Key Points
-
•
Whole genome sequencing of 36 Hodgkin lymphoma (HL) families identifies 33 coding and 11 noncoding HL-risk variants.
-
•
Recurrent damaging variants are observed in known (KDR and KLHDC8B) and novel (PAX5, GATA3, and POLR1E) predisposing loci.
Visual Abstract
Abstract
Familial aggregation of Hodgkin lymphoma (HL) has been demonstrated in large population studies, pointing to genetic predisposition to this hematological malignancy. To understand the genetic variants associated with the development of HL, we performed whole genome sequencing on 234 individuals with and without HL from 36 pedigrees that had 2 or more first-degree relatives with HL. Our pedigree selection criteria also required at least 1 affected individual aged <21 years, with the median age at diagnosis of 21.98 years (3-55 years). Family-based segregation analysis was performed for the identification of coding and noncoding variants using linkage and filtering approaches. Using our tiered variant prioritization algorithm, we identified 44 HL-risk variants in 28 pedigrees, of which 33 are coding and 11 are noncoding. The top 4 recurrent risk variants are a coding variant in KDR (rs56302315), a 5′ untranslated region variant in KLHDC8B (rs387906223), a noncoding variant in an intron of PAX5 (rs147081110), and another noncoding variant in an intron of GATA3 (rs3824666). A newly identified splice variant in KDR (c.3849-2A>C) was observed for 1 pedigree, and high-confidence stop-gain variants affecting IRF7 (p.W238∗) and EEF2KMT (p.K116∗) were also observed. Multiple truncating variants in POLR1E were found in 3 independent pedigrees as well. Whereas KDR and KLHDC8B have previously been reported, PAX5, GATA3, IRF7, EEF2KMT, and POLR1E represent novel observations. Although there may be environmental factors influencing lymphomagenesis, we observed segregation of candidate germline variants likely to predispose HL in most of the pedigrees studied.
Familial cases of Hodgkin lymphoma (HL) have long been recognized, but there is ongoing debate as to whether this reflects genetic predisposition or shared environmental factors. Flerlage et al report on whole genome sequencing of 234 individuals from 36 pedigrees with multiple first-degree relatives with HL identifying 44 HL risk variants in 28 pedigrees. Four variants were recurrent, but the others were private, occurring in only 1 family. This suggests that germline variants play a major role in familial recurrence of HL.
Introduction
Hodgkin lymphoma (HL) is a rare cancer of the lymph nodes that comprises ∼40% to 50% of all lymphomas with a unique distribution that differs geographically and ethnically.1 There is a known bimodal distribution of age at onset; both adolescents and young adults aged between 15 and 39 years and people aged >55 years are more affected.1 This signifies that there may be unique causes of disease at differing age of onset with the younger peak more likely to represent genetic predisposition than lifetime exposures.2 Known risk factors for the development of HL include age, male sex, higher socioeconomic status, smaller family size, living in westernized countries, and familial history of HL.1,3 A twofold to sixfold increased risk of developing HL has been reported for first-degree relatives of probands,4, 5, 6, 7 which is among the highest reported for all cancers.4 A twin study demonstrated an increased risk for the development of HL in monozygotic twins but no increased risk for dizygotic twins,3 pointing toward a contribution by genetics vs environmental factors. Importantly, the 20 known cases of HL in monozygotic twins occurred before the age of 50 years (mean age, 25.5 years), in alignment with the hypothesis that genetic susceptibility to developing HL may be more prevalent among individuals diagnosed in the younger adolescents and young adults peak.
In contrast with other hematological malignancies, the pathogenetic mechanisms responsible for HL formation are largely unknown and few genomic aberrations have been described thus far.8, 9, 10, 11 Genome-wide association studies (GWAS) of HL indicate a role for common genetic variation in the HLA region at 6p21.3212,13 and non-HLA loci including TCF3, REL, GATA3, and IL13.14,15 Whole exome sequencing (WES) studies from the National Cancer Institute (NCI) familial HL study 02-C-0210 identified rare coding variants segregating in families, including a missense variant in KDR16 segregating with disease in 2 pedigrees and 2 POT1 variants17 in separate families. Whole genome sequencing (WGS) identified a nonsynonymous variant in DICER1 and showed downregulation of tumor suppressor microRNAs in carriers.18,19 In addition, a homozygous 56–base pair deletion (c.2836_2892del) in ACAN was found to segregate in a Middle Eastern family.20 Furthermore, variants in KLHDC8B include a highly penetrant translocation between chromosomes 2 and 3 that removes the 5′ untranslated region (UTR) in one pedigree and a 5′ UTR single nucleotide variant (SNV) (rs387906223) found to segregate in 3 HL pedigrees, which reduces translation of KLHDC8B.21
To identify novel rare variants predisposing to HL susceptibility, we performed WGS of 36 pedigrees containing ≥2 individuals with HL, where at least 1 individual was diagnosed at the age of ≤21 years. This young age at onset is a novel approach selected to increase the probability of a genetic underpinning vs lifetime exposures for the development of HL. A subset of 23 pedigrees from NCI familial study 02-C-0210 meeting eligibility criteria were included in our study. This allowed for expansion of additional pedigree members, application of WGS, and interrogation of noncoding regions to build on previous WES efforts by Goldin, McMaster et al in the same pedigrees.16 The WGS data were analyzed with rigorous pipelines developed at St. Jude Children’s Research Hospital (SJCRH) to perform family-based segregation for the identification of coding, noncoding, and copy number variants (CNVs) using linkage and filtering approaches. Our findings and the data set amassed are from the largest cohort of familial HL with WGS to date.
Methods
Eligible study participants
We included patients diagnosed with HL at the age of ≤21 years with at least 1 first-degree relative also affected by HL regardless of age and all affected or unaffected relatives who were referred to SJCRH. The research participant or legal guardian provided informed consent for this protocol. In addition, DNA of individuals from 23 families from the Institutional Review Board–approved NCI study (NCT00039676)16,22 met the eligibility criteria and were included. The study was approved by the SJCRH IRB (NCT02795013, NCT03050268) and the Women's and Children's Health Network Human Research Ethics Committee (Adelaide, SA, Australia; 2020/HRE00981).
Sample preparation and sequencing
Samples were obtained from peripheral blood with DNA extraction per institutional protocol, and WGS was performed using a HudsonAlpha Sequencing Core. See supplementary Material for details, including quality control (QC), which are available on the Blood website.
Prioritization of coding and noncoding SNVs/indels
We developed the familial variant prioritizer (FAMVP) pipeline23 for annotation and prioritization of coding and noncoding SNVs and insertion/deletions (indels) from WGS and applied it to the 36 HL pedigrees. FAMVP checks for segregation of variants in affected individuals and obligate carriers within each pedigree were performed using SLIVAR.24 Penetrance was estimated using Bayes risk estimation (formula 2) in the study by Wang et al.25 Variants were not filtered out if they were present in unaffected individuals unless that individual was an unaffected spouse or if the estimated penetrance was negative (>50% of unaffected siblings were carriers). QC filters were applied as hard filtering thresholds (minimum genotype quality ≥ 20, minimum mean coverage depth ≥ 10, Hardy-Weinberg equilibrium P < 1 × 10−6, and maximum missing genotypes 10%).26, 27, 28, 29, 30 ANNOVAR31 was incorporated to functionally annotate variants. Splicing variants were defined as those within 4 nucleotides of an annotated splice junction and annotated based on predicted impact on splicing using SpliceAI32 and dbscSNV.33 The prioritization workflow used an investigator-derived candidate gene list comprising 160 cancer predisposition genes34, 35, 36, 37 and 242 genes associated with HL; the HL-associated gene list was formulated from the Harmonizome38 database using the search term “Hodgkin,” the Cancer GeneticsWeb database,39 published literature, and 3 investigator-selected genes (supplemental Figure 1; supplemental Table 1).
Scoring of coding and noncoding variants
Coding variants were scored into 4 priority levels C1 to C4 defined based on presence within a predefined candidate gene, annotation as loss-of-function (LOF) (nonsense, frameshift, and splice), and predicted deleteriousness from REVEL26 and CADD27 using the developer-recommended thresholds of 0.5 and 20, respectively (Figure 1A). Noncoding variants were scored into 8 priority levels NC1 to NC8 based on overlap with DNase sequencing or transcription factor chromatin immunoprecipitation sequencing clusters from Encode340 with increased weight for the GM12878 cell line, RegBase30 model scores >10, noncoding RNA overlap, and proximity to genes on our candidate list.30,41 Variants with C1 to C3 and NC1 and NC2 priority level scores were further reviewed. FIMO42 from the meme-suite of tools (version 5.1.0) was used to predict differences in occurrences of known motifs (JASPAR2018_CORE_vertebrates_non-redundant) between reference and alternative alleles with ± 30 base pairs of context sequence.
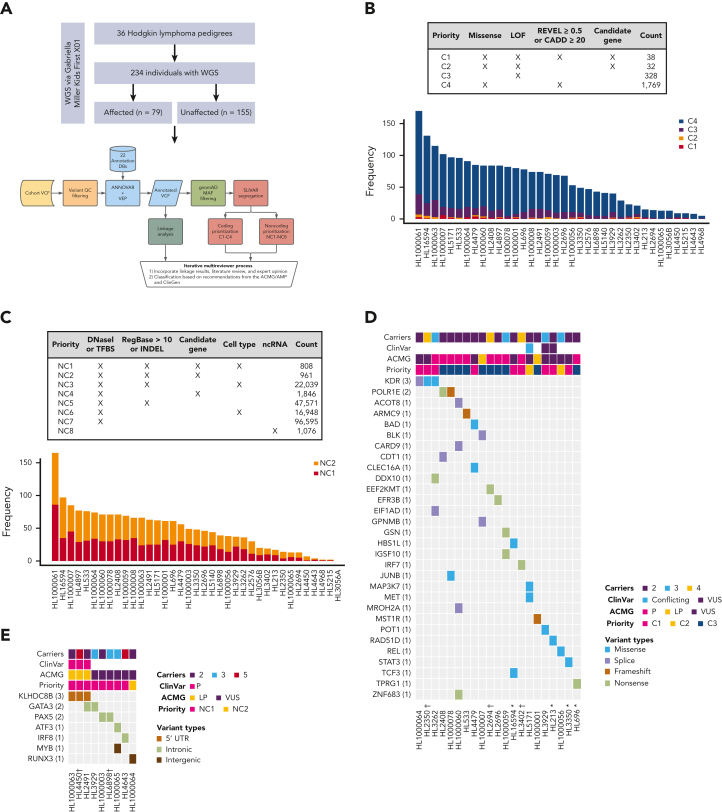
Figure 1.
Families with putative germ line variants. (A) Study overview and prioritization schema. (B) Counts of coding variants according to prioritization category overall and per pedigree. (C) Heat map of coding variants showing the number of carriers across pedigrees, the ClinVar rating, the variant classification based on American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) guidelines, and taking into account evidence of familial segregation and a possible predisposition to HL and the priority level based on our prioritization schema as described in Panel A. (D) Counts of noncoding variants according to prioritization category overall and per pedigree. (E) Heat map of noncoding variants showing recurrence across pedigrees. ncRNA, noncoding RNA; ∗pedigrees with early-onset; † the variant is within 1 logarithm of the odds (1-LOD) multipoint logarithm of the odds (MLOD) region.
Linkage and CNV analysis
Pedigree-specific multipoint parametric linkage analysis was performed using Merlin43 under an autosomal dominant inheritance model with 90% penetrance and an HL disease allele frequency of 0.00067 (ie, the prevalence of 219,128 HL cases in 2018 within the United States).44 Unaffected siblings were coded as unknown. Pedigree-specific maximum MLOD regions were defined. Segregating coding and noncoding variants falling within the 1-LOD support interval of the MLOD are indicated in Table 1.
Table 1.
Prioritized germline HL candidates
| Family | Sample count | Affected plus obligate carriers | Early-onset family | Within 1-LOD MLOD region | ACMG evidence | Pathogenicity | Priority | Gene | Type | HGVS |
|---|---|---|---|---|---|---|---|---|---|---|
| HL1000001 | 4 | 2 | PVS1∗, PP1∗ | LP∗, VUS | C3 | MST1R | Coding | NM_002447.4:c.697delinsCA (p.V233Cfs∗16) | ||
| HL1000003 | 4 | 2 | PM2_supporting, PP1_moderate∗, PP3 | VUS, VUS∗ | NC1 | PAX5 | Noncoding | NM_016734.3:c.605-2310C>T | ||
| HL1000007 | 4 | 2 | PVS1∗, PP1∗ | LP∗, VUS | C3 | GPNMB | Coding | NM_002510.3:c.367+2T>C | ||
| HL1000007 | 4 | 2 | PVS1∗, PP1∗ | LP∗, VUS | C3 | BLK | Coding | NM_001715.3:c.369-2A>G | ||
| HL1000056 | 4 | 3 | BP4, PP1∗ | VUS∗, VUS | C2 | REL | Coding | NM_001291746.2:c.920A>G (p.H307R) | ||
| HL1000059 | 4 | 3 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | IGSF10 | Coding | NM_178822.5:c.3296C>G (p.S1099∗) | ||
| HL1000059 | 4 | 3 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | GSN | Coding | NM_198252.3:c.1662G>A (p.W554∗) | ||
| HL1000060 | 4 | 2 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | CARD9 | Coding | NM_052813.5:c.184+1G>A | ||
| HL1000060 | 4 | 2 | BP4, PP1∗ | VUS∗, VUS | C3 | MROH2A | Coding | NM_001394639.1:c.4452+1G>A | ||
| HL1000060 | 4 | 2 | PVS1∗, PP1∗ | LP∗, VUS | C3 | ACOT8 | Coding | NM_005469.4:c.488+1G>A | ||
| HL1000060 | 4 | 2 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | ZNF683 | Coding | NM_001114759.3:c.103C>T (p.R35∗) | ||
| HL1000063 | 3 | 2 | PS3_supporting, PP1_strong∗, PP3 | LP∗, VUS | NC1 | KLHDC8B∗ | Noncoding | NM_173546.3:c.-158C>T | ||
| HL1000064 | 5 | 2 | PVS1_moderate, PM2_supporting, PP1 | VUS | C1 | KDR∗ | Coding | NM_002253.4:c.3849-2A>C | ||
| HL1000064 | 5 | 2 | PP1∗, PP3 | VUS∗, VUS | NC1 | RUNX3 | Noncoding | NM_004350.3:c.-61138C>T | ||
| HL1000065 | 7 | 3 | PP1∗, PP3 | VUS∗, VUS | NC1 | MYB | Noncoding | NM_001130173.2:c.-4939del | ||
| HL1000065 | 7 | 3 | PP1∗, PP3 | VUS∗, VUS | NC1 | ATF3 | Noncoding | NM_001674.4:c.-5+978A>C | ||
| HL1000078 | 5 | 2 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | POLR1E | Coding | NM_022490.4:c.847del (p.L283Sfs∗9) | ||
| HL1000078 | 5 | 2 | PM2_supporting, PP1∗, BP4 | VUS∗, VUS | C1 | JUNB | Coding | NM_002229.3:c.334C>T (p.P112S) | ||
| HL16594 | 6 | 2 | X | BP4, PP1∗ | VUS∗, VUS | C1 | TCF3 | Coding | NM_003200.5:c.1486G>A (p.E496K) | |
| HL16594 | 6 | 2 | X | BP4, PP1∗ | VUS∗, VUS | C2 | HBS1L | Coding | NM_006620.4:c.1162A>G (p.I388V) | |
| HL213 | 6 | 2 | X | PM2_supporting, PP1∗, BP4 | VUS∗, VUS | C1 | RAD51D | Coding | NM_002878.4:c.911G>A (p.G304D) | |
| HL2350 | 6 | 4 | X | PS3_supporting, PP1_strong∗ | VUS∗, VUS | C1 | KDR∗ | Coding | NM_002253.4:c.3193G>A (p.A1065T) | |
| HL2408 | 4 | 2 | PM2_supporting, PP1∗, PP3 | VUS∗, VUS | C3 | CDT1 | Coding | NM_030928.4::c.1477+3_1477+24del () | ||
| HL2408 | 4 | 2 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | POLR1E | Coding | NM_022490.4:c.445C>T (p.R149∗) | ||
| HL2491 | 6 | 2 | PS3_supporting, PP1_strong∗, PP3 | LP∗, VUS | NC1 | KLHDC8B∗ | Noncoding | NM_173546.3:c.-158C>T | ||
| HL2491 | 6 | 2 | PP1∗, PP3 | VUS∗, VUS | NC1 | GATA3 | Noncoding | NM_001002295.2:c.779-2563G>A | ||
| HL2694 | 11 | 4 | X | PVS1∗, PM2_supporting, PP1_moderate∗ | P∗, VUS | C3 | EEF2KMT | Coding | NM_201400.4:c.529A>T (p.K177∗) | |
| HL2696 | 5 | 2 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | EFR3B | Coding | NM_014971.2:c.853C>T (p.Q285∗) | ||
| HL3262 | 7 | 3 | PM2_supporting, PP1∗, PP3 | VUS∗, VUS | C3 | EIF1AD | Coding | NM_001242481.2:c.88-4C>G | ||
| HL3262 | 7 | 3 | PS3_supporting, PP1_strong∗ | VUS∗, VUS | C1 | KDR∗ | Coding | NM_002253.4:c.3193G>A (p.A1065T) | ||
| HL3262 | 7 | 3 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | DDX10 | Coding | NM_004398.4:c.2059A>T (p.K687∗) | ||
| HL3350 | 9 | 2 | X | PM2_supporting, PP1∗, PP2 | VUS∗, VUS | C1 | STAT3 | Coding | NM_139276.3:c.849A>T (p.K283N) | |
| HL3402 | 8 | 4 | X | PVS1∗, PM2_supporting, PP1_moderate∗ | P∗, VUS | C1 | IRF7 | Coding | NM_001572.5:c.396G>A (p.W132∗) | |
| HL3929 | 3 | 3 | PP1∗, PP3 | VUS∗, VUS | NC1 | GATA3 | Noncoding | NM_001002295.2:c.779-2563G>A | ||
| HL3929 | 3 | 3 | PS3_supporting, PP1_moderate, PP3 | VUS∗, VUS | C1 | POT1∗ | Coding | NM_015450.3:c.670G>A (p.D224N) | ||
| HL4450 | 7 | 5 | X | PS3_supporting, PP1_strong∗, PP3 | LP∗, VUS | NC1 | KLHDC8B∗ | Noncoding | NM_173546.3:c.-158C>T | |
| HL4479 | 7 | 2 | BP4, PP1∗ | VUS∗, VUS | C1 | BAD | Coding | NM_032989.3:c.397A>C (p.K133Q) | ||
| HL4479 | 7 | 2 | BP4, PP1∗ | VUS∗, VUS | C1 | CLEC16A | Coding | NM_015226.3:c.2578C>T (p.R860C) | ||
| HL4643 | 18 | 5 | PP1∗, PP3 | VUS∗, VUS | NC1 | IRF8 | Noncoding | NM_002163.4:c.-1-1639C>T | ||
| HL5171 | 5 | 2 | PP1∗ | VUS∗, VUS | C2 | MET | Coding | NM_000245.4:c.2318C>T (p.P773L) | ||
| HL5171 | 5 | 2 | BP4, PP1∗ | VUS∗, VUS | C1 | MAP3K7 | Coding | NM_145331.3:c.1282G>A (p.V428I) | ||
| HL533 | 6 | 2 | PVS1∗, PM2_supporting, PP1∗ | P∗, VUS | C3 | ARMC9 | Coding | NM_001352754.2:c.1268_1271del (p.K423Rfs∗29) | ||
| HL6898 | 7 | 3 | X | PM2_supporting, PP1_moderate∗, PP3 | VUS, VUS∗ | NC1 | PAX5 | Noncoding | NM_016734.3:c.605-2310C>T | |
| HL696 | 7 | 2 | X | PVS1∗, PP1_moderate∗ | P∗, VUS | C3 | TPRG1 | Coding | NM_198485.4:c.183_192del (p.Y61∗) |
Prioritized variants for each family based on segregation and predicted deleteriousness. Family specific information including sample count and number of affected and obligate carriers and counts for each priority level are provided in addition to highlighted variant details. Variants in KLHDC8B NM_173546.3:c.-158C>T, KDR NM_002253.4:c.3849-2A>C, and POT1 were previously reported based on WES analysis of the same NCI pedigrees.16,17,21
BP, benign supporting; HGVS, Human Genome Variation Society; LP, likely pathogenic; P, pathogenic; PM, pathogenic moderate; PP, pathogenic supporting; PS, pathogenic strong; PVS, pathogenic very strong; VUS, variant of unknown significance.
∗Where evidence applied based on hypothesis of genotype-phenotype correlation.
CNVnator45 was used to determine CNV genotypes for each sample. CNV calls not supported by at least 1 discordant read pair alignment were removed. SURVIVOR46 was used to merge individual sample variant call formats into pedigree-specific variant call formats and SLIVAR24 for the identification of segregating CNVs within each pedigree. We then used AnnotSV47 to automatically apply CNV-specific ranking criteria based on the ACMG and ClinGen guidelines. CNVs with ranking ≥4 (P/LP) were reviewed for their potential to be related to the HL phenotype.
Candidate variant review and classification
Coding and noncoding variant prioritization scores, linkage results, literature, and expert opinion were considered when reviewing SNV/indel candidate variants within each family (Figure 1A). This was restricted to C1 to C3 and NC1 and NC2 variants. C4 variants are low scoring in genes that have little to no previous association with HL or related phenotypes and therefore were not included. Final candidate variants were then classified based on recommendations from the ACMG, AMP, and Clinical Genome Resource. Pathogenicity was calculated using the Bayesian formulation of the ACMG/AMP.48 Refer to supplementary material for further details and thresholds.48
Results
We performed comprehensive WGS analysis on germline samples of 234 individuals (129 female and 105 male) from 36 unrelated HL pedigrees (Figure 1A and Figure 2). The cohort consisted of 79 affected and 155 unaffected individuals, with an average of 2.6 affected individuals per pedigree (range, 2-5). The median age at diagnosis was 21.98 years (range, 3-55 years). Most individuals (207, 88.5%) were of European ancestry (supplemental Figure 2).
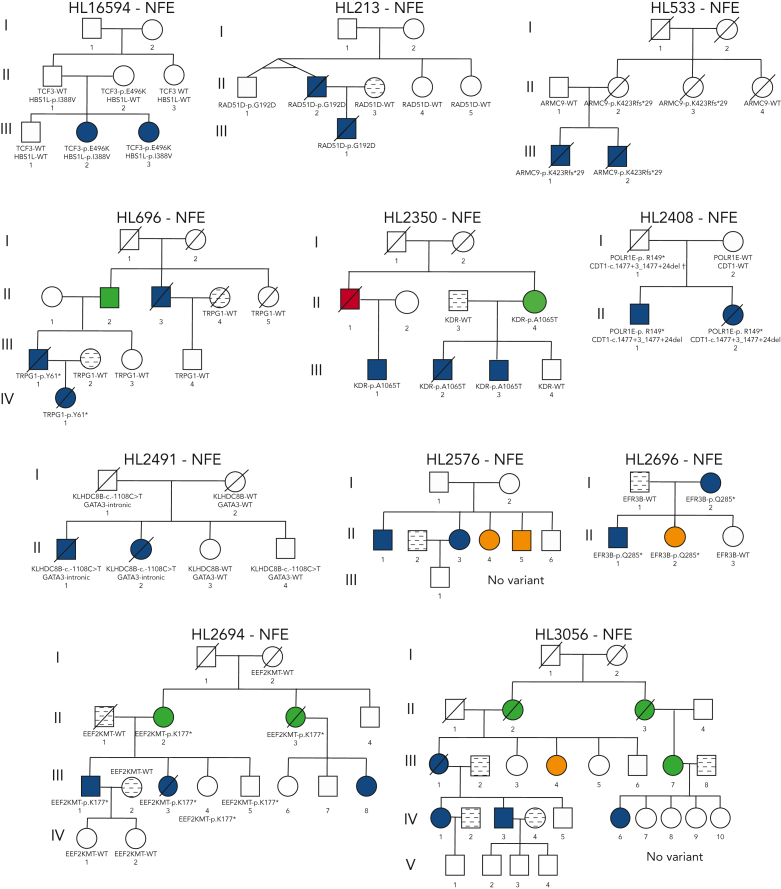
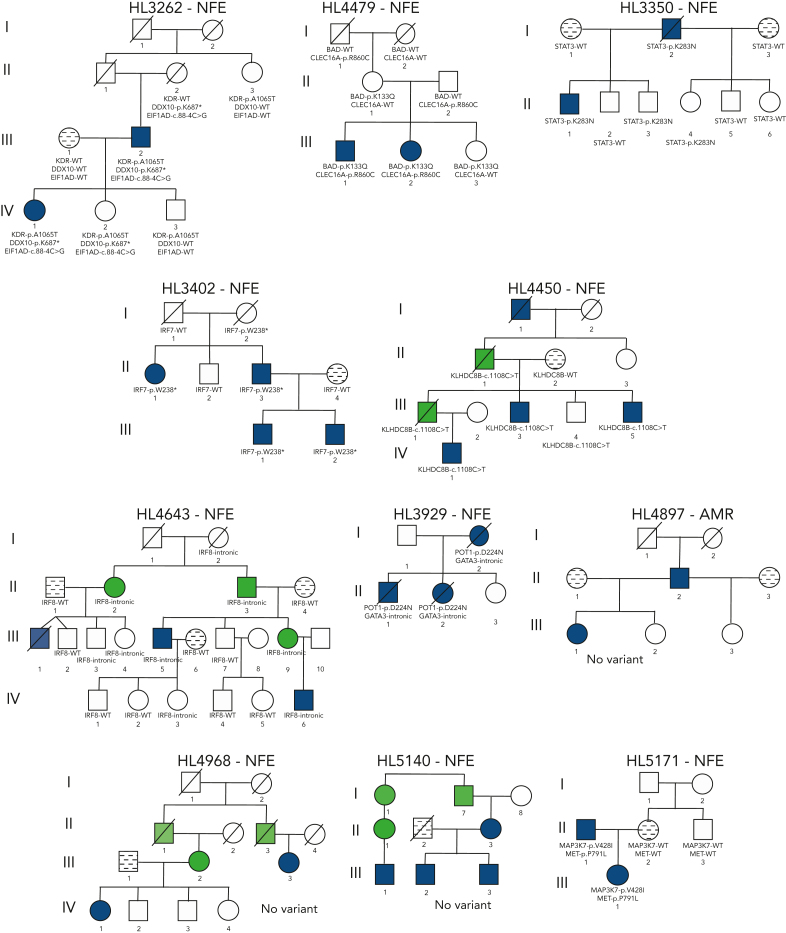
Figure 2.
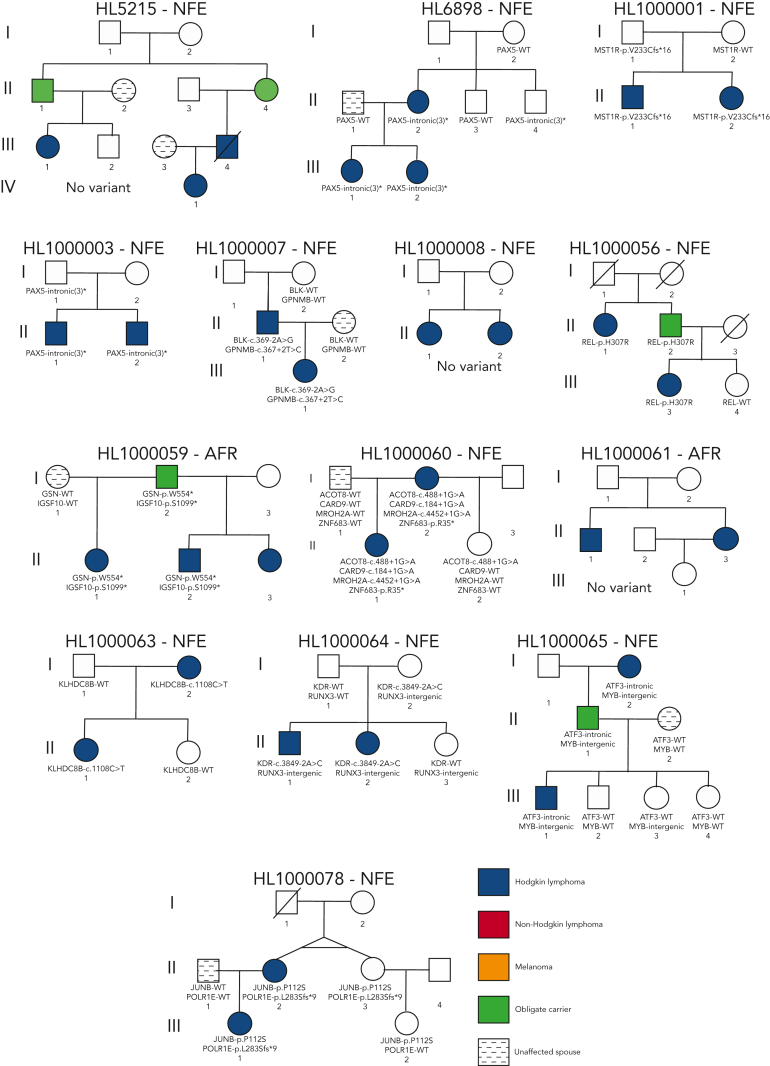
Pedigrees of families with a high frequency of HL. Pedigree representation of the 36 families included for analysis. Variants listed segregate with the disease phenotype and obligate carriers within each pedigree. HL16594: TCF3-p.E496K and HBS1L-p.I388V demonstrate potential codominant inheritance; HL213: RAD51D-p.G192D; HL533: ARMC9-p.K423Rfs∗29; HL696: TRPG1-p.Y61∗; HL2350: KDR-p.A1065T; HL2408: POLR1E-p. R149∗ and CDT1-c.1477+3_1477+24del† (limited support of alternative allele for father of proband); HL2491: KLHDC8B-c.-1108C>T and GATA3-intronic; HL2576: no variant; HL2696: EFR3B-p.Q285∗; HL2694: EEF2KMT-p.K177∗; HL3056: no variant; HL3262: KDR-p.A1065T, DDX10-p.K687∗, EIF1AD-c.88-4C>G; HL4479: BAD-p.K133Q, CLEC16A-p.R860C; HL3402: IRF7-p.W238∗; HL4450: KLHDC8B-c.1108C>T; HL4643: IRF8-intronic; HL3929: POT1-p.D224N and GATA3-intronic; HL4897: no variant; HL4968: no variant; HL5140: no variant; HL5171: MAP3K7-p.V428I and MET-p.P791L; HL5215: no variant; HL6898: PAX5-intronic(3)∗; HL1000001: MST1R-p.V233Cfs∗16; HL1000003: PAX5-intronic(3)∗; HL1000007: BLK-c.369-2A>G and GPNMB-c.367+2T>C; HL1000008: no variant; HL1000056: REL-p.H307R; HL1000059: GSN-p.W554∗ and IGSF10-p.S1099∗; HL1000060: ACOT8-c.488+1G>A, CARD9-c.184+1G>A, MROH2A-c.4452+1G>A, and ZNF683-p.R35∗; HL1000061: no variant; HL1000063: KLHDC8B-c.1108C>T; HL1000064: KDR-c.3849-2A>C and RUNX3-intergenic; HL1000065: ATF3-intronic and MYB-intergenic; and HL1000078: JUNB-p.P112S and POLR1E-p.L283Sfs∗9. WT, wild-type.
Prioritized HL-risk segregating coding and noncoding variants
On average, families carried 68 segregating C1 to C4 coding variants and 55 segregating NC1 and NC2 noncoding variants (supplemental Table 2). Of these, there were 70 high-priority coding variants (C1 and C2; Figure 1B) and 1769 high-priority noncoding variants (NC1 and NC2; Figure 1C) based on their predicted functional consequences. These prioritized coding and noncoding risk variants (C1-C2 plus C3 and NC1 and NC2) were then subjected to an iterative multireviewer process to narrow down to a list (Table 1; supplemental Table 3) of prioritized genomic candidates for 28 of the 36 pedigrees (Figure 1D-E).
Recurrence among pedigrees
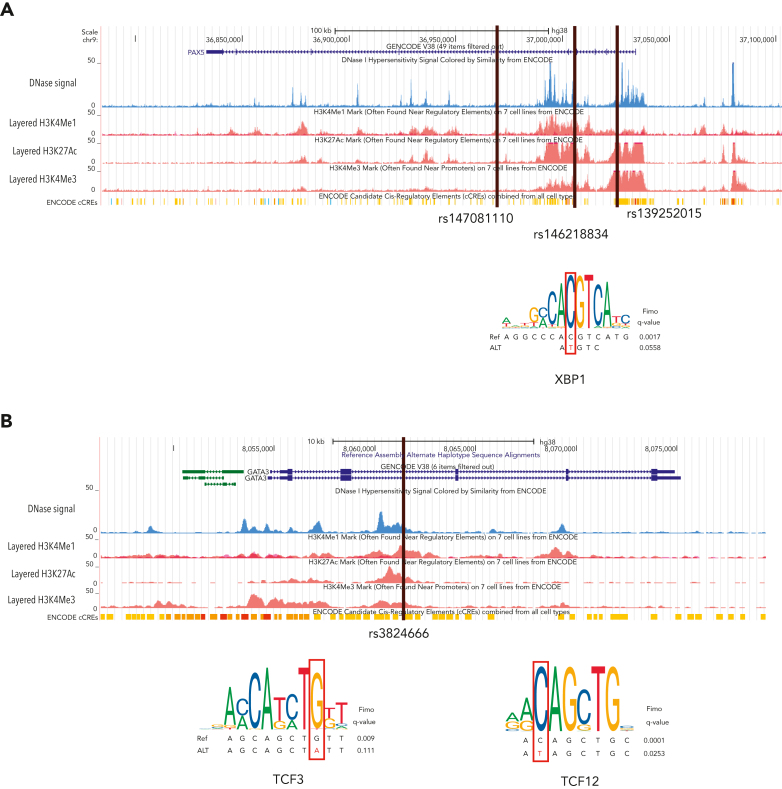
Four recurrent variants were observed (Table 1), including previously reported coding variant (rs56302315) in KDR16 and 5′ UTR variant (rs387906223) in KLHDC8B,21 along with 2 novel noncoding variants, 1 in intron 5 of PAX5 (rs147081110) and another (rs3824666) in intron 3 of GATA3 (supplemental Table 4). The PAX5 intronic variant (rs147081110, RegBase PHRED = 10.299, minor allele frequency = 3.94 × 10-5) overlaps DNase I along with transcription factor binding site (TFBS) clusters with predicted loss of XBP1 binding (Figure 3A). In addition, this variant fell within one of the maximum MLOD regions for pedigree HL6898 (MLOD = 0.30; HG38, chr9:36,833,268-37,034,267), with transmission from the affected mother to both affected children. The GATA3 intronic variant (rs3824666, RegBase PHRED = 15.39, minor allele frequency = 0.0079) overlaps DNase I and TFBS clusters with predicted loss of binding of TCF3 and TCF12 (Figure 3B). TCF3 is purported to act as a tumor suppressor in B-cell malignancies.49 TCF12 (HEB), a critical regulator of hematopoietic cell specification, has been shown to be necessary for proper B-cell and CD4 T-cell generation.50,51 We identified an additional 25 recurrent NC1 and NC2 variants (supplemental Table 4). There were 11 variants with no predicted change in transcription factor binding and 15 with complex predictions by FIMO. Although showing complex predicted binding, the recurrent variant (rs575404240) in IRF8 is of interest because it was the final prioritized candidate for HL4643, one of the largest pedigrees studied.
Figure 3.
Recurrent noncoding variants. (A) The PAX5 intronic variant (rs147081110) overlaps an ENCODE DNase I hypersensitivity peak cluster along with an ENCODE 3 transcription factor chromatin immunoprecipitation sequencing cluster and has FIMO-predicted loss of XBP1 binding. (B) The GATA3 intronic variant (rs3824666) overlaps an ENCODE DNase I hypersensitivity peak cluster along with an ENCODE 3 transcription factor chromatin immunoprecipitation sequencing cluster and has FIMO-predicted loss of binding of TCF3 and TCF12.
Gene-level recurrence of variants
Gene-level recurrence, wherein multiple unrelated families harbor different variants in the same gene, was seen for POLR1E (p.L283Sfs∗9, p.R149∗, and p.E41Dfs∗2) in 3 independent pedigrees. Although not in our candidate gene list, these POLR1E LOF variants were of interest because intrachromosomal rearrangements involving this gene have been identified in diffuse large B-cell lymphomas.52 In addition, a splice variant in KDR (c.3849-2A>C) was identified in 1 pedigree, meaning we observed gene-level recurrence of variations in KDR across 3 pedigrees. Although KDR16 and KLHDC8B21 have previously been reported, PAX5, GATA3, and POLR1E represent novel findings.
Variants under maximum MLOD peaks
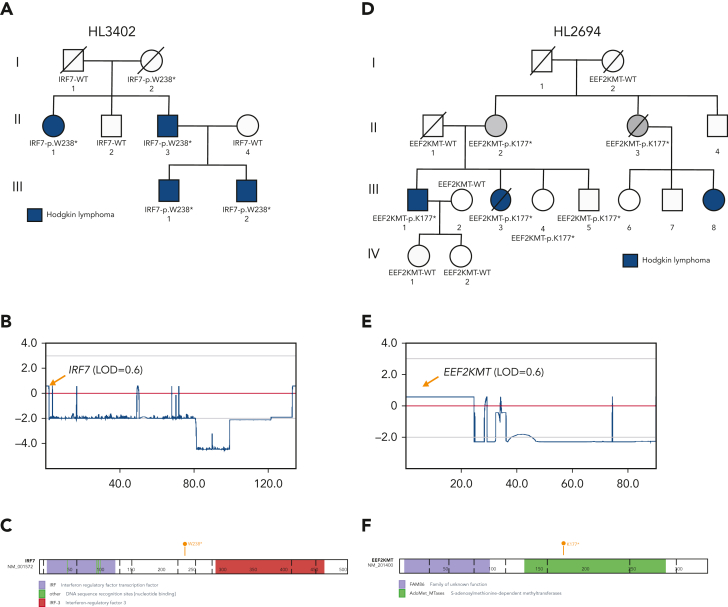
Prioritized variants in 5 genes (KDR, IRF7, EEF2KMT, KLHDC8B, and PAX5) were of increased interest given they fell under maximum MLOD peaks. KDR,16 KLHDC8B,21 and PAX5 have already been described in “Recurrence among pedigrees.” Two stop-gain variants (IRF7 p.W238∗ and EEF2KMT p.K77∗) were found to segregate within large pedigrees with 4 affected or obligate carriers and fell within or close to linkage peaks with LOD > 0.70. IRF7 p.W238∗ was the only C1 variant prioritized for HL3402 (Figure 4A-C) and showed segregation among 4 affected individuals across 2 generations. It has been classified using the ACMG criteria by InterVar as LP and resides in the interferon regulatory factor DNA-binding domain in the 5′ N-terminal region of the gene. In addition, EEF2KMT p.K77∗ is a C3 variant prioritized for HL2694, a pedigree for which no C1 or C2 variant (ie, in a candidate gene) was identified (Figure 4D-E).
Figure 4.
LOF variants falling under maximum multipoint linkage regions in large HL pedigrees. (A) Segregation of IRF7-p.W238∗ among 4 affected relatives in HL3402. (B) MLOD plot on chromosome 11 with IRF7 location indicated. (C) Protein paint diagram showing the location of stop-gain variant p.W238∗, which could result in the removal of the IRF-3 functional domain. (D) Segregation of EEF2KMT-p.K177∗ among 2 affected relatives and 2 obligate carriers in HL3402. Two unaffected siblings also carry the variant. (E) MLOD plot on chromosome 16 with the EEF2KMT location indicated. (F) Protein paint diagram showing the location of stop-gain variant p.K177∗, which could result in the removal of part of the AdoMet_MTases functional domain.
Segregating CNVs
An unbiased genome-wide screen for CNV identified 26 CNVs that segregated with affected individuals within pedigrees and were potentially P/LP based on the AnnotSV ranking (supplemental Table 5A). All 26 CNVs were observed to be common in an internal control cohort of >10 000 samples that were analyzed in a similar manner, and a majority (15/26 CNVs) also overlapped an established benign CNV region. In addition, none of the 26 CNVs overlapped a known candidate gene. We also observed 9 VUS-ranked CNVs that were annotated as affecting a candidate gene; however, only 2 were considered not common (supplemental Table 5B). These 2 rare VUS CNVs (HL1000064-CMIP and HL213-PTPRK) were intronic and shared by most unaffected individuals in each pedigree. Based on the described CNV selection criteria, we determined that none of the segregating CNVs were likely to be associated with the development of HL.
Pedigrees with very early-onset HL
Four pedigrees had a proband with an onset of HL<10 years of age, which is even more rare for this disease that has an incidence rate of 4.2% in age <10 years compared with 46.6% in ages 10 to 19 years.44 Thus, we interrogated all variants in tiers C1 to C4 for this subset of pedigrees. Pedigree HL213 had the proband with the earliest onset in the cohort (aged 3 years) and an affected father (onset at 31 years). We observed 15 coding variants for pedigree HL213 with only a single variant (rs200615280, p.G304D) in RAD51D, 1 of our candidate genes involved in DNA repair with a well-established role in breast and ovarian cancer.53 An independent study of familial HL19 found another segregating variant (rs587781813, p.R266C) in RAD51D. Both variants reside in the same RecA-like nucleoside-triphosphatase protein domain. Using our classification criteria in the context of a potential association with HL, RAD51D p.G304D is classified as VUS based on ACMG PM2, PP1, and BP4 criteria and RAD51D p.R266C as VUS.
The next youngest proband belonged to pedigree HL3350 (onset at 6 years). Their father was also affected but at a later age (onset at 51 years). There were 49 coding variants observed for pedigree HL3350, 2 of which affected study-specific candidate genes, STAT3 (p.K283N) and TP63 (rs201188464, p.P174L). STAT3 has a clear body of literature supporting its role in HL54; thus, it was prioritized over TP63.
Pedigree HL696 has a proband with an onset age of 7 years whose father had an onset at age 31 years. This pedigree also includes an affected great uncle of the proband (onset at 43 years) for whom sequencing data were not available. We observed 78 segregating coding variants for pedigree HL696, none of which affected study-specific candidate genes. However, 1 of the variants is a stop gain in TPRG1 (rs761733372, p.Y61∗), a gene whose overexpression has been associated with B-cell lymphoma.55 There were 2 affected siblings in the fourth pedigree (HL16594), one of whom was diagnosed at 10 years of age. Out of the 131 coding variants observed for this pedigree, there were 5 in candidate genes. A missense variant (rs201250905, p.E496K) affecting TCF3, a gene associated with HL risk, may be relevant in this pedigree.56 In addition, a stop-gain variant (rs758125506, p.K461∗) was observed for this pedigree in GBP5, a gene associated with chronic active Epstein-Barr virus (EBV) infection. We did not prioritize this variant given that its LOF annotation does not support an association with lymphomagenesis, which would be predicted to occur through increased expression or a gain-of-function variant in this gene.57
Discussion
HL is a rare cancer with known familial aggregation but limited understanding of its genetic predisposition. We performed WGS on the largest cohort of pedigrees with multiple occurrences of HL, thus expanding upon previous WES studies to comprehensively interrogate both coding and noncoding variation. To identify potentially causative or disease-susceptibility variants, we considered the following categories: (1) variants identified from large pedigrees with 4 or more affected relatives, particularly if they fell within linkage peaks; (2) recurrent noncoding variants segregating with HL in >1 pedigree; (3) gene-level recurrence under the assumption of genetic heterogeneity; and (4) variants prioritized in very early-onset families (onset of HL at <10 years of age).44
Findings from the largest pedigrees
Five high-confidence variants were found in each of the largest pedigrees (N = 4-5 affected relatives). Two of the variants had previously been identified by WES in the same families: KDR p.A1065T (HL2350) and KLHDC8B c.-1108C>T (HL4450)16,21,58. The remaining 3 variants (IRF7:p.W238∗; EEF2KMT:p.K116∗; rs575404240 in IRF8) are novel candidates that deserve further investigation. IRF7 plays important roles in innate immunity and immune cell differentiation and is involved in the regulation of EBV latency.59 This variant may affect the clearance of EBV in a host, which is a driver for the formation of lymphoma and may be causal in this pedigree.60 Further analyses to look for latent EBV in this pedigree would be helpful to establish causation. EEF2KMT was the best candidate for pedigree HL2694, which had no C1 or C2 variants in cancer-related candidate genes. However, the role of this gene in lymphomagenesis is unclear. IRF8 is an interferon regulatory factor that plays a role in cellular differentiation, transformation, and apoptosis.61 This gene is expressed in B cells and has been implicated in the formation of chronic lymphocytic leukemia, another B-cell malignancy. The expression of IRF8 is important for B-cell development, including pre–B-cell differentiation, and variations in this gene may be implicated in the formation of HL.61,62 Therefore, these 3 new variants are potentially pathogenic in the largest of our pedigrees and in need of further validation.
Findings in noncoding variants
WGS allowed for the identification of segregating noncoding variants that may be important for HL, particularly in pedigrees for which no obvious coding variant was found. We identified recurrent noncoding variants in PAX5 and GATA3, both well-known HL candidate genes. Predicting and testing the impact of noncoding variants can be complex depending on the tissue specificity and temporal stage of sample collection.63 The recurrent intronic GATA3 variant (rs3824666) is especially interesting because it is predicted to lead to loss of binding of tumor suppressor TCF349 and it corroborates the results from previous GWAS studies.56 Genome-wide association of single nucleotide polymorphisms within GATA3 (rs3781093, P = 9.49 × 10−13) with HL was found in a large meta-analysis of 5314 HL cases and 16 749 controls.14 In addition, meta-analysis in 1816 HL cases and 7877 controls and subsequent replication in an independent set of 1281 HL cases and 3218 controls found a significant association of common noncoding variants (rs444929) in GATA3.56 Thus, we speculate that the common and low-frequency noncoding variants in GATA3 may be related to both sporadic and familial HL. The other interesting noncoding variant was found in PAX5, which is of significant clinical interest given its association with other cancers such as leukemia and lymphoblastic lymphoma. PAX5 is needed for B-cell lineage commitment64 and given that HL is a B-cell driven process that loses its B-cell marker and aberrantly expresses CD30, it is plausible that a genetic variant in PAX5 may hinder B cells from remaining committed to the B-cell lineage and thus have a propensity to lose their classic CD20 marker.64 Moreover, the expression of PAX5 is turned on during the transition of B cells from a pre–pro-B cell to a committed pro–B cell, and in mouse models, when PAX5 is turned off, the cells return to an uncommitted progenitor cell.64 Therefore, alterations in PAX5 may explain the loss of CD20 and be related to the formation of Hodgkin Reed-Sternberg cells.
Gene-level recurrence
Recurrence at the gene level was found for POLR1E and KDR. The POLR1E gene is a polymerase (RNA) I polypeptide E programmed death ligand. Intrachromosomal alterations involving POLR1E have been described in diffuse large B-cell lymphoma tissue specimens.52 POLR1E was not on our candidate gene list but we observed 3 different segregating LOF variants in 3 independent pedigrees, which was a striking finding. Besides the recurrent published KDR coding variant, we identified a novel KDR splice variant (c.3849-2A>C) in pedigree HL1000064 that affects the splice acceptor locus before exon 30 (last exon). This splice variant is predicted to disrupt the native acceptor site. KDR, also known as VEGFR2, has been reported for its association with familial HL and validated with functional experiments.16 The KDR gene comprises 30 exons and Rotunno et al16 demonstrated that this missense variant affects the kinase domain activation loop that is important for tumor angiogenesis and cell proliferation and survival. VEGFR’s importance in angiogenesis of HL has been well described.65 This new finding of a splice variant adds to growing evidence that variants affecting KDR have a role in HL susceptibility, and the addition of a second KDR variant in familial HL suggests that this gene could be considered for clinical cancer predisposition screenings.
Variants potentially related to early onset
We were particularly interested in germline variants segregating in 4 early-onset families and hypothesized that germline predisposition may be more relevant in such families because of the presumed higher impact of genetic over environmental causes. This investigation resulted in a few candidates, including RAD51D, STAT3, TPRG1, and TCF3. However, no clear primary candidate emerged from this analysis, with the exception of RAD15D, which has also been implicated in an independent familial HL cohort.19 RAD51D is part of the homologous recombination deficiency pathway and interacts with BRCA1/2. Burden testing of germline variants in this gene was also associated with an increased risk for ovarian cancer.66 The paper reported that ∼69% of RAD51D variants contributing to ovarian cancer fell in the DNA recombination and repair protein RecA-like adenosine triphosphate–binding domain of the gene. The variant segregating with HL in our family falls within the same domain. In contrast to most nonsense and frameshift variants in RAD15D seen in patients with ovarian cancer, the HL-segregating variants from our study (p.G304D) and an independent HL familial study19 (p.R266C) are both missense variants. Somewhat surprisingly, RAD15D in HL213 segregated in 1 but not both monozygotic twins. We hypothesize that the germline susceptibility from RAD15D may be accompanied by environmentally triggered epigenetic factors including DNA methylation, which has been reported as a cause for disease in other discordant twin studies, including in HL.67, 68, 69
Strengths of this study include the highly informative pedigrees with multiple affected individuals, at least 1 proband diagnosed ≤21 years of age, and application of WGS allowed for interrogation of noncoding variants that had not previously been performed, even in the NCI families. In addition, we used several newly available annotations and computational tools for family-based screening of variants, including in-house pipelines for variant calling. Our findings validated previously published results, which served as positive controls.
Limitations include lack of updated clinical information after enrollment and reliance on unaffected married-in spouses as negative controls. Next, we based our interrogation on germline variants only and did not study the contribution of somatic mutations to the patient’s disease because few tumor blocks were available. Viral factors or cooperating mutations may also contribute, but the EBV status of the patients and tumor were unknown. Of note, we selected probands diagnosed with HL <21 years of age. These patients fit within the first bimodal age at onset peak, which has the lowest rate of association with EBV. Another limitation is the use of the GM12878 cell line for noncoding variant prioritization based on DNase I hypersensitivity and TFBS overlap, which may not be the optimal cell line for studying HL but has been used previously in GWAS studies of HL.14
Conclusions
We used WGS in 36 highly informative HL pedigrees with pediatric age probands and expanded on previous WES analysis. In the next step, these candidate risk variants should be validated for pathogenicity in a laboratory setting, in large sporadic cohorts of HL cases, and through sequencing of limited tumor samples available to compare the germline genetic changes identified with those seen in Hodgkin Reed-Sternberg cells. The genomic landscape of familial HL remains incompletely characterized. Identification of genetic predisposition variants for the development of HL may lead to novel therapeutic targets, better treatment of this rare disease, and addition of these genes to clinical germline genetic testing panels to facilitate early detection of symptoms, inform genetic counseling, and help determine risk for other family members.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors thank Bryan Roberts for his assistance with drawing of pedigrees in Figure 2 and to members of the St. Jude Children’s Research Hospital’s Center for Applied Bioinformatics for pipeline implementation and computing.
This work was supported by grants from the National Institutes of Health R03 grant (R03HD104066), National Institutes of Health Cancer Support Core grant (CA-21765), Lymphoma Research Foundation, the American Lebanese Syrian Associated Charities, and Gabriella Miller Kids First X01 grant (HL136999-01). The research activities of L.R.G., M.L. McMaster, M.R., N.C., A.V., D.F., K.W., J.L., and M.T. were supported by the Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute. Work in Australia was supported by National Health and Medical Research Council grant APP1164601.
Authorship
Contribution: J.E.F., J.L.M., and K.H. performed the clinical data preparation; J.R.M., J.E.F., J.L.M., N.O., S.R.R., T.-C.C., S.S.T., and E.R. performed the genomic analyses; and J.E.F., J.R.M., and E.R. prepared the manuscript with contributions from J.L.M., N.O., J.J.Y., Y.H., Y.-D.W., W.C., G.W., L.R.G., M.L. McMaster, M.R., N.C., A.V., D.F., K.W., J.L., M.T., C.H., A.L.B., H.S.S., C.M., K.E.N., and M.L. Metzger.
Footnotes
The results are based upon data generated by Gabriella Miller Kids First Pediatric Research Program projects phs001738.v1.p1 and accessed from the Kids First Data Resource Portal (https://kidsfirstdrc.org) and/or the database of Genotypes and Phenotypes (www.ncbi.nlm.nih.gov/gap). Code used for this publication is available at https://github.com/jrm3215/FAMVP.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Contributor Information
Jamie E. Flerlage, Email: jamie.flerlage@stjude.org.
Jun J. Yang, Email: jun.yang@stjude.org.
Evadnie Rampersaud, Email: evadnie.rampersaud@stjude.org.
Supplementary Material
References
- 1.Caporaso NE, Goldin LR, Anderson WF, Landgren O. Current insight on trends, causes, and mechanisms of Hodgkin's lymphoma. Cancer J. 2009;15(2):117–123. doi: 10.1097/PPO.0b013e3181a39585. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo J, Gasull T, Giralt M, Armario A. Brain metallothionein in stress. Biol Signals. 1994;3(4):198–210. doi: 10.1159/000109546. [DOI] [PubMed] [Google Scholar]
- 3.Mack TM, Cozen W, Shibata DK, et al. Concordance for Hodgkin's disease in identical twins suggesting genetic susceptibility to the young-adult form of the disease. N Engl J Med. 1995;332(7):413–418. doi: 10.1056/NEJM199502163320701. [DOI] [PubMed] [Google Scholar]
- 4.Kharazmi E, Fallah M, Pukkala E, et al. Risk of familial classical Hodgkin lymphoma by relationship, histology, age, and sex: a joint study from five Nordic countries. Blood. 2015;126(17):1990–1995. doi: 10.1182/blood-2015-04-639781. [DOI] [PubMed] [Google Scholar]
- 5.Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9(1):15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 6.Goldin LR, Pfeiffer RM, Gridley G, et al. Familial aggregation of Hodgkin lymphoma and related tumors. Cancer. 2004;100(9):1902–1908. doi: 10.1002/cncr.20189. [DOI] [PubMed] [Google Scholar]
- 7.Cerhan JR, Slager SL. Familial predisposition and genetic risk factors for lymphoma. Blood. 2015;126(20):2265–2273. doi: 10.1182/blood-2015-04-537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131(22):2413–2425. doi: 10.1182/blood-2017-11-812073. [DOI] [PubMed] [Google Scholar]
- 9.Camus V, Viennot M, Lequesne J, et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: a prospective study. Haematologica. 2021;106(1):154–162. doi: 10.3324/haematol.2019.237719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desch AK, Hartung K, Botzen A, et al. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia. 2020;34(1):151–166. doi: 10.1038/s41375-019-0541-6. [DOI] [PubMed] [Google Scholar]
- 11.Sobesky S, Mammadova L, Cirillo M, et al. Exhaustive circulating tumor DNA sequencing reveals the genomic landscape of Hodgkin lymphoma and facilitates ultrasensitive detection of minimal residual disease. medRxiv. https://doi.org/10.1101/2021.03.16.21253679 Preprint posted online 16 March 2021. [DOI] [PubMed]
- 12.Diepstra A, Niens M, Vellenga E, et al. Association with HLA class I in Epstein-Barr-virus-positive and with HLA class III in Epstein-Barr-virus-negative Hodgkin's lymphoma. Lancet. 2005;365(9478):2216–2224. doi: 10.1016/S0140-6736(05)66780-3. [DOI] [PubMed] [Google Scholar]
- 13.Cozen W, Li D, Best T, et al. A genome-wide meta-analysis of nodular sclerosing Hodgkin lymphoma identifies risk loci at 6p21.32. Blood. 2012;119(2):469–475. doi: 10.1182/blood-2011-03-343921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sud A, Thomsen H, Law PJ, et al. Genome-wide association study of classical Hodgkin lymphoma identifies key regulators of disease susceptibility. Nat Commun. 2017;8(1):1892. doi: 10.1038/s41467-017-00320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kushekhar K, van den Berg A, Nolte I, Hepkema B, Visser L, Diepstra A. Genetic associations in classical Hodgkin lymphoma: a systematic review and insights into susceptibility mechanisms. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2737–2747. doi: 10.1158/1055-9965.EPI-14-0683. [DOI] [PubMed] [Google Scholar]
- 16.Rotunno M, McMaster ML, Boland J, et al. Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene. Haematologica. 2016;101(7):853–860. doi: 10.3324/haematol.2015.135475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMaster ML, Sun C, Landi MT, et al. Germline mutations in Protection of Telomeres 1 in two families with Hodgkin lymphoma. Br J Haematol. 2018;181(3):372–377. doi: 10.1111/bjh.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandapalli OR, Paramasivam N, Giangiobbe S, et al. Whole genome sequencing reveals DICER1 as a candidate predisposing gene in familial Hodgkin lymphoma. Int J Cancer. 2018;143(8):2076–2078. doi: 10.1002/ijc.31576. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava A, Giangiobbe S, Kumar A, et al. Identification of familial Hodgkin lymphoma predisposing genes using whole genome sequencing. Front Bioeng Biotechnol. 2020;8:179. doi: 10.3389/fbioe.2020.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ristolainen H, Kilpivaara O, Kamper P, et al. Identification of homozygous deletion in ACAN and other candidate variants in familial classical Hodgkin lymphoma by exome sequencing. Br J Haematol. 2015;170(3):428–431. doi: 10.1111/bjh.13295. [DOI] [PubMed] [Google Scholar]
- 21.Salipante SJ, Mealiffe ME, Wechsler J, et al. Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc Natl Acad Sci U S A. 2009;106(35):14920–14925. doi: 10.1073/pnas.0904231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldin LR, McMaster ML, Ter-Minassian M, et al. A genome screen of families at high risk for Hodgkin lymphoma: evidence for a susceptibility gene on chromosome 4. J Med Genet. 2005;42(7):595–601. doi: 10.1136/jmg.2004.027433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers J ON, Flerlage J, Rashkin R, et al. FAMilial Variant Prioritizer (FAMVP): An annotation and prioritization pipeline for whole genome germline variants in family studies [abstract]. In: American Society of Human Genetics Annual Meeting. October 2020. [Google Scholar]
- 24.Pedersen BS, Brown JM, Dashnow H, et al. Effective variant filtering and expected candidate variant yield in studies of rare human disease. NPJ Genom Med. 2021;6(1):60. doi: 10.1038/s41525-021-00227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Ottman R, Rabinowitz D. A method for estimating penetrance from families sampled for linkage analysis. Biometrics. 2006;62(4):1081–1088. doi: 10.1111/j.1541-0420.2006.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch L. Exploring human genomic diversity with gnomAD. Nat Rev Genet. 2020;21(8):448. doi: 10.1038/s41576-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37(3):235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, He Y, Liu H, et al. regBase: whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants. Nucleic Acids Res. 2019;47(21):e134. doi: 10.1093/nar/gkz774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176(3):535–548 e524. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42(22):13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 35.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Wilson CL, Easton J, et al. Genetic risk for subsequent neoplasms among long-term survivors of childhood cancer. J Clin Oncol. 2018;36(20):2078–2087. doi: 10.1200/JCO.2018.77.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouillard AD, Gundersen GW, Fernandez NF, et al. Vol. 2016. Database (Oxford); 2016. (The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cotterill SJ. Home Page, Cancer Genetics Web. http://www.cancer-genetics.org/index.htm
- 40.Sloan CA, Chan ET, Davidson JM, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 44.Howlader N NA, Krapcho M, Miller D, et al., editors. SEER Cancer Statistics Review. 1975-2018. https://seer.cancer.gov/csr/1975_2018/ [Google Scholar]
- 45.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeffares DC, Jolly C, Hoti M, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017;8:14061. doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geoffroy V, Guignard T, Kress A, et al. AnnotSV and knotAnnotSV: a web server for human structural variations annotations, ranking and analysis. Nucleic Acids Res. 2021;49(W1):W21–W28. doi: 10.1093/nar/gkab402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tavtigian SV, Harrison SM, Boucher KM, Biesecker LG. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mutat. 2020;41(10):1734–1737. doi: 10.1002/humu.24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan H, Xie L, Wirth T, Ushmorov A. Repression of TCF3/E2A contributes to Hodgkin lymphomagenesis. Oncotarget. 2016;7(24):36854–36864. doi: 10.18632/oncotarget.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Brauer PM, Singh J, et al. Targeted disruption of TCF12 reveals HEB as essential in human mesodermal specification and hematopoiesis. Stem Cell Rep. 2017;9(3):779–795. doi: 10.1016/j.stemcr.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouderlique T, Pena-Perez L, Kharazi S, et al. The concerted action of E2-2 and HEB is critical for early lymphoid specification. Front Immunol. 2019;10:455. doi: 10.3389/fimmu.2019.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chong LC, Twa DD, Mottok A, et al. Comprehensive characterization of programmed death ligand structural rearrangements in B-cell non-Hodgkin lymphomas. Blood. 2016;128(9):1206–1213. doi: 10.1182/blood-2015-11-683003. [DOI] [PubMed] [Google Scholar]
- 53.Loveday C, Turnbull C, Ramsay E, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43(9):879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holtick U, Vockerodt M, Pinkert D, et al. STAT3 is essential for Hodgkin lymphoma cell proliferation and is a target of tyrphostin AG17 which confers sensitization for apoptosis. Leukemia. 2005;19(6):936–944. doi: 10.1038/sj.leu.2403750. [DOI] [PubMed] [Google Scholar]
- 55.Cornish AJ, Hoang PH, Dobbins SE, et al. Identification of recurrent noncoding mutations in B-cell lymphoma using capture Hi-C. Blood Adv. 2019;3(1):21–32. doi: 10.1182/bloodadvances.2018026419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cozen W, Timofeeva MN, Li D, et al. A meta-analysis of Hodgkin lymphoma reveals 19p13.3 TCF3 as a novel susceptibility locus. Nat Commun. 2014;5:3856. doi: 10.1038/ncomms4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito Y, Shibata-Watanabe Y, Ushijima Y, et al. Oligonucleotide microarray analysis of gene expression profiles followed by real-time reverse-transcriptase polymerase chain reaction assay in chronic active Epstein-Barr virus infection. J Infect Dis. 2008;197(5):663–666. doi: 10.1086/527330. [DOI] [PubMed] [Google Scholar]
- 58.Lawrie A, Han S, Sud A, et al. Combined linkage and association analysis of classical Hodgkin lymphoma. Oncotarget. 2018;9(29):20377–20385. doi: 10.18632/oncotarget.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Zhang J, Lambert Q, et al. Interferon regulatory factor 7 is associated with Epstein-Barr virus-transformed central nervous system lymphoma and has oncogenic properties. J Virol. 2004;78(23):12987–12995. doi: 10.1128/JVI.78.23.12987-12995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen JI. Epstein-Barr virus infection. N Engl J Med. 2000;343(7):481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 61.Slager SL, Achenbach SJ, Asmann YW, et al. Mapping of the IRF8 gene identifies a 3′UTR variant associated with risk of chronic lymphocytic leukemia but not other common non-Hodgkin lymphoma subtypes. Cancer Epidemiol Biomarkers Prev. 2013;22(3):461–466. doi: 10.1158/1055-9965.EPI-12-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shukla V, Lu R. IRF4 and IRF8: Governing the virtues of B Lymphocytes. Front Biol. 2014;9(4):269–282. doi: 10.1007/s11515-014-1318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Lee D, Dhiman V, et al. An integrative ENCODE resource for cancer genomics. Nat Commun. 2020;11(1):3696. doi: 10.1038/s41467-020-14743-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449(7161):473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 65.Marinaccio C, Nico B, Maiorano E, Specchia G, Ribatti D. Insights in Hodgkin Lymphoma angiogenesis. Leuk Res. 2014;38(8):857–861. doi: 10.1016/j.leukres.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 66.Suszynska M, Ratajska M, Kozlowski P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: mutation prevalence and precise risk estimates based on a pooled analysis of ∼30,000 cases. J Ovarian Res. 2020;13(1):50. doi: 10.1186/s13048-020-00654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Selmi C, Leung PS, Sherr DH, et al. Mechanisms of environmental influence on human autoimmunity: a National Institute of Environmental Health Sciences expert panel workshop. J Autoimmun. 2012;39(4):272–284. doi: 10.1016/j.jaut.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Ceribelli A, Selmi C. Epigenetic Methods and Twin Studies. Adv Exp Med Biol. 2020;1253:95–104. doi: 10.1007/978-981-15-3449-2_3. [DOI] [PubMed] [Google Scholar]
- 69.Wang JHA, Hwang A, Weisenberger D, et al. DNA methylation differences in twins discordant for adolescent/young adult Hodgkin lymphoma. Blood. 2015;126(23):179. [abstract] Abstract 621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.