Abstract
Optical techniques hold great potential to detect and monitor disease states as they are a fast, non-invasive toolkit. Raman spectroscopy (RS) in particular is a powerful label-free method capable of quantifying the biomolecular content of tissues. Still, spontaneous Raman scattering lacks information about tissue morphology due to its inability to rapidly assess a large field of view. Optical Coherence Tomography (OCT) is an interferometric optical method capable of fast, depth-resolved imaging of tissue morphology, but lacks detailed molecular contrast. In many cases, pairing label-free techniques into multimodal systems allows for a more diverse field of applications. Integrating RS and OCT into a single instrument allows for both structural imaging and biochemical interrogation of tissues and therefore offers a more comprehensive means for clinical diagnosis. This review summarizes the efforts made to date toward combining spontaneous RS-OCT instrumentation for biomedical analysis, including insights into primary design considerations and data interpretation.
Keywords: biomedical, label-free, multimodal, optical coherence tomography, Raman spectroscopy
Graphical Abstract
This review summarizes the efforts made to date toward combining Raman Spectroscopy (RS) and Optical Coherence Tomography (OCT) into a single instrument and aims to serve as a ‘one-stop’ guide for researchers interested in the field. It first outlines the essential features of RS and OCT, followed by an overview of how the two modalities can be integrated with instrumentation and how data from each is analyzed and combined. Lastly, a summary of reported biomedical applications of RS-OCT is provided, as well as the associated challenges of multimodal RS-OCT and future prospects.
1 |. INTRODUCTION
Label-free optical techniques have long been investigated as potential tools for clinical diagnostics. Such methods are generally non-destructive, fast, and can readily be applied in vivo because they rely on intrinsic tissue contrasts. Their ability to provide sensitivity to various pathological features without exogenous contrast agents has led to the application of optical spectroscopy and imaging in many areas of medicine [1].
The emergence of label-free optical spectroscopy for biomedical analysis has offered a clinically compatible set of methods to probe molecular changes in tissue related to disease. The interaction of light and its prescribed properties (e.g., wavelength, polarization, and phase) with primary absorbers and fluorophores allows for a quantitative measure of their changing concentration and conformation. Methods like diffuse reflectance spectroscopy, autofluorescence, and vibrational spectroscopy (i.e., infrared and Raman) exemplify the diversity of spectroscopic tools available to non-invasively probe tissue composition [2]. A large body of research has demonstrated that Raman spectroscopy (RS) provides higher specificity than any other form of label-free optical spectroscopy for disease classification [3, 4]. Unlike absorption- or fluorescence-based spectroscopy methods which are most sensitive to a small subset of the primary chromophores (e.g., hemoglobin, lipid, water, and melanin) and fluorophores (e.g., collagen, NADH, and FAD), respectively, RS provides detection of discrete molecular signals from all tissue components. Investigators have employed RS to study biochemical changes in biological specimens since the mid-1980s [5], and advances in the field have now extended its application to disease diagnostics and cell therapies [6–13]. However, the intrinsically weak signal generated by spontaneous RS requires long signal integration times and is often confined to point measurements for clinically realistic measurement speeds. This limitation introduces the potential benefit of image guidance for improved sampling accuracy. Spatially specific RS measurements can be realized using morphological features from two- or three-dimensional optical imaging to guide the position of the Raman laser within the target tissue.
Label-free optical imaging techniques such as confocal microscopy (CM) and optical coherence tomography (OCT) enable visualization of tissue morphology with micron-scale resolution, and small-form-factor designs allow integration into clinical endoscopes [14, 15]. However, imaging with CM typically uses tissue autofluorescence as the contrast mechanism, where an excitation source within the ultra-violet or visible range is scanned axially to image depth-resolved structural information. This leads to inherently slow imaging speeds, with typical frame rates of a few Hz, and limited penetration depth (<100 microns, [16]) due to the wavelength of the source beam [15]. Unlike CM, OCT relies on elastic scattering from long-wave visible or infrared (IR) sources to generate contrast and allows for faster speeds and deeper penetration depths that is more practical for clinical imaging. These benefits allow OCT to achieve frame rates of 100 Hz to a few kHz with a penetration depth of hundreds of microns to a few centimeters [17]. Another distinct advantage OCT has over other optical imaging methods is that axial resolution is determined by the coherent length of the source and decoupled from the imaging lens’ numerical aperture (NA), thereby facilitating better imaging of microstructural features and tissue layers over a wider field of view [18]. OCT has been used to assess the scattering behavior from a diverse set of tissues for identifying various disease-specific characteristics [19–24]. However, tissue pathological features can share similar morphologic and elastic scattering properties, leading to difficulty distinguishing certain disease states [25–27]. For example, basal cell carcinoma and the premalignancy of squamous cell carcinoma appeared similar in OCT images [25], pointing to a limitation of using imaging alone for pathological screening. To address this, coupling spectroscopic techniques like RS with OCT imaging adds molecular specificity and would serve to eliminate ambiguity in differentiating disease states that show analogous image features.
Characterization of both tissue morphology and biomolecular composition enables a more comprehensive analysis of disease features for diagnostic purposes. Over the past 15 years, efforts to combine RS and OCT into a single instrument derive the benefits from both techniques and mitigate their individual limitations. Multimodal RS-OCT enables microstructural imaging of the tissues, targeted RS measurement at a location of interest within the OCT image, and improved diagnostic accuracy by combining RS spectral features with image-derived OCT quantitative metrics [28–31]. Therefore, the combination of RS and OCT into a single instrument presents a more holistic means for clinical diagnosis than either modality offers alone.
This review is intended for scientists and engineers interested in optical and photonic design considerations and data analytics for RS–OCT, as it focuses on the essential features of how spontaneous RS and OCT systems are integrated into instrumentation and how data from each modality is analyzed and interpreted in the presence of the other. A summary of reported biomedical application is also provided, and the associated challenges and prospects of multimodal spontaneous and stimulated RS-OCT are also examined.
2 |. RAMAN SPECTROSCOPY OVERVIEW
Among the available optical molecular sensing techniques, RS is particularly advantageous due to its ability to probe the biomolecular content of tissue with high specificity, in both ex vivo and in vivo settings, without the need for exogenous labels [32]. It utilizes the inelastic scattering interaction to analyze the presence and abundance of biomolecules relating to sample composition. The potential for RS to be used as a clinical diagnostic tool has been extensively studied [33] and is ongoing. The following subsections will outline the basic theory, instrumentation considerations, and data processing methods relevant to biomedical applications of spontaneous RS.
2.1 |. Principles
The working principle of RS is that the specific arrangement of chemical bonds within a molecule allows certain vibrational modes to be supported, and these modes are quantized such that they can only occupy particular energy states. Photons can interact with these bond vibrations through absorption or scattering events. Vibrational excitations in RS are generated through inelastic scattering, where the coupling of a photon to a molecular bond causes an instantaneous transfer of energy between the two. This transfer results in a change in energy of the incident photon and a measurable shift in its wavelength, which depends on the specific Raman-active vibrational modes present within a molecule. This effect creates a spectral “molecular fingerprint” that represents a comprehensive chemical description of the analyte that can be used to identify the sample or characterize biochemical changes associated with disease.
2.2 |. Instrumentation
Inelastic scattering is a weak optical interaction, so collecting spectra through the spontaneous production of Raman photons requires extremely sensitive detection hardware and relatively high excitation laser power and acquisition times compared to other optical sensing methods. Furthermore, the Raman component of the measured signal must also compete with other optical interactions like tissue autofluorescence. While Raman scattering intensity increases at visible excitation wavelengths compared to near-IR, so does the likelihood of generating tissue autofluorescence. This increase in autofluorescence background impacts the signal-to-noise (SNR) of Raman peaks and spectral interpretability [34]. An established approach to reduce the effects of sample autofluorescence is to employ IR lasers (e.g., Nd:YAG, 1064 nm) where photon energies are less likely to excite fluorophores. However, a 3-fold decrease in Raman scattering power at 1064 nm relative to 785 nm excitation [35] demands higher laser powers and longer integration times. In addition, the detection of an IR Raman signal requires the use of IR-sensitive materials (e.g., Germanium, Indium–Gallium–Arsenide) that have relatively high readout noise and dark current compared to current charge-coupled device (CCD) technologies [36]. Although 1064 nm excitation improves spectral SNR in highly pigmented tissues (e.g., kidney, liver, pigmented skin, lung), absorption and heating effects become an additional challenge that limits translation to in vivo applications [35].
Spontaneous production of Raman photons is most common in biological applications of RS due to the simplicity and customizability of instrument design. In spontaneous RS systems, wavelength-stabilized diode lasers with center laser lines between 500 and 800 nm and sub-nanometer bandwidth are typically employed that balance the trade-off between Raman scattering intensity and tissue autofluorescence [37]. The backscattered light is collected with either free space optics or fiber optic probes and is then spectrally resolved with high-efficiency spectrometers that incorporate optical filters to reject the elastically scattered excitation light. High-sensitivity, thermoelectrically cooled CCD detectors are commonly used to acquire spectra due to their low-noise performance and the ability to conduct hardware binning during readout for optimal SNR of the measured signal [38].
2.3 |. Data processing
Spectral calibration and preprocessing routines are required to properly extract the Raman component of the raw optical signal. A given RS system configuration determines the instrument response and requires two types of calibration: spectral and intensity calibration. Spectral calibration converts CCD pixel number to relative Raman wavenumber, while intensity calibration uses the known emission from a calibrated source (e.g., lamp, luminescence standard) to correct for the system’s wavelength-dependent sensitivity [33].
Preprocessing encompasses the various steps needed to isolate and analyze the signal components specific to Raman scattering. This is especially important in biological RS spectra with strong autofluorescent background, often orders of magnitude stronger than Raman scattering, which impedes analysis of Raman bands and generates additional shot noise. This background is usually eliminated through software-based methods. Such methods include derivative filters, frequency domain analysis, but more popularly with fitting techniques based on polynomials or penalized least-squares [33]. Smoothing procedures suppress high-frequency noise contributions to the spectrum through a moving average window, median, derivative, or Savitzky–Golay filters [39]. Finally, normalization of the spectrum is required if direct peak heights are to be used for analysis, because variations in peak energies between measurements may be due to inconsistencies in experimental or sampling conditions rather than true spectroscopic deviations [40]. Common approaches include mean normalization, vector normalization, Standard Normal Variate, and Multiplicative Scatter Correction [39, 41].
2.4 |. Current applications
There has been considerable work in implementing spontaneous RS for pathological screening in oncology [42, 43], pathogen detection, and identification [44], and endoscopic applications in neurosurgery [45]. In most cases, using an optical approach like RS for disease screening rivals current gold-standard methods in that it is fast and has relatively simple sample preparation requirements for ex vivo applications. A unique aspect important for clinical disease screening is that RS can be applied in vivo directly to the target tissue by means of handheld optical designs that utilize fiber optics. While the U.S. Food and Drug Administration (FDA) has not filed approval for any clinical Raman device, current efforts to apply RS as a diagnostic tool is ongoing, with recent clinical trials investigating the use of benchtop Raman systems and microfluidic devices for diagnostics in areas including cancer [46–49], osteoarthritis [50], and diabetic glucose monitoring [51]. Additional clinical trials relating to probe-based RS are also evaluating direct in vivo detection of gastric dysplasia [52], esophageal cancer [53], and bone quality assessment [54].
Applications of RS for biomedical analysis have primarily relied on spontaneous generation of Raman photons, but the slow acquisition speed of RS means that collecting two-dimensional maps of spectral information is not feasible in a clinical setting. Point-wise measurement eliminates the ability to determine the spatial extent of disease and introduces the chance of sampling error in medical applications where these aspects are important; like tracking of skin lesions or tumor margin assessment. RS diagnostics can therefore be improved by the addition of an imaging modality to not only provide morphological information about the sample to complement diagnosis, but also to spatially position the RS laser to a region of interest within the imaging field.
3 |. OCT: OVERVIEW
OCT is an imaging modality commonly used in ophthalmology and is also emerging as a promising tool in cardiology, oncology, dentistry, and otolaryngology. OCT constructs depth-resolved reflectance profiles of the sample by the interference of backscattered light from the sample with a reference signal reflected from a mirror [55, 56]. The contrast seen within these images arises from differences in tissue scattering, and it provides distinctive information about the morphology of the sample with micron-scale resolution [57, 58]. In the following subsections, the fundamental principles and instrumentation of a standard OCT system are introduced.
3.1 |. Principles
In OCT, a low-coherence Michelson interferometer setup splits the light from a broadband source into two paths: a reference arm and sample arm. According to the low temporal coherence properties of a broadband light source, interference fringes are created when the optical path length difference between the two arms of the interferometer matches within the coherence length of the broadband source [59]. The backscattered light from the sample arm is combined with light reflected from a reference arm to create an interference signal, which is then captured by an optical detector and processed to obtain a depth profile of sample reflectivity (i.e., A-scan). Combining a series of A-scans at different transverse positions, by laterally scanning the probing beam, creates a cross-sectional tomogram (i.e., B-scan). Importantly, the axial resolution of an OCT image is determined by the coherence length of the broadband light source and is decoupled from the transverse resolution, which is determined by the NA of the objective lens.
3.2 |. Instrumentation
OCT instrumentation can be categorized into two primary types based on how reflectivity profiles are generated: Time-Domain OCT (TD-OCT) and Fourier-Domain OCT (FD-OCT). TD-OCT was the first implementation of OCT, but FD-OCT overcame the limitations of the former in terms of imaging speed and sensitivity [12, 51].
In TD-OCT systems, the mirror in the reference arm is translated axially to obtain depth information, and a point detector records the intensity. Interference fringes arise when the optical path lengths of scatters at various axial positions within the sample arm match that of the reference arm. The requirement for mechanical movement of the mirror in the reference arm sets the limit for image acquisition speed.
On the contrary, the reference mirror in a FD-OCT system is kept stationary. The absence of mechanical movement in the reference arm allows FD-OCT to acquire images much more rapidly; an order of magnitude faster than TD-OCT [56]. Furthermore, reflected signals from all depths of the sample are measured simultaneously so that the large number of spectral channels (N) gathering signal provides improved sensitivity by a factor of N/2 [14], providing an SNR improvement over TD-OCT. FD-OCT creates an A-scan by measuring the interference spectrum as a function of wavelength and performing a Fourier transform on this spectrum, which can be realized in two distinct ways. Spectral-Domain OCT (SD-OCT) uses a broadband light source and spectrometer to acquire the interference spectrum. Swept-source OCT (SS-OCT), also known as Optical Frequency Domain Imaging, uses a point-detector and tunable laser. Here, the wavelength-tunable laser rapidly sweeps (with a sampling rate of up to a few MHz) through a range of wavelengths, usually with a bandwidth of more than 50 nm, so that the interference fringes of different wavelengths are recorded sequentially using a single point detector.
3.3 |. Data processing
Due to the popularity of FD-OCT, an overview of the signal processing chain specific to this variant of OCT will be discussed. To generate an A-scan in either SD- or SS-OCT, the acquired spectrum must be processed and Fourier transformed. In SD-OCT, the initial step usually involves elimination of the reference power that would otherwise manifest as a background noise in the OCT image and degrade image quality. This can be done by either measuring an isolated spectrum from the reference arm, the interference spectrum with no sample in place, or by deriving one from acquired data [60], and subsequently subtracting this from the interference spectrum. In SS-OCT, balanced dual detectors allow for analog subtraction of the reference power term prior to digitization of the signal.
Resampling is essential to avoiding degradation of axial resolution in SD-OCT. Modulations in the spectrum produced by FD-OCT vary as a function of wavenumber (k), while the interference signal is often captured as a function of wavelength (λ) [61]. However, SS-OCT lasers can incorporate a k-clock which allows data to be obtained as a function of k. Following resampling, dispersion compensation is performed to recover lost axial resolution due to the difference in optical pathlengths seen by different wavelengths of light as they travel through optical components or the sample itself. Although it can be corrected for in hardware [62–64], dispersion compensation is often handled with software methods [65–67] to minimize system complexity. Once these steps are applied, the spectrum is Fast Fourier Transformed (FFT). A reflectance profile is obtained by taking the magnitude of the complex FFT output, that can then be displayed using an intensity plot. Due to high dynamic range of an OCT system (e.g., more than 30 dB), the pixel intensity range is rescaled using various compression techniques [60]. The typical workflow for an OCT processing pipeline as shown in Figure 1.
FIGURE 1.
OCT signal processing workflow, modified from reference [60]
3.4 |. Current applications
OCT has become an established medical imaging technique in ophthalmology. Compared to other imaging modalities that require direct contact with the sample, like ultrasound, OCT makes it easier to screen for ocular diseases quickly and comfortably in the clinic. In addition, near-IR light can penetrate down to the deepest layers of the eye’s fundus. These advantages, as well as the micron-level resolution offered by this modality, have made OCT a gold standard for imaging corneal and retinal pathology. Like RS, optical fibers can also be used to design endoscopic OCT probes that collect images from tissues deep inside body cavities. Endoscopic OCT is gradually gaining attention [58] due to the expansion of possible applications offered by probe-based imaging. Aside from clinical screening in ophthalmology, the FDA has given clearance for the use of OCT endoscopic probes in cardiology since 2010 [68], and has since cleared devices for applying OCT to breast cancer margin assessment [69] and otitis media diagnostics [70].
Although OCT imaging of tissue microstructure is suitable for diagnosing many conditions, in some cases qualitative analysis of OCT images lacks the specificity required to differentiate certain disease states. For example, dermatologists and pathologists trained in interpreting OCT scans had difficulty in separating basal cell carcinoma from premalignant lesions [25]. In such cases, the addition of label-free spectroscopic methods to perform biochemical characterization of tissues could improve diagnostic accuracy compared to using OCT imaging alone.
4 |. MULTIMODAL RS–OCT: OVERVIEW
The complementary information provided by RS and OCT motivation their integration into a single device. A multimodal design enhances the scalability and diagnostic accuracy beyond what is possible for either technique alone, but appropriate integration of hardware and data is first required. The following subsections will discuss aspects of multimodal instrumentation, including example RS-OCT system configurations and signal analysis methodologies toward combining RS and OCT data for diagnostics.
4.1 |. Instrumentation
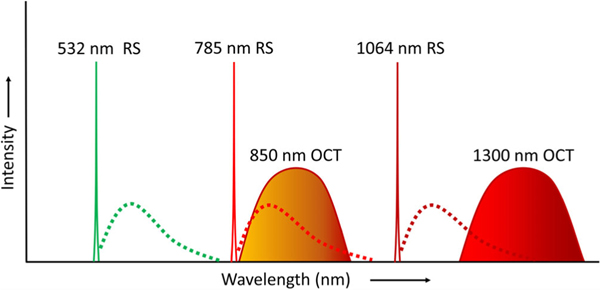
An important consideration when integrating RS and OCT into a single instrument is the unique hardware requirements for each modality. RS requires a narrowband light source because the source bandwidth sets a limit on the Raman band linewidth in the recorded spectrum [71]. On the contrary, OCT needs a broad bandwidth light source to realize low coherence interferometric signal and achieve high axial resolution. In addition, the appropriate center wavelengths of the sources are distinct. Although visible sources generate more Raman scattering compared to IR, due to the fourth-order dependency of scattering power versus wavelength (i.e., λ−4), they also increase the level of tissue autofluorescence that overwhelms the Raman components of the signal. Laser lines between 500 and 800 nm with sub-nanometer bandwidth balance these two effects for most tissues and are usually chosen for biological RS. Ultimately, the target tissue’s absorption, scattering, and fluorescent properties will determine the optimal center wavelength for the RS laser [33]. In OCT, center wavelengths that are too short reduce the penetration depth due to optical scattering inside the tissue [18] and usually increase material/waveguide dispersion. Center wavelengths that are too long can suffer from water absorption losses and deteriorate axial and lateral resolution because of the extended coherence length and enlarged spot size, respectively. So, lasers with center wavelengths between 850 and 1300 nm with ~100 nm bandwidth are typically implemented into clinical OCT systems. Accordingly, the requirements for laser sources in terms of bandwidths and center wavelength are not similar between RS and OCT. Figure 2 depicts the spectral characteristics of laser sources most commonly reported for each modality, showing that combining light sources into a fully integrated system is challenging.
FIGURE 2.
Unique light source requirements for RS and OCT in terms of laser wavelength and bandwidth. Dotted lines represent the spectral regions for Raman emissions
Optimal detector characteristics are also distinct between these two modalities. RS requires a high-sensitivity detector with low noise to capture the weak Raman signal. Because spontaneous Raman spectra must be integrated over long exposure times, speed is not a necessary detector characteristic. On the contrary, FD-OCT requires either a fast-scanning laser (with a MHz sampling rate for SS-OCT) or a high-speed spectrometer (with a kHz line speed for SD-OCT) to ensure fast imaging speeds. Another important characteristic for detecting the SD-OCT spectrum is having a large full-well capacity, as the smallest reflectivity from the sample that can be captured in an OCT scan (i.e., sensitivity) is dependent on the electron-storing capacity of pixel wells [72]. So, the ideal shared detector for RS and SD-OCT would have high sensitivity, low noise, a large full-well capacity, and fast readout; four characteristics that are seldom found in a single optical detector. The sampling density of the spectral data is also a consideration. OCT imaging depth range is dependent on the sampling density of the interference signal [17]. While sparse sampling of the RS spectrum with ~1000 pixels is sufficient for biological Raman spectra, SD-OCT typically employs line detectors with >2000 pixels. In general, it is possible to combine RS and SD-OCT detection platforms by using a common spectrometer. The caveat to this approach is that Raman emissions and OCT spectrum must occupy the same spectral window, which can be accomplished by using a narrowband 785 nm source for RS and broadband OCT source centered at 850 nm, as depicted in Figure 2. While a single detector cannot fully satisfy the unique detection requirements of both modalities, a compromise can be made in terms of sensitivity, readout speed, noise performance, and pixel array size. Due to the stringent requirements of RS source and detection characteristics for biological materials, the common detector is usually chosen toward the needs of RS. Thus, it down-scales some of the OCT performance metrics (e.g., imaging speed and depth range) [73].
Although the unique excitation and detection needs of RS and OCT makes combining such instrumentation aspects a challenge, recent advances in laser and detector technologies may help bridge this gap; as outlined in Section 6.1. Instead of sharing a common light source or detector, another option for integrating both modalities is to only employ common optics for RS and OCT beam paths in the primary sample arm. This approach allows for more flexibility in optimizing excitation and detection instrumentation decisions independently to maximize the performance for each technique, but it leads to extensive instrument components and electronic control, added cost, and increased system complexity.
Several groups have explored the integration of RS and OCT into a single instrument. Nearly all designs incorporated RS into the OCT sample arm with free-space optical designs and left the detection platforms separate, resulting in bulky benchtop systems. A diverse set of RS-OCT system configurations have been explored in this way, including miniaturized designs that are more suitable for clinical translation. To the best of our knowledge, the only attempt to detect RS and SD-OCT signals on a single spectrometer was reported by reference [73], as discussed in Section 4.1.1. Table 1 provides a summary of published RS-OCT multimodal instrumentation characteristics that compares integration strategies, light sources used, RS acquisition settings, and OCT imaging resolution. An overview of these reported system variants, and associated aspects of instrumentation like data coregistration, will now be discussed.
TABLE 1.
Overview of the reported specifications for various RS-OCT system configurations
| Multimodal integration strategy | Author | Excitation | Laser power at sample (RS) | Acquisition time (RS) | Spectral resolution (RS) | Spot size (RS) | Spatial resolution (OCT) |
|
|---|---|---|---|---|---|---|---|---|
| Axial | Lateral | |||||||
| Shared RS–OCT sample arm, separate detection systems | Patil et al. [76] | RS: 785 nm, FP region TD-OCT: 1310 ± 30 nm |
40 mW | – | 7.7 cm−1 | 75 μm | 21 μm | 25 μm |
| Evans et al. [75] | RS: 633 nm, FP region SD-OCT: 855 ± 37 nm |
4 mW | 5 s | ~6 cm−1 | 15 mm | 4.5 μm | 18 μm | |
| Patil et al. [77] | RS: 785 nm, FP region TD-OCT: 1310 ± 30 nm |
40 mW | 30 s | – | 44 μm | 14 μm | 25 μm | |
| Khan et al. [82] | RS: 785 nm, FP region TD-OCT: 1310 ± 21 nm |
60 mW | 10 s | 8 cm−1 | 12 μm | 20 μm | 15 μm | |
| Maher et al. [83] | RS: 785 nm, FP region SD-OCT: 840 ± 22 nm |
30 mW | – | 3.3 cm−1 | – | 8 μm | 40 μm | |
| Zakharov et al. [29] | RS: 785 nm, FP region SD-OCT: 840 ± 50 nm |
– | 30 s | 0.05 nm | 1 mm | 3.5 μm | 3.5 μm | |
| Wang et al. [78] | RS: 785 nm, FP/HW regions SS-OCT: 1325 ± 60 nm |
10–15 mW | 0.5 s | ~9 cm−1 | – | 12 μm | 12 μm | |
| Chen et al. [91] | RS: 785 nm, FP region SD-OCT: 850 ± 15 nm |
100 mW | 2 s | – | 15 μm | – | – | |
| Mazurenka et al. [80] | RS: 532 nm pulsed, FP region SD-OCT: 1325 nm |
2.38 mJ/cm2 | 100 s | – | Collimated beam | 5.5 μm | 13 μm | |
| Ren et al. [84] | RS: 532 nm, FP region SD-OCT: 830 ± 20 nm |
10 mW | 30–60 s | 7 cm−1 | 3 μm | 2.3 μm | 7.35 μm | |
| Shared RS-OCT sample arm and detection system | Patil, et al. [73] | RS: 785 nm, FP region SD-OCT: 855 ± 20 nm |
100 mW | 30–90 s | 7 cm−1 | 15 μm | 8 μm | 18 μm |
| Separate RS-OCT sample arms, manual or automated coregistration | Klemes, et al. [79] | RS: 632.5 nm, FP region SD-OCT: 880 ± 32 nm |
13 mW | 20–50 s | – | – | 4.3 μm | 3.6 μm |
| Placzek, et al. [86] | RS: 785 nm, FP region SS-OCT: 1304 ± 45 nm |
70 mW | 5 s | 5 cm−1 | 100 μm | 12 μm | 28 μm | |
Abbreviations: FP, fingerprint; HW, high wavenumber; CT, optical coherence tomography; RS: Raman Spectroscopy; SD, spectral domain; SS, swept source; TD, time domain.
4.1.1 |. Free space benchtop systems
The initial efforts toward combining RS and OCT data used completely separate systems for each modality. The first result of such work was reported by Ko et al. where the authors applied OCT and polarized RS to detect early dental caries ex vivo [74]. The benefits of multimodal signal detection were demonstrated with both RS microscope- and probe-based acquisition, and these results set the ground of motivation for combining RS and OCT into a single working system.
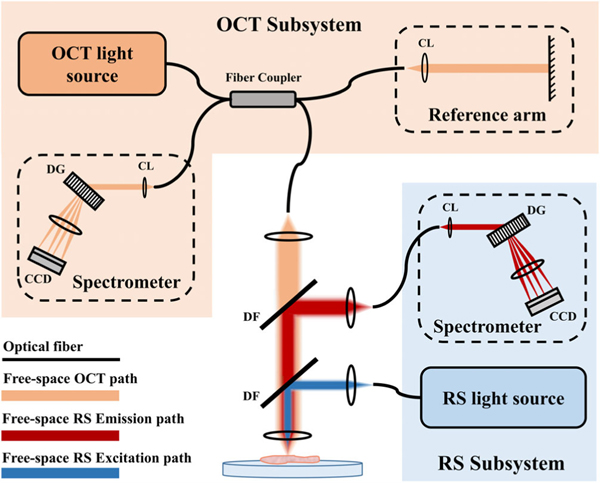
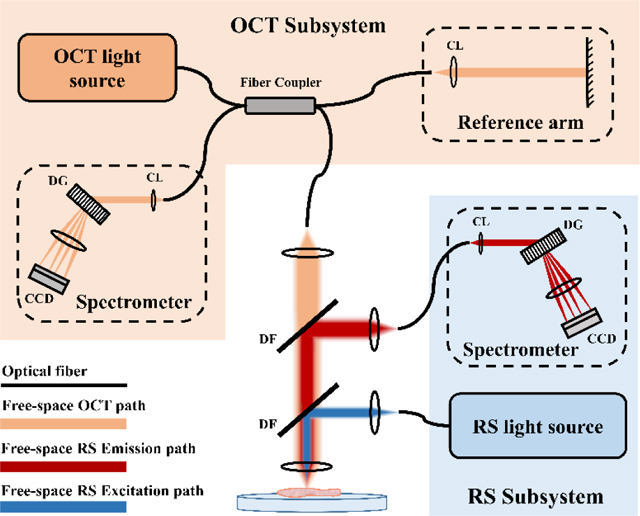
Subsequent explorations demonstrated RS-OCT using common scanning optics in a single sample arm, where both OCT and RS share the same light-guiding optics. As depicted in Figure 3, these kinds of systems involve free-space beam steering optics, dichroic filters or flip mirrors to separate optical paths, and independent laser sources and spectrometers. Evans et al. reported an example of such a system to simultaneously acquire RS and OCT signals, where Raman spectra were collected at each lateral position within an OCT scan of ex vivo retinal tissue. Collection of Raman spectra were acquired at an eye-safe laser power of 4 mW, and therefore required more exposure time (e.g., 5 s) than measuring an OCT A-scan (e.g., 50 μs). So, collecting multimodal RS-OCT maps was slow and retinal samples suffered from dehydration during these long acquisitions [75]. In scanning mode, either more sparse sampling of RS-OCT data or an increase in the Raman source power is necessary to expediate measurements. This scheme for simultaneous collection of morphological and spectroscopic maps therefore limits feasibility for in vivo implementation. For more rapid tissue interrogation with RS-OCT, OCT can first be used to perform initial real-time imaging of tissue microstructure without RS measurement. The morphological imaging can then be used to identify ambiguous lesions and subsequently apply RS point measurement to specific regions of interest to characterize lesion composition. Patil et al. was the first to report this scheme for RS-OCT data collection [76]. The authors demonstrated sequential acquisition of OCT and RS data, where OCT imaging was used to guide Raman laser positioning.
FIGURE 3.
Schematic of a multimodal RS and SD-OCT system with separate detection sub-systems, combined through a shared sample arm. This example shows spectral channels separated by dichroic filters (DF), and detected by dedicated spectrometers with collimating lens (CL), diffraction grating (DG), and charge-coupled device (CCD)
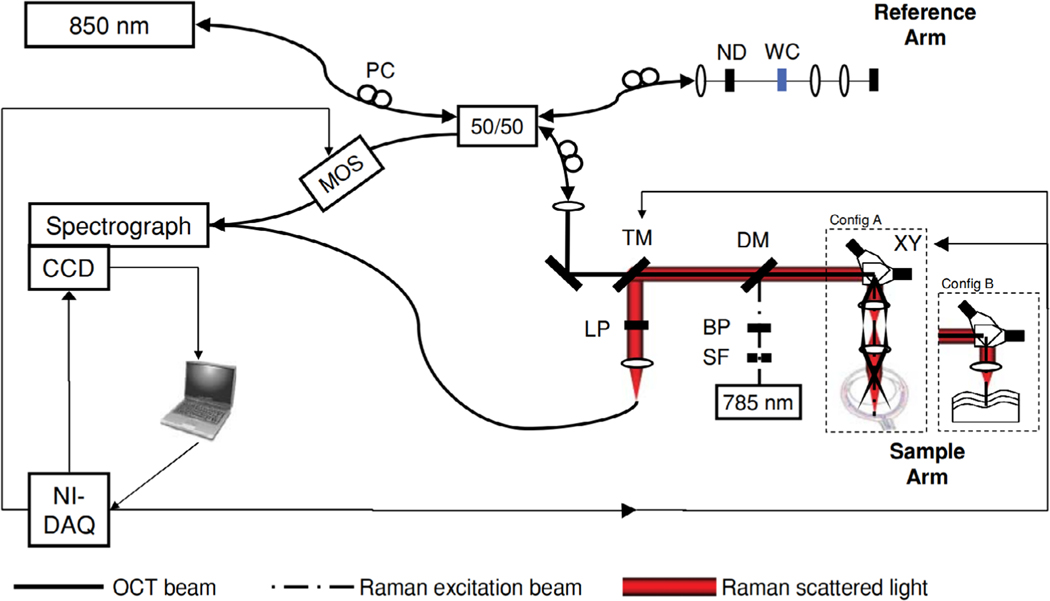
In another study, Patil et al. combined the detection platform for RS and OCT onto a single spectrometer to simplify system complexity and allowed for smaller footprint of the overall system [73], shown in Figure 4. In this design, a region of the RS spectrometer CCD was dedicated to the OCT spectrum so that both signals could be sequentially read out. Detection components were chosen that best supported the sensitivity requirements of RS while compromising OCT readout rate and dynamic range. This common detector approach required specific laser source center wavelengths (i.e., RS: 785 nm, OCT: 850 nm) so that Raman spectra and OCT source bandwidth operate within the same spectral window. However, a limited OCT imaging range of 1.27 mm and scanning rate of 2–3 Hz was reported due to the limitations imposed on using a detection platform sufficient for spontaneous RS spectral acquisition. So, RS-OCT tissue characterization on a shared detection platform requires improved spectrometer design to balance the needs of each modality, so that an optimized single-detector system can minimize RS acquisition time while maximizing OCT imaging performance.
FIGURE 4.
Multimodal RS-OCT system implemented using a shared sample arm and common detector. BP, band pass filter; DM, dichroic mirror; LP, long pass filter; MOS, MEMS optical switch; ND, neutral density filter; PC, polarization control paddles; SF, spatial filter; TM, translatable mirror; WC, water filled cuvette; XY, scanning galvanometer pair. Figure reproduced from [73] with permission
4.1.2 |. Miniature systems
Several efforts have been made toward developing miniaturized RS-OCT systems that suit particular in vivo applications [69–72]. Recent advancements in system designs utilize optical fibers that serve as light-guides to decouple the primary imaging head from excitation sources, spectral detection components, and OCT reference arm. In this way, handheld RS-OCT devices can be fabricated that offer the steric freedom to reach areas on or within the body that a benchtop system cannot. A few studies have reported small-form-factor RS-OCT probes to increase the portability and potential usability in clinical settings.
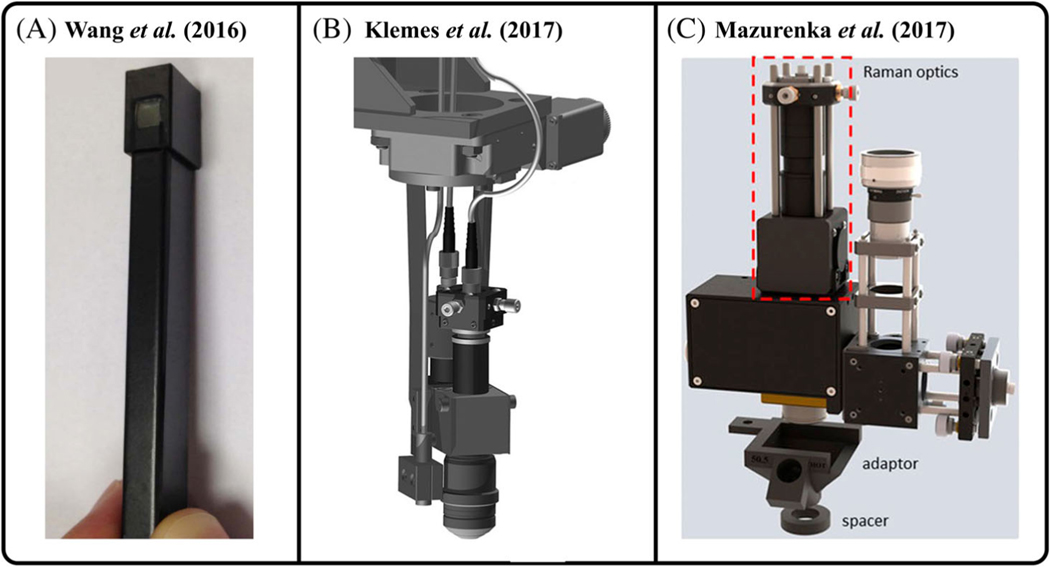
Patil et al. demonstrated a clinical probe physically separated from the source and detection systems using optical fibers to provide portability to the combined RS-OCT device and enable clinical compatibility toward identifying skin cancer. The beam paths for RS and OCT were combined within a probe that was 12.7 by 20.32 cm in size, and the laser sources and detection subsystems were set within a portable handcart to allow sufficient mobility in a hospital setting [77]. Wang et al. developed a handheld RS-OCT probe ~120 mm long and capable of OCT imaging and acquiring both fingerprint and high-wavenumber Raman spectral regions in the oral cavity (Figure 5A) [31, 78]. The OCT beam was scanned by galvo mirrors located at the back focal plane of an objective lens, and this scan path was then relayed into a half-pitch gradient index rod lens to achieve lateral scanning at the sample. A Raman fiber bundle was imaged onto the sample by a separate sapphire lens, co-aligned with the SS-OCT scan path using a long-pass dichroic mirror. This micro-optical design facilitated miniaturization of the RS-OCT probe so that it could fit within the oral cavity. In another study, Klemes et al. reported a RS–OCT probe with a length of 165 mm that consisted of separate RS and OCT fiber-coupled paths housed in a common miniature scanning head (Figure 5B) [79]. This was attached to an automated scanning system so that user-selected positions within an OCT scan could then be targeted for RS acquisition through software control of the motorized stage. Mazurenka et al. reported a portable RS–OCT probe by modifying a commercial SD-OCT device for screening melanoma (Figure 5C) [80]. The OCT scanning path was kept in place whereas the probe alignment CCD camera of the device was replaced with a Raman signal collection arm, where a fiber bundle collected and guided RS emissions to a dedicated spectrometer. The design used a separate Raman excitation fiber with beam-expanding optics that distributed the laser energy over the objective lens’ full field of view (FOV); allowing for skin irradiance to fall below Maximum Permissible Exposure (MPE) limits while maintaining adequate SNR of Raman collection. A similar attempt to incorporate dedicated RS excitation and collection fibers into a portable OCT imaging device was described by Monroy et al., where a RS fiber probe was integrated into an OCT ear speculum to determine the feasibility of applying RS-OCT within the middle ear [81].
FIGURE 5.
Miniature RS–OCT probes (A) handheld probe for real-time tissue measurements developed by Wang et al. [78]. (B) 3D technical model of the optomechanical probe developed by Klemes et al. [79] (C) 3D CAD design of the RS-OCT probe developed by Mazurenka et al. [80]. Figure reproduced from references [78–80] with permissions
4.1.3 |. Axially selective RS–OCT
The ability to retrieve Raman scattered light from specified depths within tissue is critical when the target tissue is not superficially exposed or when the assessment of disease margins is of interest to locate. The addition of axially resolved RS measurement in RS–OCT systems would expand their application space into new clinical territories. An established approach to add depth selectivity to RS measurements is to use confocal gating, termed confocal RS (CRS). Confocal optical systems spatially select emissions arising from a particular plane by using a spatial filter (i.e., pinhole) conjugate to that plane. Khan et al. demonstrated this approach with a benchtop CRS-OCT design [82]. Results showed that distinct epithelial and stromal tissue layers could be rapidly visualized with OCT imaging, and CRS could then be used to interrogate specific Raman spectral signatures from these layers to isolate their depth-dependent signals. Transitioning into fiber-based CRS, an important step toward system portability, Maher et al. [83] and Klemes et al. [79] each reported a collection geometry where the RS collection fiber was positioned at a conjugate image plane to act as a pinhole and achieve confocal detection. Ren et al. most recently demonstrated fiber confocal detection [84], which expanded the confocal capabilities of a CRS-OCT system by including a tunable lens in the RS laser path to position the Raman probing beam at any arbitrary transverse and axial position within the OCT volumetric image. While CRS facilitates axially resolved spectral information, rejection of out-of-plane emissions naturally impacts the amount of detected signal. This reduces RS collection efficiency and requires higher laser power or acquisition time.
4.1.4 |. Coregistration
In many cases, acquiring colocalized RS–OCT maps of morphologic and molecular information is helpful to understand spatial correlations of the information provided by these two modalities. To correlate such morphomolecular maps, coregistration of the OCT scan path with the Raman laser focus must first be established. If the OCT and RS optical paths share a common sample arm and primary focusing lens, coregistration is accomplished by the overlap of laser focuses. The CRS–OCT system reported by Ren et al. outlined a method to quantify coregistration error using fluorescence beads [84]. While a common focusing lens simplifies coregistration of data, the lens’ optical characteristics must be shared between RS and OCT. A high NA lens can increase collection efficiency of the weak Raman signal and reduce acquisition time, but at the cost of a shorter Rayleigh range which limits OCT imaging depth.
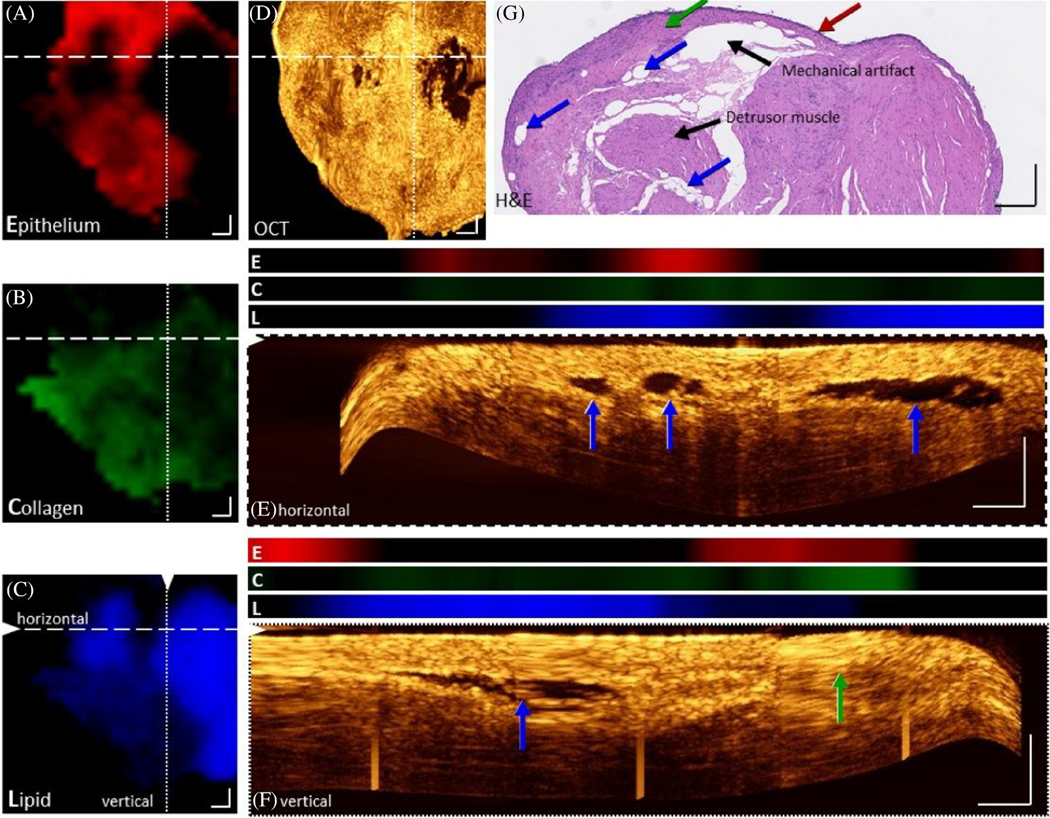
When the RS and OCT excitation/detection paths are left separate, coregistration must be achieved manually or by careful software control of sequential RS and OCT signal acquisition. Guevara et al. manually registered RS data onto the projected OCT volume of a skin specimen by using separate RS and OCT subsystems to build a mosaic image map over larger area of the sample [85]. However, the authors reported introduction of errors in marking suspicious lesions from the RS mapping due to manual registration degrading the accuracy of relating the spatial data between each modality. On the other hand, Placzek et al. described a coregistration method that utilized separate RS and SS-OCT probes with digital position control of a two-axis sample stage [86]. This system was recently used to demonstrate well-correlated morphomolecular information in bladder biopsy tissue [87] (Figure 6). OCT imaging unveiled transition zones between lamina propria and epithelium that were noted by increase in OCT scattering intensity due to the collagen-dense lamina propria tissue layer. These layer differences agreed with the coregistered RS maps fitted by a component model representing spectral contributions from epithelium, lipid, and collagen. Strong collagen versus epithelial RS component coefficients were seen in these high-scattering image zones. Importantly, areas of low contrast in the OCT images, which might be mistaken for voids caused by mechanical rupture of tissue during biopsy extraction or handling, were clearly shown to arise from homogenous low-scattering fatty regions that presented strong lipid component coefficients on the RS maps. While this approach is slow and tailored to ex vivo applications, it demonstrated that large coregistered maps of OCT and RS data can be generated with virtually infinite FOV.
FIGURE 6.
Coregistered RS-OCT morphomolecular maps of a bladder biopsy. Raman component coefficient maps of epithelium (A), collagen (B), and lipid (C) show their presence and distribution. (D) An en-face maximum intensity projection of the OCT volume between 150 and 180 μm. (e, f) Maximum intensity projections of 10 cross-sectional B-scans, positions indicated by dashed and dotted lines in the Enface OCT image with corresponding Raman component values (E: Epithelium, red, C: Collagen, green, L: Lipid, blue). An overlap of lipid Raman signals can be seen in black voids within OCT images. (g) H&E image of the biopsy. Arrows indicate lipid pools (blue arrow), epithelium (red arrow), lamina propria (green arrow), and collagen (green arrow). Scale bars: 250 μm. Figure reproduced from reference [87] with permission
4.1.5 |. Challenges in multimodal instrumentation
The key challenge of combining RS and OCT into a single instrument for clinical diagnostics is integrating the hardware for both modalities in a compact and flexible way. Integration of fully fiber-based subsystems, for example, fiber-based RS [88] and OCT fiber probe [89], could be a step toward miniaturization that would suit the requirements for clinical adoption and integration into endoscopic tools. Whether combined or left separate, a suitable choice of light sources and detection platforms is essential to addressing this challenge. A shared spectrometer minimizes cost and system size but requires that the light sources operate in the same spectral window and eliminates the ability to optimize detection for each modality or simultaneous signal acquisition. Due to the unique laser characteristics required for each technique, there are currently no reports that attempt to combine light sources within a multimodal RS–OCT system. A related challenge includes maintaining adequate Raman collection efficiency to reasonably match RS acquisition time with OCT imaging speeds. Many of the reported RS–OCT results used high RS laser powers relative to RS spot size (Table 1) that exceeded the MPE level set by American National Standards Institute to maintain useable SNR within a reasonable acquisition time. Therefore, methods to minimize laser power or distribute the RS laser power over a larger area are essential to reduce the risk of potential specimen damage, especially for in vivo applications. Finally, coregistration of the morphological and molecular information is essential if two-dimensional RS–OCT maps are desired. Optical paths can be left separate for added flexibility in integrating each modality into the platform, but this introduces the challenge of registering the laser spots for spatially correlated signals. The RS and OCT probing beams can instead be co-aligned into a shared sample arm to ensure spatial overlap of laser foci, but then the primary lens NA introduces a trade-off between RS collection efficiency and OCT imaging depth. Novel beam-shaping methods like Bessel beam formation have been explored to extend the OCT imaging depth when using high-NA objectives [90], at the expense of imaging SNR. This could serve as a potential method to maintain long OCT imaging depth and large RS collection efficiency through a shared imaging lens, but have yet to be explored for multimodal RS–OCT. Appropriate integration of instrumentation aspects for each modality should therefore consider the specific needs of the biomedical application.
4.2 |. Multimodal data analysis
Because RS–OCT systems provide unique but complementary information about sample structure and composition, it is then useful to employ appropriate data analysis procedures that extract and combine meaningful diagnostic information. Feature extraction procedures construct a comprehensive set of quantitative metrics that can be used to characterize or identify the sample. Ultimately, the employment of chemometric tools to interpret combinatorial RS–OCT feature sets allows one to explore underlying relationships between the data and pathology. This section will provide an overview of feature extraction methods for both RS and OCT, commonly used chemometric tools, and a summary of attempts to combine data from each technique to improve tissue classification accuracy.
4.2.1 |. RS feature extraction
Many molecular components within tissue are not altered during the progression of disease, and so several RS spectral features are often strongly correlated to one another and provide no discriminatory information. Feature extraction techniques are essential to overcoming the issue of spectral band correlation in RS and assist in creating more robust predictive models that can serve as diagnostic tools. They effectively reduce the number of inputs to the model, which avoids model overfitting, but also aid in determining which vibrational bands are significant for biomedical analysis. This allows the analyst to trace back to the biochemical origin of disease-related tissue alterations.
Identifying important spectral bands related to disease transformation can be performed manually through visual comparison of preprocessed spectra. Alongside direct peak analysis, it is common in RS to calculate the area-under-the-curve of a particular spectral region or perform ratiometric analysis to create spectrally derived features [92]. Principal component analysis (PCA) is a popular data transformation method used in RS to conduct feature extraction. This technique captures the primary sources of variation within the data by projecting it into a subspace whose basis vectors point in orthogonal directions of maximum variance [39]. The component “loadings” for a PC are the weights assigned to each original spectral feature. If data points are well separated along this PC axis, then high loading coefficients help to inform which spectral bands were discriminatory to disease state. Automated feature selection is also possible with the use of regularization techniques, which induce sparsity in the RS feature set by learning which spectral bands best contribute to model accuracy while the model is being trained [93, 94]. Popular regularization methods for RS like Least Absolute Shrinkage and Selection Operator (LASSO) [95–97] and Elastic Net [98, 99] add constraints to the optimization of a regression algorithm, through which the weights of irrelevant features are shrunk to zero. In summary, feature extraction methods in RS are important tools that serve to reduce dimensionality for spectral analysis and improve the performance of diagnostic models.
4.2.2 |. OCT feature extraction
Deriving quantitative metrics from OCT images, which are typically analyzed qualitatively, offers a new avenue for image-based tissue characterization. Such metrics have been widely used to build classification algorithms for pathology identification and have been explored in clinical OCT applications, including breast [100–104], ovarian [105–107], prostate [108], and urinary tract cancers [109], as well as gastrointestinal [110, 111] and arterial [112] tissue characterization.
Texture analysis is a common feature extraction approach within the OCT community. Texture is a measure of the spatial intensity variations within an OCT image that arise from speckle, which depends on the underlying optical properties of scatterers, their size and distribution, and their degree of motility [113]. Correlations between this speckle texture and sample microstructure have shown that it can be used as a metric to differentiate tissue types [114, 115] or as a tool to segment structural layers in OCT images [116–118]. These texture metrics can represent the mean, variance, or skewness of pixel histograms but often include higher-order parameters (e.g., contrast, entropy, homogeneity [119]) derived from the Gray Level Co-occurrence Matrix (GLCM) that can be calculated for different directions within the image [84].
Alternative OCT image quantification schemes attempt to derive information about the tissue directly from the intensity profile of an A-scan. Structural differences between tissue types or morphological changes associated with disease alter the degree of optical attenuation that is seen within an A-scan. So, approximations of signal attenuation represent another diagnostic feature that can be extracted from OCT images [120]. Attenuation estimation has been reported by linear [91] and exponential [92] fitting of an A-scan. More advanced approaches can retrieve depth-resolved estimates for attenuation coefficient at every pixel within an A-scan, first introduced by Vermeer et al. [121] to segment multiple tissue layers.
4.2.3 |. Chemometrics
Chemometric analysis encompasses a set of multivariate tools to discover patterns within large datasets. In the context of biomedical analysis, these tools can build predictive models to accurately classify spectroscopic or imaging data for the purpose of diagnostics. These tools can be broadly categorized into two categories: unsupervised clustering methods and supervised classification methods.
Unsupervised methods attempt to categorize data points without prior knowledge of class membership (i.e., labels). This is accomplished by calculating metrics that associate mathematical relationships between the measurements; most commonly distance metrics (e.g., Euclidean and Mahalanobis distance) or metrics of correlation (e.g., Pearson or Spearman correlation coefficients) [39]. PCA can be used for unsupervised data clustering. However, it is more commonly used as an initial dimension reduction procedure, where a subset of PC scores is used as inputs to train an alternative classification algorithm. Accuracy of prediction typically improves when only a subset of PCs that capture the majority of data variance, typically 95% of the latent variation, are used to train the model [122]. Other popular unsupervised methods include Hierarchical Cluster Analysis and K-means. In supervised methods, an algorithm trains a classification model that learns relationships between the input features and assigned class labels. K-nearest neighbor (KNN) predicts class membership of an unlabeled measurement by attributing the same class label to that of the closest neighboring labeled measurements, demonstrated in reference [108]. Support vector machine (SVM) attempts to assign measurements to their respective classes by fitting a decision boundary that optimally separates each group, reported for RS–OCT classification in references [73, 109–111]. Discriminant analysis (DA) is similar to PCA in that it projects the measured data into a subspace through linear transformation, but does so in a label-guided manner by maximizing the ratio of between-class and within-class scatter, which was utilized in references [73, 77]. A summary of reported attempts to discriminate both pathology and tissue type with RS–OCT data, and the associated feature extraction methods and chemometric tools used, is provided in Table 2.
TABLE 2.
Applications of RS–OCT for disease state and tissue identification, with associated analysis methods used and accuracies for separate and combined analysis
| Author | Sample/pathology | Modality | Feature extraction | Chemometric tool | Accuracy (%) |
|---|---|---|---|---|---|
| Bovenkamp et al. [124] | Bladder biopsy (tumor grade) | RS | PCA | KNN | 93 |
| OCT | Texture analysis | KNN | 70 | ||
| Placzek et al. [86] | Bladder biopsy (tumor stage) | RS | None | DA | 75 |
| OCT | Texture analysis | SVM | 74 | ||
| Liu et al. [125] | Ex vivo organ differentiation | RS | PCA | SVM | 100 |
| OCT | Slope, standard deviation | SVM | 84 | ||
| Zakharov et al. [29] | In vivo skin cancer | RS | Peak ratios | DA | 85 |
| OCT | Presence of “nest structures” | None | 44 | ||
| RS-OCT | Peak ratios (RS); the presence of “nest structures” (OCT) | DA | 91 | ||
| Rangaraju et al. [28] | Ex vivo skin burn wounds | RS | Peak ratios, AUC | SVM, RF, LR | 87, 85, 83 |
| OCT | Texture analysis | SVM, RF, LR | 68, 70, 67 | ||
| RS-OCT | Peak ratios, AUC (RS); texture analysis (OCT) | SVM, RF, LR | 90, 86, 84 | ||
| Ashok et al. [126] | Colon biopsy | RS | PCA | SVM | 83 |
| OCT | Texture analysis, PCA | SVM | 76 | ||
| RS-OCT | PCA (RS, OCT); texture analysis (OCT) | SVM | 94 | ||
| Wang et al. [31] | In vivo oral tissues | RS | None | DA | 70 |
| OCT | AC | DA | 66 | ||
| RS-OCT | None (RS); AC (OCT) | DA | 75 | ||
| Tamošiunas et al. [127] | Ex vivo skin cancer malignancy | RS | AUC | DA | 89 |
| OCT | Texture analysis | DA | 89 | ||
| RS-OCT | AUC (RS), texture analysis (OCT) | DA | 96 |
Abbreviations: AC, attenuation coefficient; AUC, area under the curve; DA, discriminant analysis; KNN, k-nearest neighbors; LR, logistic regression; OCT, optical coherence tomography; PCA, principal component analysis; RF, random forest; RS, Raman spectroscopy; SVM, support vector machine.
Many other chemometric tools have been employed for disease state classification of optical data, such as neural networks, decision trees, and naive Bayes, that have been recently reviewed in detail [39, 123] but have not been utilized for RS–OCT applications. Indeed, the breadth of available algorithms introduces a challenge in optimizing diagnostic models, as the ideal choice of feature extraction methods and chemometric tools for maximizing discriminatory accuracy may not be universal to all RS–OCT datasets.
4.2.4 |. Combining data in RS–OCT
Enhancing tissue classification performance is a common motivation when combining RS and OCT complimentary features. Although RS has shown promise in tissue discrimination on its own, clinical applications are impeded by inter-patient variability that affects spectral discrimination in larger-scale patient studies [128]. When classification accuracy falls below acceptable performance, combining quantitative OCT image features with spectrally derived features from RS represents one solution to enhancing classification efficiency to clinically relevant levels. For example, Ashok et al. improved discrimination accuracies between normal colon and colonic adenocarcinoma by combining texture parameters extracted from OCT images with PCA-derived RS features. The reported sensitivity and specificity for RS alone were 89% and 78%, respectively. When adding 16 texture features derived from GLCM, both sensitivity and specificity improved to 94% [126]. Tamošiunas et al. similarly showed that combining statistical metrics from OCT images with RS features increased the detection accuracy of ex vivo malignant skin lesions [127]. Independent RS and OCT datasets could discriminate healthy or benign samples from malignant samples with 89% average accuracy, while the combined RS–OCT dataset could classify malignant samples with 94% sensitivity and 98% specificity. Table 2 confirms that in all cases where RS and OCT features were combined, the trained classification model showed improved performance compared to training on RS or OCT features alone.
4.2.5 |. Challenges in multimodal data analysis
To maximize the discriminatory potential of RS–OCT classification, it is necessary to find the optimal choice of classification algorithm and feature set used for training. Data-mining software like Waikato Environment for Knowledge Analysis (WEKA) can help address this burden. This software contains data screening methods to eliminate noisy, missing, or erroneous data and includes numerous supervised and unsupervised algorithms for testing various classification models. It can also employ feature selection tools like filter and wrapper based methods, regularization methods like LASSO or elastic net [129], and stochastic search methods like swarm search [130] to assist the analyst in choosing a model and feature set that performs best for a particular task [131, 132]. Advances in this open-source machine learning platform have led to the automation of WEKA, called Auto-WELA, where the best-performing model and hyperparameter settings are found through Bayesian optimization [133]. WEKA is a powerful model optimization tool in clinical research areas like bioinformatics [134] and meta-data diagnostics [135, 136], and has recently been used in RS–OCT model optimization [28].
Similar challenges exist in preprocessing of RS–OCT data. The ideal choice of preprocessing methods for Raman spectra and OCT images is heavily dependent on data quality, and so there is little consensus in either field for a single processing pipeline that should be applied universally. Some efforts have explored the use of machine learning algorithms to find the best procedure for a given dataset, which searches for the optimal set of preprocessing steps that maximize the performance of a classification or regression model [137, 138].
5 |. APPLICATION OF MULTIMODAL RS–OCT
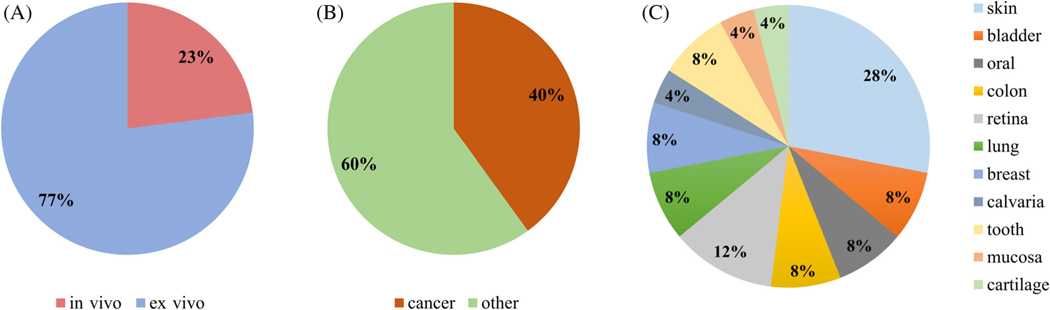
Though it is still in an early phase of exploration, multimodal RS–OCT has been used to investigate a diverse set of biological samples, including skin, dental, retinal, lung, bladder, and colonic tissues, with a large focus on cancer diagnostics. The feasibility of using RS–OCT has mainly been demonstrated in ex vivo samples, while only a small percentage of studies have been conducted in vivo. A statistical summary of the kinds of biological samples referenced within this review is provided in Figure 7. The wide application space for applying RS–OCT for biomedical analysis shows the broad utility of this multimodal approach to optical diagnostics.
FIGURE 7.
Multimodal RS-OCT application domain. (A) In vivo versus ex vivo, (B) cancer versus other applications, and (C) scope of various tissue types explored with RS–OCT.
5.1 |. Ex vivo applications
Most ex vivo studies have demonstrated the successful use of OCT to localize and visualize lesions, from which RS point measurements can be performed to probe tissue composition in a particular region. For example, in the early exploration of this multimodal tool, Chun-Te Ko et al. [74] reported OCT and polarized RS analysis of extracted teeth. Carious and normal enamel were scanned from both un-sectioned and longitudinally sectioned tooth samples. The deepest area of carious lesions was determined by OCT scan, and RS measurement was then acquired at this location to reveal differences in relative band intensities compared to healthy teeth. Demineralization of enamel caused a decrease in RS peak energies related to hydroxyapatite, which allowed for differentiation of healthy enamel from caries legions.
Other ex vivo reports of RS–OCT include application to retinal samples [73, 75], skull [73], and mucosal tissue [82]. In the study conducted by Evans et al. [75], human and porcine retinal tissues were scanned with a benchtop system to collect volumetric OCT data and RS spectral maps. Retinal layers were clearly visualized with OCT. The RS maps also revealed higher signal in the spectral range between 1500 and 1630 cm−1 at the fovea, which was thought to be indicative of Cytochrome C that is associated with mitochondria; suggesting apoptosis of the excised retina [75]. Another ex vivo study applied RS-OCT to rodent calvarial bone [73]. The OCT images allowed clear visualization of the inner and outer surfaces of the calvaria, along with notable contrast between the hyper-reflective mineralized bone tissue and the less reflective collagenous tissue that hold the plates of the skull together. The associated Raman spectra showed a corresponding signature of mineralized tissue. Similarly, Khan et al. imaged samples of goat mucosal membrane and found a clear separation between upper epithelium and lower stromal layers via OCT. The RS analysis of these stromal layers showed stronger amide-I and amide-III bands than the epithelial layer, as the stroma has a larger concentration of structural proteins [82].
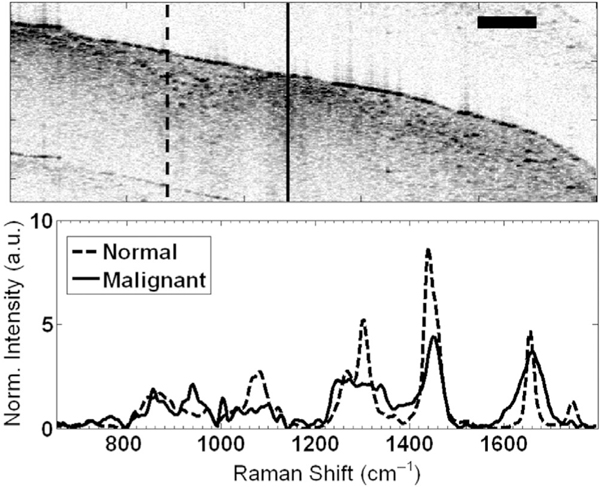
Several groups have utilized RS–OCT to investigate its potential for cancer screening and to identify tumor margins. For example, Patil et al. reported ex vivo screening of malignant breast tissue [76]. The authors concluded that OCT images could distinguish the highly scattering malignant regions from healthy tissue. Raman spectral features associated with these two zones could clear differentiate lipid-rich healthy tissue regions from malignant areas that showed higher DNA and protein content, as shown in Figure 8. Besides breast cancer detection, RS–OCT has demonstrated the potential to improve diagnosis of lung and bladder cancers [29, 86]. Zakharov et al. performed an ex vivo study on 22 lung biopsies, while Placzek et al. analyzed 119 bladder biopsies. In both cases, their results suggested that multimodal RS–OCT increases the detection accuracy compared to either modality alone.
FIGURE 8.
RS–OCT data from ex vivo breast tissues. Structural anomalies within the OCT image are spectrally characterized by RS, showing pronounced protein peaks indicative of malignancy (solid curve), while normal breast tissue exhibited a strong lipid signature (dotted curve). Scale bar represents 500 μm. Reproduced with permissions from reference [76]
Interstitial tracking of analytes within biological tissues has also been investigated with RS–OCT. Maher et al. [83] used CRS co-localized with OCT imaging to determine the concentration of Tenofovir, a microbicide drug used to prevent the transmission of HIV, in ex vivo porcine vaginal tissue. A standard-formulation gel loaded with 0% (control) or 1% Tenofovir was applied to the tissue surface, and RS measured drug concentration within this tissue at different time intervals after application. OCT images and Raman spectra were then acquired from a benchtop CRS–OCT platform that shared a common sample beam path, which utilized a motorized flip mirror to switch between the two modalities. The multimodal system determined the average depth of the interface between epithelium and stromal tissue layers with OCT imaging. Subsequently, it quantified physiologically relevant concentrations of Tenofovir in the targeted stroma with confocal RS. Building on this work, Chuchuen and Presnell et al. each used this same system to track drug transport dynamics through epithelial and stromal tissues in ex vivo vaginal tissue [139, 140].
5.2 |. In vivo applications
To date, there have been several RS–OCT reports of in vivo tissue analysis, primarily of skin. The OCT images clearly located skin lesions in each case, which was subsequently probed with RS to classify lesion type. For example, Patil et al. reported the first in vivo result from a combined RS–OCT system to analyze human skin scabs [76]. The scab was visible in the OCT image as a hyper-reflective zone. RS spectra were taken from locations within the scab and bordering regions, showing unique spectral lineshapes between the scab and the neighboring healthy tissue. Mazurenka et al. demonstrated the applicability of a miniaturized RS–OCT probe to screen for skin lesions. As a proof of concept, the device could distinguish RS spectral characteristics from volar and dorsal regions (i.e., palm and back) of the hand [80] while also imaging tissue microstructure with OCT. The spectra from the dorsal region showed lower Raman signal intensity due to higher melanin concentration, and higher carotenoid concentration was found in the volar area due to thicker stratum corneum layer. A large study was carried out by Zakharov et al. [29] that demonstrated in vivo results from 50 Caucasian subjects’ skin comprising melanomas, basal cell carcinoma, squamous cell carcinoma, pigment nevi, benign tumors, and healthy tissues. OCT images were used for tumor area localization and RS spectra were used in two-step phase analysis for tumor-grade diagnostics. An increase of 9% in sensitivity and 8% in specificity was observed by the multimodal system compared to either of the single modality systems.
In addition to skin interrogation, Wang et al. demonstrated the efficacy of RS-OCT for in vivo characterization of tissues in the oral cavity with a handheld probe. OCT showed unique tissue morphology between soft palate, blood vessels in lamina propria, and a clear contrast between the epithelium layer and lamina propria. These morphological differences were congruent with histology images of oral cavity tissues. Raman spectra of these oral tissues were also unique. When combining RS features with OCT-derived attenuation coefficients from each tissue type, discrimination between oral tissue types (e.g. hard and soft palate, floor, buccal, ventral, and dorsal tongue) was achieved with 75% accuracy [31].
6 |. FUTURE DIRECTIONS
Although the utility of multimodal RS–OCT has been demonstrated for various biomedical applications, there are still barriers that must be addressed to promote clinical adoption. The following section will outline potential technological and methodological advances that may bring RS–OCT technologies to a more clinically ready state.
6.1 |. Hardware advances
The future landscape for advancements in biomedical RS–OCT largely/mainly depends on the development of miniaturized probe designs for specific applications. This is necessary to deploy RS–OCT in clinical or remote environments. Many groups have reported miniature OCT technologies [141], which have demonstrated that more compact or cheaper OCT systems can be fabricated; for example, as stand-alone systems designed for self-OCT scanning at home [143] or even within photonic integrated circuits [142]. While there are multiple commercial companies offering portable RS technologies, like the handheld Agilent Resolve Raman (Agilent, Santa Clara, CA), the purpose of such devices is primarily for chemical identification, as they do not have the required sensitivity for tissue characterization. By a large margin, the stopgap for miniaturizing clinical RS–OCT systems rests on the instrumentation requirements for collecting biological Raman signals.
The unrealistic system cost, size, and complexity associated with using dedicated laser sources and detection platforms for RS and OCT also impedes the scalability and deployment of this technology. So, we believe next-generation RS–OCT systems will also benefit from recent advances in laser and detector technologies that may help bridge the instrumentation gaps between these two modalities. For example, a detector that can provide low noise, deep wells, long pixel arrays, and fast readout rates would allow both optical signals to be acquired efficiently on a single detection platform. Next-generation low-light detectors like the Blaze CCD (Teledyne, Waterloo, ON) are approaching the high sensitivity (98% quantum efficiency at 900 nm) and sufficiently fast spectral readout rate (215 kHz) to address the detection needs for both RS and SD–OCT signals. Swept source lasers that provide fast tunability, narrow bandwidth, and high-power output could allow for a single source to be used for both RS and SS-OCT, and custom laser designs have been reported that represent potential solutions. Strupler et al. described a modified commercial external cavity diode laser with a custom rapidly tunable filter, based on a scanning polygonal mirror, that could sweep between 761 and 798 nm with a maximal output power of 117 mW [144]. This design demonstrated a coherence length of 11 mm and instantaneous linewidth of 24 pm at a sweep rate of 30 kHz. Because the mirror position corresponds to the output laser line, such a device can park the laser output at 785 nm for standard dispersion-based RS. This provides a reasonable compromise between the source power, center wavelength, and bandwidth requirements for spontaneous RS and a sweeping rate sufficient for SS-OCT imaging, but with a low axial resolution. The authors mentioned that a more optimized design, for example with extended scan range and a faster scanning mirror, could reduce the coherence length to realize improved axial resolution while maintaining acceptable sweep rate for fast SS-OCT imaging. This configuration would require RS and OCT signals to be acquired on dedicated detectors.
An alternative solution would be to apply swept source detection for both RS and OCT using a single light source and detector for further device miniaturization. Small form-factor microelectromechanically tunable vertical-cavity surface-emitting lasers (MEMS-VCSEL) have separately demonstrated great utility for both swept-source RS (SS-RS) [145] and SS-OCT [146] miniature designs. In SS-RS, a fixed bandpass optical filter selects a narrow range of Raman scattered emissions to be collected by a point-detector. The Raman spectrum is readout by sweeping the tunable laser line, effectively eliminating the need for a dispersive spectrometer for smaller form-factor RS system designs. This collection scheme is nearly identical to SS-OCT. One could envision easily switching between SS-OCT and SS-RS spectral readout by mechanically moving the bandpass filter within the detection path.
However, combining detectors or sources for cheaper and more portable RS–OCT systems will inevitably impact system functionality and performance. Using a single detector or laser source eliminates the potential to acquire both signals simultaneously. The limited pixel array size of RS–optimized CCD detectors decreases OCT spectral sampling and, therefore, impacts the imaging range. Although MEMS-VCSEL and similar tunable sources could represent a potential solution to using a single source and detector for RS–OCT, they have a limited scanning range. Atabaki et al. reported coverage of only 400 cm−1 in their implementation of SS-RS even when integrating two MEMS-VCSEL lasers to cover a more extensive scan range [145]. Another limitation of their results was that SS-RS on biological samples required 52 s acquisitions due to the need to detect each Raman spectral channel sequentially. So, RS-OCT system designs must weigh the trade-offs associated with multimodal integration to determine if sharing common components is desirable.
6.2 |. Pump-probe RS-OCT
The long exposure times needed for spontaneous RS severely limit the potential for real-time RS–OCT morphomolecular imaging or swept-source detection of the Raman signal. Therefore, mitigation of this speed imbalance between RS and OCT is another avenue for future innovation. One way to add molecular sensitivity to OCT without the speed limitations imposed by RS is by analyzing the acquired OCT interferogram to derive compositional information, termed spectroscopic OCT (SOCT) [147]. In SOCT, wavelength-specific attenuation of the OCT spectrum can be assigned to molecular absorbers like hemoglobin and lipids [148]. Still, this approach cannot provide unambiguous molecularly-specific information that is available in RS. Instead, the integration of pump-probe optical methods like coherent anti-stokes Raman scattering (CARS) can amplify the rate of Raman scattering and enhance Raman detection speeds to approach that of OCT [149].
Vinegoni et al. proposed a CARS–OCT system design [150] to perform real-time interferometric imaging with added RS molecular contrast. Due to the need for high-powered femtosecond lasers to stimulate CARS, multiple higher-order optical processes could also be used as additional label-free contrast mechanisms, including stimulated Raman scattering (SRS), and higher-order harmonic generation. Clinical applications for CARS- and SRS-OCT have been explored for optic nerve imaging [151], characterization of human bone [152], brain tumor detection [153], and adipose tissue [154]. Although, the high cost and tissue-damaging effects of fast-pulsed lasers limit the accessibility of such devices for in vivo use. To address this issue, advances in spectral broadening techniques to lower pulse peak power are attempting to find novel optical designs better suited for clinical pump-probe RS–OCT imaging [155].
7 |. CONCLUSION
This review is intended to present the reader with design considerations and limitations of current implementations of RS–OCT but also to inspire future technical developments in this very promising area of multimodal optical diagnostics.
Hybrid RS–OCT systems demonstrate the benefits of multimodal optical systems in that combining unique optical techniques can address the limitations of one modality by offering the advantages of the other. In situations where in vivo RS acquisition is difficult due to spatial constraints (e.g., small body cavities, noncontact measurements) or requires spatially dependent probing (e.g., legion boundaries, tumor margin detection), OCT can provide feedback on tissue morphology relative to the RS probing beam. In this way, OCT scans can serve as an image guidance tool to spatially select a specific tissue region for point-wise RS measurement. When the structural information provided by OCT imaging is insufficient to characterize disease states, RS may provide the necessary molecular specificity needed for accurate diagnosis. Also, it has been shown that combining RS spectral features with OCT image-derived metrics can improve diagnostic accuracy in cases where either morphological or biochemical information is inadequare to discriminate disease. In all reported cases where quantitative features from RS and OCT were combined, the classification accuracy of diseases state or tissue types was enhanced by 1%–47% relative to their independent classification performances. Compared with either RS or OCT alone, the complementary structural and compositional information provided by multimodal RS–OCT systems expands the application space for each label-free optical method. In this way, combining these optical modalities into a single platform augments their use for tissue characterization.
While the technical challenges for integrating RS and OCT into a single device are considerable, the prospects for this field of noninvasive and label-free optical diagnostics are becoming more and more apparent. It is an active and growing field of research, and the wealth of potential biomedical applications for RS–OCT has yet to be fully realized.
ACKNOWLEDGMENTS
The authors would like to thank Andrea Locke and Guillermo Monroy for useful discussions. Jobaida Akhtar would like to acknowledge an Australian Government Research Training Program Scholarship. Jobaida Akhtar, Erik Schartner, Heike Ebendorff-Heidepriem, and Jiawen Li wish to acknowledge the Australian Research Council (CE140100003). One author would like to acknowledge funding from the National Health and Medical Research Council (GNT2008462 and APP2001646) and the National Heart Foundation of Australia Future Leader Fellowship Grant (105608). Sean Fitzgerald would like to acknowledge funding support from the National Institutes of Health (R01EB028615).
Funding information
Australian Research Council, Grant/Award Number: CE140100003; National Health and Medical Research Council, Grant/Award Numbers: GNT2008462, APP2001646; National Heart Foundation of Australia, Grant/Award Number: 105608; National Institutes of Health, Grant/Award Number: R01EB028615
Footnotes
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the creation of this review article.
CONFLICT OF INTEREST
The authors declare no financial or commercial conflict of interest.
REFERENCES
- [1].Wilson BC, Jermyn M, Leblond F, J. Biomed. Opt 2018, 23, 030901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wan Q-S, Wang T, Zhang K-H, Tumor Biol. 2017, 39, 1010428317717984. [DOI] [PubMed] [Google Scholar]
- [3].Majumder SK, Keller MD, Boulos FI, Kelley MC, Mahadevan-Jansen A, J. Biomed. Opt 2008, 13, 054009. [DOI] [PubMed] [Google Scholar]
- [4].Stone N, Kendall C, Shepherd N, Crow P, Barr H, Raman Spectrosc J. 2002, 33, 564. [Google Scholar]
- [5].H H. Mantsch, Infrared and Raman Spectroscopy of Biological Materials, CRC Press, Boca Raton, FL: 2000, p. 4. [Google Scholar]
- [6].Johnson CR, Ludwig M, O’Donnell S, Asher SA, J. Am. Chem. Soc 1984, 106, 5008. [Google Scholar]
- [7].Nie S, Bergbauer K, Ho J, Kuck J Jr., Yu N, Spectroscopy 1990, 5, 24. [DOI] [PubMed] [Google Scholar]
- [8].Pilotto S, Pacheco M, Silveira L Jr., Villaverde AB, Zangaro R, Lasers Med. Sci 2001, 16, 2. [DOI] [PubMed] [Google Scholar]
- [9].Caspers P, Lucassen G, Bruining H, Puppels G, Raman Spectrosc J. 2000, 31, 813. [Google Scholar]
- [10].Gellermann W, Ermakov IV, Ermakova MR, McClane RW, Zhao D-Y, Bernstein PS, JOSA A 2002, 19, 1172. [DOI] [PubMed] [Google Scholar]
- [11].Rangan S, Schulze HG, Vardaki MZ, Blades MW, Piret JM, Turner RF, Analyst 2020, 145, 2070. [DOI] [PubMed] [Google Scholar]
- [12].Bergholt MS, Serio A, Albro MB, Front. Bioeng. Biotechnol 2019, 7, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Choo-Smith LP, Edwards H, Endtz HP, Kros J, Heule F, Barr H, Robinson JS Jr., Bruining H, Puppels G, Biopolymers 2002, 67, 1. [DOI] [PubMed] [Google Scholar]
- [14].Yaqoob Z, Wu J, McDowell EJ, Heng X, Yang C, J. Biomed. Opt 2006, 11, 063001. [DOI] [PubMed] [Google Scholar]
- [15].Goetz M, Malek NP, Kiesslich R, Nat. Rev. Gastroenterol. Hepatol 2014, 11, 11. [DOI] [PubMed] [Google Scholar]
- [16].Smithpeter CL, Dunn AK, Welch A, Richards-Kortum R, Appl. Optics 1998, 37, 2749. [DOI] [PubMed] [Google Scholar]
- [17].Aumann S, Donner S, Fischer J, Müller F, High Resolution Imaging in Microscopy and Ophthalmology, Springer, Cham: 2019, p. 59. [Google Scholar]
- [18].Fujimoto JG, Brezinski ME, Tearney GJ, Boppart SA, Bouma B, Hee MR, Southern JF, Swanson EA, Nat. Med 1995, 1, 970. [DOI] [PubMed] [Google Scholar]
- [19].Bowd C, Zangwill LM, Berry CC, Blumenthal EZ, Vasile C, Sanchez-Galeana C, Bosworth CF, Sample PA, Weinreb RN, Invest. Ophthalmol. Vis. Sci 1993, 2001, 42. [PubMed] [Google Scholar]
- [20].Zuccaro G, Gladkova N, Vargo J, Feldchtein F, Zagaynova E, Conwell D, Falk G, Goldblum J, Dumot J, Ponsky J, Am. J. Gastroenterol Suppl. 2001, 96, 2633. [DOI] [PubMed] [Google Scholar]
- [21].Gambichler T, Orlikov A, Vasa R, Moussa G, Hoffmann K, Stücker M, Altmeyer P, Bechara FG, J. Dermatol. Sci 2007, 45, 167. [DOI] [PubMed] [Google Scholar]
- [22].Gambichler T, Regeniter P, Bechara FG, Orlikov A, Vasa R, Moussa G, Stücker M, Altmeyer P, Hoffmann K, J. Am. Acad. Dermatol 2007, 57, 629. [DOI] [PubMed] [Google Scholar]
- [23].Boppart SA, Psychophysiology 2003, 40, 529. [DOI] [PubMed] [Google Scholar]
- [24].Suter MJ, Nadkarni SK, Weisz G, Tanaka A, Jaffer FA, Bouma BE, Tearney GJ, JACC Cardiovasc. Imaging 2011, 4, 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mogensen M, Jørgensen TM, Nürnberg BM, Morsy HA, Thomsen JB, Thrane L, Jemec GB, Dermatol. Surg 2009, 35, 965. [DOI] [PubMed] [Google Scholar]
- [26].van der Meer FJ, Faber DJ, Sassoon DMB, Aalders MC, Pasterkamp G, van Leeuwen TG, IEEE Trans. Med. Imaging 2005, 24, 1369. [DOI] [PubMed] [Google Scholar]
- [27].Çilesiz I, Fockens P, Kerindongo R, Faber D, Tytgat G, ten Kate F, van Leeuwen T, Gastrointest. Endosc 2002, 56, 852. [DOI] [PubMed] [Google Scholar]
- [28].Rangaraju LP, Kunapuli G, Every D, Ayala OD, Ganapathy P, Mahadevan-Jansen A, Burns 2019, 45, 659. [DOI] [PubMed] [Google Scholar]
- [29].Zakharov VP, Bratchenko IA, Artemyev DN, Myakinin OO, Kornilin DV, Kozlov SV, Moryatov AA, J. Biomed. Opt 2015, 20, 025003. [DOI] [PubMed] [Google Scholar]
- [30].Ashok PC, Praveen BB, Bellini N, Riches A, Dholakia K, Herrington CS, in: Proceedings of SPIE BiOS: Biomedical Vibrational Spectroscopy VI: Advances in Research and Industry, California, March 2014, 89390L. [Google Scholar]
- [31].Wang J, Zheng W, Lin K, Huang Z, Biophotonics J. 2018, 11, e201700113. [DOI] [PubMed] [Google Scholar]
- [32].Balan V, Mihai C-T, Cojocaru F-D, Uritu C-M, Dodi G, Botezat D, Gardikiotis I, Materials 2019, 12, 2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pence I, Mahadevan-Jansen A, Chem. Soc. Rev 1958, 2016, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kerr LT, Byrne HJ, Hennelly BM, Anal. Methods 2015, 7, 5041. [Google Scholar]
- [35].Patil CA, Pence IJ, Lieber CA, Mahadevan-Jansen A, Opt. Lett 2014, 39, 303. [DOI] [PubMed] [Google Scholar]
- [36].Santos IP, Caspers PJ, Bakker Schut T, van Doorn R, Koljenović S, Puppels GJ, J. Raman Spectrosc 2015, 46, 652. [Google Scholar]
- [37].Tuschel D, Spectroscopy 2016, 31, 14. [Google Scholar]
- [38].Jahn IJ, Grjasnow A, John H, Weber K, Popp J, Hauswald W, Sensors 2021, 21, 5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gautam R, Vanga S, Ariese F, Umapathy S, EPJ Tech. Instrum 2015, 2, 1.26146600 [Google Scholar]
- [40].Lasch P, Chemom. Intel. Lab. Syst 2012, 117, 100. [Google Scholar]
- [41].Afseth NK, Segtnan VH, Wold JP, Appl. Spectrosc 2006, 60, 1358. [DOI] [PubMed] [Google Scholar]
- [42].Jermyn M, Desroches J, Aubertin K, St-Arnaud K, Madore W-J, De Montigny E, Guiot M-C, Trudel D, Wilson BC, Petrecca K, Phys. Med. Biol 2016, 61, R370. [DOI] [PubMed] [Google Scholar]
- [43].Auner GW, Koya SK, Huang C, Broadbent B, Trexler M, Auner Z, Elias A, Mehne KC, Brusatori MA, Cancer Metastasis Rev. 2018, 37, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pahlow S, Meisel S, Cialla-May D, Weber K, Rösch P, Popp J, Adv. Drug Deliv. Rev 2015, 89, 105. [DOI] [PubMed] [Google Scholar]
- [45].DePaoli D, Lemoine É, Ember K, Parent M, Prud’homme M, Cantin L, Petrecca K, Leblond F, Côté DC, J. Biomed. Opt 2020, 25, 050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Clinicaltrials.org. U.S. National Library of Medicine, NCT04239105; c2020–01. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [47].Clinicaltrials.org. U.S. National Library of Medicine, NCT04215861; c2020–01. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [48].Clinicaltrials.org. U.S. National Library of Medicine, NCT03482622; c2018–03. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [49].Clinicaltrials.org. U.S. National Library of Medicine, NCT02758678; c2016–05. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [50].Clinicaltrials.org. U.S. National Library of Medicine, NCT04931121; c2021–06. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [51].Clinicaltrials.org. U.S. National Library of Medicine, NCT03781232; c2018–12. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [52].v. U.S. National Library of Medicine, NCT04869618; c2021–05. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [53].Clinicaltrials.org. U.S. National Library of Medicine, NCT03468634; c2018–03. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/https://clinicaltrials.gov/.
- [54].Clinicaltrials.org. U.S. National Library of Medicine, NCT02814591; c2019–04. [Cited March 3, 2022]. Available from: https://clinicaltrials.gov/.
- [55].Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Science 1991, 254, 1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].McLaughlin RA, Sampson DD, Opt. Fiber Technol 2010, 16, 467. [Google Scholar]
- [57].Popescu DP, Flueraru C, Mao Y, Chang S, Disano J, Sherif S, Sowa MG, Biophys. Rev 2011, 3, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gora MJ, Suter MJ, Tearney GJ, Li X, Biomed. Opt. Express 2017, 8, 2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Swanson EA, Fujimoto JG, Biomed. Opt. Express 2017, 8, 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ali M, Parlapalli R, Texas Instruments, SPRABB9–June, 2010. [Google Scholar]
- [61].Uribe-Patarroyo N, Kassani SH, Villiger M, Bouma BE, Opt. Express 2018, 26, 9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tearney G, Bouma B, Fujimoto J, Opt. Lett 1811, 1997, 22. [DOI] [PubMed] [Google Scholar]
- [63].Iyer S, Coen S, Vanholsbeeck F, Opt. Lett 2009, 34, 2903. [DOI] [PubMed] [Google Scholar]
- [64].Xie T, Wang Z, Pan Y, Appl. Optics 2005, 44, 4272. [DOI] [PubMed] [Google Scholar]
- [65].Wojtkowski M, Srinivasan VJ, Ko TH, Fujimoto JG, Kowalczyk A, Duker JS, Opt. Express 2004, 12, 2404. [DOI] [PubMed] [Google Scholar]
- [66].Cense B, Nassif NA, Chen TC, Pierce MC, Yun S-H, Park BH, Bouma BE, Tearney GJ, De Boer JF, Opt. Express 2004, 12, 2435. [DOI] [PubMed] [Google Scholar]
- [67].Liu D, Ge C, Xin Y, Li Q, Tao R, Opt. Express 2020, 28, 5919. [DOI] [PubMed] [Google Scholar]
- [68].Dicardiology.com. Diagnostic and Interventional Cardiology; c2010–05. [Cited March 3, 2022]. Available from: https://www.dicardiology.com/product/fda-clears-first-coronary-oct-system-united-states.
- [69].investorintel.com. InvestorIntel Corporation; c2021–05. [Cited March 4, 2022]. Available from: https://investorintel.com/markets/biotech-healthcare/biotech-news/perimeter-medical-imaging-ai-receives-u-s-fda-breakthrough-device-designation-for-its-optical-coherence-tomography-oct-imaging-system-with-imgassist-ai/.
- [70].Fiercebiotech.com. c2020–01. [Cited March 3, 2022]. Available from: https://www.fiercebiotech.com/medtech/fda-clears-photonicare-s-handheld-oct-scanner-for-checking-ear-infections.
- [71].Cordero E, Latka I, Matthäus C, Schie IW, Popp J, J. Biomed. Opt 2018, 23, 071210. [DOI] [PubMed] [Google Scholar]
- [72].Kufcsak A, Bagnaninchi P, Erdogan A, Henderson RK, Krstajić N, Opt. Express 2021, 29, 18720. [DOI] [PubMed] [Google Scholar]
- [73].Patil C, Kalkman J, Faber DJ, Nyman JS, van Leeuwen TG, Mahadevan-Jansen A, J. Biomed. Opt 2011, 16, 011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chun-Te Ko A, Hewko MD, Leonardi L, Sowa MG, Dong CC, Williams P, Cleghorn B, J. Biomed. Opt 2005, 10, 031118. [DOI] [PubMed] [Google Scholar]
- [75].Evans JW, Zawadzki RJ, Liu R, Chan JW, Lane SM, Werner JS, Biophotonics J. 2009, 2, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Patil CA, Bosschaart N, Keller MD, van Leeuwen TG, Mahadevan-Jansen A, Opt. Lett 2008, 33, 1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Patil CA, Kirshnamoorthi H, Ellis DL, van Leeuwen TG, Mahadevan-Jansen A, Lasers Surg. Med 2011, 43, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang J, Zheng W, Lin K, Huang Z, Opt. Lett 2016, 41, 3045. [DOI] [PubMed] [Google Scholar]
- [79].Klemes J, Kotzianova A, Pokorny M, Mojzes P, Novak J, Sukova L, Demuth J, Vesely J, Sasek L, Velebny V, Biophotonics J. 2017, 10, 1442. [DOI] [PubMed] [Google Scholar]
- [80].Mazurenka M, Behrendt L, Meinhardt-Wollweber M, Morgner U, Roth B, Rev. Sci. Instrum 2017, 88, 105103. [DOI] [PubMed] [Google Scholar]
- [81].Monroy GL, Fitzgerald ST, Locke A, Won J, Spillman DR Jr., Ho A, Zaki FR, Choi H, Chaney EJ, Werkhaven JA, Front. Photon 2022, 3, 929574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Khan KM, Krishna H, Majumder SK, Rao KD, Gupta PK, Biophotonics J. 2014, 7, 77. [DOI] [PubMed] [Google Scholar]
- [83].Maher JR, Chuchuen O, Henderson MH, Kim S, Rinehart MT, Kashuba AD, Wax A, Katz DF, Biomed. Opt. Express 2022, 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ren X, Lin K, Hsieh C-M, Liu L, Ge X, Liu Q, Biomed. Opt. Express 2022, 13, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Guevara E, Gutierrez-Hernandez JM, Castonguay A, Lesage F, Moncada B, González FJ, Biomed. Res 2017, 28, 1664. [Google Scholar]
- [86].Placzek F, Bautista EC, Kretschmer S, Wurster LM, Knorr F, González-Cerdas G, Erkkilä MT, Stein P, Ataman Ç, Hermann GG, Analyst 2020, 145, 1445. [DOI] [PubMed] [Google Scholar]
- [87].Schie IW, Placzek F, Knorr F, Cordero E, Wurster LM, Hermann GG, Mogensen K, Hasselager T, Drexler W, Popp J, Sci. Rep 2021, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Desroches J, Lemoine É, Pinto M, Marple E, Urmey K, Diaz R, Guiot MC, Wilson BC, Petrecca K, Leblond F, Biophotonics J. 2019, 12, e201800396. [DOI] [PubMed] [Google Scholar]
- [89].Li J, Schartner E, Musolino S, Quirk BC, Kirk RW, Ebendorff-Heidepriem H, McLaughlin RA, Opt. Lett 2018, 43, 1682. [DOI] [PubMed] [Google Scholar]
- [90].Lorenser D, Singe CC, Curatolo A, Sampson DD, Opt. Lett 2014, 39, 548. [DOI] [PubMed] [Google Scholar]
- [91].Chen M, Mas J, Forbes LH, Andrews MR, Dholakia K, Biophotonics J. 2018, 11, e201700129. [DOI] [PubMed] [Google Scholar]
- [92].Kumar S, Verma T, Mukherjee R, Ariese F, Somasundaram K, Umapathy S, Chem. Soc. Rev 1879, 2016, 45. [DOI] [PubMed] [Google Scholar]
- [93].Kemmler M, Denzler J, in: Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Stockholm, Sweden, 2012, 1823. [Google Scholar]
- [94].Andries E, Martin S, Appl. Spectrosc 2013, 67, 579. [DOI] [PubMed] [Google Scholar]
- [95].Chen G, Lin X, Lin D, Ge X, Feng S, Pan J, Lin J, Huang Z, Huang X, Chen R, RSC Adv. 2016, 6, 7760. [Google Scholar]
- [96].Zhao J, Zeng H, Kalia S, Lui H, Analyst 2016, 141, 1034. [DOI] [PubMed] [Google Scholar]
- [97].Gautam R, Peoples D, Jansen K, O’Connor M, Thomas G, Vanga S, Pence IJ, Mahadevan-Jansen A, Appl. Spectrosc 2020, 74, 1238. [DOI] [PubMed] [Google Scholar]
- [98].Bi X, Rexer B, Arteaga CL, Guo M, Mahadevan-Jansen A, J. Biomed. Opt 2014, 19, 025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zeng HT, Hou MH, Ni YP, Fang Z, Fan XQ, Lu HM, Zhang ZM, Chemometr J. 2020, 34, e3293. [Google Scholar]
- [100].Zysk AM, Boppart SA, J. Biomed. Opt 2006, 11, 054015. [DOI] [PubMed] [Google Scholar]
- [101].Bhattacharjee M, Ashok P, Rao KD, Majumder S, Verma Y, Gupta P, J. Innov. Opt. Heal. Sci 2011, 4, 59. [Google Scholar]
- [102].Sullivan AC, Hunt JP, Oldenburg AL, J. Biomed. Opt 2011, 16, 066010. [DOI] [PubMed] [Google Scholar]
- [103].Wan S, Lee H-C, Huang X, Xu T, Xu T, Zeng X, Zhang Z, Sheikine Y, Connolly JL, Fujimoto JG, Med. Image Anal 2017, 38, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Butola A, Ahmad A, Dubey V, Srivastava V, Qaiser D, Srivastava A, Senthilkumaran P, Mehta DS, Appl. Optics 2019, 58, A135. [DOI] [PubMed] [Google Scholar]
- [105].Nandy S, Sanders M, Zhu Q, Biomed. Opt. Express 2016, 7, 5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Pierre CS, Madore W-J, De Montigny É, Trudel D, Boudoux C, Godbout N, Mes-Masson A-M, Rahimi K, Leblond F, Med J. Imaging 2017, 4, 041306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Sawyer TW, Chandra S, Rice PF, Koevary JW, Barton JK, Phys. Med. Biol 2018, 63, 235020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].D’Amico AV, Weinstein M, Li X, Richie JP, Fujimoto J, Urology 2000, 55, 783. [DOI] [PubMed] [Google Scholar]
- [109].Freund JE, Faber DJ, Bus MT, van Leeuwen TG, de Bruin DM, Lasers Surg. Med 2019, 51, 399. [DOI] [PubMed] [Google Scholar]
- [110].Qi X, Pan Y, Sivak MV, Willis JE, Isenberg G, Rollins AM, Biomed. Opt. Express 2010, 1, 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Garcia-Allende PB, Amygdalos I, Dhanapala H, Goldin RD, Hanna GB, Elson DS, Biomed. Opt. Express 2011, 2, 2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gan Y, Tsay D, Amir SB, Marboe CC, Hendon CP, J. Biomed. Opt 2016, 21, 101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Schmitt JM, Xiang S, Yung KM, J. Biomed. Opt 1999, 4, 95. [DOI] [PubMed] [Google Scholar]
- [114].Gossage KW, Tkaczyk TS, Rodriguez JJ, Barton JK, J. Biomed. Opt 2003, 8, 570. [DOI] [PubMed] [Google Scholar]
- [115].Karamata B, Hassler K, Laubscher M, Lasser T, JOSA A 2005, 22, 593. [DOI] [PubMed] [Google Scholar]
- [116].Baroni M, Diciotti S, Evangelisti A, Fortunato P, La Torre A, in: Proceedings of 11th Mediterranean Conference on Medical and Biomedical Engineering and Computing, Ljubljana, Slovenia, 2007, 847. [Google Scholar]
- [117].Mishra A, Wong A, Bizheva K, Clausi DA, Opt. Express 2009, 17, 23719. [DOI] [PubMed] [Google Scholar]
- [118].Manfredi M, Grana C, Pellacani G, in: Proceedings of 11th International Conference on Computer Vision Theory and Applications, Rome, Italy, February 2016: 234. [Google Scholar]
- [119].Rao CN, Sastry SS, Mallika K, Tiong HS, Mahalakshmi K, Int. J. Innov. Res. Sci. Eng. Technol 2013, 2, 4531. [Google Scholar]
- [120].Chang S, Bowden AK, J. Biomed. Opt 2019, 24, 090901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vermeer KA, Mo J, Weda JJ, Lemij HG, de Boer JF, Biomed. Opt. Express 2014, 5, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Howley T, Madden MG, O’Connell M-L, Ryder AG, AICS’05, 2005, 277. [Google Scholar]
- [123].Sen PC, Hajra M, Ghosh M, Emerging Technology in Modelling and Graphics, Springer, Singapore: 2020, p. 99. [Google Scholar]
- [124].Bovenkamp D, Sentosa R, Rank E, Erkkilä MT, Placzek F, Püls J, Drexler W, Leitgeb RA, Garstka N, Shariat SF, Stiebing C, Schie IW, Popp J, Andreana M, Unterhuber A, in: Proceedings of European Conferences on Biomedical Optics: Optical Coherence Imaging Techniques and Imaging in Scattering Media III, Munich, Germany, July 2019, 11078_25. [Google Scholar]
- [125].Liu C-H, Qi J, Lu J, Wang S, Wu C, Shih W-C, Larin KV, in: Proceedings of SPIE BiOS: Dynamics and Fluctuations in Biomedical Photonics XI, California, February 2014, 894208. [Google Scholar]
- [126].Ashok PC, Praveen BB, Bellini N, Riches A, Dholakia K, Herrington CS, Biomed. Opt. Express 2013, 4, 2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tamošiūnas M, Čiževskis O, Viškere D, Melderis M, Rubins U, Cugmas B, Cancers (Basel) 2022, 14, 2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Pence IJ, Vargis E, Mahadevan-Jansen A, Appl. Spectrosc 2013, 67, 789. [DOI] [PubMed] [Google Scholar]
- [129].Srivastava S, Int. J. Comput. Appl 2014, 88, 26. [Google Scholar]
- [130].Fong S, Biuk-Aghai RP, Millham RC, in: Proceedings of the 10th International Conference on Machine Learning and Computing, New York, February 2018, 122. [Google Scholar]
- [131].Ahmed Z, Zeeshan S, Data Mining J, Genomics Proteomics 2014, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Smith TC, Frank E, Statistical genomics, Springer, New York, NY: 2016, p. 353. [Google Scholar]
- [133].Kotthoff L, Thornton C, Hoos HH, Hutter F, Leyton-Brown K, Automated Machine Learning, Springer, Cham: 2019, p. 81. [Google Scholar]
- [134].David SK, Saeb A, Al Rubeaan K, Comput. Eng. Intell. Syst 2013, 4, 28. [Google Scholar]
- [135].Aličković E, Subasi A, Neural Comput. Applic 2017, 28, 753. [Google Scholar]
- [136].Álvarez JD, Matias-Guiu JA, Cabrera-Martín MN, Risco-Martín JL, Ayala JL, BMC Bioinformatics 2019, 20, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Guo S, Bocklitz T, Popp J, Analyst 2016, 141, 2396. [DOI] [PubMed] [Google Scholar]
- [138].Martyna A, Menżyk A, Damin A, Michalska A, Martra G, Alladio E, Zadora G, Chemom. Intel. Lab. Syst 2020, 202, 104029. [Google Scholar]
- [139].Chuchuen O, Maher JR, Henderson MH, Desoto M, Rohan LC, Wax A, Katz DF, PLoS One 2017, 12, e0185633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Presnell AL, Chuchuen O, Simons MG, Maher JR, Katz DF, Drug Deliv. Transl. Res 2018, 8, 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Rank EA, Agneter A, Schmoll T, Leitgeb RA, Drexler W, Transl. Biophoton 2022, 4, e202100007. [Google Scholar]
- [142].Akca B, Považay B, Alex A, Wörhoff K, De Ridder R, Drexler W, Pollnau M, Opt. Express 2013, 21, 16648. [DOI] [PubMed] [Google Scholar]
- [143].Keenan TD, Goldstein M, Goldenberg D, Zur D, Shulman S, Loewenstein A, Ophthalmol. Sci 2021, 1, 100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Strupler M, Goulamhoussen N, De Montigny E, Boudoux C, IEEE Photon. Technol. Lett 2010, 23, 197. [Google Scholar]
- [145].Atabaki AH, Herrington WF, Burgner C, Jayaraman V, Ram RJ, Opt. Express 2021, 29, 24723. [DOI] [PubMed] [Google Scholar]
- [146].John DD, Lee B, Potsaid B, Kennedy AC, Robertson ME, Burgner CB, Cable AE, Fujimoto JG, Jayaraman V, in: Proceedings of Advanced Solid State Lasers, Massachusetts, November 2016, ATh5A.2. [Google Scholar]
- [147].Morgner U, Drexler W, Kärtner F, Li X, Pitris C, Ippen E, Fujimoto J, Opt. Lett 2000, 25, 111. [DOI] [PubMed] [Google Scholar]
- [148].Fleming CP, Eckert J, Halpern EF, Gardecki JA, Tearney GJ, Biomed. Opt. Express 2013, 4, 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Rao KD, Choma MA, Yazdanfar S, Rollins AM, Izatt JA, Opt. Lett 2003, 28, 340. [DOI] [PubMed] [Google Scholar]
- [150].Vinegoni C, Bredfeldt JS, Marks DL, Boppart SA, Opt. Express 2004, 12, 331. [DOI] [PubMed] [Google Scholar]
- [151].Hoffmann C, Hofer B, Unterhuber A, Poavzay B, Morgner U, Drexler W, in: Proceedings of SPIE BiOS: Multimodal Biomedical Imaging VI, California, February 2011, 78920B. [Google Scholar]
- [152].Kamali T, Považay B, Kumar S, Silberberg Y, Hermann B, Werkmeister R, Drexler W, Unterhuber A, Opt. Lett 2014, 39, 5709. [DOI] [PubMed] [Google Scholar]
- [153].Soltani S, Guang Z, Zhang Z, Olson JJ, Robles FE, J. Biomed. Opt 2021, 26, 076004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Robles FE, in: Proceedings of SPIE BiOS: Label-free Biomedical Imaging and Sensing (LBIS), California, March 2019, 108900N. [Google Scholar]
- [155].Robles FE, Linnenbank H, Mörz F, Ledwig P, Steinle T, Giessen H, Opt. Lett 2019, 44, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]