Abstract
Polychlorinated biphenyls (PCBs) accumulate in adipose tissue and are linked to obesity and diabetes. The congener, PCB52 (2,2’,5,5’-tetrachorobiphenyl), is found at high levels in school air. Hydroxylation of PCB52 to 4-OH-PCB52 (4-hydroxy-2,2’,5,5’-tetrachorobiphenyl) may increase its toxicity. To understand PCB52’s role in causing adipose dysfunction, we exposed human preadipocytes to PCB52 or 4-OH-PCB52 across a time course and assessed transcript changes using RNAseq. 4-OH-PCB52 caused considerably more changes in the number of differentially expressed genes as compared to PCB52. Both PCB52 and 4-OH-PCB52 upregulated transcript levels of the sulfotransferase SULT1E1 at early time points, but cytochrome P450 genes were generally not affected. A set of genes known to be transcriptionally regulated by PPARα were consistently downregulated by PCB52 at all time points. In contrast, 4-OH-PCB52 affected a variety of pathways, including those involving cytokine responses, hormone responses, focal adhesion, Hippo, and Wnt signaling. Sets of genes known to be transcriptionally regulated by IL17A or parathyroid hormone (PTH) were found to be consistently downregulated by 4-OH-PCB52. Most of the genes affected by PCB52 and 4-OH-PCB52 were different and, of those that were the same, many were changed in an opposite direction. These studies provide insight into how PCB52 or its metabolites may cause adipose dysfunction to cause disease.
Keywords: PCB52, preadipocytes, adipose, RNAseq, inflammation, diabetes, obesity
INTRODUCTION
Polychlorinated biphenyls (PCBs) were used extensively in building materials such as caulking, electrical light ballast fluid, and paints. While their production was banned several decades ago, high levels of PCBs are persistent in food and air and are inadvertently produced in in the making of silicone for kitchen cabinetry and colorants in paint (Jahnke and Hornbuckle, 2019; Jahnke et al., 2022; Saktrakulkla et al., 2020; Schettgen et al., 2022). PCBs are persistent organic pollutants and there is increasing evidence that exposure is associated with neurotoxicity, cancer, and cardiometabolic disease, including diabetes (Clair et al., 2018; Dirinck et al., 2011; Everett et al., 2011). There are 209 different congeners of PCBs with varying numbers of chlorines attached to a biphenyl structure (Grimm et al., 2015). The congeners can exhibit vastly different biological properties. Individual PCB congeners are present at different levels as contaminants depending mainly on their original source. Mixtures of PCB congeners, referred to as Aroclors, were used for different industrial purposes. Studies from the Iowa Superfund Research Program have demonstrated that the signature of Aroclor 1254 is present at very high levels in school air (Bannavti et al., 2021; Marek et al., 2017; Wang et al., 2020). The highest inhalation exposure in children was experienced by the most active children in schools with the highest air concentrations (Ampleman et al., 2015; Koh et al., 2016). Thus, PCB exposure, particularly in children, is a major concern.
One of the most prominent congeners found in school air is PCB52 (2,2’,5,5’-tetrachorobiphenyl)(Bannavti et al., 2021). The different Aroclors that were commonly used for building materials and for other industrial purposes contain different percentages of PCB52. For example, as assessed by gas chromatography-mass spectrometry, Aroclor 1260 was found to contain 0.24% PCB52 of the total PCB, whereas Aroclor 1254 contained 0.78%, Aroclor 3.2%, and Aroclor 1016 4.1% (Frame, 1997). Interestingly, school air has high levels of PCB52 of approximately 10.5% of the total PCB mass detected (Wang et al., 2022). This over-representation in school air is likely due to higher levels of emission compared to other congeners present in the Aroclors used in the building materials. Despite these high levels, very little is known about the toxicity of PCB52, particularly regarding its effects on disease. PCB52 is known to cause hepatoxicity in rodents and in liver cell lines (Xie et al., 2019). Toxic effects on mitochondria in liver have been noted (Mildaziene et al., 2002a). PCB52 can be converted by the liver into hydroxylated and sulfated forms (Espandiari et al., 2003; Robertson et al., 2000; Shimada et al., 2016). Accumulating evidence indicates that PCB52, among other PCB congeners, can also be hydroxylated through environmental mechanisms, likely microbial activity in sediments, raising the concern that humans are exposed to more soluble and toxic breakdown products of this prevalent PCB congener (Marek et al., 2013; Saktrakulkla et al., 2022).
It has been demonstrated that people who are exposed to PCBs accumulate them to high levels in fat and PCB52 is no exception (Beyer and Biziuk, 2009; Jackson et al., 2017). PCB52 was found to accumulate to exceedingly high levels in adipose tissue in studies in which school air mixtures were administered to rodents and naturally in animals that are exposed to it in the wild (Wang et al., 2020; Wang et al., 2022). PCB52’s high presence in adipose tissue makes it a candidate for altering adipose tissue function and potential development of metabolic disease, including diabetes. One study found that PCB52 exposure was associated with the development of gestational diabetes, specifically (Zhang et al., 2018). Another report from Germany demonstrated a significant association between higher levels of HbA1c and PCB52 (Esser et al., 2016). These studies suggest a link between PCB52 exposure and disruption of metabolism, but more research is needed.
Adipose tissue is critical for regulation of metabolism. The main cells found in adipose tissue, adipocytes, not only accumulate and store lipid, they also secrete important adipokines that can regulate other cells and tissues (Cohen and Spiegelman, 2016; Rosen and Spiegelman, 2014). Adipocytes are also capable of thermogenesis through upregulation of the uncoupling protein, UCP1 (Cohen and Spiegelman, 2015; Gourronc et al., 2020). Adipocytes are generated through differentiation of preadipocytes. Disruption of the process of adipogenesis is known to lead to aberrant adipocyte function, causing insulin resistance, reduction in secretion of “good” adipokines, increase in proinflammatory factors, and fatty acids that can then accumulate in other tissues such as muscle and liver (Kohlgruber and Lynch, 2015). In previous studies, we found that preadipocytes, the precursors to adipocytes, are particularly sensitive to toxicants such as PCB126 (Gadupudi et al., 2015; Gourronc et al., 2022; Gourronc et al., 2018). These cells are derived from the stromal vascular fraction of adipose tissue and largely resemble mesenchymal stem cells (MSCs). We propose that preadipocytes, more so than adipocytes themselves, are major targets for toxicants that are associated with obesity and diabetes.
While there are reports that indicate that PCB52 causes cytotoxicity in the liver (Espandiari et al., 2003; Xie et al., 2019; Zhou et al., 2020), nothing is known about how PCB52 affects adipose tissue, specifically, preadipocytes. In this study, we use RNAseq to assess how PCB52 causes changes in transcript levels over a time course in human preadipocytes that may be associated with the development of cardiometabolic disease. Because PCB52 is metabolized to hydroxylated forms by the liver and in the environment, we also assessed the effects of the main hydroxylated form of PCB, 4-OH-PCB52 (4-hydroxy-2,2’,5,5’-tetrachorobiphenyl), on human preadipocytes. Our results indicate that oxidation of PCB52 to an hydroxylated form significantly changes how it affects gene expression in this cell type with 4-OH-PCB52 causing both more changes and changes that are more consistent over a time course of exposure as compared to PCB52. Many of the genes and pathways altered by PCB52 and 4-OH-PCB52 would be expected to have significant biological consequences for adipose function, providing insight into how exposure to these compounds could lead to metabolic disease.
MATERIALS AND METHODS
Cell culture and treatments
The human preadipocyte cell line, NPAD (Normal PreADipocyte), has been described previously (Gadupudi et al., 2015; Vu et al., 2013). It was derived from subcutaneous adipose of a non-diabetic female donor. The cell line is immortal and can differentiate into mature adipocytes that can be induced to beige.(Gourronc et al., 2020; Vu et al., 2013). Early passage cells were cultured in PGM2 medium (Lonza). PGM2 medium contains 10% FBS.
PCB52 was synthesized by reduction of 2,2’,5,5’-tetrachlorobenzidine with hypophosphorous acid (Espandiari et al., 2003) and authenticated as described previously (Li et al., 2018; Sethi et al., 2019). 4-OH-PCB52 was synthesized by Suzuki-coupling reaction of 2,5-dichloro-4-iodoanisole and 2,5-dichlorobenzene boronic acid followed by deprotection of the methoxy group with boron tribromide (Rodriguez et al., 2016). Both study compounds were provided in the Synthesis core of the Iowa Superfund Research Program.
PCB52 or 4-OH-PCB52 were dissolved in DMSO and used at a concentration of 10 μM in PGM2 medium for treatments. Cells that were approximately 90% confluent were treated with PCB or vehicle (same concentration of DMSO as treatments) with complete medium change. To determine how changes in gene expression occur over a time, treatments were allowed to proceed for 9 hours, 1 day, or 3 days, a range of time points post-exposure that we have previously shown to result in progressive changes in gene expression by PCB126 treatment (Gourronc et al., 2022), before the collection of RNA. Four biological replicates were prepared for each timepoint and treatment.
RNA isolation and processing for RNAseq
RNA was isolated and prepared as previously described (Gourronc et al., 2022). Briefly, cells were homogenized in 1 ml of TRIzol Reagent (Invitrogen). Total RNA from the aqueous phase was further purified using RNeasy Columns (Qiagen). Transcription profiling using RNAseq was performed by the University of Iowa Genomics Division using manufacturer recommended protocols. Briefly, 500 ng of Dnase I-treated total RNA was used to enrich for polyA-containing transcripts using beads coated with oligo(dT) primers. The enriched RNA pool was then fragmented, converted to cDNA and ligated to sequencing adaptors containing indexes using the Illumina TruSeq stranded mRNA sample preparation kit (Cat. #RS-122–2101, Illumina, Inc., San Diego, CA). The molar concentrations of the indexed libraries were measured using the 2100 Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) and combined equally into pools for sequencing. The RNA concentration of the pools was measured using the Illumina Library Quantification Kit (KAPA Biosystems, Wilmington, MA) and sequenced on the Illumina NovaSeq 6000 genome sequencer using 50 bp paired-end SBS chemistry.
RNAseq bioinformatic analysis
Bioinformatic analysis was performed as previously described (Gourronc et al., 2022). Briefly, four biological replicates were prepared per treatment and timepoint. Barcoded samples were pooled and sequenced on an Illumina NovaSeq6000 located in the Iowa Institute of Human Genetics (IIHG) Genomics Division. A minimum of 25 million paired-end 50 bp reads were obtained per sample. Reads were converted to fastq format with bcl2fastq. The bcbio-nextgen pipeline (v 1.2.2; https://github.com/chapmanb/bcbio-nextgen) was run in “RNA-seq” mode to align reads against the hg19 reference genome using the splice-aware hisat aligner. Alignments were stored as indexed BAM files (Kim et al., 2015; Kim et al., 2019). Quantification of reads against the human transcriptome (GENCODE 19) was also performed using the salmon aligner.(Patro et al., 2017) Analysis of BAM files showed that for all samples, ~95% of RNA-seq reads were uniquely mapped to the reference, with ~90% of mapped reads originating in exonic regions. Additional QC was performed with MultiQC (Ewels et al., 2016). All samples passed the QC thresholds. Each treatment and time point had a minimum of 3 replicates.
Salmon transcript expression values were summarized to the gene level using tximport (Soneson et al., 2015). Estimated, non-normalized gene-level counts from this procedure were used for differential gene expression analysis with DESeq2 (Love et al., 2014). The sets of differentially expressed genes (DEGs) between PCB52- or 4-OH-PCB52-treated and DMSO-treated cells at the same time points were converted to Excel files and are provided in the GEO deposit. Code used to obtain the DE results in this analysis is available online (https://github.com/mchimenti/klingelhutz_rnaseq_july2020_pcb126/blob/main/rnaseq_analysis_DEseq2.Rmd). Raw and processed RNAseq data were deposited in NCBI Gene Expression Omnibus (GEO) and are available for review (GSE205813).
Genes were considered differentially expressed (DE) if they exhibited an average log2fold change of an absolute value of 0.3 compared to vehicle alone at the same time point and a threshold of p=0.05 (FDR adjusted p-value of 0.05) for statistical significance. Meta-Analysis in the iPathwayGuide software was used to determine what DE genes overlapped between treatments and timepoints.
Sets of the DEGs were imported into iPathwayGuide commercial pathway enrichment software (https://advaitabio.com/ipathwayguide) to perform proprietary “impact analysis” to generate reports describing enriched pathways. Both the direction and type of all signals on a pathway along with the position, role and type of each gene are considered (Draghici et al., 2007; Khatri and Draghici, 2005; Nguyen et al., 2019). Briefly, these analyses include two types of evidence to determine pathways: 1) the over-representation of DE genes in a given pathway and 2) the perturbation of that pathway computed by propagating the measured expression changes across the pathway topology. These aspects are captured by two independent probability values, pORA (perturbation over-representation) and pAcc (perturbation accumulation), that are then combined in a unique pathway-specific p-value. Only pathways with a p-value of less than or equal to 0.05 were included in the pathway lists. Meta-Analysis in the iPathwayGuide software was used to determine what pathways overlapped between treatments and time points.
For prediction of upstream regulator genes in the iPathwayGuide software, the prediction is based on information of two types which include the enrichment of differentially expressed genes with consideration of direction of change and the network of regulatory interactions from a proprietary knowledge base. For each upstream regulator, the number of consistent DE genes downstream of the regulator is compared to the number of measured target genes expected to be both consistent and DE just by chance. The iPathwayGuide software uses an over-representation approach to compute the statistical significance of observing at least the given number of consistent DE genes. The p-value of the prediction of an activated or inhibited upstream regulator is calculated using the iPathwayGuide software. Upstream regulators with a p-value of 0.05 or less were considered significant. Meta-Analysis in the iPathwayGuide software was used to determine upstream regulators that overlapped between treatments and timepoints.
Cytotoxicity Assay
To assess cytotoxicity of PCB52 and 4-OH-PCB52, NPAD cells were plated using PGM2 in a manner to result in subconfluent cultures 1 day after plating (8,000 cells/well in 96-well format). The following day, the media was replaced with vehicle or varying concentrations of PCB52 or 4-OH-PCB52, all with the same concentration of DMSO in PGM2 media containing 10% FBS. Cells were allowed to grow for the indicated times before assessment of ATP production using Cell Titer-Glo (Promega). Luminescence units were converted to percent of control for graphing purposes.
Quantitative RT-PCR
RNA was extracted by using TRIzol followed by Qiagen RNA columns and real-time quantitative-PCR (RT-qPCR) was performed as described previously using at least three replicates (Gourronc et al., 2018). The following primers were used for RT-qPCR:
GAPDH Forward 5’- AAGGTCATCCATGACAACTTTG, Reverse 5’- GTAGAGGCAGGGATGATGTTCT
STMN2 Forward 5’-GGTGGCTTATTTGTGGATGCC, Reverse 5’-AGCTTGAGTGAAAGTCCGCA
Statistical Analyses
Statistics on results of cytotoxicity assays were performed using GraphPrism. One-way ANOVA using the Dunnett’s multi-comparisons test comparing results from at least 3 biological replicates was performed to compare percent viability at the different concentrations with vehicle control at the same time point. Similar statistical analyses were performed for the RT-qPCR assays. Statistical analyses of the RNAseq data that were used to determine DE genes, affected biological pathways, and upstream regulators were performed as described above.
RESULTS
To determine how PCB52 or 4-OH-PCB52 affected gene expression in human preadipocytes over a time course, we first wanted to determine a sublethal concentration that didn’t elicit significant cytotoxic effects at the time points being assessed. We reasoned that significant cytotoxicity would cause changes in gene expression related to cell death pathways and obscure transcriptional changes caused by these compounds that might be important in modulating biological function. Standard culture conditions (10% FBS, PGM2 media) and a range of concentrations were used over a time course of 9 hours (9h), 1 day (1d), 2 days (2d), and 3 days (3d). Viability was assessed by measuring ATP production (Cell Titer Glo, Promega). PCB52 did not elicit a cytotoxic response at any of the concentrations used, (1, 5, 10, or 20 μM), or any of the time points, whereas 4-OH-PCB52 exhibited cytotoxicity that was statistically significant only at the 20 μM concentration at the 1 day, 2 day and 3 day treatment time points (Supplemental figure 1). Since the 10 μM concentration did not elicit significant cytotoxicity (i.e., was a sublethal concentration), we used this concentration for our RNAseq studies. Since our previous studies using another PCB congener, PCB126, demonstrated a progressive change in gene expression over a time-course of what can be considered early (9h), middle (1d), and late (3d) post-exposure (Gourronc et al., 2022), we decided to also use these three time points for PCB52 or 4-OH-PCB52 exposure.
Preadipocytes were exposed to vehicle, PCB52 or 4-OH-PCB52 for the indicated time points, RNA was collected, and RNAseq analysis was performed (Figure 1). Transcripts that exhibited statistically significant (adjusted p-value≤0.05) differences in expression of PCB-treated cells compared to vehicle control-treated cells at the same time point were determined. Excel files containing all the genes that met the p≤0.05 criteria have been deposited with GEO, along with raw data files (GSE205813). The software program iPathwayGuide (Advaita) was used to further process and analyze the data. Using an average absolute value of log2 fold change of at least 0.3 and an adjusted p≤0.05, we found that exposure to PCB52 caused changes in a relatively small number of genes at all time points with only 70 gene transcript changes at the 9h timepoint, 59 at the 1d timepoint, and 133 at the 3d timepoint (Table 1). As compared to PCB52, the hydroxylated form of PCB52, 4-OH-PCB52 caused many more genes to be differentially expressed compared to control with 445 gene transcripts at the 9h time point, 418 at the 1d time point, and 335 at the 3d time point (Table 1). In addition to more genes being changed, the magnitude of change was greater in 4-OH-PCB52-treated cells compared to PCB52-treated cells (Figure 2). After sorting by log2 fold change, the top 50 up- or down-regulated genes (or less if that number was not reached) are presented in Supplemental Tables 1–6.
Figure 1:
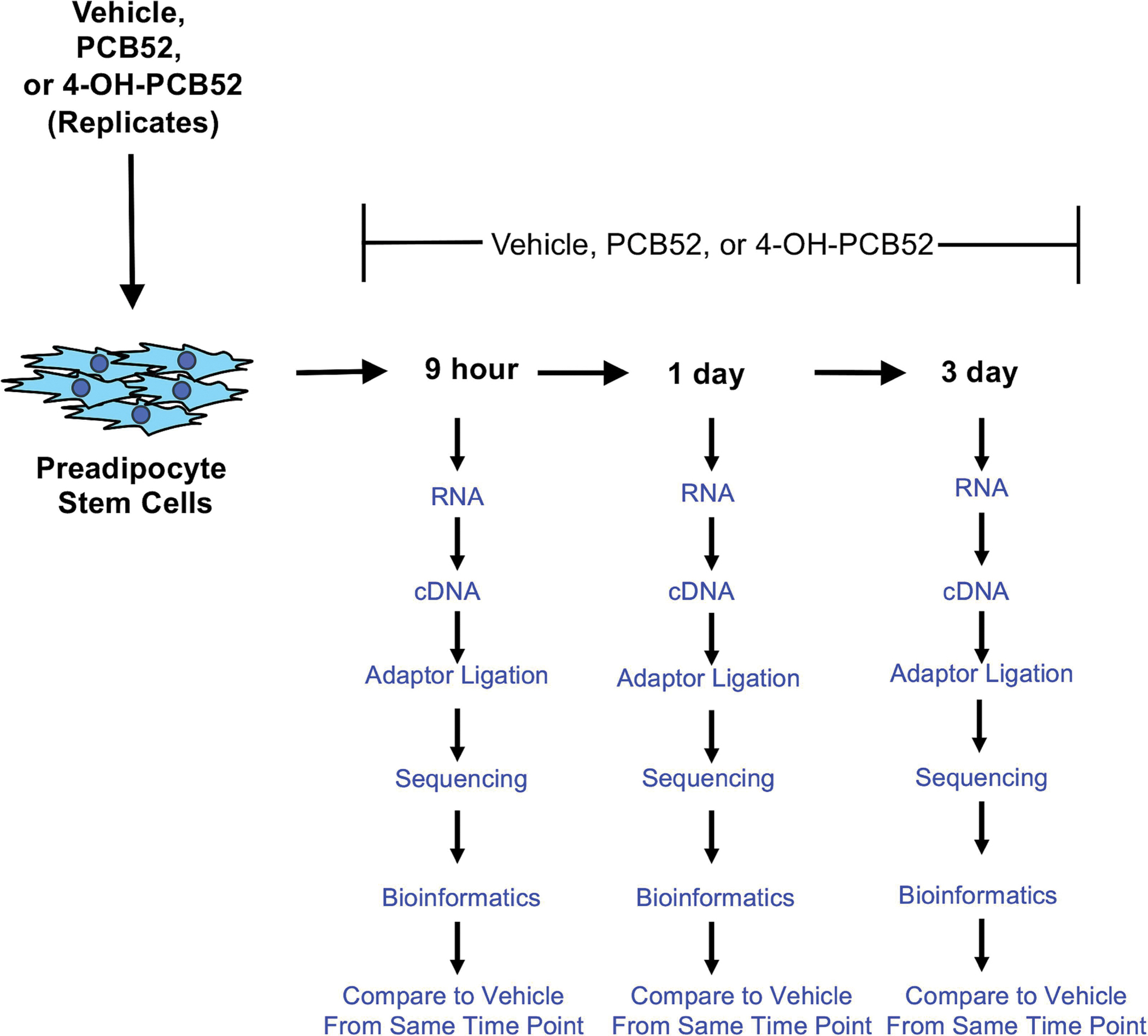
Scheme of PCB52 and 4-OH-PCB52 treatments of preadipocytes over a time course.
Table 1:
Number of genes altered by PCB52 or 4-OH-PCB52 at different time points
| 9 hrs Number of genes changed |
1 d Number of genes changed |
3d Number of genes changed |
Common genes at 2 of 3 time pointsa | Common genes at all time pointsa | |
|---|---|---|---|---|---|
| PCB52 | 70 | 59 | 133 | 32 | 8 |
| 4-OH-PCB52 | 445 | 418 | 335 | 228 | 90 |
| Common genes between PCB52 and 4-OH-PCB52a | 21 | 12 | 30 | 3 | 1 |
Figure 2:
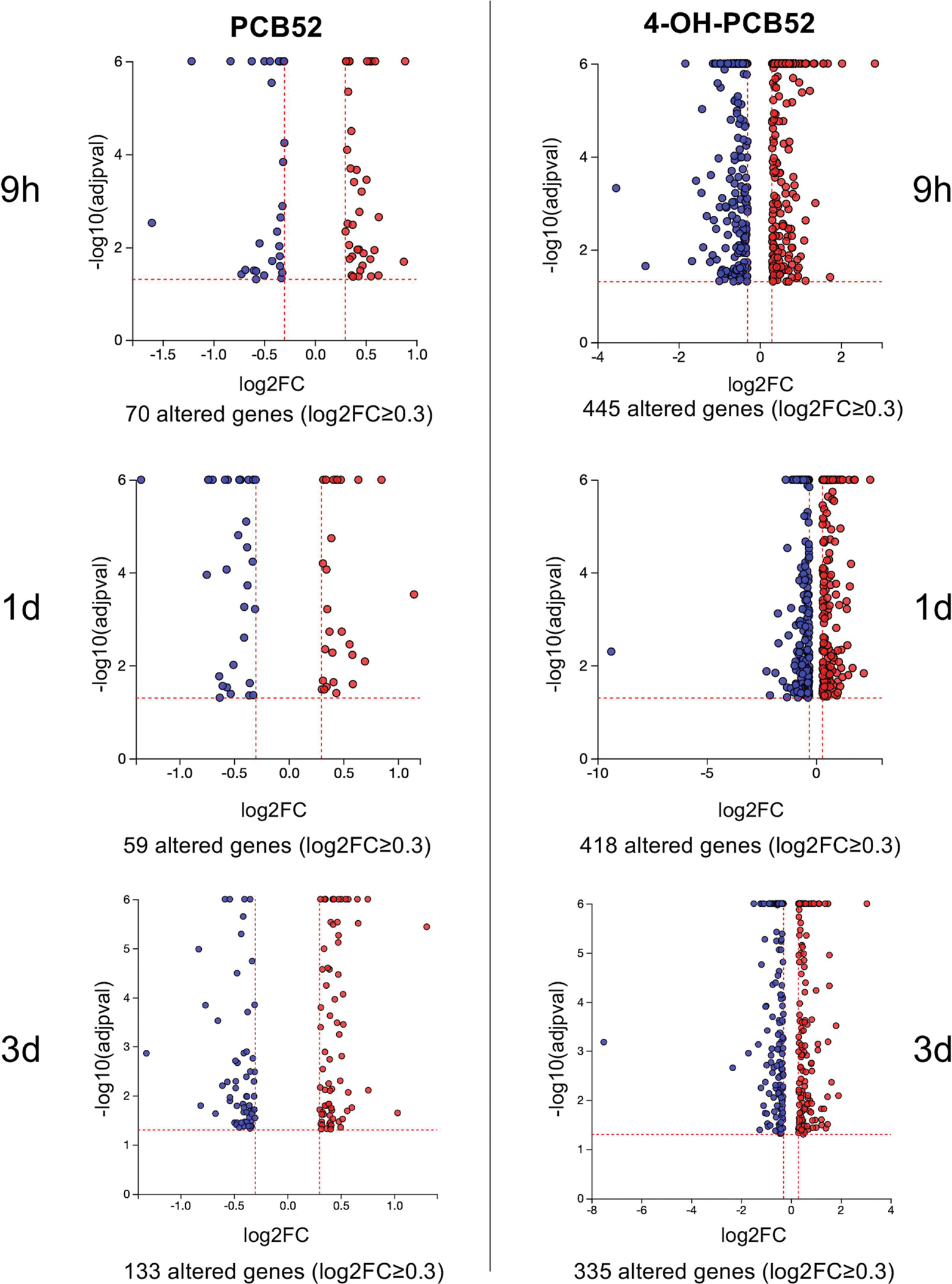
Example volcano plots showing significantly altered genes compared to control at all time points. p≤0.05, log2FC≤0.3
PCBs have been shown to induce the expression of Phase I (e.g., cytochrome P450s) and Phase II (e.g., sulfotransferases) enzymes, usually in the liver, that catalyze the metabolism of PCBs leading to the formation of metabolites that could be more biologically active (Routti et al., 2008). We did not observe upregulation of any of cytochrome P450 (CYP) enzyme genes by PCB52 or 4-OH-PCB52 following exposure of preadipocytes. For other genes that code for enzymes involved in the metabolism of organic compounds, the RNAseq data demonstrated upregulation of the sulfotransferase, SULT1E1, at the early 9-hour and 1-day time points for both PCB52 and 4-OH-PCB52 (Supplemental Tables 1–4, Figure 3A and B). 4-OH-PCB52, but not PCB52, upregulated another sulfotransferase, SULT1A1, at all three time points (Supplemental Tables 2, 4, 6, Figure 3B). Hydroxylated PCB52 also upregulated the arylacetamide deacetylase, AADAC, an enzyme that catalyzes one of the initial biotransformation pathways for arylamine and heterocyclic amine carcinogens (Figure 3B) (Zhang et al., 2012). Interestingly, the parent PCB52 inhibited transcript levels of this gene (Figure 3A). PCB52 also downregulated CYP26B1, a P450 gene that encodes an enzyme involved in the specific inactivation of all-trans-retinoic acid to hydroxylated forms, such as 4-oxo-, 4-OH-, and 18-OH-all-trans-retinoic acid (Figure 3A) (Isoherranen and Zhong, 2019).
Figure 3:
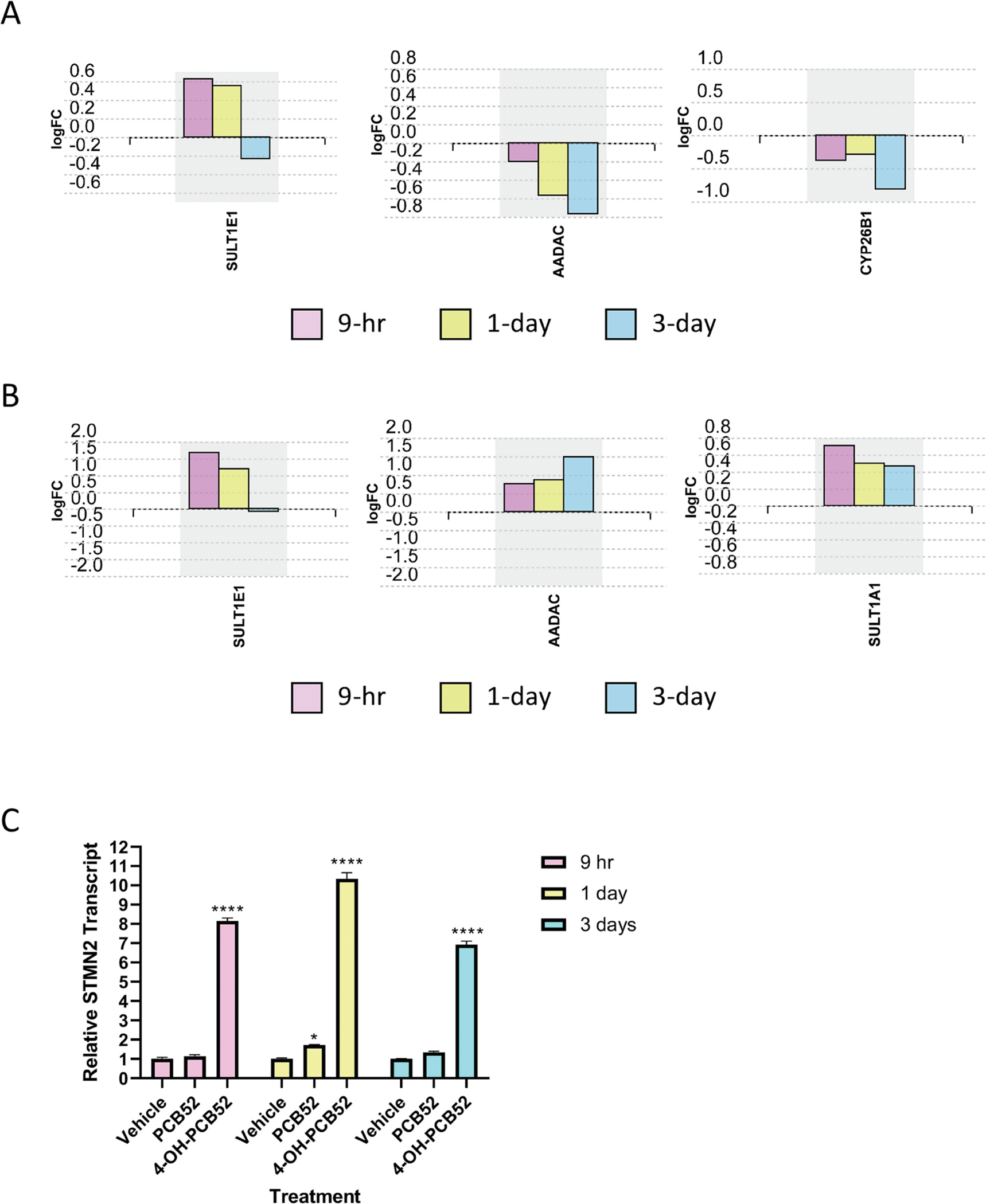
Expression levels of individual genes across time points of PCB52 or 4-OH-PCB52 treatments A. PCB52, B. 4-OH-PCB52, C. Stathmin-2(STMN2) transcript levels measured by RT-qPCR. *p≤−.05, ****p≤0.001
In addition to 4-OH-PCB52 altering expression of more genes than PCB52, changes associated with exposure were also more consistent across time points for 4-OH-PCB52 as compared to PCB52. Using the same stringency as above (log2 fold change cutoff of at least 0.3 and p≤0.05), there were only 8 genes that were shared across all time points for PCB52 treatment, all in the same direction of change (Figure 4A, Supplemental Table 7). In contrast, 91 genes were shared across all time points for 4-OH-PCB52 treatment, with all in the same direction of change (Figure 4B, Supplemental Table 8). One gene that stands out is STMN2 (stathmin 2). This gene had the highest fold-change and the lowest p-value across all three time-points (Supplemental Tables 2A, 4A, 6A). For direct comparison across PCB52 and 4-OH-PCB52 treatments and time points, RT-qPCR was performed. This verified a 7- to 10-fold upregulation of STMN2 as compared to vehicle at all time points but little, if any, upregulation caused by PCB52 (Figure 3C). This gene codes for a phosphoprotein that has been shown to be involved in microtubule dynamics (Chiellini et al., 2008).
Figure 4:
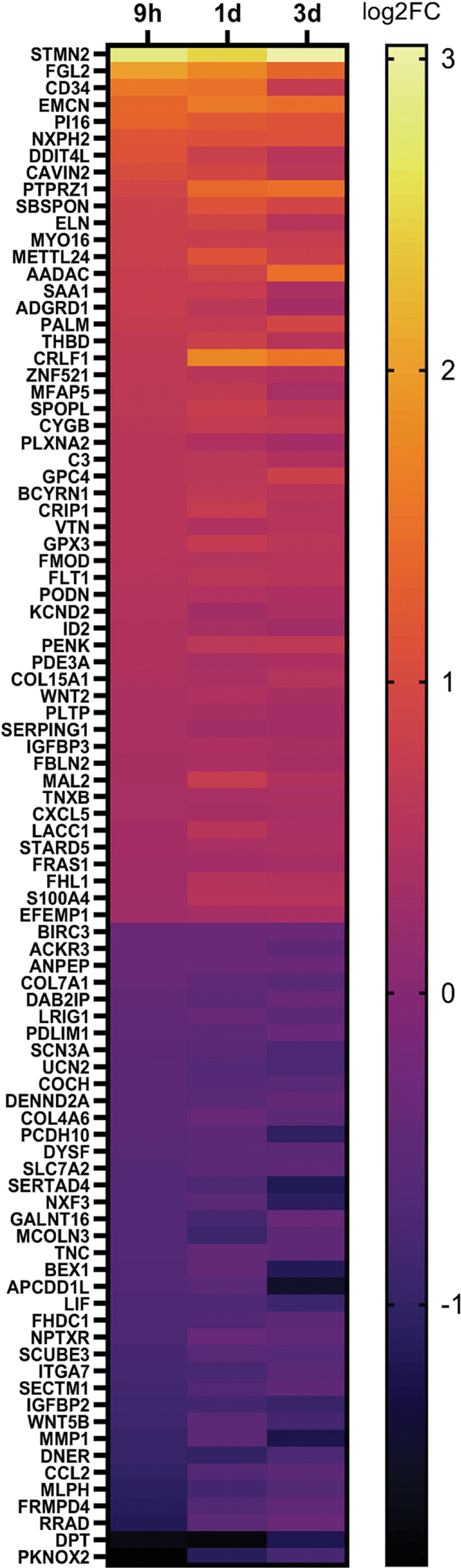
Common genes altered by PCB52 or 4-OH-PCB52 compared to vehicle alone across all time points A. PCB52 common gene changes at all time points. p≤0.05, log2FC≤0.3. B. 4-OH-PCB52 Common Changes at all Time Points. p≤0.05, log2FC≤0.3
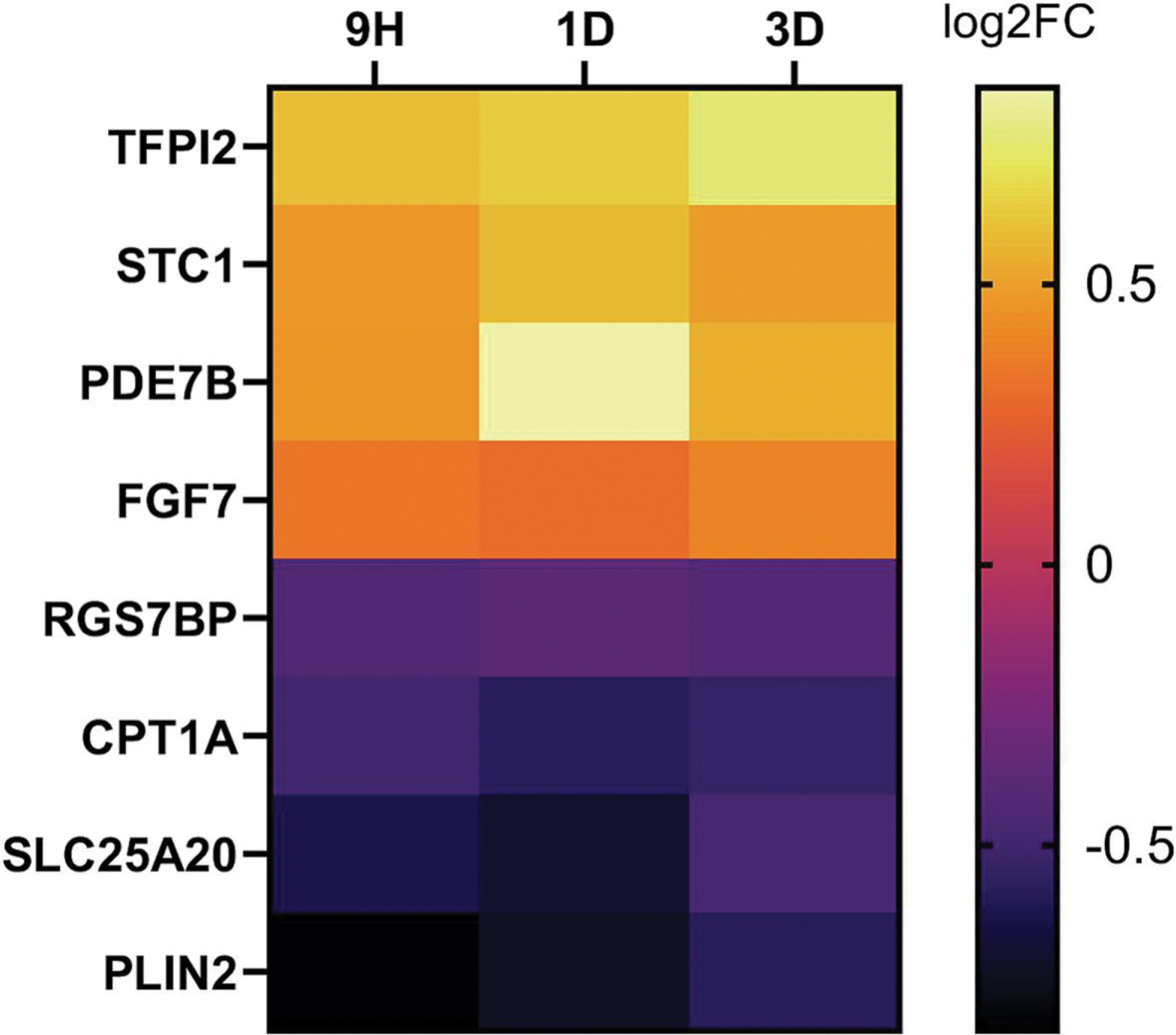
Very few gene changes were shared between PCB52 and 4-OH-PCB52 (Table 1). While there was some overlap in genes that were affected by both PCB52 and 4-OH-PCB52 at single time points, the direction of change in the gene transcripts was more often opposite in the two groups than in the same direction. An example heatmap for genes that were shared at the day 3 time point is shown (Figure 5A). Looking at all the time points, very few genes (only 4) were commonly changed when comparing PCB52 and 4-OH-PCB52 treatments and only one, FGF7 was changed in the same direction (Figure 5B).
Figure 5:
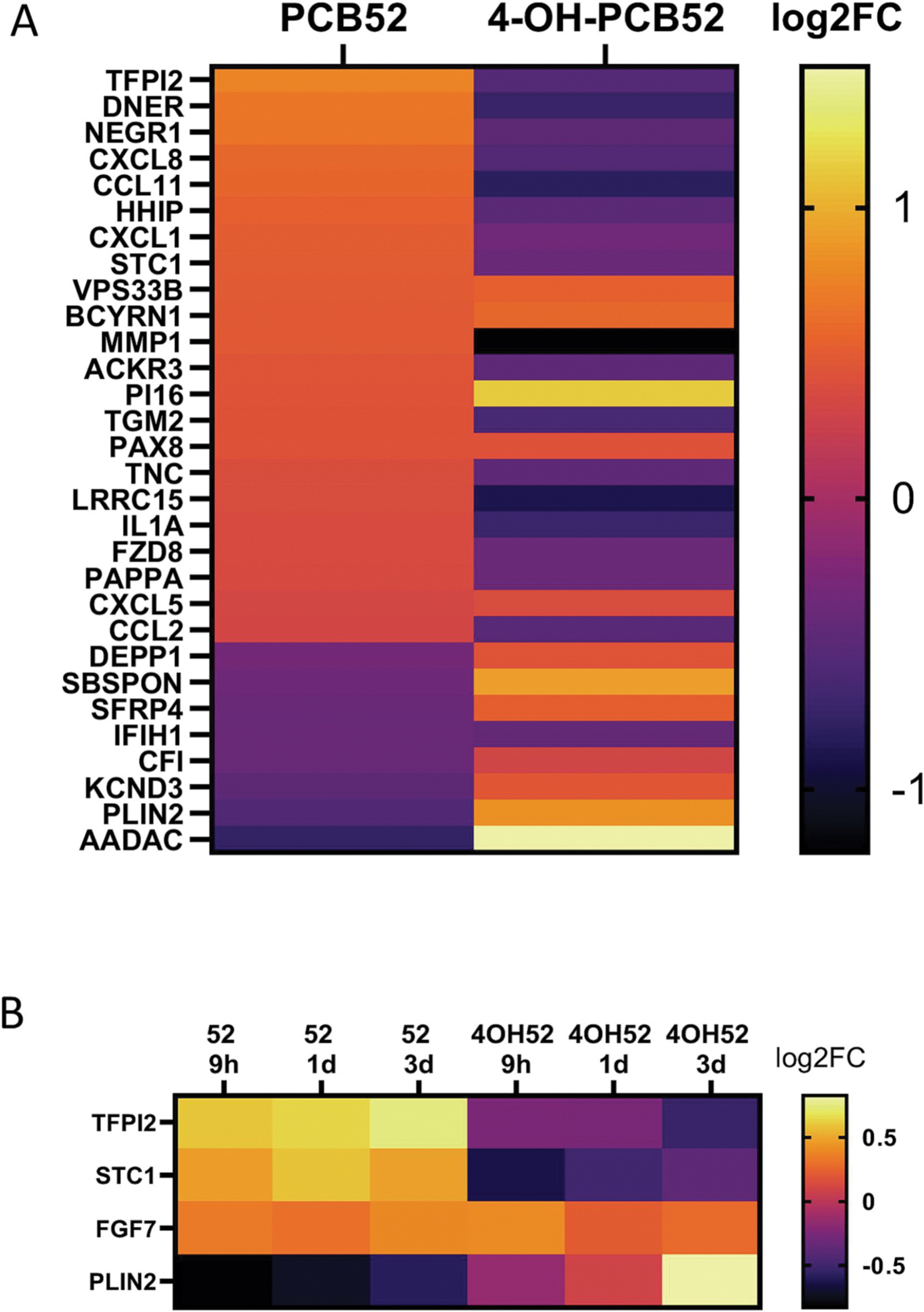
Overlap between PCB52 and 4-OH-PCB52 treatments. A. Genes considered to be altered in common between PCB52 and 4-OH-PCB52 treatments at day 3 post-treatment. Those genes altered by PCB52 were sorted according to log2fold change, largest to smallest. B. Genes altered in common between PCB52 and 4-OH-PCB52 across at least 2 of 3 time points.
Pathway analysis was used to place the gene transcript alterations into a biological context. At the early time point of 9 hours, three pathways were found to be significantly altered by PCB52 (Supplemental Table 9). These were the PPAR signaling pathway, the insulin resistance pathway, and the cytokine-cytokine receptor interaction pathway. For 4-OH-PCB52, many more pathways were altered including cytokine-cytokine receptor interaction, complement and coagulation cascade, TNF signaling, Hippo, NF-kappa B and IL-17 signaling pathways, among others (Supplemental Table 10). Similar but not identical pathways were altered by PCB52 or 4-OH-PCB52 at the 1d-time point (Supplementary Tables 11 and 12).
With the idea that a longer exposure time would provide a better representation of stable changes induced by the compounds, we focused on the longest exposure time point of 3 days. Twenty-two biological pathways that were altered by PCB52 were mainly associated with inflammation and included chemokine signaling, cytokine-cytokine receptor interaction, IL-17A signaling, PPAR signaling, and AGE-RAGE signaling (Supplemental Table 13, Figure 6A). For 4-OH-PCB52, 51 pathways were predicted to be affected on day 3, with some similar inflammatory pathways shared between PCB52 and 4-OH-PCB52 (Supplemental Table 14, Figure 6B). These include changes in pathways associated with rheumatoid arthritis, cytokine-cytokine receptor interactions, AGE-RAGE signaling, NF-kappaB signaling, IL17A signaling and PI3K-Akt signaling. Other pathways predicted to be significantly changed by 3-day exposure to 4-OH-PCB52 included ECM-receptor interaction, focal adhesion, and Wnt signaling. Interestingly, when comparing genes in pathways that were similar between PCB52 and 4-OH-PCB52 treatments, many of the changes were in opposing directions. For example, for the IL17A pathway, which was considered altered for both PCB52 and 4-OH-PCB52, most of the genes in the pathway were upregulated with PCB52 treatment but were downregulated for 4-OH-PCB52 treatment (Figure 7).
Figure 6:
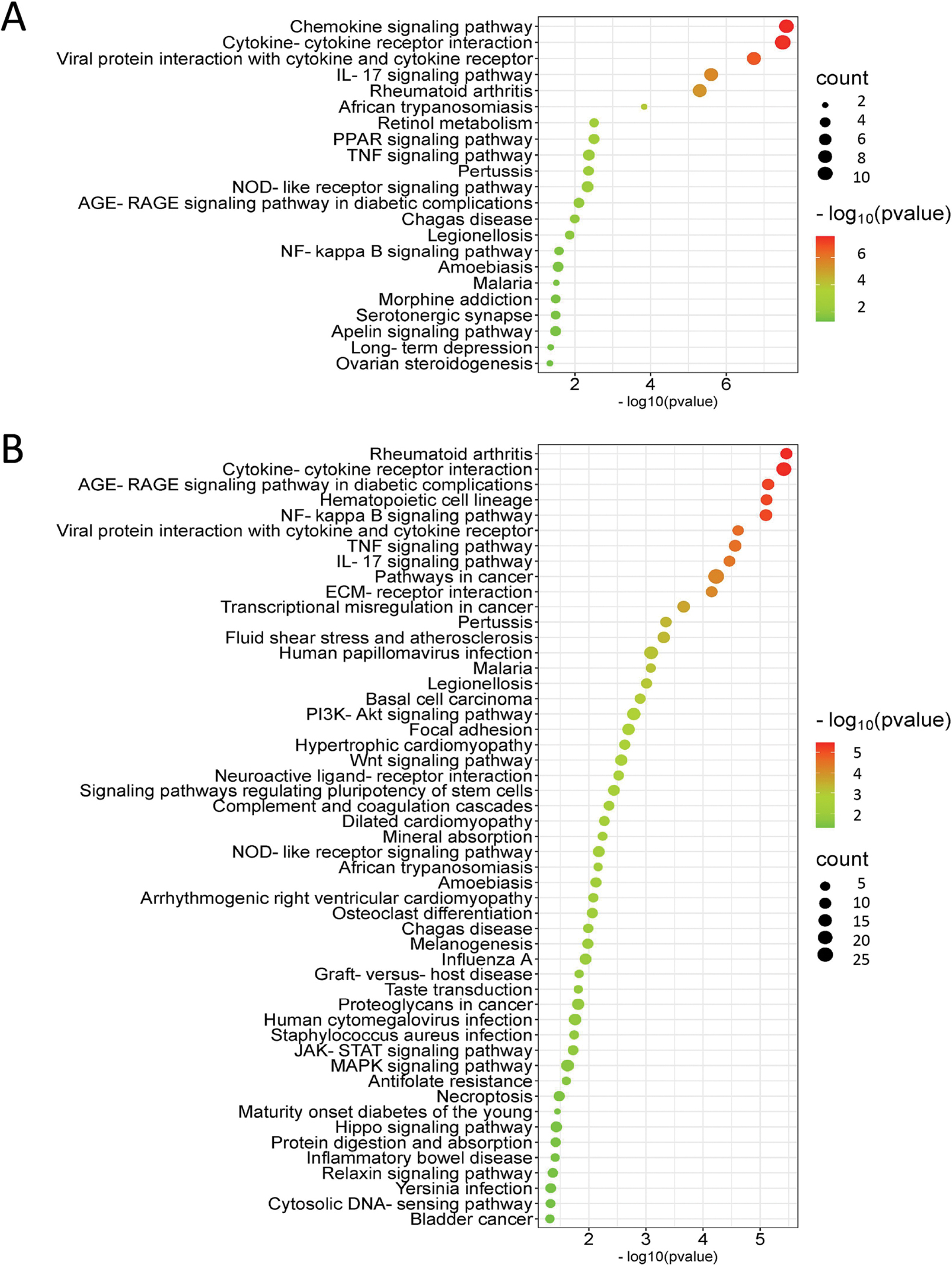
Pathways considered to be altered by PCB52 (A) or 4-OH-PCB52 (B) at the day 3 time point. The affected pathways were determined by iPathwayGuide analysis and a bubble plot was generated using SRPLOT (bioinformatics.com.cn/en). The size of the bubble represents how many genes in the pathway were affected and the color indicates significance (as calculated by iPathwayGuide).
Figure 7:
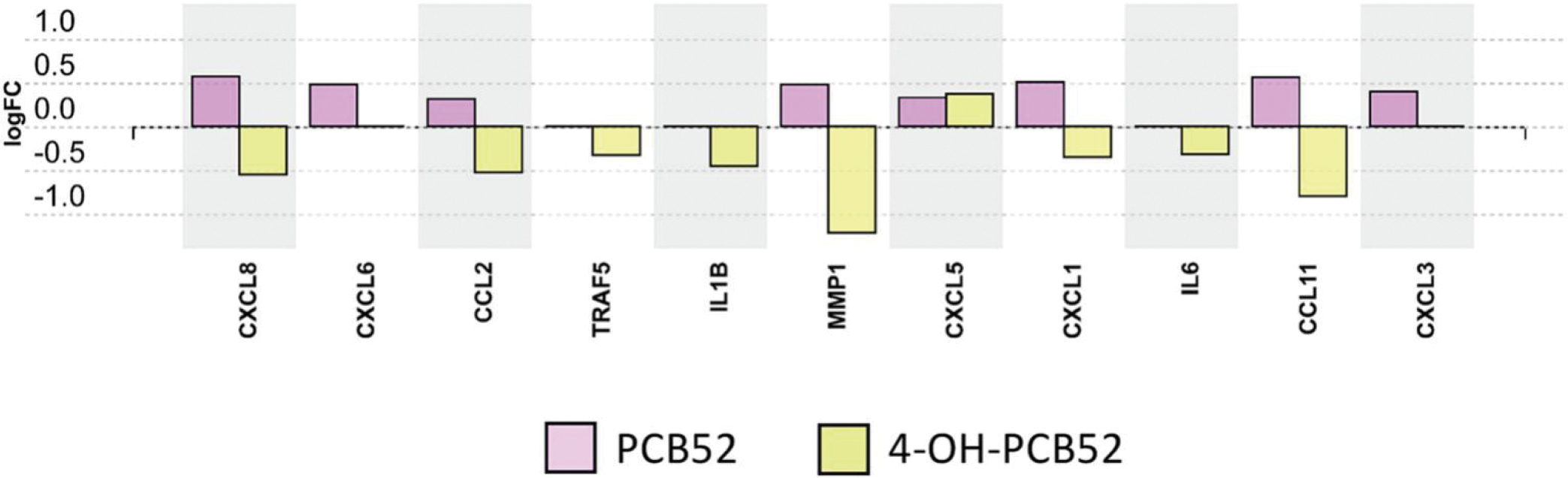
Differentially expressed genes in the IL17A pathway at day 3 for PCB52 and 4-OH-PCB52 treated preadipocytes. Values represent log2fold change over vehicle control.
Looking at pathways that were consistently altered across time points, the PPAR signaling pathway was the only pathway considered to be commonly affected at all 3 time points for PCB52 treatment (Table 2). In contrast, pathways in common across all time points for 4-OH-PCB52 treatment included complement and coagulation, cytokine-cytokine receptor interaction, ECM-receptor, Hippo, stem cell pluripotency and Wnt signaling pathways (Table 2).
Table 2:
Biological pathways altered by PCB52 or 4-OH-PCB52 across all time points
| PCB52 | |||
| Biological Pathway | pv 9h | pv 1d | pv 3d |
| PPAR signaling pathway | 0.000277 | 0.029122 | 0.003131 |
| 4-OH-PCB52 | |||
| Biological Pathway | pv 9h | pv 1d | pv 3d |
| Complement and coagulation cascades | 1.41E-05 | 0.001733 | 0.004425 |
| Cytokine-cytokine receptor interaction | 1.05E-06 | 0.004758 | 3.86E-06 |
| ECM-receptor interaction | 0.000477 | 0.000703 | 7.08E-05 |
| Focal adhesion | 0.017636 | 0.019932 | 0.002017 |
| Hippo signaling pathway | 0.000854 | 0.027494 | 0.037106 |
| Human papillomavirus infection | 0.021398 | 0.029324 | 0.000811 |
| Neuroactive ligand-receptor interaction | 0.000013 | 0.000763 | 0.003003 |
| Protein digestion and absorption | 0.001687 | 0.023737 | 0.038139 |
| Rheumatoid arthritis | 0.001185 | 0.003154 | 3.47E-06 |
| Signaling pathways regulating pluripotency of stem cells | 0.000609 | 0.013898 | 0.003633 |
| Transcriptional misregulation in cancer | 0.000827 | 0.012527 | 0.000219 |
| Wnt signaling pathway | 0.021619 | 0.015331 | 0.002704 |
A useful tool in iPathwayGuide is the upstream gene analysis that allows for a prediction of upstream regulator genes based on the downstream genes that are altered and the direction they are altered. The function gives a prediction as to whether the specific upstream regulator is inhibited or activated. A pathway diagram is provided that shows the measured and predicted relationships between the genes and the predicted upstream regulator. We used this analysis to predict upstream regulators affected by PCB52 or 4-OH-PCB52. We used the criteria that at least 2 of the 3 time points had to have the same predicted and significant (FDR-adjusted p-value<0.05) upstream regulator. For PCB52, PPARα although itself not downregulated at the transcript level, was predicted to be inhibited at all three time points (Supplemental table 15, Figure 8A). This prediction is based on the lower transcript levels of genes that PPARα is known to upregulate and the higher transcript levels of genes that PPARα is known to suppress (Figure 8B). The genes in this group include CPT1A, PLIN2, SLC25A20, and ANGPTL4, all of which have been implicated in the development of obesity and diabetes, mainly through their effects on fatty acid metabolism (Calderon-Dominguez et al., 2016; DiDonna et al., 2022; MacPherson and Peters, 2015). Two other upstream regulators, CREBBP and EP300, were predicted by this analysis to be inhibited by PCB52 at two of the three time points (Supplemental table 15, Supplemental Figure 2). For 4-OH-PCB52 treatment, IL17A, a cytokine, and PTH, a gene that encodes for parathyroid hormone, were predicted by upstream regulator analysis to be inhibited at the later time points (Supplemental Table 15, Figure 9A and 9B). IL17A has also been recently implicated in adipogenesis and the development of obesity and diabetes (Teijeiro et al., 2021). PTH has been shown to be intricately involved in adipocyte browning (Breining et al., 2021; Hedesan et al., 2019; Le et al., 2021).
Figure 8:
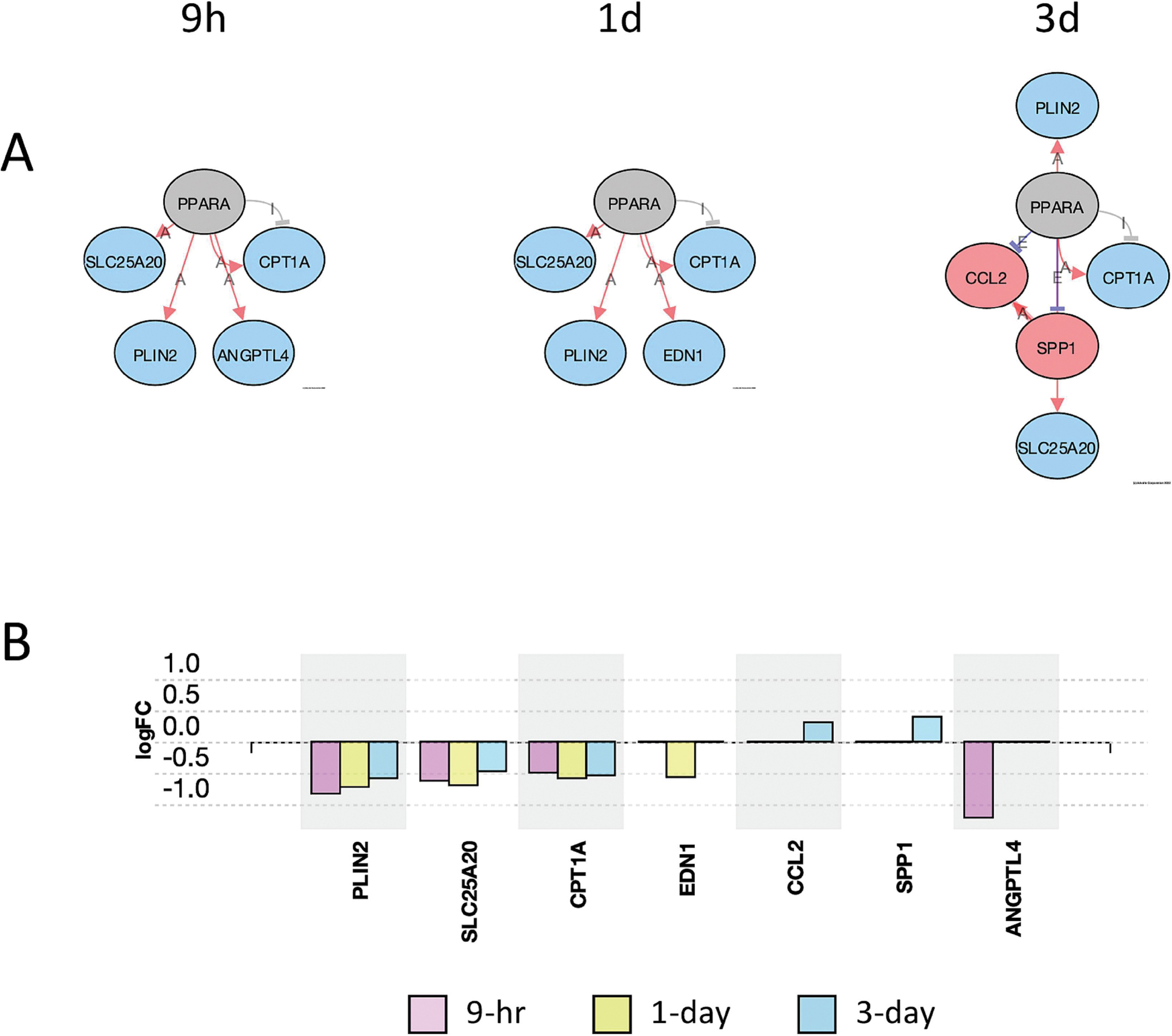
Inhibition of PPARα pathway genes by PCB52 at different time points. A. Upstream regulator analysis showing actual and predicted relationships between genes in the PPARα pathway. Red Oval: Upregulated (actual); Blue Oval: Downregulated (actual); Gray or White oval: Unchanged or undetected (actual); “A”: Activated (predicted); “E”: Expression Inhibited (predicted); “I”: Inhibited (predicted); →Activation (predicted); ┤Inhibition (predicted). All time points were considered significant (p≤0.05, FDR≤0.05). B. Differentially expressed genes in the PPARα pathway at all time points for PCB52-treated preadipocytes. Values represent log2fold change over vehicle control.
Figure 9:
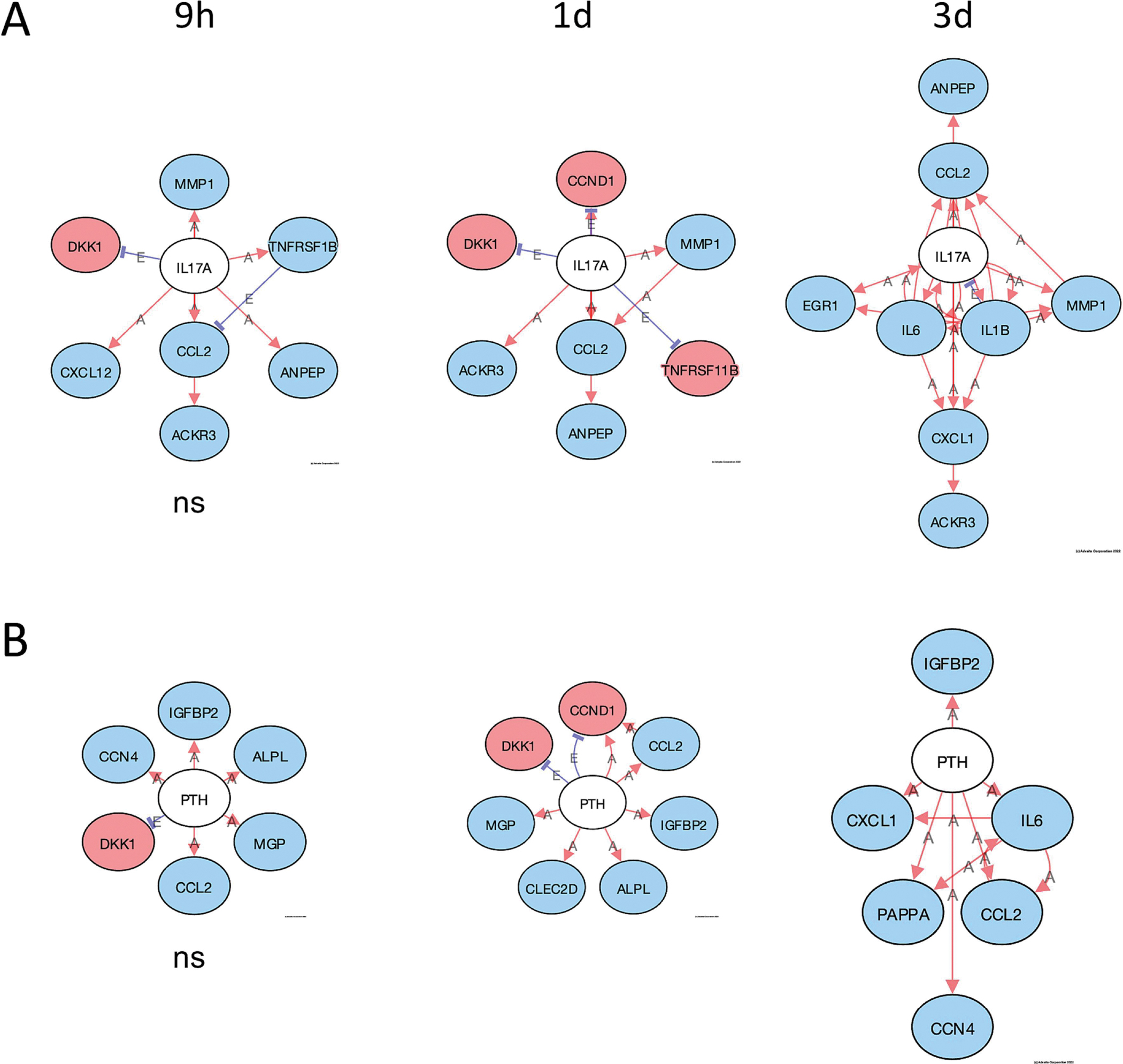
Predicted upstream regulators affected by 4-OH-PCB52. A. Inhibition of IL17A by 4-OH-PCB52 at different time points, B. Inhibition of PTH at different time points by 4-OH-PCB52. Red Oval: Upregulated (actual); Blue Oval: Downregulated (actual); Gray or White oval: Unchanged or undetected (actual); “A”: Activated (predicted); “E”: Expression Inhibited (predicted); “I”: Inhibited (predicted); →Activation (predicted); ┤Inhibition (predicted). ). Except where indicated, all pathways are significant (p≤0.05, FDR≤0.05). ns=nonsignificant
DISCUSSION
The effects of PCB52, one of the most prevalent PCB congeners found in school air, on the biology of human cells have not been extensively studied. To begin to understand how PCB52 and this hydroxylated metabolite might affect human health, particularly the development of metabolic syndrome through effects on adipose tissue, we performed RNAseq studies on PCB52- and 4-OH-PCB52-treated human preadipocytes. We found that while both PCB52 and 4-OH-PCB52 affect genes involved adipocyte biology, they affect gene transcription differently. Unexpectedly, there was little overlap in the pathways and direction of transcript level changes. Oxidation of PCB52 to the hydroxylated metabolite not only caused many more changes in human preadipocytes than the parent compound, but the direction of the changes was often different, demonstrating the very different biological activity of these two compounds. These findings demonstrate that hydroxylation of PCB52 significantly alters its biological properties, which has important implications for how PCB52 may cause disease.
Most metabolism of PCBs, including PCB52, would be expected to mainly take place in the liver (Grimm et al., 2015). It is unknown if cells in adipose tissue, such as preadipocytes, are capable of metabolizing PCB52 to 4-OH-PCB52 or any of its other metabolites. The dramatic differences caused by 4-OH-PCB52 as compared to PCB52 would suggest that preadipocytes do not readily convert PCB52 to 4-OH-PCB52 in cell culture. If there was conversion, 4-OH-PCB52 would be generated in the PCB52-treated cultures, and it might be expected that there would be more overlap in the genes and pathways and similar changes in direction when comparing groups. Certain immortalized human cells such as the liver cell line HepG2 are capable of metabolizing certain PCB congeners, particularly those that are lower-chlorinated, such as PCB11, but not others (Rodriguez et al., 2018; Zhang et al., 2020). P450 enzyme genes necessary for metabolism of these congeners are often shut down in cultured cell lines (Takemura et al., 2021). In our studies using immortal preadipocytes, we did not observe PCB52-associated upregulation of any P450 enzyme genes such as CYP2A6 that have been proposed to be needed for the formation of 4-OH-PCB52 (Richardson and Schlenk, 2011; Shimada et al., 2016). PCB52 can also be metabolized to 4-PCB52 Sulfate by sulfotransferases (SULTs). This sulfation depends on initial oxidation of PCB52 to 4-OH-PCB52. Interestingly, we did detect upregulation of the sulfotransferase gene SULT1E1 by both PCB52 and 4-OH-PCB52 and SULT1A1 by 4-OH-PCB52, suggesting that these enzymes might be active in preadipocytes. It is therefore possible that 4-PCB52 Sulfate can be produced in preadipocytes, particularly in the 4-OH-PCB52-treated cultures. Generation of sulfated metabolites could alter what genes are affected, as it has been demonstrated that hydroxylated and sulfated metabolites cause very different cytotoxic responses in cell culture (Rodriguez et al., 2018). Further studies will be needed for addressing whether and how PCB52 is metabolized in cells, such as preadipocytes and adipocytes, that are found in adipose tissue.
It is interesting that PCB52 metabolites can be found in sediments where high-level PCB contamination has occurred (Marek et al., 2013; Saktrakulkla et al., 2022), indicating biotransformation occurs in the environment. From both liver-specific and environmental conversion of PCB52 to its metabolites, it is possible that humans are exposed to significant levels of the more biologically active PCB52 and other congener metabolites, and this warrants further investigation.
The most consistent pathways found to be affected by PCB52 was the PPARα pathway. PPARα is a ligand-activated transcription factor of a nuclear hormone receptor superfamily comprising PPARα, PPARγ, and PPARβ/δ (Issemann and Green, 1990). Activation of PPARα reduces triglyceride levels, is involved in the regulation of fatty acid beta-oxidation, and is a major regulator of energy homeostasis (Tsuchida et al., 2005). Indeed, genes in the carnitine pathway were altered, mainly inhibited, by PCB52 at all time points that were assessed. PPARα inhibition would be expected to affect mitochondrial function which would likely lead to changes in the ability of adipocytes to regulate energy metabolism (Li et al., 2005). PCB52 has been demonstrated to dysregulate mitochondrial function in rat liver (Mildaziene et al., 2002a; Mildaziene et al., 2002b). While there have been no studies directly linking PCBs in regulating PPARα levels, it has been demonstrated previously that PPARα is inhibited in liver HepG2 cells by treatment with the PCB Aroclor 1260 mixture (Wahlang et al., 2014). However, it is unclear if the PCB52 present in this complex Aroclor mixture was responsible for the inhibition of PPARα. Our studies indicate that PPARα was itself not downregulated by PCB52 at the transcript level. This would suggest that PCB52 is somehow inhibiting the PPARα protein. PPARs are similar to steroid or thyroid hormone receptor and are stimulated in response to small lipophilic ligands. PPARs bind to co-activator complex to stimulate gene transcription. PCB52 could be inhibiting PPARα directly or blocking the ability of another factor to stimulate PPARα. PCBs have been previously implicated in binding to and inhibiting or activating steroid hormones (Wahlang et al., 2016). The mechanism by which PCB52 inhibits PPARα is unclear and will be the subject of future studies.
The various pathways that were consistently affected by 4-OH-PCB52 were quite diverse and included complement and coagulation, cytokine-cytokine receptor interaction, ECM-receptor, Hippo, stem cell pluripotency and Wnt signaling pathways. Individual time points such as the day 3 exposure also included AGE-RAGE signaling in diabetic complications as well as the NFκB pathway. These changes all point to potential mechanisms by which 4-OH-PCB52 could alter proinflammatory responses and differentiation in preadipocytes and adipocytes. Upstream regulators predicted to be inhibited by 4-OH-PCB52 included IL17A and PTH. Alterations in all these genes in preadipocytes could lead to potential problems in adipocyte function and differentiation and play a role in the development of cardiometabolic disease. For example, the IL17A axis has recently been shown to act in adipocytes to suppress diet-induced obesity and metabolic disorders in mice (Teijeiro et al., 2021). PTH has been shown to be important in adipocyte lipolysis and browning (Breining et al., 2021; Hedesan et al., 2019). Transcript levels of IL17A or PTH were not changed by 4-OH-PCB52 exposure, indicating that the compound is affecting these pathways through a mechanism that does not involve transcription regulation of IL17A or PTH.
One gene of interest that was found to be consistently upregulated by 4-OH-PCB52 at all time points (highest fold-change and lowest p-value of any genes) was STMN2 (stathmin2). This gene codes for a phosphoprotein that has been implicated in the microtubule regulatory network as an important part of cytoskeletal regulation (Chiellini et al., 2008). Intriguingly, upregulation of this gene was found to be associated with age-related changes in preadipocytes specifically (Cartwright et al., 2010), potentially implicating 4-OH-PCB52 in causing age-associated changes in preadipocytes. Further studies will be needed to determine the relevance of STMN2 upregulation in these cells.
As noted above, both PCB52 and 4-OH-PCB52 upregulated transcript levels of the sulfotransferase SULT1E1. SULT1E1 has been previously implicated in sulfating estrogens to inactivate them (Barbosa et al., 2019; Yi et al., 2021). Intriguingly, it has been shown in previous studies that PCB52 inhibits activity of the SULT1E1 protein (Parker et al., 2018). It is possible that transcription regulation is different. Several nuclear receptors, including PPARα, CAR (constitutive androstane receptor), and ERα (estrogen receptor alpha) have been implicated in SULT1E1 transcriptional regulation (Yi et al., 2021) and the activity of these may be affected by PCB52 and 4-OH-PCB52. How regulation of SULT1E1 by PCB52 or its hydroxylated form affects adipocyte function and differentiation is currently unknown. Another xenobiotic processing gene that was affected by both PCB52 and 4-OH-PCB52 was AADAC (arylacetamide deacetylase), an enzyme that catalyzes one of the initial biotransformation pathways for arylamine and heterocyclic amine carcinogens (Zhang et al., 2012). The gene has been implicated in regulating triglyceride homeostasis (Konstandi et al., 2019). Interestingly, PCB52 was found to downregulate AADAC whereas 4-OH-PCB52 had the opposite effect and downregulated it. What this means for preadipocyte or adipocyte function is unknown.
Some caveats of the present study should be noted. One is that only one concentration of PCB52 or 4-OH-PCB52 was used in the exposures of preadipocytes over the time course. The 10 μM concentration was chosen because this was a sublethal concentration did not cause significant cytotoxicity in the conditions of the experiment. It is a level that people would not be exposed to. We used this high dose to ensure that the compounds would cause robust changes in gene expression that could be detected. It is possible that lower concentrations have differential effects and/or no effects at all, except for over a longer-term exposure. The present findings provide motivation to perform further studies with a wider range of concentrations over different lengths of time. We also should note that our studies were performed in conditions of 10% FBS in the media. We used FBS to ensure good cell growth. It is possible and likely that much of the PCB52 is bound up by the FBS, making it unavailable to act on the cells. In fact, the concentration of 10 μM PCB52 would be expected to be above the solubility limit in water. However, the PCBs are partitioned to albumin in serum making their solubility in culture media appear much higher than that of pure water (Zhang et al., 2020). Simulations based on octanol/water partitioning coefficients show that with the use of 10% FBS media, ~99% of the PCBs partition to the albumin leaving only a small fraction left soluble to interact with the cells in culture (unpublished). Thus, having this amount of FBS requires higher levels of PCB52. Further, oxidation of PCB52 to an hydroxylated form might not only change its biological activity but also make it more available because the hydroxylated form may bind less to FBS. Future studies are needed to precisely assess levels of PCB52 and metabolites in the cells and cell culture media (Zhang et al., 2020). Further, experiments using less, or no, serum would be needed to address how serum levels might change PCB availability, with the risk, though, of changing cell growth or function. Finally, it should be noted that the various pathway analyses and upstream regulator results are based on predictions derived from available data in the literature and in publicly available databases. Some of this information may not be completely accurate or may not be applicable to preadipocytes. Thus, the predictions may be considered relatively speculative. To draw causative associations and more definitive biological conclusions, it will be necessary to perform additional in vitro and in vivo experiments.
Regardless of these caveats, our studies provide a framework for understanding how PCB52 exposure leads to cell dysfunction to cause disease. The rich dataset that we have generated will be useful to other researchers who are studying how environmental toxins cause cardiometabolic disease through effects on adipose tissue.
Supplementary Material
HIGHLIGHTS.
RNAseq was performed on human preadipocytes exposed to PCB52 or 4-OH-PCB52
4-OH-PCB52 caused considerably more changes in gene expression than PCB52
PCB52 or 4-OH-PCB52 often caused gene expression changes in opposite direction
PCB52 inhibited expression of genes involved in the PPARα pathway
4-OH-PCB52 altered genes in cytokine, hormone, focal adhesion, and Wnt pathways
Acknowledgments and Funding
The authors thank Dr. Xueshue Li for synthesizing and authenticating the PCB52 and 4-OH-PCB52 used in these studies. The RNAseq data and analyses were obtained at the Genomics and Bioinformatics Divisions of the Iowa Institute of Human Genetics which is supported, in part, by the University of Iowa Carver College of Medicine and the Holden Comprehensive Cancer Center (National Cancer Institute of the National Institutes of Health under Award Number P30 CA086862). This study was supported by NIH P42 ES013661 (AJK, JAA, HJL) and a pilot grant from the Environmental Health Research Center P30 ES005605 (AJK, JAA).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ampleman MD, Martinez A, DeWall J, Rawn DF, Hornbuckle KC, Thorne PS, 2015. Inhalation and dietary exposure to PCBs in urban and rural cohorts via congener-specific measurements. Environ Sci Technol 49, 1156–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannavti MK, Jahnke JC, Marek RF, Just CL, Hornbuckle KC, 2021. Room-to-Room Variability of Airborne Polychlorinated Biphenyls in Schools and the Application of Air Sampling for Targeted Source Evaluation. Environ Sci Technol 55, 9460–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa ACS, Feng Y, Yu C, Huang M, Xie W, 2019. Estrogen sulfotransferase in the metabolism of estrogenic drugs and in the pathogenesis of diseases. Expert Opin Drug Metab Toxicol 15, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A, Biziuk M, 2009. Environmental fate and global distribution of polychlorinated biphenyls. Rev Environ Contam Toxicol 201, 137–158. [DOI] [PubMed] [Google Scholar]
- Breining P, Pedersen SB, Kjolby M, Hansen JB, Jessen N, Richelsen B, 2021. Parathyroid hormone receptor stimulation induces human adipocyte lipolysis and browning. Eur J Endocrinol 184, 687–697. [DOI] [PubMed] [Google Scholar]
- Calderon-Dominguez M, Sebastian D, Fucho R, Weber M, Mir JF, Garcia-Casarrubios E, Obregon MJ, Zorzano A, Valverde AM, Serra D, Herrero L, 2016. Carnitine Palmitoyltransferase 1 Increases Lipolysis, UCP1 Protein Expression and Mitochondrial Activity in Brown Adipocytes. PLoS One 11, e0159399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright MJ, Schlauch K, Lenburg ME, Tchkonia T, Pirtskhalava T, Cartwright A, Thomou T, Kirkland JL, 2010. Aging, depot origin, and preadipocyte gene expression. J Gerontol A Biol Sci Med Sci 65, 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiellini C, Grenningloh G, Cochet O, Scheideler M, Trajanoski Z, Ailhaud G, Dani C, Amri EZ, 2008. Stathmin-like 2, a developmentally-associated neuronal marker, is expressed and modulated during osteogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun 374, 64–68. [DOI] [PubMed] [Google Scholar]
- Clair HB, Pinkston CM, Rai SN, Pavuk M, Dutton ND, Brock GN, Prough RA, Falkner KC, McClain CJ, Cave MC, 2018. Liver Disease in a Residential Cohort With Elevated Polychlorinated Biphenyl Exposures. Toxicol Sci 164, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Spiegelman BM, 2015. Brown and Beige Fat: Molecular Parts of a Thermogenic Machine. Diabetes 64, 2346–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Spiegelman BM, 2016. Cell biology of fat storage. Mol Biol Cell 27, 2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonna NM, Chen YQ, Konrad RJ, 2022. Angiopoietin-like proteins and postprandial partitioning of fatty acids. Curr Opin Lipidol 33, 39–46. [DOI] [PubMed] [Google Scholar]
- Dirinck E, Jorens PG, Covaci A, Geens T, Roosens L, Neels H, Mertens I, Van Gaal L, 2011. Obesity and persistent organic pollutants: possible obesogenic effect of organochlorine pesticides and polychlorinated biphenyls. Obesity (Silver Spring) 19, 709–714. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R, 2007. A systems biology approach for pathway level analysis. Genome Res 17, 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espandiari P, Glauert HP, Lehmler HJ, Lee EY, Srinivasan C, Robertson LW, 2003. Polychlorinated biphenyls as initiators in liver carcinogenesis: resistant hepatocyte model. Toxicol Appl Pharmacol 186, 55–62. [DOI] [PubMed] [Google Scholar]
- Esser A, Schettgen T, Gube M, Koch A, Kraus T, 2016. Association between polychlorinated biphenyls and diabetes mellitus in the German HELPcB cohort. Int J Hyg Environ Health 219, 557–565. [DOI] [PubMed] [Google Scholar]
- Everett CJ, Frithsen I, Player M, 2011. Relationship of polychlorinated biphenyls with type 2 diabetes and hypertension. J Environ Monit 13, 241–251. [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Kaller M, 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame GM, 1997. A collaborative study of 209 PCB congeners and 6 Aroclors on 20 different HRGC columns .2. Semi-quantitative Aroclor congener distributions. Fresenius Journal of Analytical Chemistry 357, 714–722. [Google Scholar]
- Gadupudi G, Gourronc FA, Ludewig G, Robertson LW, Klingelhutz AJ, 2015. PCB126 inhibits adipogenesis of human preadipocytes. Toxicol In Vitro 29, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourronc FA, Helm BK, Robertson LW, Chimenti MS, Joachim-Lehmler H, Ankrum JA, Klingelhutz AJ, 2022. Transcriptome sequencing of 3,3’,4,4’,5-Pentachlorobiphenyl (PCB126)-treated human preadipocytes demonstrates progressive changes in pathways associated with inflammation and diabetes. Toxicol In Vitro 83, 105396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourronc FA, Perdew GH, Robertson LW, Klingelhutz AJ, 2020. PCB126 blocks the thermogenic beiging response of adipocytes. Environ Sci Pollut Res Int 27, 8897–8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourronc FA, Robertson LW, Klingelhutz AJ, 2018. A delayed proinflammatory response of human preadipocytes to PCB126 is dependent on the aryl hydrocarbon receptor. Environ Sci Pollut Res Int 25, 16481–16492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm FA, Hu D, Kania-Korwel I, Lehmler HJ, Ludewig G, Hornbuckle KC, Duffel MW, Bergman A, Robertson LW, 2015. Metabolism and metabolites of polychlorinated biphenyls. Crit Rev Toxicol 45, 245–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedesan OC, Fenzl A, Digruber A, Spirk K, Baumgartner-Parzer S, Bilban M, Kenner L, Vierhapper M, Elbe-Burger A, Kiefer FW, 2019. Parathyroid hormone induces a browning program in human white adipocytes. Int J Obes (Lond) 43, 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoherranen N, Zhong G, 2019. Biochemical and physiological importance of the CYP26 retinoic acid hydroxylases. Pharmacol Ther 204, 107400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issemann I, Green S, 1990. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347, 645–650. [DOI] [PubMed] [Google Scholar]
- Jackson E, Shoemaker R, Larian N, Cassis L, 2017. Adipose Tissue as a Site of Toxin Accumulation. Compr Physiol 7, 1085–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke JC, Hornbuckle KC, 2019. PCB Emissions from Paint Colorants. Environ Sci Technol 53, 5187–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke JC, Martinez A, Hornbuckle KC, 2022. Distinguishing Aroclor and non-Aroclor sources to Chicago Air. Sci Total Environ 823, 153263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Draghici S, 2005. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics 21, 3587–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL, 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL, 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh WX, Hornbuckle KC, Wang K, Thorne PS, 2016. Serum polychlorinated biphenyls and their hydroxylated metabolites are associated with demographic and behavioral factors in children and mothers. Environ Int 94, 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlgruber A, Lynch L, 2015. Adipose tissue inflammation in the pathogenesis of type 2 diabetes. Curr Diab Rep 15, 92. [DOI] [PubMed] [Google Scholar]
- Konstandi M, Kypreos KE, Matsubara T, Xepapadaki E, Shah YM, Krausz K, Andriopoulou CE, Kofinas A, Gonzalez FJ, 2019. Adrenoceptor-related decrease in serum triglycerides is independent of PPARalpha activation. FEBS J 286, 4328–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le PT, Liu H, Alabdulaaly L, Vegting Y, Calle IL, Gori F, Lanske B, Baron R, Rosen CJ, 2021. The role of Zfp467 in mediating the pro-osteogenic and anti-adipogenic effects on bone and bone marrow niche. Bone 144, 115832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhu Z, Lu Y, Granneman JG, 2005. Metabolic and cellular plasticity in white adipose tissue II: role of peroxisome proliferator-activated receptor-alpha. Am J Physiol Endocrinol Metab 289, E617–626. [DOI] [PubMed] [Google Scholar]
- Li X, Holland EB, Feng W, Zheng J, Dong Y, Pessah IN, Duffel MW, Robertson LW, Lehmler HJ, 2018. Authentication of synthetic environmental contaminants and their (bio)transformation products in toxicology: polychlorinated biphenyls as an example. Environ Sci Pollut Res Int 25, 16508–16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson RE, Peters SJ, 2015. Piecing together the puzzle of perilipin proteins and skeletal muscle lipolysis. Appl Physiol Nutr Metab 40, 641–651. [DOI] [PubMed] [Google Scholar]
- Marek RF, Martinez A, Hornbuckle KC, 2013. Discovery of hydroxylated polychlorinated biphenyls (OH-PCBs) in sediment from a lake Michigan waterway and original commercial aroclors. Environ Sci Technol 47, 8204–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek RF, Thorne PS, Herkert NJ, Awad AM, Hornbuckle KC, 2017. Airborne PCBs and OH-PCBs Inside and Outside Urban and Rural U.S. Schools. Environ Sci Technol 51, 7853–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildaziene V, Nauciene Z, Baniene R, Demin O, Krab K, 2002a. Analysis of effects of 2,2’,5,5’-tetrachlorobiphenyl on the flux control in oxidative phosphorylation system in rat liver mitochondria. Mol Biol Rep 29, 35–40. [DOI] [PubMed] [Google Scholar]
- Mildaziene V, Nauciene Z, Krab K, 2002b. The targets of 2,2’,5,5’-tetrachlorobiphenyl in the respiratory chain of rat liver mitochondria revealed by modular kinetic analysis. Mol Biol Rep 29, 31–34. [DOI] [PubMed] [Google Scholar]
- Nguyen TM, Shafi A, Nguyen T, Draghici S, 2019. Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biol 20, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker VS, Squirewell EJ, Lehmler HJ, Robertson LW, Duffel MW, 2018. Hydroxylated and sulfated metabolites of commonly occurring airborne polychlorinated biphenyls inhibit human steroid sulfotransferases SULT1E1 and SULT2A1. Environ Toxicol Pharmacol 58, 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson KL, Schlenk D, 2011. Biotransformation of 2,2’,5,5’-tetrachlorobiphenyl (PCB 52) and 3,3’,4,4’-tetrachlorobiphenyl (PCB 77) by liver microsomes from four species of sea turtles. Chem Res Toxicol 24, 718–725. [DOI] [PubMed] [Google Scholar]
- Robertson LW, Espandiari P, Lehmler HJ, Pereg D, Srinivasan A, Tampal N, Twaroski T, Ludewig G, Glauert HP, Arif J, Gupta R, 2000. Metabolism and activation of polychlorinated biphenyls (PCBs). Cent Eur J Public Health 8 Suppl, 14–15. [PubMed] [Google Scholar]
- Rodriguez EA, Li X, Lehmler HJ, Robertson LW, Duffel MW, 2016. Sulfation of Lower Chlorinated Polychlorinated Biphenyls Increases Their Affinity for the Major Drug-Binding Sites of Human Serum Albumin. Environ Sci Technol 50, 5320–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez EA, Vanle BC, Doorn JA, Lehmler HJ, Robertson LW, Duffel MW, 2018. Hydroxylated and sulfated metabolites of commonly observed airborne polychlorinated biphenyls display selective uptake and toxicity in N27, SH-SY5Y, and HepG2 cells. Environ Toxicol Pharmacol 62, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM, 2014. What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routti H, Letcher RJ, Arukwe A, Van Bavel B, Yoccoz NG, Chu S, Gabrielsen GW, 2008. Biotransformation of PCBs in relation to phase I and II xenobiotic-metabolizing enzyme activities in ringed seals (Phoca hispida) from Svalbard and the Baltic Sea. Environ Sci Technol 42, 8952–8958. [DOI] [PubMed] [Google Scholar]
- Saktrakulkla P, Lan T, Hua J, Marek RF, Thorne PS, Hornbuckle KC, 2020. Polychlorinated Biphenyls in Food. Environ Sci Technol 54, 11443–11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saktrakulkla P, Li X, Martinez A, Lehmler HJ, Hornbuckle KC, 2022. Hydroxylated Polychlorinated Biphenyls Are Emerging Legacy Pollutants in Contaminated Sediments. Environ Sci Technol 56, 2269–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schettgen T, Esser A, Alt A, Randerath I, Kraus T, Ziegler P, 2022. Decomposition Products of the Initiator Bis(2,4-dichlorobenzoyl)peroxide in the Silicone Industry: Human Biomonitoring in Plasma and Urine of Workers. Environ Sci Technol. [DOI] [PubMed] [Google Scholar]
- Sethi S, Morgan RK, Feng W, Lin Y, Li X, Luna C, Koch M, Bansal R, Duffel MW, Puschner B, Zoeller RT, Lehmler HJ, Pessah IN, Lein PJ, 2019. Comparative Analyses of the 12 Most Abundant PCB Congeners Detected in Human Maternal Serum for Activity at the Thyroid Hormone Receptor and Ryanodine Receptor. Environ Sci Technol 53, 3948–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Kakimoto K, Takenaka S, Koga N, Uehara S, Murayama N, Yamazaki H, Kim D, Guengerich FP, Komori M, 2016. Roles of Human CYP2A6 and Monkey CYP2A24 and 2A26 Cytochrome P450 Enzymes in the Oxidation of 2,5,2’,5’-Tetrachlorobiphenyl. Drug Metab Dispos 44, 1899–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD, 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura A, Gong S, Sato T, Kawaguchi M, Sekine S, Kazuki Y, Horie T, Ito K, 2021. Evaluation of Parent- and Metabolite-Induced Mitochondrial Toxicities Using CYP-Introduced HepG2 cells. J Pharm Sci 110, 3306–3312. [DOI] [PubMed] [Google Scholar]
- Teijeiro A, Garrido A, Ferre A, Perna C, Djouder N, 2021. Inhibition of the IL-17A axis in adipocytes suppresses diet-induced obesity and metabolic disorders in mice. Nat Metab 3, 496–512. [DOI] [PubMed] [Google Scholar]
- Tsuchida A, Yamauchi T, Takekawa S, Hada Y, Ito Y, Maki T, Kadowaki T, 2005. Peroxisome proliferator-activated receptor (PPAR)alpha activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: comparison of activation of PPARalpha, PPARgamma, and their combination. Diabetes 54, 3358–3370. [DOI] [PubMed] [Google Scholar]
- Vu BG, Gourronc FA, Bernlohr DA, Schlievert PM, Klingelhutz AJ, 2013. Staphylococcal superantigens stimulate immortalized human adipocytes to produce chemokines. PLoS One 8, e77988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlang B, Falkner KC, Clair HB, Al-Eryani L, Prough RA, States JC, Coslo DM, Omiecinski CJ, Cave MC, 2014. Human receptor activation by aroclor 1260, a polychlorinated biphenyl mixture. Toxicol Sci 140, 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlang B, Prough RA, Falkner KC, Hardesty JE, Song M, Clair HB, Clark BJ, States JC, Arteel GE, Cave MC, 2016. Polychlorinated Biphenyl-Xenobiotic Nuclear Receptor Interactions Regulate Energy Metabolism, Behavior, and Inflammation in Non-alcoholic-Steatohepatitis. Toxicol Sci 149, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Adamcakova-Dodd A, Flor S, Gosse L, Klenov VE, Stolwijk JM, Lehmler HJ, Hornbuckle KC, Ludewig G, Robertson LW, Thorne PS, 2020. Comprehensive Subchronic Inhalation Toxicity Assessment of an Indoor School Air Mixture of PCBs. Environ Sci Technol 54, 15976–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Adamcakova-Dodd A, Lehmler HJ, Hornbuckle KC, Thorne PS, 2022. Toxicity Assessment of 91-Day Repeated Inhalation Exposure to an Indoor School Air Mixture of PCBs. Environ Sci Technol 56, 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XL, Zhou WT, Zhang KK, Yuan Y, Qiu EM, Shen YW, Wang Q, 2019. PCB52 induces hepatotoxicity in male offspring through aggravating loss of clearance capacity and activating the apoptosis: Sex-biased effects on rats. Chemosphere 227, 389–400. [DOI] [PubMed] [Google Scholar]
- Yi M, Negishi M, Lee SJ, 2021. Estrogen Sulfotransferase (SULT1E1): Its Molecular Regulation, Polymorphisms, and Clinical Perspectives. J Pers Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CY, Flor S, Ludewig G, Lehmler HJ, 2020. Atropselective Partitioning of Polychlorinated Biphenyls in a HepG2 Cell Culture System: Experimental and Modeling Results. Environ Sci Technol 54, 13817–13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu X, Meng G, Chi M, Li J, Yin S, Zhao Y, Wu Y, 2018. Non-dioxin-like polychlorinated biphenyls in early pregnancy and risk of gestational diabetes mellitus. Environ Int 115, 127–132. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng X, Aleksunes L, Klaassen CD, 2012. Transcription factor-mediated regulation of carboxylesterase enzymes in livers of mice. Drug Metab Dispos 40, 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WT, Wang LB, Yu H, Zhang KK, Chen LJ, Wang Q, Xie XL, 2020. N-acetylcysteine alleviates PCB52-induced hepatotoxicity by repressing oxidative stress and inflammatory responses. PeerJ 8, e9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


