ABSTRACT
The production of β-lactamases is a prevalent mechanism that poses serious pressure on the control of bacterial resistance. Furthermore, the unavoidable and alarming increase in the transmission of bacteria producing extended-spectrum β-lactamases complicates treatment alternatives with existing drugs and/or approaches. Class D β-lactamases, designated as OXA enzymes, are characterized by their activity specifically towards oxacillins. They are widely distributed among the ESKAPE bugs that are associated with antibiotic resistance and life-threatening hospital infections. The inadequacy of current β-lactamase inhibitors for conventional treatments of ‘OXA’ mediated infections confirms the necessity of new approaches. Here, the focus is on the mechanistic details of OXA-10, OXA-23, and OXA-48, commonly found in highly virulent and antibiotic-resistant pathogens Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Enterobacter spp. to describe their similarities and differences. Furthermore, this review contains a specific emphasis on structural and computational perspectives, which will be valuable to guide efforts in the design/discovery of a common single-molecule drug against ESKAPE pathogens.
KEYWORDS: Β-lactamase, OXA-10, OXA-23, OXA-48, ESKAPE bugs
Introduction
β-lactam antibiotics are life-saving drugs that kill bacterial cells by irreversibly preventing the proper formation of the peptidoglycan polymers in their cell walls through the inactivation of the peptidoglycan transpeptidases. Unfortunately, bacteria evolve different resistance mechanisms to counteract the action of these wonder drugs [1–3]. The production of β-lactamases is one of the most prevalent resistance mechanisms encountered in clinical settings. Just like the peptidoglycan transpeptidases, β-lactamases form an intermediate complex with β-lactams and then render these drugs ineffective by cleaving their β-lactam rings, the chemical moiety with the amidic function [4,5]. The use of β-lactamase inhibitors in combinatorial treatments is one major strategy to restore the efficacy of β-lactam antibiotics to prevent the antibiotic from being hydrolyzed by the enzyme [3].
There exist two different schemes for the classification of β-lactamases. Bush-Jacoby-Medeiros classification divides the enzymes into groups based on substrate and inhibitor profiles [6]. The more widely used Ambler classification divides the enzymes based on the similarities of the catalytic site residues and the motifs in their primary sequences [7,8]. Originally, Ambler specified only the A and B classes. For catalysis, while class A enzymes had a serine residue in the active site, class B enzymes required the metal zinc in the active site. Later, two more classes with active-site serine residues but with little sequence similarity to class A enzymes [9] were discovered; ‘Amp C’ β-lactamases, designated as the class C [10], and ‘OXA’ β-lactamases, designated as the class D [11,12].
Most OXA-enzymes have the ability to hydrolyze the so-called ‘last resort’ β-lactams, such as carbapenems and oxyimino-substituted agents as ceftazidime and cefotaxime [13]. For the management of antibiotic resistance, in-depth knowledge of mechanistic details of these enzymes, especially those with carbapenemase activity, is crucial. Hence, in order to guide drug and inhibitor development efforts, this review will focus on three OXA enzymes widely distributed in highly virulent and antibiotic-resistant bacterial pathogens associated with life-threatening hospital infections. There will be specific emphasis on the OXA enzymes of ESKAPE bugs [14], Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Enterobacter spp., from structural and computational perspectives to guide efforts in the design/discovery of a single-molecule targeting all four pathogens.
Class D β-lactamases of ‘ESKAPE’ bugs
Class D β-lactamases are commonly known as oxacillinases based on their strong hydrolytic activity against the semisynthetic penicillin oxacillin. Thus, they are conventionally named using the ‘OXA’ nomenclature [15]. Later studies have confirmed their activities also against cephalosporins and carbapenems. Within β-lactamases, this class has a unique mechanism for catalysis, which is featured by a carboxylated lysine [16].
Most genes encoding OXA-type β-lactamases are located on large, transferable plasmids in Gram-negative bacteria, constituting a danger for their wide dispersal. More recent studies report that they can be encoded by both chromosomal and plasmid-mediated genes in Gram-negative and Gram-positive bacteria [17,18]. Currently, there are more than 750 types of OXA-enzymes displaying heterogeneous substrate profiles; consequently, they are categorized as narrow- or extended-spectrum enzymes [13,19]. Generally, OXA-enzymes resist inhibition by common β-lactam inhibitors (i.e. clavulanate, sulbactam, and tazobactam) and confer resistance to the amino-, carboxy-, and ureidopenicillins [20]. Not surprisingly, these enzymes are widely distributed among the clinically challenging ESKAPE bugs [21]. Hence, OXA-enzymes deserve special attention under the topic of β-lactomics, the term introduced by Khan [22]. Here the focus will be on the OXA-10, OXA-23, and OXA-48 β-lactamases, commonly found in highly virulent and antibiotic-resistant pathogens A. baumannii, K. pneumoniae, P. aeruginosa, and Enterobacter spp.
P. aeruginosa primarily produces the class D β-lactamases of the OXA-10 group [23,24]. The blaOXA-10 gene was first characterized from a P. aeruginosa plasmid and its product has been initially named as PSE-2 [25]. Later, it was transferred to the OXA group and renamed as OXA-10 [26]. Due to horizontal gene transfer, they are also encountered in different bacterial species [27–29]. Many enzymes within this group are generally narrow-spectrum with weak carbapenemase activity; however, natural variants with point mutations on OXA-10 such as OXA-11, OXA-14, OXA-16, OXA-17, and OXA-35 have been shown to be extended-spectrum class D β-lactamases [13,30]. This demonstrates how these enzymes have the potential to evolve into clinically important variants under selective pressure [31].
The general drug resistance associated with A. baumannii is through the production of OXA-type-β-lactamases with carbapenem hydrolyzing activities. These enzymes could be intrinsic like the chromosomal OXA-51 group enzymes or acquired (e.g. OXA-23 group, OXA-40 group, OXA-58 group, etc.) [13]. Among the plasmid-borne enzymes, the OXA-23 group is the most prevalent element [32]. All around the world, outbreaks of A. baumannii synthesizing OXA-23 have been reported [33–39]. The blaOXA-23 gene was first characterized from an A. baumannii strain resistant to imipenem, penicillins, and all classes of cephalosporins. Its product has been initially named as ARI-1 [40] and later renamed as OXA-23 [41]. This was the first carbapenem-hydrolyzing class D β-lactamase (CHDL) on a self-transferable plasmid [42].
Different K. pneumoniae strains are capable of synthesizing β-lactamases of all classes from A to D; however, drug resistance to multiple antibiotics is mostly associated with the synthesis of extended-spectrum β-lactamases of the Ambler class A (NMCA, IMI, SME, GES, and KPC), of extended-spectrum metallo β-lactamases (IMP, VIM), and of extended-spectrum oxacillinase, OXA-48 [43–50]. The first blaOXA-48 gene was identified in a K. pneumoniae strain [50], but now surveillance studies identify the OXA-48 group enzymes as the 2nd or 3rd most common carbapenemases among global Enterobacteriaceae [20].
Structural and catalytic features of OXA-10, OXA-23, and OXA-48
Despite having varying amino acid sequences, class D β-lactamases assume very similar secondary structures [51–53]. In Figure 1, the aligned OXA-10, OXA-23, and OXA-48 enzymes are presented to provide an insight into their sequential and structural similarities. The limited sequence identity of 33.4–45.6 % (Figure 1 -a-c) yielded Cα RMSD values between 0.782 and 1.092 Å, which indicates quite similar secondary structures (Figure 1 -b-d). Similar folding patterns in these OXA structures might be associated with the motifs present in their primary sequences. In particular, all three OXA enzymes have the major conserved motifs of class D enzymes, as well as the Ser-x-x-Lys motif, which is the minimal requirement for serine acylation [51,54–58]. Multiple sequence alignment of OXA-10, OXA-23, and OXA-48 further revealed additional shared motifs, which are summarized in Table 1.
Figure 1.
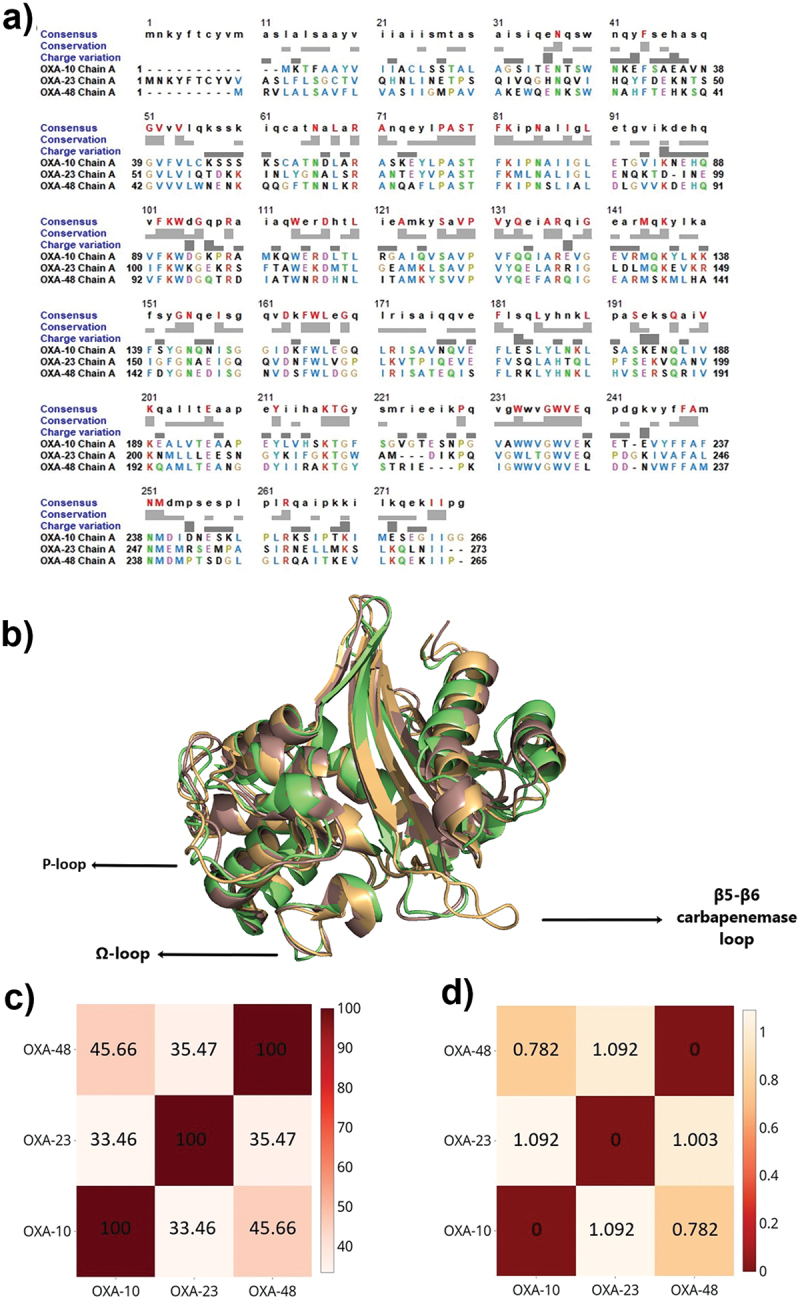
a) Multiple sequence alignment of OXA-10 (PDB ID: 1FOF), OXA-23 (PDB ID: 4K0X), and OXA-48 (PDB ID: 3HBR). Consensus residues are colored and indicated in the upper case at the top row. The degree of conservation is presented in light grey. b) Aligned 3D structures of OXA-10 (light Orange), OXA-23 (pale green), and OXA-48 (dark salmon) highlighting the differences occurring especially on important loops surrounding active site cleft. c) Heatmap of sequential identities on a scale of 0 to 100, where 100 denotes the highest similarity. d) Heatmap of RMSD differences, where 0 denotes the highest similarity.
Table 1.
Conserved motifs of OXA-10, OXA-23, and OXA-48 enzymes.
| |
OXA-10 |
OXA-23 |
OXA-48 |
|---|---|---|---|
| Conserved motifs with DBL numbering [59] | |||
|
SER70-x-x-LYS |
Ser67-Thr-Phe-Lys |
Ser79-Thr-Phe-Lys |
Ser70-Thr-Phe-Lys |
|
SER118-x-VAL |
Ser115-Ala-Val |
Ser126-Ala-Val |
Ser118-Val-Val |
|
LYS216-THR-GLY |
Lys205-Thr-Gly |
Lys216-Thr-Gly |
Lys208-Thr-Gly |
|
Additional common motifs with DBL numbering [59] | |||
|
GLY40-VAL-x-VAL |
Gly39-Val-x-Val |
Gly51-Val-x-Val |
Gly42-Val-x-Val |
|
PHE87-LYS-TRP-x-GLY |
Phe90-Lys-Trp-x-Gly |
Phe101-Lys-Trp-x-Gly |
Phe93-Lys-Trp-x-Gly |
| PHE163-TRP-LEU | Phe153-Trp-Leu | Phe164-Trp-Leu | Phe156-Trp-Leu |
| GLY235-TRP-VAL-GLU | Gly224-Trp-Val-Glu | Gly232-Trp-Val-Glu | Gly224-Trp-Val-Glu |
Class D β-lactamases are commonly found in monomeric and/or dimeric forms. In accordance with this, OXA-23 primarily exists as a monomer, while OXA-10 and OXA-48 usually exist as homodimers [52]. Their dimerization is stabilized via cation-binding sites, chloride ions, and hydrogen bonding, as cations are hydrogen-bonded to water molecules in their vicinity. For OXA-10, an additional β-strand has been shown to mediate the dimer formation [58]. Size-wise, the dimerization surface area of OXA-48 is narrower compared to OXA-10. When this dimeric structure is disrupted, a decrease in enzyme activity against antibiotics such as oxacillin and penicillin is observed in OXA-10, whereas this is not the case for OXA-48 [9,53,56,60].
OXA-10, OXA-23, and OXA-48 fold as two-domain proteins, like most other β-lactamases (Figure 2). The first domain (D1) is a mixed α/β domain containing the β5-β6 carbapenemase loop, the Lys[SerThr]Gly motif, and N- and C-termini; whereas the second domain (D2) is a helix-only domain, containing the active site serine acylation motif, the Ω-loop, and other active site stabilizing elements [51,55–58]. The active sites of these OXA enzymes are positioned in the cleft between the two domains like a transition zone, and the conserved motifs (Table 1) present in this region are important for activity. Active site residues of OXA-10 [58,60–63], OXA-23 [57,64–66], and OXA-48 [51,61,62,67–69] are summarized in Table 2 and presented in Figure 3. Overall, the total number of active site residues (marked in bold in Table 2) is highest in OXA-10, while the least number of active site residues is in OXA-48. For the most part, many of the residues are conserved in the primary sequences and on the superimposed structures. The strictly conserved residues Ser70, Lys73, Ser118, Lys216, and Gly218 (given in DBL numbering) [70] are overlaid when superimposed, though the last three are present in the active site of OXA-23 only, none of these three residues contribute to the active site in OXA-48. Found within these residues are the active-site Ser70 and the carboxylated Lys73 [16]. This carboxylysine residue acts as the general base in the hydrolysis mechanism, playing a particularly essential role in the deacylation step of catalysis [71,72]. The residues 102, 105, 120, 165, 219, and 260 (DBL numbering) [59] confer hydrophobic character to the active site region. The residues 120 and 165 (DBL numbering) are key elements for carbapenem binding [51–53,57,73,74]. These conserved residues interact with the antibiotics through hydrophobic interactions, thus, contribute to the adoption of different tautomerizations [52,53].
Figure 2.
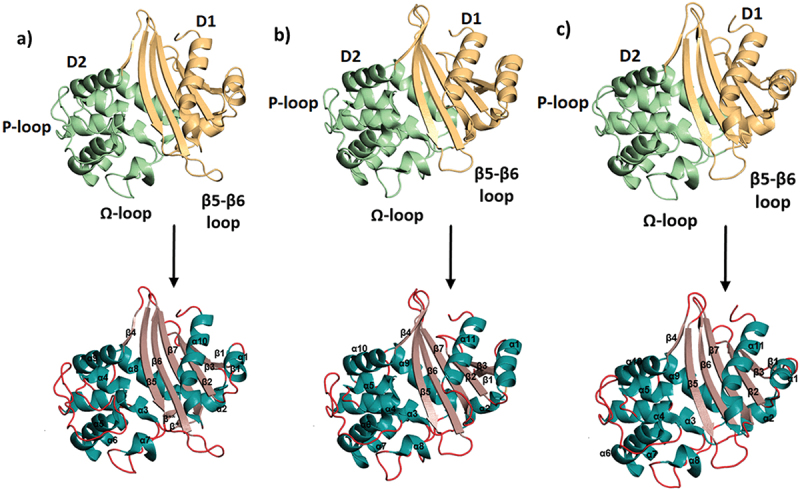
Domains (top row) and secondary structural elements (bottom row) of OXA-10, OXA-23, and OXA-48. First domain (D1) is shown in yellow, the second domain (D2) is shown in lime green, α-helices are shown in dark green, β-strands are shown in pink and the loops are shown in red. a) OXA-10 (PDB ID: 1FOF) α1 (28–34), α2 (55–60), α3 (65–80), α4 (107–114), α5 (116–128), α6 (127–139), α7 (152–157), α8 (163–175), α9 (181–192), α10 (243–261): β1 (21–24), β2 (39–45), β3 (50–53), β* (62–63), β**(162–163), β4 (193–196), β5 (200–208), β6 (218–228), β7 (231–241). b) OXA-23 (PDB ID: 4K0X),: α1 (37–47), α2 (67–73), α3 (77–92), α4 (110–114), α5 (118–126), α6 (127–139), α7 (139–151), α8 (163–168), α9 (173–186), α10 (192–203) α11 (257–270); β1 (32–34), β2 (51–56), β3 (61–65), β4 (204–208), β5, (211–220), β6 (226–235), β7 (241–250). c) OXA-48 (PDB ID: 3HBR) α1 (31–37), α2 (58–63), α3 (68–83), α4 (102–106), α5 (110–117), α6 (119–131), α7 (131–143), α8 (155–160), α9 (166–178), α10 (184–195), α11 (243–261); β1 (26–27), β2 (42–48), β3 (53–56), β* (65–66), β** (164–165), β4 (196–199), β5 (204–212), β6 (219–227), β7 (232–240) (β* and β** not shown).
Table 2.
| DBL numbering [59] | OXA-10 | OXA-23 | OXA-48 |
|---|---|---|---|
| 69 | Ala66 | Ala78 | Ala69 |
| 70 | Ser67 | Ser79 | Ser70 |
| 73 | Lys70CX | Lys82CX | Lys73CX |
| 76 | Asn73 | Asn85 | Asn76 |
| 102 | : Met99 | : Phe110 | : Ile102 |
| 105 | Trp102 | Trp113 | Trp105 |
| 118 | Ser115 | Ser126 | Ser118 |
| 120 | Val117 | Val128 | Val120 |
| 123 | : Phe120 | : Tyr131 | : Tyr123 |
| 124 | Gln121 | Gln132 | Gln124 |
| 164 | Trp154 | Trp165 | Trp157 |
| 165 | Leu155 | Leu166 | Leu158 |
| 216 | Lys205 | Lys216 | Lys208 |
| 217 | Thr206 | Thr217 | Thr209 |
| 218 | Gly207 | Gly218 | Gly210 |
| 219 | : PHE208 | : TRP219 | : TYR211 |
| 220 | Ser209 | : Ala220 | Ser212 |
| 221 | Gly210 | Met221 | Thr213 |
| 225 | Glu214 | Asp222 | Arg214 |
| 257 | Glu244 | . Glu253 | . Ser242 |
| 260 | Leu247 | Ala256 | Leu247 |
| 263 | Arg250 | Arg259 | Arg250 |
* Amino acids in bold are active site residues. Residues stabilizing the oxyanion cleft of the active region are capitalized, residues connecting the conserved active Ser and Lys residues are italicized, and carboxylated Lys residues are labeled with ‘CX’.
Figure 3.
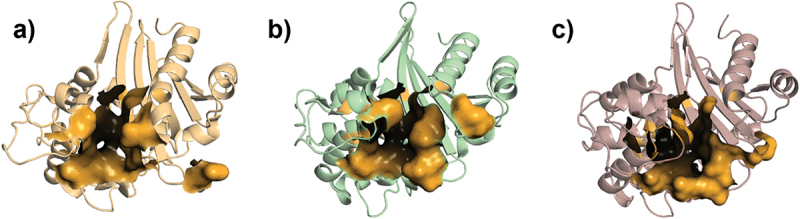
Active site regions of a) OXA-10, b) OXA-23, and c) OXA-48 are shown on the 3D structures.
In the active sites of the enzymes, there is an ‘oxyanion hole’ (Figure 4), which is responsible for the tetrahedral oxyanion intermediate stabilization (Table 3). Of the residues stabilizing the oxyanion hole, the first one is the conserved active-site serine residue while the second one is an aromatic amino acid. The identity of the aromatic amino acid is different in all three OXA enzymes. These aromatic residues are found as a part of a nonpolar patch important for binding substrate side-chains [15]. A water molecule resides in the oxyanion hole of all three enzymes, OXA-10 [16], OXA-23 [75], and OXA-48 [51]:, in their apo structures.
Figure 4.
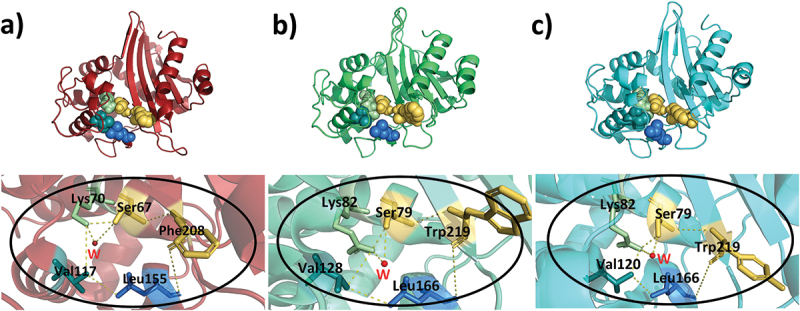
Oxyanion hole of a) OXA-10, b) OXA-23, and c) OXA-48. In the above panels, residues surrounding and stabilizing the oxyanion holes are represented with spheres. In the below panels, the zoomed views of the oxyanion holes with stick representation of the stabilizing residues which form the crevice are given. The water molecules are shown in red and the stabilizing residues are shown in yellow-orange spheres.
Table 3.
Residues correlated with the oxyanion hole.
| Residues around the oxyanion hole |
Residues stabilizing the oxyanion hole |
|
|---|---|---|
| OXA-10 | Ser67, Lys70, Val117, Leu155 | Ser67, Phe208 |
| OXA-23 | Ser79, Lys82, Val128, Leu166 | Ser79, Trp219 |
| OXA-48 | Ser70, Lys73, Val120, Leu158 | Ser70, Tyr211 |
The active sites of the enzymes are surrounded mainly by the two characteristic loops, the Ω-loop, residing in D2, and the carbapenemase loop, connecting β5 and β6 in D1 (β5-β6 loop) (Figure 3). Variations in these loops are responsible for the differences in observed activities and substrate profiles. The Ω-loop, which has a variable length, forms one side of the active site [15]. It adopts similar conformations in all three enzymes. While in OXA-10 this loop connects α6 and α7; in OXA-23 and OXA-48, it connects α7 and α8. It is also shorter in size and has a more compact conformation in OXA-10 [58]. The absence of this loop is found to affect not only antibiotic recognition profiles but also acylation by third-generation cephalosporins [76]. The β5-β6 loop residing close to the active site connects two β-strands and delimits one side of the active site in OXA-23 and OXA-48, while folds in the opposite direction in OXA-10, pointing outwards to the active site [51,56,58]. Insertions and deletions within this loop have been shown to affect functional properties [52] and the substrate profiles, particularly for carbapenems [62]. In OXA-23 and OXA-48, this loop has similar folds and extends towards the outer portion of the active site crevice, changing its charge and narrowing its width. In OXA-10, it adopts an open conformation easing the passage of water molecules, which can be related to the weaker carbapenemase activity but stronger activity against bulky chain antibiotics, such as penicillin due to the larger entrance region [1,2]. An additional study showed that OXA-10 adopted carbapenemase activity when its β5-β6 loop has been replaced with the β5-β6 loop of OXA-48 or OXA-24. However, oxacillinase, ampicillinase, and cephalotinase activities were not altered in the same variants, pointing out their independence from this loop [62].
OXA-23 displays carbapenemase activity as a result of mutations and mechanisms it has evolved [57,66]. A conformational change in OXA-23 characterized by turning of the Ω-loop element, Leu166, away from the Val128 side chain upon substrate binding, plays a key role in providing access for the hydrolytic water molecule to the N-carboxylated Lys for efficient deacylation of the bound substrate. In OXA-23, there is a tunnel-like entrance to the active site forming a hydrophobic barrier to give access only to certain substrates [52,57]. The hydrophobic bridge that forms between Phe110 and Met221 of OXA-23 narrows its active site significantly. Hydrophobic-bridge deficient OXA-23 mutants exhibit lower carbapenemase activity [57]. This bridge was initially hypothesized to be responsible for carbapenemase activity in class D β-lactamases but later it was found not to be a universal feature for activity [70,77], e.g. it is not found in OXA-10 and OXA-48 [53]. Due to the absence of the hydrophobic bridge, OXA-48 has a more open active site [78]. OXA-48 has enhanced hydrolytic activity against carbapenems like OXA-23, yet its functional mechanism is almost completely different [53]. The carboxylate group of the Lys73 (DBL numbering) is responsible for activation of the water molecule which leads to deacylation and it is protected by a hydrophobic patch formed by juxtaposed aliphatic residues, Val120 and Leu165 (DBL numbering) that form the ‘deacylating water channel’. In OXA-23, the surface is fully closed over the lysine and this channel opens via the movement of residues of the hydrophobic patch upon substrate binding, while in OXA-48 there is a preexisting channel between the hydrophobic patch and the catalytic serine, which slightly widens upon substrate binding [57,77]. On the other hand, OXA-48 has lower activity against the antibiotics with bulkier side-chain substituents [52] compared to OXA-10, which is consistent with its higher carbapenemase activity and narrower active site [78,79].
Substrate and inhibitor profiles of OXA-10, OXA-23, and OXA-48
Substrate profiles of OXA-10, OXA-23, and OXA-48 can be determined by measuring the kinetic parameters such as kcat and Km. As a general rule, the kcat/Km ratio provides an idea about the catalytic efficiency of the enzymes, e.g. a high kcat/Km value would indicate high catalytic efficiency despite low kcat and Km values. Thus, the actual catalytic efficiency of a β-lactamase against a β-lactam antibiotic is commonly determined by evaluating kcat and kcat/Km values together [80]. Following this, the kinetic values presented in Tables 4, 5, and 6 clearly show that OXA-10, OXA-23, and OXA-48 display diverse substrate specificities. However, overall, they hydrolyze penicillins more efficiently when compared to other classes of β-lactams. Despite their higher affinity for carbapenems (in the nanomolar range for selected antibiotics), their hydrolysis is very slow. This demonstrates that hydrolysis of cephalosporins and monobactams is not as efficient as penicillins.
Table 4.
Kinetic values of OXA-10 β-lactamase against β-lactam antibiotics.
| kcat (s−1) | Km (µM) | kcat/Km (mM−1·s−1) | |
|---|---|---|---|
| Carbapenems | |||
| Imipenem | <0.1 [62] 0.041 ± 0.001 [30] 0.047 ± 0.001 [31] |
0.04 ± 0.007 [62] 2.0 ± 0.1 [30] >2 [31] |
ND [62] 21 ± 3 [30] >23 [31] |
| Meropenem | 0.039 ± 0.001 [30] 0.023 ± 0.001[31] |
5.6 ± 0.8[30] >2 [31] |
7 ± 0.2 [30] >12 [31] |
| Ertapenem | 0.022 ± 0.001 [30] | 4.1 ± 0.5 [30] | 5.4 ± 0.7 [30] |
| Doripenem | 0.037 ± 0.001 [30] | 4.8 ± 0.8 [30] | 8 ± 1 [30] |
| Penicillins | |||
| Benxylpenicillin (Penicillin G) |
89 ± 10 [60] 109 ± 3 [16] 120 ± 10 [104] 120 ± 5 [105] 91 ± 1 [31] |
63 ± 6 [60] 23 ± 0.4 [16] 20 ± 1 [104] 20 ± 1 [105] 14 ± 1 [31] |
1.41x103 [60] (5 ± 1)x103 [16] (6.0 ± 0.6)x103 [104] (6 ± 0.4)x103 [105] 6.7x103 [31] |
| Oxacillin | 608 ± 10 [60] (1.26 ± 0.03)x103 [16] 300 ± 10 [104] 300 ± 4 [105] 660 [62] 530 ± 10 [30] 346 ± 3 [31] |
222 ± 16 [60] 29 ± 2 [16] 100 ± 20 [104] 100 ± 20 [105] 96 [62] 87 ± 5 [30] 65 ± 5 [31] |
2.74x103 [60] (43 ± 3)x103 [16] (3 ± 0.6)x103 [104] (3 ± 0.6)x103 [105] 6.9x103 [62] (6.1 ± 0.3)x103 [30] 5.3x103 [31] |
| Ampicillin | (5.59 ± 0.03)x103 [60] 143 ± 7 [16] 220 ± 20 [104] 220 ± 10 [105] 530 [62] |
235 ± 30 [60] 34 ± 4 [16] 35 ± 5 [104] 35 ± 5 [105] 77 [62] |
2.5x103 [60] (4.2 ± 0.6)x103 [16] (6.02 ± 0.1)x103 [104] (6.3 ± 0.9)x103 [105] 6.9x103 [62] |
| Carbenicillin | 31 ± 1 [60] 112 ± 14 [16] |
195 ± 13 [60] 92 ± 16 [16] |
159 [60] (1.2 ± 0.3)x103 [16] |
| Cloxacillin | 530 ± 36 [60] (1.53 ± 0.02)x103 [16] 120 ± 10 [104] |
(2.64 ± 0.3)x103 [60] 114 ± 22 [16] 110 ± 10 [104] |
196 [60] (13 ± 3)x103 [16] (1 ± 0.1)x103 [104] |
| Cephalosporins | |||
| Cephalothin | 6 ± 0.1 [60] 8.3 ± 0.1 [16] 2.5 ± 0.1 [104] 10 ± 1 [105] 2.9 ± 0.1 [62] 1.07 ± 0.03 [31] |
38 ± 2 [60] 32 ± 2 [16] 7 ± 1 [104] 35 ± 2 [105] 11 ± 1.4 [62] 4.4 ± 1.4 [31] |
158 [60] 260 ± 10 [16] 380 ± 50 [104] 300 ± 30 [105] 260 [62] 250 [31] |
| Cephaloridine | 79 ± 8 [60] 57 ± 14 [16] 70 ± 20 [104] |
(2.34 ± 0.3)x103 [60] 374 ± 94 [16] 400 ± 100 [104] |
33 [60] 150 ± 50 [16] 180 ± 60 [104] |
| Cefoxitin | >0.07 [31] | >200 [31] | 0.38 ± 0.012 [31] |
| Cefotaxime | 9 ± 0.2 [60] 2.06 ± 0.19 [31] |
346 ± 19 [60] 104 ± 17 [31] |
26 [60] 19 [31] |
| Ceftriaxone | 3 ± 0.3 [60] | 55 ± 2 [60] | 54 [60] |
| Ceftazidime | ND [60] | ND [60] | ND [60] |
ND: Hydrolysis not detected.
Table 5.
Kinetic values of OXA-23 β-lactamase against β-lactam antibiotics.
| kcat (s−1) | Km (µM) | kcat/Km (mM−1·s−1) | |
|---|---|---|---|
| Carbapenems | |||
| Imipenem | 0.35 ± 0.01 [57] 2.8 [65] 0.490 ± 0.01 [66] 0.35 ± 0.01 [30] 0.5 [106] |
4.8 ± 0.3 [57] 6.26 [65] 0.204 ± 0.023 [66] ≤2.0 [30] 80 [106] |
74 ± 4 [57] 450 [65] (2.4 ± 0.3)x103 [66] ≥180 [30] 6 [106] |
| Meropenem | 0.068 ± 0.001 [57] 0.7 [65] 0.068 ± 0.001 [30] |
≤1 [57] 17 [65] ≤2.0 [30] |
≤68 [57] 40 [65] ≥34 [30] |
| Ertapenem | 0.021 ± 0.001 [57] 0.021 ± 0.001 [30] |
0.50 ± 0.10 [57] ≤2.0 [30] |
42 ± 8 [57] ≥ 11 [30] |
| Doripenem | 0.036 ± 0.001 [57] 0.028 ± 0.003 [66] 0.036 ± 0.001 [30] |
0.70 ± 0.09 [57] 0.018 ± 0.002 [66] ≤2.0 [30] |
52 ± 7 [57] (1.5 ± 0.3)x103 [66] ≥18 [30] |
| Penicillins | |||
| Benzylpenicillin (Penicillin G) |
78 [65] 40 [106] |
188 [65] 60 [106] |
400 [65] 670 [106] |
| Oxacillin | 320 ± 10 [30] | 110 ± 10 [30] | (3.1 ± 0.2)x103 [30] |
| Ampicillin | 460 ± 10 [66] 31 [65] <0.1 [106] |
82 ± 9 [66] 161 [65] ND* [106] |
(5.7 ± 0.6)x103 [66] 200 [65] ND* [106] |
| Ticarcillin | 7 [106] | 70 [106] | 100 [106] |
| Piperacillin | 47 [65] | 302 [65] | 150 [65] |
| Cephalosporins | |||
| Cefotaxime | 5.5 ± 0.1 [66] | 340 ± 30 [66] | 16 ± 2 [66] |
| Ceftriaxone | 0.016 ± 0.001 [66] | 3.7 ± 0.5 [66] | 4.4 ± 0.7 [66] |
| Ceftazidime | <0.01 [66] | ||
| Cefepime | 3 [65] 20 ± 1 [73] |
173 [65] (1.1 ± 0.2)x103 [73] |
18 [65] 18 ± 3 [73] |
| Cefiderocol | <0.1 [106] | ND [106] | ND [106] |
| Monobactams | |||
| Aztreonam | 0.24 ± 0.01 [66] | (2.4 ± 0.14)x103 [66] | 0.10 ± 0.01 [66] |
ND*: not determined due to instability of the hydrolysis complex.
ND: Hydrolysis not detected.
Table 6.
Kinetic values of OXA-48 β-lactamase against β-lactam antibiotics.
| kcat (s−1) | Km (µM) | kcat/Km (mM−1·s−1) | |
|---|---|---|---|
| Carbapenems | |||
| Imipenem | 2 [50] 4.8 [51] 22.48 [69] 6.7 ± 0.2 [30] 2.7 ± 0.2 [107] 4.5 ± 0.8 [108] 1.5 ± 0.1 [109] 1 [106] 4.8 ± 0.2 [79] 11.4 ± 0.5 [110] 5 ± 0.3 [111] |
14 [50] 13 [51] 28.3 [69] 5.3 ± 0.6 [30] 3.7 ± 0.7 [107] 7.9 ± 0.1 [108] 60.3 ± 12.4 [109] 15 [106] 13 ± 2 [79] 57.7 ± 9.3 [110] 2.3 ± 0.07 [111] |
145 [50] 370 [51] 794 [69] (1.3 ± 0.2)x103 [30] 900 [107] 570 [108] 25 [109] 70 [106] 365 ± 71 [79] 200 ± 30 [110] (2.0 ± 0.2)x103 [111] |
| Meropenem | 0.1 [50] 0.07 [51] 0.112 [69] 0.16 ± 0.01 [30] 0.11 ± 0.01 [107] 0.098 ± 0.005 [108] 0.71 ± 0.02 [79] 0.087 ± 0.01 [110] 0.12 ± 0.001 [111] |
200 [50] 11 [51] 5.5 [69] ≤2.0 [30] 6.0 ± 1.2 [107] 1.0 ± 0.2 [108] 4 ± 1 [79] <1.9 [110] 0.06 ± 0.002 [111] |
0.5 [50] 6.2 [51] 20.4 [69] ≥80 [30] 17 [107] 98 [108] 177 ± 50 [79] <45 [110] (1.9 ± 0.1)x103 [111] |
| Ertapenem | 0.13 [51] 0.112 [69] 0.067 ± 0.001 [30] 0.3 ± 0.02 [109] 0.03 ± 0.002 [111] |
100 [51] 2.4 [69] ≤2.0 [30] 123.7 ± 36.2 [109] 0.2 ± 0.003 [111] |
1.3 [51] 46.7 [69] ≥34 [30] 2 [109] 200 ± 30 [111] |
| Doripenem | ND [69] 0.14 ± 0.01 [30] 0.066 ± 0.002 [107] |
≤2.0 [30] 4.1 ± 0.6 [107] |
≥70 [30] 16 [107] |
| Panipenem | 1.4 [51] | 14 [51] | 100 [51] |
| Faropenem | 0.038 [51] | 13 [51] | 2.9 [51] |
| Penicillins | |||
| Benzylpenicillin (Penicillin G) |
245 [50] 446 [112] (8.0 ± 1.1)x103 [108] 1750 [106] |
40 [50] 79 [112] 700 ± 300 [108] 200 [106] |
6.1x103 [50] 5.6x103 [112] 1.1x104 [108] 8.75x103 [106] |
| Oxacillin | 25 [50] 130 [51] 160 ± 10 [30] |
30 [50] 95 [51] ≤30 [30] |
850 [50] 1.4x103 [51] ≥6x103 [30] |
| Ampicillin | 340 [50] 955 [51] 1349 [69] 560 ± 30 [108] 370 [106] 608 ± 53 [79] |
5200 [50] 395 [51] 572.7 [69] 150 ± 30 [108] 1.1x103 [106] 370 ± 70 [79] |
65 [50] 2.4x103 [51] 2.36x103 [69] 3.7x103 [108] 340 [106] (1.64 ± 0.46)x103 [79] |
| Ticarcillin | 45 [50] 70 [106] |
55 [50] 90 [106] |
820 [50] 780 [106] |
| Piperacillin | 75 [50] 3.9 ± 0.5 [79] |
410 [50] 898 ± 155 [79] |
180 [50] 4 ± 1 [79] |
| Temocillin | 0.3 [51] | 45 [51] | 6.6 [51] |
| Cephalosporins | |||
| Cephalothin | 3 [50] 44 [51] 2.8 ± 0.1 [107] |
20 [50] 195 [51] 140 ± 10 [107] |
150 [50] 230 [51] 20 [107] |
| Cephaloridine | 2 [50] | 27 [50] | 75 [50] |
| Cefoxitin | >0.05 [51] | >200 [51] | 0.26 [51] |
| Cefotaxime | 10 [50] >9 [51] ND [107] <0.001 [111] |
190 [50] >900 [51] >1.0x103 [107] 174 ± 14 [111] |
60 [50] 10 [51] 4.7 [107] <0.006 [111] |
| Ceftazidime | 4 [50] ND [69] ND [107] 3.0 ± 0.8 [79] ND [111] ND [51] |
5.1x103[50] ND [107] 300 ± 150 [79] (9.9 ± 0.74)x103 [111] |
1 [50] ND [107] 10 ± 8 [79] ND [111] |
| Cefepime | 1 [50] >0.6 [51] 7.87 [69] 9 ± 2 [108] 1.7 ± 0.6 [79] |
160 [50] >550 [51] 2514.5 [69] 300 ± 110 [108] (1.68 ± 0.7)x103 [79] |
6 [50] 1.1 [51] 3.13 [69] 30 [108] 1 ± 0.8 [79] |
| Cefpirome | 8 [50] | 390 [50] | 20 [50] |
| Cefiderocol | <0.1 [106] | ND [106] | ND [106] |
| Monobactams | |||
| Aztreonam | ND [50] | ND [50] | ND [50] |
ND: Hydrolysis not detected.
In literature, reports on the kinetic parameters for different class D β-lactamases are available. However, there exist variations in the reported values for OXA-10, OXA-23, and OXA-48, even with the same substrate. The major reason for this discrepancy is whether or not a CO2 source (such as NaHCO3) is present during kinetic measurements. Class D β-lactamases require a CO2 source for the N-carboxylation of the catalytic lysine [16]. In the absence of a CO2 source, the enzyme may be inactive, which in turn significantly alters kinetic measurements. Some other studies also explain this discrepancy among the reports by the difference in the enzyme formulations employed, which could be crude extracts, partially purified enzymes, or enzymes that lose part of their activity upon purification [30].
Following the discovery of β-lactamase inhibitors, which can be coadministered with β-lactam antibiotics to restore drug efficacies, much research was directed towards finding new inhibitor molecules to be used in combinatorial therapies. The evaluation of the efficiency of an inhibitor molecule requires the measurement of additional kinetic parameters, IC50 and/or Ki. In Table 7 presented are the kinetic parameters of different inhibitors tested against OXA-10, OXA-23, and OXA-48.
Table 7.
Kinetic parameters of different inhibitors tested against OXA-10, OXA-23, and OXA-48.
| OXA-10 | OXA-23 | OXA-48 | ||
|---|---|---|---|---|
| FIRST GENERATION INHIBITORS | ||||
| Clavulanic acid | IC50 (nM) | 810 [113] | 2.1x108 [65] | 1.6x104 [114] 2.85x104 [115] |
| Ki (nM) | (1.04 ± 0.02)x105 [116] | |||
| Sulbactam | IC50 (nM) | 3.7x104 [113] | 3.3x109 [65] | 5x104 [114] |
| Ki (nM) | 1.3x105 [117] | |||
| kinact/Ki (M−1 s−1) | 22 [117] | 4 [117] | ||
| Tazobactam | IC50 (nM) | 9.4x102 [113] | (1.5 ± 0.2)x103 [118] (2.13 ± 0.80)x103 [90] 3.1x107 [65] |
550 [119] (1.5 ± 0.5)x103 [109] 1.7x103 [114] 2.0x104 [115] |
| Ki (nM) | (1.7 ±0.17)x105 [116] |
(1.14 ± 0.25)x104 [90] | (3 ± 0.3)x104 [109] | |
| kinact/Ki (M−1 s−1) | (1.18 ± 0.078)x103 [90] | (3 ± 0.5)x103 [109] | ||
| k2/K (M−1 s−1) | 193.53 ± 39.10 [90] | |||
| NEW GENERATION INHIBITORS | ||||
| Diazabicyclooctane derivatives | ||||
| Avibactam (NXL104) |
IC50 (nM) | 445 [61] | 1.78x103 [61] (3.1 ± 0.6)x103 [120] (8.93 ± 0.99)x103 [90] |
180 ± 50 [120] 550 [119] 593 [61] 880 [121] (1.7 ± 0.2)x103 [118] |
| Ki (nM) | (1.7 ± 0.4)x103 [122] >1.0x105 [123] |
27 ± 15 [122] 260 ± 5 [124] (3 ± 0.3)x104 [123] |
||
| kinact/Ki (M−1 s−1) | 70 [125] | 100 [125] 286.60 ± 61.78 [90] |
5x103 [125] | |
| k2/K (M−1 s−1) | 11 ± 1 [126] | 300 ± 20 [127] | (1.4 ± 0.1)x103 [126] | |
| Relebactam (MK-7655) |
IC50 (nM) | (9 ± 0.3)x104 [120] | ||
| Ki (nM) | >1.0x105 [123] | >1.0x105 [123] | ||
| kinact/Ki (M−1 s−1) | 44 [128] | 6 [128] | 39 [128] | |
| Zidebactam (WCK-5107) |
IC50 (nM) | |||
| Ki (nM) | >1.0x105 [123] | >1.0x105 [123] | ||
| Durlobactam (EXT2514) |
IC50 (nM) | |||
| Ki (nM) | ||||
| kinact/Ki (M−1 s−1) | (9 ± 2)×103 [125] | (5.1 ± 0.2)x103 [125] | (8 ± 2)x105 [125] | |
| ETX1317 | IC50 (nM) | 77 [121] | ||
| Ki (nM) | ||||
| kinact/Ki (M−1 s−1) | 680 ± 30 [125] 8.6x103 [128] |
(1.54 ± 0.06)x103 [125] 5.1x103 [128] |
(5.3 ± 0.2)x104 [125] 8.3x105 [128] |
|
| WCK4234 | IC50 (nM) | |||
| Ki (nM) | 8.0x103 [123] | 290 [123] | ||
| kinact/Ki (M−1 s−1) | 9.5x103 [128] | 1.4x104 [128] | 6x105 [128] | |
| Boronic acid derivatives | ||||
| Vaborbactam (RPX7009) |
IC50 (nM) | >4x105 [129] | (1.2 ± 0.2)x105 [120] | (6.9 ± 2.3)x103 [120] 3.2x104 [129] 3.88x104 [119] |
| Ki (nM) | > 4.0 × 104 [122] | (1.4 ± 0.5)x104 [122] 350 ± 7 [124] |
||
| Taniborbactam (VNRX-5133) |
IC50 (nM) | 234−/645+ [130] | 537−/2390+ [130] 420 [119] |
|
| Ki (nM) | 350 ± 7 [124] | |||
| QPX7728 | IC50 (nM) | 1.2 ± 0.4 [120] | 1.1 ± 0.4 [120] | |
| Ki (nM) | 0.74 [122] | 0.28 [122] | ||
| k2/K (M−1 s−1) | (9.9 ± 0.6)×105 [120] | (2.75 ± 0.09)x106 [120] | ||
| β-lactam derivatives | ||||
| Enmetazobactam (AAI101) |
IC50 (nM) | (11 ± 1)x103 [118] | ||
| Ki (nM) | ||||
| LN-1-255 | IC50 (nM) | 12±8 [90] | 3 ± 0.3 [109] | |
| Ki (nM) | 88 ± 6 [90] | 170 ± 10 [109] | ||
| kinact/Ki (M−1 s−1) | (1.39 ± 0.29)x105 [90] | |||
| k2/K (M−1 s−1) | (2.90 ± 0.03)x104 [90] | (10 ± 1)×104 [109] | ||
−/+: in the absence/presence of 100 mM aqueous sodium bicarbonate.
First-generation inhibitors (clavulanic acid, sulbactam, and tazobactam) are effective against class A serine β-lactamases, while they are proven to be futile against class C and class D serine β-lactamases. The limited spectrum of first-generation inhibitors necessitated the development of novel β-lactamase inhibitors [81]. Currently, as new generation inhibitors, different inhibitor scaffolds are available; such as diazabicyclooctane (DBO) derivatives, boronic acid derivatives, and β-lactam derivatives. Unlike first-generation inhibitors, DBO and boronic acid derivatives have non-β-lactam scaffolds that also target the active site of the enzyme [81]. The DBO inhibitors display different activities against OXA-10, OXA-23, and OXA-48. For example; OXA-48 is more susceptible to avibactam with IC50 values in the nanomolar range and with a k2/K value of (1.4 ± 0.1)x103 M−1 s−1, while OXA-10 is not susceptible at all. On the other hand, the same DBO moderately inhibits OXA-23. Other DBO-derived inhibitors, durlobactam, ETX1317, and WCK4234 are potent inhibitors for all three enzymes. Among the boronic acid inhibitors; taniborbactam shows inhibitory effect against OXA-10 and OXA-48. QPX7728 shows a high affinity for OXA-23 and OXA-48 with very low Ki values. IC50 values of this inhibitor are very low, which indicates its great potential for clinical use. LN-1-255 is a β-lactam inhibitor among these new generation inhibitors with higher or broader activity than the first-generation inhibitors. Kinetic values show that this inhibitor has a significant inhibitory effect on OXA-23 and OXA-48 enzymes (Table 7). Other novel inhibitors such as ANT2681 (Antabio), J-110,441, and J-111,225 have displayed potency against other classes of β-lactamases but information on their activities against OXA-10, OXA-23, and OXA-48 is not available.
Computational studies on OXA-10, OXA-23, and OXA-48
Computational studies gain increasing attention since they not only shed light on the mechanistic details at the structural level but also support experimental findings. To this end, several approaches, e.g. molecular docking and molecular dynamics (MD), are commonly employed to study enzymes (Figure 5). The primary goal in such studies is usually to elucidate catalytic mechanisms, to find potential inhibitors, and to understand inhibition mechanisms at the molecular level. Computational docking and MD simulation studies of OXA enzymes with a specific emphasis on OXA-10, OXA-23, and OXA-48 structures and their selected variants are briefly summarized below.
Figure 5.
Schematic representation of computational workflow for drug discovery.
In one of the earlier studies of OXA-48, the crystallized structure of the protein (PDB ID: 3HBR) was used for computations to get an insight into the mechanism of carbapenem hydrolysis [51]. Molecular docking followed by 10 ns MD simulations were conducted with the inhibitor (meropenem) bound structure to identify critical residues for carbapenem hydrolysis. The structural basis for different functional properties of the OXA enzymes toward carbapenems and oxacillins was investigated and a new catalytic mechanism, which relied on the nature and conformation of the β5-β6 loop residues was proposed. A comparative docking study by Stojanoski et al. (2015) on OXA-48 and OXA-163 structures undertook an effort to understand the underlying mechanism in their different substrate specificities [107]. Different than OXA-48 by four amino acid deletions (214-Arg-Ile-Glu-Pro-217) in the loop region connecting β5 and β6 strands and one substitution on the β5-strand (Ser212Asp), OXA-163 displays increased cephalosporinase and decreased carbapenemase activities due to an enlargement in the active site. A later study by Pina Vaz et al. (2016) tried to understand the affinity of different drugs to different types of enzymes produced by Enterobacteriaceae using experimental methods complemented with computational approaches [82]. Their results showed that among ertapenem, doripenem, meropenem, and imipenem; ertapenem had the highest affinity, while imipenem had the lowest. A similar combined approach of experimental and molecular docking techniques to investigate OXA enzymes was by Fröhlich et al. (2019), which focused on the effects of ceftazidime (CAZ) and CAZ-AVI (Ceftazidime-Avibactam) on OXA-48 and the epidemic OXA-48 plasmid in Escherichia coli [79]. Based on their experiments with both wild-type and mutated OXA-48 structures (OXA-48: Pro68Ala, Tyr211Ser), they concluded that CAZ hydrolysis is mechanistically infeasible in OXA-48 and that Pro68Ala mutation results in increased activity towards CAZ, whereas Pro68Ala and Tyr211Ser mutations together lead to decreased activity. Since crystallization of the structure with these two mutations (OXA-48: Pro68Ala, Tyr211Ser) was unsuccessful, computational modeling was helpful to get a deeper understanding of the underlying mechanism. More recently, Hirvonen et al. (2020) [78] used QM/MM and MD simulations to understand cephalosporin breakdown by OXA-48 and OXA-163 variants. They reported that in OXA-163, extra water molecules were able to enter the active site accelerating ceftazidime breakdown, which suggested that the differences in the variants can be related to changes in solvation. In another study, molecular docking calculations were performed with imipenem and temocillin to shed light on the observation of different hydrolytic parameters [83]. They suggested alternative binding modes and conformations. Together with their experimental observations, which were consistent with computational results, their mutational analysis showed that residue 214 in OXA-48-like β-lactamases is critical for carbapenemase activity. More recently, Pestana-Nobles et al. (2022) [84] conducted a computational study based on molecular docking and MD simulations aiming to find new inhibitors from microalgal metabolites against six β-lactamase enzymes including OXA-48. Their computational studies revealed that metabolites belonging to the same structural families, (phenylacridine (4-Ph), quercetin (Qn), and cryptophycin (Cryp)), exhibit better performance with β-lactamase than the existing commercial inhibitors (clavulanic acid, sulbactam, and tazobactam). To this end, they suggested the usage of these metabolites as novel inhibitors as well as possible structural templates to be used in further studies.
One of the first modeling studies with OXA-10 was conducted by Johnson et al. (2010) [85]. As they re-evaluated the cyclobutanone analogs as potential inhibitor compounds, they modeled enzyme-inhibitor complexes to gain insight into specific active site interactions and provide support for the inhibition observed with the synthesized molecules. Though their inhibition was modest, with molecular modeling studies they were able to suggest modifications on the inhibitor compounds to increase their affinity with the enzymes. Malathi et al. (2016) conducted molecular docking and MD simulations with imipenem and imipenem-like drugs retrieved from the ZINC database [86] not only to propose new non-hydrolyzing inhibitors for P. aeruginosa OXA-10 (PDB: 1FOF) but also to understand the resistance mechanism at a structural level [87]. They came up with the molecule ZINC44672480 possessing ideal characteristics to be a potential inhibitor. In another study, Singh et al. (2017) [88] focused on an OXA-10 from an A. baumannii strain. They first constructed the 3D structure of OXA-10 from A. baumannii by homology modeling and then carried out molecular docking with different protease inhibitor compounds of cyanobacterial origin. They suggested kempopeptin as a potential inhibitor to be tested for its in vivo inhibitory activities. In the studies by Kotsakis et al. (2019) and Leiros et al. (2020), the focus was on the differences among OXA-10, OXA-656, and OXA-655. An experimental study was complemented with 10 ns MD simulations to elucidate interaction profiles on a residue-based level [31,89]. Simulations revealed Val117Leu to affect internal interactions, which in turn has a role in carbapenem alignment through the structural shifts in the active site.
As for OXA-23, computational studies are very limited. In a study by Smith et al. (2013), MD simulations were conducted to complement experimental observations, which included the crystallization of OXA-23 structures of wild type strains, under varying pH conditions and in complex with meropenem (PDB ID: 4JF4, 4JF5, and 4JF6) [57]. The molecular mechanism of carbapenemase activity was investigated in detail and it was observed that the binding of the carbapenem substrate causes a substantial movement in the conserved sites, which is stabilized by the special fold of the β6/β7 loop allowing a water molecule to enter the enzyme’s active site. In another study, the inhibitory activity of LN-1-255 was characterized and compared with avibactam and tazobactam against class D carbapenemases in A. baumannii (OXA-23, OXA-24/40, OXA-58, OXA-143, OXA-235, and OXA-51) [90]. Experimental studies followed by computational docking studies showed that LN-1-255 was effective and is a potential new inhibitor that might have a significant role against infections. A recent computational biology study aimed to find novel inhibitors for OXA enzymes (OXA-1, OXA-10, OXA-23, OXA-24/40, OXA-48, OXA-51, and OXA-58) [91] using virtual screening followed by 50 ns MD simulations. Two known structures (cilastatin and meropenem) and two newly identified potential inhibitors were docked and binding energies and bond formations were examined in detail. As a result, two new novel inhibitors were proposed (M1593 and M2680) for OXA variants that can be used as reversible competitive inhibitors.
Future prospects in novel inhibitor search
The alarming increase in the transmission of multidrug-resistant (MDR) pathogenic bacteria, especially those expressing extended-spectrum β-lactamases, makes it very difficult to cope with such bacteria using existing drugs and approaches. Furthermore, the most widely used β-lactamase inhibitors of combinatorial treatments in the clinic are commonly ineffective against class B, C, and D enzymes. Prudent support for the development of new antibiotics and alternative solutions are required to avoid antibacterial agents exhibiting cross-resistance [92]. In this context, the focus is turned to nature, which serves as a rich source for untapped novel bioactive compounds. Indeed, clavulanic acid, the first β-lactamase inhibitor introduced into clinical medicine, is a natural product [3]. These compounds can be administered alone or in combinatorial therapies to fight against increased incidences of antibiotic resistance. Thus, natural products obtained from different sources, which are not limited to microbial and plant cells, play key roles in the discovery of new molecules with distinct antimicrobial mechanisms and multi-target properties [93].
Several experimental studies report the potential of natural compounds to inhibit the different classes of β-lactamases, also providing limited mechanistic details [94–99]. However, these studies primarily focus on enzyme classes other than D or work with a collection of strains that may synthesize OXA types along with other enzyme classes. Therefore, the specificity of the inhibitor molecule under question remains vague. Among the available experimental studies specific to OXAs, Somboro et al. (2019) [100] and Nishimura et al. (2021) [101] have reported natural product based inhibitors for OXA-48, tannic acid (a polyphenolic biomolecule) and JBIR-155 (a molecule from a Streptomyces polymachus strain). A few recent studies report the antibacterial and synergistic activities of natural products against OXA-48 producing K. pneumoniae [102] and OXA-23 producing A. baumannii [103], but no details on their mechanisms are available; thus it is not clear if these molecules can function as inhibitors.
Present findings strongly suggest the importance of molecules of natural origin for the identification of new compounds and medicinal leads in β-lactamase inhibition. Despite this potential, studies with natural products targeting OXA enzymes of ESKAPE bugs are very limited. The increase in the availability of structural and mechanistic details of these enzymes, combined with the development of novel computational tools, is valuable to push this rate in the discovery of new inhibitor molecules. The implementation of the power of computers and bioinformatic tools not only to find ligands targeting active sites but also to find ligands targeting allosteric sites will further increase the pace of the fight against this worldwide health crisis. Here, a challenging approach would be to design/discover a candidate molecule to target multiple pathogens.
Acknowledgements
BSA acknowledges TUBITAK-118Z572.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Meziane-Cherif D, Bonnet R, Haouz A, et al. Structural insights into the loss of penicillinase and the gain of ceftazidimase activities by OXA-145 β-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother. 2016;71(2):395–402. [DOI] [PubMed] [Google Scholar]
- [2].Hocquet D, Colomb M, Dehecq B, et al. Ceftazidime-hydrolysing -lactamase OXA-145 with impaired hydrolysis of penicillins in Pseudomonas aeruginosa. J Antimicrob Chemother. 2011;66(8):1745–1750. [DOI] [PubMed] [Google Scholar]
- [3].Drawz SM, Bonomo RA.. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23(1):160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264(5157):375–382. [DOI] [PubMed] [Google Scholar]
- [5].Green DW. The bacterial cell wall as a source of antibacterial targets. Expert Opin Ther Targets. 2002;6(1):1–19. [DOI] [PubMed] [Google Scholar]
- [6].Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39(6):1211–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ambler RP. The Structure of Beta-lactamases. Philos Trans R Soc B Biol Sci. 1980;289:321–331. [DOI] [PubMed] [Google Scholar]
- [8].Hall BG, Barlow M. Revised Ambler classification of β-lactamases. J Antimicrob Chemother. 2005;55(6):1050–1051. [DOI] [PubMed] [Google Scholar]
- [9].Naas T, Nordmann P. OXA-type beta-lactamases. Curr Pharm Des. 1999;5(11):865–879. [PubMed] [Google Scholar]
- [10].Jaurin B, Grundstrom T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci. 1981;78(8):4897–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Medeiros AA. Beta-Lactamases. Br Med Bull. 1984;40(1):18–27. [DOI] [PubMed] [Google Scholar]
- [12].Ouellette M, Bissonnette L, Roy PH. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc Natl Acad Sci. 1987;84(21):7378–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother. 2010;54(1):24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2008;197(8):1079–1081. [DOI] [PubMed] [Google Scholar]
- [15].Leonard DA, Bonomo RA, Powers RA. Class D β‑lactamases: a reappraisal after five decades. Acc Chem Res. 2013;46(11):2407–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Golemi D, Maveyraud L, Vakulenko S, et al. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc Natl Acad Sci U S A. 2001;98(25):14280–14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Toth M, Antunes NT, Stewart NK, et al. Class D β-lactamases do exist in gram-positive bacteria. Nat Chem Biol. 2016;12(1):9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Beyrouthy R, Robin F, Cougnoux A, et al. Chromosome-mediated OXA-48 carbapenemase in highly virulent Escherichia coli. J Antimicrob Chemother. 2013;68(7):1558–1561. [DOI] [PubMed] [Google Scholar]
- [19].Antunes NT, Fisher JF. Acquired class D β-Lactamases. Antibiotics. 2014;3(3):398–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pitout JDD, Peirano G, Kock MM, et al. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2020;33:1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pandey D, Singhal N, Kumar M. Investigating the OXA variants of ESKAPE pathogens. Antibiotics. 2021;10(12):1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Khan AU. Beta-lactomics: a new term coined in “OMICS. J Proteomics Bioinform. 2014;7(10):10000e27. [Google Scholar]
- [23].Huovinen P, Huovinen S, Jacoby GA. Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother. 1988;32(1):134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Philippon AM, Paul GC, Jacoby GA. Properties of PSE-2 beta-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1983;24(3):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Matthew M, Sykes RB. Properties of the Beta-lactamase specified by the Pseudomonas Plasmid RPL11. J Bacteriol. 1977;132(1):341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hall LMC, Livermore DM, Gur D, et al. Oxa-11, an extended-spectrum variant of Oxa-10 (Pse-2) Beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37(8):1637–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Aibinu IE, Pfeifer Y, Ogunsola F, et al. Emergence of -lactamases OXA-10, VEB-1 and CMY in providencia spp. from Nigeria. J Antimicrob Chemother. 2011;66(8):1931–1932. [DOI] [PubMed] [Google Scholar]
- [28].Maurya AP, Dhar D, Basumatary MK, et al. Expansion of highly stable bla OXA-10 β-lactamase family within diverse host range among nosocomial isolates of gram-negative bacilli within a tertiary referral hospital of Northeast India. BMC Res Notes. 2017;10(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kiratisin P, Apisarnthanarak A, Laesripa C, et al. Molecular characterization and epidemiology of extended-spectrum- β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008;52(8):2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Antunes NT, Lamoureaux TL, Toth M, et al. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother. 2014;58(4):2119–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kotsakis SD, Flach CF, Razavi M, et al. Characterization of the first OXA-10 natural variant with increased carbapenemase activity. Antimicrob Agents Chemother. 2019;63(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mugnier PD, Poirel L, Naas T, et al. Worldwide dissemination of the bla OXA-23 carbapenemase gene of Acinetobacter baumannii 1. Emerg Infect Dis. 2010;16(1):35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ning NZ, Liu X, Bao CM, et al. Molecular epidemiology of bla OXA-23 -producing carbapenem-resistant Acinetobacter baumannii in a single institution over a 65-month period in north China. BMC Infect Dis. 2017;17(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Goic-Barisic I, Kovacic A, Medic D, et al. Endemicity of OXA-23 and OXA-72 in clinical isolates of Acinetobacter baumannii from three neighbouring countries in Southeast Europe. J Appl Genet. 2021;62(2):353–359. [DOI] [PubMed] [Google Scholar]
- [35].Principe L, Piazza A, Giani T, et al. Epidemic diffusion of OXA-23-producing Acinetobacter baumannii isolates in Italy: results of the first cross-sectional countrywide survey. J Clin Microbiol. 2014;52(8):3004–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Merino M, Poza M, Roca I, et al. Nosocomial Outbreak of a Multiresistant Acinetobacter baumannii Expressing OXA-23 Carbapenemase in Spain. Microb Drug Resist. 2014;20(4):259–263. [DOI] [PubMed] [Google Scholar]
- [37].Lee Y, Kim YR, Kim J, et al. Increasing prevalence of blaOXA-23-carrying Acinetobacter baumannii and the emergence of blaOXA-182-carrying Acinetobacter nosocomialis in Korea. Diagn Microbiol Infect Dis. 2013;77(2):160–163. [DOI] [PubMed] [Google Scholar]
- [38].Opazo A, Domínguez M, Bello H, et al. OXA-type carbapenemases in Acinetobacter baumannii in South America. J Infect Dev Ctries. 2012;6(4):311–316. [DOI] [PubMed] [Google Scholar]
- [39].Neves FC, Clemente WT, Lincopan N, et al. Clinical and microbiological characteristics of OXA-23- and OXA-143-producing Acinetobacter baumannii in ICU patients at a teaching hospital, Brazil. Brazilian J Infect Dis. 2016;20(6):556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Paton R, Miles RS, Hood J, et al. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2(2):81–87. [DOI] [PubMed] [Google Scholar]
- [41].Donald HM, Scaife W, Amyes SGB, et al. Sequence Analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44(1):196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Scaife W, Young HK, Paton RH, et al. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36(3):585–586. [DOI] [PubMed] [Google Scholar]
- [43].Nordmann P, Cuzon G, Naas T. The real threat of KPC carbapenemase– producing bacteria. Lancet Infect Dis. 2009;9(4):228–236. [DOI] [PubMed] [Google Scholar]
- [44].Cuzon G, Naas T, Truong H, et al. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase bla KPC-2 gene1. Emerg Infect Dis. 2010;16(9):1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Paterson DL, Hujer KM, Hujer AM, et al. Extended-Spectrum β-Lactamases in Klebsiella pneumoniae Bloodstream Isolates from Seven Countries: dominance and Widespread Prevalence of SHV- and CTX-M-Type β-Lactamases. Antimicrob Agents Chemother. 2003;47(11):3554–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Daikos GL, Karabinis A, Syriopoulou VP, et al. VIM-1-producing Klebsiella pneumoniae bloodstream infections: analysis of 28 cases. Int J Antimicrob Agents. 2007;29(4):471–483. [DOI] [PubMed] [Google Scholar]
- [47].Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhu YL, Zhang XN, Gao F, et al. ACT-6, a novel plasmid-encoded class C Β-lactamase in a Klebsiella pneumoniae isolate from China. J Antibiot (Tokyo). 2011;64(4):317–320. [DOI] [PubMed] [Google Scholar]
- [49].Bauernfeind A, Schneider I, Jungwirth R, et al. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother. 1999;43(8):1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Poirel L, Héritier C, Tolün V, et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Docquier JD, Calderone V, De Luca F, et al. Crystal Structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol. 2009;16(5):540–547. [DOI] [PubMed] [Google Scholar]
- [52].Docquier JD, Mangani S. Structure-function relationships of class D carbapenemases. Curr Drug Targets. 2016;17(9):1061–1071. [DOI] [PubMed] [Google Scholar]
- [53].Smith CA, Stewart NK, Toth M, et al. Structural insights into the mechanism of carbapenemase activity of the OXA-48 β-lactamase. Antimicrob Agents Chemother. 2019;63(10):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schneider KD, Ortega CJ, Renck NA, et al. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J Mol Biol. 2011;406(4):583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Paetzel M, Danel F, De Castro L, et al. Crystal structure of the class D β-lactamase OXA-10. Nat Struct Biol. 2000;7(10):918–925. [DOI] [PubMed] [Google Scholar]
- [57].Smith CA, Antunes NT, Stewart NK, et al. Structural basis for carbapenemase activity of the OXA-23 β-Lactamase from Acinetobacter baumannii. Chem Biol. 2013;20(9):1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Maveyraud L, Golemi D, Kotra LP, et al. Insights into class D β-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa. Structure. 2000;8(12):1289–1298. [DOI] [PubMed] [Google Scholar]
- [59].Couture F, Lachapelle J, Levesque RC. Phylogeny of LCR‐1 and OXA‐5 with class A and class D β‐lactamases. Mol Microbiol. 1992;6(12):1693–1705. [DOI] [PubMed] [Google Scholar]
- [60].Danel F, Hall LMC, Gur D, et al. OXA-16, a further extended-spectrum variant of OXA-10 β-lactamase, from two pseudomonas aeruginosa Isolates. Antimicrob Agents Chemother. 1998;42(12):3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lohans CT, Wang DY, Jorgensen C, et al. 13 C-Carbamylation as a mechanistic probe for the inhibition of class D β-lactamases by avibactam and halide ions. Org Biomol Chem. 2017;15(28):6024–6032. [DOI] [PubMed] [Google Scholar]
- [62].De Luca F, Benvenuti M, Carboni F, et al. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D β-lactamase OXA-10 by rational protein design. Proc Natl Acad Sci U S A. 2011;108(45):18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schneider KD, Bethel CR, Distler AM, et al. Mutation of the active site carboxy-lysine (K70) of OXA-1 β-lactamase results in a deacylation-deficient enzyme. Biochemistry. 2009;48(26):6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pratap S, Katiki M, Gill P, et al. Active-site plasticity is essential to carbapenem hydrolysis by OXA-58 class D β-lactamase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60(1):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Torol S, Kasap M. Purification and characterization of OXA-23 from Acinetobacter baumannii. J Enzyme Inhib Med Chem. 2013;28(4):836–842. [DOI] [PubMed] [Google Scholar]
- [66].Kaitany KCJ, V KN, June CM, et al. Structures of the class D carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob Agents Chemother. 2013;57(10):4848–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liapis E, Pantel A, Robert J, et al. Molecular epidemiology of OXA-48-producing Klebsiella pneumoniae in France. Clin Microbiol Infect. 2014;20(12):O1121–O1123. [DOI] [PubMed] [Google Scholar]
- [68].Cuzon G, Ouanich J, Gondret R, et al. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother. 2011;55(5):2420–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kasap M, Torol S, Kolayli F, et al. OXA-162, a novel variant of OXA-48 displays extended hydrolytic activity towards imipenem, meropenem and doripenem. J Enzyme Inhib Med Chem. 2013;28(5):990–996. [DOI] [PubMed] [Google Scholar]
- [70].Jeon JH, Lee JH, Lee JJ, et al. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci. 2015;16(12):9654–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schneider KD, Karpen ME, Bonomo RA, et al. The 1.4 Å crystal structure of the class D β-Lactamase OXA-1 complexed with doripenem. Biochemistry. 2009;48(50):11840–11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Stewart NK, Smith CA, Antunes NT, et al. Role of the hydrophobic bridge in the carbapenemase activity of class D beta-lactamases. Antimicrob Agents Chemother. 2019;63(2):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Harper TM, June CM, Taracila MA, et al. Multiple substitutions lead to increased loop flexibility and expanded specificity in Acinetobacter baumannii carbapenemase OXA-239. Biochem J. 2018;475(1):273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Selim S, Faried OA, Almuhayawi MS, et al. Dynamic gene clusters mediating carbapenem-resistant Acinetobacter baumannii clinical isolates. Antibiotics. 2022;11(2):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Stewart NK, Toth M, Alqurafi MA, et al. C6 hydroxymethyl-substituted carbapenem MA-1-206 inhibits the major Acinetobacter baumannii carbapenemase oxa-23 by impeding deacylation. MBio. 2022;e0036722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Banerjee S, Pieper U, Kapadia G, et al. Role of the Ω-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry. 1998;37(10):3286–3296. [DOI] [PubMed] [Google Scholar]
- [77].Hirvonen VHA, Spencer J, van der Kamp MW. Antimicrobial resistance conferred by OXA-48 β-lactamases: towards a detailed mechanistic understanding. Antimicrob Agents Chemother. 2021;65(6):e00184–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hirvonen VHA, Mulholland AJ, Spencer J, et al. Small Changes in Hydration Determine Cephalosporinase Activity of OXA-48 β-Lactamases. ACS Catal. 2020;10(11):6188–6196. [Google Scholar]
- [79].Fröhlich C, Sørum V, Thomassen AM, et al. OXA-48-mediated ceftazidime-avibactam resistance is associated with evolutionary trade-offs. mSphere. 2019;4(2):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Matagne A, Lamotte-Brasseur J, Frère JM. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330(2):581–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vázquez-Ucha JC, Arca-Suárez J, Bou G, et al. New carbapenemase inhibitors: clearing the way for the β-lactams. Int J Mol Sci. 2020;21(23):1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Pina-Vaz C, Silva AP, Faria-Ramos I, et al. A flow cytometric and computational approaches to carbapenems affinity to the different types of carbapenemases. Front Microbiol. 2016;7:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Oueslati S, Retailleau P, Marchini L, et al. Role of arginine 214 in the substrate specificity of OXA-48. Antimicrob Agents Chemother. 2020;64(5):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pestana-Nobles R, Aranguren-Díaz Y, Machado-Sierra E, et al. Docking and molecular dynamic of microalgae compounds as potential inhibitors of Beta-lactamase. Int J Mol Sci. 2022;23(3):1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Johnson JW, Gretes M, Goodfellow VJ, et al. Cyclobutanone analogues of β-lactams revisited: insights into conformational requirements for inhibition of serine- and metallo-β-lactamases. J Am Chem Soc. 2010;132(8):2558–2560. [DOI] [PubMed] [Google Scholar]
- [86].Sterling T, Irwin JJ. ZINC 15 – ligand Discovery for Everyone. J Chem Inf Model. 2015;55(11):2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Malathi K, Ramaiah S. Molecular docking and molecular dynamics studies to identify potential OXA-10 extended spectrum β-lactamase non-hydrolysing inhibitors for Pseudomonas aeruginosa. Cell Biochem Biophys. 2016;74(2):141–155. [DOI] [PubMed] [Google Scholar]
- [88].Singh RK, Mishra S, Singh VK, et al. Molecular modeling and docking studies on OXA-10 enzyme of Acinetobacter baumannii. J Pharm Res. 2017;11:352–358. [Google Scholar]
- [89].Leiros HKS, Thomassen AM, Samuelsen Ø, et al. Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10. FEBS Open Bio. 2020;10(9):1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Vázquez-Ucha J, Maneiro M, Martínez-Guitián M, et al. Activity of the β-lactamase inhibitor LN-1-255 against carbapenem-hydrolyzing class D β-lactamases from Acinetobacter baumannii. Antimicrob Agents Chemother. 2017;61(11):e01172–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gupta D, Singh A, Somvanshi P, et al. Structure-based screening of non-β-lactam inhibitors against class D β-lactamases: an approach of docking and molecular dynamics. ACS Omega. 2020;5(16):9356–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Docquier JD, Mangani S. An update on β-lactamase inhibitor discovery and development. Drug Resist Updat. 2018;36:13–29. [DOI] [PubMed] [Google Scholar]
- [93].Prakash B, Kujur A, Yadav A. Drug synthesis from natural products: a historical overview and future perspective. In: Tewari A, Tiwari S, editors. Synthesis of Medicinal Agents from Plants. Elsevier; 2018. p. 25–46. doi: 10.1016/C2016-0-01140-3. [DOI] [Google Scholar]
- [94].King AM, Reid-Yu SA, Wang W, et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature. 2014;510(7506):503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sychantha D, Rotondo CM, Tehrani KHME, et al. Aspergillomarasmine A inhibits metallo-β-lactamases by selectively sequestering Zn 2. J Biol Chem. 2021;297(2):100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Liu S, Zhou Y, Niu X, et al. Magnolol restores the activity of meropenem against NDM-1-producing Escherichia coli by inhibiting the activity of metallo-beta-lactamase. Cell Death Discov. 2018;4(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Vinod N, Shijina R, Dileep K, et al. Inhibition of Beta-lactamase by 1,4-naphthalenedione from the plant Holoptelea integrifolia. Appl Biochem Biotechnol. 2010;160(6):1752–1759. [DOI] [PubMed] [Google Scholar]
- [98].Payne DJ, Hueso-Rodríguez JA, Boyd H, et al. Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-β-lactamases. Antimicrob Agents Chemother. 2002;46(6):1880–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Aydemir E, Sarıyer E, Akyıldız E, et al. In vitro and in silico evaluation of some plant extracts and phytocompounds against multidrug-resistant Gram-negative bacteria. Adv Tradit Med. 2021;1–11. [Google Scholar]
- [100].Somboro AM, Osei Sekyere J, Amoako DG, et al. In vitro potentiation of carbapenems with tannic acid against carbapenemase-producing enterobacteriaceae: exploring natural products as potential carbapenemase inhibitors. J Appl Microbiol. 2019;126(2):452–467. [DOI] [PubMed] [Google Scholar]
- [101].Nishimura T, Kawahara T, Kagaya N, et al. JBIR-155, a specific class D β-lactamase inhibitor of microbial Origin. Org Lett. 2021;23(11):4415–4419. [DOI] [PubMed] [Google Scholar]
- [102].Shakib P, Taherikalani M, Ramazanzadeh R, et al. Chemical Composition, Genotoxicity and Antimicrobial Activities of Dracocephalum kotschyi Boiss against OXA-48 Producing Klebsiella pneumoniae Isolated from Major Hospitals of Kurdistan Province, Iran. Microbiol Res J Int. 2018;24(3):1–8. [Google Scholar]
- [103].Vasconcelos NG, Mallmann V, Costa ÉR, et al. Antibacterial activity and synergism of the essential oil of Nectandra megapotamica (L.) flowers against OXA-23-producing Acinetobacter baumannii. J Essent Oil Res. 2020;32(3):260–268. [Google Scholar]
- [104].Baurin S, Vercheval L, Bouillenne F, et al. Critical role of Tryptophan 154 for the Activity and Stability of Class D β-Lactamases. Biochemistry. 2009;48(47):11252–11263. [DOI] [PubMed] [Google Scholar]
- [105].Vercheval L, Bauvois C, Di Paolo A, et al. Three factors that modulate the activity of class D β-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem J. 2010;432(3):495–504. [DOI] [PubMed] [Google Scholar]
- [106].Poirel L, Kieffer N, Nordmann P. Stability of cefiderocol against clinically significant broad-spectrum oxacillinases. Int J Antimicrob Agents. 2018;52(6):866–867. [DOI] [PubMed] [Google Scholar]
- [107].Stojanoski V, Chow DC, Fryszczyn B, et al. Structural basis for different substrate profiles of two closely related class D β-lactamases and their inhibition by halogens. Biochemistry. 2015;54(21):3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lund BA, Christopeit T, Guttormsen Y, et al. Screening and design of inhibitor scaffolds for the antibiotic resistance oxacillinase-48 (OXA-48) through surface plasmon resonance screening. J Med Chem. 2016;59(11):5542–5554. [DOI] [PubMed] [Google Scholar]
- [109].Vallejo JA, Martínez-Guitián M, Vázquez-Ucha JC, et al. LN-1-255, a penicillanic acid sulfone able to inhibit the class D carbapenemase OXA-48. J Antimicrob Chemother. 2016;71(8):2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Aertker KMJ, Chan HTH, Lohans CT, et al. Analysis of β-lactone formation by clinically observed carbapenemases informs on a novel antibiotic resistance mechanism. J Biol Chem. 2020;295(49):16604–16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].De Belder D, Ghiglione B, Pasteran F, et al. Comparative kinetic analysis of OXA-438 with related OXA-48-Type carbapenem-hydrolyzing class d β-lactamases. ACS Infect Dis. 2020;6(11):3026–3033. [DOI] [PubMed] [Google Scholar]
- [112].Antonelli A, D’Andrea MM, Vaggelli G, et al. OXA-372, a novel carbapenem-hydrolysing class D β-lactamase from a Citrobacter freundii isolated from a hospital wastewater plant. J Antimicrob Chemother. 2015;70(10):2749–2756. [DOI] [PubMed] [Google Scholar]
- [113].Payne DJ, Cramp R, Winstanley DJ, et al. Comparative activities of clavulanic acid, sulbactam, and tazobactam against clinically important Beta-lactamases. Antimicrob Agents Chemother. 1994;38(4):767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Potron A, Rondinaud E, Poirel L, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem- hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. 2013;41(4):325–329. [DOI] [PubMed] [Google Scholar]
- [115].Oueslati S, Retailleau P, Marchini L, et al. Biochemical and structural characterization of OXA-405, an OXA-48 Variant with Extended- spectrum β -lactamase activity. Microorganisms. 2020;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Drawz SM, Bethel CR, Doppalapudi VR, et al. Penicillin sulfone inhibitors of class D β-lactamases. Antimicrob Agents Chemother. 2010;54(4):1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Shapiro A. Kinetics of Sulbactam Hydrolysis by Beta- Lactamases, and Kinetics of Beta-Lactamase Inhibition by Sulbactam. Antimicrob Agents Chemother. 2017;61(12):e01612–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Papp-Wallace KM, Bethel CR, Caillon J, et al. Beyond piperacillin-tazobactam: cefepime and AAI101 as a potent -β-lactam-β-lactamase inhibitor combination. Antimicrob Agents Chemother. 2019;63(5):e00105–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Liu B, Trout REL, Chu G, et al. Discovery of Taniborbactam (VNRX-5133): a broad-spectrum serine- and metallo- β -lactamase inhibitor for carbapenem-resistant bacterial infections. J Med Chem. 2020;63(6):2789–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Tsivkovski R, Totrov M, Lomovskaya O. Biochemical characterization of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases. Antimicrob Agents Chemother. 2020;64(6):e00130–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Miller AA, Shapiro AB, Mcleod SM, et al. In Vitro characterization of ETX1317, a broad-spectrum β-lactamase inhibitor that restores and enhances β-lactam activity against multi-drug-resistant enterobacteriales, including carbapenem-resistant strains. ACS Infect Dis. 2020;6(6):1389–1397. [DOI] [PubMed] [Google Scholar]
- [122].Hecker SJ, Reddy KR, Lomovskaya O, et al. Discovery of cyclic boronic acid QPX7728, an ultrabroad-spectrum inhibitor of serine and metallo- β -Lactamases. J Med Chem. 2020;63(14):7491–7507. [DOI] [PubMed] [Google Scholar]
- [123].Papp-Wallace KM, Nguyen NQ, Jacobs MR, et al. Strategic approaches to overcome resistance against gram-negative pathogens using β-lactamase inhibitors and β-lactam enhancers: activity of three novel diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234. J Med Chem. 2018;61(9):4067–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Hamrick JC, Docquier JD, Uehara T, et al. VNRX-5133 (taniborbactam), a broad-spectrum inhibitor of serine- and metallo-β-lactamases, restores activity of cefepime in enterobacterales and pseudomonas aeruginosa. Antimicrob Agents Chemother. 2020;64(3):e01963–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Durand-Réville TF, Comita-Prevoir J, Zhang J, et al. Discovery of an orally available diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine β-Lactamases. J Med Chem. 2020;63(21):12511–12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ehmann DE, Jahić H, Ross PL, et al. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem. 2013;288(39):27960–27971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lahiri SD, Mangani S, Jahić H, et al. Molecular basis of selective inhibition and slow reversibility of avibactam against class D carbapenemases: a structure-guided study of OXA-24 and OXA-48. ACS Chem Biol. 2015;10(2):591–600. [DOI] [PubMed] [Google Scholar]
- [128].Shapiro A, Guler S, Carter N, et al. ETX2514, a Novel, Rationally Designed Inhibitor of Class A, C and D β-lactamases, for the Treatment of Gram-negative Infections. ASM Microbe 2016 Boston, MA. doi: 10.1080/14656566.2019.1660772. [DOI] [Google Scholar]
- [129].Langley GW, Cain R, Tyrrell JM, et al. Profiling interactions of vaborbactam with metallo-β-lactamases. Bioorganic Med Chem Lett. 2019;29(15):1981–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Krajnc A, Brem J, Hinchliffe P, et al. Bicyclic Boronate VNRX-5133 Inhibits Metallo- And Serine-β-Lactamases. J Med Chem. 2019;62(18):8544–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]


