Abstract
The 2022 Metrics of the Human Proteome from the HUPO Human Proteome Project (HPP) illustrates that protein expression has now been credibly detected (neXtProt PE1 level) for 18,407 (93.2%) of the 19,750 predicted proteins coded in the human genome, a net gain of 50 since 2021 from datasets generated around the world and reanalyzed by the HPP. Conversely, the number of neXtProt PE2, PE3, and PE4 missing proteins has been reduced by 78 from 1421 to 1343. This represents continuing experimental progress on the proteome parts list across all the chromosomes, as well as significant reclassifications. Meanwhile, applying proteomics in a vast array of biological and clinical studies continues to yield significant findings and growing integration with other omics platforms. We present highlights from the Chromosome-Centric HPP, Biology and Disease-driven HPP, and HPP Resource Pillars, compare features of mass spectrometry and Olink and Somalogic platforms, note the emergence of translation products from ribosome profiling of small open reading frames, and discuss the launch of the initial HPP Grand Challenge Project, “A Function for Each Protein”.
Keywords: Human Proteome Project (HPP), neXtProt protein existence (PE metrics), missing proteins (MP), non-MS PE1 proteins, uncharacterized protein existence 1 (uPE1), Chromosome-centric HPP (C-HPP), Biology and Disease-HPP (B/D-HPP), PeptideAtlas, Mass Spectrometry Interactive Virtual Environment (MassIVE), Human Protein Atlas, small open reading frames (smORFs), Ribo-Seq, Grand Challenge Project
Graphical Abstract
PROGRESS ON THE HUMAN PROTEOME PARTS LIST
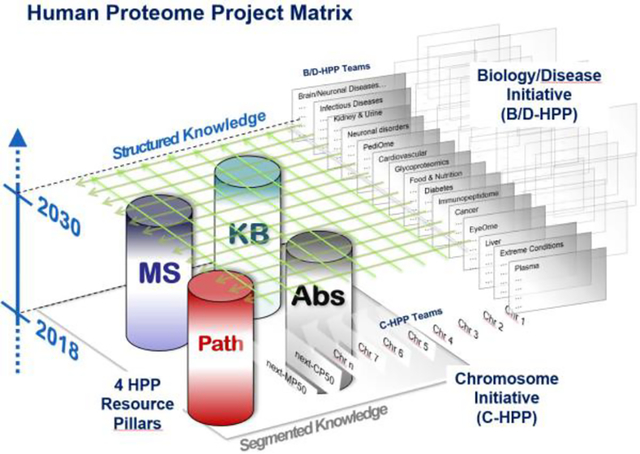
Since 2010, the Human Proteome Project (HPP)1, the flagship initiative of the global Human Proteome Organization (HUPO)2, has pursued two goals: (1) to credibly identify the protein “parts list”, primarily but not entirely by mass spectrometry, and (2) to make proteomics an integral part of multi-omics studies of human health and disease3–5. Through international collaboration, data sharing, standardized reanalysis of datasets, and guidelines for quality assurance, the HPP consortium has stimulated progress in building and utilizing proteomics knowledge globally. The HPP is organized into 25 teams by chromosome and mitochondria, 19 teams by biological and disease categories, and four resource pillars for antibody-based protein localization, mass spectrometry, knowledgebases, and pathology. This work is supported by advances in developing data formats and standards by the HUPO Proteomics Standards Initiative (HUPO-PSI)6 and the concomitant growth of PRIDE, PeptideAtlas, and other data repositories.
The human proteome database neXtProt curates and documents the proteins identified or predicted in a chromosome-specific manner for ease of organization7. Table 1 shows the annual progress in identifying proteins with credible protein-level evidence (PE1) from 13,975 proteins in 2012–02 to 18,407 PE1 proteins in the neXtProt release of 2022–02. The total represents 93.2% of the 19,750 PE1,2,3,4 predicted proteins from protein-coding genes. These proteins constitute the main gene-based translation products, but the total number of protein species, including sequence variants, splice variants, and post-translationally modified proteoforms, is a substantial multiple of that figure. Table 1 further documents the reduction of PE2,3,4 “missing proteins”, classified based on detectable levels of transcripts in human specimens (PE2), homologous protein expression in other species (PE3), or gene models predicted to be translated (PE4), from 5511 in 2012–02 to 1343 in 2022–02, a reduction of 78 in the past year, and now representing just 6.8% of the PE1,2,3,4 total of 19,750 predicted human proteins. Figure 1 shows the very complex dynamics of changes in the past year.
Table 1.
Numbers of Proteins by neXtProt Protein Existence Evidence Levels from 2012–02 to 2022–02 Showing Progress in Identifying PE2,3,4 Missing Proteins to Become PE1 Proteins and PeptideAtlas Canonical Proteins by Mass Spectrometry a,b,c
| PE Level | 2012–02 | 2013–09 | 2014–10 | 2016–01 | 2017–01 | 2018–01 | 2019–01 | 2020–01 | 2021–02 | 2022–02 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1: Evidence at protein level | 13,975 | 15,646 | 16,491 | 16,518 | 17,008 | 17,470 | 17,694 | 17,874 | 18,357 | 18,407 |
| 2: Evidence at transcript level | 5205 | 3570 | 2647 | 2290 | 1939 | 1660 | 1548 | 1596 | 1265 | 1135 |
| 3: Inferred from homology | 218 | 187 | 214 | 565 | 563 | 452 | 510 | 253 | 147 | 195 |
| 4: Predicted | 88 | 87 | 87 | 94 | 77 | 74 | 71 | 50 | 9 | 13 |
| MP = PE2 + PE3 + PE4 | 5511 | 3844 | 2948 | 2949 | 2579 | 2186 | 2129 | 1899 | 1421 | 1343 |
| Human PeptideAtlas canonical proteins | 12,509 | 13,377 | 14,928 | 14,569 | 15,173 | 15,798 | 16,293 | 16,655 | 16,702 | 16,957 |
Figure 1.
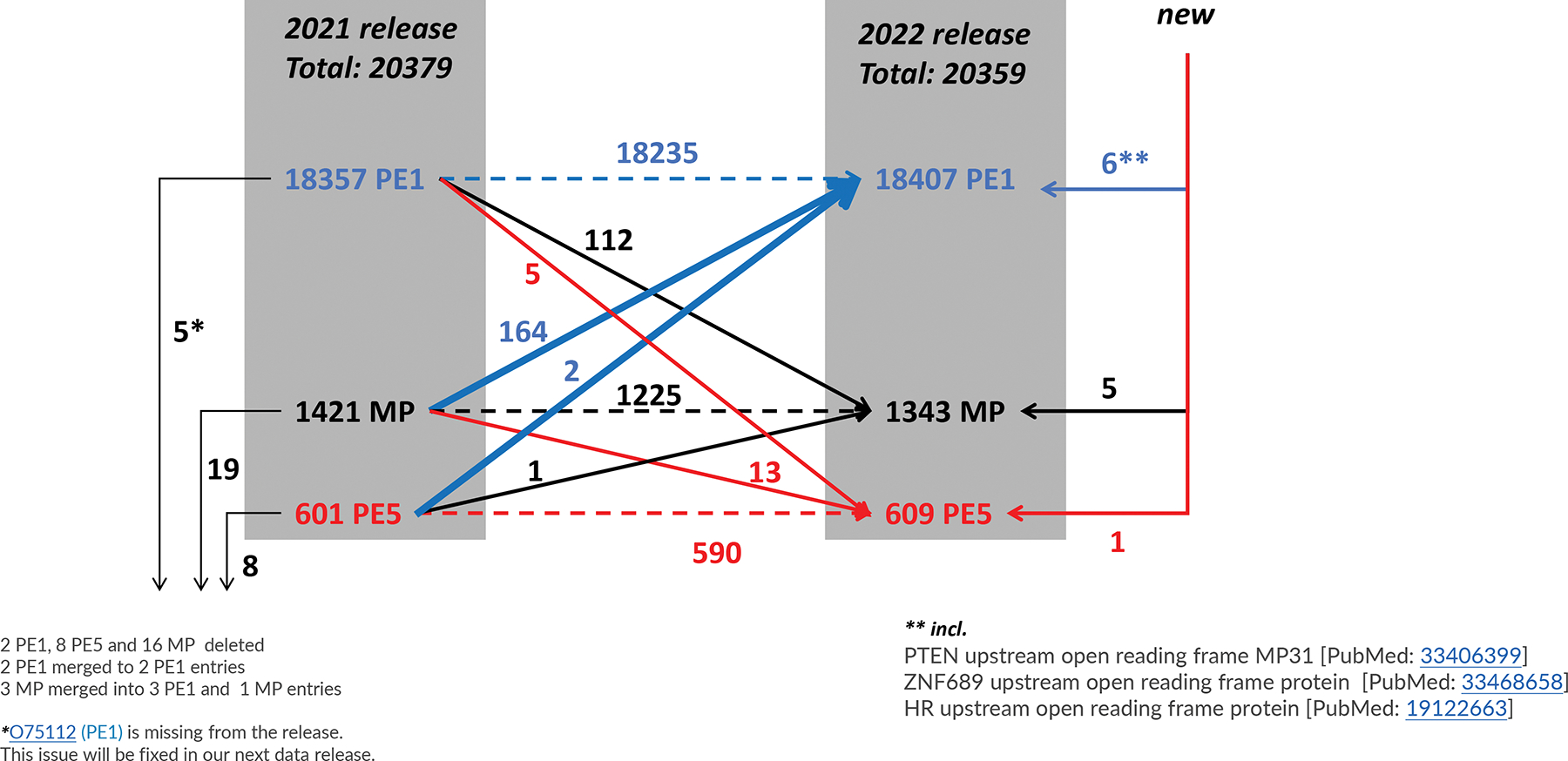
Flow diagram of the changes in numbers of PE1, PE2, PE3, PE4 (and PE5) classes of predicted proteins from 2021–02 to 2022–02. Blue solid arrows indicate increases to PE1; red arrows indicate demotions from PE1 or PE2,3,4 MPs, to PE5; black arrows show increases to MPs. Dashed arrows indicate continuation in the same status. In the margins we show examples of the 12 new entries to and 32 removals from neXtProt (see discussion later about uORFs and smORFs). Note, PE5 represents a category of dubious or uncertain genes, mostly pseudogenes, which we have excluded from the HPP metrics since 2013.
For MS-based PE1 decisions, neXtProt incorporates peptide identifications from PeptideAtlas8, 9 and MassIVE10 and applies guidelines for protein validation (at least two uniquely-mapping, non-nested peptides of 9 or more amino acids covering at least 18 amino acids) based on the HPP Mass Spectrometry Data Interpretation Guidelines v3.011. Of the 1343 PE2,3,4 missing proteins, 310 have some MS data which are insufficient to meet the Guidelines. Of the 18,407 PE1 proteins, 17,539 are based on mass spectrometry evidence (Figure 2). Of the MS-based PE1 proteins, 17,174 resulted from PeptideAtlas reanalysis of raw data from published datasets, and 17,033 resulted from similar reanalysis by MassIVE and submitted to neXtProt; 16,668 proteins met the guideline requirements in both PeptideAtlas and MassIVE; 506 proteins met the guidelines only in PeptideAtlas, whereas 365 only met the guidelines in MassIVE. Peptide evidence must meet the guidelines either in PeptideAtlas or MassIVE (or both); partial evidence from the two sources cannot be combined to create sufficient proof11.
Figure 2.
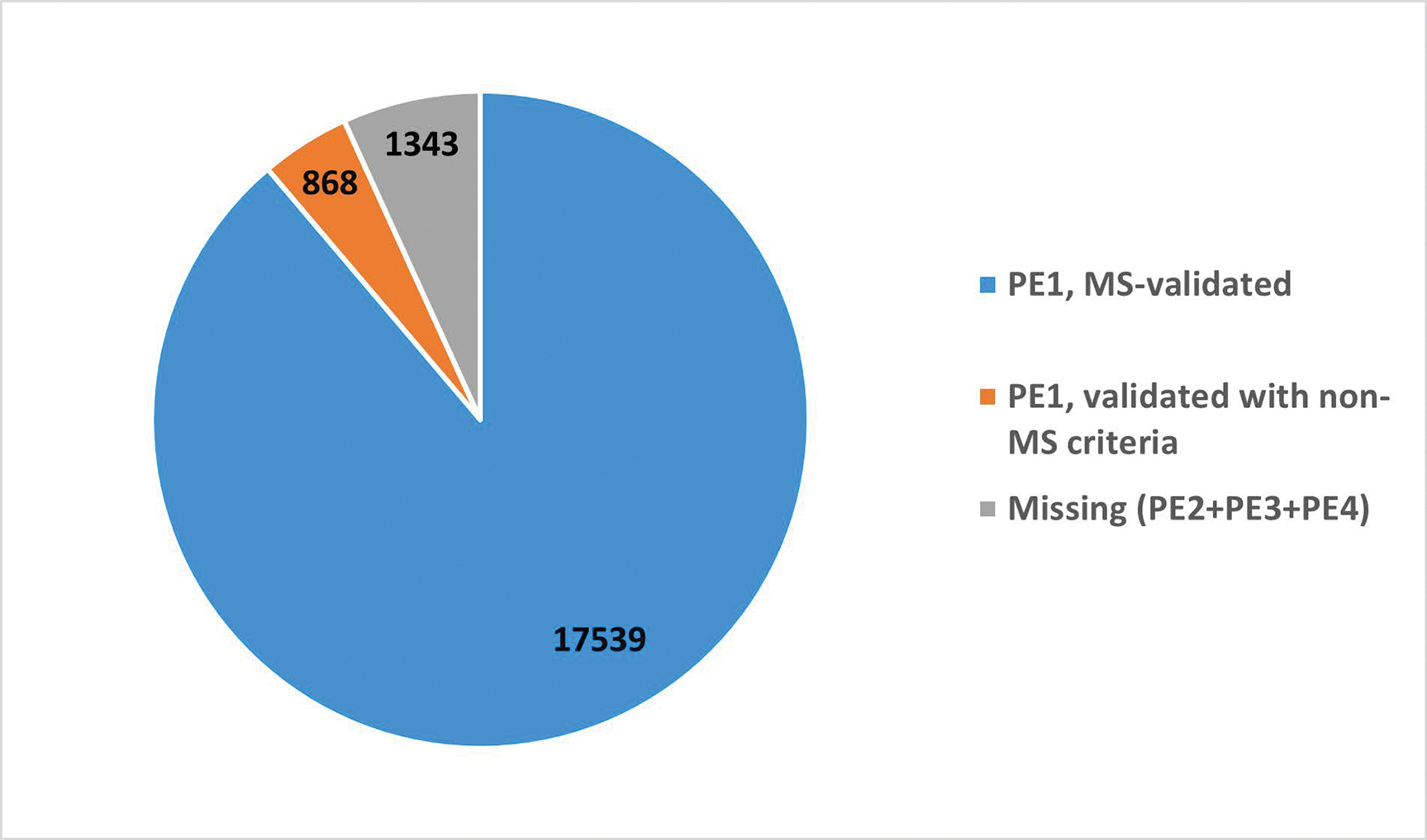
Pie chart depicting the 2022 status of proteins in the human proteome. PE1 proteins that have MS validation are shown in blue. PE1 validated via means other than MS are shown in orange. Missing proteins (PE2+PE3+PE4) are shown in gray.
Of the 868 non-MS-based PE1 proteins, 47 are primarily based on protein sequencing by N-terminal sequencing by Edman degradation, 18 on 3D structures of natural proteins (not recombinant) in the Protein Data Bank, 453 on protein-protein interactions, 31 on antibody studies, 87 on PTMs and proteolytic processing, 69 on genetic mutations, and 163 from biochemical studies. Many are supported by multiple types of studies, as shown in Figure 3; some have MS data insufficient to meet the HPP MS Data Interpretation Guidelines v3.0.
Figure 3.
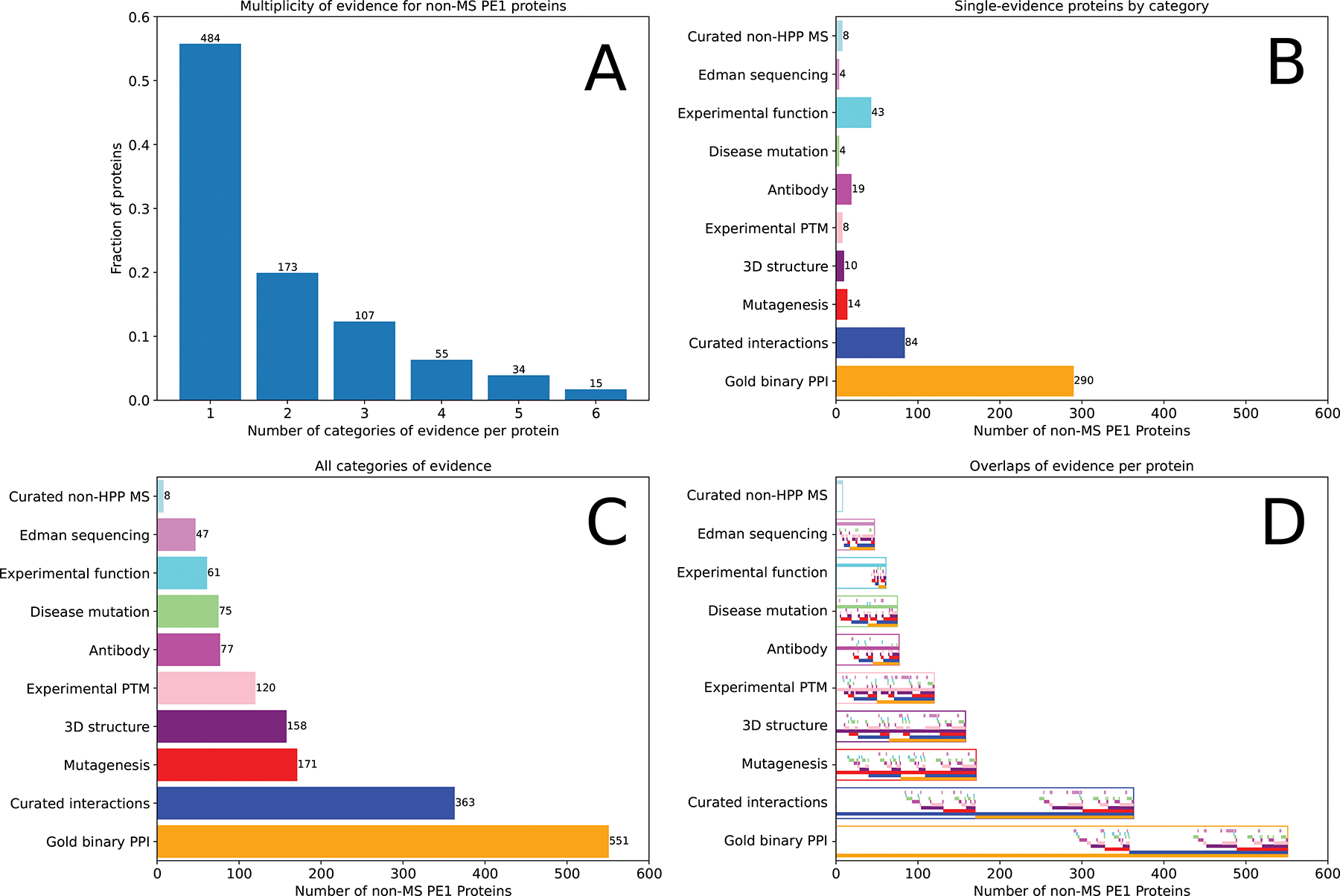
Bar charts summarizing the 868 PE1 proteins that have MS evidence in neither PeptideAtlas nor MassIVE meeting HPP Guidelines (designated non-MS PE1). A) Number of different categories of evidence for these proteins, with 484 proteins having only a single type. B) Distribution of the 484 proteins with only a single category of evidence across the 10 different categories. Protein-protein interactions (PPI) from IntAct far outweigh all other categories. C) Distribution of all evidence categories (many proteins have multiple categories of evidence). D) Similar to panel C but within each major type (depicted with the outline of the appropriate color), each of the 10 categories is depicted by a narrow bar, thereby showing the overlap between categories. Of the 551 proteins with “Gold Binary PPI” evidence, 194 also have “Curated Interactions” (blue), and, of those, 62 have “Mutagenesis” evidence as well (red). A complete listing of the individual proteins is provided in Supplementary Table 1.
The quality criteria for protein-protein interaction annotations in neXtProt have been changed so that only interactions shown by two independent sources of evidence are considered Gold level. As a result, 112 entries that had been promoted during 2020/2021 to PE1 based on previous Gold protein-protein interactions were demoted between 2021 and 2022, reducing the net gain in PE1 to 50 entries.
A complementary depiction of the progress since the application of the HPP MS Data Interpretation Guidelines v2.112 in 2016 is shown in Figure 4. As documented in Table 1, there continues to be remarkable progress on credibly identifying PE2,3,4 missing proteins, thereby increasing PE1 numbers while simultaneously reducing PE2,3,4 numbers (now only 6.8% of the total PE1,2,3,4 proteins.
Figure 4.
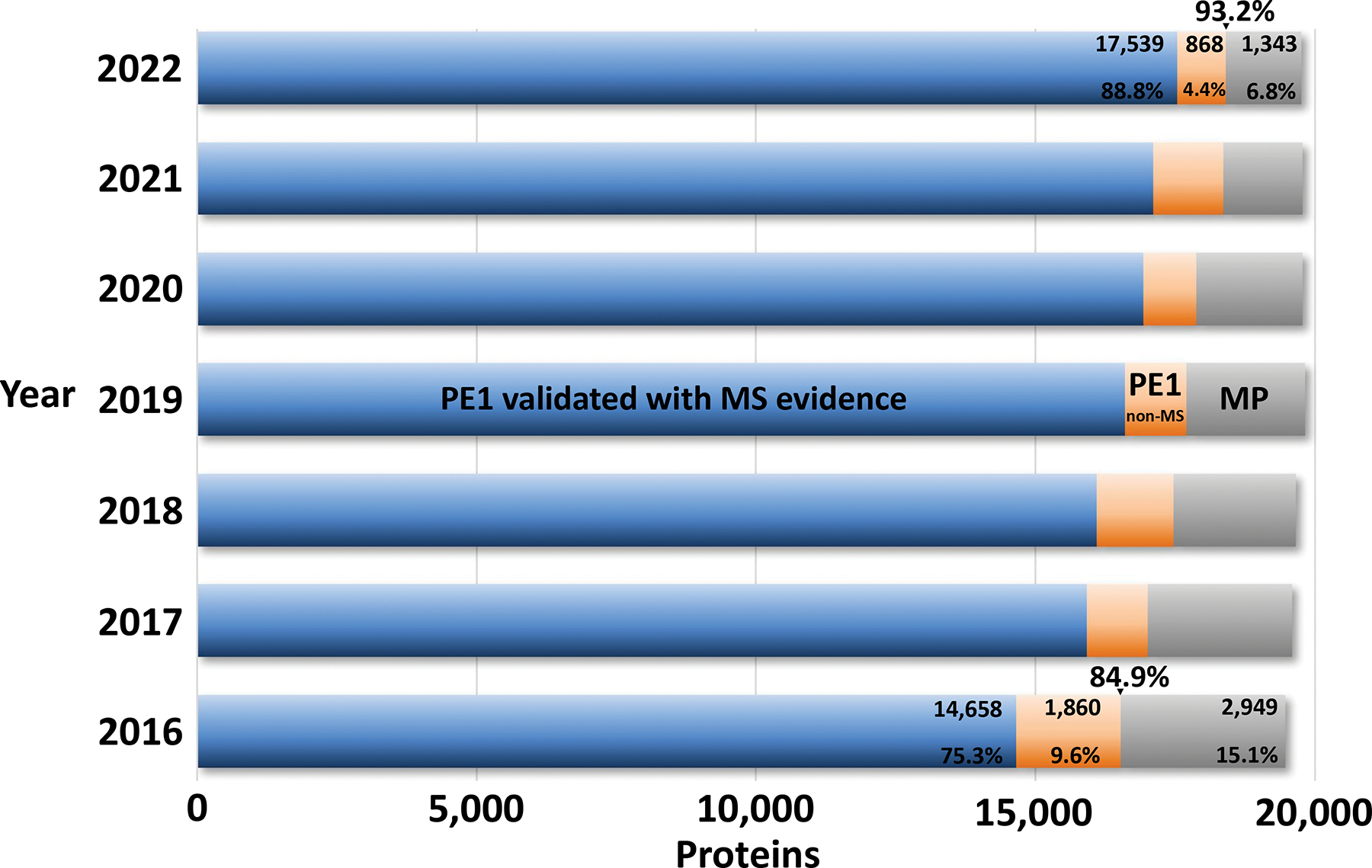
This bar chart documents progress in reducing PE2,3,4 missing proteins (grey) and increasing PE1 proteins (blue + orange). Within the PE1 proteins, there is a continuing shift of non-MS PE1 proteins (orange) to MS-based PE1 proteins (blue). The bulge in 2021 for non-MS reflects the inclusion and then removal of 112 PPI-based entries, as noted above.
Simple linear extrapolation based on the historical progress since 2016 shows annual increments of ~300 MS-based PE1 proteins and ~280 total PE1 proteins per year. If the pace were to continue, this would yield ~18,700 PE1 proteins in 2023, 19,000 PE1 in 2024, and 19,250 in 2025. However, there are multiple reasons why progress would likely slow in coming years due to certain classes of proteins likely to remain difficult to detect; these include negligible transcription for up to 800 predicted PE2,3,4 proteins (based on HPA, GTEx, FANTOM5)13; membrane-embedded proteins not well solubilized; expression only in understudied tissues or cell types; and proteins lacking two tryptic digestion sites 9–40 aa apart (though these proteins may be detected with alternative proteases, missed cleavages, or terminal peptides).
Finally, Figure 5 shows the contributions of the 8 top new data sets in PeptideAtlas that generated 160 of the increment of 255 canonical proteins in 2022 (Table 1).
Figure 5.
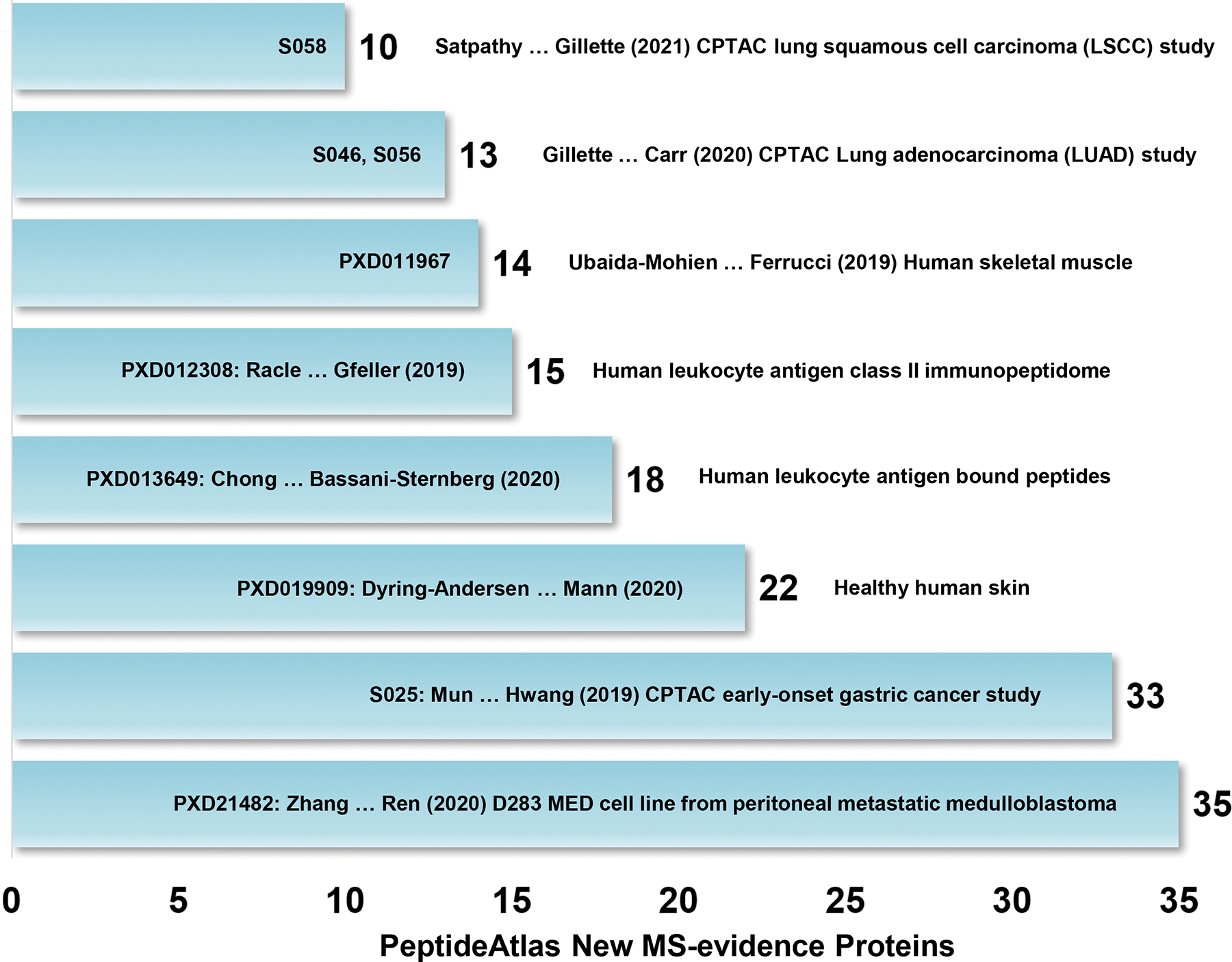
Primary papers that generated ten or more new PE1 proteins in Protein Atlas-2022-01 and neXtProt-2022-02. These eight reports14–22 include CPTAC proteogenomic studies of gastric, lung adeno, and lung squamous carcinomas14, 15, 21; proteins associated with inflammation, proteolysis, and splicing in aging skeletal muscle16; hundreds of non-canonical shared and tumor-specific HLA peptides from non-coding regions18, 19; 10,701 proteins identified across 4 layers and 9 cell types in skin20; and 12 missing proteins from a medulloblastoma cell line confirmed with synthetic peptides in PRM, enhanced to 35 MPs upon review by PeptideAtlas22.
Supplementary Table 2 lists the eleven major datasets that contributed 10+ MPs each, a total of 551, to the big increase of PE1 entries in MassIVE, many confirming canonical PE1 entries in PeptideAtlas.
HIGHLIGHTS FROM THE CHROMOSOME-CENTRIC HPP (C-HPP)
The C-HPP is organized as 25 teams by chromosome and mitochondria and are geographically dispersed. Table 2 shows the current status of identified and predicted proteins by chromosome. For all but 5 highlighted chromosomes, between 92 and 98% of predicted proteins have been confidently detected; only 2–8% are classified as missing proteins. In contrast, 20% of chr Y, 17% of chr 11, and 11% of the predicted proteins encoded on chromosomes 7, 14, and 21 still lack credible detection. Among the credibly identified PE1 proteins, between 3% (chromosome Y) and 11% (chromosome X) lack functional annotation (uPE1). The total of uPE1 proteins lacking functional annotation (direct or by homology) is down from 1260 in 2017 to 1191 in 2022.
Table 2.
Chromosome-by-Chromosome Status of Predicted Proteins in neXtProt 2022–02, showing Missing Proteins (MP) and Unannotated Proteins (uPE1). Note: The total from neXtProt (18,407) differs from the sum of all PE1 proteins (18,413), which includes six whose genes are duplicated on a second chromosome.
| chr | PE1 | PE2 | PE3 | PE4 | PE1-PE4 | MP | uPE1 | %MP/PE1–4 | %uPE1/PE1 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1864 | 120 | 33 | 2 | 2019 | 155 | 145 | 7.68 | 7.78 |
| 2 | 1231 | 43 | 3 | 0 | 1277 | 46 | 74 | 3.60 | 6.01 |
| 3 | 1011 | 44 | 5 | 0 | 1060 | 49 | 57 | 4.62 | 5.64 |
| 4 | 706 | 29 | 12 | 0 | 747 | 41 | 47 | 5.49 | 6.66 |
| 5 | 834 | 27 | 2 | 0 | 863 | 29 | 53 | 3.36 | 6.35 |
| 6 | 940 | 53 | 4 | 1 | 998 | 58 | 58 | 5.81 | 6.17 |
| 7 | 867 | 100 | 5 | 3 | 975 | 108 | 48 | 11.08 | 5.54 |
| 8 | 626 | 32 | 5 | 0 | 663 | 37 | 36 | 5.58 | 5.75 |
| 9 | 705 | 50 | 8 | 2 | 765 | 60 | 65 | 7.84 | 9.22 |
| 10 | 686 | 40 | 0 | 1 | 727 | 41 | 43 | 5.64 | 6.27 |
| 11 | 1074 | 149 | 68 | 0 | 1291 | 217 | 70 | 16.81 | 6.52 |
| 12 | 962 | 42 | 7 | 0 | 1011 | 49 | 39 | 4.85 | 4.05 |
| 13 | 311 | 9 | 2 | 0 | 322 | 11 | 26 | 3.42 | 8.36 |
| 14 | 642 | 60 | 17 | 2 | 721 | 79 | 37 | 10.96 | 5.76 |
| 15 | 537 | 37 | 0 | 0 | 574 | 37 | 35 | 6.45 | 6.52 |
| 16 | 773 | 41 | 1 | 1 | 816 | 43 | 45 | 5.27 | 5.82 |
| 17 | 1085 | 57 | 3 | 0 | 1145 | 60 | 62 | 5.24 | 5.71 |
| 18 | 257 | 7 | 0 | 0 | 264 | 7 | 12 | 2.65 | 4.67 |
| 19 | 1327 | 67 | 10 | 1 | 1405 | 78 | 73 | 5.55 | 5.50 |
| 20 | 512 | 21 | 1 | 0 | 534 | 22 | 37 | 4.12 | 7.23 |
| 21 | 199 | 21 | 4 | 0 | 224 | 25 | 11 | 11.16 | 5.53 |
| 22 | 452 | 25 | 4 | 0 | 481 | 29 | 32 | 6.03 | 7.08 |
| X | 762 | 51 | 1 | 0 | 814 | 52 | 85 | 6.39 | 11.15 |
| Y | 32 | 8 | 0 | 0 | 40 | 8 | 1 | 20.00 | 3.13 |
| MT | 15 | 0 | 0 | 0 | 15 | 0 | 0 | 0.00 | 0.00 |
| unknown | 3 | 2 | 0 | 0 | 5 | 2 | 0 | ||
| All | 18407 | 1135 | 195 | 13 | 19756 | 1343 | 1191 |
Like most scientific activities, those of the C-HPP teams were curtailed by the COVID-19 pandemic; with widespread vaccination, laboratories and meetings have reopened. A highlight was the first in-person and hybrid workshop of the C-HPP, held in Faro, Portugal, as a satellite of the Proteo Villamoura combined Portuguese, Spanish, and French Proteomic Societies on May 13, 2022. More than 75 attendees contributed to a White Paper on implementing the HPP Grand Challenge Project (see later section). Speakers included Gong Zhang on the hidden human proteome: the protein-coding “non-coding” RNAs; Xuejuan Gao on uPE1 protein functionalization of ZSWIMI and FAM210B; Wanting Liu on protein function prediction; Marina Gay on top-down proteomics applied to the HPP; and Lydie Lane with an update to the neXtProt release 2022–02.
Many C-HPP labs turned their attention to Covid-19 biology and assays during the pandemic. A notable example from the Chromosome 6 team utilized the N-TAILS (Terminal Amine Isotope Labeling of Substrates) approach to generate an atlas of 101 human host cell substrates of the SARS-CoV-2 3-chymotrypsin-like protease, 3CL pro23, a remarkable landscape of human proteins that are inactivated or co-opted for viral replication and transmission by precise proteolytic cleavage. A new intracellular sensor of viruses was discovered in the hand-off from the viral receptor, ACE2, to galectin-8, targeting the virus for destruction in the lysosomal pathway, which 3CLpro inactivated.
Meanwhile, the Chromosome 14 team initiated a search for missing proteins in several cell lines derived from induced pluripotent stem cells, identifying several MPs by MS in naive and trophoblastic stem cells that met the HPP Data Interpretation Guidelines v3.024.
We warmly acknowledge the retirement of Young-Ki Paik who, with Sam Hanash, coordinated and pioneered the launch of HUPO in 2000–2002 when many scientists did not know what proteomics was. When he was President of HUPO, Young-Ki played a key role in developing the C-HPP with investigators in individual countries taking ownership of characterizing the protein-coding genes on a selected chromosome. This concept facilitated a global initiative with funding of individual projects, clear country leadership, and a strong sense of local achievements. Over 20 years Young-Ki guided the C-HPP, hosted semi-annual workshops, brought in new teams and leadership, and nurtured the continuing growth of the C-HPP with humility.
HIGHLIGHTS FROM THE B/D-HPP
The Biology and Disease-driven Human Proteome Project (B/D-HPP) focuses on determining the biological functions of human proteins to apply disease-related proteins to developing preventive and therapeutic interventions. The B/D-HPP branch comprises international teams of scientists in 19 areas of biological and clinical relevance (see https://hupo.org/B/D-HPP). The teams develop and disseminate state-of-the-art proteomics methods and software to the broader scientific community and implement proteomics discovery in multidisciplinary biological and clinical studies. Increasingly, the B/D-HPP teams are collaborating across the various initiatives. Here we highlight recent accomplishments of several B/D-HPP teams.
Cardiovascular
This very active initiative investigates many heart diseases cutting-edge proteomics and other omics technologies to map the dynamic cardiac and vascular proteomes, elucidate disease mechanisms, identify candidate therapeutic targets, and provide clinically useful diagnoses and risk predictions. There is emphasis on single-cell and proteoform resolution. The Van Eyk group shared their insights on how proteomics helped elucidate hallmark proteins associated with COVID-19 infections with respect to both short- and long-term diagnosis and prognosis25. They utilized phosphoproteomics to investigate the effects of cardiosphere-derived cell treatments in hyperphosphorylated heart failure, especially HFpEF26. The Gundry group developed a method for preparing isolated cardiomyocytes and whole cardiac tissue homogenates for bottom-up proteomic analyses27; they applied glycomics to characterize sickle cell disease and hPSC-derived cardiomyocytes28. The Lau group utilized multi-omics to compare skeletal muscle and cardiac muscle in young and aged mice, revealing new details of how aging reshapes gene expression at the transcript and protein levels29. Members of the CV team worked closely with the PediOME, Rheumatic Disorders, EyeOme, and Liver Initiatives to generate popular proteins lists. Lam further developed the PUBPULAR website (https://heart.shinyapps.io/PubPular/) to integrate data on intensively investigated proteins.
Cancers
The ultimate goal here is to map the proteome of all types of human cancers to reveal tumor biology and drive improved diagnostics, treatments, and management of cancers. The members collaborate closely with the Pathology Pillar to source clinical samples with full medical history in order to identify cancer-specific proteins, proteoforms, or protein networks through data deposition, sharing, and analysis. Large-scale clinical cancer proteogenomics studies published in 2021 include pancreatic ductal adenocarcinoma30, lung squamous cell carcinoma14, glioblastoma31, and head and neck squamous cell carcinoma32. Guo published a review on high-throughput proteomics and AI for cancer biomarker discovery33. Jimenez published a phosphoproteomics study of AML focused on response to Flt3 inhibitors34 and bench-marked a protocol that enabled (phospho)proteomics on the same sample35. Further analyses were performed on the DIA-MS-based pan-cancer atlas of 1200+ cancers representing 22 cancer types (The Cancer Proteome Atlas, www.cancerproteome.org) led by the Jimenez and Guo laboratories. They plan to expand to a larger Cancer HPP community effort.
Glycoproteomics
The Human Glycoproteomics Initiative (HGI) (@HumanGlycoprot) aims to increase understanding of the functional significance of the extensive post-translational modification of proteins by glycans. This requires well-integrated analytical and informatics tools to more easily enable the determination of site-, protein-, cell- and tissue-specific glycoform structural heterogeneity in complex biological systems. The HGI is a project/study-centric initiative. The modus operandi is to carefully assemble experts and field-leaders to complete discrete studies of particular glycoproteomic analytical methods and software of interest to the community that increasingly is becoming interested in the role of glycosylation in diseases. The first HGI large community study, which compared glycoinformatics solutions for intact glycopeptide analysis from LC-MS/MS data, a recognized bottleneck in the field36, was completed in 2021 with 55 participants (many HUPO members) across 22 teams from 11 different countries. The work was published in Nature Methods35 and presented at international meetings by Thaysen-Andersen (HUPO 2021, Australian Glycoscience Symposium), Kolarich (Queensland MS Symposium, Lorne Proteomics), and Packer (Microscale Separations and Bioanalysis, B/D HPP Webinar), and at the Dagstuhl Seminar on Computational Proteomics in order to directly help the community create the toolboxes required to address unexplored glycobiology-focused fundamental and applied research questions in human health and disease.
Human Immuno-Peptidome
The Human Immuno-Peptidome Project (HiPP) with Chair Michal Bassani-Sternberg and over 200 members, was very busy in 2021. The long-term aims of HiPP are to map the entire repertoire of peptides presented by HLA molecules using mass spectrometry technologies, and make its robust analysis accessible to immunologists, clinical investigators, and other researchers. Members of this team published three definitive articles on the peptidome of SARS-COV-2 in Cell, Cell Reports, and Nature Immunology37–39 plus other landmark immunology publications identifying specific immuno-epitopes in Nature Communications, Journal of Clinical Investigation, and Frontiers in Immunology and Immunity40–43.
Pillars of the HUPO-HIPP program are method and technology development, standardization, effective data sharing, and education. They have organized a biennial HUPO-HIPP summer school to expand the immunopeptidomics community; organized quarterly virtual webinars/panels to share and discuss the latest advances in immunopeptidome research and technology development; designed multi-laboratory studies to benchmark protocols and MS technologies; and established partnerships with MS developers to improve detection and analysis of immunopeptidomes. To promote the visibility of HUPO-HIPP, the members include their affiliation in publications, conferences, and workshops, a good practice for everyone significantly involved in the HPP. They are working on establishing partnerships with appropriate journal editors and funding agencies to enforce sharing of immunopeptidomic data.
Food and Nutrition
With the growing understanding that there is a link between the health of humans, other animals within our ecosystem, and the broader environment, there is great interest in using proteomics to characterize dietary proteins and proteome alterations linked to nutritional issues, food safety, and food security. A collaborative manuscript from members of the Food and Nutrition (FAN) team, led by Paola Roncada, offered an overview of advanced methods available for studying food allergenic proteins, which include cross-linking combined with mass spectrometry and thermal proximity profiling among the mass spectrometry-based approaches44. The FAN team is linked to other consortia of scientists that aim to promote the use of omics technologies within dietary and nutrition studies, such as the FoodOmicsGR_RI consortium45. With a goal to understand plant survival and crop losses connected to high-temperature stress, the group of Subhra Chakraborty found a role for 2-Cys peroxiredoxin in wheat thermotolerance46. Characterizing the human gut microbiota is another important focus of the FAN, exemplified by the Figeys group’s report of an optimized experimental and bioinformatics workflow for defining lysine acylations in microbial species within the human gut47.
Rheumatic and Autoimmune Diseases
The overarching goal of the Rheumatic and Autoimmune Diseases (RAD) team is to discover and characterize human proteins that can be used in the prognosis or therapy of rheumatologic disorders, including age-related pathologies, inflammatory diseases, and systemic autoimmune diseases. This large collaborative effort among proteomics groups, basic research scientists, and clinicians last year completed the Applied Public-Private Research enabling OsteoArthritis Clinical Headway (APPROACH) project48, 49. Connected to this effort, the Blanco and Nilsson labs monitored protein biomarkers to identify predictors of radiographic knee osteoarthritis50. The Blanco group further implemented targeted mass spectrometry or imaging mass spectrometry, in collaboration with either the Ruiz-Romero or Heeren group, to uncover signature lipidomic profiles within the synovial fluid or membranes of individuals with osteoarthritis51, 52. These findings and protocols established for characterizing proteins as biomarkers for patient stratification and therapeutic management were disseminated by RAD team members at the European Congress of Rheumatology, the Congress on Osteoarthritis, and through organized symposia.
Infectious Disease
Dedicated to understanding the biology and pathogenesis of infection with microbes, including human viruses and bacteria, members of the infectious disease (ID) team have advanced MS-based methods for the detection of pathogenic proteins, virus-host protein interactions, and infection-induced alterations in protein abundance or posttranslational modifications53–55. A targeted mass spectrometry assay, TRUSTED, was developed by the Cristea group for the specific detection of viral proteins from the three Herpesviridae subfamilies, demonstrating its use for monitoring the effects of clinical therapeutic agents and anti- or pro-viral factors56. Targeted mass spectrometry also defined the impact of diverse human viruses on membrane contact site proteins that link organelles in dynamic functional networks, leading to the discovery of a previously unrecognized mitochondria-ER encapsulation structure that aids virus production57. The group of Nita-Lazar integrated metabolomics with secretome analyses, establishing a link between bacterial lipopolysaccharide (LPS) tolerance, immunometabolism, and secretome changes in macrophages58. Searching for pathogen-specific tuning of innate immune responses, they further characterized LPS structures from different bacterial pathogens and the resulting inflammatory actions and protein secretion59. The group of Concha Gil determined proteome alterations in Candida albicans in response to stress, finding a role for Prn1 in oxidative stress and possible targets for future antifungal drugs60, as well as characterizing released extracellular vesicles containing virulence-related proteins61. The Malmstrom group used quantitative and structural mass spectrometry to demonstrate that Streptococcus pyogenes, a human-specific Gram-positive bacterium, establishes serotype-specific interactions with human plasma proteins as a likely means to inhibit immune responses during mucosal and systemic infections62. Integrating information from protein microarray data, the LaBaer group identified a set of common autoantibodies present in healthy individuals, without gender preference, thereby providing critical information for determining possible false positives within reported disease-related biomarkers63. In response to the ongoing pandemic, the ID team joined members of other B/D-HPP teams (noted above) to elucidate the biology and pathology of SARS-CoV-2 infection. The Srivastava, Schmidt, and Volker groups, among others, established robust mass spectrometry methods for detection of SARS-CoV-2 proteins, defining protein-metabolite signature profiles and crosstalk that may predict clinical outcomes and characterize vaccine-induced complications64–66. The LaBaer group joined the SARS-CoV-2 Serological Sciences Network (SeroNet)67.
Lastly, to continue the dissemination of results and newly developed methodologies of interest during the pandemic-imposed virtual setting in 2021, the B/D-HPP held a well-received webinar, entitled “SARS-CoV-2 through the lens of proteomics”, in which members from different B/D-HPP teams presented their research contributions in response to the COVID-19 pandemic from the specific scientific lens of their individual teams (https://www.hupo.org/Webinars-and-Virtual-Presentations).
HIGHLIGHTS FROM THE KNOWLEDGEBASE RESOURCE PILLAR
UniProtKB/Swiss-Prot curators continue to focus on improving both the sequence quality and annotation content of the human proteome. This work includes the re-curation of sequences that have been shown to be incorrectly represented, the removal of entries for proteins which are now believed not to be expressed, such as putative products of what are now regarded as pseudogenes, and the creation of records describing the products of newly identified protein-coding genes. New isoforms are added when verified by experimental data or removed when shown to be experimental artifacts or the result of erroneous gene model predictions. About 93% of UniProtKB/Swiss-Prot canonical sequences are now identical to their corresponding Ensembl protein sequences translated from the reference human genome; work is ongoing to understand the differences between entries, including reference sequences, for the remaining 7%. Efforts are also focused on ensuring consistency with the Matched Annotation from NCBI and EMBL-EBI (MANE)68. MANE is a collaboration between EMBL-EBI and NCBI to converge on human gene and transcript annotation and to jointly define a high-value set of transcripts and corresponding proteins. Functional information is continually being added to UniProtKB/Swiss-Prot entries by expert curators reading and evaluating the scientific literature and, in parallel, annotating the same proteins in the Gene Ontology annotation project69 and adding to the protein interaction data in the IMEx databases70. In both cases, this represents computationally accessible information which is then imported back into the appropriate UniProt entries. Large-scale datasets, including peptides identified by MS, are increasingly imported into UniProt, visualized on the website, and available for downloading via the API.
neXtProt automatically carries forward curation decisions by UniProtKB/Swiss-Prot, which includes certain anomalies, such as 15 MP entries with sequences 100% identical to non-MS-PE1 entries, 31 entries 100% identical to other MPs, 17 entries for highly-similar USP17-like peptides (chr 4 and chr 8), 13 entries for putative nuclear-encoded peptides of 24–28 aa very similar to a peptide from the mitochondrial genome, 85 entries for T cell receptors or immunoglobulin regions, and numerous lncRNAs, including 17 with LINCXXX as the gene name. These are candidates for exclusion from the MP target list (the denominator for the HPP parts list), which will be discussed in the coming year.
Potential Polypeptides Translated from Ribo-seq Open Reading Frame (ORF) Sequences
A strategy for standardized annotation of Ribo-seq ORFs led by Jonathan Mudge (EMBL-EBI), with participation of the HPP, was announced in a July 2022 article in Nature Biotechnology71. The group’s aim is to find a “home” like GENCODE for these data on upstream uORFs or small smORFs that will stimulate a broad range of studies about their potential functions. PeptideAtlas will perform a reanalysis of public human datasets with its process based on the Trans-Proteomic Pipeline72, 73 to search for translation evidence for the 7264 ORFs, some as short as 16 aa in length, with substantial evidence of ribosome translation activity based on Ribo-seq experiments. The intent is not to annotate them in the same manner as the current set of canonical human proteins – and thus very substantially increase the number of human proteins – if there is no compelling evidence that they have functions, but rather to annotate them in a manner that allows them to be identified and studied as a separate class. We note from Figure 1 above, that UniProtKB/Swiss-Prot has recently incorporated three new uORFs, which were added to neXtProt during 2021, based on literature evidence of potential function.
This focus on short polypeptides raises the question not whether peptides can be biologically active (which is well known) but whether the community should distinguish polypeptides shorter than some threshold length as separate from “proteins”, and what quality assurance guidelines should be applied if not the HPP Guidelines v3.0. The 2005 Human Plasma Proteome Project papers included one on the “low MW proteome” or “peptidome”74 with MW <15 kDa. neXtProt and UniProtKB do not make a distinction between proteins and peptides based on a minimum size limit.
HIGHLIGHTS FROM THE ANTIBODY PILLAR/HUMAN PROTEIN ATLAS
The HPP Antibody Resource Pillar, based on the Human Protein Atlas (HPA) project, focuses on mapping the human proteome using spatial proteomics and antibody-based imaging. In version 21.1 of the HPA (release date 31 May 2022), a significant update of the structure and content of the database was introduced, dividing the data into ten main sections, each focusing on particular aspects of the human proteome and genome. HPA v21 builds upon the Ensembl version 103 genome release for annotation of all protein-coding genes and introduces a new normalization scheme for all datasets. The Single Cell Type section75, introduced in v20 (released 19 November 2020), was expanded to include single-cell transcriptomics data from 25 tissue types, providing a body-wide overview of expression in human single-cell types. The dataset constitutes an excellent resource for comparing antibody-based proteomics data using immunohistochemistry to study expression in single-cell types across platforms. This also facilitates more stringent antibody validation, one of the main objectives of the Antibody Resource Pillar.
A complementary section of the HPA, added in v21, is the Tissue Cell Type section. In this novel section, deconvolution and integrated network analysis of publicly available bulk RNA-seq data were used to predict cell type expression specificity for all human protein-coding genes. This allows for determining which genes are enriched in each cell type within tissues and detailed studies of core cell types expressed across several tissues, such as endothelial cells. The Brain section, extensively updated in 2021, focuses on expression profiles in different brain regions; internally generated RNA-seq data from >1300 human brain samples, including 200 regions, areas, and nuclei, were added. The Subcellular section provides insights into the spatiotemporal subcellular distribution of proteins; an in-depth analysis of cell cycle-dependent genes76 has been expanded in v21 to include 129 additional genes with cell cycle-dependent transcripts.
The Blood protein section77, which presents estimated concentrations of proteins detected in human blood plasma, also had major updates; 850 additional proteins now have plasma concentrations based on mass spectrometry data from the PeptideAtlas Build 2021–01. The data for longitudinal plasma expression levels were updated and are now based on the Olink Explore platform for an additional 708 proteins. The human Secretome was updated, with a new category of Immunoglobulin genes added. Altogether, the total number of predicted secreted proteins is now 2739, of which the number of proteins annotated as secreted in the blood is 784. Yet another recent section addresses Adipose tissues78.
In addition to the updates to the HPA database, the HPA workflow and antibodies have been utilized for several major research studies, including several projects linked to the COVID-19 pandemics79–82 and efforts for advanced imaging. One example is 3D imaging of sympathetic innervations, leading to further understanding of nonalcoholic fatty liver disease83. Future objectives of the Antibody Resource Pillar include a continuous close collaboration with the HPA project and other groups of the HPP to study and identify missing proteins using antibody-based proteomics. Possible efforts include integrated projects utilizing various methods for mRNA and protein detection in the same samples and multiplexed imaging, which will allow for determining protein expression patterns in rare cell types not distinguishable by the human eye.
HIGHLIGHTS FROM THE PATHOLOGY RESOURCE PILLAR
The Pathology Pillar (PP) was launched at the HUPO World Congress in Orlando, USA, in 2018 to recognize the important role that pathology must play in the translation and delivery of proteomics-driven biomarker discovery and next generation diagnostics and theranostics84. This Pillar aims at coordinating the identification of key unmet needs in clinical medicine, stimulating guidelines and standards for the development of fit-for-purpose validated clinical assays, promoting awareness of best practices, and coordinating access to bioresources of clinical samples with their associated data. One of the most significant contributions of proteomics will be the improvement of human health85. An important example is the Clinical Proteomic Tumor Analysis Consortium (CPTAC) coordinated by the NCI, noted above. CPTAC working groups have elucidated unique and powerful proteogenomics landscapes for a total of 14 common and less common cancers (e.g., pancreas30, lung14, acute myeloid leukemia86, glioblastoma31, head and neck32, 87). A new round characterizing 10 additional tumor types will start in late 2022 (including hepatocellular, biliary, and gastric cancers). In the current CPTAC approach, only untreated primary cancers are being analyzed. Better understanding the metastatic processes and the outcomes of targeted chemo- and immune-therapies on the proteomic evolution of cancers will require large-scale, longitudinal studies.
Other interesting PP-related publications include the role of redox signaling pathways in cancers88, drug resistance89, 90, and the emerging role of tumor cell plasticity in modifying therapeutic responses91. Martens et al developed a molecular pathway model of malignant pleural mesothelioma (MPM), a visual, interactive overview of interactions and connections between proteins and molecular pathways known to be related to MPM92. Extracellular vesicles, small membrane-enclosed particles released by cells and able to traffic information between them, are seeing increasing application, e.g., proteomic alterations in extracellular vesicles from activating mutations in PIK3CA, one of the most frequently mutated genes in human cancers93. At the technical level, a surfactant-assisted one-pot sample preparation for label-free single-cell proteomics improves detection sensitivity for single-cell proteomics94 to address intra-tumoral heterogeneity. There were also reviews on proteomics, personalized medicine, and cancers85, 95, and roles of separation sciences in proteomics95, 96.
The identification of significant medical needs and specific diseases or applications for which proteomics will make a difference over conventional DNA- and RNA-based molecular diagnostics is an important goal of the Pathology Pillar (Figure 6). Over the past year, several studies have shown new protein markers for risk prediction of cancers97, 98. This is particularly significant for early-stage cancers, such as colorectal adenocarcinoma, because there currently exist no robust predictive markers to direct additional adjuvant therapy to high-risk patients after curative-intent surgery, while protecting low-risk patients from harmful, debilitating, and costly overtreatment. A human melanoma proteome atlas focused on in-depth histopathology coupled to proteomic characterization defines the molecular pathology of melanomas99. A pan-cancer proteomic map of 949 human cell lines across 28 tissue types (https://cellmodelpassports.sanger.ac.uk) revealed principles of protein regulation with important implications for future clinical studies.
Figure 6.
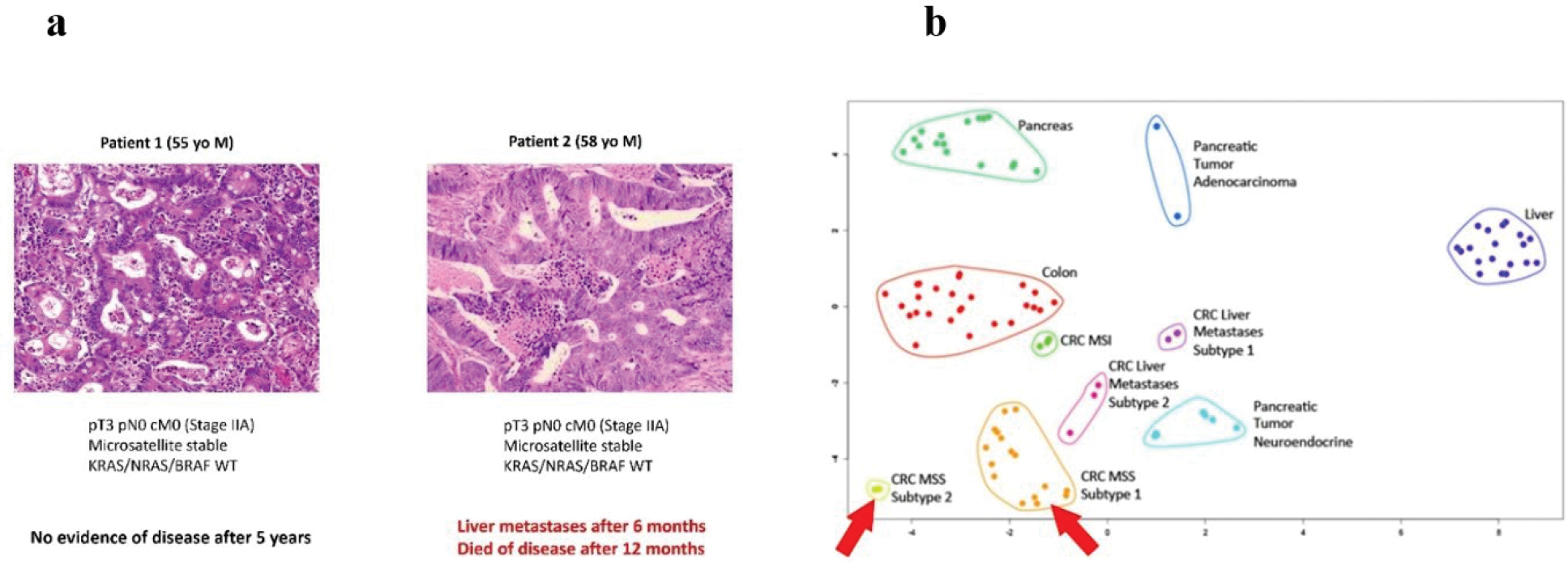
Personalized outcome risk prediction of colon cancers is a specific example of high value “clinical need” with possible proteomics solutions. (a) Two patients with colon cancers appear – to the best of today’s diagnostic capabilities – identical by demographics, histo-morphology, stage, and genomics, yet have vastly different survival outcomes (patient on left cured by surgery vs. patient on right died of progressive disease). (b) Proteomics analyses may reveal subtypes of previously indistinguishable cancers (as in CPTAC). The patients from (a) fall into two different proteomic subtypes of microsatellite stable (MSS) colon cancer (green and orange clusters, see arrows), illustrating that proteomics “sees” more than traditional molecular diagnostics. For comparison, clusters of other cancer types (pancreatic adenocarcinoma and neuroendocrine tumor, microsatellite unstable (MSI) colon cancer, metastatic colon cancer) and benign parenchyma (colon, pancreas, liver) are shown.
Meanwhile, proteomics has also made significant contributions to understanding SARS-CoV-2 and COVID-19, including “long COVID”, with persistent cough, fatigue, muscle and joint pain, memory loss, brain fog, or depression. Several papers show a striking relationship to human autoimmunity100–103; an analysis of human lung cells showed that a large number of COVID-dysregulated proteins during acute infection are known human autoantigens104.
Finally, the HPP Pathology mission is to enhance education and outreach for physicians and healthcare providers worldwide, raise awareness about proteomics, provide education through meetings and tutorials, and increasingly bring international pathology and laboratory medicine societies into close collaborative contact with HUPO and HPP. A pilot proteomics education program for pathologists and clinicians has been established with The Royal College of Pathologists of Australasia and the Biobanks and Complex Data Management MSc Program at the Université Côte d’Azur (Nice, France). We aim to extend these initiatives to other Pathology and Clinical Diagnosis groups around the world.
COMPLEMENTARY OMICS TECHNOLOGIES FOR BLOOD PROTEIN DETECTION
The continued development of affinity-based assays for protein identification in blood has accelerated the adoption of these technologies for high-throughput plasma and serum proteomics. A recent review of these non-mass spectrometry-based blood proteomic technologies describes the advantages and disadvantages of each and highlights the need for quantitative assessment and comparison of each technology to validate each approach105. These plate-based affinity assays can take the form of small arrays of protein targets marketed by a number of companies, to large, ordered arrays with considerable depth in coverage of unique proteins in blood samples. Most prominent of these targeted assay developments are the Proximity Extension Assays (PEA by Olink) constructed as paired antibodies with either quantitative polymerase chain reaction (qPCR)106–108 or sequencing as a readout109, or the large library of modified slow off-rate DNA aptamers (SomaScan by SomaLogic)110, 111. Currently, Olink provides 2940 (3K system) unique assays and SomaLogic provides 6377 (7K system) targeted assays to human plasma proteins. Between these plate-based affinity platforms, there is quite a high overlap in protein targets, with 2033 unique proteins detectable across both targeted affinity platforms. However, compared with current mass spectrometry-based approaches to analyze blood proteins, there are only 1484 unique proteins detected by all three technologies based on the 2021 Human Plasma PeptideAtlas. Combining all three platforms, >8000 proteins can be targeted for potential detection of plasma proteins, as recently reviewed in the 2021 HUPO Human Proteome Project paper, “Advances and utility of the human plasma proteome”96.
As interest in plate-based affinity assays towards blood proteins has increased markedly over the last few years, several studies have compared the different targeted assay approaches and the correlations in their readout. Because each of these large-scale plate-based targeted assays reports only relative quantitative values, there is considerable difficulty in correlating responses between the assays due to the different approaches to target protein-binding molecules, binding epitopes of each protein assayed, and detection modalities112–121. The correlations varied widely from being very closely correlated (r=0.99) for very few assays between the large-scale platform readouts to very low correlated (r=0.02) for a majority of the assays, and to even heavily negatively correlated (r=−0.48). The explanation for the difficulty in correlating findings across platforms stems from the differences in the protein detection and quantitative readout of each platform causing differences in signal conversion; confounding each protein detection are the differing binding site epitopes on each target protein. These differences greatly complicate clinical chemistry applications, where accurate identification and quantitation are needed. For example, Baird and Hoofnagle reported that aptamer-based approaches fail to detect major structural proteins (Apo-A1) in high-density lipoproteins (HDL) associated with HDL cholesterol, a stark contrast to validated clinical immunoassays to Apo-A1 in HDL cholesterol determination122. Other examples include the failure to associate retinol binding protein-4, a protein secreted by adipocytes and increased in insulin-resistant states in diabetes patients, again in contrast to other studies123.
As with any new technology, much work is needed to convince researchers and clinical pathologists of the specificity and accuracy of measurements made by the novel technology. Somalogic has assessed the specificity of their aptamer-based affinity reagents through a project conducted by researchers at Novartis124. A library of 4783 SOMAmers was multiplexed in sets of 8 and used for enrichment of target proteins from cell lysates, cell culture conditioned media, human plasma, and human serum, with a subset also screened in urine. Mass spectrometry analyses were performed (736 by DDA and 104 by MRM) alongside 779 SOMAmer reagents binding their endogenous targets. The raw DDA data were deposited to the HUPO Proteomics Standards Initiative ProteomeXchange Consortium via PRIDE74, 125, with five dataset identifiers PXD008819-PXD008823. The raw MRM data were deposited to ProteomeXchange via the PeptideAtlas PASSEL repository with the dataset identifier PASS01145126. Although this dataset is the first to provide some evidence of specificity, the experimental design does not allow an investigation of the specificity of each SOMAmer because they were grouped as a collective target recovery experiment with additional contaminants detected, making it impossible to assign targets and contaminants identified to a specific SOMAmer. While accuracy of quantitation with each reagent remains to be investigated, this dataset provides the first evidence toward characterizing the core reagents of one of these novel technologies.
High-throughput analyses utilizing standardized 96-well plate-based targeted proteomics measurements serve the purpose of rapidly quantifying differences of analytes detected within and across experiments. Comparison of results within a technology is both feasible and expansive as shown by the development of analysis consortia such as SCALLOP by Anders Mälarstig127–129. Within SCALLOP, researchers with Olink data wishing to compare results across different cohorts can apply for membership, with the obligation to make their own data available to the consortia, increasing the total pool of searchable results. Currently, SCALLOP has over 70,000 sample results available to members and continues to add members. Another large consortium is the Human Pharma Proteomics Project (HPPP), a collaboration between the UK Biobank (UKB) and 13 biopharmaceutical companies, headed by Chris Whelan of Biogen, which has recently completed the initial characterization of 54,306 plasma proteomic profiles from UK Biobank participants. Using the Olink 1.5K panel (1463 unique proteins) as a first phase analysis and including protein quantitative trait loci (pQTL) mapping, they identified 10,248 primary genetic associations, of which 85% are newly discovered130. The data identified independent secondary associations in 92% of cis and 29% of trans loci, expanding the catalogue of genetic instruments for downstream analyses. This study has provided an updated characterization of the genetic architecture of the plasma proteome, leveraging population-scale proteomics to provide novel extensive insights into trans pQTLs across multiple biological domains. This large study with a singular technology identified numerous pharmacological actionable results such as identifying genetic influences on ligand-receptor interactions, pathway perturbations, and novel drug targets, illustrated by the genetic proxied effect of PCSK9 levels on lipid concentrations and cardio- and cerebro-vascular diseases. These data will be enhanced by additional efforts of the HPPP to extend the Olink analysis to the Olink 3K panel as well as performing various pilot scale mass spectrometry-based analyses on a smaller cohort of ~2,000 of the same UKB subjects. Adding to the public plasma proteome knowledgebase as an open-access proteomics resource will help enable the elucidation of biological mechanisms underlying genetic discoveries and accelerate the development of novel biomarkers and therapeutics.
To correlate quantitative responses between platforms, mass spectrometry has been compared to quantitative readouts with Olink assays in a few systematic studies. Price, et al., described high correlation for some proteins when comparing targeted Olink and selected reaction monitoring assays (SRM-MS) for C-reactive protein, but for other targets the quantitative correlation was lower131. Ruffieux, et al., reported two sample cohorts analyzed by SomaScan, detecting 1096 proteins in both cohorts, and MS-based assays using immunodepleted and 6-plex isobaric labeling,132 detecting 133 proteins in both cohorts133. In the common set of 72 proteins detected by both methods, they found agreement between some of the reported protein levels (e.g., SHBG, CAH1, CXCL7) and another 25 proteins with concordant effects associated with pQTL (e.g., CFB, CO7, FETUA).
As highlighted in the above technologies, mass spectrometry-based approaches do not escape the difficulty in correlating quantitative readouts. The vast majority of protein detection and quantitation in mass spectrometry is derived from bottom–up approaches where proteins are enzymically cleaved to short peptides, identified, and quantitated in the mass spectrometer, and then finally aggregated to a protein level quantitative value. Given there are estimated to be ~100 different proteoforms per gene product133 the averaging of peptide signals discounts the contributions of post-translational modifications and could hamper the discovery of important biological differences134. The effort to define proteoforms for every human protein135 is a worthwhile initiative that complements the effort by the Human Proteome Grand Challenges Project to define function(s) for every human protein. Consequently, until we understand the context of each protein identified, the quantitative values of each protein afforded by any technology need to be validated by orthogonal approaches to account for the biological diversity. With ever-increasing technology development, we need to utilize each technology for its strengths and complement them with cross-platform capabilities to arrive at conclusions that explain the biological differences represented.
THE HPP GRAND CHALLENGE PROJECT: “A FUNCTION FOR EVERY PROTEIN”
The Human Proteome Organization, through the HPP, has developed over the past 20 years a framework to generate and disseminate focused biological knowledge to better understand various human biology and disease. Based on the milestone of >90% of predicted human proteins credibly detected by 20203–5, the vast high-quality human proteome databases, and the C-HPP neXt_CP50 initiative136 to functionally annotate the then 1260 unannotated PE1 proteins (uPE1) in neXtProt, the HPP has developed a framework to integrate forces and resources from its various initiatives and pillars, and collectively contribute to a large-scale “Grand Challenge Project” important to human health and well-being. The mandate came from a critical review in 2020 by the HPP Scientific Advisory Board chaired by Ruedi Aebersold and an open community discussion. During the 2020 HUPO virtual annual meeting, the HPP Executive Committee led by Robert Moritz announced the HPP Grand Challenge Project framework and subsequently posted a White Paper on the HPP website (www.hupo.org). The goal is to advance our understanding of complex biological processes in an open community framework where individuals and groups can propose various fundable work plans to contribute towards the aims of the Project. The first of these is the HPP Grand Challenge Project “A Function for Every Protein”.
The Project will enable interrogation of the often-multiple functions of any protein through profiling and modeling of proteomic changes after the expression or deletion of a particular protein in a cell. In essence, the functions of a protein are revealed by perturbing the protein in question within the molecular networks of the cell. The Project includes the following aspects:
Quantitative status of proteins (expression level, isoforms, PTMs, localization, interactions) in the context of networks and interactomes that can be perturbed by variation amongst individuals, specific binding interactions, and protein complex formation, influencing their functional relevance. Computational predictions92,93 may provide Gene Ontology clues for experimental design. Measurements of proteins can be based on MS, antibodies, and any other technologies with rigorous technical quality control. These molecular measurements could be performed in cell line models, or in cells or tissue specimens from animal models or patients with specific diseases such as infectious diseases, cancers, cardiovascular and metabolic diseases, and neurodegenerative disorders. Analyses might be conducted at the single cell level. Both bottom up and top-down approaches could be adopted.
Illumination of the functional basis of a disease by building off the principles of disease manifestation from a genomic viewpoint and applying proteomics knowledge to define protein interactions and their roles. Clinical proteomics studies can be rich sources of data for this Project. In the disease-centric design of clinical cohorts, various subtypes of clinical phenotypes arising from patient heterogeneity are conceptually equivalent to biological perturbation experiments.
Measurements of the initiation and progression of a disease through a pathology-focused view, including spatial and temporal measurements, disease stratification, aberration of disease-related protein networks after intervention, and identification of novel therapeutic targets through affiliated resources such as the Human Protein Atlas antibody resource headed by Mathias Uhlén (SciLifeLab, Stockholm) and the Target2035 small molecule binder resource headed by Cheryl Arrowsmith and Aled Edwards (U. Toronto). Both address the entire human proteome. In 2011 Edwards had estimated that 90% of the literature is focused on 10% of the proteins137.
Generation of databases and informational resources to collect, organize, and disseminate the contextual network states after perturbation of the specific proteins. Following the experience of the HPP, we will generate guidelines and metrics for the credible annotation of function(s) of specific proteins in network and cell//tissue/organ context. An open, user-friendly knowledge base with effective visualization tools will be critical to sharing the information with the broader public and with complementary initiatives, such as the PharmaProteomics Project with the UK Biobank85, the Understudied Proteins Initiative with the Wellcome Trust94, and the π-HuB Proteomic Navigator of the Human Body in China.
The HPP Grand Challenge Project “A Function for Every Protein” was presented to the proteomics public at the HUPO 2021 virtual Congress, the 2022 Lorne Australian Proteomics Symposium, and other national and international meetings by members of the HPP EC. A task force has been announced within the HPP Executive Committee to provide comments and perspective on emerging proposals. This task force does not aim to evaluate or rank grant applications but will advise and provide collective support to project leaders on their objectives, milestones, and deliverables and fit with other components of the HPP Grand Project. The first countries to express significant interest in developing programs towards this focus are France, through the current HPP Chair, Charles Pineau, and China through the efforts of Tiannan Guo and Fuchu He.
Specifically, Pineau presented the project at several universities and to the French National Alliance for Life Sciences and Health (Aviesan) and the French National Research Agency (ANR). These groups together constitute the main stakeholders of life and health sciences in France. It was agreed that French research teams could apply for funding in connection with the HPP during the next annual call for proposals in October 2022. A similar nationally focused presentation strategy is being organized with the German Research Foundation (Deutsche Forschungsgemeinschaft; DFG) and will be extended to other European countries by their scientists.
In China, Guo proposed the Westlake Pilot Project in Hangzhou; an initial 50 representative proteins were selected based on their relevance to breast cancers for functional analysis in HEK293T human embryonic kidney cells. Most of these proteins have known functions to build a training model, while some proteins with unclear function(s) are to be investigated by interrogating the proteome perturbation for each protein using CRISPR-Cas9 experiments. This pilot will set up a workflow for data quality control, storage, analysis, and visualization, with extensive communication among HPP members and be the initial contribution from China.
The HPP Grand Challenges Projects will serve as a coordination point to reach out and connect such initiatives to complete the ambitious goals of decoding the function of every human protein and realizing the aims of precision medicine.
Supplementary Material
Table S2: MassIVE data sets with 10+ new proteins with MS evidence for this latest release
Table S1: The 868 PE1 proteins WITHOUT mass spectrometry evidence from the PeptideAtlas-MassIVE-neXtProt pipeline
ACKNOWLEDGMENTS
We appreciate the guidance from the HPP Executive Committee and the participation of all HPP investigators. We thank the UniProt groups at SIB, EBI, and PIR for providing high-quality annotations for the human proteins in UniProtKB/Swiss-Prot. The neXtProt server is hosted at SIB Swiss Institute of Bioinformatics in Switzerland, ProteomeXchange and PRIDE at the European Bioinformatics Institute in Cambridge, UK, PeptideAtlas at the Institute for Systems Biology in Seattle, and MassIVE at the University of California San Diego. G.S.O. acknowledges support from National Institutes of Health Grants P30ES017885-01A1 and U24CA210967; E.W.D. and R.L.M. from National Institutes of Health Grants R01GM087221, R24GM127667, U19AG023122, S10OD026936, and from National Science Foundation Grant DBI-1933311; C.M.O. by Canadian Institutes of Health Research Foundation Grant 148408 and a Canada Research Chair in Protease Proteomics and Systems Biology; N.B. from NIH grant 1R01LM013115, and NSF grant ABI 1759980; M.S.B. by Australian Research Council (CE140100003); C.L. by the Knut and Alice Wallenberg Foundation for the Human Protein Atlas; M.H.R by National Institutes of Health Grants R21 CA263262, U01 CA253217, R21 CA251992, P30 CA008748 (MSKCC CCSG, Pathology Component), NIH-Leidos CPTAC contract 17X173, and Farmer Family Foundation; and I.M.C. from National Institutes of Health Grant R01GM114141 and Stand Up To Cancer Convergence 3.1416.
Footnotes
SUPPORTING INFORMATION
The Supporting Information is available free of charge.
REFERENCES
- 1.Legrain P; Aebersold R; Archakov A; Bairoch A; Bala K; Beretta L; Bergeron J; Borchers CH; Corthals GL; Costello CE; Deutsch EW; Domon B; Hancock W; He F; Hochstrasser D; Marko-Varga G; Salekdeh GH; Sechi S; Snyder M; Srivastava S; Uhlen M; Wu CH; Yamamoto T; Paik YK; Omenn GS, The Human Proteome Project: Current state and future direction. Mol Cell Proteomics 2011, 10 (7), M111 009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanash S; Celis JE, The Human Proteome Organization: a mission to advance proteome knowledge. Mol Cell Proteomics 2002, 1 (6), 413–4. [DOI] [PubMed] [Google Scholar]
- 3.Adhikari S; Nice EC; Deutsch EW; Lane L; Omenn GS; Pennington SR; Paik YK; Overall CM; Corrales FJ; Cristea IM; Van Eyk JE; Uhlen M; Lindskog C; Chan DW; Bairoch A; Waddington JC; Justice JL; LaBaer J; Rodriguez H; He F; Kostrzewa M; Ping P; Gundry RL; Stewart P; Srivastava S; Srivastava S; Nogueira FCS; Domont GB; Vandenbrouck Y; Lam MPY; Wennersten S; Vizcaino JA; Wilkins M; Schwenk JM; Lundberg E; Bandeira N; Marko-Varga G; Weintraub ST; Pineau C; Kusebauch U; Moritz RL; Ahn SB; Palmblad M; Snyder MP; Aebersold R; Baker MS, A high-stringency blueprint of the human proteome. Nat Commun 2020, 11 (1), 5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omenn GS; Lane L; Overall CM; Paik YK; Cristea IM; Corrales FJ; Lindskog C; Weintraub S; Roehrl MHA; Liu S; Bandeira N; Srivastava S; Chen YJ; Aebersold R; Moritz RL; Deutsch EW, Progress identifying and analyzing the human proteome: 2021 Metrics from the HUPO Human Proteome Project. J Proteome Res 2021, 20 (12), 5227–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omenn GS, Reflections on the HUPO Human Proteome Project, the flagship project of the Human Proteome Organization, at 10 Years. Mol Cell Proteomics 2021, 20, 100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutsch EW; Orchard S; Binz PA; Bittremieux W; Eisenacher M; Hermjakob H; Kawano S; Lam H; Mayer G; Menschaert G; Perez-Riverol Y; Salek RM; Tabb DL; Tenzer S; Vizcaino JA; Walzer M; Jones AR, Proteomics standards initiative: Fifteen years of progress and future work. J Proteome Res 2017, 16 (12), 4288–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahn-Zabal M; Michel PA; Gateau A; Nikitin F; Schaeffer M; Audot E; Gaudet P; Duek PD; Teixeira D; Rech de Laval V; Samarasinghe K; Bairoch A; Lane L, The neXtProt knowledgebase in 2020: data, tools and usability improvements. Nucleic Acids Res 2020, 48 (D1), D328–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desiere F; Deutsch EW; King NL; Nesvizhskii AI; Mallick P; Eng J; Chen S; Eddes J; Loevenich SN; Aebersold R, The PeptideAtlas project. Nucleic Acids Res 2006, 34 (Database issue), D655–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutsch EW; Sun Z; Campbell D; Kusebauch U; Chu CS; Mendoza L; Shteynberg D; Omenn GS; Moritz RL, State of the human proteome in 2014/2015 as viewed through PeptideAtlas: Enhancing accuracy and coverage through the AtlasProphet. J Proteome Res 2015, 14 (9), 3461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M; Wang J; Carver J; Pullman BS; Cha SW; Bandeira N, Assembling the community-scale discoverable human proteome. Cell Syst 2018, 7 (4), 412–421 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch EW; Lane L; Overall CM; Bandeira N; Baker MS; Pineau C; Moritz RL; Corrales F; Orchard S; Van Eyk JE; Paik YK; Weintraub ST; Vandenbrouck Y; Omenn GS, Human Proteome Project mass spectrometry data interpretation guidelines 3.0. J Proteome Res 2019, 18 (12), 4108–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutsch EW; Overall CM; Van Eyk JE; Baker MS; Paik YK; Weintraub ST; Lane L; Martens L; Vandenbrouck Y; Kusebauch U; Hancock WS; Hermjakob H; Aebersold R; Moritz RL; Omenn GS, Human proteome project mass spectrometry data interpretation guidelines 2.1. J Proteome Res 2016, 15 (11), 3961–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omenn GS; Lane L; Overall CM; Corrales FJ; Schwenk JM; Paik YK; Van Eyk JE; Liu S; Snyder M; Baker MS; Deutsch EW, Progress on identifying and characterizing the human proteome: 2018 metrics from the HUPO Human Proteome Project. J Proteome Res 2018, 17 (12), 4031–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satpathy S; Krug K; Jean Beltran PM; Savage SR; Petralia F; Kumar-Sinha C; Dou Y; Reva B; Kane MH; Avanessian SC; Vasaikar SV; Krek A; Lei JT; Jaehnig EJ; Omelchenko T; Geffen Y; Bergstrom EJ; Stathias V; Christianson KE; Heiman DI; Cieslik MP; Cao S; Song X; Ji J; Liu W; Li K; Wen B; Li Y; Gumus ZH; Selvan ME; Soundararajan R; Visal TH; Raso MG; Parra ER; Babur O; Vats P; Anand S; Schraink T; Cornwell M; Rodrigues FM; Zhu H; Mo CK; Zhang Y; da Veiga Leprevost F; Huang C; Chinnaiyan AM; Wyczalkowski MA; Omenn GS; Newton CJ; Schurer S; Ruggles KV; Fenyo D; Jewell SD; Thiagarajan M; Mesri M; Rodriguez H; Mani SA; Udeshi ND; Getz G; Suh J; Li QK; Hostetter G; Paik PK; Dhanasekaran SM; Govindan R; Ding L; Robles AI; Clauser KR; Nesvizhskii AI; Wang P; Carr SA; Zhang B; Mani DR; Gillette MA; Clinical Proteomic Tumor Analysis Consortium, A proteogenomic portrait of lung squamous cell carcinoma. Cell 2021, 184 (16), 4348–4371 e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillette MA; Satpathy S; Cao S; Dhanasekaran SM; Vasaikar SV; Krug K; Petralia F; Li Y; Liang WW; Reva B; Krek A; Ji J; Song X; Liu W; Hong R; Yao L; Blumenberg L; Savage SR; Wendl MC; Wen B; Li K; Tang LC; MacMullan MA; Avanessian SC; Kane MH; Newton CJ; Cornwell M; Kothadia RB; Ma W; Yoo S; Mannan R; Vats P; Kumar-Sinha C; Kawaler EA; Omelchenko T; Colaprico A; Geffen Y; Maruvka YE; da Veiga Leprevost F; Wiznerowicz M; Gumus ZH; Veluswamy RR; Hostetter G; Heiman DI; Wyczalkowski MA; Hiltke T; Mesri M; Kinsinger CR; Boja ES; Omenn GS; Chinnaiyan AM; Rodriguez H; Li QK; Jewell SD; Thiagarajan M; Getz G; Zhang B; Fenyo D; Ruggles KV; Cieslik MP; Robles AI; Clauser KR; Govindan R; Wang P; Nesvizhskii AI; Ding L; Mani DR; Carr SA; Clinical Proteomic Tumor Analysis Consortium, Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 2020, 182 (1), 200–225 e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ubaida-Mohien C; Lyashkov A; Gonzalez-Freire M; Tharakan R; Shardell M; Moaddel R; Semba RD; Chia CW; Gorospe M; Sen R; Ferrucci L, Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ubaida-Mohien C; Gonzalez-Freire M; Lyashkov A; Moaddel R; Chia CW; Simonsick EM; Sen R; Ferrucci L, Physical activity associated proteomics of skeletal muscle: Being physically active in daily life may protect skeletal muscle from aging. Front Physiol 2019, 10, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Racle J; Michaux J; Rockinger GA; Arnaud M; Bobisse S; Chong C; Guillaume P; Coukos G; Harari A; Jandus C; Bassani-Sternberg M; Gfeller D, Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol 2019, 37 (11), 1283–1286. [DOI] [PubMed] [Google Scholar]
- 19.Chong C; Muller M; Pak H; Harnett D; Huber F; Grun D; Leleu M; Auger A; Arnaud M; Stevenson BJ; Michaux J; Bilic I; Hirsekorn A; Calviello L; Simo-Riudalbas L; Planet E; Lubinski J; Bryskiewicz M; Wiznerowicz M; Xenarios I; Zhang L; Trono D; Harari A; Ohler U; Coukos G; Bassani-Sternberg M, Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun 2020, 11 (1), 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyring-Andersen B; Lovendorf MB; Coscia F; Santos A; Moller LBP; Colaco AR; Niu L; Bzorek M; Doll S; Andersen JL; Clark RA; Skov L; Teunissen MBM; Mann M, Spatially and cell-type resolved quantitative proteomic atlas of healthy human skin. Nat Commun 2020, 11 (1), 5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mun DG; Bhin J; Kim S; Kim H; Jung JH; Jung Y; Jang YE; Park JM; Kim H; Jung Y; Lee H; Bae J; Back S; Kim SJ; Kim J; Park H; Li H; Hwang KB; Park YS; Yook JH; Kim BS; Kwon SY; Ryu SW; Park DY; Jeon TY; Kim DH; Lee JH; Han SU; Song KS; Park D; Park JW; Rodriguez H; Kim J; Lee H; Kim KP; Yang EG; Kim HK; Paek E; Lee S; Lee SW; Hwang D, Proteogenomic characterization of human early-onset gastric cancer. Cancer cell 2019, 35 (1), 111–124 e10. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y; Zhang K; Bu F; Hao P; Yang H; Liu S; Ren Y, D283 Med, a cell line derived from peritoneal metastatic medulloblastoma: A good choice for missing protein discovery. J Proteome Res 2020, 19 (12), 4857–4866. [DOI] [PubMed] [Google Scholar]
- 23.Vellios N; van der Zee K, Dataset on cigarette smokers in six South African townships. Data Brief 2020, 32, 106260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girard et al. , Naive Pluripotent and Trophoblastic Stem Cell Lines as a Model for Detecting Missing Proteins in the Context of the Chromosome-Centric Human Proteome Project [Manuscript submitted to Journal of Proteome Research for publication] August 2022 [DOI] [PubMed] [Google Scholar]
- 25.McArdle A; Washington KE; Chazarin Orgel B; Binek A; Manalo DM; Rivas A; Ayres M; Pandey R; Phebus C; Raedschelders K; Fert-Bober J; Van Eyk JE, Discovery proteomics for COVID-19: Where we are now. J Proteome Res 2021, 20 (10), 4627–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soetkamp D; Gallet R; Parker SJ; Holewinski R; Venkatraman V; Peck K; Goldhaber JI; Marban E; Van Eyk JE, Myofilament phosphorylation in stem cell treated diastolic heart failure. Circ Res 2021, 129 (12), 1125–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wojtkiewicz M; Berg Luecke L; Castro C; Burkovetskaya M; Mesidor R; Gundry RL, Bottom-up proteomic analysis of human adult cardiac tissue and isolated cardiomyocytes. J Mol Cell Cardiol 2022, 162, 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashwood HE; Ashwood C; Schmidt AP; Gundry RL; Hoffmeister KM; Anani WQ, Characterization and statistical modeling of glycosylation changes in sickle cell disease. Blood Adv 2021, 5 (5), 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y; Li LZ; Kastury NL; Thomas CT; Lam MPY; Lau E, Transcriptome features of striated muscle aging and predictability of protein level changes. Mol Omics 2021, 17 (5), 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao L; Huang C; Cui Zhou D; Hu Y; Lih TM; Savage SR; Krug K; Clark DJ; Schnaubelt M; Chen L; da Veiga Leprevost F; Eguez RV; Yang W; Pan J; Wen B; Dou Y; Jiang W; Liao Y; Shi Z; Terekhanova NV; Cao S; Lu RJ; Li Y; Liu R; Zhu H; Ronning P; Wu Y; Wyczalkowski MA; Easwaran H; Danilova L; Mer AS; Yoo S; Wang JM; Liu W; Haibe-Kains B; Thiagarajan M; Jewell SD; Hostetter G; Newton CJ; Li QK; Roehrl MH; Fenyo D; Wang P; Nesvizhskii AI; Mani DR; Omenn GS; Boja ES; Mesri M; Robles AI; Rodriguez H; Bathe OF; Chan DW; Hruban RH; Ding L; Zhang B; Zhang H; Clinical Proteomic Tumor Analysis Consortium, Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 2021, 184 (19), 5031–5052 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang LB; Karpova A; Gritsenko MA; Kyle JE; Cao S; Li Y; Rykunov D; Colaprico A; Rothstein JH; Hong R; Stathias V; Cornwell M; Petralia F; Wu Y; Reva B; Krug K; Pugliese P; Kawaler E; Olsen LK; Liang WW; Song X; Dou Y; Wendl MC; Caravan W; Liu W; Cui Zhou D; Ji J; Tsai CF; Petyuk VA; Moon J; Ma W; Chu RK; Weitz KK; Moore RJ; Monroe ME; Zhao R; Yang X; Yoo S; Krek A; Demopoulos A; Zhu H; Wyczalkowski MA; McMichael JF; Henderson BL; Lindgren CM; Boekweg H; Lu S; Baral J; Yao L; Stratton KG; Bramer LM; Zink E; Couvillion SP; Bloodsworth KJ; Satpathy S; Sieh W; Boca SM; Schurer S; Chen F; Wiznerowicz M; Ketchum KA; Boja ES; Kinsinger CR; Robles AI; Hiltke T; Thiagarajan M; Nesvizhskii AI; Zhang B; Mani DR; Ceccarelli M; Chen XS; Cottingham SL; Li QK; Kim AH; Fenyo D; Ruggles KV; Rodriguez H; Mesri M; Payne SH; Resnick AC; Wang P; Smith RD; Iavarone A; Chheda MG; Barnholtz-Sloan JS; Rodland KD; Liu T; Ding L; Clinical Proteomic Tumor Analysis Consortium, Proteogenomic and metabolomic characterization of human glioblastoma. Cancer cell 2021, 39 (4), 509–528 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C; Chen L; Savage SR; Eguez RV; Dou Y; Li Y; da Veiga Leprevost F; Jaehnig EJ; Lei JT; Wen B; Schnaubelt M; Krug K; Song X; Cieslik M; Chang HY; Wyczalkowski MA; Li K; Colaprico A; Li QK; Clark DJ; Hu Y; Cao L; Pan J; Wang Y; Cho KC; Shi Z; Liao Y; Jiang W; Anurag M; Ji J; Yoo S; Zhou DC; Liang WW; Wendl M; Vats P; Carr SA; Mani DR; Zhang Z; Qian J; Chen XS; Pico AR; Wang P; Chinnaiyan AM; Ketchum KA; Kinsinger CR; Robles AI; An E; Hiltke T; Mesri M; Thiagarajan M; Weaver AM; Sikora AG; Lubinski J; Wierzbicka M; Wiznerowicz M; Satpathy S; Gillette MA; Miles G; Ellis MJ; Omenn GS; Rodriguez H; Boja ES; Dhanasekaran SM; Ding L; Nesvizhskii AI; El-Naggar AK; Chan DW; Zhang H; Zhang B; Clinical Proteomic Tumor Analysis Consortium, Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer cell 2021, 39 (3), 361–379 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Q; Zhang F; Xu L; Yue L; Kon OL; Zhu Y; Guo T, High-throughput proteomics and AI for cancer biomarker discovery. Adv Drug Deliv Rev 2021, 176, 113844. [DOI] [PubMed] [Google Scholar]
- 34.Cucchi DGJ; Van Alphen C; Zweegman S; Van Kuijk B; Kwidama ZJ; Al Hinai A; Henneman AA; Knol JC; Piersma SR; Pham TV; Jimenez CR; Cloos J; Janssen J, Phosphoproteomic characterization of primary AML samples and relevance for response toward FLT3-inhibitors. Hemasphere 2021, 5 (7), e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rolfs F; Piersma SR; Dias MP; Jonkers J; Jimenez CR, Feasibility of phosphoproteomics on leftover samples after RNA extraction with guanidinium thiocyanate. Mol Cell Proteomics 2021, 20, 100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawahara R; Chernykh A; Alagesan K; Bern M; Cao W; Chalkley RJ; Cheng K; Choo MS; Edwards N; Goldman R; Hoffmann M; Hu Y; Huang Y; Kim JY; Kletter D; Liquet B; Liu M; Mechref Y; Meng B; Neelamegham S; Nguyen-Khuong T; Nilsson J; Pap A; Park GW; Parker BL; Pegg CL; Penninger JM; Phung TK; Pioch M; Rapp E; Sakalli E; Sanda M; Schulz BL; Scott NE; Sofronov G; Stadlmann J; Vakhrushev SY; Woo CM; Wu HY; Yang P; Ying W; Zhang H; Zhang Y; Zhao J; Zaia J; Haslam SM; Palmisano G; Yoo JS; Larson G; Khoo KH; Medzihradszky KF; Kolarich D; Packer NH; Thaysen-Andersen M, Community evaluation of glycoproteomics informatics solutions reveals high-performance search strategies for serum glycopeptide analysis. Nat Methods 2021, 18 (11), 1304–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weingarten-Gabbay S; Klaeger S; Sarkizova S; Pearlman LR; Chen DY; Gallagher KME; Bauer MR; Taylor HB; Dunn WA; Tarr C; Sidney J; Rachimi S; Conway HL; Katsis K; Wang Y; Leistritz-Edwards D; Durkin MR; Tomkins-Tinch CH; Finkel Y; Nachshon A; Gentili M; Rivera KD; Carulli IP; Chea VA; Chandrashekar A; Bozkus CC; Carrington M; MGH COVID-19 Collection & Processing Team; Bhardwaj N; Barouch DH; Sette A; Maus MV; Rice CM; Clauser KR; Keskin DB; Pregibon DC; Hacohen N; Carr SA; Abelin JG; Saeed M; Sabeti PC, Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell 2021, 184 (15), 3962–3980 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker R; Partridge T; Wormald C; Kawahara R; Stalls V; Aggelakopoulou M; Parker J; Powell Doherty R; Ariosa Morejon Y; Lee E; Saunders K; Haynes BF; Acharya P; Thaysen-Andersen M; Borrow P; Ternette N, Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell reports 2021, 35 (8), 109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelde A; Bilich T; Heitmann JS; Maringer Y; Salih HR; Roerden M; Lubke M; Bauer J; Rieth J; Wacker M; Peter A; Horber S; Traenkle B; Kaiser PD; Rothbauer U; Becker M; Junker D; Krause G; Strengert M; Schneiderhan-Marra N; Templin MF; Joos TO; Kowalewski DJ; Stos-Zweifel V; Fehr M; Rabsteyn A; Mirakaj V; Karbach J; Jager E; Graf M; Gruber LC; Rachfalski D; Preuss B; Hagelstein I; Marklin M; Bakchoul T; Gouttefangeas C; Kohlbacher O; Klein R; Stevanovic S; Rammensee HG; Walz JS, SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol 2021, 22 (1), 74–85. [DOI] [PubMed] [Google Scholar]
- 40.Ebrahimi-Nik H; Moussa M; Englander RP; Singhaviranon S; Michaux J; Pak H; Miyadera H; Corwin WL; Keller GLJ; Hagymasi AT; Shcheglova TV; Coukos G; Baker BM; Mandoiu II; Bassani-Sternberg M; Srivastava PK, Reversion analysis reveals the in vivo immunogenicity of a poorly MHC I-binding cancer neoepitope. Nat Commun 2021, 12 (1), 6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son ET; Faridi P; Paul-Heng M; Leong ML; English K; Ramarathinam SH; Braun A; Dudek NL; Alexander IE; Lisowski L; Bertolino P; Bowen DG; Purcell AW; Mifsud NA; Sharland AF, The self-peptide repertoire plays a critical role in transplant tolerance induction. J Clin Invest 2021, 131 (21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee S; Sanchez-Bernabeu A; Demmers LC; Wu W; Heck AJR, The HLA ligandome comprises a limited repertoire of O-GlcNAcylated antigens preferentially associated with HLA-B*07:02. Front Immunol 2021, 12, 796584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehx G; Larouche JD; Durette C; Laverdure JP; Hesnard L; Vincent K; Hardy MP; Theriault C; Rulleau C; Lanoix J; Bonneil E; Feghaly A; Apavaloaei A; Noronha N; Laumont CM; Delisle JS; Vago L; Hebert J; Sauvageau G; Lemieux S; Thibault P; Perreault C, Atypical acute myeloid leukemia-specific transcripts generate shared and immunogenic MHC class-I-associated epitopes. Immunity 2021, 54 (4), 737–752 e10. [DOI] [PubMed] [Google Scholar]
- 44.Benede S; Lozano-Ojalvo D; Cristobal S; Costa J; D’Auria E; Velickovic TC; Garrido-Arandia M; Karakaya S; Mafra I; Mazzucchelli G; Picariello G; Romero-Sahagun A; Villa C; Roncada P; Molina E, New applications of advanced instrumental techniques for the characterization of food allergenic proteins. Crit Rev Food Sci Nutr 2021, 1–17. [DOI] [PubMed] [Google Scholar]
- 45.Theodoridis G; Pechlivanis A; Thomaidis NS; Spyros A; Georgiou CA; Albanis T; Skoufos I; Kalogiannis S; Tsangaris GT; Stasinakis AS; Konstantinou I; Triantafyllidis A; Gkagkavouzis K; Kritikou AS; Dasenaki ME; Gika H; Virgiliou C; Kodra D; Nenadis N; Sampsonidis I; Arsenos G; Halabalaki M; Mikros E; On Behalf of The FoodOmicsGR Ri Consortium, FoodOmicsGR_RI. A consortium for comprehensive molecular characterisation of food products. Metabolites 2021, 11 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishra D; Shekhar S; Chakraborty S; Chakraborty N, Wheat 2-Cys peroxiredoxin plays a dual role in chlorophyll biosynthesis and adaptation to high temperature. Plant J 2021, 105 (5), 1374–1389. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X; Cheng K; Ning Z; Mayne J; Walker K; Chi H; Farnsworth CL; Lee K; Figeys D, Exploring the microbiome-wide lysine acetylation, succinylation, and propionylation in human gut microbiota. Anal Chem 2021, 93 (17), 6594–6598. [DOI] [PubMed] [Google Scholar]
- 48.van Helvoort EM; van Spil WE; Jansen MP; Welsing PMJ; Kloppenburg M; Loef M; Blanco FJ; Haugen IK; Berenbaum F; Bacardit J; Ladel CH; Loughlin J; Bay-Jensen AC; Mobasheri A; Larkin J; Boere J; Weinans HH; Lalande A; Marijnissen ACA; Lafeber F, Cohort profile: The applied public-private research enabling OsteoArthritis clinical headway (IMI-APPROACH) study: A 2-year, European, cohort study to describe, validate and predict phenotypes of osteoarthritis using clinical, imaging and biochemical markers. BMJ Open 2020, 10 (7), e035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Helvoort EM; Ladel C; Mastbergen S; Kloppenburg M; Blanco FJ; Haugen IK; Berenbaum F; Bacardit J; Widera P; Welsing PMJ; Lafeber F, Baseline clinical characteristics of predicted structural and pain progressors in the IMI-APPROACH knee OA cohort. RMD Open 2021, 7 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lourido L; Balboa-Barreiro V; Ruiz-Romero C; Rego-Perez I; Camacho-Encina M; Paz-Gonzalez R; Calamia V; Oreiro N; Nilsson P; Blanco FJ, A clinical model including protein biomarkers predicts radiographic knee osteoarthritis: A prospective study using data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2021, 29 (8), 1147–1154. [DOI] [PubMed] [Google Scholar]
- 51.Rocha B; Cillero-Pastor B; Ruiz-Romero C; Paine MRL; Canete JD; Heeren RMA; Blanco FJ, Identification of a distinct lipidomic profile in the osteoarthritic synovial membrane by mass spectrometry imaging. Osteoarthritis Cartilage 2021, 29 (5), 750–761. [DOI] [PubMed] [Google Scholar]
- 52.Rocha B; Illiano A; Calamia V; Pinto G; Amoresano A; Ruiz-Romero C; Blanco FJ, Targeted phospholipidomic analysis of synovial fluid as a tool for osteoarthritis deep phenotyping. Osteoarthritis and Cartilage Open 2021, 3 (4), 100219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acharjee A; Stephen Kingsly J; Kamat M; Kurlawala V; Chakraborty A; Vyas P; Vaishnav R; Srivastava S, Rise of the SARS-CoV-2 variants: Can proteomics be the silver bullet? Expert Rev Proteomics 2022, 1–16. [DOI] [PubMed] [Google Scholar]
- 54.Justice JL; Kennedy MA; Hutton JE; Liu D; Song B; Phelan B; Cristea IM, Systematic profiling of protein complex dynamics reveals DNA-PK phosphorylation of IFI16 en route to herpesvirus immunity. Sci Adv 2021, 7 (25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Duran C; Martinez-Lopez R; Monteoliva L; Gil C, Sample processing for metaproteomic analysis of human gut microbiota. Methods Mol Biol 2022, 2420, 53–61. [DOI] [PubMed] [Google Scholar]
- 56.Kennedy MA; Tyl MD; Betsinger CN; Federspiel JD; Sheng X; Arbuckle JH; Kristie TM; Cristea IM, A TRUSTED targeted mass spectrometry assay for pan-herpesvirus protein detection. Cell Rep 2022, 39 (6), 110810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cook KC; Tsopurashvili E; Needham JM; Thompson SR; Cristea IM, Restructured membrane contacts rewire organelles for human cytomegalovirus infection. Nat Commun 2022, 13 (1), 4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillen J; Ondee T; Gurusamy D; Issara-Amphorn J; Manes NP; Yoon SH; Leelahavanichkul A; Nita-Lazar A, LPS tolerance inhibits cellular respiration and induces global changes in the macrophage secretome. Biomolecules 2021, 11 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ernst O; Khan MM; Oyler BL; Yoon SH; Sun J; Lin FY; Manes NP; MacKerell AD Jr.; Fraser IDC; Ernst RK; Goodlett DR; Nita-Lazar A, Species-specific endotoxin stimulus determines toll-like Receptor 4- and Caspase 11-mediated pathway activation characteristics. mSystems 2021, 6 (4), e0030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amador-Garcia A; Zapico I; Borrajo A; Malmstrom J; Monteoliva L; Gil C, Extending the proteomic characterization of Candida albicans exposed to stress and apoptotic inducers through data-independent acquisition mass spectrometry. mSystems 2021, 6 (5), e0094621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinez-Lopez R; Hernaez ML; Redondo E; Calvo G; Radau S; Pardo M; Gil C; Monteoliva L, Candida albicans hyphal extracellular vesicles are different from yeast ones, carrying an active proteasome complex and showing a different role in host immune response. Microbiol Spectr 2022, 10 (3), e0069822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chowdhury S; Khakzad H; Bergdahl GE; Lood R; Ekstrom S; Linke D; Malmstrom L; Happonen L; Malmstrom J, Streptococcus pyogenes forms serotype- and local environment-dependent interspecies protein complexes. mSystems 2021, 6 (5), e0027121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shome M; Chung Y; Chavan R; Park JG; Qiu J; LaBaer J, Serum autoantibodyome reveals that healthy individuals share common autoantibodies. Cell Rep 2022, 39 (9), 110873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banerjee A; Pai MGJ; Singh A; Nissa MU; Srivastava S, Mass spectrometry and proteome analysis to identify SARS-CoV-2 protein from COVID-19 patient swab samples. STAR Protoc 2022, 3 (1), 101177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buyukozkan M; Alvarez-Mulett S; Racanelli AC; Schmidt F; Batra R; Hoffman KL; Sarwath H; Engelke R; Gomez-Escobar L; Simmons W; Benedetti E; Chetnik K; Zhang G; Schenck E; Suhre K; Choi JJ; Zhao Z; Racine-Brzostek S; Yang HS; Choi ME; Choi AMK; Cho SJ; Krumsiek J, Integrative metabolomic and proteomic signatures define clinical outcomes in severe COVID-19. iScience 2022, 25 (7), 104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greinacher A; Selleng K; Palankar R; Wesche J; Handtke S; Wolff M; Aurich K; Lalk M; Methling K; Volker U; Hentschker C; Michalik S; Steil L; Reder A; Schonborn L; Beer M; Franzke K; Buttner A; Fehse B; Stavrou EX; Rangaswamy C; Mailer RK; Englert H; Frye M; Thiele T; Kochanek S; Krutzke L; Siegerist F; Endlich N; Warkentin TE; Renne T, Insights in ChAdOx1 nCoV-19 vaccine-induced immune thrombotic thrombocytopenia. Blood 2021, 138 (22), 2256–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figueiredo JC; Hirsch FR; Kushi LH; Nembhard WN; Crawford JM; Mantis N; Finster L; Merin NM; Merchant A; Reckamp KL; Melmed GY; Braun J; McGovern D; Parekh S; Corley DA; Zohoori N; Amick BC; Du R; Gregersen PK; Diamond B; Taioli E; Sariol C; Espino A; Weiskopf D; Gifoni A; Brien J; Hanege W; Lipsitch M; Zidar DA; Scheck McAlearney A; Wajnberg A; LaBaer J; Yvonne Lewis E; Binder RA; Moormann AM; Forconi C; Forrester S; Batista J; Schieffelin J; Kim D; Biancon G; VanOudenhove J; Halene S; Fan R; Barouch DH; Alter G; Pinninti S; Boppana SB; Pati SK; Latting M; Karaba AH; Roback J; Sekaly R; Neish A; Brincks AM; Granger DA; Karger AB; Thyagarajan B; Thomas SN; Klein SL; Cox AL; Lucas T; Furr-Holden D; Key K; Jones N; Wrammerr J; Suthar M; Yu Wong S; Bowman NM; Simon V; Richardson LD; McBride R; Krammer F; Rana M; Kennedy J; Boehme K; Forrest C; Granger SW; Heaney CD; Knight Lapinski M; Wallet S; Baric RS; Schifanella L; Lopez M; Fernandez S; Kenah E; Panchal AR; Britt WJ; Sanz I; Dhodapkar M; Ahmed R; Bartelt LA; Markmann AJ; Lin JT; Hagan RS; Wolfgang MC; Skarbinski J, Mission, organization, and future direction of the serological sciences network for COVID-19 (SeroNet) epidemiologic cohort studies. Open Forum Infect Dis 2022, 9 (6), ofac171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morales J; Pujar S; Loveland JE; Astashyn A; Bennett R; Berry A; Cox E; Davidson C; Ermolaeva O; Farrell CM; Fatima R; Gil L; Goldfarb T; Gonzalez JM; Haddad D; Hardy M; Hunt T; Jackson J; Joardar VS; Kay M; Kodali VK; McGarvey KM; McMahon A; Mudge JM; Murphy DN; Murphy MR; Rajput B; Rangwala SH; Riddick LD; Thibaud-Nissen F; Threadgold G; Vatsan AR; Wallin C; Webb D; Flicek P; Birney E; Pruitt KD; Frankish A; Cunningham F; Murphy TD, A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature 2022, 604 (7905), 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gene Ontology Consortium, The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 2021, 49 (D1), D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porras P; Barrera E; Bridge A; Del-Toro N; Cesareni G; Duesbury M; Hermjakob H; Iannuccelli M; Jurisica I; Kotlyar M; Licata L; Lovering RC; Lynn DJ; Meldal B; Nanduri B; Paneerselvam K; Panni S; Pastrello C; Pellegrini M; Perfetto L; Rahimzadeh N; Ratan P; Ricard-Blum S; Salwinski L; Shirodkar G; Shrivastava A; Orchard S, Towards a unified open access dataset of molecular interactions. Nat Commun 2020, 11 (1), 6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mudge JM; Ruiz-Orera J; Prensner JR; Brunet MA; Calvet F; Jungreis I; Gonzalez JM; Magrane M; Martinez TF; Schulz JF; Yang YT; Alba MM; Aspden JL; Baranov PV; Bazzini AA; Bruford E; Martin MJ; Calviello L; Carvunis AR; Chen J; Couso JP; Deutsch EW; Flicek P; Frankish A; Gerstein M; Hubner N; Ingolia NT; Kellis M; Menschaert G; Moritz RL; Ohler U; Roucou X; Saghatelian A; Weissman JS; van Heesch S, Standardized annotation of translated open reading frames. Nat Biotechnol 2022, 40 (7), 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deutsch EW; Mendoza L; Shteynberg D; Farrah T; Lam H; Tasman N; Sun Z; Nilsson E; Pratt B; Prazen B; Eng JK; Martin DB; Nesvizhskii AI; Aebersold R, A guided tour of the Trans-Proteomic Pipeline. Proteomics 2010, 10 (6), 1150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deutsch EW; Mendoza L; Shteynberg D; Slagel J; Sun Z; Moritz RL, Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin Appl 2015, 9 (7–8), 745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Omenn GS; States DJ; Adamski M; Blackwell TW; Menon R; Hermjakob H; Apweiler R; Haab BB; Simpson RJ; Eddes JS; Kapp EA; Moritz RL; Chan DW; Rai AJ; Admon A; Aebersold R; Eng J; Hancock WS; Hefta SA; Meyer H; Paik YK; Yoo JS; Ping P; Pounds J; Adkins J; Qian X; Wang R; Wasinger V; Wu CY; Zhao X; Zeng R; Archakov A; Tsugita A; Beer I; Pandey A; Pisano M; Andrews P; Tammen H; Speicher DW; Hanash SM, Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 2005, 5 (13), 3226–45. [DOI] [PubMed] [Google Scholar]
- 75.Karlsson M; Zhang C; Mear L; Zhong W; Digre A; Katona B; Sjostedt E; Butler L; Odeberg J; Dusart P; Edfors F; Oksvold P; von Feilitzen K; Zwahlen M; Arif M; Altay O; Li X; Ozcan M; Mardinoglu A; Fagerberg L; Mulder J; Luo Y; Ponten F; Uhlen M; Lindskog C, A single-cell type transcriptomics map of human tissues. Sci Adv 2021, 7 (31). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verma M; Sharma N; Rashi; Arora V; Bashar MA; Nath B; Kalra S, Diabetic foot care knowledge and practices in rural North India: Insights for preventive podiatry. J Assoc Physicians India 2021, 69 (2), 30–34. [PubMed] [Google Scholar]
- 77.Uhlen M; Karlsson MJ; Hober A; Svensson AS; Scheffel J; Kotol D; Zhong W; Tebani A; Strandberg L; Edfors F; Sjostedt E; Mulder J; Mardinoglu A; Berling A; Ekblad S; Dannemeyer M; Kanje S; Rockberg J; Lundqvist M; Malm M; Volk AL; Nilsson P; Manberg A; Dodig-Crnkovic T; Pin E; Zwahlen M; Oksvold P; von Feilitzen K; Haussler RS; Hong MG; Lindskog C; Ponten F; Katona B; Vuu J; Lindstrom E; Nielsen J; Robinson J; Ayoglu B; Mahdessian D; Sullivan D; Thul P; Danielsson F; Stadler C; Lundberg E; Bergstrom G; Gummesson A; Voldborg BG; Tegel H; Hober S; Forsstrom B; Schwenk JM; Fagerberg L; Sivertsson A, The human secretome. Sci Signal 2019, 12 (609). [DOI] [PubMed] [Google Scholar]
- 78.Norreen-Thorsen M; Struck EC; Oling S; Zwahlen M; Von Feilitzen K; Odeberg J; Lindskog C; Ponten F; Uhlen M; Dusart PJ; Butler LM, A human adipose tissue cell-type transcriptome atlas. Cell reports 2022, 40 (2), 111046. [DOI] [PubMed] [Google Scholar]
- 79.Mulder J; Lindqvist I; Rasmusson AJ; Husen E; Ronnelid J; Kumlien E; Rostami E; Virhammar J; Cunningham JL, Indirect immunofluorescence for detecting anti-neuronal autoimmunity in CSF after COVID-19 - Possibilities and pitfalls. Brain Behav Immun 2021, 94, 473–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong W; Altay O; Arif M; Edfors F; Doganay L; Mardinoglu A; Uhlen M; Fagerberg L, Next generation plasma proteome profiling of COVID-19 patients with mild to moderate symptoms. EBioMedicine 2021, 74, 103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roxhed N; Bendes A; Dale M; Mattsson C; Hanke L; Dodig-Crnkovic T; Christian M; Meineke B; Elsasser S; Andrell J; Havervall S; Thalin C; Eklund C; Dillner J; Beck O; Thomas CE; McInerney G; Hong MG; Murrell B; Fredolini C; Schwenk JM, Multianalyte serology in home-sampled blood enables an unbiased assessment of the immune response against SARS-CoV-2. Nat Commun 2021, 12 (1), 3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), Canadian Interventional Radiology Association (CIRA), Congress of Neurological Surgeons (CNS), European Society of Minimally Invasive Neurological Therapy (ESMINT), European Society of Neuroradiology (ESNR), European Stroke Organization (ESO), Society for Cardiovascular Angiography and Interventions (SCAI), Society of Interventional Radiology (SIR), Society of NeuroInterventional Surgery (SNIS), and World Stroke Organization (WSO), Sacks D, Baxter B, Campbell BCV, Carpenter JS, Cognard C, Dippel D, Eesa M, Fischer U, Hausegger K, Hirsch JA, Shazam Hussain M, Jansen O, Jayaraman MV, Khalessi AA, Kluck BW, Lavine S, Meyers PM, Ramee S, Rüfenacht DA, Schirmer CM, Vorwerk D. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke 2018, 13 (6), 612–632.29786478 [Google Scholar]
- 83.Adori C; Daraio T; Kuiper R; Barde S; Horvathova L; Yoshitake T; Ihnatko R; Valladolid-Acebes I; Vercruysse P; Wellendorf AM; Gramignoli R; Bozoky B; Kehr J; Theodorsson E; Cancelas JA; Mravec B; Jorns C; Ellis E; Mulder J; Uhlen M; Bark C; Hokfelt T, Disorganization and degeneration of liver sympathetic innervations in nonalcoholic fatty liver disease revealed by 3D imaging. Sci Adv 2021, 7 (30). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He B; Huang Z; Huang C; Nice EC, Clinical applications of plasma proteomics and peptidomics: Towards precision medicine. Proteomics Clin Appl. 2022. May 1:e2100097. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 85.Roehrl MH; Roehrl VB; Wang JY, Proteome-based pathology: the next frontier in precision medicine. Expert Rev Precis Med Drug Dev 2021, 6 (1), 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Joshi SK; Nechiporuk T; Bottomly D; Piehowski PD; Reisz JA; Pittsenbarger J; Kaempf A; Gosline SJC; Wang YT; Hansen JR; Gritsenko MA; Hutchinson C; Weitz KK; Moon J; Cendali F; Fillmore TL; Tsai CF; Schepmoes AA; Shi T; Arshad OA; McDermott JE; Babur O; Watanabe-Smith K; Demir E; D’Alessandro A; Liu T; Tognon CE; Tyner JW; McWeeney SK; Rodland KD; Druker BJ; Traer E, The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer cell 2021, 39 (7), 999–1014 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez H; Zenklusen JC; Staudt LM; Doroshow JH; Lowy DR, The next horizon in precision oncology: Proteogenomics to inform cancer diagnosis and treatment. Cell 2021, 184 (7), 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao W; Huang Z; Duan J; Nice EC; Lin J; Huang C, Elesclomol induces copper-dependent ferroptosis in colorectal cancer cells via degradation of ATP7A. Mol Oncol 2021, 15 (12), 3527–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luo M; Yang X; Chen HN; Nice EC; Huang C, Drug resistance in colorectal cancer: An epigenetic overview. Biochim Biophys Acta Rev Cancer 2021, 1876 (2), 188623. [DOI] [PubMed] [Google Scholar]
- 90.Wu P; Gao W; Su M; Nice EC; Zhang W; Lin J; Xie N, Adaptive mechanisms of tumor therapy resistance driven by tumor microenvironment. Front Cell Dev Biol 2021, 9, 641469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin S; Jiang J; Lu Y; Nice EC; Huang C; Zhang J; He W, Emerging role of tumor cell plasticity in modifying therapeutic response. Signal Transduct Target Ther 2020, 5 (1), 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martens M; Kreidl F; Ehrhart F; Jean D; Mei M; Mortensen HM; Nash A; Nymark P; Evelo CT; Cerciello F, A community-driven, openly accessible molecular pathway integrating knowledge on malignant pleural mesothelioma. Front Oncol 2022, 12, 849640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saraswat M; Garapati K; Kim J; Budhraja R; Pandey A, Proteomic alterations in extracellular vesicles induced by oncogenic PIK3CA mutations. Proteomics. 2022. Jun 11:e2200077. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 94.Tsai CF; Zhang P; Scholten D; Martin K; Wang YT; Zhao R; Chrisler WB; Patel DB; Dou M; Jia Y; Reduzzi C; Liu X; Moore RJ; Burnum-Johnson KE; Lin MH; Hsu CC; Jacobs JM; Kagan J; Srivastava S; Rodland KD; Steven Wiley H; Qian WJ; Smith RD; Zhu Y; Cristofanilli M; Liu T; Liu H; Shi T, Surfactant-assisted one-pot sample preparation for label-free single-cell proteomics. Commun Biol 2021, 4 (1), 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Su M; Zhang Z; Zhou L; Han C; Huang C; Nice EC, Proteomics, personalized medicine and cancer. Cancers (Basel) 2021, 13 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deutsch EW; Omenn GS; Sun Z; Maes M; Pernemalm M; Palaniappan KK; Letunica N; Vandenbrouck Y; Brun V; Tao SC; Yu X; Geyer PE; Ignjatovic V; Moritz RL; Schwenk JM, Advances and utility of the human plasma proteome. J Proteome Res 2021, 20 (12), 5241–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ogawa M; Tanaka A; Namba K; Shia J; Wang JY; Roehrl MHA, Tumor stromal nicotinamide N-methyltransferase overexpression as a prognostic biomarker for poor clinical outcome in early-stage colorectal cancer. Scientific reports 2022, 12 (1), 2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ogawa M; Tanaka A; Namba K; Shia J; Wang JY; Roehrl MH, Early-stage loss of GALNT6 predicts poor clinical outcome in colorectal cancer. Front Oncol 2022, 12, 802548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Betancourt LH; Gil J; Kim Y; Doma V; Cakir U; Sanchez A; Murillo JR; Kuras M; Parada IP; Sugihara Y; Appelqvist R; Wieslander E; Welinder C; Velasquez E; de Almeida NP; Woldmar N; Marko-Varga M; Pawlowski K; Eriksson J; Szeitz B; Baldetorp B; Ingvar C; Olsson H; Lundgren L; Lindberg H; Oskolas H; Lee B; Berge E; Sjogren M; Eriksson C; Kim D; Kwon HJ; Knudsen B; Rezeli M; Hong R; Horvatovich P; Miliotis T; Nishimura T; Kato H; Steinfelder E; Oppermann M; Miller K; Florindi F; Zhou Q; Domont GB; Pizzatti L; Nogueira FCS; Horvath P; Szadai L; Timar J; Karpati S; Szasz AM; Malm J; Fenyo D; Ekedahl H; Nemeth IB; Marko-Varga G, The human melanoma proteome atlas-Defining the molecular pathology. Clin Transl Med 2021, 11 (7), e473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang JY; Zhang W; Roehrl MW; Roehrl VB; Roehrl MH, An autoantigen profile of human A549 lung cells reveals viral and host etiologic molecular attributes of autoimmunity in COVID-19. J Autoimmun 2021, 120, 102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang JY; Zhang W; Roehrl VB; Roehrl MW; Roehrl MH, An autoantigen-ome from HS-Sultan B-Lymphoblasts offers a molecular map for investigating autoimmune sequelae of COVID-19. bioRxiv [Preprint]. 2021. Apr 6:2021.04.05.438500. [Google Scholar]
- 102.Wang JY; Zhang W; Roehrl MW; Roehrl VB; Roehrl MH, An autoantigen profile from Jurkat T-Lymphoblasts provides a molecular guide for investigating autoimmune sequelae of COVID-19. bioRxiv [Preprint]. 2021. Jul 7:2021.07.05.451199. [Google Scholar]
- 103.Wang JY; Roehrl MW; Roehrl VB; Roehrl MH, A master autoantigen-ome links alternative splicing, female predilection, and COVID-19 to autoimmune diseases. J Transl Autoimmun 2022, 5, 100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang JY; Zhang W; Roehrl VB; Roehrl MW; Roehrl MH, An autoantigen atlas from human lung HFL1 cells offers clues to neurological and diverse autoimmune manifestations of COVID-19. Front Immunol 2022, 13, 831849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ren AH; Diamandis EP; Kulasingam V, Uncovering the depths of the human proteome: Antibody-based technologies for ultrasensitive multiplexed protein detection and quantification. Mol Cell Proteomics 2021, 20, 100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Assarsson E; Lundberg M; Holmquist G; Bjorkesten J; Thorsen SB; Ekman D; Eriksson A; Rennel Dickens E; Ohlsson S; Edfeldt G; Andersson AC; Lindstedt P; Stenvang J; Gullberg M; Fredriksson S, Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PloS one 2014, 9 (4), e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gullberg M; Gustafsdottir SM; Schallmeiner E; Jarvius J; Bjarnegard M; Betsholtz C; Landegren U; Fredriksson S, Cytokine detection by antibody-based proximity ligation. Proceedings of the National Academy of Sciences of the United States of America 2004, 101 (22), 8420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blokzijl A; Nong R; Darmanis S; Hertz E; Landegren U; Kamali-Moghaddam M, Protein biomarker validation via proximity ligation assays. Biochim Biophys Acta 2014, 1844 (5), 933–9. [DOI] [PubMed] [Google Scholar]
- 109.Zhong W; Edfors F; Gummesson A; Bergstrom G; Fagerberg L; Uhlen M, Next generation plasma proteome profiling to monitor health and disease. Nat Commun 2021, 12 (1), 2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rohloff JC; Gelinas AD; Jarvis TC; Ochsner UA; Schneider DJ; Gold L; Janjic N, Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids 2014, 3, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brody EN; Willis MC; Smith JD; Jayasena S; Zichi D; Gold L, The use of aptamers in large arrays for molecular diagnostics. Mol Diagn 1999, 4 (4), 381–8. [DOI] [PubMed] [Google Scholar]
- 112.Lim SY; Lee JH; Welsh SJ; Ahn SB; Breen E; Khan A; Carlino MS; Menzies AM; Kefford RF; Scolyer RA; Long GV; Rizos H, Evaluation of two high-throughput proteomic technologies for plasma biomarker discovery in immunotherapy-treated melanoma patients. Biomark Res 2017, 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Finkernagel F; Reinartz S; Schuldner M; Malz A; Jansen JM; Wagner U; Worzfeld T; Graumann J; von Strandmann EP; Muller R, Dual-platform affinity proteomics identifies links between the recurrence of ovarian carcinoma and proteins released into the tumor microenvironment. Theranostics 2019, 9 (22), 6601–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Graumann J; Finkernagel F; Reinartz S; Stief T; Brodje D; Renz H; Jansen JM; Wagner U; Worzfeld T; Pogge von Strandmann E; Muller R, Multi-platform affinity proteomics identify proteins linked to metastasis and immune suppression in ovarian cancer plasma. Front Oncol 2019, 9, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Raffield LM; Dang H; Pratte KA; Jacobson S; Gillenwater LA; Ampleford E; Barjaktarevic I; Basta P; Clish CB; Comellas AP; Cornell E; Curtis JL; Doerschuk C; Durda P; Emson C; Freeman CM; Guo X; Hastie AT; Hawkins GA; Herrera J; Johnson WC; Labaki WW; Liu Y; Masters B; Miller M; Ortega VE; Papanicolaou G; Peters S; Taylor KD; Rich SS; Rotter JI; Auer P; Reiner AP; Tracy RP; Ngo D; Gerszten RE; O’Neal WK; Bowler RP; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, Comparison of proteomic assessment methods in multiple cohort studies. Proteomics 2020, 20 (12), e1900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Petrera A; von Toerne C; Behler J; Huth C; Thorand B; Hilgendorff A; Hauck SM, Multiplatform approach for plasma proteomics: Complementarity of Olink proximity extension assay technology to mass spectrometry-based protein profiling. J Proteome Res 2021, 20 (1), 751–762. [DOI] [PubMed] [Google Scholar]
- 117.Pietzner M; Wheeler E; Carrasco-Zanini J; Kerrison ND; Oerton E; Koprulu M; Luan J; Hingorani AD; Williams SA; Wareham NJ; Langenberg C, Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat Commun 2021, 12 (1), 6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Daniels JR; Ma JZ; Cao Z; Beger RD; Sun J; Schnackenberg L; Pence L; Choudhury D; Palevsky PM; Portilla D; Yu LR, Discovery of novel proteomic biomarkers for the prediction of kidney recovery from dialysis-dependent AKI patients. Kidney360 2021, 2 (11), 1716–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Haslam DE; Li J; Dillon ST; Gu X; Cao Y; Zeleznik OA; Sasamoto N; Zhang X; Eliassen AH; Liang L; Stampfer MJ; Mora S; Chen ZZ; Terry KL; Gerszten RE; Hu FB; Chan AT; Libermann TA; Bhupathiraju SN, Stability and reproducibility of proteomic profiles in epidemiological studies: Comparing the Olink and SOMAscan platforms. Proteomics 2022, 22 (13–14), e2100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pietzner M; Wheeler E; Carrasco-Zanini J; Kerrison ND; Oerton E; Koprulu M; Luan J. a.; Hingorani AD; Williams SA; Wareham NJ; Langenberg C, Cross-platform proteomics to advance genetic prioritisation strategies. 2021. bioRxiv 2021.03.18.435919; doi: 10.1101/2021.03.18.435919 [DOI] [Google Scholar]
- 121.Eldjarn GH; Ferkingstad E; Lund SH; Helgason H; Magnusson OT; Olafsdottir TA; Halldorsson BV; Olason PI; Zink F; Gudjonsson SA; Sveinbjornsson G; Magnusson MI; Helgason A; Oddsson A; Halldorsson GH; Magnusson MK; Saevarsdottir S; Eiriksdottir T; Masson G; Stefansson H; Jonsdottir I; Holm H; Rafnar T; Melsted P; Saemundsdottir J; Norddahl GL; Thorleifsson G; Ulfarsson MO; Gudbjartsson DF; Thorsteinsdottir U; Sulem P; Stefansson K, Large-scale comparison of immunoassay- and aptamer-based plasma proteomics through genetics and disease. bioRxiv 2022.02.18.481034; doi: 10.1101/2022.02.18.481034. [DOI] [Google Scholar]
- 122.Baird GS; Hoofnagle AN, A novel discovery platform: Aptamers for the quantification of human proteins. Clin Chem 2017, 63 (6), 1061–1062. [DOI] [PubMed] [Google Scholar]
- 123.Graham TE; Yang Q; Bluher M; Hammarstedt A; Ciaraldi TP; Henry RR; Wason CJ; Oberbach A; Jansson PA; Smith U; Kahn BB, Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med 2006, 354 (24), 2552–63. [DOI] [PubMed] [Google Scholar]
- 124.Emilsson V; Ilkov M; Lamb JR; Finkel N; Gudmundsson EF; Pitts R; Hoover H; Gudmundsdottir V; Horman SR; Aspelund T; Shu L; Trifonov V; Sigurdsson S; Manolescu A; Zhu J; Olafsson O; Jakobsdottir J; Lesley SA; To J; Zhang J; Harris TB; Launer LJ; Zhang B; Eiriksdottir G; Yang X; Orth AP; Jennings LL; Gudnason V, Co-regulatory networks of human serum proteins link genetics to disease. Science 2018, 361 (6404), 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martens L; Hermjakob H; Jones P; Adamski M; Taylor C; States D; Gevaert K; Vandekerckhove J; Apweiler R, PRIDE: the proteomics identifications database. Proteomics 2005, 5 (13), 3537–45. [DOI] [PubMed] [Google Scholar]
- 126.Farrah T; Deutsch EW; Kreisberg R; Sun Z; Campbell DS; Mendoza L; Kusebauch U; Brusniak MY; Huttenhain R; Schiess R; Selevsek N; Aebersold R; Moritz RL, PASSEL: the PeptideAtlas SRMexperiment library. Proteomics 2012, 12 (8), 1170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Folkersen L; Fauman E; Sabater-Lleal M; Strawbridge RJ; Franberg M; Sennblad B; Baldassarre D; Veglia F; Humphries SE; Rauramaa R; de Faire U; Smit AJ; Giral P; Kurl S; Mannarino E; Enroth S; Johansson A; Enroth SB; Gustafsson S; Lind L; Lindgren C; Morris AP; Giedraitis V; Silveira A; Franco-Cereceda A; Tremoli E; IMPROVE study group; Gyllensten U; Ingelsson E; Brunak S; Eriksson P; Ziemek D; Hamsten A; Malarstig A, Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet 2017, 13 (4), e1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Folkersen L; Gustafsson S; Wang Q; Hansen DH; Hedman AK; Schork A; Page K; Zhernakova DV; Wu Y; Peters J; Eriksson N; Bergen SE; Boutin TS; Bretherick AD; Enroth S; Kalnapenkis A; Gadin JR; Suur BE; Chen Y; Matic L; Gale JD; Lee J; Zhang W; Quazi A; Ala-Korpela M; Choi SH; Claringbould A; Danesh J; Davey Smith G; de Masi F; Elmstahl S; Engstrom G; Fauman E; Fernandez C; Franke L; Franks PW; Giedraitis V; Haley C; Hamsten A; Ingason A; Johansson A; Joshi PK; Lind L; Lindgren CM; Lubitz S; Palmer T; Macdonald-Dunlop E; Magnusson M; Melander O; Michaelsson K; Morris AP; Magi R; Nagle MW; Nilsson PM; Nilsson J; Orho-Melander M; Polasek O; Prins B; Palsson E; Qi T; Sjogren M; Sundstrom J; Surendran P; Vosa U; Werge T; Wernersson R; Westra HJ; Yang J; Zhernakova A; Arnlov J; Fu J; Smith JG; Esko T; Hayward C; Gyllensten U; Landen M; Siegbahn A; Wilson JF; Wallentin L; Butterworth AS; Holmes MV; Ingelsson E; Malarstig A, Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab 2020, 2 (10), 1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Klaric L; Gisby JS; Papadaki A; Muckian MD; Macdonald-Dunlop E; Zhao JH; Tokolyi A; Persyn E; Pairo-Castineira E; Morris AP; Kalnapenkis A; Richmond A; Landini A; Hedman AK; Prins B; Zanetti D; Wheeler E; Kooperberg C; Yao C; Petrie JR; Fu J; Folkersen L; Walker M; Magnusson M; Eriksson N; Mattsson-Carlgren N; Timmers P; Hwang SJ; Enroth S; Gustafsson S; Vosa U; Chen Y; Siegbahn A; Reiner A; Johansson A; Thorand B; Gigante B; Hayward C; Herder C; Gieger C; Langenberg C; Levy D; Zhernakova DV; Smith JG; Campbell H; Sundstrom J; Danesh J; Michaelsson K; Suhre K; Lind L; Wallentin L; Padyukov L; Landen M; Wareham NJ; Goteson A; Hansson O; Eriksson P; Strawbridge RJ; Assimes TL; Esko T; Gyllensten U; Baillie JK; Paul DS; Joshi PK; Butterworth AS; Malarstig A; Pirastu N; Wilson JF; Peters JE, Mendelian randomisation identifies alternative splicing of the FAS death receptor as a mediator of severe COVID-19. medRxiv [Preprint]. 2021. Apr 7:2021.04.01.21254789. [Google Scholar]
- 130.Sun BB; Chiou J; Traylor M; Benner C; Hsu Y-H; Richardson TG; Surendran P; Mahajan A; Robins C; Vasquez-Grinnell SG; Hou L; Kvikstad EM; Burren OS; Cule M; Davitte J; Ferber KL; Gillies CE; Hedman ÅK; Hu S; Lin T; Mikkilineni R; Pendergrass RK; Pickering C; Prins B; Raj A; Robinson J; Sethi A; Ward LD; Welsh S; Willis CM; Alnylam Human Genetics, AstraZeneca Genomics Initiative, Biogen Biobank Team, Bristol Myers Squibb, Genentech Human Genetics, GlaxoSmithKline Genomic Sciences, Pfizer Integrative Biology, Population Analytics of Janssen Data Sciences, Regeneron Genetics Center; Burkitt-Gray L; Black MH; Fauman EB; Howson JMM; Kang HM; McCarthy MI; Melamud E; Nioi P; Petrovski S; Scott RA; Smith EN; Szalma S; Waterworth DM; Mitnaul LJ; Szustakowski J; Gibson BW; Miller MR; Whelan CD, Genetic regulation of the human plasma proteome in 54,306 UK Biobank participants. 2022, bioRxiv 2022.06.17.496443; doi: 10.1101/2022.06.17.496443. [DOI] [Google Scholar]
- 131.Price ND; Magis AT; Earls JC; Glusman G; Levy R; Lausted C; McDonald DT; Kusebauch U; Moss CL; Zhou Y; Qin S; Moritz RL; Brogaard K; Omenn GS; Lovejoy JC; Hood L, A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol 2017, 35 (8), 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oller Moreno S; Cominetti O; Nunez Galindo A; Irincheeva I; Corthesy J; Astrup A; Saris WHM; Hager J; Kussmann M; Dayon L, The differential plasma proteome of obese and overweight individuals undergoing a nutritional weight loss and maintenance intervention. Proteomics. Clinical applications 2018, 12 (1). [DOI] [PubMed] [Google Scholar]
- 133.Ruffieux H; Carayol J; Popescu R; Harper ME; Dent R; Saris WHM; Astrup A; Hager J; Davison AC; Valsesia A, A fully joint Bayesian quantitative trait locus mapping of human protein abundance in plasma. PLoS computational biology 2020, 16 (6), e1007882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Plubell DL; Kall L; Webb-Robertson BJ; Bramer LM; Ives A; Kelleher NL; Smith LM; Montine TJ; Wu CC; MacCoss MJ, Putting Humpty Dumpty back together again: What does protein quantification mean in bottom-up proteomics? J Proteome Res 2022, 21 (4), 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smith LM; Agar JN; Chamot-Rooke J; Danis PO; Ge Y; Loo JA; Pasa-Tolic L; Tsybin YO; Kelleher NL; Consortium for Top-Down Proteomics, The Human Proteoform Project: Defining the human proteome. Sci Adv 2021, 7 (46), eabk0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Omenn GS; Lane L; Overall CM; Corrales FJ; Schwenk JM; Paik YK; Van Eyk JE; Liu S; Pennington S; Snyder MP; Baker MS; Deutsch EW, Progress on identifying and characterizing the human proteome: 2019 metrics from the HUPO Human Proteome Project. J Proteome Res 2019, 18 (12), 4098–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Edwards AM; Isserlin R; Bader GD; Frye SV; Willson TM; Yu FH, Too many roads not taken. Nature 2011, 470 (7333), 163–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2: MassIVE data sets with 10+ new proteins with MS evidence for this latest release
Table S1: The 868 PE1 proteins WITHOUT mass spectrometry evidence from the PeptideAtlas-MassIVE-neXtProt pipeline


