Fig. 3.
Merging, splitting, and querying BLOW5 files. a Normalized execution times for merging all individual batches of reads produced by MinKNOW into a single BLOW5 file for a typical ONT dataset (~9M reads), when using increasing numbers of parallel CPU threads. Data merging was evaluated on three separate machines with different disk systems (SSD, HDD, Distributed). For each system, execution times are normalized relative to the execution time for a single thread run on that system. Ideal curves (dashed lines) model the expected performance under optimal parallelism. b The same comparison for splitting a typical BLOW5 file into separate files for each read_group with slow5tools split. c The same comparison for retrieving a set of records from a BLOW5 file based on their read_ids with slow5tools get
The slow5tools cat command provides a lightweight alternative to merge, which can be used to quickly combine batches of reads without assigning read_groups. Similar to the Unix “cat” command, this tool is intended for concatenating the small batches of reads produced by ONT’s MinKNOW software into a single BLOW5 file for a given sequencing run and does so faster than possible with merge (Additional File 1: Fig. S2b) Cat does not permit the user to combine files with different metadata (e.g., reads from different flow cells) in their headers.
The user may also wish to split a dataset into multiple separate files. This can be achieved using the split command, which offers the user a choice to direct reads from each read_group to a single output file, or alternatively split the input into a specific number of output files or a specific number of reads per file:
# split a BLOW5 file into separate BLOW5 files based on read groups
slow5tools split file.blow5 -d blow5_dir -g
# split a BLOW5 file (single read group) into separate BLOW5 files such that there are 4000 reads per file
slow5tools split file.blow5 -d blow5_dir -r 4000
# split a BLOW5 file (single read group) into 100 separate BLOW5 files
slow5tools split file.blow5 -d blow5_dir -f 100
When splitting by read_group, split is accelerated via a multi-threading strategy similar to merge (see the “Methods” section) and also exhibits near-optimal parallelism on different machines (Fig. 3b). Together, the merge, cat, and split tools provide the user an efficient, flexible framework for reorganizing ONT raw signal datasets into the most convenient structure for any use case.
Indexing SLOW5/BLOW5 files
Efficient access to a SLOW5/BLOW5 file is facilitated by an accompanying binary index file that specifies the position of each read (in bytes) within the main file (see the “Methods” section). This file should be created using the slow5tools index command:
# index a SLOW5/BLOW5 file
slow5tools index file.blow5
Creating an index file for a BLOW5 file involves decompressing and parsing each record to get the position of each read_id. Decompressing and parsing an entire record is time-consuming. However, the zlib compression algorithm, used by default in slow5tools, supports partial decompression. Index is hence accelerated through a partial decompression strategy, where only the read_id is decompressed for a given record. This method enjoys substantial performance benefits during index creation, compared to complete decompression (Additional File 1: Fig. S2c).
Viewing and querying SLOW5/BLOW5 files
SLOW5 datasets can be stored in human-readable ASCII SLOW5 format, or the more compact binary BLOW5 format, which is preferable in most scenarios. The view tool allows the user to view the contents of a BLOW5 file or convert between SLOW5/BLOW5:
# to view a BLOW5 file in SLOW5 ASCII on standard output
slow5tools view file.blow5
# Convert a BLOW5 file into SLOW5 ASCII
slow5tools view file.blow5 -o file.slow5
# convert a SLOW5 file to BLOW5 (default compression)
slow5tools view file.slow5 -o file.blow5
The view command can also be used to specify different compression methods available for BLOW5 format, with various combinations available across two compression layers (see the “Methods” section).
Rather than viewing the whole file, many use-cases require retrieval of a specific record/s from a dataset, based on their read_ids. This can be performed with the slow5tools get command, which enables efficient random access on BLOW5 files, as follows:
# extract records from a SLOW5/BLOW5 file corresponding to given read ids
slow5tools get file.blow5 readid1 readid2 -o output.slow5
Get employs a similar multi-threading strategy as above and exhibits optimal parallelism on all systems tested (Fig. 3c). Together, these tools allow users to efficiently view and query datasets in SLOW5/BLOW5 format.
POD5 conversion
POD5 is a prototype file format for raw signal data that is currently under active development by ONT. It is anticipated that POD5 will eventually replace FAST5 as the native file format on ONT devices. It is therefore essential to develop capabilities for POD5-to-SLOW5 and SLOW5-to-POD5 conversion. We have implemented and validated alpha-version tools for POD5 conversion under a repository that is currently separate from slow5tools (https://github.com/Psy-Fer/project_blue_crab). If the development of POD5 and its associated API for writing POD5 files continues to a point of maturity and stability, we intend to integrate the above tools into the slow5tools main code base, as core utilities s2p and p2s, ensuring ongoing compatibility of the SLOW5 ecosystem with ONT data frameworks.
Other tools
In addition to the commands outlined above, slow5tools also currently includes two tools for checking the integrity and/or specifications of a dataset in SLOW5/BLOW5 format. The slow5tools stats command generates simple summary statistics including the read count, number of read_groups, etc. A lightweight alternative, quickcheck simply checks a SLOW5/BLOW5 file to make sure it is not malformed. The slow5tools skim command allows the user to quickly skim through a SLOW5/BLOW5 file, printing only specified components to the standard output, rather than the entire file. The modular architecture of slow5tools will allow new tools to be readily added as new use cases arise. Similarly, the slow5tools command structure enables multiple operations to be combined in efficient one-line BASH commands familiar to the bioinformatics community (https://hasindu2008.github.io/slow5tools/oneliners.html).
Discussion
SLOW5 format was developed as an open-source, community-centric file format that addresses several inherent design flaws in ONT’s FAST5 format, on which the nanopore community was previously dependent [9]. Performance, compatibility, usability, and transparency are central principles of the ongoing SLOW5 initiative.
In addition to the slow5lib and pyslow5 APIs for reading and writing SLOW5 files, slow5tools is critical for the usability of SLOW5 format. With ONT devices currently limited to write data in FAST5 format, the slow5tools f2s utility for FAST5-to-SLOW5 conversion will be the first step in any workflow that integrates SLOW5. We also provide a method for live data conversion during sequencing, which easily keeps pace with a PromethION P48 device at maximum load. This allows the user to obtain their data in analysis-ready BLOW5 format with no additional cost to the overall workflow time, or to more efficiently move raw data off the sequencing machine for real-time analysis [10]. This also provides a base for future implementation of real-time base-calling with SLOW5 input, which we expect will enjoy significant performance benefits over FAST5.
Following data conversion, slow5tools provides a fast and flexible framework for managing and interacting with SLOW5/BLOW5 files. We recommend the use of binary BLOW5 for almost all applications, because it is more compact and efficient than ASCII SLOW5. BLOW5 supports multiple compression methods, or no compression, at the user’s discretion. Zlib was chosen as the default compression method due to its near-universal compatibility; however, svb+zstd is more compact and provides superior file-reading performance. In some cases, such as when writing temporary BLOW5 files during an analysis, no compression may also be preferable. SLOW5 was designed to allow future compression strategies to be easily incorporated, and we anticipate domain-specific compression algorithms will deliver future improvements.
Since its beta release alongside the SLOW5 format [9], slow5tools has undergone numerous performance optimizations, such as multi-threading and multi-processing accelerations, and various new utilities and options have been added. This includes fast and flexible methods for combining SLOW5/BLOW5 files (merge, cat); splitting by various different specifications (split); efficiently retrieving specific reads, groups of reads, or specifics fields from reads within a file (get, skim); modular, customizable file compression (view); and basic summary stats or QC checks (stats, quickcheck) (Additional File 1: Table S1). Slow5tools will continue to grow and evolve with community needs. For example, an early-stage tool for POD5-to-SLOW5 conversion is now available and will mature in parallel to this prototype file format being developed by ONT.
Conclusions
Slow5tools greatly simplifies the basic processes involved in structuring, querying, and interacting with nanopore raw signal data files, which are at the heart of all data management and analysis workflows. Slow5tools is the latest addition to the SLOW5 ecosystem, which already includes (i) the SLOW5/BLOW5 file format and accompanying design specifications; (ii) the slow5lib (C/C++), pyslow5 (python), and slow5-rs (rust) software libraries for reading and writing SLOW5/BLOW5 files; (iii) the Buttery-eel client wrapper that enables Guppy base-calling with SLOW5 data input [11]; and (iv) a suite of open source bioinformatics software packages with which SLOW5 is now integrated [3, 7, 12–15]. We provide slow5tools as a free, open-source software package to facilitate the adoption of the SLOW5 format by nanopore users: https://github.com/hasindu2008/slow5tools.
Methods
Basic architecture/implementation of slow5tools
Slowtools is written in C/C++ and links with two file format-specific libraries. Hdf5lib and slow5lib are used to read and write files in FAST5 and SLOW5/BLOW5 formats, respectively (Additional File 1: Fig. S1a). There are different FAST5 versions (v2.2 is the latest at the time of submission), and all current/previous versions can be converted to SLOW5 format using slow5tools. When converting back to FAST5 format, slow5tools conforms to the latest FAST5 version (currently v2.2). Slow5tools will provide support for future FAST5 versions. SLOW5 file format specification (https://hasindu2008.github.io/slow5specs/) is maintained separately from slow5lib and slow5tools. Backwards compatibility will be ensured in future SLOW5 versions and a compatibility table is maintained here: https://hasindu2008.github.io/slow5tools/compatibility.html
Slow5tools multi-processing strategy
All slow5tools commands are optimized to run efficiently on modern computer CPUs. The key optimization is to exploit parallelization. Both the f2s and s2f commands require hdf5lib to read and write FAST5 files, respectively. Hdf5lib internally uses global locks to serialize parallel thread-based I/O operations. Therefore, to parallelize the conversion from FAST5 to SLOW5 and vice versa, we employ a multi-processing strategy (Additional File 1: Fig. S1b). In this approach, each process keeps a separate memory instance of the library. Firstly, the program recursively searches the input directories to list the files to be converted based on the file extension (e.g., f2s lists all files with .fast5 extensions). Then, using the C function fork(), a pool of processes is created. The user can determine the number of processes to spawn (default is 8). Next, the number of files is equally distributed among the processes. Inside a process, an input file is opened, an output file is created, the content of the input file is written to the output file as per the output file format, and lastly, both files are closed. Then, the process iterates to the next allotted input file, if any. When all the processes terminate, all the files are converted successfully.
Slow5tools multi-threading strategy
To exploit parallelization in the rest of the programs (where FAST5 files are not involved), a multithreading model is adopted (Fig. S1c), which uses less resources than a similar multiprocessing model. This is possible as these programs only depend on slow5lib that supports C/C++ POSIX threads. A SLOW5 record has a finite set of primary attributes—read_id, read_group, digitisation, offset, range, sampling rate, len_raw_signal, and the raw signal. It can optionally have auxiliary attributes. By default, the raw signal is compressed using the stream variable byte (svb) encoding algorithm and the whole record is compressed using standard zlib compression (see below). When reading a SLOW5 file, first the compressed read should be read from the storage disk, decompressed, and then parsed into structures in the memory. These steps can exploit parallelism to speed up the I/O operations. That is, a single-threaded application (main program) fetches a batch of compressed records (using slow5_get_next_mem()) from the file on the disk and spawns a set of worker threads to decompress and parse (using slow5_rec_depress_parse()) them. Later, the necessary processing on the records is also carried out on the threads. The output records are written to a buffer (using slow5_rec_to_mem()) that is accessible by the main program’s thread before the worker threads exit. Eventually, after returning to the main thread (after worker threads are joined by the main thread), the content in the buffer is written to the output stream. This workflow is carried out until the end of the file is reached. The number of records to fetch at a time is determined by the batch size parameter of the programs. The user can also set the number of threads to match the available compute resources. This method ensures the order of the records of the input is maintained in the output.
Single-thread implementations
Slow5tools stats gives a summary of the slow5 file. In addition to the details about the primary and auxiliary attributes, it counts the number of reads in the file. Since no processing is done on the records, a single-threaded program that continuously calls slow5_get_next_mem() on the file is the optimal method to count the records. However, the buffer size of the standard C function fread() determines how many bytes are fetched from the file through system calls. This affects the performance of slow5_get_next_mem() and thus slow5tools. Hence, the buffer size was set to a larger buffer size of 128k bytes using setvbuf() in stdio. To split a SLOW5/BLOW5 file into a given number of reads or files, split also uses a single-threaded implementation unless the user sets a different output format from the input format. That is because this requires no processing on a record but just writing as it is to an output file.
BLOW5 compression strategies
There are two compression layers applied on a SLOW5 record. By default, BLOW5 format has zlib compression and svb-zd encoding [zlib, svb-zd] on the entire record and on the signal respectively. Slow5tools view can be used to create BLOW5 files with different compression combinations. View also adopts the multithreading model explained above (Additional File 1: Fig. S1c). For example, to convert a BLOW5 record from [zlib, svb-zd] to [zlib, none] (i.e., no signal encoding), the record should be first decompressed using zlib followed by a svb-zd decoding of the signal. Then, the records are compressed back using zlib. On average, this process is ~6X times slower than the conversion from [zlib, svb-zd] to [zstd, svb-zd], because zlib compression is slower than zstd compression. Despite this disparity, zlib compression is used as the default BLOW5 record compression method for the sake of compatibility. Zstd library is not yet widely available by default, especially on academic HPC servers and the user may face associated dependency and version issues. However, we advise the users to use zstd compression where this is possible on their specific system.
Indexing
Slow5tools index creates a binary index file that stores the read_ids and the byte offset of the records. Loading the index to memory allows bioinformatics programs to quickly fetch the necessary records from a SLOW5/BLOW5 file in a random access pattern. Creating an index file from a BLOW5 file involves decompressing and parsing each record to get the read_ids. Decompressing and parsing an entire record just to fetch the read_id is time-consuming. The default SLOW5 record compression method, zlib, supports partial decompression. This enables quick access to the read_id by only decompressing up to the read_id of a record. The other compression method, zstd, does not support partial decompression. Therefore, index first determines which compression is used, and then, either partially or completely decompresses the BLOW5 record to fetch the read_id. Using this partial decompression method, indexing is ~15 times faster than with the default zlib decompression and, despite the fact that zlib is slower than zstd, indexing is ~3 times faster with zlib partial decompression than zstd (which must fully decompress each record; Additional File 1: Fig. S2c).
Live data conversion
Live FAST5-to-SLOW5 data conversion can be carried out using the realf2s.sh script provided under the slow5tools repository at https://github.com/hasindu2008/slow5tools/tree/master/scripts/realtime-f2s. This is a BASH script that takes the sequencing data directory as an argument (e.g., /data/sample_id on a standard PromethION architecture). The script continuously monitors this specified directory for newly created FAST5 files using the inotifywait tool available under Linux’s inotify interface. As soon as a newly created FAST5 file is detected (the default in MinKNOW is such that one FAST5 file contains 4000 reads), the slow5tools f2s command is invoked unless the maximum number of processes is already reached. If the maximum number of processes has been reached, the newly detected FAST5 files are queued until the ongoing conversions are complete. This strategy prevents the system being overloaded if hundreds of files are generated by MinKNOW at once.
To test the capacity of real-time data conversion on a PromethION computer tower, we simulated 48 parallel sequencing runs using the playback feature in MinKNOW. The bulk-FAST5 file for MinKNOW playback was saved during the sequencing of a typical human genome sample (~10–20-kb reads, R9.4.1 flow cell, LSK110 kit). The simulation was performed by modifying the relevant MinKNOW TOML file (TOML file named sequencing_PRO002_DNA.toml under /opt/ont/minknow/conf/package/sequencing was modified to include the “simulation = /path/to/bulk/fast5” field under “[custom_settings]” section) to point to this bulk-FAST5 file. The simulation was run for a period of 1 h with live base-calling disabled. The rate of data generation is highest during the first hour of a sequencing run; therefore, this simulation emulates the maximum load one could expect to encounter. Live base-calling was disabled during the simulation so that completed FAST5 files are released at the sequencing rate—this would otherwise be tapered by MinKNOW because live base-calling cannot keep up with the sequencing rate. For this experiment, the maximum number of processes was capped at 56 (half of the CPU threads available on the PromethION compute-node), thus limiting the maximum number of FAST5 files simultaneously converted to be 56.
Supplementary Information
Additional file 1. Additional supporting information. Supplementary tables and figures supporting the work presented in this article.
Acknowledgements
We thank our colleague Derrick Lin for providing excellent technical support and freedom to use the institute’s HPC system in some quite exotic ways. Resources from the Australian National Computational Infrastructure (NCI) were used during benchmarking experiments.
Peer review information
Andrew Cosgrove was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional File 2.
Authors’ contributions
All authors (H.S., J.M.F., S.P.J., T.G.A., S.P., H.G., and I.W.D.) contributed to the conception, design, and testing of slow5tools. H.S., J.M.F., S.P.J., and H.G. implemented core slow5tools software. H.S. and H.G. performed benchmarking experiments. H.S. and I.W.D. prepared the figures. H.S., H.G., and I.W.D prepared the manuscript, with support from all authors. The authors read and approved the final manuscript.
Funding
We acknowledge the following funding support: Australian Medical Research Futures Fund Investigator Grants MRF1173594, MRF2023126, and MRF2016008 (to I.W.D.) and ARC Discovery Early Career Researcher Award (DECRA) DE230100178 to H.G.
Availability of data and materials
SLOW5 format and all associated software are free and open source under a standard MIT license. Datasets used in benchmarking experiments are described in Additional File 1: Table S2 and are available on NCBI Sequence Read Archive [16].
Slow5tools [17], Slow5lib & pyslow5 [18], and SLOW5 format specification documents [19] are all available on GitHub and specific source code versions used in this manuscript are available on Zenodo [20].
Declarations
Ethics approval and consent to participate
This study did not require ethical approval.
Competing interests
I.W.D. manages a fee-for-service sequencing facility at the Garvan Institute of Medical Research that is a customer of Oxford Nanopore Technologies but has no further financial relationship. H.G. and J.M.F. have previously received travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences. The authors declare no other competing financial or non-financial interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hasindu Gamaarachchi and Ira W. Deveson are joint senior authors.
Contributor Information
Hasindu Gamaarachchi, Email: hasindu@garvan.org.au.
Ira W. Deveson, Email: i.deveson@garvan.org.au
References
- 1.Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, et al. Real-time mapping of nanopore raw signals. Bioinformatics. 2021;37:i477–i483. doi: 10.1093/bioinformatics/btab264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang L, et al. DeepRepeat: direct quantification of short tandem repeats on signal data from nanopore sequencing. Genome Biol. 2022;23:108. doi: 10.1186/s13059-022-02670-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begik O, et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00915-6. [DOI] [PubMed] [Google Scholar]
- 6.Lee I, et al. Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. Nat Methods. 2020;17:1191–1199. doi: 10.1038/s41592-020-01000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson JT, et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 8.Aw JGA, et al. Determination of isoform-specific RNA structure with nanopore long reads. Nat Biotechnol. 2021;39:336–346. doi: 10.1038/s41587-020-0712-z. [DOI] [PubMed] [Google Scholar]
- 9.Gamaarachchi H, et al. Fast nanopore sequencing data analysis with SLOW5. Nat Biotechnol. 2022 doi: 10.1038/s41587-021-01147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goenka SD, et al. Accelerated identification of disease-causing variants with ultra-rapid nanopore genome sequencing. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samarakoon H, et al. Accelerated nanopore basecalling with SLOW5 data format. bioRxiv. 2023 doi: 10.1101/2023.02.06.527365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao Y, et al. SquiggleNet: real-time, direct classification of nanopore signals. Genome Biol. 2021;22:298. doi: 10.1186/s13059-021-02511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamaarachchi H, et al. GPU accelerated adaptive banded event alignment for rapid comparative nanopore signal analysis. BMC Bioinformatics. 2020;21:343. doi: 10.1186/s12859-020-03697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih PJ, et al. Efficient real-time selective genome sequencing on resource-constrained devices. arXiv. 2022 doi: 10.48550/arXiv.2211.07340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senanayake A, et al. DeepSelectNet: deep neural network based selective sequencing for oxford nanopore sequencing. BMC Bioinformatics. 2023;24:31. doi: 10.1186/s12859-023-05151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamaarachchi H, et al. SLOW5: a new file format enables massive acceleration of nanopore sequencing data analysis. PRJNA744329. Sequence Read Archive. 2023. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA744329/
- 17.Gamaarachchi H, et al. Slow5tools. Github. 2023. https://hasindu2008.github.io/slow5tools/
- 18.Gamaarachchi H, et al. Slow5lib. Github. 2023. https://hasindu2008.github.io/slow5lib/
- 19.Gamaarachchi H, et al. Slow5spec. Github. 2023. https://hasindu2008.github.io/slow5specs
- 20.Gamaarachchi H, et al. Permanent source code - Flexible and efficient handling of nanopore sequencing signal data with slow5tools. 2023. Zenodo. 10.5281/zenodo.7742923. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional supporting information. Supplementary tables and figures supporting the work presented in this article.
Data Availability Statement
SLOW5 format and all associated software are free and open source under a standard MIT license. Datasets used in benchmarking experiments are described in Additional File 1: Table S2 and are available on NCBI Sequence Read Archive [16].
Slow5tools [17], Slow5lib & pyslow5 [18], and SLOW5 format specification documents [19] are all available on GitHub and specific source code versions used in this manuscript are available on Zenodo [20].