ABSTRACT
In recent years, researchers have begun to realize the importance of the role of non-coding RNAs in the treatment of cancer and cardiovascular and neurological diseases. LncRNAs and miRNAs are important non-coding RNAs, which regulate gene expression and activate mRNA translation through binding to diverse target sites. Their involvement in the regulation of protein function and the modulation of physiological and pathological conditions continues to be investigated. Sirtuins, especially Sirt1, have a critical function in regulating a variety of physiological processes such as oxidative stress, inflammation, apoptosis, and autophagy. The lncRNAs/miRNAs/Sirt1 axis may be a novel regulatory mechanism, which is involved in the progression and/or prevention of numerous diseases. This review focuses on recent findings on the crosstalk between non-coding RNAs and Sirt1 in myocardial and cerebral injuries and may provide some insight into the development of novel approaches in the treatment of these disorders.
Abbreviation: BMECs, brain microvascular endothelial cells; C2dat1, calcium/calmodulin-dependent protein kinase type II subunit delta (CAMK2D)-associated transcript 1; EPCs, endothelial progenitor cells; FOXOs, forkhead transcription factors; GAS5, growth arrest-specific 5; HAECs, human aortic endothelial cells; HAND2-AS1, HAND2 Antisense RNA 1; HIF-1α, hypoxia-inducible factor-1α; ILF3-AS1, interleukin enhancer-binding factor 3-antisense RNA 1; KLF3-AS1, KLF3 antisense RNA 1; LncRNA, long noncoding RNA; LUADT1, Lung Adenocarcinoma Associated Transcript 1; MALAT1, Metastasis-associated lung adenocarcinoma transcript 1; miRNA, microRNA; NEAT1, nuclear enriched abundant transcript 1; NF-κB, nuclear factor kappa B; OIP5-AS1, Opa-interacting protein 5-antisense transcript 1; Sirt1-AS, Sirt1 Antisense RNA; SNHG7, small nucleolar RNA host gene 7; SNHG8, small nucleolar RNA host gene 8; SNHG12, small nucleolar RNA host gene 12; SNHG15, small nucleolar RNA host gene 15; STAT3, signal transducers and activators of transcription 3; TUG1, taurine up-regulated gene 1; VSMCs, vascular smooth muscle cells; XIST, X inactive specific transcript; ZFAS1, ZNFX1 Antisense RNA 1.
KEYWORDS: Non-coding RNA, microRNA, MALAT1, ANRIL, ischemia, atherosclerosis
1. Introduction
1.1. The importance of myocardial and cerebral injuries
Myocardial and cerebral injuries are among the most prevalent and costly health problems, the leading cause of severe illness and long-term disability and deaths globally [1]. Nearly 15 million people died from heart attack and stroke in 2019 [2]. The mortality from myocardial diseases is expected to decrease by about 25% in the coming years, partly due to the development of new diagnostic and treatment techniques [3]. Similarly, ischemic stroke (IS) deaths which account for nearly 87% of total strokes manifesting with complex pathogenesis [4], have also diminished over the past several decades because of advances in early diagnosis and treatment management [5].
Myocardial and cerebral injuries occur because of arterial occlusion due to thrombosis after the rupture of atherosclerotic plaques. Unfortunately, the molecular mechanism of these injuries has not been fully elucidated. For this reason, a better understanding of the key molecules involved in these disorders is needed to develop more effective preventive and therapeutic measures. The current treatment regime, while effective, has a narrow therapeutic window with numerous contraindications [6]. An understanding of the intricate cellular and molecular mechanisms underlying these injuries is essential for the development of more efficient and safer therapeutic approaches to enhance current treatment strategies.
1.2. The non-coding RNAs
Protein synthesis and the process of translation are tightly controlled in both prokaryotic and eukaryotic organisms. Translation is a key regulatory point in post-transcriptional control pathways in normal homeostasis and adverse outcomes including cancer and neurodegenerative and heart diseases [7,8]. The regulation of mRNAs translation can occur during initiation, elongation, and/or termination through RNA-binding proteins (RBPs), translation initiation factors, and non-coding RNAs such as long non-coding RNAs (lncRNAs) or microRNAs (miRNAs) [9]. The stability of mRNAs is essential for protein translation.
Improvements in RNA-sequencing methods have resulted in the identification of numerous non-coding transcripts often without understanding their functions in contrast to mRNAs, which are responsible for protein coding [10]. Recently, several kinds of non-coding RNAs with different lengths including long non-coding RNAs (lncRNAs), short interfering RNAs (siRNAs), microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), and circular RNAs (circRNA) with a closed continuous loop have been identified [11,12]. Among these non-coding RNAs, lncRNAs and miRNAs are two primary families of non-protein-coding transcripts, which play essential roles in the adjustment of numerous molecular mechanisms including gene expression [13]. Growing evidence has confirmed the importance and impact of non-coding RNAs on diverse molecular mechanisms. The non-coding RNA content correlates with an organism’s complexity, which suggests a particular involvement of non-coding RNAs in the development and organization of higher structured animals [11].
1.2.1. Long non-coding RNAs
LncRNAs the major class of linear non-coding RNAs with over 200 nucleotides in length and no protein-coding potential, were initially detected in mice [14]. LncRNAs have structural similarities with mRNA and like mRNAs, lncRNAs can make secondary structures, which undergo splicing and post-transcriptional processing such as 5′-cap structure and poly-adenylation [15]. These linear non-coding RNAs are found in the nucleus, nucleolus, mitochondria, cytoplasm, and even in some exosomes with tissue-specific expression patterns [16,17]. Their cellular location is responsible for the diverse functions of the lncRNAs [18]. For example, the nuclear lncRNAs are related to chromatin structures. Transcriptional regulation of specific genes, as well as pre-mRNA processing, are only a few of the diverse processes that nuclear lncRNAs have in controlling gene expression [19]. In contrast, cytoplasmic lncRNAs predominantly regulate the stability and translation of mRNAs [20].
Despite the low level of lncRNAs compared to protein-coding genes, increasing evidence has confirmed that lncRNAs strictly regulate cellular processes, including mRNA translation, splicing, editing, transport, and stability as well as post-translational modification [21,22]. Moreover, lncRNAs are associated with protein translation through the interaction with ribosomal RNAs or eukaryotic initiation factors (eIFs) [21]. The regulation of cellular processes involves lncRNAs binding to DNAs, mRNAs, miRNA, and/or proteins which can occur in physiological situations and disease conditions such as neurological and heart diseases, diabetes, and cancer [7,13,23]. It is shown that lncRNAs have different regulatory relationships with miRNAs through sponging miRNAs, the mutual suppression of lncRNAs and miRNAs, and the coexpression of lncRNAs with miRNAs, which positively regulates the level of miRNAs [7]. Besides, the role of miRNAs were reported as negative regulators of lncRNAs [7]. Examples include the role of lncRNA H19 in cervical cancer [24], lncRNA taurine up-regulated gene 1 (TUG1) in liver injury and obesity [25,26], and lncRNA X inactive specific transcript (XIST) and Lung Adenocarcinoma Associated Transcript 1 (LUADT1) in osteoarthritis [27,28].
1.2.2. MicroRNAs
MiRNAs, single-strand RNA with approximately 22 nucleotides in length, are another class of conserved and endogenous non-coding RNAs family, which have been reported to control more than 30% of genes [29]. Initially, RNA polymerase II or III is transcribed from DNA to form primary-miRNAs (pri-miRNAs) and then processed into precursor-miRNAs (pre-miRNAs) in the cell nucleus. The mature miRNAs are further processed from pre-miRNAs in the cytoplasm [30]. The post-transcriptional regulation mechanism of miRNAs is mediated by RISC (RNA-induced silencing complex) proteins, which is embedded one functional miRNA strand in Argonaute protein family as a central to the RISC function [31]. The interaction between mature miRNAs and target mRNAs by RISC complex through binding to 3′- or 5′-UTRs of mRNAs leads to inhibiting mRNA translation or stimulating mRNA degradation. This binding can be perfect or imperfect complementarity, resulting in the regulation of protein synthesis, for example [13,32,33]. However, a few miRNAs can activate the translation of target mRNAs [30].
In these cases, miRNAs function may be involved in certain human diseases. The involvement of miR‐29b and miR-653-5p in age-related hearing loss [34,35], miR-34a in liver injury [36], miR-204 in prostate cancer [37], and miR-204-5p in type 2 diabetes [38] have been reported. Both lncRNAs and miRNAs, therefore, have potential as novel therapeutic targets.
1.3. Sirtuin 1 (Sirt1)
Proteins carry out a wide range of biological functions according to the information encoded in their respective genes, including DNA replication, transporting molecules, catalyzing metabolic reactions, providing structure to cells, and responding to stimuli [39]. Among the intracellular proteins, the sirtuin (silent mating-type information regulation 2 homolog) family of proteins is involved in several roles essential for maintaining cellular homeostasis [40]. The sirtuins comprise the histone deacetylase class III family of enzymes, which are reliant on nicotinamide adenine dinucleotide (NAD+) for their deacetylase activity. Seven members (Sirt1 to Sirt7) of the sirtuin family have been described in mammals [41]. Sirt1 is classified as a nuclear protein and is the best characterized of the sirtuin family of proteins [42]. However, Sirt1 has been observed in both the nucleus and the cytoplasm and its movement between these two cellular compartments has been reported in several cell types and under different circumstances [43,44].
Sirt1 is activated in response to the disruption of intracellular redox potential and is involved in the regulation of several physiological processes essential for maintaining homeostasis and cell survival like senescence, inflammation, cell growth, redox homeostasis, apoptosis, DNA repair, stress resistance, and metabolism [45]. Sirt1 functions through the deacetylation of histones and nonhistone proteins such as p53, forkhead transcription factors (FOXOs), hypoxia-inducible factor-1α (HIF-1α), nuclear factor kappa B (NF-κB), and signal transducers and activators of transcription 3 (STAT3) in the lysine residues [46–50]. Additionally, the presence of Sirt1 in multiple mice and human organs such as the heart, liver, brain, lung, and kidney has been reported [51,52]. Interestingly, high expression of Sirt1 has been reported in the heart, vascular endothelial cells, brain, and spinal cord [51,53].
1.4. The lncRNAs/miRNAs/Sirt1 axis
A deficiency of Sirt1 has been reported to be crucial in the development of several cardiovascular and neuroinflammation diseases [54–56]. Sirt1 improved cerebral ischemic injury in mice by deacetylation of p53 and NF-κB and subsequently prevented or greatly reduced the activity of both the inflammatory and apoptotic pathways [57]. In another study, the cardiomyocyte protective effect of Sirt1 was reported in ischemia-reperfusion oxidative injury and cell death through a positive effect on the MAPK pathway [58]. Sirt1 function is regulated by several lncRNAs and miRNAs. Furthermore, it is demonstrated that one lncRNA can adjust diverse miRNAs, while simultaneously, the function of one miRNA can be controlled by several lncRNAs. For example, the Sirt1 expression can be affected by Sirt1 AS lncRNA through competing with different miRNAs such as miR-34a, miR-22, and miR-29c [59,60], while miR-34a also negatively regulate the Sirt1 mRNA translation via C2dat1, OIP5-AS1, and HOTAIR [61–63]. It is reported that several microRNAs such as miR-93, miR-204, miR-132, miR-22, miR-141, and miR-543 are involved in the regulation of Sirt1 expression and the repression of its synthesis [64]. On the other hand, for instance, miR-204 can be regulated by lncRNAs TUG1, UCA1, HCG11, and MEG3 in different situations [26,37,65,66]. Interestingly, the above lncRNAs such as TUG1, in addition to regulating miR-204 can cause the adjustment of other miRNAs including miR-194, miR-29, miR-200, and miR-138, all of which play a major role in the management of Sirt1 expression [25,67–69]. As a result, Sirt1 should be considered a potential therapeutic target for the treatment of such diseases.
Recent studies have suggested that lncRNAs, miRNAs, and mRNAs are associated with the initiation and progression of myocardial and cerebral injuries [7,70]. Sirt1 is involved in neuroinflammation and neurodegenerative disorders as well as ischemia/reperfusion injury in the brain and heart [54,55,71]. In this review, we discuss recent information on the relationship between these two non-coding RNAs, lncRNAs and miRNAs, and Sirt1 regulation in myocardial and cerebral injury (Figure 1).
Figure 1.
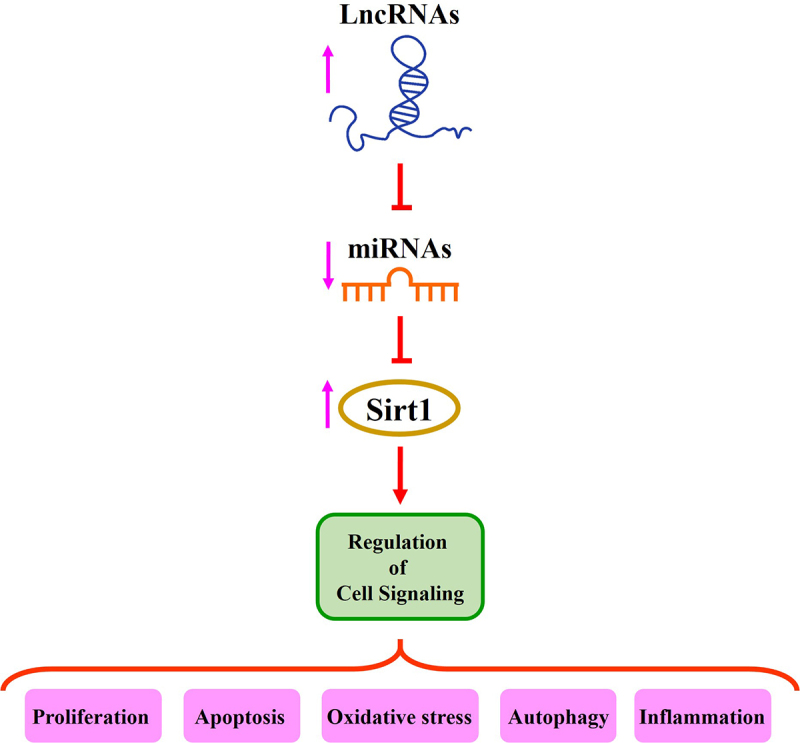
The role of lncRNAs/miRNAs/Sirt1 axis in the regulation of different signaling pathways.
2. The role of lncRNAs/miRNAs/Sirt1 axis in myocardial and cerebral injury
2.1. Myocardial injuries
2.1.1. Myocardial infarction
Myocardial infarction (MI) results from the sudden ischemic death of myocardial tissue and is usually caused by thrombotic occlusion of a coronary vessel following the rupture of an atherosclerotic plaque. Ischemia induces profound metabolic and ionic perturbations in the affected myocardium and causes a depression of systolic function. Myocardial infarction (MI) is a severe cardiac disease with high incidence and mortality worldwide [72]. The overexpression of Sirt1 has been reported to reduce the infarcted area, decreasing cardiomyocyte apoptosis as well as improving myocardial injury and heart failure caused by MI [73].
An important relationship exists between lncRNAs and miRNAs and the expression of Sirt1 in MI [74,75]. Mao et al. have reported that the overexpression of lncRNA KLF3 antisense RNA 1 (KLF3-AS1) in exosomes decreased cell apoptosis, reduced the area of the MI, and attenuated MI progression. Sirt1 expression was increased following the binding of KLF3-AS1 to miR-138-5p (ceRNA) as a miR-138-5p target in MI and hypoxic H9c2 cells, leading to an inhibition of cardiomyocytes’ pyroptosis after MI [74]. An elevated level of lncRNA ANRIL was observed in hypoxic H9c2 cells, a cellular model of MI. ANRIL competitively binds to miR‐7‐5p and reduced its function [75]. Consequently, Sirt1 increased and attenuated the hypoxia‐induced H9c2 cell injury. ANRIL positively regulated Sirt1 in this model [75].
Interleukin enhancer-binding factor 3-antisense RNA 1 (ILF3-AS1) is another lncRNA involved in the regulation of Sirt1 in MI. Hypoxia decreases ILF3-AS1 expression and induces damage to H9c2 cells by reduction of cell viability and enhancement of apoptosis. miR-212-3p expression increases and could negatively regulate Sirt1 expression under hypoxia conditions. So, the overexpression of ILF3-AS1 alleviates hypoxia damage and attenuates cardiomyocytes apoptosis by the downregulation of miR-212-3p [76].
Recently, the use of cardiac progenitor cells (CPCs) for myocardial regeneration has been suggested as an alternative to MI therapy. However, the proliferation of CPCs can decline because of the lack of oxygen in the infarcted myocardium. In the study by Li et al., the molecular mechanism involved in CPCs proliferation was investigated. Their results showed that under hypoxia stress, the decreased H19 expression resulted in attenuation of CPCs proliferation and migration through mediating the Sirt1 expression as a target of miR-200a-3p. miR-200a-3p, which is a target of H19, significantly increased after hypoxia stimulus in CPCs. These results demonstrated the protective role of the H19/miR-200a-3p/Sirt1 signaling axis in CPCs in response to hypoxia, suggesting a new therapeutic approach for heart ischemia damage and MI [77].
These studies showed that the lncRNAs such as KLF3-AS1, ANRIL, ILF3-AS1, and H19 decrease the hypoxia‐induced injury in MI model systems, suggesting them as possible targets in MI therapy.
2.1.2. Myocardial ischemia/reperfusion
Myocardial ischemia/reperfusion (MI/R) injury occurs when the blood supply returns to previously ischemic tissues. Under I/R conditions, the restored blood circulation can cause additional damage by inducing oxidative damage, calcium overload, mitochondrial dysfunction, and inflammation leading to apoptosis and cell death as well as increased infarct size in ischemic tissues [78]. However, timely myocardial reperfusion is the most effective approach to improving MI injury and reducing MI size [79]. In this regard, understanding the mechanisms involved in MI/R would be beneficial in improving MI treatment.
Mechanisms implicated in MI/R were investigated in both an in vivo (rat heart with MI/R) and an in vitro (H9c2 cells treated with oxygen-glucose deprivation/reoxygenation; OGD/R) model by Niu and colleagues. Their results showed that the overexpression of lncRNA Opa-interacting protein 5-antisense transcript 1 (OIP5-AS1) reduced reactive oxygen species (ROS) production, mitochondrial membrane depolarization, infarct size, and subsequently decreased apoptosis in both models. It was illustrated that OIP5-AS1 contains a conserved binding site for miR-29a, which can detect the 3′‐UTR of Sirt1 mRNA. OIP5-AS1 regulated Sirt1 expression through suppression of miR-29a and as a result, diminished the apoptosis that was triggered by OGD/R [80]. The positive regulation of Sirt1 expression involved the sponging of miR‑181a by ANRIL in H9c2 cardiomyocytes exposed to hypoxia/reoxygenation (H/R). In this study, ANRIL inhibited lactate dehydrogenase release and prevented H/R‑induced H9c2 cardiomyocyte apoptosis by mediating miR‑181a and Sirt1, suggesting a potential therapeutic role of the ANRIL/miR‑181a/SIRT1 axis in mitigating MI/R injury [81]. The overexpression of MALAT1 lncRNA has been shown to improve the hypoxia-induced H9c2 cell injury through the increment of Sirt1 by suppression of miR-217. The overexpression of Sirt1 also increased the activity of the PI3K/AKT and Notch pathways thus alleviating H9c2 cell injury [82]. lncRNAs such as OIP5-AS1, ANRIL, and MALAT1 may have a protective effect on the MI/R injury by suppressing miRNAs and increasing Sirt1.
2.1.3. Atherosclerosis
Atherosclerosis (AS) is a progressive arterial disorder initiated by vascular endothelial injury involving focal aggregation of lipoproteins, foam cells, and fatty streaks on the artery luminal surface. These events eventually lead to the form of atherosclerotic plaques, which can obstruct blood flow and if the plaques rupture, can result in thrombogenesis, coronary thrombosis, ischemia, myocardial infarction, and ischemic stroke [83]. Finally, the development of necrotic cores is stimulated in advanced atherosclerotic plaque by inflammatory cytokine release and macrophage apoptosis [84]. One frequently seen aspect of the development of atherosclerosis is the deregulated proliferation of VSMCs. The study by Wang et al. showed that the overexpression of novel lncRNA CAMK2D-associated transcript 1 (C2dat1) stimulated VSMC growth, proliferation and migration by inhibiting the expression level of miR-34a and promoting Sirt1 expression [63]. LncRNA MALAT1 also activated macrophage autophagy through the upregulation of Sirt1 and inhibited macrophage apoptotic rate, so it improved atherosclerosis [84].
The expression of miR-221 was markedly enhanced in carotid tissue in an AS mouse model in comparison to normal mice. The overexpression of miR-221 prevented the proliferation, migration, and tube formation of human aortic endothelial cells (HAECs) in ox-LDL-treated HAECs, while lncRNA p21 as a sponge for miR-221 and Sirt1 as a downstream target of miR-221 was poorly expressed in this HAECs model of AS. The decline of Sirt1 increased the acetylation of pro-protein convertase subtilisin/kexin type 9 (Pcsk9), which participates in the regulation and promotion of AS [85].
In another study, Ming et al. investigated the mechanisms involved in the migration and proliferation of endothelial progenitor cells (EPCs) for the possible treatment of AS. They found that nicotinamide phosphoribosyl-transferase (NAMPT) prevented EPCs senescence and stimulated EPCs migration and proliferation through the regulation of the Sirt1-AS lncRNA/miR-22/Sirt1 pathway. They also confirmed that Sirt1 expression, as well as Sirt1 Antisense RNA (Sirt1-AS) lncRNA, increased in EPCs-treated with NAMPT, while miR-22 expression decreased in these cells. NAMPT as an endogenous enzyme may play an important role in the prevention and treatment of AS and other CVDs [60]. The expression of HAND2 Antisense RNA 1 (HAND2-AS1) and Sirt1 diminished in the plasma of patients with atherosclerotic plaques. HAND2-AS1 suppressed miR-128 activity through a sequence of complementation, whereby the expression of Sirt1 intensified in macrophages originating from THP-1. HAND2-AS1 and lncRNAs such as p21, and Sirt1-AS may have potential as therapeutic targets for AS [86].
2.1.4. Cardiomyopathy
Chen et al. reported that the expression of lncRNA ZNFX1 Antisense RNA 1 (ZFAS1), as well as a decline in Sirt1, occurred while miR-34b-5p was elevated in LPS-induced myocardial injury in rats and H9C2 cells. The incremental expression of ZFAS1 or the inhibition of miR-34b-5p improved inflammation and cardiomyocyte apoptosis in cardiomyopathy [87]. In addition, the downregulation of various lncRNAs such as OIP5-AS1 and HOTAIR along with low Sirt1 levels in cardiomyopathy has been reported in previous studies [61,62]. Meanwhile, the miRNA expression of the target of these lncRNAs, miR-34a, is increased. Interestingly, the overexpression of OIP5-AS1 and HOTAIR lead to a decrease in the level of miR-34a followed by an increase in Sirt1 expression. So, these lncRNAs result in improved cardiac function and decreased oxidative stress and inflammation, suggesting the role of lncRNAs/miRNAs/Sirt1 in the treatment of cardiomyopathy [61,62].
2.2. Cerebral injuries
2.2.1. Ischemic stroke
Ischemic stroke (IS) occurs when a blood clot blocks or narrows an artery leading to the brain and can result in long-term disability and high mortality, especially in the elderly [88]. The potential role of lncRNAs, miRNAs, and Sirt1 in IS has been investigated by several groups [89,90]. Tian et al. have reported that lncRNA small nucleolar RNA host gene 8 (SNHG8) overexpression inhibited cell apoptosis and reduced neuronal damage, cerebral edema, and neurological function loss in mice following middle cerebral artery occlusion (MCAO) surgery. Additionally, following SNHG8 overexpression, the miR‐425‐5p expression was repressed and Sirt1 increased, indicating the role of SNHG8 as a competitive endogenous RNA through sponge of miR‐425‐5p by targeting Sirt1 to improve microglial inflammation and brain microvascular endothelial cells (BMECs) injury [91]. It has also been demonstrated that SNHG8 regulated the Sirt1 expression by sponging miR-449c-5p and as a result, inhibited microglia activation and improved brain tissue damage and inflammation in IS [92].
In another study by Zhou et al., it was demonstrated that the expression of lncRNA SNHG7 and Sirt1 decreased, while the level of miR-9 was enhanced in mice brain tissue with IS. The overexpression of SNHG7 diminished the ROS and MDA levels, enhanced cell viability, and reduced apoptosis in the IS PC12 cells [93]. Wang et al. also reported the relation between MALAT1, miR-200c-3p, and Sirt1 with autophagy and cell survival in OGD-induced ischemia in BEMCs cells by showing that MALAT1 binds to miR-200c-3p, and subsequently can upregulate Sirt1 expression, activate autophagy, and stimulate cell survival in an OGD model of IS [94]. In contrast, it has been reported that the H19 overexpression decreases cell viability, enhances inflammatory cytokine, and induces apoptosis in the MCAO mouse model and HT22 cells. Interestingly, H19 knockdown augments the miR-29b and Sirt1 levels in the HT22 cells, which results in cell viability improvement as well as a decline in inflammation and apoptosis [95]. These studies showed that lncRNAs such as SNHG8, SNHG7, MALAT1, and H19 decreased the injury induced by IS through their regulatory effect on the miRNAs/Sirt1 pathway.
2.2.2. Cerebral ischemia/reperfusion
Cerebral ischemia and reperfusion can lead to severe disability, microvascular dysfunction, and even neurological injury [96,97]. The role of 1ncRNAs, miRNAs, and Sirt1 in cerebral ischemia/reperfusion has been investigated by several groups. The expression of lncRNA nuclear enriched abundant transcript 1 (NEAT1), vascular endothelial growth factor (VEGF), Sirt1, and BCL-XL increased leading to angiogenesis and inhibition of apoptosis following OGD exposure in BMECs. Meanwhile, miR-377 expression was downregulated because of targeting 5′-UTR of miR-377 by NEAT1 [98]. SNHG12 increased cell proliferation and inhibited apoptosis by interacting with miR-199a and activating Sirt1 expression and the AMPK pathway thereby attenuating cerebral IR injury [99]. The knockdown of lncRNA SNHG15 resulted in increased miR-141 expression but a decrease in its target (Sirt1) in SH-SY5Y cells exposed to OGD/R. miR-141 also increased oxidative stress markers by reducing the expression level of Sirt1 [100]. Meng et al. reported that in OGD/R-induced PC12 cells, MALAT1 and Sirt1 levels diminished, but the expression of miR-142-3p increased. On the other hand, the overexpression of MALAT1 reduced oxidative stress markers and enhanced antioxidant activity and as a result repressed OGD/R-induced cell apoptosis and stimulated cell proliferation by regulating the miR-142-3p/SIRT1 axis [101].
These studies indicated that lncRNAs such as NEAT1, SNHG12, SNHG15, and MALAT1 decrease cerebral ischemia/reperfusion injury by reducing oxidative stress and inhibiting apoptosis through the enhancement of the level of Sir1.
3. Conclusion and perspective
This review highlights the crosstalk between lncRNAs, miRNAs, and Sirt1 in myocardial and cerebral disorders (Table 1). The majority of lncRNAs such as MALAT1, H19, SHNG, and ANRIL function as sponges of target miRNAs, which is related to Sirt1 regulation (Figure 2). Although there is growing evidence that ncRNAs play distinct roles in a wide range of gene regulation processes, this field is relatively infancy, necessitating the development of fresh strategies for the identification and annotation of the complex properties of the lncRNAs/miRNAs/mRNA axis. LncRNAs play a protective role in myocardial and cerebral disorder through the regulation of miRNAs/Sirt1 and subsequently modulate many biological processes including oxidative stress, apoptosis, and autophagy. The significance of lncRNAs, miRNAs, and Sirt1 in the pathogenesis of human diseases as possible therapeutic targets or strong prognosis indicators offers a potential strategy for MI, atherosclerosis, and ischemic stroke therapy. Therefore, a better understanding of the lncRNAs/miRNAs/Sirt1 axis may well provide fresh approaches for maximizing the effects of pharmacotherapy in the treatment of myocardial and cerebral disorders. Though little is known about whether the dynamic regulation of the this axis might be considered as the disease development marker or as the effect of medication therapy or due to the paucity of large-scale clinical investigations.
Table 1.
Examples of lncRNAs and their miRNA targets involved in myocardial and cerebral injuries.
| Human Disease | LncRNA | miRNA target | Function | Examinated cell type | References |
|---|---|---|---|---|---|
| Myocardial Infarction | KLF3-AS1 | miR-138-5p | Inhibit cardiomyocytes pyroptosis after MI | H9c2 | [74] |
| ANRIL | miR‐7‐5p | Protect H9c2 cells from hypoxia‐induced injury | H9c2 | [75] | |
| ILF3-AS1 | miR-212-3p | Relieve hypoxia damage, attenuate cardiomyocytes apoptosis | H9c2 | [76] | |
| H19 | miR-200a-3p | Increase CPCs proliferation and migration | CPCs | [77] | |
| Myocardial Ischemia/Reperfusion | OIP5-AS1 | miR-29a | Reduce reactive oxygen species, decreased cardiomyocytes apoptosis | H9c2 | [80] |
| ANRIL | miR‑181a | Inhibite lactate dehydrogenase release, prevent cardiomyocytes apoptosis | H9c2 | [81] | |
| MALAT1 | miR-217 | Alleviate H9c2 cell injury | H9c2 | [82] | |
| Atherosclerosis | p21 | miR-221 | Increase HAECs proliferation and migration | HAECs | [85] |
| Sirt1-AS | miR-22 | Increase EPCs proliferation and migration | EPCs | [60] | |
| HAND2-AS1 | miR-128 | Alleviate the formation of foam cells in atherosclerosis | THP-1 | [86] | |
| Ischemic Stroke | SNHG8 | miR‐425‐5p | Improve microglial inflammation | BMECs | [91] |
| miR-449c-5p | Inhibit microglia activation, improve inflammation | Microglia | [92] | ||
| SNHG7 | miR-9 | Enhance cell viability, reduce ROS and apoptosis | PC12 | [93] | |
| MALAT1 | miR-200c-3p | Stimulate cell survival, activate autophagy | BEMCs | [94] | |
| H19 | miR-29b | Increase cell apoptosis and inflammatory cytokine | HT22 | [95] | |
| Cerebral Ischemia/Reperfusion | NEAT1 | miR-377 | Stimulate angiogenesis, inhibit apoptosis | BMECs | [98] |
| SNHG12 | miR-199a | Increased cell proliferation, suppress apoptosis | primary neuronal and N2a cells | [99] | |
| SNHG15 | miR-141 | Reduce oxidative stress markers | SH-SY5Y | [100] | |
| MALAT1 | miR-142-3p | Stimulate cell proliferation, reduce oxidative stress, enhance antioxidant activities | PC12 | [101] |
Figure 2.
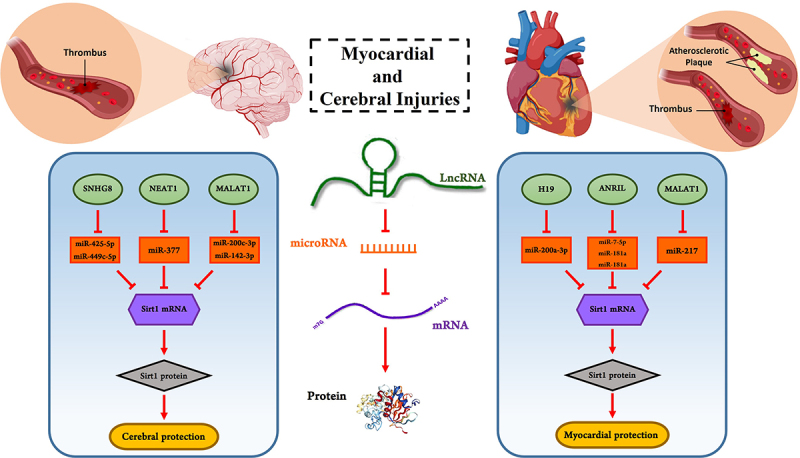
Examples of lncRNAs and miRNAs involved in myocardial and cerebral injury by influencing Sirt1. LncRNAs MALAT1, ANRIL, H19, NEAT1, and SNHG8 can bind to target miRNAs as a competitive endogenous RNA and prevent from miRNAs binding to mRNA. This function of lncRNAs leads to increase mRNA translation and as a result, the level of target protein such as Sirt1 enhance. Thus, lncRNAs can protect cells against injuries.
Funding Statement
The work was supported by the Mashhad University of Medical Sciences [484]
CRediT authorship contribution statement
SB and GK designed the project. SB wrote the first draft of the manuscript and designed the figure. GK and AWH reviewed and edited the manuscript. All authors approved the final version of the manuscript.
Data availability statement
No data was used for the research described in the article.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation. 2016. Jan 26;133:e38–360. [DOI] [PubMed] [Google Scholar]
- [2].Virani SS, Alonso A, Benjamin EJ, et al. heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation. 2020. Mar 3;141:e139–596. [DOI] [PubMed] [Google Scholar]
- [3].Joseph P, Leong D, McKee M, et al. Reducing the global burden of cardiovascular disease, Part 1: the epidemiology and risk factors. Circ Res. 2017. Sep 1;121:677–694. [DOI] [PubMed] [Google Scholar]
- [4].Radu RA, Terecoasă EO, Băjenaru OA, et al. Etiologic classification of ischemic stroke: where do we stand? Clin Neurol Neurosurg. 2017. Aug;159:93–106. [DOI] [PubMed] [Google Scholar]
- [5].Herpich F, Rincon F.. Management of acute ischemic stroke. Crit Care Med. 2020. Nov;48:1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rzemieniec J, Castiglioni L, Gelosa P, et al. Nuclear receptors in myocardial and cerebral ischemia—mechanisms of action and therapeutic strategies. Int J Mol Sci. 2021. Nov 15; 22;22:12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018. Dec;22:5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Knight JRP, Garland G, Pöyry T, et al. Control of translation elongation in health and disease. Dis Model Mech. 2020. Mar 26;13. doi: 10.1242/dmm.043208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Song P, Yang F, Jin H, et al. The regulation of protein translation and its implications for cancer. Signal Transduct Target Ther. 2021. Feb 18;6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li J, Liu C. Coding or noncoding, the converging concepts of RNAs. Front Genet. 2019;10:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016. Oct;96:1297–1325. [DOI] [PubMed] [Google Scholar]
- [12].Barangi S, Hayes AW, Reiter R, et al. The therapeutic role of long non-coding RNAs in human diseases: a focus on the recent insights into autophagy. Pharmacol Res. 2019. Apr;142:22–29. [DOI] [PubMed] [Google Scholar]
- [13].Xue M, Zhuo Y, Shan B. MicroRNAs, long noncoding RNAs, and their functions in human disease. Methods Mol Biol. 2017;1617:1–25. [DOI] [PubMed] [Google Scholar]
- [14].Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDnas. Nature. 2002. Dec 5;420:563–573. [DOI] [PubMed] [Google Scholar]
- [15].Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016. Jan;17:47–62. [DOI] [PubMed] [Google Scholar]
- [16].Fernandes JCR, Acuña SM, Aoki JI, et al. Long noncoding RNAs in the regulation of gene expression: physiology and disease. Noncoding RNA. 2019. Feb 17;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aillaud M, Schulte LN. Emerging roles of long noncoding RNAs in the cytoplasmic Milieu. Noncoding RNA. 2020. Nov 9;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Karakas D, Ozpolat B. The Role of LncRNAs in Translation. Noncoding RNA. 2021. Feb 20;7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Singh DK, Prasanth KV. Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res. 2013. Dec;21:695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Noh JH, Kim KM, McClusky WG, et al. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018. May;9:e1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Z, Zhao W, Wang M, et al. The role of long noncoding RNAs in gene expression regulation. In: Vlachakis D, editor. Gene expression profiling in cancer. London: IntechOpen; 2019. pp. 1–17. [Google Scholar]
- [22].Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019. May;21:542–551. [DOI] [PubMed] [Google Scholar]
- [23].DiStefano JK. The emerging role of long noncoding RNAs in human disease. Methods Mol Biol. 2018;1706:91–110. [DOI] [PubMed] [Google Scholar]
- [24].Ou L, Wang D, Zhang H, et al. Decreased expression of miR-138-5p by lncRNA H19 in cervical cancer promotes tumor proliferation. Oncol Res. 2018. Apr 10;26:401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gu XX, Xu XX, Liao HH, et al. Dexmedetomidine hydrochloride inhibits hepatocyte apoptosis and inflammation by activating the lncRNA TUG1/miR-194/SIRT1 signaling pathway. J Inflamm (Lond). 2021. May 26;18:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang Y, Gu M, Ma Y, et al. LncRNA TUG1 reduces inflammation and enhances insulin sensitivity in white adipose tissue by regulating miR-204/SIRT1 axis in obesity mice. Mol Cell Biochem. 2020. Dec;475:171–183. [DOI] [PubMed] [Google Scholar]
- [27].Lian LP, Xi XY. Long non-coding RNA XIST protects chondrocytes ATDC5 and CHON-001 from IL-1β-induced injury via regulating miR-653-5p/sirt1 axis. J Biol Regul Homeost Agents. 2020. Mar-Apr;34:379–391. [DOI] [PubMed] [Google Scholar]
- [28].Ni S, Xu C, Zhuang C, et al. LncRNA LUADT1 regulates miR-34a/sirt1 to participate in chondrocyte apoptosis. J Cell Biochem. 2020. Feb 7;122:1003–1008. [DOI] [PubMed] [Google Scholar]
- [29].Rothman AM, Arnold ND, Pickworth JA, et al. MicroRNA-140-5p and SMURF1 regulate pulmonary arterial hypertension. J Clin Invest. 2016. Jul 1;126:2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ni WJ, Leng XM. miRNA-Dependent Activation of mRNA Translation. Microrna. 2016;5:83–86. [DOI] [PubMed] [Google Scholar]
- [31].Da Sacco L, Masotti A. Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5’ untranslated region. Int J Mol Sci. 2012. Dec 27;14: 480–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Poller W, Dimmeler S, Heymans S, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. 2018. Aug 1;39:2704–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- [34].Hao S, Wang L, Zhao K, et al. Rs1894720 polymorphism in MIAT increased susceptibility to age-related hearing loss by modulating the activation of miR-29b/sirt1/pgc-1α signaling. J Cell Biochem. 2019. Apr;120:4975–4986. [DOI] [PubMed] [Google Scholar]
- [35].Xie W, Shu T, Peng H, et al. LncRNA H19 inhibits oxidative stress injury of cochlear hair cells by regulating miR-653-5p/sirt1 axis. Acta Biochim Biophys Sin (Shanghai). 2022. Mar 25;54:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Barangi S, Mehri S, Moosavi Z, et al. Melatonin inhibits Benzo(a)pyrene-Induced apoptosis through activation of the Mir-34a/sirt1/autophagy pathway in mouse liver. Ecotoxicol Environ Saf. 2020. Jun 15;196:110556. [DOI] [PubMed] [Google Scholar]
- [37].Wang X, Yang B, Ma B. The UCA1/miR-204/sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Cancer Chemother Pharmacol. 2016. Nov;78:1025–1031. [DOI] [PubMed] [Google Scholar]
- [38].Zhu X, Liu Y, Cui J, et al. LncRNA LYPLAL1-DT screening from type 2 diabetes with macrovascular complication contributes protective effects on human umbilical vein endothelial cells via regulating the miR-204-5p/sirt1 axis. Cell Death Discov. 2022. May 4;8:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lodish UH, Lodish H, Berk A, et al. Molecular cell biology, Chapter 3. New York: W. H. Freeman; 2008. [Google Scholar]
- [40].Sosnowska B, Mazidi M, Penson P, et al. The sirtuin family members SIRT1, SIRT3 and SIRT6: their role in vascular biology and atherogenesis. Atherosclerosis. 2017. Oct;265:275–282. [DOI] [PubMed] [Google Scholar]
- [41].Singh CK, Chhabra G, Ndiaye MA, et al. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal. 2018. Mar 10;28:643–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Donmez G. The neurobiology of sirtuins and their role in neurodegeneration. Trends Pharmacol Sci. 2012. Sep;33:494–501. [DOI] [PubMed] [Google Scholar]
- [43].Jin Q, Yan T, Ge X, et al. Cytoplasm-localized SIRT1 enhances apoptosis. J Cell Physiol. 2007. Oct;213:88–97. [DOI] [PubMed] [Google Scholar]
- [44].Yanagisawa S, Baker JR, Vuppusetty C, et al. The dynamic shuttling of SIRT1 between cytoplasm and nuclei in bronchial epithelial cells by single and repeated cigarette smoke exposure. PLoS ONE. 2018;13:e0193921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hwang JW, Yao H, Caito S, et al. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med. 2013. Aug;61:95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001. Oct 19;107:149–159. [DOI] [PubMed] [Google Scholar]
- [47].Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004. Jun 16;23:2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Joo HY, Yun M, Jeong J, et al. SIRT1 deacetylates and stabilizes hypoxia-inducible factor-1α (HIF-1α) via direct interactions during hypoxia. Biochem Biophys Res Commun. 2015. Jul 10;462:294–300. [DOI] [PubMed] [Google Scholar]
- [49].Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004. Mar 26;303:2011–2015. [DOI] [PubMed] [Google Scholar]
- [50].Nie Y, Erion DM, Yuan Z, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009. Apr;11:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sakamoto J, Miura T, Shimamoto K, et al. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004. Jan 2;556:281–286. [DOI] [PubMed] [Google Scholar]
- [52].Ogawa T, Wakai C, Saito T, et al. Distribution of the longevity gene product, SIRT1, in developing mouse organs. Congenit Anom (Kyoto). 2011. Jun;51:70–79. [DOI] [PubMed] [Google Scholar]
- [53].Potente M, Ghaeni L, Baldessari D, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007. Oct 15;21:2644–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jiao F, Gong Z. The beneficial roles of SIRT1 in neuroinflammation-related diseases. Oxid Med Cell Longev. 2020;2020:6782872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Meng X, Tan J, Li M, et al. Sirt1: role under the condition of ischemia/hypoxia. Cell Mol Neurobiol. 2017. Jan;37:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Winnik S, Auwerx J, Sinclair DA, et al. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J. 2015. Dec 21;36:3404–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hernández-Jiménez M, Hurtado O, Cuartero MI, et al. Silent information regulator 1 protects the brain against cerebral ischemic damage. Stroke. 2013. Aug;44:2333–2337. [DOI] [PubMed] [Google Scholar]
- [58].Becatti M, Taddei N, Cecchi C, et al. SIRT1 modulates MAPK pathways in ischemic-reperfused cardiomyocytes. Cell Mol Life Sci. 2012. Jul;69:2245–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang GQ, Wang Y, Xiong Y, et al. Sirt1 as lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. 2016. Feb 23;6:21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ming GF, Wu K, Hu K, et al. NAMPT regulates senescence, proliferation, and migration of endothelial progenitor cells through the SIRT1 as lncRna/mir-22/SIRT1 pathway. Biochem Biophys Res Commun. 2016. Sep 23;478:1382–1388. [DOI] [PubMed] [Google Scholar]
- [61].Gao L, Wang X, Guo S, et al. LncRNA HOTAIR functions as a competing endogenous RNA to upregulate SIRT1 by sponging miR-34a in diabetic cardiomyopathy. J Cell Physiol. 2019. Apr;234:4944–4958. [DOI] [PubMed] [Google Scholar]
- [62].Sun H, Wang C, Zhou Y, et al. Long noncoding RNA OIP5-AS1 overexpression promotes viability and inhibits high glucose-induced oxidative stress of cardiomyocytes by targeting MicroRNA-34a/sirt1 axis in diabetic cardiomyopathy. Endocr Metab Immune Disord Drug Targets. 2021;21:2017–2027. [DOI] [PubMed] [Google Scholar]
- [63].Wang H, Jin Z, Pei T, et al. Long noncoding RNAs C2dat1 enhances vascular smooth muscle cell proliferation and migration by targeting MiR-34a-5p. J Cell Biochem. 2019. Mar;120:3001–3008. [DOI] [PubMed] [Google Scholar]
- [64].Shahgaldi S, Kahmini FR. A comprehensive review of Sirtuins: with a major focus on redox homeostasis and metabolism. Life Sci. 2021. Oct 1;282: 119803. [DOI] [PubMed] [Google Scholar]
- [65].Li D, Liu Y, Gao W, et al. LncRNA HCG11 inhibits adipocyte differentiation in human adipose-derived mesenchymal stem cells by sponging miR-204-5p to upregulate SIRT1. Cell Transplant. 2020. Jan-Dec;29:963689720968090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tu Y, Song E, Wang Z, et al. Melatonin attenuates oxidative stress and inflammation of Müller cells in diabetic retinopathy via activating the Sirt1 pathway. Biomed Pharmacother. 2021. May;137:111274. [DOI] [PubMed] [Google Scholar]
- [67].Wang S, Yi P, Wang N, et al. LncRNA TUG1/miR-29c-3p/sirt1 axis regulates endoplasmic reticulum stress-mediated renal epithelial cells injury in diabetic nephropathy model in vitro. PLoS ONE. 2021;16:e0252761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wu P, Yu X, Peng Y, et al. Ginsenoside Rg3 alleviates septic liver injury by regulating the lncRNA TUG1/miR-200c-3p/sirt1 axis. J Inflamm (Lond). 2021. Dec 20;18:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhu J, Shi H, Liu H, et al. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017. Sep 12;8:65253–65264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yin KJ, Vemuganti R. Long noncoding RNAs and CNS disorders. Neurochem Int. 2021. Nov;150:105176. [DOI] [PubMed] [Google Scholar]
- [71].Zhang Y, Anoopkumar-Dukie S, Arora D, et al. Review of the anti-inflammatory effect of SIRT1 and SIRT2 modulators on neurodegenerative diseases. Eur J Pharmacol. 2020. Jan 15;867:172847. [DOI] [PubMed] [Google Scholar]
- [72].Koenen RR, Binder CJ. Platelets and coagulation factors: established and novel roles in atherosclerosis and atherothrombosis. Atherosclerosis. 2020. Aug;307:78–79. [DOI] [PubMed] [Google Scholar]
- [73].Chen L, Li S, Zhu J, et al. Mangiferin prevents myocardial infarction-induced apoptosis and heart failure in mice by activating the Sirt1/FoxO3a pathway. J Cell Mol Med. 2021. Mar;25:2944–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mao Q, Liang XL, Zhang CL, et al. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138-5p/sirt1 axis. Stem Cell Res Ther. 2019. Dec 17;10:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shu L, Zhang W, Huang C, et al. lncRNA ANRIL protects H9c2 cells against hypoxia-induced injury through targeting the miR-7-5p/sirt1 axis. J Cell Physiol. 2020. Feb;235:1175–1183. [DOI] [PubMed] [Google Scholar]
- [76].Zhang JY, Yang Z, Fang K, et al. Long noncoding RNA ILF3-AS1 regulates myocardial infarction via the miR-212-3p/sirt1 axis and PI3K/Akt signaling pathway. Eur Rev Med Pharmacol Sci. 2020. Mar;24:2647–2658. [DOI] [PubMed] [Google Scholar]
- [77].Li L, Wang Q, Yuan Z, et al. Long non-coding RNA H19 contributes to hypoxia-induced CPC injury by suppressing Sirt1 through miR-200a-3p. Acta Biochim Biophys Sin (Shanghai). 2018. Oct 1;50:950–959. [DOI] [PubMed] [Google Scholar]
- [78].Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med. 2007. Sep 13;357: 1121–1135. [DOI] [PubMed] [Google Scholar]
- [79].Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest. 2013. Jan;123:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Niu X, Pu S, Ling C, et al. lncRNA Oip5-as1 attenuates myocardial ischaemia/reperfusion injury by sponging miR-29a to activate the SIRT1/AMPK/PGC1α pathway. Cell Prolif. 2020. Jun;53:e12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Song B, Wei D, Yin G, et al. Critical role of SIRT1 upregulation on the protective effect of lncRNA ANRIL against hypoxia/reoxygenation injury in H9c2 cardiomyocytes. Mol Med Rep. 2021. Aug 24;24. DOI: 10.3892/mmr.2021.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yao Y, Fan X, Yu B, et al. Knockdown of long noncoding RNA Malat1 aggravates hypoxia-induced cardiomyocyte injury by targeting miR-217. Adv Clin Exp Med. 2019. Jun;28:719–728. [DOI] [PubMed] [Google Scholar]
- [83].Bentzon JF, Otsuka F, Virmani R, et al. Mechanisms of plaque formation and rupture. Circ Res. 2014. Jun 6;114:1852–1866. [DOI] [PubMed] [Google Scholar]
- [84].Yang J, Lin X, Wang L, et al. LncRNA MALAT1 enhances ox-LDL-induced autophagy through the SIRT1/MAPK/NF-κB pathway in macrophages. Curr Vasc Pharmacol. 2020;18:652–662. [DOI] [PubMed] [Google Scholar]
- [85].Wang H, He F, Liang B, et al. LincRNA-p21 alleviates atherosclerosis progression through regulating the miR-221/sirt1/pcsk9 axis. J Cell Mol Med. 2021. Oct;25:9141–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ma L, He S, Li H, et al. HAND2-AS1 targeting miR-1208/SIRT1 axis alleviates foam cell formation in atherosclerosis. Int J Cardiol. 2022. Jan 1;346:53–61. [DOI] [PubMed] [Google Scholar]
- [87].Chen DD, Wang HW, Cai XJ. Long non-coding RNA ZFAS1 alleviates sepsis-induced myocardial injury via target miR-34b-5p/sirt1. Innate Immun. 2021. Jul;27:377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rodrigo R, Fernández-Gajardo R, Gutiérrez R, et al. Oxidative stress and pathophysiology of ischemic stroke: novel therapeutic opportunities. CNS Neurol Disord Drug Targets. 2013. Aug;12:698–714. [DOI] [PubMed] [Google Scholar]
- [89].Bao MH, Szeto V, Yang BB, et al. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018. Feb 15;9:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Teertam SK, Jha S, Prakash Babu P. Up-regulation of Sirt1/miR-149-5p signaling may play a role in resveratrol induced protection against ischemia via p53 in rat brain. J Clin Neurosci. 2020. Feb;72:402–411. [DOI] [PubMed] [Google Scholar]
- [91].Tian J, Liu Y, Wang Z, et al. LncRNA Snhg8 attenuates microglial inflammation response and blood-brain barrier damage in ischemic stroke through regulating miR-425-5p mediated SIRT1/NF-κB signaling. J Biochem Mol Toxicol. 2021. May;35:e22724. [DOI] [PubMed] [Google Scholar]
- [92].Zhang D, Pan N, Jiang C, et al. LncRNA SNHG8 sponges miR-449c-5p and regulates the SIRT1/FoxO1 pathway to affect microglia activation and blood-brain barrier permeability in ischemic stroke. J Leukocyte Biol. 2022. May;111:953–966. [DOI] [PubMed] [Google Scholar]
- [93].Zhou T, Wang S, Lu K, et al. Long non-coding RNA SNHG7 alleviates oxygen and glucose deprivation/reoxygenation-induced neuronal injury by modulating miR-9/SIRT1 axis in PC12 cells: potential role in ischemic stroke. Neuropsychiatr Dis Treat. 2020;16:2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wang S, Han X, Mao Z, et al. MALAT1 lncRNA induces autophagy and protects brain microvascular endothelial cells against oxygen-glucose deprivation by binding to miR-200c-3p and upregulating SIRT1 expression. Neuroscience. 2019. Jan 15;397:116–126. [DOI] [PubMed] [Google Scholar]
- [95].Xu J, Wang C, Meng F, et al. Long non‑coding RNA H19 inhibition ameliorates oxygen‑glucose deprivation‑induced cell apoptosis and inflammatory cytokine expression by regulating the microRna‑29b/SIRT1/PGC‑1α axis. Mol Med Rep. 2021. Feb 23;23. DOI: 10.3892/mmr.2020.11770 [DOI] [PubMed] [Google Scholar]
- [96].Ishikawa M, Zhang JH, Nanda A, et al. Inflammatory responses to ischemia and reperfusion in the cerebral microcirculation. Front Biosci. 2004. May 1;9:1339–1347. [DOI] [PubMed] [Google Scholar]
- [97].Mehta SL, Manhas N, Raghubir R. Molecular targets in cerebral ischemia for developing novel therapeutics. Brain Res Rev. 2007. Apr;54:34–66. [DOI] [PubMed] [Google Scholar]
- [98].Zhou ZW, Zheng LJ, Ren X, et al. LncRNA NEAT1 facilitates survival and angiogenesis in oxygen-glucose deprivation (OGD)-induced brain microvascular endothelial cells (BMECs) via targeting miR-377 and upregulating SIRT1, VEGFA, and BCL-XL. Brain Res. 2019. Mar 15;1707:90–98. [DOI] [PubMed] [Google Scholar]
- [99].Yin WL, Yin WG, Huang BS, et al. LncRNA SNHG12 inhibits miR-199a to upregulate SIRT1 to attenuate cerebral ischemia/reperfusion injury through activating AMPK signaling pathway. Neurosci Lett. 2019. Jan 18;690:188–195. [DOI] [PubMed] [Google Scholar]
- [100].Kang M, Ji F, Sun X, et al. LncRNA SNHG15 promotes oxidative stress damage to regulate the occurrence and development of cerebral ischemia/reperfusion injury by targeting the miR-141/SIRT1 axis. J Healthc Eng. 2021;2021:6577799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Meng S, Wang B, Li W. LncRNA MALAT1 improves cerebral ischemia-reperfusion injury and cognitive dysfunction by regulating miR-142-3p/sirt1 axis. Int J Neurosci. 2021. Aug;31:1–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.


