Abstract
Background and Aim:
Slaughterhouses and their effluents could serve as a “hotspot” for the occurrence and distribution of antibiotic-resistant bacteria in the environment. This study aimed to understand the distribution of tetracycline resistance genes in Escherichia coli isolated from the floor surface and effluent samples of pig slaughterhouses in Banten Province, Indonesia.
Materials and Methods:
Ten samples, each from floor surface swabs and effluents, were collected from 10 pig slaughterhouses in Banten Province. Escherichia coli strains were isolated and identified by referring to the protocol of the Global Tricycle Surveillance extended-spectrum beta-lactamase E. coli from the WHO (2021). Quantitative real-time polymerase chain reaction (qPCR) was used to detect the tet genes.
Results:
The tetA, tetB, tetC, tetM, tetO, and tetX genes were distributed in the isolates from the floor surface samples, and the tetA, tetC, tetE, tetM, tetO, and tetX genes were distributed in the isolates from the effluent samples. The tetO gene (60%) was the most dominant gene in the isolates from floor surface samples, while the tetA gene was the dominant one in the isolates from the effluent samples (50%). The tetA + tetO gene combination was the dominant pattern (15%) in the E. coli isolates.
Conclusion:
The high prevalence and diversity of the tet genes in floor surface and effluent samples from pig slaughterhouses in Banten Province indicated that the transmission of the tet genes had occurred from pigs to the environment; thus, this situation should be considered a serious threat to public health.
Keywords: effluent, Escherichia coli, pig slaughterhouse, tet gene, tetracycline resistance
Introduction
The emergence and spread of antibiotic resistance due to the overuse of antibiotics in the farming industry pose a serious global threat to public health. Some studies have reported that both resistant pathogenic and commensal bacteria can be transmitted through the consumption of animal-based food products and environmental contamination [1]. Deaths due to infections caused by drug-resistant bacteria are estimated to increase yearly and reach 700,000–10,00,000 persons annually by 2050. Furthermore, global economic loss due to antibiotic resistance is likely to reach 100 trillion USD by 2050 [1]. Infection with multidrug-resistant bacteria renders the treatment ineffective; this eventually increases material loss, diminishes the quality of life, causes death of the infected individuals, and reduces the success rate of health improvement programs [2].
The crossover use of medically important antibiotics in livestock is considered one of the reasons for the emergence of antibiotic-resistant bacteria and their transmission to humans and the environment. Tetracycline is a commonly used antibiotic in humans because of its broad spectrum of activity, easy availability, and low cost. According to Jurnalis et al. [3], tetracycline is the most common antibiotic class available in public healthcare centers. Kallau et al. [4] and Detha et al. [5] reported that tetracycline is the common antibiotic used to treat pigs in Indonesia. Some studies have shown that bacterial species resist tetracycline through their tet genes [6, 7].
Effluents from slaughterhouses are known to be “hotspots” for the development and spread of antibiotic-resistant bacteria, resulting in the transfer of antibiotic resistance genes (ARGs) to other bacterial species [8]. The animal slaughtering process produces abundant liquid waste (1000 L/1000 kg live animal mass), and this liquid waste is potentially contaminated with antibiotic-resistant bacteria [8, 9]. The incomplete removal of antibiotic-resistant bacteria from wastewater treatment plants leads to the transportation of these bacteria along with the liquid waste to larger water bodies and terrestrial areas, resulting in environmental pollution, especially of aquatic systems, and eventually reaching humans through the contaminated food chain [10–12].
Escherichia coli, Gram-negative enteric commensal bacteria, are commonly found in humans and animals. Escherichia coli is one of the 12 enlisted critical priority pathogens. It can transmit ARGs to other bacteria, regardless of the species, horizontally through mobile genetic elements (MGEs) or vertically through replication [13, 14]. Escherichia coli is an indicator organism mostly used to determine the level of microbial contamination of water [15] and as an indicator to monitor antimicrobial resistance [13, 14].
Banten Province in Indonesia has several pig slaughterhouses centered in Tangerang City. The untreated effluent from pig slaughterhouses has a high potential to pollute the environment and ecosystem and poses a serious threat to public health. The contaminated floor where slaughtering is performed also acts as a source of contamination of the pork by drug-resistant bacteria. At present, limited data are available regarding the presence of antibiotic-resistant E. coli and ARGs, particularly tetracycline resistance genes, in pig slaughterhouses.
This study aimed to understand the distribution of tetracycline resistance genes in E. coli isolated from floor surface and effluent samples of pig slaughterhouses in Banten Province, Indonesia. The study findings can be used as a base to develop appropriate strategies to control antibiotic resistance.
Materials and Methods
Ethical approval
Ethical approval was not required for this study. The samples were collected in accordance with the guidelines of the standards for sample collection procedure (SNI 6989.59-2008 and ISO 19458:2006) [16, 17].
Study period and location
The study was conducted from July to October 2022. The isolation and identification of E. coli from the samples were performed at the microbiology laboratory of SKHB IPB. Tetracycline resistance genes were detected by quantitative real-time polymerase chain reaction (qPCR) at the Quality Control Laboratory and Certification of Animal Products, Ministry of Agriculture, Republic of Indonesia.
Sample collection
All of the pig slaughterhouses in Banten Province were taken as samples (as many as 10 pig slaughterhouses). Ten samples each of floor surface swab and effluent were collected. The effluent was sampled in accordance with the standards SNI 6989.59-2008 for the wastewater sampling method [16] and ISO 19458:2006 for sample collection for microbiological analysis of water quality [17]. Samples were collected aseptically and transported to the laboratory while being preserved at 4°C. The effluent at pig slaughterhouses was collected at two different time points: During the slaughtering process and after completion of the process; 500 mL sample was collected at each time point (total sample volume: 1 L).
Isolation and identification of E. coli
Escherichia coli was isolated and identified in accordance with the protocol of the Global Tricycle Surveillance extended-spectrum beta-lactamase E. coli from WHO [18]. All samples were serially diluted up to 10-5 dilution in duplicate by using sterile phosphate-buffered saline (PBS; pH 7.4) in a ratio of 1:9. Next, 0.1 mL of the sample from each dilution was added to a petri dish containing tryptone bile X-glucuronide (TBX) agar (Merck, Germany) and plated on the surface using the spread plate method. Bluish-green colonies on the TBX agar plate were suspected to be of E. coli. Petri dishes with a colony count of ≤100 colony-forming unit (CFU)/mL were used for subsequent analysis. Five bluish-green colonies from each TBX agar plate were inoculated into MacConkey agar (MCA; Oxoid, UK) plates. The suspected E. coli colonies on MCA plates appeared as morphologically flat, dry, pink, and nonmucoid colonies with a surrounding darker pink area of precipitated bile salts. The suspected E. coli colony was cultured on a tryptic soy agar medium (Oxoid) and then cultured on sulfide indole motility medium (Oxoid) to perform the indole test to confirm E. coli. The formation of a cherry red ring in the indole test was considered to be a positive confirmation of the E. coli isolate. Escherichia coli ATCC 25922 was used as a positive control.
DNA extraction
DNA from the E. coli isolate was extracted using the Mericon DNA Bacteria Kit (Qiagen, Germany) in accordance with the manufacturer’s protocol. The pure E. coli isolate was transferred using an inoculating loop from the culture medium into a microtube containing 1 mL of sterile PBS to achieve a turbidity of 0.5 McFarland standard or more (depending on the availability of the isolate). The suspension was centrifuged at 13,000× g for 5 min. The supernatant was discarded using a pipette, and 200 μL of sterile PBS was added to the bacterial pellet; the mixture was homogenized using a vortex mixer. Subsequently, the suspension was centrifuged at 13.000× g for 5 min. The bacterial pellet was washed several times to obtain a colorless suspension. Next, 200 μL of Fast Lysis Buffer was added, and the mixture was placed in a ThermoMixer (Eppendorf, Germany) for heating at 100°C at the rotation speed of 122× g for 10 min. The suspension was then incubated at room temperature (TM) for 2 min. The resulting suspension was centrifuged at 13,000× g for 5 min. Subsequently, 100 μL DNA-containing supernatant was transferred into a 2 mL microtube and incubated at –20°C or –80°C until further analysis.
Quality control of the extracted DNA
DNA concentration and purity were tested using a nanodrop spectrophotometer. The DNA purity ratio assessed by nanodrop was considered appropriate when it matched the set value of 1.8–2.0 (A260/A280). The amount of DNA concentration needed for the qPCR test was >36 ng/μL.
Detection of tetracycline resistance genes
Tetracycline resistance genes were detected using the qPCR SYBR Green method with primers of the target genes (Table-1 [19–21]). A real-time PCR thermal cycler (Rotor-Gene Q, Germany) was used for this method. The reagents were added to microtubes according to the required experimental design as follows: 12 μL of SYBR select master mix, 2 μL of 10 μM reverse primer, 2 μL of 10 μM forward primer, 3.5 μL of nuclease-free water, and 5 μL of DNA sample. The total reaction volume was 25 μL. Each microtube was placed on a PCR plate cooler to keep the reagent at low TM. In the qPCR SYBR Green method, melting was performed using Q-Rex software (Qiagen). The tetA, tetM, tetO, and tetX genes were amplified with the procedure proposed by Li et al. [22] using a two-step qPCR program with the following protocol: 3 min of initial heating at 95°C followed by 40 cycles of denaturation for 10 s at 95°C, 60 s of annealing at the TM adjusted for the primers listed in Table-1, and extension at 72°C for 1 min. The tetB, tetC, and tetE genes were amplified by referring to the procedure of Jia et al. [23] as follows: Initial heating at 94°C for 5 min, followed by 40 cycles of denaturation for 60 s at 94°C, 30 s of annealing at the TM adjusted for the primers listed in Table-1 [19–21], and extension at 72°C for 90 s. The specificity of the amplified product was analyzed using the melting curve (95°C for 10 s, 65°C–95°C with 0.5°C increment every 0.05 s).
Table-1.
Details of the primers used to detect tetracycline resistance genes.
| Gene | Primer | Primer sequence (5'–3') | Temperature annealing (°C) | Reference |
|---|---|---|---|---|
| tetA | tetA-F | GCTACATCCTGCTTGCCTTC | 57 | [19] |
| tetA-R | CATAGATCGCCGTGAAGAGG | |||
| tetB | tetB-F | TTG GTT AGG GGC AAG TTT TG | 56 | [19] |
| tetB-R | GTA ATG GGC CAA TAA CAC CG | |||
| tetC | tetC-F | CTTGAGAGCCTTCAACCCAG | 55 | [19] |
| tetC-R | ATGGTCGTCATCTACCTGCC | |||
| tetE | tetE-F | AAACCACATCCTCCATACGC | 57 | [19] |
| tetE-F | AAATAGGCCACAACCGTCAG | |||
| tetO | tetO-F | ACGGARAGTTTATTGTATACC | 57 | [20] |
| tetO-R | TGGCGTATCTATAATGTTGAC | |||
| tetM | tetM-F | ACAGAAAGCTTATTATATAAC | 52 | [20] |
| tetM-R | TGGCGTGTCTATGATGTTCAC | |||
| tetX | tetX-F | AGCCTTACCAATGGGTGTAAA | 57 | [21] |
| tetX-R | TTCTTACCTTGGACATCCCG |
The result was considered positive if the cycle threshold (CT) value was <40 with the amplification curve and a single melt peak was formed with melting TM equal to or the tolerance value of TM <2°C. The result was considered negative/undetectable if the CT value was >40 without the amplification curve. The result was considered indeterminate if the CT value was between >36 and <40.
Statistical analysis
The experimental data were presented as tables and figures and analyzed using a descriptive approach.
Results
Isolation and identification of E. coli
All the tested samples (ten floor surface samples and ten effluent samples) were positive for E. coli. Table-2 shows the complete results of the isolation and identification of E. coli.
Table-2.
Results of E. coli isolation and identification.
| Sample type | Amount isolate culture | Testing stages | Positive E. coli (%) | ||
|---|---|---|---|---|---|
|
| |||||
| TBX media culture (%) | MCA media culture (%) | Indole test (%) | |||
| Floor surface | 10 | 10 (100) | 10 (100) | 10 (100) | 100% |
| Effluent | 10 | 10 (100) | 10 (100) | 10 (100) | 100% |
E. coli=Escherichia coli, TBX=Tryptone bile X-glucuronide, MCA=MacConkey agar
Distribution of tetracycline resistance genes
The tetA, tetB, tetC, tetM, tetO, and tetX genes were present in the E. coli isolates from floor surface samples, while the tetA, tetC, tetE, tetM, tetO, and tetX genes were detected in the E. coli isolates from effluent samples (Table-3). Figure-1 shows the amplification curve and melting curve from the test result of tetracycline resistance genes determined using qPCR.
Table-3.
Cycle threshold and melt peak values of the tet genes detected in the floor surface and effluent samples using qPCR.
| Floor surface sample code | tet genes | CT value | Melt peak (°C) | Effluent sample code | tet genes | CT value | Melt peak (°C) |
|---|---|---|---|---|---|---|---|
| 1U | tetM | 12.27 | 85.0 | 2U | tetM | 10.90 | 85.0 |
| tetX | 20.15 | 91.3 | tetX | 19.99 | 91.3 | ||
| 4U | tetA | 11.16 | 90.4 | 5B | tetX | 20.29 | 93.0 |
| tetB | 29.99 | 82.1 | 8U | tetA | 11.89 | 90.6 | |
| tetO | 11.88 | 84.3 | tetO | 12.09 | 84.3 | ||
| tetX | 20.32 | 91.3 | 11U | tetA | 11.29 | 90.9 | |
| 9U | tetC | 31.78 | 87.9 | 14B | tetC | 33.94 | 85.8 |
| tetO | 12.01 | 84.3 | 16B | tetA | 11.10 | 90.5 | |
| 10U | tetA | 23.73 | 90.6 | tetX | 20.15 | 93.0 | |
| tetO | 12.24 | 84.8 | 19B | tetA | 33.22 | 90.5 | |
| 15B | tetA | 11.73 | 90.9 | 26U | tetA | 24.76 | 90.8 |
| tetO | 12.03 | 84.5 | tetE | 15.78 | 83.5 | ||
| 18U | tetO | 11.93 | 84.8 | 30B | tetE | 29.42 | 84.1 |
| tetX | 19.99 | 91.3 | tetO | 11.96 | 85.0 | ||
| 21B | tetO | 11.96 | 84.5 | ||||
| tetX | 20.29 | 93.0 | |||||
| 24B | tetC | 33.25 | 87.0 | ||||
| 25B | tetB | 30.48 | 82.1 |
qPCR=Quantitative real-time polymerase chain reaction, CT=Cycle threshold
Figure-1.
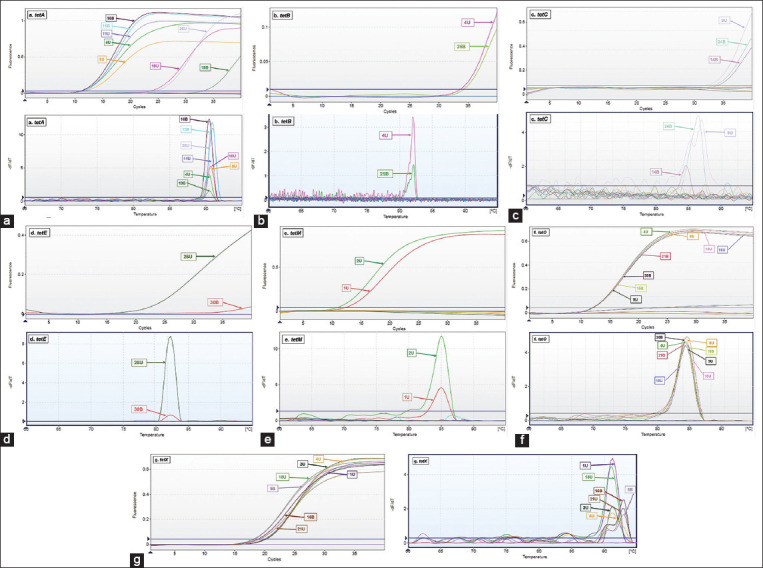
Test results showing the detection of tet genes in floor surface and effluent samples by quantitative real-time polymerase chain reaction. (a) Amplification curve and melting curve of tetA; (b) amplification curve and melting curve of tetB; (c) amplification curve and melting curve of tetC; (d) amplification curve and melting curve of tetE; (e) amplification curve and melting curve of tetM; (f) amplification curve and melting curve of tetO; (g) amplification curve and melting curve of tetX.
The tetO gene was the most dominant one and detected in 60% of floor surface samples, followed by tetX (40%), tetA (30%), tetB and tetC (20%), and tetM (10%), while the tetE gene was not detected (0%). In the effluent samples, the tetA gene was the most dominant one and detected in 50% of samples, followed by tetX (30%), tetE and tetO (20%), and tetC and tetM (10%), while the tetB gene was not detected (0%) (Figure-2). Twelve tet gene patterns were distributed between the samples from the pig slaughterhouses; the dominant pattern was tetA + tetO (15%), followed by tetA, tetC, tetM + tetX, and tetO + tetX (10%) and tetB, tetX, tetA + tetX, tetC + tetO, tetA + tetE, tetE + tetO, and tetA + tetB + tetO + tetX (5%) (Figure-3).
Figure-2.
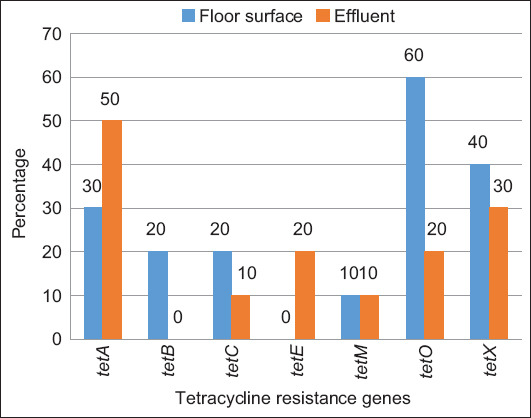
Prevalence percentage of tetracycline resistance genes in floor surface and effluent samples from pig slaughterhouses.
Figure-3.
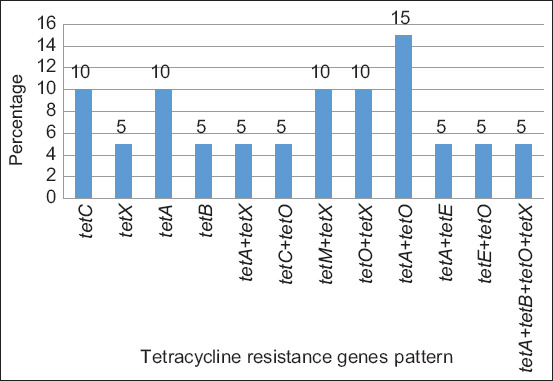
Pattern of tetracycline resistance genes in samples from pig slaughterhouses.
Discussion
Isolation and identification of E. coli
The floor surface and effluent samples from the ten pig slaughterhouses showed a high prevalence (100%) of E. coli. Researchers from several countries have also reported a high prevalence of E. coli in pig slaughterhouses. Savin et al. [24] reported a high prevalence (85.1%) of E. coli in wastewater treatment plants of pig slaughterhouses in Germany. A high prevalence of E. coli was also observed in the waste of pig slaughterhouses in Portugal [25] and in the effluent of slaughterhouses in Addis Ababa, Ethiopia [26]. Blaak et al. [27] showed that the percentage of E. coli in several slaughterhouses varied depending on different factors, such as the condition of the sampling area, fecal contamination level, sampling period, effluent flow rate, and disposal conditions.
In this study, the high prevalence of E. coli (100%) in floor surface and effluent samples from pig slaughterhouses was caused by a high rate of fecal contamination from the intestines of the slaughtered animals and poor health condition of the slaughtered animals [28]. The large amount of E. coli detected in the samples of floor surfaces and effluent of pig slaughterhouses might be caused by the abundance of blood traces (high protein medium), which could be an ideal source of nutrition for E. coli to grow and reproduce [29]. The presence of E. coli also suggests the possibility of the presence of other pathogens critical to public health safety [30]. Poor hygiene and sanitation conditions at pig slaughterhouses led to the spread of E. coli from animals, humans, the environment, and ecosystems through contaminated waste. In Banten Province, most of the effluent from pig slaughterhouses is disposed to Cisandane River, without pretreatment of the waste; this practice poses a serious risk to people who use the river water for their daily needs. A study on Cisandane River by Purwati et al. [31] showed a high level of E. coli contamination between 2 × 102 and 3 × 105 CFU/100 mL.
Distribution of tetracycline resistance genes
The qPCR SYBR Green method used in the present study showed positive results if the CT value was <40, an amplification curve was formed, and there was a single melt peak with equal TM value or the tolerance of TM value was <2°C. This concept was based on the studies of Ririe et al. [32] and Dortmans et al. [33] who reported that melting curve analysis could differentiate amplification products with the same length but have different GC/AT ratios with TM values <2°C. Melting point is generally acceptable in the range of 0.5–2°C. The narrow range (0.5°C–2°C) of melting point usually indicates the purity of the testing materials. The melting peak values of all the tested tet genes were as follows: tetA, 90.53°C; tetB, 82.0°C; tetC, 87.0°C; tetE, 84.11°C; tetM, 85.04°C; tetO, 84.45°C; and tetX, 91.28°C.
Melting curve analysis using high-resolution DNA melting analysis (HRM) generates a DNA melt curve profile that enables DNA characterization based on sequence length, guanine and cytosine as the main components, and complementarity of the DNA sequence. This method is very sensitive for classification based on variations in nucleic acid sequences or for the detection of changes in a single-nucleotide base that would form a melting curve with different patterns; thus, this method is useful to detect mutations or identify other genetic variants [34]. The difference in the melting curve pattern in the HRM method is a reflection of various DNA sequences of the target genes in each isolate [35]. This method can detect different nucleic acid compositions without sequencing, and thus, it is relatively cheaper than other conventional techniques [36, 37].
The present study tested seven types of tet genes that induce resistance to tetracycline through different mechanisms: Efflux pump (tetA, tetB, tetC, and tetE), ribosomal protection (tetM and tetO), and enzymatic inactivation (tetX). Tet genes are responsible for the emergence of resistance to tetracycline antibiotics in E. coli. Among the seven tet genes tested, almost all of them, except the tetE gene, were detected in the floor surface samples, and the tetB gene was detected in the effluent samples. The tetO gene (60%) was the dominant gene in the floor surface samples, whereas tetA was the dominant one in the effluent samples (50%). Savin et al. [38] demonstrated the presence of the tetA, tetB, and tetM genes in the effluent of pig slaughterhouses in Germany, but the tetX gene was not detected. Similar results have been reported in Portugal [25], wherein the tetA, tetB, tetM, and tetK genes were detected in the waste of a pig farm, and the tetA, tetB, tetK, tetL, tetM, tetO, and tetA(P) genes were detected in the effluent of slaughterhouses; the tetX gene was not found in both pig farm waste and slaughterhouse effluent in Portugal.
The tetA, tetB, tetC, tetD, tetE, and tetG genes are the dominant genes that induce tetracycline resistance in E. coli through the efflux pump mechanism that actively pumps out tetracycline to the periplasm during proton (H+) exchange by active transport, resulting in resistance due to inadequate concentration of the antibiotic, which subsequently prevents the binding of tRNA to the A-site of the ribosome 30S subunit and inhibits protein synthesis [39]. The tetA gene is the dominant efflux pump gene, followed tetC, and the least dominant ones are tetB and tetE. This finding agrees with the results of most other studies which stated that the tetA gene was indeed the most dominant one to be present in Gram-negative bacteria [40] and was often detected in pigs [41] or other animals [42, 43], meats, processed meat products [44], and various environmental samples [25, 45]. The tetA gene is present in the conjugation plasmid, from where it can easily transmit resistance genes to other bacterial species throughout conjugative horizontal gene transfer. The genes tetC and tetE are also present in the bacterial plasmid, while the tetB gene is present in the transposon and integrative and conjugative elements (ICE) [46]. The tetE gene is often associated with the nonconjugative plasmid [21] because of its limited transmission. In the present study, the percentage occurrence of the tetE gene was much lower than that of the other efflux pump genes, and it was even undetectable in the floor surface samples.
The tetO and tetM genes were the tet genes responsible for the ribosomal protection resistance mechanism, which is known as ribosomal protection proteins or RPPs that protect the ribosome by disrupting the main binding site of tetracycline [47, 48]. The tetO gene was the dominant gene in the floor surface samples (60%), but it showed a low occurrence (20%) in the effluent samples. Similarly, the tetM gene showed a low percentage of occurrences in both floor surface and effluent samples (10%). Amador et al. [25] detected tetO in E. coli isolated from slaughterhouse effluent, even though it had a low percentage of occurrences (12.5%), while the gene tetM, in contrast, was detected in a quite high percentage (43.8%).
The tetO gene is mostly associated with the conjugation plasmid in Campylobacter spp., and an inter-isolate transfer of tetO in Campylobacter jejuni has also been reported [49]. The tetO gene is also associated with the hot plasmid that was mediated within Enterococcus and Streptococcus spp. [50]. A recent study revealed that the tetO gene was integrated into transposons carrying the macrolide-resistant efflux genes mefA and msrD. This transposon can be transferred conjugatively to different strains of Streptococcus pyogenes and the unrelated Enterococcus faecalis [51, 52]. The tetO gene is rarely found in E. coli; however, in the present study, a high prevalence of this gene was detected, particularly in floor surface samples. This most likely occurred because the conjugative transposon carrying the tetO gene had a wider transmission among other unrelated bacteria, such as E. coli. As reported by Roberts [48], tetO found on the conjugative transposon could possibly cause much wider transmission of this gene among various unrelated bacteria in the future. Antibiotic resistance genes associated with a conjugative transposon can be easily transmitted to other bacteria, even those that are unrelated, as compared to ARGs associated with a nonconjugative plasmid [53].
The tetX genes responsible for the enzymatic inactivation resistance mechanism can modify the enzymatic activity, resulting in the inactivation of tetracycline. The tetX gene encodes a cytoplasmic protein that chemically transmutes tetracycline using oxygen and NADPH through the addition of a –OH cluster on C-11 of the tetracycline molecule [47]. At present, tetX has been identified in E. coli, although the enzymatic inactivation resistance mechanism has not been deeply observed [54]. Some studies have detected tetX in the effluent of pig slaughterhouses [55], manure made of pig feces [56], well water in close proximity to a pig farm [57], and river water [58].
The tet genes have been discovered on various MGEs, such as transposons, plasmids, integrons, and ICEs [59–61]. The abundance of MGEs carrying the tet genes has significantly contributed to the transmission of tetracycline-resistant bacteria to a wider environment through gene transfer from one bacterial species to another. Plasmids have contributed in transmitting multidrug resistance genes not only among bacteria of the same species but also among bacteria of unrelated species [62]. Integrons are often found on the plasmid and/or transposon that increase the transmission rate of ARGs. Class-1 integron is known to distribute ARGs among Gram-negative and Gram-positive bacteria [63]. Moredo et al. [64] reported that approximately 17.5% of E. coli isolates from pig slaughterhouses carried integrons as the transmitter of resistant genes.
In the present study, most E. coli isolates carried 2 tet genes, while some isolates had 4 tet genes. The dominant pattern among the combination of the tet genes was tetA + tetO (15%) (Figure-3). The high rate of prevalence and the variety of the tet genes detected in either floor surface samples or effluent samples from the 10 pig slaughterhouses in Banten Province indicated the occurrence of pig-to-environment transmission due to the overuse and uncontrolled application of tetracycline antibiotics in pig farms. Kallau et al. [4] showed that antibiotics are mostly used in pig farms for medication (55.21%), disease prevention (42.71%), and enhanced production (2.08%). Antibiotics of tetracycline class (oxytetracycline and tetracycline) are frequently used in pig farms [4, 65, 66].
The effluent of slaughterhouses could function as a “hotspot” for the growth and reproduction of different bacterial species as well as for the transmission and exchange of ARGs at a high level among different bacterial cells, thereby resulting in various combinations of such genes. The E. coli strain carrying multiple tetracycline resistance genes might increase the probability of forming a new combination or pattern of the tet genes, if this condition persists. The occurrence of new tet gene combinations is a very serious concern with unpredictable consequences on human health and environment [67].
The presence of antibiotic-resistant E. coli in pigs might cause contamination of the pork [66], processed meat products [68], and pig slaughterhouse environment and its waste [38], in addition to having the potential to transmit to a wider area and ecosystem [57, 58, 69]. Marshall and Levy [70] and Sanders et al. [11] showed that ARGs could be distributed to agricultural lands by direct and indirect contact through food, water, and animal feces.
Conclusion
The tetO gene (60%) was the most dominantly distributed gene in floor surface samples, while tetA (50%) was the dominant one in the effluent samples. The dominant pattern in the combination of the tet genes in the E. coli isolates was tetA + tetO (15%). The high prevalence and diversity of the tet genes in floor surface and effluent samples from pig slaughterhouses in Banten Province indicated that the transmission of the tet genes had occurred from pigs to the environment. In addition, poor waste management could be a source of transmission of E. coli that carries ARGs to the environment and may pose a serious threat to public health.
Authors’ Contributions
DFP: Coordinated the research, performed sample and data collection, conducted sample testing, performed data analysis, and wrote the manuscript. HL and IWTW: Involved in coordinating the research, data interpretation, and manuscript preparation and review. CB and PR: Involved in coordinating the research, data analysis, and manuscript preparation. All authors have read, reviewed, and approved the final manuscript.
Acknowledgments
The study was funded by Doctoral Research of the Ministry of Education, Culture, Research, and Technology, Indonesia with contract number: 082/E5/PG.02.00.PT/2022. Also, thanks to the Head of Agriculture and Food Security Service of Tangerang Regency and staff, as well as the Head of Food Security Service of Tangerang City and staff for the facilities provided during the research. Moreover, thanks to Quality Control Laboratory and Certification of Animal Products (QCLCAP/BPMSPH) Bogor and staff for the assistance and laboratory facility provided during research.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.O'Neill J. Antimicrobial Resistance:Tackling a Crisis for Health and Wealth of Nations. HM Government and Wellcome Trust, London, UK. 2014 [Google Scholar]
- 2.WHO. Regional Strategy on Prevention and Containment of Antimicrobial Resistance. New Delhi: SEARO Publication; 2010. [Google Scholar]
- 3.Jurnalis Y.D, Sayoeti Y, Aslinar A. Pattern of resistance of bacteria that cause diarrhea to antibiotics [Pola resistensi kuman penyebab diare terhadap antibiotika] Andalas Med. Magazine [Majalah Kedokt Andalas] 2009;33(1):41–46. [Google Scholar]
- 4.Kallau N.H.G, Wibawan I.W.T, Lukman D.W, Sudarwanto M.B. Analysis of relationship between knowledge and attitudes towards the practice of using antibiotics by pig farms in the city of Kupang, East Nusa Tenggara province [Analisis hubungan antara pengetahuan dan sikap terhadap praktik penggunaan antibiotik oleh peternakan babi di kota kupang provinsi nusa tenggara timur] J. Sain Vet. 2018;36(2):200–212. [Google Scholar]
- 5.Detha A, Wuri D.A, Ramos F, Biru D, Meha M.M, Lakapu A. Inappropriate use of antibiotics in pig farms in Kupang City, East Nusa Tenggara [Penggunaan antibiotik yang kurang tepat pada peternakan babi di Kota kupang, nusa tenggara timur] J. Vet. 2021;22(2):162–167. [Google Scholar]
- 6.Wang J, Ben W, Yang M, Zhang Y, Qiang Z. Dissemination of veterinary antibiotics and corresponding resistance genes from a concentrated swine feedlot along the waste treatment paths. Environ. Int. 2016;92–93:317–23. doi: 10.1016/j.envint.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Zhang P, Shen Z, Zhang C, Song L, Wang B, Shang J, Yue X, Qu Z, Li X, Wu L, Zheng Y, Aditya A, Wang Y, Xu S, Wu C. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet. Microbiol. 2017;203:49–55. doi: 10.1016/j.vetmic.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Savin M, Bierbaum G, Hammerl J.A, Heinemann C, Parcina M, Sib E, Voigt A, Kreyenschmidt J. Eskape bacteria and extended-spectrum-beta-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl. Environ. Microbiol. 2020;86(8):e02748–19. doi: 10.1128/AEM.02748-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal G.S. Characterization of the effluent wastewater from abattoirs for land application. Food Rev. Int. 2004;20(3):229–256. [Google Scholar]
- 10.Ivanov V, Stabnikov V, Zhuang W.Q, Tay J.H, Tay S.T.L. Phosphate removal from the returned liquor of municipal wastewater treatment plant using iron-reducing bacteria. J. Appl. Microbiol. 2005;98(5):1152–1161. doi: 10.1111/j.1365-2672.2005.02567.x. [DOI] [PubMed] [Google Scholar]
- 11.Sanders E.C, Yuan Y, Pitchford A. Fecal coliform and E. coli concentrations in effluent-dominated streams of the upper Santa Cruz watershed. Water. 2013;5(1):243–261. [Google Scholar]
- 12.Wadepohl K, Müller A, Seinige D, Rohn K, Blaha T, Meemken D, Kehrenberg C. Association of intestinal colonization of ESBL producing Enterobacteriaceae in poultry slaughterhouse workers with occupational exposure-a German pilot study. PLoS One. 2020;15(6):e0232326. doi: 10.1371/journal.pone.0232326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gekenidis M.T, Qi W, Hummerjohann J, Zbinden R, Walsh F, Drissner D. Antibiotic-resistant indicator bacteria in irrigation water:High prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS One. 2018;13(11):e0207857. doi: 10.1371/journal.pone.0207857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Xiao S, Jiang X, Li Y, Fan Z, Yu Y, Wang P, Li D, Zhao X, Liu C. Genomic characterization of Escherichia coli LCT-EC001, an extremely multidrug-resistant strain with an amazing number of resistance genes. Gut Pathog. 2019;11(1):25. doi: 10.1186/s13099-019-0298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He L.Y, Ying G.G, Liu Y.S, Su H.C, Chen J, Liu S.S, Zhao J.L. Discharge of swine wastes risks water quality and food safety:Antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 2016;92–93:210–219. doi: 10.1016/j.envint.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 16.BSN. SNI 6989.59-2008Concerning Wastewater Sampling Methods [Tentang Metoda Pengambilan Contoh Air Limbah]. BSN, Jakarta, ID. 2008 [Google Scholar]
- 17.ISO. ISO 19458:2006 Water Quality-Sampling for Microbiological. Geneva: International Organization for Standardization; 2006. [Google Scholar]
- 18.World Health Organization. WHO Integrated Global Surveillance on ESBL-Producing E. coli using a “One Health”Approach:Implementation and Opportunities. Geneva: World Health Organization; 2021. [Google Scholar]
- 19.Ng L.K, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline-resistant genes. Mol. Cell. Probes. 2001;15(4):209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 20.Aminov R.I, Garrigues-Jeanjean N, Mackie R.I. Molecular ecology of tetracycline resistance:Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 2001;67(1):22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh S, Sadowsky M.J, Roberts M.C, Gralnick J.A, LaPara T.M. Sphingobacterium spp. strain PM2-P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 2009;106(4):1336–1342. doi: 10.1111/j.1365-2672.2008.04101.x. [DOI] [PubMed] [Google Scholar]
- 22.Li N, Chen J, Liu C, Li B, Zhu C, Li H. Fate of antibiotic resistance genes in abandoned swine feedlots in China:Seasonal variation. Environ. Sci. Eur. 2021;33:121. [Google Scholar]
- 23.Jia S, He X, Bu Y, Shi P, Miao Y, Zhou H, Shan Z, Zhang X. Environmental fate of tetracycline resistance genes originating from swine feedlots in river water. J. Environ. Sci. Health B. 2014;49(8):624–631. doi: 10.1080/03601234.2014.911594. [DOI] [PubMed] [Google Scholar]
- 24.Savin M, Bierbaum G, Hammerl J.A, Heinemann C, Parcina M, Sib E, Voigt A, Kreyenschmidt J. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020;727:138788. doi: 10.1016/j.scitotenv.2020.138788. [DOI] [PubMed] [Google Scholar]
- 25.Amador P, Fernandes R, Prudêncio C, Duarte I. Prevalence of antibiotic resistance genes in multidrug-resistant Enterobacteriaceae on Portuguese livestock manure. Antibiotics (Basel) 2019;8(1):23. doi: 10.3390/antibiotics8010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tesfaye H, Alemayehu H, Desta A.F, Eguale T. Antimicrobial susceptibility profile of selected Enterobacteriaceae in wastewater samples from health facilities, abattoir, downstream rivers and a WWTP in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 2019;8:134. doi: 10.1186/s13756-019-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaak H, Lynch G, Italiaander R, Hamidjaja R.A, Schets F.M, De Husman A.M.R. Multidrug-resistant and extended-spectrum beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLoS One. 2015;10(6):e0127752. doi: 10.1371/journal.pone.0127752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onuoha S.C, Eluu S.C, Okata M.O. In-vitro antimicrobial resistance of Shigella and Salmonella species recovered from abattoir effluent in Afikpo, South Eastern Nigeria. Int. J. Curr. Microbiol. Appl. Sci. 2016;5(4):488–497. [Google Scholar]
- 29.Dankaka S.M, Farouq A.A, Bagega A, Abubakar U. Microbiological and physicochemical analysis of old Sokoto abattoir wastewater (sewage) contaminated with blacksmith activities. Bioremediation Sci. Technol. Res. 2018;6(2):9–13. [Google Scholar]
- 30.Atuanya E.I, Nwogu N.A, Orah C.U. Antibiotic resistance and plasmid profiles of bacteria isolated from Abattoir effluents around Ikpoba river in Benin City, Nigeria. J. Appl. Sci. Environ. Man. 2018;22(11):1749–1755. [Google Scholar]
- 31.Purwati S.U, Lestari N.S, Nasution E.L. Water quality assessment of Cisadane River using pollution indicator parameters. IOP Conf. Seri. Earth Environ. Sci. 2019;407:012009. [Google Scholar]
- 32.Ririe K.M, Rasmussen R.P, Wittwer C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245(2):154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 33.Dortmans J.C.F.M, Koch G, Rottier P.J.M, Peeters B.P.H. Virulence of pigeon paramyxovirus Type 1 does not always correlate with the cleavability of its fusion protein. J. Gen. Virol. 2009;90(11):2746–2750. doi: 10.1099/vir.0.014118-0. [DOI] [PubMed] [Google Scholar]
- 34.Wittwer C.T. High-resolution DNA melting analysis:Advancements and limitations. Hum. Mutat. 2009;30(6):857–859. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- 35.Banowary B, Dang T.V, Sarker S, Connolly J.H, Chenu J, Groves P, Ayton M, Raidal S, Devi A, Vanniasinkam T, Ghorashi S.A. Differentiation of Campylobacter jejuni and Campylobacter coli using multiplex-PCR and high-resolution melt curve analysis. PLoS One. 2015;10(9):e0138808. doi: 10.1371/journal.pone.0138808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong S.Y.C, Xie S, Richardson L.J, Ballard S.A, Dakh F, Grabsch E.A, Grayson M.L, Howden B.P, Johnson P.D.R. High-resolution melting genotyping of Enterococcus faecium based on multilocus sequence typing derived single nucleotide polymorphisms. PLoS One. 2011;6(12):e29189. doi: 10.1371/journal.pone.0029189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price E.P, Smith H, Huygens F, Giffard P.M. High-resolution DNA melt curve analysis of the clustered, regularly interspaced short-palindromic-repeat locus of Campylobacter jejuni. Appl. Environ. Microbiol. 2007;73(10):3431–3436. doi: 10.1128/AEM.02702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savin M, Bierbaum G, Kreyenschmidt J, Schmithausen R.M, Sib E, Schmoger S, Käsbohrer A, Hammerl J.A. Clinically relevant Escherichia coli isolates from process waters and wastewater of poultry and pig slaughterhouses in Germany. Microorganisms. 2021;9(4):698. doi: 10.3390/microorganisms9040698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen F, Starosta A.L, Arenz S, Sohmen D, Dönhöfer A, Wilson D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014;395(5):559–575. doi: 10.1515/hsz-2013-0292. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Zhu Z, Chen Y, Wang W, He F. The Plasmid-borne tetA gene is an important factor causing tigecycline resistance in ST11 carbapenem-resistant Klebsiella pneumoniae under selective pressure. Front. Microbiol. 2021;12:644949. doi: 10.3389/fmicb.2021.644949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurado-Rabadán S, de la Fuente R, Ruiz-Santa-Quiteria J.A, Orden J.A, de Vries L.E, Agersø Y. Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coli isolates from pigs. BMC Vet. Res. 2014;10:155. doi: 10.1186/1746-6148-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karczmarczyk M, Walsh C, Slowey R, Leonard N, Fanning S. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. Appl. Environ. Microbiol. 2011;77(20):7121–7127. doi: 10.1128/AEM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abo-Amer A.E, Shobrak M.Y, Altalhi A.D. Isolation and antimicrobial resistance of Escherichia coli isolated from farm chickens in Taif, Saudi Arabia. J. Glob. Antimicrob. Resist. 2018;15:65–68. doi: 10.1016/j.jgar.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Koo H.J, Woo G.J. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 2011;145(2):407–413. doi: 10.1016/j.ijfoodmicro.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Ling Z, Yang Y, Huang Y, Zou S, Luan T. A preliminary investigation on the occurrence and distribution of antibiotic resistance genes in the Beijiang River, South China. J. Environ. Sci. 2013;25(8):1656–1661. doi: 10.1016/s1001-0742(12)60223-x. [DOI] [PubMed] [Google Scholar]
- 46.Pazda M, Kumirska J, Stepnowski P, Mulkiewicz E. Antibiotic resistance genes identified in wastewater treatment plant systems-a review. Sci. Total. Environ. 2019;697:134023. doi: 10.1016/j.scitotenv.2019.134023. [DOI] [PubMed] [Google Scholar]
- 47.Chopra I, Roberts M. Tetracycline antibiotics:Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts M.C. Update on acquired tetracycline resistance genes. FEMS. Microbiol. Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 49.Avrain L, Vernozy-Rozand C, Kempf I. Evidence for natural horizontal transfer of tetO gene between Campylobacter jejuni strains in chickens. J. Appl. Microbiol. 2004;97(1):134–140. doi: 10.1111/j.1365-2672.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 50.Luna V.A, Roberts M.C. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J. Antimicrob. Chemother. 1998;42(5):613–619. doi: 10.1093/jac/42.5.613. [DOI] [PubMed] [Google Scholar]
- 51.Giovanetti E, Brenciani A, Lupidi R, Roberts M.C, Varaldo P.E. The presence of the tet(O) gene in erythromycin and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 2003;47(9):2844–2849. doi: 10.1128/AAC.47.9.2844-2849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenciani A, Ojo K.K, Monachetti A, Menzo S, Roberts M.C, Varaldo P.E, Giovanetti E. A new genetic element, carrying tet(O) and mef(A) genes. J. Antimicrob. Chemother. 2004;54:991–998. doi: 10.1093/jac/dkh481. [DOI] [PubMed] [Google Scholar]
- 53.Roberts M.C. Ciba Foundation Symposium 207. Chichester, UK: Wiley; 1997. Genetic Mobility and Distribution of Tetracycline Resistance Determinants Antibiotic Resistance:Origins, Evolution, Selection and Spread; pp. 206–218. [PubMed] [Google Scholar]
- 54.Poirel L, Madec J, Lupo A, Schink A, Kieffer N, Nordmann P, Schwarz S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectrum. 2018;6(4):1–27. doi: 10.1128/microbiolspec.arba-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He L.Y, He L.K, Liu Y.S, Zhang M, Zhao J.L, Zhang Q.Q, Ying G.G. Microbial diversity and antibiotic resistome in swine farm environments. Sci. Total Environ. 2019;685:197–207. doi: 10.1016/j.scitotenv.2019.05.369. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Y, Johnson T.A, Su J, Qiao M, Guo G, Stedtfeld R.D, Hashsham S.A, Tiedje J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U S A. 2013;110(9):3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang L, Xu Y, Xu J, Ling J, Zheng L, Zhou X, Xie G. Dissemination of antibiotic resistance genes (ARGs) by rainfall on a cyclic economic breeding livestock farm. Int. Biodeterior Biodegradation. 2019;138:114–121. [Google Scholar]
- 58.Shi W, Zhang H, Li J, Liu Y, Shi R, Du H, Chen J. Occurrence and spatial variation of antibiotic resistance genes (ARGs) in the Hetao Irrigation District, China. Environ. Pollut. 2019;251:792–801. doi: 10.1016/j.envpol.2019.04.119. [DOI] [PubMed] [Google Scholar]
- 59.de Barbeyrac B, Dupon M, Rodriguez P, Renaudin H, Bébéar C. A Tn1545-like transposon carries the tet(M) gene in tetracycline-resistant strains of Bacteroides ureolyticus as well as Ureaplasma urealyticum but not Neisseria gonorrhoeae. J. Antimicrob. Chemother. 1996;37(2):223–232. doi: 10.1093/jac/37.2.223. [DOI] [PubMed] [Google Scholar]
- 60.Randall L.P, Cooles S.W, Osborn M.K, Piddock L.J, Woodward M.J. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004;53(2):208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 61.Dasti J.I, Gross U, Pohl S, Lugert R, Weig M, Schmidt-Ott R. Role of the plasmid-encoded tet(O) gene in tetracycline-resistant clinical isolates of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 2007;56(Pt6):833–837. doi: 10.1099/jmm.0.47103-0. [DOI] [PubMed] [Google Scholar]
- 62.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, Rocha E.P.C, Garcillán-Barcia M.P, de la Cruz F. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 2020;11(1):3602. doi: 10.1038/s41467-020-17278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu K, Wang F, Sun J, Wang Q, Chen Q, Yu S, Rui Y. Class 1 integron gene cassettes in multidrug-resistant gram-negative bacteria in Southern China. Int. J. Antimicrob. Agents. 2012;40(3):264–267. doi: 10.1016/j.ijantimicag.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 64.Moredo F.A, Pineyro P.E, Marquez G.C, Sanz M, Colello R, Etcheverria A, Padola N.L, Quiroga M.A, Perfumo C.J, Galli L, Leotta G.A. Enterotoxigenic Escherichia coli subclinical infection in pigs:Bacteriological and genotypic characterization and antimicrobial resistance profiles. Foodborne Pathog. Dis. 2015;12(8):704–711. doi: 10.1089/fpd.2015.1959. [DOI] [PubMed] [Google Scholar]
- 65.Arief R.A, Darmawan R.D, Sunandar Widyastuti M.D.W, Nugroho E, Jatikusumah A, Putra G.A.A, Basuno E, Kuniawati A, Suwandono A, Willyanto I, Suandy I, Latif H. The use of Antibiotics on Farms Pigs in the Province of Central Java, Indonesia. In:Scientific Proceedings of the 14th, September 2016 22–25. Tangerang, Indonesia. CIVAS, Tangerang. 2016:161–163. [Google Scholar]
- 66.Rizaldi A, Lukman D.W, Pisestyani H. Antibiotic resistance of Escherichia coli in pork sold at Tamiang Layang Market, East Barito District. Adv. Anim. Vet. Sci. 2019;7(9):791–797. [Google Scholar]
- 67.Al-Bahry S, Al-Sharji N, Yaish M, Al-Musharafi S, Mahmoud I. Diversity of tetracycline-resistant genes in Escherichia coli from human and environmental sources. Open Biotechnol. J. 2016;10(Suppl-2, M2):289–300. [Google Scholar]
- 68.Amalo G.F, Purnawarman T, Pisestyani H. Escherichia coli contamination and resistance to antibiotics in se'i meat. JKH. 2021;15(1):27–30. [Google Scholar]
- 69.Casanova L.M, Sobsey M.D. Antibiotic-resistant enteric bacteria in environmental waters. Water. 2016;8(12):561. [Google Scholar]
- 70.Marshall B.M, Levy S.B. Food animals and antibiotics:Impacts on human health. Clin. Microbiol. Rev. 2011;24(4):718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


