Abstract
Myocardial Brg1 is essential for heart regeneration in zebrafish, but it remains unknown whether and how endothelial Brg1 plays a role in heart regeneration. Here, we found that both brg1 mRNA and protein were induced in cardiac endothelial cells after ventricular resection and endothelium-specific overexpression of dominant-negative Xenopus Brg1 (dn-xbrg1) inhibited myocardial proliferation and heart regeneration and increased cardiac fibrosis. RNA-seq and ChIP-seq analysis revealed that endothelium-specific overexpression of dn-xbrg1 changed the levels of H3K4me3 modifications in the promoter regions of the zebrafish genome and induced abnormal activation of Notch family genes upon injury. Mechanistically, Brg1 interacted with lysine demethylase 7aa (Kdm7aa) to fine-tune the level of H3K4me3 within the promoter regions of Notch family genes and thus regulated notch gene transcription. Together, this work demonstrates that the Brg1-Kdm7aa-Notch axis in cardiac endothelial cells, including the endocardium, regulates myocardial proliferation and regeneration via modulating the H3K4me3 of the notch promoters in zebrafish.
Subject terms: Regeneration, Epigenetics
Introduction
The high mortality and morbidity of myocardial infarction are of public concern worldwide. The loss of cardiomyocytes following myocardial infarction and the inadequate self-repair capability of the mammalian heart make it difficult to treat cardiac diseases1. As one of the least regenerative organs in the human body, the heart replaces the infarcted myocardium with a noncontractile scar instead of new muscles, which is initially beneficial but eventually leads to loss of contraction and function. Although various cell-based and cell-free strategies have been explored to restore infarcted heart function, the efficacy and side effects, such as arrhythmia and immune rejection, currently prevent translation to the clinic. The neonatal mouse can regenerate its heart, but this ability is lost after 7 postnatal days2–4. A number of elegant studies have provided evidence for the underlying mechanisms, but how to efficiently stimulate mammalian heart regeneration remains largely unknown. Unlike mammals, some lower vertebrates, such as zebrafish, can fully regenerate the heart after injury throughout life5. Dissecting the cellular and molecular mechanisms of zebrafish heart regeneration may provide clues for promoting heart regeneration in mammals.
It is conceivable that cardiomyocyte dedifferentiation and proliferation contribute to heart regeneration in zebrafish6,7. Over the past decades, a number of signaling pathways and transcription factors have been reported to regulate myocardial proliferation and regeneration in zebrafish, including fibroblast growth factor, sonic hedgehog, retinoic acid, insulin-like growth factor, Notch, GATA4, Hand2, NF-kB, and Stat38–13. Retinaldehyde dehydrogenase 2, which produces retinoic acid, is activated in the epicardium and endocardium within hours after injury, and transgenic inhibition of retinoic acid receptors impairs myocardial proliferation8. Conditional inhibition of Notch signaling via overexpression of dominant-negative Notch transcriptional co-activator Master-mind like-1 (MAML) in endothelial cells (including the endocardium) decreases myocardial proliferation12,14. These studies suggest an essential role of endocardial signaling in regulating myocardial proliferation, but it remains to be addressed how endocardial Notch components are regulated or how endocardial signals regulate myocardial proliferation and regeneration upon injury.
Epigenetic regulation plays an important role in gene expression in various cellular processes such as differentiation, proliferation, fate determination, as well as organ regeneration15–18. Epigenetic regulation is, in general, defined as controlling gene expression beyond the DNA sequence itself, consisting of histone modifications, DNA/RNA modifications, noncoding RNAs, and chromatin-remodeling complexes19. The SWI/SNF (SWItch/Sucrose Non-Fermentable)-like complex, a member of the ATP-dependent chromatin-remodeling complex family, uses energy from ATP hydrolysis, regulates gene transcription by rearranging nucleosome positions and histone-DNA interactions, and thus facilitates the transcriptional activation or repression of targeted genes20. We previously reported that its central subunit, brahma-related gene 1 (brg1 or smarca4), had a critical function in zebrafish heart regeneration by interacting with DNA (cytosine-5)-methyltransferase 3 alpha b to modify DNA methylation of the cyclin-dependent kinase inhibitor 1C promoter21. We found that brg1 was not only induced in cardiomyocytes but also in cardiac endothelial cells, including the endocardium, during myocardial regeneration21. In this work, we investigated how endothelial Brg1 played a role in zebrafish heart regeneration. Inhibition of Brg1 via dominant-negative (DN)-xBrg1 in cardiac endothelial/endocardial cells decreased myocardial proliferation and heart regeneration, and Brg1 interacted with the histone demethylase Kdm7aa (lysine (K)-specific demethylase 7Aa) to regulate notch receptor gene expression upon injury. Together, this work supports the notion that the Brg1-Kdm7aa axis fine-tunes Notch signaling in cardiac endothelium and endocardium during heart regeneration.
Results
Endothelial Brg1 is required for heart regeneration in zebrafish
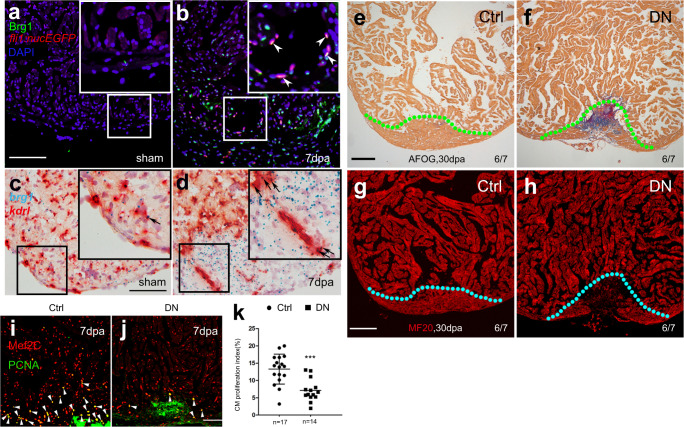
Our previous work has shown that both global and myocardium-specific inhibition of Brg1 results in impaired myocardial proliferation and regeneration, while global inhibition of Brg1 leads to more severe cardiac fibrosis than its myocardium-specific inhibition21. In addition to elevated expression in the injured myocardium, Brg1 was also induced in other cardiac cells, including endothelial cells, during heart regeneration. To evaluate Brg1 expression in endothelial cells during zebrafish heart regeneration, we used immunofluorescence staining (Fig. 1a, b) and RNAscope in situ hybridization (Fig. 1c, d) to determine whether Brg1 was induced in endothelial cells upon ventricular amputation. Consistent with our previous report, Brg1 protein was co-localized with Tg(fli1:nucEGFP)-positive endothelial cells in the injury site at 7 days post-amputation (dpa) (Fig. 1a, b). Moreover, RNAscope staining revealed that brg1 mRNA was elevated and partially overlapped with kdrl-positive endothelium at 3 dpa (Fig. 1c, d). We then turned to tamoxifen-induced endothelium-specific inhibition of Brg1 with the transgenic strains Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:CreER)21,22 to address whether Brg1 had a function in endothelial cells during regeneration. To validate the endothelial-specific expression of Tg(kdrl:CreER), we created compound transgenic zebrafish line Tg(ubi:LoxP-DsRed-STOP-LoxP-EGFP; kdrl:CreER), where endothelial-specific expression of GFP is driven by the kdrl promoter after Cre-mediated recombination. The immunostaining with GFP antibody showed that GFP was expressed in the endocardium/coronary vessels in the double transgenic fish hearts treated with 4-HT (Supplementary Fig. 1a). Furthermore, RNA in situ analysis with either CreER or dn-xbrg1 probes revealed expression of dn-xbrg1 and CreER mRNA in the endocardium/coronary vessels on frozen heart sections from Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:CreER) [DN] hearts but not Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1) [Ctrl] hearts treated with tamoxifen (Supplementary Fig. 1b). Interestingly, endothelium-specific overexpression of dn-xbrg1 resulted in abnormal cardiac fibrosis (Fig. 1e, f) and compromised myocardial regeneration (Fig. 1g, h) at 30 dpa as well as decreased the number of proliferating cardiomyocytes at 7 dpa (Fig. 1i–k). Using RNAscope in situ hybridization, we also found that endothelium-specific inhibition of Brg1 decreased the number of kdrl-positive endothelial cells (Supplementary Fig. 2a–c) and coronin1a-positive leukocytes (Supplementary Fig. 2d–f) while having no effect on tcf21-positive epicardium (Supplementary Fig. 2g–i) in DN hearts compared with Ctrl sibling hearts at 7 dpa in the presence of 4-hydroxytamoxifen (4-HT). Taken together, these data demonstrate that endothelial Brg1 is required for myocardial proliferation during heart regeneration.
Fig. 1. Inhibition of endothelial Brg1 impairs myocardial proliferation and regeneration.
a, b Immunofluorescence staining of Brg1 and EGFP on paraffin sections of Tg(fli1:nucEGFP) transgenic hearts from sham-operated (a) and injured zebrafish hearts (b) at 7 dpa (arrowheads, Brg1- and EGFP-positive endothelial cell nuclei). c, d RNAscope in situ hybridization of brg1 and kdrl probes in frozen sections from sham-operated (c) and injured hearts (d) at 3 dpa (arrows, brg1- and kdrl-positive endothelial cells). The upper right corner presented in (a–d) is a high-magnification image of the framed area. e–h Representative images of Acid fuchsin orange G (AFOG) staining (e, f) and immunofluorescence with anti-myosin heavy chain (MF20) (g, h) of heart sections from control siblings Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1) (Ctrl) and endothelium-specific dominant-negative brg1 mutants Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:CreER) (DN) at 30 dpa, noting that, compared with robust regenerated myocardium and rare cardiac fibrosis in Ctrl group (e, g), the DN group failed to regenerate the myocardium (h) and had evident fibrin (red) and collagen (blue) deposition (f). Dashed lines mark the resection traces. N numbers indicate biological replicates. i, j Immunostaining of representative heart sections at 7 dpa identified cardiomyocyte nuclei (Mef2C+) and nuclei undergoing DNA replication (PCNA+). Noting fewer proliferative cardiomyocytes (Mef2C+/PCNA+) in the DN group than in the Ctrl group. Arrowheads, Mef2C+/PCNA+ proliferating cardiomyocytes. k Statistical analysis of experiments as in (i) and (j) (CM, cardiomyocyte; n = 17 or 14 biological replications; data were the mean percentage ± s.e.m.; ***p < 0.001, unpaired t-test). Scale bars, 100 μm.
Endothelium-specific inhibition of Brg1 changes the levels of H3K4me3 in the promoter regions of zebrafish genome
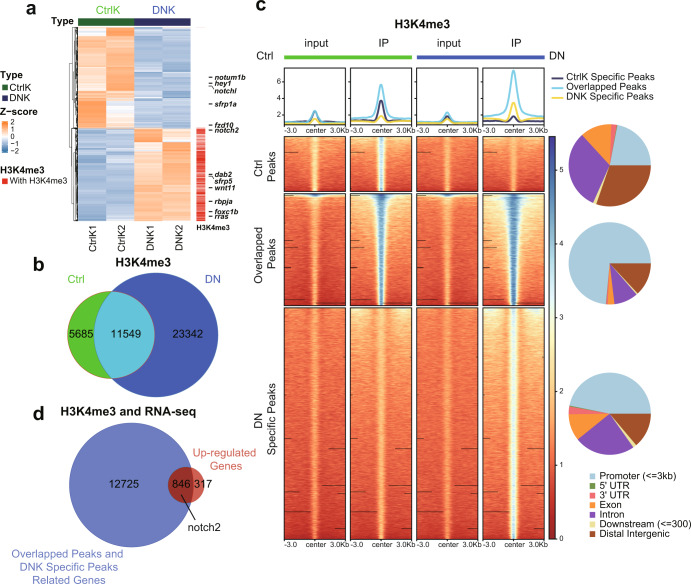
To decipher the molecular action of endothelial Brg1, we used RNA-seq analysis to search for Brg1-regulated genes during heart regeneration. We applied Tg(kdrl:EGFP) to label cardiac endothelial cells, including the endocardium, and achieved endothelium-specific overexpression of dn-xbrg1 by using the compound zebrafish line consisting of Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:CreER; kdrl:EGFP) (defined as DNK), while we used Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:EGFP) as control (CtrlK) in the presence of 4-HT starting at 3 days before ventricular resection. The kdrl:EGFP endothelial cells, which were sorted by fluorescence-activated cell sorting (FACS) from CtrlK and DNK hearts at 7 dpa (Supplementary Fig. 3), were subjected to RNA-seq analysis, and differentially expressed genes were identified (Fig. 2a). Compared with CtrlK group, we found 1163 upregulated genes and 1266 downregulated genes in DNK group (Fig. 2a; Supplementary Table 3). Gene ontology (GO) enrichment analysis of these genes revealed that receptor activity-related genes were among the top-affected leads, in which the Notch signaling component notch2 was strongly induced in the DNK group (Fig. 2a; Supplementary Fig. 4a, b; Supplementary Table 4). Some other biological processes, such as cell adhesion, response to wounding, extracellular matrix organization, cell migration, wound healing, and immune response, were also significantly affected after the inhibition of Brg1, specifically in endothelial cells (Supplementary Fig. 4a; Supplementary Table 4).
Fig. 2. Endothelium-specific inhibition of Brg1 changes the levels of H3K4me3 in the promoter regions of the zebrafish genome.
a Heatmap displaying Z-score normalized gene expression for differentially expressed genes between FACS-sorted kdrl:EGFP endothelial cells from dominant-negative Brg1 groups (DNK1 and DNK2) and control groups (CtrlK1 and CtrlK2). FPKM value of each gene was normalized using Z-scores across samples. Columns represent individual samples (two biological replicates for each group); rows represent differentially expressed genes ordered by hierarchical clustering. Labeled genes are part of the differentially expressed Notch signaling genes. The upregulated genes in the DNK group that were labeled with ‘red color’ had H3K4me3 peaks in their promoters. b Venn plot representing the intersection of H3K4me3 peaks between Ctrl and DN groups. c Heatmaps and summary plots displaying the signal profile of normalized read coverage around three categories of H3K4me3 peaks across different samples. The read coverage was normalized to 1x sequencing depth in all samples. Each row of the heatmap represents one peak, with coverage plotted across the 3 kb surrounding the peak summit. H3K4me3 peaks were classified into three categories: Ctrl-Specific Peaks represent peaks specifically in the Ctrl group; Overlapped Peaks represent peaks overlapped between the Ctrl and DN groups; DN-Specific Peaks represent peaks specifically in the DN group. The genomic distribution for three types of peaks is presented with pie charts on the right side. d Venn plot representing the intersection between genes with promoters marked by Overlapped Peaks and DN-Specific Peaks and genes that were differentially upregulated in the DNK group.
It is well recognized that Brg1 is involved in both gene activation and repression through interacting with epigenetic modifiers and influencing histone modifications at the targeted gene promoters23. And previous studies have established that the nucleosomes with histone H3 Lysine 4 trimethylation (H3K4me3) are mainly associated with the promoter regions of active transcription24,25. Therefore, we examine whether endothelial-specific overexpression of dn-xbrg1 has an effect on the level of the histone marker H3K4me3 in the zebrafish genome. Genome-wide ChIP-seq analyses of Ctrl and DN amputated ventricles at 7 dpa using H3K4me3 antibody revealed that, in addition to 11,549 overlapping H3K4me3 peaks between the Ctrl and DN groups, more H3K4me3 peaks emerged in the DN group, suggesting that inhibition of Brg1 enhanced H3K4me3 modifications (Fig. 2b). Peaks were then divided into three categories according to the Venn plot, namely Ctrl-Specific Peaks in Ctrl group, Overlapped Peaks representing peaks overlapped between Ctrl and DN groups, and DN-Specific Peaks representing peaks specifically in DN group. Heatmaps and summary plots of H3K4me3 ChIP-seq signals in 3 kb surrounding the peak summits displayed slightly stronger Ctrl-Specific Peaks signals in the Ctrl group, while increased Overlapped Peaks signals and DNK-Specific Peaks signals in the DN group (Fig. 2c). Moreover, genomic distribution analysis for three categories of peaks revealed that peaks with increased signals in DN group were more concentrated in the promoter region than that with decreased peak signals (Fig. 2c), suggesting that endothelial Brg1 inhibition led to elevated levels of H3K4me3 in the promoter regions. Apart from basic biological processes, we also found that GO terms about “Notch signaling pathway” were highly enriched near the overlapped peaks between Ctrl and DN groups (Supplementary Fig. 5b), and GO terms about “chromatin remodeling” were enriched near both DN-Specific Peaks and Overlapped Peaks (Supplementary Fig. 5). We then examined the correlation of differentially expressed genes from RNA-seq and H3K4me3 modification levels. We analyzed the overlapping genes by comparing upregulated genes in the DNK group with the genes whose promoters were marked by Overlapped Peaks and DN-Specific Peaks (Fig. 2d), as well as comparing downregulated genes in the DNK group with the genes whose promoters were marked by Overlapped Peaks and Ctrl-Specific Peaks. Venn plot identified 846 of the 1163 upregulated genes in the DNK group, which consisting of receptor activity-related Notch signaling component notch2 are occupied with Overlapped Peaks and DN-Specific H3K4me3 Peaks in the promoter regions (Fig. 2d, Supplementary Fig. 4c). These data suggest that endothelial-specific inhibition of Brg1 results in increased H3K4me3 modification levels in the promoter region of genes, which in turn leads to upregulation of genes expression, including notch2, in DN hearts.
Endothelium-specific inhibition of Brg1 induces upregulation of Notch signaling by increasing the level of H3K4me3 in the promoters
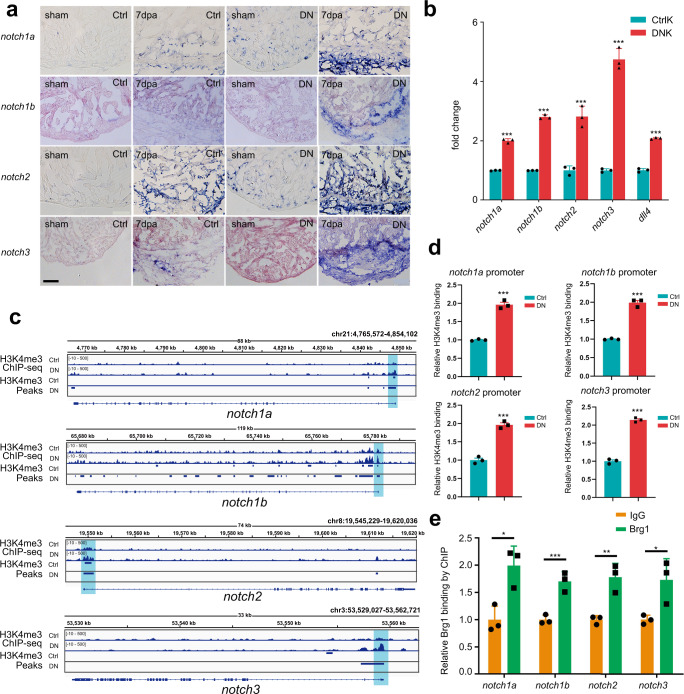
Previous works by Zhao and colleagues have found that endocardial expression of all four notch receptors was induced after injury, while hyperactivation of Notch signaling blocked zebrafish heart regeneration11. However, how notch receptors were transcriptionally regulated during zebrafish heart regeneration is yet unknown. Importantly, from the above RNA- and ChIP-seq data, we found overexpression of dn-xbrg1 increased the levels of H3K4me3 modifications and mRNA expression of notch2, and GO terms about “Notch signaling pathway” were highly enriched near the overlapped peaks between Ctrl and DN group, we then ask how Brg1 regulates notch receptor genes expression during heart regeneration. By performing RNA in situ analysis on frozen heart sections using either notch1a, notch1b, notch2, or notch3 probes, we found that inhibition of Brg1 in endothelial cells (DN) resulted in slight upregulation in sham-operated hearts, but had strong induction of notch1a, notch1b, notch2, and notch3 in injured hearts at 7 dpa compared with control sibling hearts (Ctrl) (Fig. 3a). Furthermore, RNAscope in situ hybridization showed that notch1b overlapped with kdrl-positive endothelial cells but rarely with tcf21-positive epicardial cells in Ctrl hearts (Supplementary Fig. 6a, c) and DN hearts (Supplementary Fig. 6b, d). Also, notch2 was induced in both kdrl- and tcf21-positive cells of DN hearts compared with Ctrl hearts (Supplementary Fig. 6e–h). In addition, qRT-PCR of FACS-sorted kdrl:EGFP-positive endothelial cells from CtrlK and DNK hearts at 7 dpa showed that, compared with CtrlK group, the expression levels of notch1a, notch1b, notch2, and notch3, as well as Notch ligands dll4, significantly increased in DNK group (Fig. 3b) in the presence of 4-HT. Together, these data suggest an inhibitory effect of Brg1 on the expression of notch genes during heart regeneration.
Fig. 3. Endothelium-specific inhibition of Brg1 induces abnormal upregulation of Notch signaling via the increased levels of H3K4me3 in their promoters.
a Representative images of RNA in situ hybridization with notch1a, notch1b, notch2, and notch3 probes on frozen sections of sham-operated Ctrl hearts, injured Ctrl hearts, sham-operated DN hearts, and injured DN hearts at 7 dpa. Scale bar, 100 μm. b Quantitative RT-PCR analysis showing that the expression of notch receptors and ligand in FACS-sorted kdrl:EGFP endothelial cells from the DNK group was higher than those from the CtrlK group. Data represented one of three independent experiments. Data were mean fold changes ± s.e.m., ***p < 0.005, unpaired t-test. c H3K4me3 ChIP-seq showing the traces and peak intervals of representative genomic loci from Ctrl and DN hearts. H3K4me3 peaks in both Ctrl and DN groups were shown as bars. Putative promoter regions were indicated in blue color. d Anti-H3K4me3 ChIP and quantitative PCR in Ctrl and DN hearts at 7 dpa (primers designed from notch receptor genomic regions: notch1a, −171/+3 bp; notch1b, −41/+58 bp; notch2, −263/−115 bp; notch3, +394/+504 bp; ATG site designed as +1 bp). Data represented one of three independent experiments. Data were the mean fold changes ± s.e.m.; ***p < 0.005, unpaired t-test. e Anti-Brg1 ChIP and quantitative PCR in wild-type hearts at 3 dpa. Data represented one of two independent experiments. Data were the mean fold change ± s.e.m.; *p < 0.05, **p < 0.01, ***p < 0.005; unpaired t-test. n number shown here (b, d, e) indicated technical replicates.
We then investigated how Brg1 regulated notch receptor genes. Genome-wide H3K4me3 ChIP-seq data showed that the H3K4me3 levels and peaks were increased in the promoters of notch1a, notch1b, notch2, and notch3 genomic loci in the DN group compared with those in the Ctrl group (Fig. 3c). Particularly, the promoter regions of notch1a, notch1b, and notch2 occupied with the Overlapped Peaks and the peaks signals were stronger in the DN group compared with Ctrl group; and the promoter region of notch3 had an H3K4me3 peak in DN group that was not in Ctrl group (Fig. 3c). We then used ChIP-qPCR to further confirm the levels of H3K4me3 modification in each of the notch promoter regions. ChIP with H3K4me3 antibody and quantitative PCR (ChIP-qPCR) showed that the levels of H3K4me3 of all four notch promoter regions were higher in DN hearts than in Ctrl hearts at 7 dpa in the presence of 5 μM 4-HT for 3 days before surgery (Fig. 3d), which was consistent with the elevated expression levels of these genes upon endothelial Brg1 inhibition (Fig. 3a, b). Furthermore, ChIP-qPCR with Brg1 antibody showed that Brg1 bound to the promoter regions of notch1a, notch1b, notch2, and notch3 (Fig. 3e), suggesting that Brg1 is involved in regulating the H3K4me3 modifications in the notch promoters.
Abnormally activated Notch signaling is responsible for the reduced cardiomyocyte proliferation in DN-xBrg1 hearts
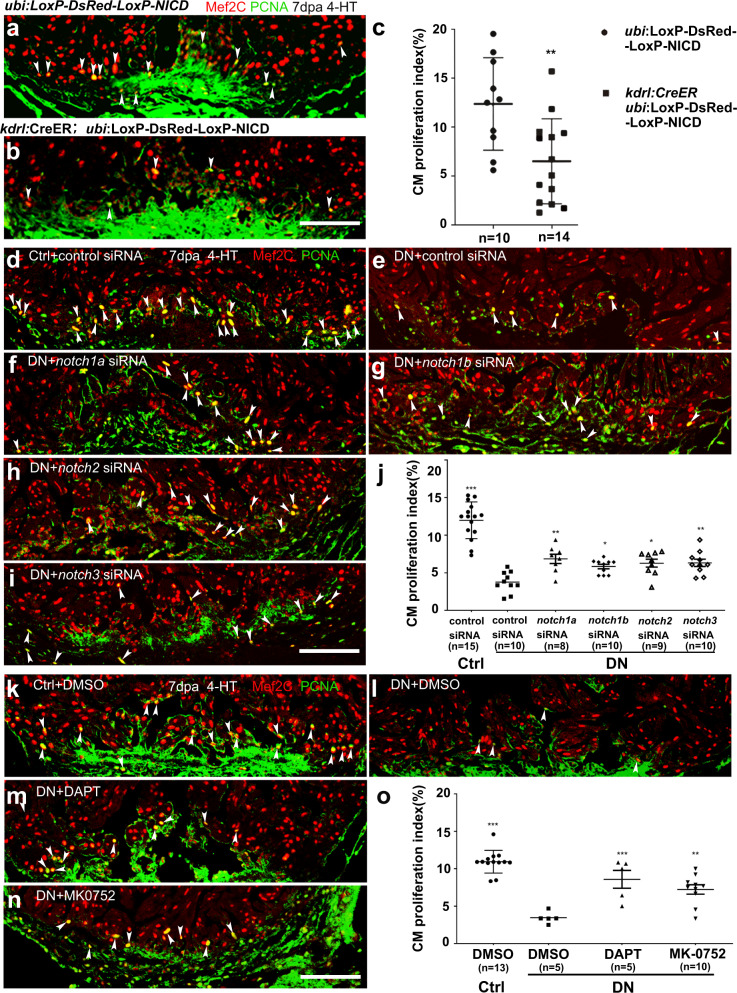
Since a previous study has shown that hyperactivation of Notch signaling impairs cardiomyocyte proliferation and heart regeneration11, we suspected that abnormally activated Notch signaling might contribute to defects of cardiomyocyte proliferation and regeneration in the endothelium-specific DN-xBrg1 hearts. We generated Tg(ubi:LoxP-DsRed-STOP-LoxP-NICD; kdrl:CreER) transgenic fish line to carry out tamoxifen-inducible overexpression of NICD (zebrafish notch1b intracellular domain) that specifically activated Notch signaling in endothelial cells. Compared with control hearts at 7 dpa, we found that hyperactivation of Notch signaling in endothelial cells decreased the numbers of PCNA+/Mef2C+ proliferating cardiomyocytes (Fig. 4a–c), which was consistent with the previous report11. We then asked whether simultaneous knockdown of notch receptors could rescue the numbers of proliferating cardiomyocytes in DN-xBrg1 mutant hearts. To validate the efficacy of nanoparticle-encapsulated siRNAs mediated gene knockdown, we first confirmed that siRNA could be delivered into endothelial/endocardial cells, epicardial cells, and cardiomyocytes (Supplementary Figs. 7 and 8). We then injected either notch1a, notch1b, notch2, notch3, or control siRNA at 1 dpa, analyzed expression levels of the target genes at 2 dpa by performing qRT-PCR, PCR, and RNAscope or RNA in situ hybridization (Supplementary Figs. 9a, c, 10, and 11). All the data showed that the siRNAs used here could decrease expression levels of the corresponding genes. As described above, control and DN zebrafish were infused with 5 μM 4-hydroxytamoxifen (4-HT) for 3 days before surgery, and nanoparticle-encapsulated notch1a, notch1b, notch2, notch3, or control siRNA was, respectively, injected every day after surgery until the hearts were harvested at 7 dpa. With control siRNA injection, we found that the PCNA+/Mef2C+ proliferating cardiomyocytes were fewer in DN hearts than in Ctrl hearts (Fig. 4d, e, j). Interestingly, either notch1a, notch1b, notch2, or notch3 siRNA was able to partially rescue the numbers of PCNA+/Mef2C+ proliferating cardiomyocytes in DN zebrafish hearts at 7 dpa, but was unable to return them to the control level (Fig. 4d–j), suggesting that hyperactivation of Notch signaling contributes to defects of myocardial proliferation in DN mutant hearts. In addition, we also chose two chemical inhibitors, DAPT and MK-0752, to interfere with Notch signaling. Compared with DN hearts injected with control DMSO (Fig. 4l), we found more PCNA+/Mef2C+ proliferating cardiomyocytes in the DN hearts injected with either of the Notch inhibitors (Fig. 4m, n), which was similar to the effect of delivering siRNAs targeting all four notches (Supplementary Fig. 12), but fewer than those in Ctrl hearts injected with DMSO (Fig. 4k, o). Thus, these results suggest that abnormally activated Notch signaling is partially responsible for the cardiomyocyte-proliferation defects in the DN mutant hearts. Together, our data suggest that injury-induced endothelial Brg1 negatively regulates the level of H3K4me3 in the promoter regions of notch1b, notch2, and notch3 and thus prevents the overactivation of Notch signaling during heart regeneration. When this suppression is released, such as in DN hearts, the level of H3K4me3 modifications in the notch promoter regions is abnormally upregulated, resulting in the overactivation of Notch signaling and thus inhibiting regeneration.
Fig. 4. Endothelium-specific expression of NICD or dn-xbrg1 decreases cardiomyocyte proliferation that is partially rescued by inhibition of Notch signaling.
a, b Immunostaining showing that Mef2C+ and PCNA+ proliferating cardiomyocytes of control (a) and endothelial NICD-overexpressing heart sections (b) at 7 dpa after 4-HT induction. c Statistics of panels (a) and (b) (data are the mean fold change ± s.e.m.; **p < 0.01, unpaired t-test). d–i Representative images of immunostaining showing that, compared with control siRNA treatment (d), PCNA+/Mef2C+ proliferating cardiomyocytes decreased at 7 dpa in DN-xBrg1 hearts (DN) treated with control siRNA (e), which were partially rescued by either notch1a (f), notch1b (g), notch2 (h), or notch3 (i) siRNA treatment in the presence of 4-HT. Scale bar, 100 μm. j Statistics of panels (d–i) (data are the mean ± s.e.m.; *p < 0.05; **p < 0.01; ***p < 0.005; one-way analysis of variance [F = 32.5] followed by Dunnett’s multiple comparison test). k–n Representative images of immunostaining at 7 dpa showing that, compared with DMSO treatment (k), PCNA+/Mef2C+ proliferating cardiomyocytes in DN mutant hearts decreased (l), which were partially rescued by either DAPT (m) or MK-0752 treatment (n) in the presence of 4-HT. Scale bar, 100 μm. o Statistics of panels (k–n) (data were the mean ± s.e.m.; ***p < 0.005; one-way analysis of variance [F = 22.4] followed by Dunnett’s multiple comparison test). n number shown here (c, j, o) indicated biological replicates.
Brg1 interacts with Kdm7aa to fine-tune Notch signaling
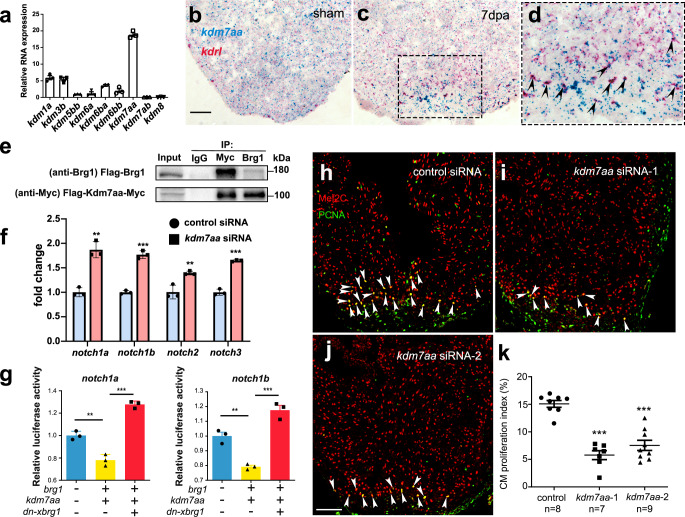
We then asked how Brg1 negatively regulated H3K4me3 modifications in the promoter regions and had its function in regulating notch receptor gene expression. It has been reported that Brg1 and histone demethylase (lysine demethylases, KDMs) jointly regulated gene expression in other organs26–28. To determine whether KDMs were involved in the regulation of the levels of H3K4me3 by Brg1, we first examined the expression pattern of KDMs during zebrafish heart regeneration. RT-PCR data revealed that kdm7aa had the strongest expression while kdm1a, kdm3b, kdm5bb, kdm6a, kdm6ba, and kdm6bb, but not kdm7ab and kdm8, were weakly expressed in injured hearts at 2 dpa (Fig. 5a). Kdm7aa has been shown to be responsible for histone demethylation at multiple sites, including H3K9, H3K27, H3K36, and H3K2029. Interestingly, we also found that kdm7aa was induced and enriched in cardiac endothelial cells upon injury using RNAscope with kdrl and kdm7aa probes (Fig. 5b–d). Therefore, we further examined the interaction between Brg1 and Kdm7aa using immunoprecipitation (IP) in 293T cells. Lysates of cells overexpressing both Flag-kdm7aa-Myc and Flag-brg1 were precipitated by either Myc or Brg1 antibodies. Western blots revealed that IP with either Myc antibody (Myc-tagged Kdm7aa) or Brg1 antibody was able to pull down both Flag-tagged Brg1 (~180 kD) and Myc-tagged Kdm7aa (~100 kD), suggesting that Brg1 physically interacted with Kdm7aa (Fig. 5e, Supplementary Fig. 13a). To examine whether Kdm7aa is involved in Brg1-regulated notch receptor gene expression, we utilized nanoparticle-mediated gene-silencing30,31 to knock down kdm7aa (Supplementary Figs. 7j, k, 8e, 9b, c, 10, and 11c, h). RT-PCR, RNAscope, and RNA in situ hybridization results showed that the downregulation of kdm7aa significantly upregulated the expression levels of notch1a, notch1b, notch2, and notch3 (Fig. 5f, Supplementary Fig. 13b–e). The IP assays performed in the 293T cells indicated the interaction between Kdm7aa and H3K4me3 (Supplementary Figs. 13f and 14a). Western blot data of H3K4me3 in zebrafish hearts revealed that H3K4me3 protein level increased after kdm7aa siRNA knockdown compared with control siRNA (Supplementary Figs. 13g and 14b). Consistently, overexpressed zebrafish kdm7aa in 293T cells decreased H3K4me3 protein level (Supplementary Figs. 13h and 14c). ChIP Q-PCR analysis showed that H3K4me3 modifications at all four notches promoter regions of hearts increased after kdm7aa siRNA knockdown compared with control siRNA (Supplementary Fig. 13i). We also used the luciferase reporter system that was driven by the notch1a or notch1b promoters, and made stable 293T cell lines expressing each of the luciferase reporters. Luciferase assays showed that overexpression of zebrafish brg1 and kdm7aa decreased notch1a and notch1b reporter activity, while overexpression of dn-xbrg1 increased the activity of notch1a and notch1b reporters (Fig. 5g), suggesting a synergistic role of Brg1 and Kdm7aa in controlling the expression levels of notch reporter genes. We finally set out to address whether kdm7aa was directly involved in regulating zebrafish heart regeneration. We found that the knockdown of kdm7aa with two independent siRNAs decreased the numbers of PCNA+/Mef2C+ proliferating cardiomyocytes compared with control siRNA (Fig. 5h–k). Together, our data suggest that endothelial cell Brg1 interacts with Kdm7aa to maintain the normal activity of notch gene promoters, and Kdm7aa modulates the level of H3K4me3 to fine-tune notch gene expression during heart regeneration.
Fig. 5. Endothelial Brg1 regulates notch receptor expression and myocardial proliferation via interaction with Kdm7aa.
a Quantitative RT-PCR of kdm genes expression, normalized by gapdh. b–d Representative images of RNAscope in situ hybridization with kdrl and kdm7aa probes (scale bar, 100 μm) and high-magnification image of boxed region in (c) (arrowheads, double kdrl- and kdm7aa-positive endothelial cells). e Immunoprecipitation (IP) assays with either anti-Myc or anti-Brg1 antibody in 293T cells. Inputs were used as loading controls and IgG as negative controls. The blots were derived from the same experiment, and they were processed in parallel. f Quantitative RT-PCR analysis showing that the expression levels of notch1a, notch1b, notch2, and notch3 from hearts at 7 dpa injected with encapsulated kdm7aa siRNA was higher compared with the control siRNA group. Data represented one of three independent experiments. Data were mean fold changes ± s.e.m., ***p < 0.005, unpaired t-test. g Luciferase reporter assays in 293T cells stably expressing the notch promoter-luciferase reporter in the pGL4.26 vector. Expression plasmid clones containing kdm7aa, brg1, or dn-xbrg1 were co-transfected into cells stably expressing each notch reporter. Data represented one of two independent experiments, ***p < 0.005, one-way analysis of variance (F = 110.6 for the left panel and 60.3 for the right panel) followed by the Bonferroni test. n number shown here (f and g) indicated technical replicates. h–j Representative images of immunostaining showing the numbers of Mef2C+/PCNA+ proliferating cardiomyocytes (arrowheads, Mef2C+/PCNA+ proliferating cardiomyocytes; scale bar, 100 μm). k Statistics of panels (h–k) (n numbers indicated biological replicates; data were the mean ± s.e.m.; ***p < 0.005; one-way analysis of variance [F = 36.79] followed by Dunnett’s multiple comparison test).
Discussion
In this study, we showed that endothelial Brg1 was required for myocardial proliferation and regeneration in zebrafish; Brg1 interacted with Kdm7aa to fine-tune the level of H3K4me3 in the notch receptor promoters and negatively regulated notch genes expression during heart regeneration; and Kdm7aa was induced in cardiac endothelial cells and was required for myocardial proliferation. Therefore, our data reveal the role of the endothelial Brg1-Kdm7aa axis in regulating notch gene transcription and the essential role of histone methylation via Kdm7aa in myocardial proliferation and regeneration in zebrafish.
Previous studies have shown that Brg1 plays an important role in oocyte genome activation, erythropoiesis, T-cell generation, erythropoiesis, vascular development, nerve development, heart development, and regeneration21,32–39. We here demonstrated that conditional inhibition of Brg1 function in endothelial cells, including the endocardium, led to increased cardiac fibrosis and compromised myocardial proliferation and regeneration. Either hypo- or hyperactivation of Notch signaling has been reported to impair cardiomyocyte proliferation and heart regeneration10–12,40, suggesting that the precise modulation of Notch family expression is essential for cardiac regeneration. Here, we present several layers of evidence to demonstrate that injury-induced Brg1 and Kdm7aa regulate notch gene expression in cardiac endothelium and endocardium. Brg1 and Kdm7aa normally fine-tune the level of the histone marker H3K4me3 in the notch gene promoters, thus preventing the abnormal hyperactivation of notch receptors after injury. When Brg1 was inhibited in cardiac endothelial cells, the H3K4me3 level increased in the notch promoter regions, and notch genes were abnormally overexpressed, leading to enhanced cardiac fibrosis and compromised myocardial proliferation and regeneration. Injury-induced expression of brg1 and kdm7aa was evident in cardiac endothelial cells that were consistent with their function, which was further supported by our data on the physical interaction between Brg1 and Kdm7aa, and their function in regulating notch promoter activities. Furthermore, either encapsulated siRNA knockdown of notch receptors, or chemical Notch inhibitors, partially rescued the phenotype of myocardial proliferation in DN-xBrg1 hearts, further suggesting an important role of Brg1 in regulating notch gene expression during heart regeneration. At the same time, how hyperactivated Notch signaling in cardiac endothelium and endocardium represses myocardial proliferation via endocardium–myocardium interaction warrants future investigations.
Chromatin remodeling has been reported to be essential for tissue/organ regeneration in urodeles and zebrafish17,18. Brg1 is the major subunit of the SWI/SNF complex and is also an important component of the trithorax group, both of which play essential roles in histone modification, such as the histone markers H3K4me3 (active) and H3K27me3 (repressive). Although data on genome-wide histone acetylation and methylation during organ regeneration are still limited, recent studies suggest that a more open chromatin state is adopted during early fin, retina, and heart regeneration in zebrafish41–43. The level of the histone marker H3K4me3 is influenced and catalyzed by lysine methyltransferases of the MLL2 complex and KDMs. Although the MLL2 complex does not provide selective specificity in a particular organ or biological process, it is believed that ATP-dependent chromatin-remodeling proteins such as Brg1 may specifically regulate the “bivalency” state of H3K4me3 and H3K27me344. KDM7 has been reported to act as a dual KDM for histone silencing markers H3K9 and H3K27 in brain development and germ cell genome stability29,45, but it is unknown whether it also works for the active histone marker H3K4. We found that brg1 and kdm7aa co-expressed in cardiac endothelial cells upon injury in zebrafish, and they formed a protein complex and functioned synergistically to regulate notch receptor gene promoters in mammalian cells. Inhibition of Brg1 function via DN-xBrg1 mutant proteins increased the notch promoter activity, suggesting that DN-xBrg1 might replace and/or inhibit Kdm7aa function, and so increased the level of H3K4me3. Our IP data suggest protein interaction between Kdm7aa and H3K4me3, and kdm7aa siRNA knockdown or overexpression respectively increased or decreased H3K4me3 modifications. Furthermore, the data on nanoparticle-mediated kdm7aa siRNA knockdown supported its function in myocardial proliferation and regeneration. Thus, this work reveals an interesting mechanism on the selective modulation of H3K4me3 by Brg1 and Kdm7aa and their essential function in zebrafish heart regeneration, even though we cannot rule out the possibility that Brg1-Kdm7aa affect the expression of the notch receptors in ways beyond H3K4me3 modifications.
Methods
Animal models
Male and female zebrafish were raised and handled according to a zebrafish protocol (IMM-XiongJW-3) approved by the Institutional Animal Care and Use Committee at Peking University, which is fully accredited by The Association for Assessment and Accreditation of Laboratory Animal Care International. Wild-type TU, Tg(kdrl:EGFP)46, Tg(kdrl:CreER)22, Tg(fli1:nucEGFP)47, Tg(tcf21:DsRed)21, Tg(myl7:cypher-EGFP)21, Tg(kdrl:mCherry)48, and Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1) zebrafish21 were maintained at 28 °C at a density of four fish per liter. Adult zebrafish were anesthetized in standard E3 medium containing 0.4% tricaine (ethyl 3-aminobenzoate methanesulfonate salt; E10521; Sigma-Aldrich) before ventricular resection as described previously21. Animals were randomized into groups for each experiment.
Wild-type C57BL/6 mice were purchased from Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). The mice used in this study were raised and handled according to the animal protocol IMM-XiongJW-4 approved by the Peking University Institutional Animal Care and Use Committee, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The mice were maintained at the humidity ranges from 40 to 70% and temperature from 21 to 23 °C and with the 12-h light/12-h dark cycle.
Construction of Tg(ubi:LoxP-DsRed-LoxP-NICD) transgenic zebrafish line
To generate the Tg(ubi:LoxP-DsRed-STOP-LoxP-NICD) zebrafish line that overexpresses NICD, a homologous recombination reaction was conducted with ubi:LoxP-DsRed-STOP-LoxP-EGFP plasmid (kindly provided by Dr. C. Geoffrey Burns at Harvard Medical School)49 by replacing EGFP with zebrafish notch1b-NICD cDNA (N1bICD). The function of N1bICD has been validated as described previously48. This Tol2-NICD plasmid was made and injected into one-cell stage wild-type embryos together with Tol2 transposase mRNA as described previously50. Heterozygous transgenic zebrafish were raised and genotyped for all experiments.
4-hydroxytamoxifen (4-HT) treatment
We generated Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:CreER) mutant (DN) and Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1) control sibling (Ctrl) adult zebrafish by crossing Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1) with Tg(kdrl:CreER) zebrafish. To induce Cre recombination, adult DN mutant and Ctrl sibling zebrafish were bathed for 24 h in the presence of 5 μM 4-HT (H7904; Sigma) made from a 10 mM stock solution dissolved in 100% ethanol at room temperature. These zebrafish were treated with 4-HT at a density of 3–4 zebrafish per 150 ml system water. Ventricular resection was performed 3 days after 4-HT treatment. Transgenic zebrafish were confirmed by PCR-based genotyping and were randomly selected for all experiments.
Ventricular resection in adult zebrafish
The ventricular resection was performed according to a well-established procedure31,51,52. Briefly, zebrafish were anesthetized with 0.4% tricaine and placed in the groove of a sponge. The pericardial sac was exposed by removing surface scales and a small piece of skin, and the ventricle apex was gently pulled up and removed with Vannas scissors. The zebrafish was quickly placed back into a system water tank, and water was puffed over the gills with a plastic pipette until it breathed and swam regularly. The surface opening sealed automatically within a few days.
Fluorescence-activated cell sorting (FACS) of cardiac endothelial cells and flow cytometry
Cardiac endothelial cells from Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:EGFP) control (CtrlK) and Tg(ubi:LoxP-DsRed-STOP-LoxP-dn-xbrg1; kdrl:CreER; kdrl:EGFP) mutant (DNK) ventricles at 7 dpa with 4-HT treatment were isolated according to an established protocol53. Briefly, ~15 adult zebrafish hearts were isolated and washed in cold PBS with 10 U/ml heparin (H8060; Solarbio). After the atrium and bulbus were removed, the ventricles were carefully cut into small pieces using forceps and collected into 1.5-ml centrifuge tubes containing cold PBS with 5 mM glucose. The sliced tissue was then transferred to a glass tube along with a magnetic stir bar and 1.5 ml digestion buffer in Dulbecco’s modified Eagle’s medium containing collagenase type II (250 U/ml) (17101015; Gibco), collagenase type IV (300 U/ml) (17104019; Gibco), and DNase I (30 μg/ml) (A3778; AppliChem). The tube was then transferred to a 32 °C water bath with stirring and incubated for 1 min. After incubation, the tube was removed from the water bath and left at room temperature until the tissue settled on the bottom. The supernatant was discarded to remove blood cells, followed by washing once with cold PBS. This was followed by a series of digestion steps with 1.5 ml digestion buffer. Each step consisted of 10 min of digestion followed by 3 min of sedimentation. The supernatants were collected in a 15-ml falcon tube containing 2 ml ice-cold PBS. The cell suspensions were centrifuged at 300 g for 5 min at 4 °C, and the cell pellets were gently resuspended in 1 ml PBS. The resuspended cells were strained using a 70-µm cell strainer and kept on ice for FACS. Cardiac endothelial cells were sorted through the GFP channel and were collected into a tube containing 0.1 ml Lysis Buffer from Magen RNA Nano Kit (R4125; Magen) for RNA isolation. Cell sorting was carried out on the Beckman Coulter MoFlo XDP, and flow cytometry was carried out on the Beckman Cytoflex EX S. Data analysis was performed by FlowJo.
RNA-seq of cardiac endothelial cells
The RNA of heart endothelial cells from CtrlK sibling and DNK mutant ventricles at 7 dpa was purified using a Magen RNA Nano Kit (R4125; Magen). About 30 ng of total RNA was used for next-generation library preparation under the guidelines of the NEBNext Ultra DNA Library Prep Kit for Illumina (E7370; NEB). The libraries were loaded for 2 × 150 bp pair-end sequencing using Illumina Hiseq 2500. Raw reads were preprocessed and quality controlled with FastQC (The Bioinformatics Group, Babraham Institute; Version: 0.11.9). Reads for each library were mapped using HISAT2 (Version: 2.2.1)54 against the zebrafish reference genome assembly GRCz11 with default parameters. Uniquely mapped reads were extracted to calculate the read counts of each gene, using the matching gene annotation (http://ftp.ensembl.org/pub/release-103/gtf/danio_rerio/Danio_rerio.GRCz11.103.gtf.gz; v. 103) from Ensembl with FeatureCounts (Version: 2.0.1)55. Genes were further filtered, and those with low expression in all samples (FPKM < 0.5 in all samples) were removed from differential gene expression analysis. Differential analysis was conducted with DEseq256. Genes with an adjusted p-value <0.05 were taken as significantly differentially expressed genes in the DNK condition compared with CtrlK. FPKM values were calculated with Stringtie (Version: 2.1.5)57, and normalized Z-score values were used to draw heatmaps using the ComplexHeatmap R package58. Gene ontology enrichment analysis for the differentially expressed genes was performed with clusterProfiler tool (R package, version: 3.18.1)59,60. All expressed genes in either CtrlK or DNK condition were used as the background. GO terms with BH adjusted p-value <0.01 were taken as enriched GO terms. The top ten enriched GO terms were shown as barplot for biological process, cellular component, and molecular function. Sequencing data have been deposited in GEO under accession code GSE200936.
ChIP-seq
ChIP-seq libraries were prepared using the VAHTS Universal Pro DNA Library Prep Kit (ND608; Vazyme) for Illumina. Five nanograms of DNA was used as starting material for input and IP samples. Libraries were amplified using 13 cycles on the thermocycler. Post-amplification libraries were size selected at 250–450 bp in length using Agencourt AMPure XP beads (A63880; Beckman Coulter). Libraries were validated using the High Sensitivity DNA Kit (5067-4626; Agilent) and loaded for pair-end sequencing using Illumina NovaSeq 6000. Trimmomatic tool (https://github.com/usadellab/Trimmomatic; Version: 0.39) was used to trim reads with a quality drop below a mean of Q15 in a window of 5 nucleotides and reads with length below 15 nucleotides were filtered out. After the quality control step, the trimmed and filtered reads were aligned to the Zebrafish reference genome GRCz11 using STAR (https://github.com/alexdobin/STAR; Version: 2.7.8a) with the parameters “--outFilterMismatchNoverLmax 0.2-outFilterMatchNmin 20 --alignIntronMax 1 --outFilterMultimapNmax 1” to retain only unique alignments. Reads were deduplicated using Picard (Broad Institute; Version: 2.25.0) to remove PCR artifacts. Since the numbers of H3K4me3 peaks may be affected by the sequencing depths, we used the same number of reads (17.5 million pairs) randomly selected from samples of each condition for downstream analysis. The MACS2 peak caller (https://github.com/macs3-project/MACS; Version: 2.2.7.1) was employed for each condition with parameters “-q 0.0001 -broad -nomodel -nolambda”. Peaks not located in defined chromosomes were further removed. The filtered peaks were used to do the downstream analysis. Intersection between peaks in CtrlK and DNK conditions was performed with Bedtools toolkit (Version: 2.30.0)61. Normalized read coverages and subtraction of read coverage were calculated with deeptools toolkit (Version: 2.5.3)62. ChIPseeker63 was performed to display the genomic distribution of H3K4me3 peaks based on the matching gene annotation (v. 103) from Ensembl. Peaks for different groups were annotated with ChIPseeker package (version: 1.26.2)63. GO enrichment analysis was further performed for genes closely located near peaks of each group with clusterProfiler package (version: 3.18.1). The H3K4me3 ChIP-seq traces were represented in IGV (Integrative Genomics Viewer) browser64. Sequencing data have been deposited in GEO under accession code GSE200937.
Quantitative RT-PCR analysis
For FACS-sorted cardiac endothelial cells, RNA from CtrlK sibling and DNK mutant ventricles at 7 dpa was purified using a Magen RNA Nano Kit (R4125; Magen). About 20 ng RNA was used for reverse transcription with MALBAC RNA amplification Kit (KT110700424; YIKON GENOMICS)65. For RNA extraction from whole hearts, an RNeasy Mini Kit (74106; Qiagen) was used to purify RNA, and 500 ng RNA was used for reverse transcription with a Prime Script RT Reagent Kit (RR037A; Takara). Quantitative PCR was performed using TB Green Premix DimerEraser Kit (RR091A; Takara) or ChamQ Universal SYBR qPCR Master Mix (Q711; Vazyme). The primer sequences are listed in Supplementary Table 1.
Delivery of chemical Notch inhibitors and siRNAs into adult zebrafish heart
siRNAs were encapsulated in nanoparticles and then injected into the pericardial sac as described previously30,31,66,67. To evaluate the effect of siRNA-mediated rescue on cardiomyocyte proliferation, 10 μl polyethylene glycol-polylactic acid nanoparticle-encapsulated siRNAs was injected into the pericardial sac daily from 2 to 7 dpa. The Notch inhibitors MK-0752 (HY-10974; MCE) and DAPT (A07D5942; Sigma) were first dissolved in DMSO to make a 20 mM stock solution. Before injection, the stock was diluted to the working concentration (30 μM), and 10 μl of diluted inhibitor was injected daily from 4 to 6 dpa. The injected hearts at 7 dpa were then collected for subsequent experiments. siRNA sequences for notch1a, notch1b, notch2, notch3, and kdm7aa are listed in Supplementary Table 2.
RNAscope and RNA in situ hybridization, immunostaining, and histology
RNAscope (Advanced Cell Diagnostics, Hayward, CA) was applied to 10-μm sections from freshly frozen hearts embedded in Optimal Cutting Temperature (OCT) compound (4583; Sakura). Fresh tissue was fixed in 10% prechilled neutral buffered formalin in 1 × PBS at 4 °C, followed by dehydration, and then treated with RNAscope® hydrogen peroxide (in RNAscope Universal Pretreatment Kit; 322380; ACD) for 10 min at room temperature. The slides were washed with water and incubated with RNAscope Protease IV (in RNAscope Universal Pretreatment Kit; 322380; ACD) for 30 min at room temperature. Then, they were washed five times in 1 × PBS, and the RNAscope® 2.5 HD Duplex Detection Kit (322430; ACD) was applied to visualize hybridization signals. The RNAscope Probes used in this work included RNAscope Probe-Dr-smarca4a (457431; ACD), RNAscope Probe-Dr-kdm7aa (822391; ACD), RNAscope Probe-Dr-notch1b (431941; ACD), RNAscope Probe-Dr-notch2 (590431, ACD), RNAscope Probe-Dr-kdrl-C2 (416611-C2; ACD), RNAscope Probe-Dr-tcf21-C2 (485341-C2; ACD), and RNAscope Probe-Dr-coro1a-C2 (496571-C2; ACD). Three injured and sham-operated hearts were used for each RNAscope in situ hybridization.
RNA in situ hybridization was performed on 10-μm sections from fixed frozen hearts embedded in OCT compound. To generate RNA probes, we amplified notch1a, notch1b, notch2, and notch3 cDNA from regenerating hearts at 7 dpa, blunt-ligated cDNA into a pEASy-Blunt vector, and generated digoxigenin-labeled RNA probes using T7 RNA polymerases (M0251; NEB). In situ hybridization was performed on cryosections of 4% paraformaldehyde-fixed hearts as previously described68.
For immunofluorescence staining, adult zebrafish hearts were fixed in 4% paraformaldehyde at room temperature for 2 h, dehydrated, and embedded in paraffin and sectioned at 5 μm. The sections were dewaxed, rehydrated, and washed in 1 × PBS. The antigens were repaired with the citric acid buffer (CW0128S; CWBIO). After washing, the sections were blocked in 10% FBS in PBST (1% Tween 20 in PBS) and then incubated with diluted primary antibodies (in PBST containing 10% FBS) overnight at 4 °C. The primary antibodies used for immunofluorescence were anti-Mef2c (HPA005533; Sigma; 1:200), anti-GFP (A-11122; Invitrogen; 1:500), anti-PCNA (P8825; Sigma; 1:300), anti-myosin heavy-chain monoclonal antibody (14-6503-82; eBioscience; 1:500), anti-DsRed (BE3307; EASYBIO; 1:300) and the Brg1 antibody (1:250), which was raised against a glutathione S-transferase-BRG1 fusion protein (human BRG1 amino-acids 1086–1307)69,70. After washing, the sections were incubated with secondary antibodies for 2 h at room temperature. The secondary antibodies (1:300 diluted in PBST containing 10% FBS) were Alexa Fluor 488 goat anti-mouse IgG (A21121; Invitrogen; 1:300), Alexa Fluor 488 goat anti-rabbit IgG (A11034; Invitrogen; 1:300), Alexa Fluor 555 goat anti-mouse IgG (A21424; Invitrogen; 1:300), and Alexa Fluor 555 goat anti-rabbit IgG (A21428; Invitrogen; 1:300).
RNA and RNAscope in situ hybridization was examined under a DM5000B microscope (Leica, Germany); immunofluorescence images were captured on a confocal microscope (LSM510 or LSM980; Carl Zeiss, Germany). The signal intensity or area was quantified using MBF ImageJ (https://imagej.nih.gov/ij/)71.
Acid fuchsin orange G staining (AFOG)
AFOG staining was performed on paraffin sections following the manufacturer’s instructions51. The sections were incubated in Bouin’s solution (HT10132; Sigma) at 56 °C for 2.5 h, and at room temperature for 1 h, washed in tap water, incubated in 1% phosphomolybdic acid (P4869; Sigma) for 5 min, washed with water, and then stained with AFOG solution consisting of 3 g acid fuchsin (F8129; Sigma), 2 g orange G (O3756, Sigma), and 1 g aniline blue (AB0083; BBI) dissolved in 200 ml acidified distilled water (pH 1.1) for 10 min. The sections were rinsed with distilled water, dehydrated, mounted, and staining was photographed under a DM5000B microscope (Leica, Germany).
Chromatin immunoprecipitation (ChIP) and quantitative ChIP (qChIP)
About 25 zebrafish hearts were pooled for each ChIP experiment. The hearts were dissected from adult zebrafish, and the outflow tract and atrium were removed. Chromatin isolation and ChIP assays were performed using a Pierce Magnetic ChIP Kit (26157; Pierce). Anti-Brg1 (5 µl for each reaction) and anti-H3K4me3 (Ab8580; Abcam; 5 µl for each reaction) antibodies were used for the ChIP assays. The Brg1 antibody used here was a mouse polyclonal antibody raised against glutathione S-transferase (GST)-tagged zebrafish Brg1 amino acid 1098–1286 that was made in our laboratory. The purified antigen protein was provided by HUABIO, HangZhou. Mice were inoculated with purified antigens, and their serum was collected as polyclonal antibodies. The DNA bound by ChIP was used for library construction and quantitative PCR. The primer sequences are listed in Supplementary Table 2.
Immunoprecipitation (IP)
The full-length coding cDNA of zebrafish kdm7aa was isolated from the regenerating heart cDNA library and cloned into the pcDNA3.1 vector. For co-IP, 293T cells (CRL-1573; American Type Culture Collection) were transfected with pcDNA3.1-Flag-kdm7aa-Myc and pcDNA3.1-Flag-brg1 using Fugen HD Transfection Reagents (2311; Promega), and after 48 h the transfected cells were lysed in NP-40 lysis buffer (P0013F; Beyotime). After brief centrifugation, the supernatants were collected for immunoprecipitation, while a protein extraction fraction was set aside for input controls. Equal volumes of supernatants were incubated overnight with either anti-Myc (AT0023; Engibody; 5 µg for each reaction), anti-Flag (F3165; Sigma; 5 µg for each reaction), anti-IgG (PI31160; Thermo Fisher Scientific; 5 µg for each reaction), and anti-Brg1 (10 µl for each reaction). The next morning, 25 μl of Pierce Protein A/G Magnetic Beads (88802; Pierce) were added and incubated with the IP mixture for 2 h at room temperature. The beads were then washed for 5 min and repeated three times in IP wash buffer (30 mM HEPES, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, pH 7.5), and were subsequently eluted with 1× loading buffer with heating at 100 °C for 10 min. The Brg1 antibody used here was a mouse polyclonal antibody raised against GST-tagged zebrafish Brg1 amino acid 1098–1286, as described above.
Zebrafish notch promoter-luciferase assays
The promoter sequences of notch receptors were cloned into the luciferase reporter vector pGL4.26, with the notch1a promoter (from 171 bp to +3 bp) and notch1b promoter (from −41 bp to +58 bp), of which the ATG was considered to be +1 bp. Stable 293T cell lines (CRL-1573; American Type Culture Collection) for each of the four notch reporters were generated in the presence of 150 μg/ml hygromycin B. Isolated reporter cells for each of the notch receptors were co-transfected with pcDNA3.1-brg1, pcDNA3.1-kdm7aa, pcDNA3.1-dn-xbrg1, and pREP4-Renilla. Luciferase assays were carried out at 48 h after infection following the manufacturer’s instructions with the Dual-luciferase Reporter Assay System (E1910; Promega). Firefly luciferase activity was normalized by Renilla luciferase activity.
Statistical analysis
All statistics were calculated using Statistical Product and Service Solutions (SPSS) software from IBM or GraphPad Prism. The statistical significance of differences between the two groups was determined using the independent unpaired t-test, with two-tailed p-values, and the data are reported as the mean ± s.e.m. Among three or more groups, one-way analysis of variance followed by Bonferroni’s multiple comparison test or Dunnett’s multiple comparison test was used for comparisons.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors thank Dr. I.C. Bruce (Guest Professor at the Institute of Molecular Medicine, Peking University) for commenting and revising the manuscript, and members of Dr. Jing-Wei Xiong’s and Dr. Yan-Yi Huang’s laboratories for helpful discussions and technical assistance. The authors acknowledge Chenyang Geng, Yun Zhang, Jing Sun, and Yang Xu for technical assistance at the Beijing Advanced Innovation Center of Genomics and the High-Throughput Sequencing Center at Peking University; Fangjin Chen, Ting Fang, and Wenzhong Zhang for technical help at the Computing Platform of the Center for Life Sciences of Peking University; Li-Ying Du and Hong-Xia Lv for technical help at the Flow Cytometry Core at the National Center for Protein Sciences at Peking University; Liqin Fu for technical help at the Optical Imaging Group of Core Facility, School of Life Sciences and National Biomedical Imaging Center of Peking University. This work was supported by the National Key R&D Program of China (2019YFA0801602 and 2018YFA0800501) and grants from the National Natural Science Foundation of China (32230032, 31701272, 31730061, 31430059, and 81870198). C.X. was supported in part by postdoctoral fellowships from Peking University Boya Program and Peking-Tsinghua Center for Life Sciences.
Author contributions
C.X. and J.H. performed most of the experiments, analyzed the data, and wrote the manuscript; W.D. analyzed RNA-seq and ChIP-seq data and contributed manuscript writing; F.W. made encapsulated siRNAs and analyzed the data; J.Z., Y.S., and J.W. contributed to design and analysis of RNA-seq experiments; L.L. contributed essential transgenic zebrafish lines; X.Z. supervised this work, analyzed data, and contributed manuscript writing; and J.-W.X. conceived this work, supervised this work, analyzed data, and wrote the manuscript.
Data availability
Sequencing data that support the findings of this study were deposited into the GEO with the accession codes GSE200936 and GSE200937. The non-sequencing data and/or materials generated during this study are available from J.-W.X. upon request.
Code availability
Only public software and custom code were used to generate or process datasets in this work. The software version, references, and parameters were well described in the “Methods” section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chenglu Xiao, Junjie Hou.
Contributor Information
Wanqiu Ding, Email: dingwq@pku.edu.cn.
Xiaojun Zhu, Email: zhuxiaojun@pku.edu.cn.
Jing-Wei Xiong, Email: jingwei_xiong@pku.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41536-023-00293-4.
References
- 1.Hesse M, Welz A, Fleischmann BK. Heart regeneration and the cardiomyocyte cell cycle. Pflugers Arch. 2018;470:241–248. doi: 10.1007/s00424-017-2061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadek H, Olson EN. Toward the goal of human heart regeneration. Cell Stem Cell. 2020;26:7–16. doi: 10.1016/j.stem.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzahor E, Poss KD. Cardiac regeneration strategies: staying young at heart. Science. 2017;356:1035–1039. doi: 10.1126/science.aam5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611–620. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pronobis MI, Poss KD. Signals for cardiomyocyte proliferation during zebrafish heart regeneration. Curr. Opin. Physiol. 2020;14:78–85. doi: 10.1016/j.cophys.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl Acad. Sci. USA. 2003;100:11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L, et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl Acad. Sci. USA. 2014;111:1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Ben-Yair R, Burns CE, Burns CG. Endocardial Notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through Wnt pathway antagonism. Cell Rep. 2019;26:546–554.e545. doi: 10.1016/j.celrep.2018.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng L, et al. Molecular regulation of myocardial proliferation and regeneration. Cell Regen. 2021;10:13. doi: 10.1186/s13619-021-00075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Fan L, Zhao L, Su Y. The interaction of Notch and Wnt signaling pathways in vertebrate regeneration. Cell Regen. 2021;10:11. doi: 10.1186/s13619-020-00072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan EM, Sanchez Alvarado A. Regulation of genomic output and (pluri)potency in regeneration. Annu. Rev. Genet. 2019;53:327–346. doi: 10.1146/annurev-genet-112618-043733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Redondo P, Izpisua Belmonte JC. Tailored chromatin modulation to promote tissue regeneration. Semin. Cell Dev. Biol. 2020;97:3–15. doi: 10.1016/j.semcdb.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, Xiao C, Xiong JW. Epigenetic regulation of organ regeneration in zebrafish. J. Cardiovasc. Dev. Dis. 2018;5:57. doi: 10.3390/jcdd5040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oyama K, El-Nachef D, Zhang Y, Sdek P, MacLellan WR. Epigenetic regulation of cardiac myocyte differentiation. Front. Genet. 2014;5:375. doi: 10.3389/fgene.2014.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao C, et al. Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat. Commun. 2016;7:13787. doi: 10.1038/ncomms13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan Y, et al. The caudal dorsal artery generates hematopoietic stem and progenitor cells via the endothelial-to-hematopoietic transition in zebrafish. J. Genet. Genomics. 2018;45:315–324. doi: 10.1016/j.jgg.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Menon DU, Shibata Y, Mu W, Magnuson T. Mammalian SWI/SNF collaborates with a polycomb-associated protein to regulate male germline transcription in the mouse. Development. 2019;146:dev174094. doi: 10.1242/dev.174094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu W, Xu X, Wang X, Liu J. Reprogramming histone modification patterns to coordinate gene expression in early zebrafish embryos. BMC Genomics. 2019;20:248. doi: 10.1186/s12864-019-5611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vastenhouw NL, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, et al. Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/beta-catenin pathway in mice. FASEB J. 2019;33:327–338. doi: 10.1096/fj.201800197R. [DOI] [PubMed] [Google Scholar]
- 27.Liu CC, et al. Regulation of KDM2B and Brg1 on Inflammatory response of nasal mucosa in CRSwNP. Inflammation. 2019;42:1389–1400. doi: 10.1007/s10753-019-01000-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. An interaction between BRG1 and histone modifying enzymes mediates lipopolysaccharide-induced proinflammatory cytokines in vascular endothelial cells. J. Cell. Biochem. 2019;120:13216–13225. doi: 10.1002/jcb.28595. [DOI] [PubMed] [Google Scholar]
- 29.Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–437. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diao J, et al. PEG-PLA nanoparticles facilitate siRNA knockdown in adult zebrafish heart. Dev. Biol. 2015;406:196–202. doi: 10.1016/j.ydbio.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Xiao C, et al. Nanoparticle-mediated siRNA gene-silencing in adult zebrafish heart. J. Vis. Exp. 2018;137:e58054. doi: 10.3791/58054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bultman SJ, et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi TH, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/S1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 35.Eroglu B, Wang G, Tu N, Sun X, Mivechi NF. Critical role of Brg1 member of the SWI/SNF chromatin remodeling complex during neurogenesis and neural crest induction in zebrafish. Dev. Dyn. 2006;235:2722–2735. doi: 10.1002/dvdy.20911. [DOI] [PubMed] [Google Scholar]
- 36.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 39.Stankunas K, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munch J, Grivas D, Gonzalez-Rajal A, Torregrosa-Carrion R, de la Pompa JL. Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development. 2017;144:1425–1440. doi: 10.1242/dev.143362. [DOI] [PubMed] [Google Scholar]
- 41.Goldman JA, et al. Resolving heart regeneration by replacement histone profiling. Dev. Cell. 2017;40:392–404.e395. doi: 10.1016/j.devcel.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart S, Tsun ZY, Izpisua Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl Acad. Sci. USA. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, et al. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science. 2020;369:eaaz3090. doi: 10.1126/science.aaz3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harikumar A, Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015;16:1609–1619. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myers TR, Amendola PG, Lussi YC, Salcini AE. JMJD-1.2 controls multiple histone post-translational modifications in germ cells and protects the genome from replication stress. Sci. Rep. 2018;8:3765. doi: 10.1038/s41598-018-21914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 47.Roman BL, et al. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development. 2002;129:3009–3019. doi: 10.1242/dev.129.12.3009. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Q, et al. Proprotein convertase furina is required for heart development in zebrafish. J. Cell Sci. 2021;134:jcs258432. doi: 10.1242/jcs.258432. [DOI] [PubMed] [Google Scholar]
- 49.Mosimann C, et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Han P, et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 2014;24:1091–1107. doi: 10.1038/cr.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 53.Patra C, et al. The zebrafish ventricle: a hub of cardiac endothelial cells for in vitro cell behavior studies. Sci. Rep. 2017;7:2687. doi: 10.1038/s41598-017-02461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 59.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu T, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 64.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chapman AR, et al. Single cell transcriptome amplification with MALBAC. PLoS ONE. 2015;10:e0120889. doi: 10.1371/journal.pone.0120889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, et al. Functionalized dendrimer-based delivery of angiotensin type 1 receptor siRNA for preserving cardiac function following infarction. Biomaterials. 2013;34:3729–3736. doi: 10.1016/j.biomaterials.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang XZ, et al. Systemic delivery of siRNA with cationic lipid assisted PEG-PLA nanoparticles for cancer therapy. J. Control. Release. 2011;156:203–211. doi: 10.1016/j.jconrel.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 68.Liu KL, Wang XM, Li ZL, He RQ, Liu Y. In situ hybridization and immunostaining of Xenopus brain. Methods Mol. Biol. 2014;1082:129–141. doi: 10.1007/978-1-62703-655-9_9. [DOI] [PubMed] [Google Scholar]
- 69.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 70.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. doi: 10.1002/j.1460-2075.1996.tb00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data that support the findings of this study were deposited into the GEO with the accession codes GSE200936 and GSE200937. The non-sequencing data and/or materials generated during this study are available from J.-W.X. upon request.
Only public software and custom code were used to generate or process datasets in this work. The software version, references, and parameters were well described in the “Methods” section.