Abstract
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-based genome editing technology is an emerging RNA-guided nuclease system initially identified from the microbial adaptive immune systems. In recent years, the CRISPR-Cas system has been reprogrammed to target specific regions of the eukaryotic genome and become a powerful tool for genetic engineering. Researchers have explored many approaches to improve the genome editing activity of the CRISPR-Cas system and deliver its components both ex vivo and in vivo. Moreover, these strategies have been applied to genome editing in preclinical research and clinical trials. In this review, we focus on representative strategies for regulation and delivery of the CRISPR-Cas system, and outline current therapeutic applications in their clinical translation.
Keywords: CRISPR, gene editing, CRISPR delivery, CRISPR-based therapeutics
CRISPR: From a Prokaryotic Immune System to a Genome Editing Tool
In the battle against foreign genetic elements such as viruses and DNA plasmids, bacteria and archaea have evolved a wide array of immune systems such as the CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats) (see Glossary) system [1–6]. Many bacteria and archaea possess this unique system to protect themselves from invading viruses and DNA plasmids [5–8]. The microbial CRISPR-Cas system has been extensively used as a genome editing tool because of its ease of use and high precision [1–8]. The CRISPR-Cas system has been divided into two classes: class I with multisubunit Cas effector proteins and class II with a single Cas effector protein [5, 6]. Although some of class I CRISPR-Cas systems have been reprogrammed for eukaryotic genome editing, their complexity has limited their applications [5, 6, 9]. The class II systems employ a single Cas protein to fulfill target DNA or RNA cleavage, and show great potential for genome engineering (Figure 1) [1–6, 10, 11]. More recently, many new class II CRISPR-Cas systems like Cas12b, Cas12c, Cas12d (CasY), Cas12e (CasX), Cas12g, Cas12h, Cas12i, and Cas14a have been characterized, expanding the capability of the CRIPSR technology [11–14].
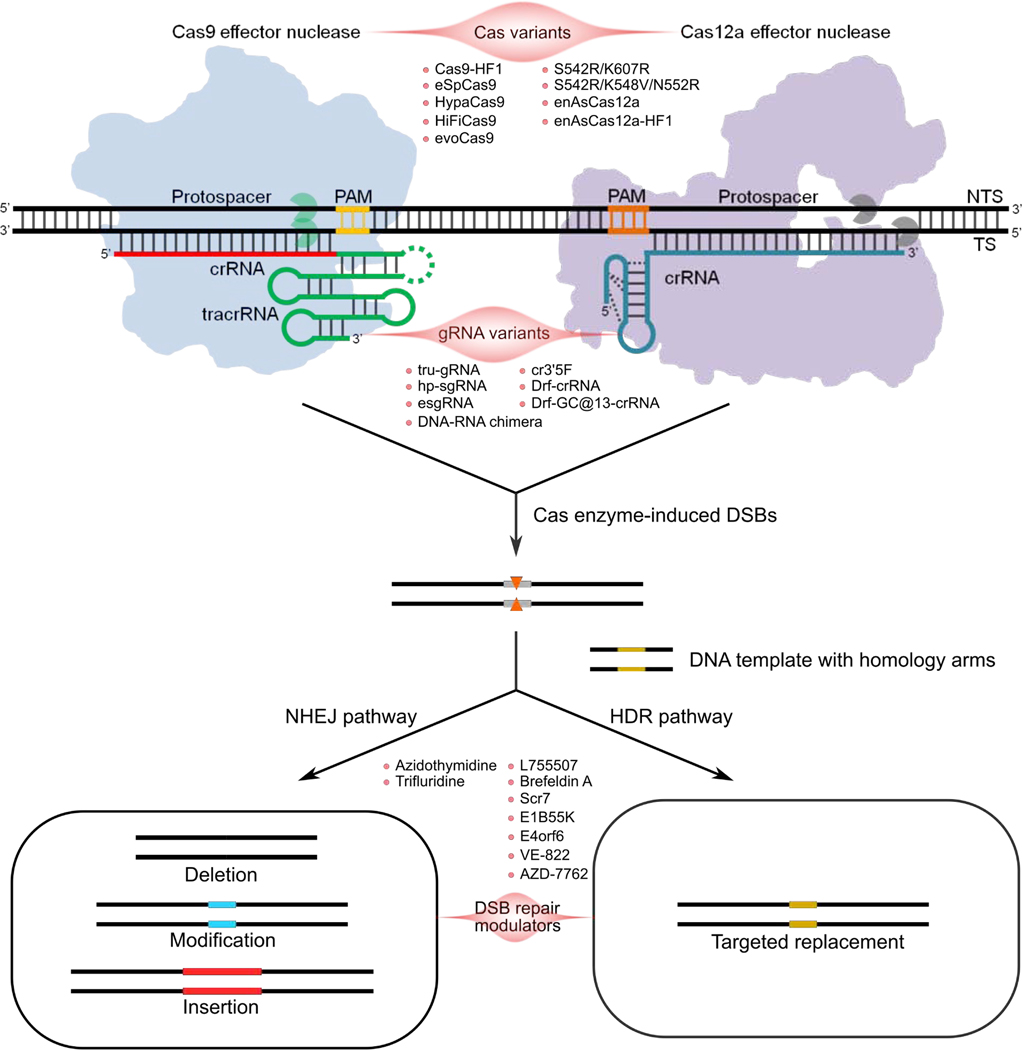
Figure 1.
Modulations of the CRISPR-Cas9 and CRISPR-Cas12a Systems. Cas and gRNA variants have been created to modulate the CRISPR system with improved efficiency, specificity or altered PAM. Biological molecules can regulate DSBs repair pathways after the CRISPR-Cas-mediated genome editing. PAM, protospacer-adjacent motif. TS, target strand. NTS, nontarget strand. crRNA and tracrRNA of the CRISPR-Cas9 system can be truncated and connected via a loop to form a single gRNA. The RuvC and HNH domains of Cas9 are indicated with green symbols. For the CRISPR-Cas12a system, crRNA is also called gRNA in some cases. The RuvC and Nuc domains of Cas12a are indicated with gray symbols.
The widely used class II Cas9 and Cas12a (also known as Cpf1) protein recognize and cleave the target double-stranded DNA under the guidance of guide RNA (gRNA) (Figure 1), [1–4, 10]. Briefly, gRNA-bound Cas complex (ribonucleoprotein, RNP) scan targets, recognize the protospacer adjacent motif (PAM), and initiate R-loop formation [8]. The process leads to PAM-dependent Cas nuclease activation that generates DNA breaks and eventual DNA repair and genome editing capability [8]. However, there are some differences between two systems (Figure 1) [1–4, 10]. First, the size of Cas12a and its gRNA is smaller than Cas9’s. Second, Cas12a recognizes a PAM sequence of TTTN, whereas the PAM for the Cas9 is NGG. Third, Cas12a make staggered cut at a distal site with RuvC and Nuc domains, but Cas9 produces a blunt end directly adjacent to the PAM with RuvC and HNH domains [1–4, 10]. Both systems are now increasingly being applied to preclinical studies and clinical trials (Table 1) for treating severe genetic diseases, angiogenesis-related eye diseases, cardiac diseases, neural diseases, cancers, infectious diseases and orphan diseases [15–19]. Below, we mainly focus on multiple therapeutic applications of the Cas9 and Cas12a systems.
Table 1.
The CRISPR-based therapy in clinical trials. The information is from ClinicalTrials.govii, accessed in September 2019.
| Target gene | Disease indication | Delivery system | Phase | Status | ClinicalTrials.gov identifier i | Sponsor |
|---|---|---|---|---|---|---|
| BCL11 A | Sickle Cell Disease, Hematological Diseases, Hemoglobinopathies | Infusions of CRISPR-Cas9 treated autologous CD34+ hematopoietic stem and progenitor cells (CTX001) (ex vivo) | Phase I/II | Recruiting | NCT03745287 | Vertex Pharmaceuticals Incorporated, USA |
| BCL11 A | Beta-Thalassemia, Thalassemia, Genetic Diseases, Inborn Hematologic Diseases, Hemoglobinopathies | Infusions of CRISPR-Cas9 treated autologous CD34+ hematopoietic stem and progenitor cells (CTX001) (ex vivo) | Phase I/II | Recruiting | NCT03655678 | Vertex Pharmaceuticals Incorporated, USA |
| CEP290 | Leber Congenital Amaurosis 10 | Subretinal injection of AGN-151587 (EDIT-101) (in vivo) | Phase I/II | Recruiting | NCT03872479 | Allergan, USA |
| Unknown | B-cell Malignancy, Non-Hodgkin Lymphoma, B-cell Lymphoma | Infusions of CRISPR-Cas9 treated CD19-directed T cells (CTX110) (ex vivo) | Phase I/II | Recruiting | NCT04035434 | CRISPR Therapeutics AG, USA |
| TCR and B2M | B Cell Leukemia, B Cell Lymphoma | Infusions of CRISPR-Cas9 treated CD19-directed CAR-T cells (ex vivo) | Phase I/II | Recruiting | NCT03166878 | Chinese PLA General Hospital, China |
| Unknown | B Cell Leukemia, B Cell Lymphoma | Infusions of CRISPR-Cas9 treated CD19- and CD20- (or CD19- and CD22-) directed CAR-T cells (ex vivo) | Phase I/II | Recruiting | NCT03398967 | Chinese PLA General Hospital, China |
| PD-1 | Gastric Carcinoma, Nasopharyngeal Carcinoma, T-Cell Lymphoma, Adult Hodgkin Lymphoma, Diffuse Large B-Cell Lymphoma | Infusions of CRISPR-Cas9 treated cytotoxic T lymphocytes (ex vivo) | Phase I/II | Recruiting | NCT03044743 | The Affiliated Nanjing Drum Tower Hospital of Nanjing University Medical School, China |
| HPV E6/E7 | HPV-related Cervical Intraepithelial Neoplasia | Plasmid in gel (in vivo) | Phase I | Not yet recruiting | NCT03057912 | First Affiliated Hospital, Sun Yat-Sen University, China |
| PD-1and TCR | Solid Tumor, Adult | CRISPR-Cas9 treated CAR-T cells infusions (ex vivo) | Phase I | Recruiting | NCT03545815 | Chinese PLA General Hospital, China |
| T cell receptor and PD-1 | Multiple Myeloma, Melanoma, Synovial Sarcoma, Myxoid/Round Cell Liposarcoma | Infusions of CRISPR-Cas9 treated autologous T cells (ex vivo), combined with chemotherapy agents | Phase I | Recruiting | NCT03399448 | University of Pennsylvania, USA |
| HPK1 | CD19 Positive Leukemia, Lymphoma | Infusions of CRISPR-Cas9 treated autologous CD19-directed T cells (transfection with a lentiviral vector and electroporation) (ex vivo), combined with chemothe rapy agents | Phase I | Recruiting | NCT04037566 | Xijing Hospital, China |
| PD-1 | Solid Tumor, Adult | CRISPR-Cas9 treated CAR-T cells infusions (ex vivo), combined with chemotherapy agents | Phase I | Recruiting | NCT03747965 | Chinese PLA General Hospital, China |
| PD-1 | Metastatic Non-small Cell Lung Cancer | Infusions of CRISPR-Cas9 treated T cells (ex vivo) | Phase I | Active, not recruiting | NCT02793856 | Sichuan University, China |
| HBB | Thalassemia | Infusions of CRISPR-Cas9 treated induced hematopoietic stem cells (ex vivo) | Early Phase I | Not yet recruiting | NCT03728322 | Allife Medical Science and Technology Co., Ltd., China |
| CCR5 | HIV-1-infection | Transplantation of CRISPR-Cas9 treated CD34+ hematopoietic stem and progenitor cells (ex vivo) | Not Applicable | Recruiting | NCT03164135 | Affiliated Hospital to Academy of Military Medical Sciences, China |
| β-globin | Sickle Cell Disease | Unknown | Not Applicable | Suspended | NCT03167450 | National Human Genome Research Institute, USA |
| KMT2 D | Kabuki Syndrome 1 | Unknown (ex vivo) | Not Applicable | Active, not recruiting | NCT03855631 | University Hospital, Montpellier, France |
| PD-1 | Esophageal Cancer | Infusions of CRISPR-Cas9 treated T cells (ex vivo) | Not Applicable | Completed | NCT03081715 | Hangzhou Cancer Hospital, China |
| NF1 | Neurofibromatosis Type 1 with Tumors of the Central Nervous System | Unknown (ex vivo) | Not Applicable | Completed | NCT03332030 | Children’s Research Institute, USA |
| KMT2 D | Kabuki Syndrome 1 | Unknown (ex vivo) | Not Applicable | Active, not recruiting | NCT03855631 | University Hospital, Montpellier, France |
Modulation of the CRISPR System for Improved Genome Editing
On the basis of crystal structure information of the CRISPR-Cas system, many Cas9 and Cas12a variants have been developed to improve the performance of CRISPR-mediated genome editing (Figure 1) [18, 20, 21]. For instance, structure-guided mutagenesis screen is being explored to identify new Cas9 and Cas12a variants with improved DNA specificity or targeting range [18, 20, 21]. Phage-assisted continuous evolution is also used to rapidly generate Cas9 variants [22]. Evolved Cas9 not only possesses greater DNA specificity than the wild-type Cas9, but also recognizes noncanonical PAMs such as NG, GAA and GAT [22]. Moreover, catalytically inactive Cas (dCas) variants (dCas9 and dCas12a) have been generated to increase the system’s specificity [1, 10]. Fusion of deaminases to dCas9 or dCas12a makes it possible to accomplish mutual conversion among four bases, which expands the potential applications of the CRISPR-Cas system for correcting disease-associated single nucleotide polymorphisms (SNPs) [23–27]. More recently, prime editing has been developed by fusing dCas9 to an engineered reverse transcriptase for genome engineering without introducing double-strand breaks or donor DNA [28].
Both the Cas9 and Cas12a systems have been reprogrammed to cleave targeted DNA sequences by rational design of gRNA sequences, enabling researchers to easily edit DNA sequences [1–4, 10]. Introduction of chemical modifications to synthetic gRNA has a critical effect on the activity of the CRISPR-Cas system [29–35]. It has also been found that co-delivery of Cas9 mRNA and gRNAs with 2’-O-methyl-3’-phosphorothioate modifications at both termini enhances Cas9-mediated genome editing efficiency from the background level to detectable frequency in multiple human primary cells [29]. Also, the combination of chemically modified Cas12a mRNA and gRNA containing five 2’-fluoro ribose at the 3’ terminus augments editing efficiency at least 3-fold [31, 35]. To further improve the potency of Cas9, the Anderson and Morrissey group respectively constructed gRNAs with massive modifications at hairpins [32, 33]. When co-delivered with Cas9 mRNA, these modified gRNAs induce dramatic knockout of serum protein encoded by the edited gene in the mouse liver [32, 33]. Additionally, modification of the 5’-triphosphate group of in vitro transcribed gRNA with phosphatase reduces T cell mediated immune response [36]. Apart from chemical modifications, engineering of the secondary structure of gRNA has been reported to enhance the CRISPR efficiency or specificity (Figure 1) [37–44]. Truncation of gRNA, partial DNA replacement, and 5’ hairpin addition has led to remarkable improvement in the CRISPR specificity [37–39]. Additionally, extension of the either end of Cas12a gRNA and optimization of hairpin structure are two important strategies to enhance on-target activity of the CRISPR-Cas12a system [40–44].
Repair of the CRISPR-induced double-stranded breaks (DSBs) involves in two types of pathways, including homology-directed repair (HDR) and non-homologous end joining (NHEJ) (Figure 1). A series of biological molecules have been found to increase the CRISPR-mediated HDR efficiency, the process that is desired for efficient gene knockin [45–48]. These HDR enhancers include the β3-adrenergic receptor agonist L755507 [45], the protein transport inhibitor Brefeldin A [45], ligase IV inhibitors Scr7 [46, 47], E1B55K and E4orf6 [47], kinase inhibitors VE-822 and AZD-7762 [48], and so on. Among them, Scr7 dramatically promotes the HDR efficiency in both cells and mice [46]. The improved knockin efficiency attributes to the suppression of the NHEJ pathway [45–48]. By contrast, repair with NHEJ can cause DNA alterations including deletions, insertions and modifications, which is beneficial to gene knockout. Thymidine analogs such as azidothymidine and trifluridine, were found to increase the CRISPR-mediated NHEJ knockout efficiency by impairing the HDR pathway [45].
Delivery of the CRISPR Components
Ex vivo and in vivo genome editing are two approaches for the CRISPR-mediated therapy in clinical development (Figure 2) where the CRISPR components can be delivered into mammalian cells in the form of DNA, RNA, or RNP [49, 50]. However, delivery of the CRISPR components is one of the challenges to overcome for successful clinical applications of the CRISPR technologies [49–51]. In most current clinical trials, electroporation remains one of the widely used methods for delivery of CRISPR ex vivo genome editing [49–51]. In recent years, new electroporation-based technologies such as electroporation of mouse zygotes with pre-assembled Cas9 RNP complex (CRISPR-EZ) and combination of adeno-associated virus (AAV)-mediated donor infection with Cas9 RNP electroporation (CRISPR-READI) have been developed to codeliver the CRISPR components [52, 53]. CRISPR-EZ causes 88% gene disruption and 42% HDR-mediated editing[52]. Further optimization of electroporation conditions enables complete gene disruption and up to 62.5% HDR efficiency [52]. CRISPR-READI allows site-specific integration of AAV donors with lengths up to 4.9 kb [53].
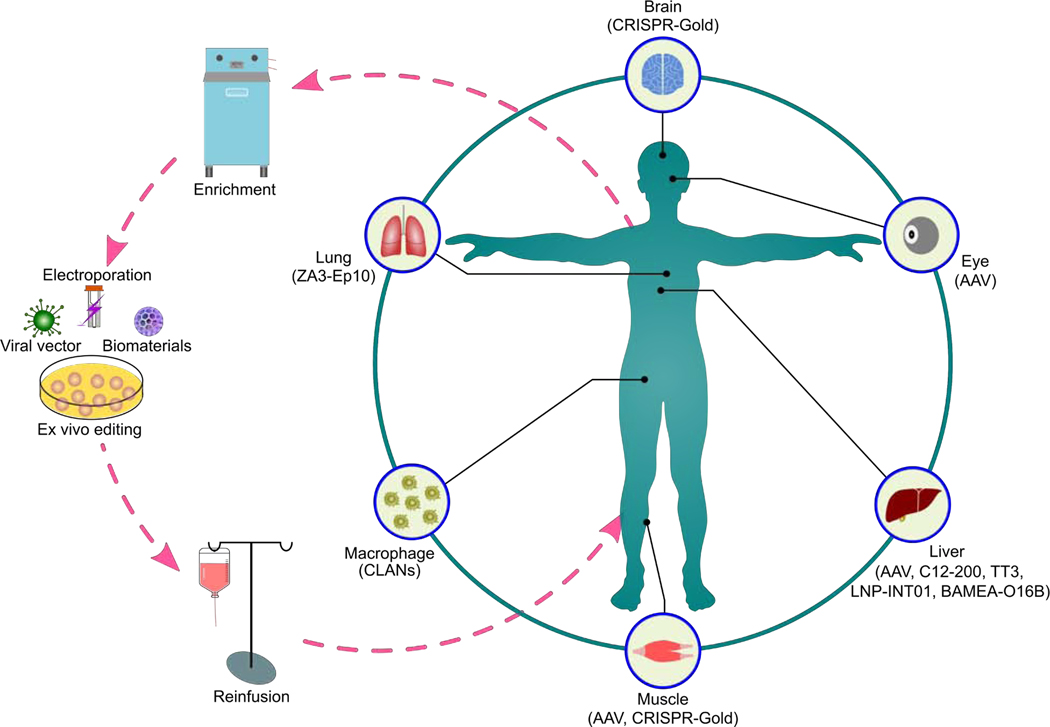
Figure 2.
Graphic Overview of the CRISPR-based Ex Vivo and In Vivo Therapeutics. The CRISPR systems can be delivered ex vivo and in vivo by physical methods, viral vectors, biomaterials or their combinations. The dashed pink circle indicates procedures for the CRISPR-mediated genome editing ex vivo. The light green circle indicates in vivo therapeutic applications of the CRISPR system.
In the case of systemic delivery in vivo, multiple factors including absorption, distribution, metabolism, and excretion (ADME) need to be carefully studied in order to ensure effective and safe genome editing in target cells and organs. Moreover, when selecting delivery vehicles for the CRISPR system, the delivery strategy should match the need of specific diseases. An appropriate in vivo delivery method should possess favorable pharmaceutical profiles like high delivery efficiency, high tissue specificity, and low side effects [49]. AAVs are the commonly used viral vectors for CRISPR delivery in vivo [54]. AAV serotypesi with different tropisms have the ability to target different organs [54]. Apart from AAVs, lentivirus has also been explored for CRISPR delivery in preclinical studies [51, 55].
Recently, a number of biomaterials such as lipid, polymeric, and inorganic nanoparticles (NPs) are developed for transient expression of the CRISPR systems in vivo [49, 50, 56]. For example, zwitterionic amino lipid, ZA3-Ep10 has been formulated for co-delivery of Cas9 mRNA and sgRNA in mice [57]. In vivo bioluminescence imaging indicates that ZA3-Ep10 NPs mainly distribute in the lung (Figure 2) [57]. Cationic lipid-assisted NPs (CLANs) have been constructed as NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3) inflammasome-targeting vehicles in order to deliver Cas9 mRNA and gRNA to inflammasome in macrophages (Figure 2) [58]. Intravenous injection of CLANs containing Cas9 mRNA and gRNA targeting NLRP3 alleviates NLRP3-dependent acute inflammation and adipose inflammation in mouse models [58].
Further, PEGylated helical polypeptide nanoparticles (P-HNPs) loaded with Cas9 plasmid- and gRNA targeting polo-like kinase 1 have displayed more than 71% tumor inhibition efficacy in HeLa xenograft tumor-bearing mice [59]. Recent studies show that the CRISPR-Gold NPs composed of Cas9 RNP, gold nanoparticles, DNA, and cationic polymers not only mediate genome editing in neurons, astrocytes, and microglia, but also alleviate the exaggerated repetitive behaviors of fragile X syndrome by reduction of the metabotropic glutamate receptor 5 (mGluR5) protein in the brain striatum of a mouse model with an intracranial injection (Figure 2) [60]. Meanwhile, CRISPR-Gold NPs enable local delivery of the CRISPR for treating muscle degeneration disease (Figure 2) [61]. Collectively, these biomaterials represent promising delivery platforms for effective in vivo genome editing.
Ex Vivo Therapeutic Applications of the CRISPR System
As stated earlier, ex vivo remains the mainstream method for the CRISPR-based therapeutics in current clinical translations (Table 1). Editing of hematopoietic stem and progenitor cells (HSPCs) with the CRISPR-Cas system provides a potential treatment option for hematological diseases, as evidenced by preclinical and clinical studies [62–65]. CTX001, a CRISPR-mediated autologous HSPCs therapy, is currently in a Phase I/II clinical trial for the treatment of severe sickle cell disease and transfusion-dependent β-thalassemia (ClinicalTrials.gov Identifier: NCT03745287 and NCT03655678, Table 1). Preclinical studies show that disruption of the BCL11A erythroid enhancer with the CRISPR technology increase therapeutic levels of hemoglobin [64]. This strategy can also be used to restore globin chain balance of hematopoietic stem cells with β-thalassemia [64]. For the sickle cell disease, the CRISPR-mediated genome editing of the mutation in the β-globin gene in patient-derived HSPCs not only effectively reduces the number of sickle cells but also induces stable expression of normal hemoglobin in mice post-transplantation [63]. In addition to hematological diseases, CRISPR-mediated gene editing of HSPCs is applicable to X-linked chronic granulomatous disease (X-CGD). Electroporation of Cas9 mRNA, gRNA targeting the CYBB gene, and ssDNA template into CD34+ HSPCs from X-CGD patients enables more than 20% reparation of the mutant CYBB gene. Moreover, production of functional mature human myeloid and lymphoid cells lasts up to 5 months when corrected X-CGD HSPCs are transplanted into mouse models [62].
In addition, ex vivo genome editing of induced pluripotent stem cells (iPSCs) holds promises for the treatment of Duchenne muscular dystrophy (DMD, a severe muscle-degenerative disease caused by small deletions, exon duplications, or loss of exons in the dystrophin gene) [66]. Different strategies are needed for different type of mutations. Electroporation of DMD-patient-derived iPSCs with CRISPR-Cas9 system for exon knockin is a useful strategy for correcting iPSCs from DMD patients who suffer from loss of exons in the dystrophin gene [66]. Full-length dystrophin protein expression can be detected after DMD-patient-derived iPSCs were corrected and differentiated into skeletal muscle cells [66]. Similar outcomes are observed when CRISPR-based deletion strategy is utilized to restore the reading frame of DMD-patient-derived iPSCs with frameshift mutations [67]. After engrafting these iPSCs into the animal model of DMD, these cells result in the functional dystrophin glycoprotein complex in vivo [67].
CRISPR-based ex vivo therapeutics have also been applied to other orphan diseases. For example, recessive dystrophic epidermolysis bullosa (RDEB), a rare hereditary skin disorder, is caused by mutations in the gene of COL7A1-encoding type VII collagen [68]. Grafting of ex vivo CRISPR-edited RDEB keratinocytes or fibroblasts onto immunodeficient mice leads to functional collagen VII expression and right localization [68, 69]. Another example of application of CRISPR-based therapeutics in orphan diseases is in hereditary tyrosinemia type 1, a rare metabolic disease. Recent studies show that transplantation of the CRISPR-edited hepatocytes improves the metabolic liver injury caused by the disease [70, 71].
In Vivo Therapeutic Applications of the CRISPR System
Muscle degeneration disease
One of the most successful examples of in vivo applications of CRISPR is in the muscle degeneration disease DMD (Figure 2). In 2016, three separate studies described use of AAV9-mediated CRISPR-Cas9 technology to treat DMD in mouse models [72–74].
Although distinct administration routes including intraperitoneal, intramuscular, and systemic administration were employed to deliver the CRISPR-Cas9 using AAV9 vectors, researchers are able to detect corrected dystrophin expression, and thereby partially recover skeletal or cardiac muscle functional in the mouse model of DMD [72–74]. The common strategy among these studies is to correct the point mutation in exon 23 of the mouse model of DMD [72–74]. Recently, the Olson group reported that AAV9-Cas9-mediated correction of exon 44 deletion mutations is also an efficient strategy for the treatment of DMD in human cells and transgenic mice harboring the same deletion mutation [75, 76].
Eye-related genetic diseases
In early 2019, a single ascending dose clinical trial (Phase I/II, ClinicalTrials.gov Identifier: NCT03872479, Table 1) was initiated for the treatment of Leber Congenital Amaurosis type 10 (LCA10, a retinal degenerative disease caused by a mutation in the CEP290 gene). Preclinical studies show that subretinal injection of EDIT-101 (also called AGN-151587, Table 1), AAV5 packaged CRISPR-Cas9, restores vision loss in humanized CEP290 mice [77]. In addition, vascular endothelial growth factor 2 (VEGFR2) is an important therapeutic target for angiogenesis-associated diseases such as proliferative diabetic retinopathy and neovascular age-related macular degeneration [78]. To disrupt genomic VEGFR2 locus, a recent work uses recombinant AAV1 to pack Cas9 and gRNA [78]. Intravitreal injection of AAV1-mediated Cas9/gRNA leads to remarkable elimination of angiogenesis, as demonstrated in two mouse models of eye diseases: oxygen-induced retinopathy and laser-induced choroid neovascularization (Figure 2) [78]. This study suggests that AAV1-mediated CRISPR-Cas9 targeting aberrant VEGFR2 gene not only inhibits pathological angiogenesis in mouse models, but also provides a method for the treatment of other VEGF-induced neovascularization like inhibiting tumor growth and tumor metastasis by reduction of the formation of angiogenesis. Further, to edit angiogenesis-associated vascular endothelial growth factor A (Vegfa) and hypoxia inducing factor 1a (Hif1a) genes, DNA sequences encoding Cas12a and crRNA targeting Vegfa and Hif1a are incorporated into an AAV9 vector. A single intravitreal administration of these AAV into the mouse retina gives rise to a long-term reduction of the area of laser-induced choroidal neovascularization [79].
Liver-related genetic diseases
Gene correction of pathogenic mutations in the liver with the CRISPR systems offers a treatment option for liver-related genetic diseases such as hereditary tyrosinemia. Systemic delivery of Cas9 mRNA with C12–200 lipid NPs and gRNA/HDR template with AAV vector that is able to correct 6% of hepatocytes in a mouse model of human hereditary tyrosinemia (Figure 2) [80]. Another example is alpha-1 antitrypsin deficiency (AATD). Delivery of dual AAV vectors expressing Cas9 and gRNA/HDR template into mouse models of AATD is able to correct mutant allele in the liver and restore serum AAT level to within normal ranges (Figure 2) [81]. Proprotein convertase subtilisin/kexin type 9 (PCSK9) related to hypercholesterolemia, plays an important role in metabolism of the low-density lipoprotein. Administration of Cas9/gRNA packaged in AAV2/8 targeting the Pcsk9 gene in the mouse liver with AAV titer of 0.5×1011 to 4×1011 leads to a 95% decrease in serum Pcsk9 and a 40% decrease in total cholesterol in mice for a month (Figure 2) [82].
Several groups have also developed biomaterials to deliver the CRISPR components to the liver [83, 84]. For instance, TT3 and BAMEA-O16B are lipid NPs are used for systemic co-delivery of Cas9 mRNA and gRNA to the mice liver, allowing effectively Pcsk9 gene editing and down-regulation of Pcsk9 protein level (Figure 2) [83–85]. LNP-INT01 is also a lipid NPs which can be used for CRISPR delivery to the liver to treat transthyretin amyloidosis [33]. Administration of single dosed LNP-INT01 containing Cas9 mRNA and chemically modified gRNA targeting the transthyretin (Ttr) gene in the mouse liver decreases over 97% of serum TTR protein in both mice and rats. Moreover, the low systemic TTR levels is observed for at least one year (Figure 2) [33].
Cancers
NP-mediated delivery of the CRISPR system has also shown potential for cancer therapy. Co-delivery of the CRISPR system and paclitaxel (an anti-cancer drug) to tumor tissues with R8-dGR (a cell penetrating peptide)-modified cationic liposome is capable of inhibiting the metastasis of pancreatic tumor cells and prolonged survival time without inducing severe toxicity [85]. Encapsulating plasmids encoding Cas9 and gRNA targeting VEGFA into a lipopolymer display tumor targeting properties [86]. Intravenous administration of this formulation results in significant accumulation in the tumor, thus effectively inhibiting osteosarcoma malignancy and lung metastasis by down-regulation of the angiogenesis [86]. Targeting of oncogenes or tumor suppressor genes using the CRISPR technology is another possible method for the treatment of cancer [87]. Editing of endogenous activated oncogenes such as epidermal growth factor receptor (EGFR), HRAS, and BRAF that are recurrently found in various cancer types, via the CRISPR-Cas9, may disrupt gain-of-function mutations [87]. Similar concepts have been applied to target the viral oncogenes. For instance, the CRISPR-mediated disruption of HPV E6 and /or E7 oncogene, responsible for cervical cancer, is currently in a phase I clinical trial (ClinicalTrials.gov Identifier: NCT03057912, Table 1) [87].
Conclusions and Future Perspectives
The bacteria derived CRISPR-Cas system enables precise and effective genome engineering in eukaryotic organisms. Recent developments in the CRISPR technology have witnessed the continuous progresses in the field of genome editing. Rational design of the CRISPR delivery platforms make it possible to achieve tissue-specific genome editing in vivo. A number of CRISPR-based ex vivo and in vivo therapeutics are currently in clinical trials (Table 1). Moreover, the structure-guided mutagenesis screening or directed evolution of the CRISPR-Cas system results in the next generation Cas endonucleases with enhanced target specificity and expanded targeting ranges [18].
While massive advances have been made to facilitate clinical translations of the CRISPR, there are some limitations that need to be solved before the CRISPR technology become a prevailing genome editing tool for modern medicine (see Outstanding Questions) [18, 19, 88]. Systemic delivery of the CRISPR components remains a key challenge. Many types of cells such as neurons, cardiomyocytes, and immune cells demand in vivo delivery vehicles for efficient genome editing. Another concerns is to minimize off-target effects (undesired genome editing in other gene loci) of the system, so as to prevent large deletions and complex rearrangements [89]. Standard and quantitative methods should be established to analyze the genome editing data. Additionally, clinical translation of the CRISPR system may encounter multiple concerns such as the p53-mediated response of DNA damage [90, 91] and pre-existing antibodies to Cas proteins [92–94]. Comprehensive studies are needed to carefully assess the impact of these findings. By gaining more insights from preclinical and clinical data, researchers and clinicians will have a profound understanding of the CRIPSR system. With these new knowledge and experience, we envision that the CRISPR technology may be exploited to treat broad human diseases in the future.
Outstanding Questions.
What would be a gold standard method to assess off-target mutations induced by the CRISPR-Cas system?
How could we rationally design delivery vehicles for the CRISPR-Cas system to address various delivery challenges including specificity, efficiency, and safety?
How to evaluate the safety of the CRISPR technology in addition to standard clinical toxicity studies?
What would be the appropriate applications of the CRISPR technology in compliance with ethical and social concerns?
Highlights.
The CRISPR-Cas system, a RNA-guided endonuclease, capable of sequence-specific cleavage of target DNA, has been widely used as a genome editing toolbox.
Multiple strategies have been developed to modulate the activity of the CRISPR-Cas system, thus enabling precise control of the CRISPR-mediated genome editing.
The CRISPR-Cas system has been applied to edit eukaryotic genome both in vitro and in vivo, and has been intensively investigated in preclinical studies and clinical trials.
ACKNOWLEDGEMENTS
Y.N. and W.J. acknowledge the National Key Research and Development Program (2016YFA0101401). Y.D. acknowledges the support of the Maximizing Investigators’ Research Award R35GM119679 from the National Institute of General Medical Sciences.
GLOSSARY
- Catalytically inactive Cas (dCas) variants
Cas variants retain DNA binding affinity without their cleavage activity
- CRISPR-Cas
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated genes) refers to both the CRISPR locus and an array of the CRISPR associated genes (cas) located in the genomes of bacteria and archaea. The CRISPR locus consists of short repeated sequences separated by spacers, which is transcribed and processed into guide RNA (gRNA) that can direct Cas nucleases encoded by cas genes to recognize and cut target nucleic acids sequences
- Electroporation
a physical approach that is being widely explored for delivery of exogenous nucleic acids into a variety of cell types ex vivo especially for hard-to-transfect cells, due to the increased cell permeability induced by controlled electrical pulses
- Guide RNA (gRNA)
The CRISPR-Cas system is directed by the CRISPR RNA (crRNA) or a base-paired precursor crRNA and trans-activating crRNA (tracrRNA). To simplify the dual-RNA components, crRNA and tracrRNA are truncated and connected via a loop to form a single guide RNA (gRNA)
- Hematopoietic stem and progenitor cells (HSPCs)
Cells possess the ability to selfrenewal and differentiation into blood cells
- Homology-directed repair (HDR)
a DNA repair pathway that repairs double-stranded DNA breaks (as caused by the CRISPR-Cas system) in the presence of a homologous DNA sequence. This repair process leads to targeted gene replacement
- Induced pluripotent stem cells (iPSCs)
Cells, derived from non-pluripotent adult cells, possess embryonic stem cell-like properties
- Intravitreous injection
an administration route via the vitreous of the eye
- Non-homologous end joining (NHEJ)
a DNA repair pathway that repairs double-stranded DNA breaks in the absence of a homologous DNA sequence. Repair with NHEJ can cause DNA alterations including deletions, insertions and modifications
- Orphan disease
also called rare disease. The incidence of such disease is low in the population (< 200,000 people in the US)
- Protospacer adjacent motif (PAM)
a short and specific DNA sequence adjacent to the target DNA region (protospacer), which is required for target DNA recognition and cleavage by Cas endonuclease
- R-loop
a triple-stranded structure comprises a gRNA-DNA hybrid and a DNA strand which is paired with the DNA strand in the hybrid before unwinding
- Subretinal injection
an administration route via the subretinal space of the eye
- X-linked chronic granulomatous disease (X-CGD)
an chronic immunodeficiency disease caused by mutations in the CYBB gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jinek M, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasiunas G, et al. (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109, E2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, et al. (2013) RNA-Guided Human Genome Engineering via Cas9. Science 339, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, et al. (2015) An updated evolutionary classification of CRISPR–Cas systems. Nature Reviews Microbiology 13, 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shmakov S, et al. (2017) Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Oost J, et al. (2014) Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol 12, 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JS and Doudna JA (2017) The chemistry of Cas9 and its CRISPR colleagues. Nature Reviews Chemistry 1, 0078 [Google Scholar]
- 9.Pickar-Oliver A, et al. (2019) Targeted transcriptional modulation with type I CRISPR–Cas systems in human cells. Nature Biotechnology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zetsche B, et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S, et al. (2015) Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 60, 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burstein D, et al. (2017) New CRISPR-Cas systems from uncultivated microbes. Nature 542, 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington LB, et al. (2018) Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan WX, et al. (2019) Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chertow DS. (2018) Next-generation diagnostics with CRISPR. Science 360, 381–382 [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. (2019) CRISPR/Cas Systems towards Next-Generation Biosensing. Trends in Biotechnology 37, 730–743 [DOI] [PubMed] [Google Scholar]
- 17.Porteus MH (2019) A New Class of Medicines through DNA Editing. N Engl J Med 380, 947–959 [DOI] [PubMed] [Google Scholar]
- 18.Pickar-Oliver A. and Gersbach CA (2019) The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 20, 490–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fellmann C, et al. (2017) Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov 16, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L, et al. (2017) Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol 35, 789–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinstiver BP, et al. (2019) Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nature Biotechnology 37, 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu JH, et al. (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komor AC, et al. (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudelli NM, et al. (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. (2018) Base editing with a Cpf1–cytidine deaminase fusion. Nature Biotechnology 36, 324. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, et al. (2018) Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nature Biotechnology 36, 946. [DOI] [PubMed] [Google Scholar]
- 27.Kim HS, et al. (2019) Adenine base editors catalyze cytosine conversions in human cells. Nature Biotechnology 37, 1145–1148 [DOI] [PubMed] [Google Scholar]
- 28.Anzalone AV, et al. (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendel A, et al. (2015) Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahdar M, et al. (2015) Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc Natl Acad Sci U S A 112, E7110–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, et al. (2017) Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat Biomed Eng 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin H., et al. (2017) Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol 35, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn JD, et al. (2018) A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep 22, 2227–2235 [DOI] [PubMed] [Google Scholar]
- 34.McMahon MA, et al. (2018) Chemically Modified Cpf1-CRISPR RNAs Mediate Efficient Genome Editing in Mammalian Cells. Mol Ther 26, 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, et al. (2018) Design and assessment of engineered CRISPR-Cpf1 and its use for genome editing. Nat Protoc 13, 899–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, et al. (2018) CRISPR RNAs trigger innate immune responses in human cells. Genome Research 28, 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y, et al. (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology 32, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin H, et al. (2018) Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nature Chemical Biology 14, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kocak DD, et al. (2019) Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol 37, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park HM, et al. (2018) Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nature Communications 9, 3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bin Moon S, et al. (2018) Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang. Nature Communications 9, 3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L, et al. (2018) Engineering the Direct Repeat Sequence of crRNA for Optimization of FnCpf1-Mediated Genome Editing in Human Cells. Molecular Therapy 26, 2650–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng F, et al. (2019) Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biology 20, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu P, et al. (2019) Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res 47, 4169–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu C, et al. (2015) Small Molecules Enhance CRISPR Genome Editing in Pluripotent Stem Cells. Cell Stem Cell 16, 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maruyama T, et al. (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature Biotechnology 33, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu VT, et al. (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature Biotechnology 33, 543. [DOI] [PubMed] [Google Scholar]
- 48.Ma X, et al. (2018) Small molecules promote CRISPR-Cpf1-mediated genome editing in human pluripotent stem cells. Nature Communications 9, 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glass Z, et al. (2018) Engineering the Delivery System for CRISPR-Based Genome Editing. Trends in Biotechnology 36, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H-X, et al. (2017) CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chemical Reviews 117, 9874–9906 [DOI] [PubMed] [Google Scholar]
- 51.Yin H, et al. (2017) Delivery technologies for genome editing. Nature Reviews Drug Discovery 16, 387. [DOI] [PubMed] [Google Scholar]
- 52.Modzelewski AJ, et al. (2018) Efficient mouse genome engineering by CRISPR-EZ technology. Nat Protoc 13, 1253–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, et al. (2019) CRISPR-READI: Efficient Generation of Knockin Mice by CRISPR RNP Electroporation and AAV Donor Infection. Cell Rep 27, 3780–3789 e3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D, et al. (2019) Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 18, 358–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilbie D, et al. (2019) Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc Chem Res 52, 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan T, et al. (2019) Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Materials Today 26, 40–66 [Google Scholar]
- 57.Miller JB, et al. (2017) Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl 56, 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu C, et al. (2018) Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases. Nat Commun 9, 4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang HX, et al. (2018) Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc Natl Acad Sci U S A 115, 4903–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee B, et al. (2018) Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nature Biomedical Engineering 2, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee K, et al. (2017) Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nature Biomedical Engineering 1, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Ravin SS, et al. (2017) CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Science Translational Medicine 9, eaah3480 [DOI] [PubMed] [Google Scholar]
- 63.Park SH, et al. (2019) Highly efficient editing of the beta-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Y, et al. (2019) Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med 25, 776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humbert O, et al. (2019) Therapeutically relevant engraftment of a CRISPR-Cas9– edited HSC-enriched population with HbF reactivation in nonhuman primates. Science Translational Medicine 11, eaaw3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li HL, et al. (2015) Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 4, 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young Courtney S., et al. (2016) A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell 18, 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Izmiryan A, et al. (2018) Ex Vivo COL7A1 Correction for Recessive Dystrophic Epidermolysis Bullosa Using CRISPR/Cas9 and Homology-Directed Repair. Mol Ther Nucleic Acids 12, 554–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonafont J, et al. (2019) Clinically Relevant Correction of Recessive Dystrophic Epidermolysis Bullosa by Dual sgRNA CRISPR/Cas9-Mediated Gene Editing. Mol Ther 27, 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.VanLith C, et al. (2018) Curative Ex Vivo Hepatocyte-Directed Gene Editing in a Mouse Model of Hereditary Tyrosinemia Type 1. Hum Gene Ther 29, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.VanLith CJ, et al. (2019) Ex Vivo Hepatocyte Reprograming Promotes Homology-Directed DNA Repair to Correct Metabolic Disease in Mice After Transplantation. Hepatol Commun 3, 558–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tabebordbar M, et al. (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nelson CE, et al. (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Long C, et al. (2016) Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Min YL and Li H. (2019) CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Science Advances 5, eaav4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dash PK, et al. (2019) Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat Commun 10, 2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maeder ML, et al. (2019) Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nature Medicine 25, 229–233 [DOI] [PubMed] [Google Scholar]
- 78.Huang X, et al. (2017) Genome editing abrogates angiogenesis in vivo. Nat Commun 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koo T, et al. (2018) CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Nat Commun 9, 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin H, et al. (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 34, 328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song CQ, et al. (2018) In Vivo Genome Editing Partially Restores Alpha1-Antitrypsin in a Murine Model of AAT Deficiency. Hum Gene Ther 29, 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ran FA, et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li B, et al. (2015) An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo. Nano Lett 15, 8099–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang C, et al. (2017) A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res 27, 440–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu J, et al. (2019) Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv Mater 31, e1902575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liang C, et al. (2017) Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 147, 68–85 [DOI] [PubMed] [Google Scholar]
- 87.Oppel F, et al. (2018) Specific Targeting of Oncogenes Using CRISPR Technology. Cancer Res 78, 5506–5512 [DOI] [PubMed] [Google Scholar]
- 88.Rossant J. (2018) Gene editing in human development: ethical concerns and practical applications. Development 145, dev150888 [DOI] [PubMed] [Google Scholar]
- 89.Kosicki M, et al. (2018) Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36, 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haapaniemi E, et al. (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24, 927–930 [DOI] [PubMed] [Google Scholar]
- 91.Ihry RJ, et al. (2018) p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24, 939–946 [DOI] [PubMed] [Google Scholar]
- 92.Simhadri VL, et al. (2018) Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol Ther Methods Clin Dev 10, 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagner DL, et al. (2019) High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med 25, 242–248 [DOI] [PubMed] [Google Scholar]
- 94.Charlesworth CT, et al. (2019) Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 25, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]