Abstract
Coronavirus Disease‐19 (COVID‐19) is an infectious disease caused by severe acute respiratory syndrome‐coronaviruses‐2 (SARS‐CoV‐2), a highly pathogenic and transmissible coronavirus. Most cases of COVID‐19 have mild to moderate symptoms, including cough, fever, myalgias, and headache. On the other hand, this coronavirus can lead to severe complications and death in some cases. Therefore, vaccination is the most effective tool to prevent and eradicate COVID‐19 disease. Also, rapid and effective diagnostic tests are critical in identifying cases of COVID‐19. The COVID‐19 pandemic has a dynamic structure on the agenda and contains up‐to‐date developments. This article has comprehensively discussed the most up‐to‐date pandemic situation since it first appeared. For the first time, not only the structure, replication mechanism, and variants of SARS‐CoV‐2 (Alpha, Beta, Gamma, Omicron, Delta, Epsilon, Kappa, Mu, Eta, Zeta, Theta, lota, Lambda) but also all the details of the pandemic, such as how it came out, how it spread, current cases, what precautions should be taken, prevention strategies, the vaccines produced, the tests developed, and the drugs used are reviewed in every aspect. Herein, the comparison of diagnostic tests for SARS‐CoV‐2 in terms of procedure, accuracy, cost, and time has been presented. The mechanism, safety, efficacy, and effectiveness of COVID‐19 vaccines against SARS‐CoV‐2 variants have been evaluated. Drug studies, therapeutic targets, various immunomodulators, and antiviral molecules applied to patients with COVID‐19 have been reviewed.
Keywords: COVID‐19 infection, diagnosis tests, drug, SARS‐CoV‐2, vaccine type, variants
This paper reviews not only the structure, replication mechanism, and variants of severe acute respiratory syndrome (SARS)‐CoV‐2 but also exposes all the details of the pandemic, such as how it came out, how it spread, what precautions should be taken, and prevention strategies in every aspect. It also compares the molecular and serological assays used for SARS‐CoV‐2 diagnosis and lists the COVID‐19 vaccines approved by the countries.
1. INTRODUCTION
Respiratory viral diseases such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) have posed a threat to humans worldwide. 1 Coronaviruses (CoVs), which cause these diseases, have a size of nearly 65–125 nm in diameter and pertain to the Coronaviridae family. 2 This family, together with the Roniviridae, the Arteriviridae, and the Mesoniviridae, belong to the Nidovirales order. 3 The Coronaviridae family includes a single‐stranded ribonucleic acid (ssRNA) that changes size between 20 and 32 kbs in length. Therefore, they are described as large‐genome Nidoviruses. 4 On the other hand, this family divides into four subgroups that are beta (β), alpha (α), delta (δ), and gamma (γ) CoVs. Only β and α CoVs are recognized to affect humans. 2 The Coronaviridae family is known to be transmitted from animals to humans or zoonotic. 5 These contain SARS‐CoV and MERS‐CoV that were first recognized in 2003 and 2012, respectively. According to the epidemiological data before 2019, there were six CoVs members infecting humans: MERS‐CoV, SARS‐CoV, NL63 (α coronavirus), OC43 (β coronavirus), HKU1 (β coronavirus), and 229E (α coronavirus). 4 , 6 , 7
For an unknown reason, pneumonia was diagnosed in a few patients in Hubei province, China, in 2019. Several laboratories in China carried out sequencing and etiological investigations for a causative agent of pneumonia. As a result of these investigations, the causative agent of pneumonia was verified as a novel coronavirus. According to the sequence analysis, this new virus pertains to β coronavirus, which contains both SARS‐CoV and MERS‐CoV. 1
In this review, the status of COVID‐19 in early 2023 has been summarized. First, how COVID‐19 came out, how it spread, precautions to be taken, and prevention strategies have been mentioned, and current cases have been given. The variants of SARS‐CoV‐2 have been given, and it has been mentioned that the Omicron variant and especially its subvariant XBB 1.5 was the most transmissible form. The structure and replication mechanism of SARS‐CoV‐2 have been detailed. The procedure, accuracy rate, cost, sample sources, detection region, and result time of tests used to detect SARS‐CoV‐2 have been compared. The general mechanism of vaccines, including protein‐based, viral‐vector, whole virus, and nucleic acid vaccines, have been explained. The safety, efficacy, and effectiveness of COVID‐19 vaccines against SARS‐CoV‐2 variants have been reviewed, and approved COVID‐19 vaccines have been listed. Various immunomodulators and antiviral molecules applied to patients with COVID‐19 have been evaluated. Viral polymerase, the main protease (Mpro), and the papain‐like protease (PLpro) have been discussed as therapeutic targets. Drug resistance and convalescent plasma and monoclonal antibodies (mAbs) used for COVID‐19 therapies have been detailed.
2. CURRENT CASES OF COVID‐19
World Health Organization (WHO) was informed about the cases of pneumonia for unknown reasons in Wuhan City of China on December 31, 2019. 8 On February 4, 2021, 103,989,900 cases and 2,260,259 deaths were reported worldwide. As of January 21, 2023, 663,640,386 total cases and 6,713,093 total deaths have been reported worldwide. These data showed that the cumulative number of cases had increased more than six times and deaths had increased almost three times in the last 2 years. The current number of cases, deaths, and persons fully vaccinated with the last dose of primary series per 100 population based on continent and country is given in Table 1. 9
TABLE 1.
The current numbers based on the continent and country (as of January 21, 2023).
| Cases a | Deaths a | Vaccinated persons (%) b | |
|---|---|---|---|
| Country | |||
| United States of America | 100,304,472 | 1,088,854 | 68.42 |
| India | 44,681,650 | 530,728 | 68.95 |
| France | 38,372,611 | 160,051 | 78.93 |
| Germany | 37,659,518 | 164,585 | 76.4 |
| Brazil | 36,677,844 | 695,615 | 79.52 |
| Japan | 31,819,310 | 64,220 | 81.55 |
| Republic of Korea | 29,955,366 | 33,134 | 86.7 |
| Italy | 25,363,742 | 185,993 | 82.96 |
| The United Kingdom | 24,259,240 | 203,229 | 74.59 |
| Russian Federation | 21,882,414 | 394,610 | 54.09 |
| Türkiye | 17,004,677 | 101,419 | 60.93 |
| Spain | 13,711,251 | 117,759 | 79.17 |
| Viet Nam | 11,526,284 | 43,186 | 88.2 |
| Australia | 11,269,081 | 16,989 | 84,92 |
| China | 11,001,439 | 34,375 | 86.82 |
| Argentina | 10,024,095 | 130,338 | 83.72 |
| The Netherlands | 8,578,818 | 22,992 | 68.69 |
| Iran | 7,562,998 | 144,728 | 69.68 |
| Mexico | 7,314,891 | 331,595 | 63.48 |
| Indonesia | 6,727,609 | 160,772 | 63.14 |
| Poland | 6,374,976 | 118,678 | 59.72 |
| Colombia | 6,354,791 | 142,385 | 72.56 |
| Austria | 5,749,735 | 21,595 | 75.04 |
| Greece | 5,683,897 | 35,392 | 72.22 |
| Portugal | 5,562,214 | 25,942 | 86.33 |
| Ukraine | 5,367,032 | 110,978 | 34.65 |
| Chile | 5,096,886 | 63,599 | 92.57 |
| Malaysia | 5,033,943 | 36,923 | 85.07 |
| Continent | |||
| Europe | 271,197,645 | 2,174,093 | 64.49 |
| Americas | 187,862,206 | 2,902,348 | 70.79 |
| Western Pacific | 111,125,686 | 308,060 | 84.91 |
| South‐East Asia | 60,751,075 | 803,537 | 68.11 |
| Eastern Mediterranean | 23,236,089 | 349,221 | 48.24 |
| Africa | 9,466,921 | 175,177 | 27.85 |
| GLOBAL | 663,640,386 | 6,713,093 | 64.61 |
Cumulative total.
Persons fully vaccinated with the last dose of primary series per 100 population.
3. VARIANTS OF SARS‐CoV‐2
All viruses, including SARS‐CoV‐2, naturally change or mutate over time. Most of these changes have minimal impact on the virus's properties, while some could affect the virus's characteristics. As a result of mutations, change in the spreading rate or the severity of the associated disease could be observed. Also, the efficacy of therapeutic drugs and vaccines could change. 10 Variants of concern (VOC) and variants of interest (VOI) are two main classifications used to assess the virological and clinical relevance of different variants. 11 As of May 15, 2022, SARS‐CoV‐2 variants that meet the definition of a VOC are the Alpha variant (Lineage B.1.1.7), Beta variant (Lineage B.1.351), Gamma variant (Lineage P1), Omicron variant (Lineage B.1.1.529), and Delta variant (Lineage B.1.617.2). The variants that meet the definition of a VOI are the Epsilon variant (Lineage B.1.427 and B.1.429), Kappa variant (B.1.617.1), Mu variant (Lineage B.1.621), Eta variant (B.1.525), Zeta variant (Lineage P.2), Theta variant (P.3), lota variant (B.1.526), and Lambda variant (Lineage C.37). 12 Currently, circulating VOC are only the Omicron variant, while the Beta, Alpha, Gamma, and Delta variants are previously circulating variants. Also, there is no currently circulating variant of interest. The Omicron variant's transmission, reported on November 11, 2021, is hugely higher than previously identified SARS‐CoV‐2 variants. 11 This is due to the greater ability of this variant to evade the immune system rather than higher viral loads. 13 As of October 12, 2022, Omicron subvariants under monitoring were BA.5, BA.2.75, BJ.1, BA.4.6, XBB, and BA.2.3.20. On the other hand, as of January 13, 2023, Omicron subvariants under monitoring are BF.7, BQ.1, BA.2.75, and XBB. XBB, which is a recombinant of BJ.1 and BA.2.75, is currently being seen across many countries. Especially its subvariant XBB 1.5 is thought to be the most transmissible variant to date. 12 This variant is a less severe disease (risk of hospitalization and death), compared with the others owing to less lower respiratory tract replication. 14 A study reported that the Alpha variant carries a 48% higher risk of serious disease than wild‐type variants. On the other hand, it was stated that the Beta variant has a 24% higher risk of serious disease, compared to the Alpha variant, and 57% of COVID‐19‐related deaths were caused by this variant. 15
4. STRUCTURE AND REPLICATION MECHANISM OF SARS‐CoV‐2
SARS‐CoV‐2 is a spherical enveloped particle involving ssRNA—positive sense—associated with a nucleoprotein within a capsid that consists of matrix protein. The surface of the envelope is covered by spikes with a glycoprotein structure. Also, some CoVs involve a hemagglutinin‐esterase protein. CoVs guanine+cytosine contents change from 32% to 43%. 16 This virus, which shows the typical β coronavirus organization, has nonstructural proteins (nsp) and structural proteins. The replication enzyme coding region encodes open reading frames (ORFs): ORF1a and ORF1b genes. All CoVs include specific genes in ORF1 downstream regions that encode proteins for nucleocapsid, spikes formation, and viral replication. Sixteen nsp, which are highly conserved throughout the coronavirus and have proceeded from pp1a and pp1b, are encoded by ORF1a/b at the 5′ end, while envelope protein (E), spike protein (S), nucleocapsid protein (N), and membrane protein (M) are encoded by ORFs at the 3′ end 16 , 17 , 18 , 19 , 20 as demonstrated in Figure 1A,B. The envelope involves three proteins: M, E, and S. The M protein binds the nucleocapsid (nucleoprotein core) to host membranes and increases budding and viral assembly. The E proteins play multiple roles during infection, including pathogenesis, release, and viral morphogenesis. The S protein is a homotrimer on the virus surface that recognize the cell receptor and is essential for infectivity. 1
FIGURE 1.
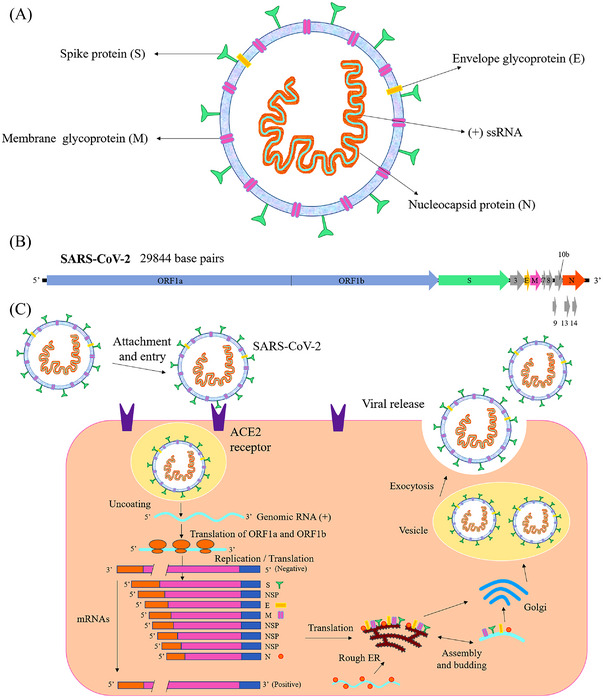
Schematic of the SARS‐CoV‐2 structure (A) and genome (B), the life cycle of SARS‐CoV‐2 in the host cell (C). Reproduced with permission from Shereen et al. 2 (2020), Copyright 2020 Elsevier. ACE2, angiotensin‐converting enzyme 2; ER, endoplasmic reticulum; mRNA, messenger RNA; ORF, open reading frame; RNA, ribonucleic acid; SARS‐CoV‐2, severe acute respiratory syndrome‐coronaviruses‐2; ssRNA, single‐stranded RNA.
MERS‐coronavirus utilizes dipeptidyl peptidase 4 as a key receptor, while SARS‐coronavirus needs the angiotensin‐converting enzyme 2 (ACE2) to enter the host cell. 21 The S protein found on the surface of the SARS‐CoV‐2 virus involves a receptor‐binding domain (RBD), similar to SARS‐CoV, which binds to the ACE2 receptor on the host cell for penetrating virus 2 , 22 as demonstrated in Figure 1C. After that, the virus reveals its RNA, translates its RNA replicase, and then constructs the RNA replicase–transcriptase complex. This complex creates RNA‐negative strands, which will later be translated into relevant viral proteins via replication and transcription. 19 Afterward, the RNA and structural proteins are assembled into virions in the rough endoplasmic reticulum and Golgi, then transported by vesicles and finally released outside the infected cells through exocytosis in an attempt to infect other cells. 2 As a result, one infected cell can release thousands of new viral particles. These particles spread to the bronchi, ultimately achieving the alveoli and extrapulmonary organs, and may cause pneumonia. 19
5. PROTECTION STRATEGIES AGAINST COVID‐19
The most common symptoms of COVID‐19 are dry cough, fever, tiredness, and loss of smell and taste. The less common symptoms are diarrhea, sore throat, headache, pains and aches, irritated or red eyes, and a skin rash or discoloration of toes or fingers. If people have any of these symptoms or have tested positive, they are advised to isolate, take care of themselves, and protect others. In addition, they must stay in touch with their doctor and seek emergency medical care if needed. After isolation period, they can return to their routine. 23
According to WHO, the management of COVID‐19 has focused chiefly on the prevention of infection, detection and monitoring of cases, and supportive care. 24 WHO listed simple protective preventions against the COVID‐19 as follows: (i) washing hands with water and soap for at least 40 s, (ii) using disinfectant or cologne including alcohol, (iii) stop rubbing your mouth, nose, and eyes, (iv) keeping the distance at least 1 m with the other person, and (v) using the mask. The person not in a particular risk group can wear a fabric mask, while the risk group should wear a medical/surgical mask. The mask's utilization and disposal after the first usage are crucial. 23 Mild and moderate COVID‐19 patients and asymptomatic cases should isolate themselves in a well‐ventilated room. If the caregiver enters the room, the patient and the caregiver may consider using an N95 mask. The patients should drink fluids to maintain adequate hydration and rest. 25
SARS‐CoV‐2 infects various cells in the human body including intestinal cells, lung cells, vascular cells, olfactory epithelial cells, and so forth. The damage caused by the infections of these cells, as well as enduring immune response, lead to long COVID. 26 Indeed, the crucial and essential protection strategy against the COVID‐19 is developing effective and rapid diagnostic tests and vaccine studies. Several molecular and serological assays have been designed to determine the presence of SARS‐CoV‐2. 27 Besides diagnostic tests, numerous vaccine and drug development studies have been carried out, and some have even been approved for use. Different strategies, including immunotherapy, cellular therapy, and antiviral therapy, have been applied to treat COVID‐19. Various bioinformatics strategies have also been utilized in this process. 28
6. DIAGNOSTIC TESTS FOR COVID‐19 DETECTION
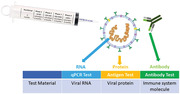
Early diagnosis is a critical step that allows timely intervention for patients suffering from COVID‐19. More complicated diagnostic tests have become significant in revealing the presence of the SARS‐CoV‐2 virus. Nowadays, commercially produced COVID‐19 tests are classified into two main groups. The first group involves molecular assay techniques based on the polymerase chain reaction (PCR) or strategies related to nucleic acid hybridization to detect SARS‐CoV‐2 viral RNA. 29 These assays, which use the virus's genetic material, are far more sensitive than other available assays and allow to identify the virus much earlier in clinical samples. 30
On the other hand, false‐negative results may occur because of improper collection and handling of the sample or improper extraction of nucleic acid from the sample. Also, false‐negative results are possible due to insufficient amount of virus molecules, cross‐contamination, or inhibitors of amplification. 27 Due to their high accuracy rate and sensitivity (≈ 100%) in the early stages (between 2 and 3 days to 20 days after exposure), PCR‐based methods are used primarily in hospitals (Figure 2). Moreover, these tests cannot be used at home due to the need for special tools, equipment, laboratories, and specialist personnel. 27 , 31 , 32
FIGURE 2.
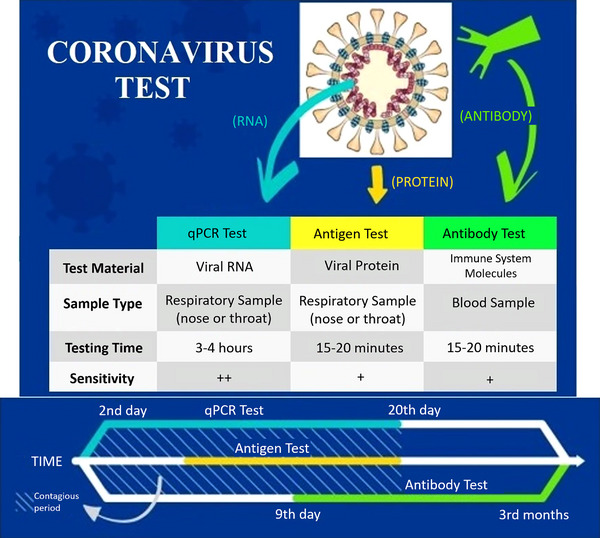
The comparison of COVID‐19 tests. PCR, polymerase chain reaction; RNA, ribonucleic acid.
The second group involves serological and immunological analyses based on the identification of either antibodies generated by the immune system of infected people or the detection of the antigenic protein 29 (Figure 2). Both past and current SARS‐CoV‐2 infections can be detected using serological methods. In addition, the progress of the disease periods and immune response could be monitored using these assays. Serological methods are cheaper and take less time to diagnose, and the steps needed to perform the tests are less complicated. 30 However, there are also some disadvantages: They cannot be used in the early stages of the infection (up to the sixth day) and do not provide information about the viral load. They can make the diagnosis in the later stages of the disease (from the ninth day to the third month). 33 In addition, the reliability rate of serological kits is limited (89%–99%) due to false‐positive results in patients infected with other human coronavirus types such as HCoV‐OC43 and HCoV‐229E because of cross‐reactivity and false‐negative results in immunocompromised individuals including congenital patients, chronic patients, cancer patients, and elderly patients. 34 That is why they are not preferred as the primary test in hospitals but allow individuals to test themselves based on the presence–absence of the virus at home.
6.1. Molecular assays for diagnosis of SARS‐CoV‐2
In recent years, progress in molecular technology has made these methods one of the most widely used approaches for laboratory diagnosis. It has allowed a better understanding of the epidemiology and pathogenesis of infectious diseases. The virus can be identified and diagnosed in molecular assays by evaluating the genome. Changes in the expression of those genes can be detected over time, especially after the infection. There are three main issues in molecular diagnosis: (i) a high capacity to test large numbers of samples accurately and quickly, (ii) decreasing the number of false‐negative results through detecting small amounts of viral RNA, (iii) preventing false‐positive results via the correct differentiation of positive signals between different pathogens. 35 Molecular assays such as reverse transcription‐PCR (RT‐PCR), RT loop‐mediated isothermal amplification (RT‐LAMP), clustered regularly interspaced short palindromic repeats (CRISPR), amplicon‐based metagenomic sequencing, and nucleic acid hybridization using microarray provide the accurate diagnosis for SARS‐CoV‐2. 36
6.1.1. Detection with RT‐PCR
RT‐PCR is based on its potential to amplify minimal amounts of the virus's genetic material in a sample and is regarded as a gold standard for identifying the SARS‐CoV‐2 virus. In this method, it is possible to analyze the samples of potential SARS‐CoV‐2 patients picked up from the feces, blood, and the lower or upper respiratory tract. Swabs have been usually used to obtain specimens from the upper respiratory system in the RT‐PCR COVID‐19 test. 29 Generally, two sequence regions (N and ORF1b) are selected from Genbank for primer and probe designs that enable in vitro identification of new coronavirus infection with RT‐PCR. The N gene‐based assay is about 10 times more sensitive than the ORF1b gene‐based assay to detect the positive sample. The actual PCR methods have high specificity; however, it is possible to obtain false‐positive results. 1 Therefore, this system used as a molecular diagnostic test by clinical laboratories needs improvement. The WHO recommended that at least two distinct targets on the SARS‐CoV‐2 virus genome be detected. 37
RT‐PCR assay begins with the extraction and conversion of viral RNA into deoxyribonucleic acid (DNA) using RNA‐dependent DNA polymerase, also called reverse transcriptase. RT‐PCR reaction is based on the primers, specifically designed for complementary sequences on the virus's genetic material, and reverse transcriptase enzyme in addition to deoxynucleotide triphosphates and cofactor to produce complementary DNA (cDNA) of the viral RNA as shown in Figure S1. In real‐time RT‐PCR, DNA amplification can be monitored using a sequence‐specific probe labeled with a fluorescent dye or a fluorescent molecule. In the automated system, the amplification step is repeated for approximately 40 cycles until the viral cDNA is detected. 29 Table 2 includes some of the market's RT‐PCR diagnostic tests used to detect SARS‐CoV‐2.
TABLE 2.
Molecular and serological assays used for SARS‐CoV‐2 diagnosis.
| Test Name | Manufacturer name | Sample source | Detected region | Total result time | Ref. |
|---|---|---|---|---|---|
| Real‐time reverse transcription‐polymerase chain reaction | |||||
| CDC 2019‐Novel Coronavirus Real‐Time RT‐PCR Diagnostic Panel | CDC | Sputum, nasal aspirate, NS, OS | N gene | – | 67 |
| Simplexa COVID‐19 Direct Kit | DiaSorin Molecular | NS | ORF1ab and S gene | ~ 1 h | 68 |
| COVID‐19 RT‐PCR Test | Laboratory Corporation of America | Sputum, nasal aspirate, NS, OS | N gene | 48–96 h | 69 |
| ARIES SARS‐CoV‐2 Assay | Luminex Corporation | NS | ORF1ab and N gene | ~ 2 h | 70 |
| Novel Coronavirus (2019‐nCoV) Nucleic Acid Diagnostic Kit | Sansure Biotech Inc. | NS, OS, alveolar lavage fluid, sputum, whole blood, feces | ORF1ab and N genes | 30 min | 71 |
| COVID‐19 Coronavirus Real Time PCR Kit | Eagle Biosciences | NS, OS, anterior nasal swabs, mid‐turbinate nasal swabs, nasal aspirates, nasal washes, BAL fluid, sputum | ORF1ab and N genes | 72 min | 72 |
| COVID‐19 RT‐PCR PNA Kit | BioTNS | NS, OS, anterior nasal swabs, mid‐turbinate nasal swabs, BAL, nasopharyngeal wash/aspirates, nasal aspirates specimens | N and RdRP genes | 140 min | 73 |
| STANDARD M nCoV Real‐Time Detection Kit | SD Biosensor Inc. | NS, OS, nasal mid‐turbinate, anterior nares specimen, sputum | N and RdRP genes | 90 min | 74 |
| Bio‐Speedy Direct RT‐qPCR SARS‐CoV‐2 | Bioeksen R&D Technologies Inc. | NS, OS, BAL, nasopharyngeal aspirate, sputum, saliva, gargle | ORF1ab | 65 min | 75 |
| RT‐LAMP | |||||
| ID NOW COVID‐19 | Abbot Diagnostics Scarborough, Inc. | Nasal, throat, NS | RdRp gene | 5–13 min | 76 |
| Isopollo COVID‐19 Detection Kit | M Monitor Inc. | NS, OS, sputum, BAL | N and RdRP genes | 30 min | 77 |
| Loopamp™ SARS‐CoV‐2 Detection Kit | Eiken Chemical Co. Ltd. | NS, OS, saliva | N and RdRP genes | 60 min | 78 |
| MobileDetect Bio BCC19 Test Kit | Detectachem Inc. | Nasal swab, NS, OS | E and N genes | 30 min | 79 |
| Detect COVID‐19 Test | Detect Inc. | Anterior nasal swabs | ORF1ab | 1 h | 80 |
| Lucira™ COVID‐19 All‐In‐One Test Kit | Lucira Health | Nasal swab | N gene | 30 min | 81 |
| Clustered regularly interspaced short palindromic repeats (CRISPR) | |||||
| SARS‐CoV‐2 DETECTR | Mammoth Biosciences | Respiratory samples | E and N genes | 30–40 min | 82 |
| Sherlock CRISPR SARS‐CoV‐2 Kit | Sherlock Biosciences | Anterior nasal swabs, NS, OS, nasopharyngeal wash/aspirate, nasal aspirate, BAL | ORF1ab, N gene | ~ 1 h | 83 |
| TATA MD CHECK CRISPR SARS‐COV‐2 KIT 1.0 | Tata Medical and Diagnostics | NS, OS | S gene | 1.67 h | 84 |
| Amplicon‐based metagenomic sequencing | |||||
| ISARIC 4C (scientific work) | ISARIC 4C Consortium | NS | ORF1ab | 8 h | 43 |
| Nucleic acid hybridization using microarray + RT‐PCR | |||||
| DetectX‐Rv | PathogenDx, Inc. | NS, OS, mid‐turbinate nasal swabs, anterior nasal swabs, nasal aspirates, nasopharyngeal wash/aspirates, BAL | N1, N2 genes | – | 85 |
| CovidArray | Italy (scientific work) | NS | N1, N2 genes | 125 min | 86 |
| Enzyme‐linked immunosorbent assay (ELISA) | |||||
| SARS‐CoV‐2 IgG ELISA Kit | Creative Diagnostics | Serum, plasma | IgG | – | 87 |
| EDI Novel Coronavirus COVID‐19 IgM ELISA Kit | Epitope Diagnostics, Inc. | Serum, plasma | IgM | 120 min | 88 |
| AMP ELISA TEST SARS‐COV‐2 AB | AMEDA Labordiagnostik GmbH | Serum, plasma | Total Antibody | 70 min | 89 |
| Anti‐SARS‐CoV‐2 QuantiVac ELISA (IgG) | EUROIMMUN AG | Fingerprick blood, serum, plasma | IgG | 120 min | 90 |
| GB SARS‐CoV‐2 Ab ELISA | General Biologicals Corporation | Serum, plasma | Antibody | 120 min | 91 |
| Lateral flow test | |||||
| COVID‐19 IgM/IgG Rapid Test | BioMedomics Inc. | Fingerprick or whole blood, serum, plasma samples | IgG/IgM | 10 min | 51 |
| COVID‐19 Ag Respi‐Strip | Coris Bioconcept | NS | N protein antigen | 15–30 min | 92 |
| CRT COVID‐19 Rapid Test | Oncosem Onkolojik Sistemler San. ve Tic. A.S. | Blood, plasma, serum | IgG/IgM | 10–15 min | 93 |
| Rapid COVID‐19 IgM/IgG Combo Test Kit | Megna Health Inc. | Serum, ACD plasma, fingerstick whole blood | IgG/IgM | 15 min | 94 |
| Pilot™ COVID‐19 At‐Home Test | SD Biosensor Inc. | Anterior nasal swabs | N protein antigen | 20 min | 95 |
| INDICAID COVID‐19 Rapid Antigen At‐Home Test | PHASE Scientific International, Ltd. | Nasal swabs, NS | N protein antigen | 20 min | 96 |
| VivaDiag™ SARS‐CoV‐2 IgM/IgG Rapid Test | VivaChek Biotech Co., Ltd. | Plasma, serum, whole blood | IgG/IgM | 15 min | 97 |
| SCoV‐2 Ag Detect Rapid Self‐Test | InBios International Inc. | Anterior nasal swabs | N protein antigen | 25 min | 98 |
| BinaxNOW COVID‐19 Ag Card Home Test | Abbott Diagnostics Scarborough, Inc. | Anterior nasal swabs | N protein antigen | 15 min | 99 |
| iHealth COVID‐19 Antigen Rapid Test Pro | iHealth Labs, Inc. | Anterior nasal swabs | N protein antigen | 15 min | 100 |
| SPERA COVID‐19 Ag Test | Xtrava Health | Anterior nasal swabs | N protein antigen | 15 min | 101 |
| NIDS COVID‐19 Antigen Rapid Test Kit | ANP Technologies, Inc. | Mid‐turbinate nasal swabs | N protein antigen | 15 min | 102 |
Abbreviations: ACD, acid citrate dextrose; BAL, bronchoalveolar lavage; CDC, Centers for Disease Control and Prevention; DETECTR, deoxyribonucleic acid (DNA) endonuclease‐targeted CRISPR trans reporter; IgG, immunoglobulin G; IgM, immunoglobulin M; NS, nasopharyngeal swabs; OS, oropharyngeal swabs; RdRp, ribonucleic acid (RNA)‐dependent RNA polymerase.
6.1.2. RT‐LAMP assay
The LAMP is a nucleic acid amplification method that amplifies DNA under isothermal conditions with high selectivity, specificity, sensitivity, and rapidity, thus eliminating the need for a thermal cycler. 38 The absence of a thermal cycler reduces the diagnostic device's complexity and cost. 39 The amplification efficiency of this method is very high, and due to its isothermal reaction, there is no time loss for thermal change. 38 In order to increase the sensitivity, RT‐LAMP requires a set of 4–6 primers specific for 6–8 different target DNA regions and combines LAMP with the RT step as demonstrated in Figure S2. 40 A strand‐displacing DNA polymerase initiates synthesis. Also, two primers generate loop structures to facilitate subsequent amplification steps. As a byproduct of amplification, magnesium pyrophosphate precipitate forms in the solution, and this precipitate causes turbidity that can be detected by photometry. Also, this reaction could be followed in real time by fluorescence using intercalating dyes and measuring turbidity. Since the RT‐LAMP diagnostic test requires only real‐time heating and visual inspection, the sensitivity and simplicity of this tool make it a hopeful candidate for virus detection. 29 However, this method has some difficulties, such as optimizing reaction conditions and designing sequence‐specific primers. It is possible to overcome these drawbacks using the CRISPR method. 39 Table 2 includes some of the RT‐LAMP assays used to detect SARS‐CoV‐2 in the market.
6.1.3. Assay based on CRISPR
CRISPR, one of the effective gene‐editing methods, performs the recognition and cutting of targets in DNA or RNA sequences using bacterial enzymes such as Cas12 or Cas13. This method is used not only as a gene‐editing tool but also as a molecular diagnostic tool for detecting infectious diseases, including COVID‐19. CRISPR has high sensitivity and high selectivity and offers fast and specific read‐out. It could be run several times on the same sample, reducing the possibility of false‐negative results. DNA endonuclease‐targeted CRISPR trans reporter (DETECTR) technique and the specific high sensitivity enzymatic reporter unlocking (SHERLOCK) technique are two different CRISPR‐based assays used to detect viral RNA. 39
SHERLOCK technique based on CRISPR can ultra‐sensitively detect RNA or DNA, as demonstrated in Figure S3. This method uses Cas13, which can excise reporter RNA sequences in response to activation via the SARS‐CoV‐2‐specific guide RNA. Viral RNA is converted to double‐stranded DNA (dsDNA) by recombinase polymerase amplification (RT‐RPA), and then complementary RNA is generated from the dsDNA template with T7 transcription. The Cas13‐tracrRNA (i.e., trans‐activating CRISPR RNA) complex binds to the target sequence, and thus the nuclease enzyme activity of Cas13 is activated to cleave the target sequence and the fluorescent RNA reporter. The DETECTR technique allows cleavage of the reporter RNA via Cas12a for specific detection of viral RNA sequences—the N and E genes. After that, the isothermal amplification of the target takes place and finally leads to a visual read‐out with a fluorophore. These tests have a low cost and short response time (almost 1 h), which is very important for point‐of‐care (PoC) diagnosis. 29 , 41 Table 2 includes some assays based on the CRISPR used to detect SARS‐CoV‐2.
6.1.4. Detection with amplicon‐based metagenomic sequencing
Amplicon sequencing is a highly targeted technique that enables genetic variation to be analyzed in specific genomic regions. The ultra‐deep sequencing of PCR products (amplicons) provides efficient variant identification and characterization. This technique helps detect complex samples of uncommon somatic mutations. It enables researchers to sequence targets ranging from a few to 100 genes in a single process. 42 On the other hand, the metagenomic sequencing technique is used to tackle the background microbiome of those infected. 43 This approach allows the rapid detection of the SARS‐CoV‐2 virus and other pathogens that lead to secondary infections. In addition, metagenomic methods such as sequence‐independent single primer amplification ensure additional control on sequence divergence. This dual technique can evaluate the mutation rate and identify possible recombination with other human CoVs. These issues are crucial regarding antiviral efficacy and vaccine development. 29 Nanopore sequencing has excellent potential as an emerging technology that allows a single DNA or RNA molecule to be sequenced in real‐time. The MinION, the first commercial sequencer based on the nanopore technology released by Oxford Nanopore Technologies, is a small and powerful sequencing instrument suitable for the genetic analysis of pathogens. 44 Amplicon‐based MinION sequencing has been used to rapidly (∼8 h) sequence the SARS‐CoV‐2 genome and the nasopharyngeal swab microbiome collected from COVID‐19 patients 29 , 43 (Table 2).
6.1.5. Nucleic acid hybridization using microarray
Microarray is a highly efficient diagnostic method used to detect SARS‐CoV‐2 nucleic acids. This method produces cDNAs from viral RNA and reference RNA via RT and then labeled with specific probes. In the next step, viral cDNA and reference cDNA, both of which are with different fluorescent labels, are mixed. These labeled cDNAs are hybridized with solid‐phase oligonucleotides fixed on the microarray by loading into each well as demonstrated in Figure S4. 29 After a series of washing steps to remove the free DNA, coronavirus RNA can be detected by following the specific probes. 45 In the literature, this technology has detected various single nucleotide polymorphisms in the S gene of SARS‐CoV‐2 with 100% accuracy. 46 Table 2 includes some of the microarrays that are used to detect SARS‐CoV‐2.
6.2. Serological assays for diagnosis of SARS‐CoV‐2
Serological assays have been used to test the existence of immunoglobulin G (IgG) and immunoglobulin M (IgM) and antibodies produced by the immune system. They detect the presence of antibodies to the virus in the patient's blood, serum, saliva, plasma, sputum, and other biological fluids instead of detecting viral antigens. 39 , 47 Serological tests play a significant role in the development of vaccines and epidemiology, offering an evaluation of both long‐term (years or permanence) and short‐term (days to weeks) trajectories of antibody response, abundance, and diversity. 29 Following the SARS‐CoV‐2 infection, IgM becomes detectable in serum after a few days, peaks within 2 weeks, and is reduced to near‐background levels. On the other hand, IgG is produced after a week, peaks 3 weeks, and is maintained at a high level for an extended period. The serological tests can be used as a confirmative method in detecting SARS‐CoV‐2 to check the accuracy of the nucleic acid tests. 29 , 48 These tests, such as detection with enzyme‐linked immunosorbent assay (ELISA) and detection with lateral flow test, provide a rapid diagnosis for SARS‐CoV‐2 infections.
6.2.1. Detection with ELISA
ELISA is a plate‐based assay designed to identify and quantify substances such as antibodies, peptides, proteins, and hormones within typically 1−5 h. 29 This assay, which could be quantitative or qualitative, is quite sensitive to IgG from the 10 days following the initial symptoms. Therefore, the ELISA method, targeting the nucleocapsid or S protein of the virus, has been used for SARS‐CoV‐2 specific antibody responses and the identification of infection. 49 , 50 In this method, the plate wells are coated with a viral protein to ensure specific binding of antiviral antibodies in the samples of patients if they exist. Then secondary enzyme‐labeled antibody binds to the patient's antibody. Finally, a colorimetric or fluorescent‐based read‐out can be obtained as a result of enzymatic conversion of the substrate as demonstrated in Figure S5. 29 Table 2 includes some of the ELISA assays used to detect SARS‐CoV‐2 in the market.
6.2.2. Detection with lateral flow test
Lateral flow test is a qualitative chromatographic assay, which is portable, small, and used for PoC testing. This cheap test does not require trained personnel and gives qualitative results. This test is a kind of rapid diagnostic test because of getting results in 10 to 30 min. Lateral flow tests can detect SARS‐CoV‐2 viral antigens, including the S protein or N protein. In this assay, liquid samples such as blood, plasma, and serum are applied to a substrate, allowing the sample to flow through a band of immobilized viral antigens. If they exist, anti‐SARS‐CoV‐2 antibodies are gathered at the band, where, along with co‐collected tracer antibodies, a color develops to demonstrate to test result. A diagnosis of infection could be possible when used in combination with symptomology. 29 Various companies developed and launched rapid PoC lateral immunoassays to diagnose coronavirus infection. One of these tests has been developed by BioMedomics Inc., which can detect early and late markers in serum, plasma, whole venous blood, or fingerprick. The test card includes colloidal gold‐labeled recombinant novel coronavirus antigen, two detection lines—M line and G line—and one quality control line—C line—fixed on a nitrocellulose membrane. When an adequate quantity of the test material is inserted into the sample well, it will move forward along with the test card through capillary action. The antibody will bind to the colloidal gold‐labeled recombinant coronavirus antigen if the sample includes IgM. The antigen/antibody complex will be captured via the antihuman IgM antibody, immobilized on the membrane, generating a red M line that suggests a positive outcome for the IgM antibody. Likewise, the same process will perform with IgG antibodies. A negative result is indicated if no antibodies are present. 51 This workflow is shown in Figure S6. Table 2 includes some lateral flow tests used to detect SARS‐CoV‐2 in the market.
6.3. Comparison of accuracy rate and cost of different detection assays
Assays used in the diagnosis of COVID‐19 have different advantages and disadvantages. RT‐PCR techniques are costly, that is why many countries—especially low‐income countries—cannot afford enough number of tests in order to screen a large population. 52 The assay time can vary between 30–120 min for RT‐PCR. The sensitivity and specificity are high. 53 According to a recent meta‐analysis, the sensitivity of RT‐PCR for the detection of SARS‐CoV‐2 in sputum and NS samples was 97.2% and 73.3%, respectively. 54 Also RT‐LAMP has high sensitivity and specificity. 52 According to a systematic review, the sensitivity of RT‐LAMP was 78% in crude samples and 94% in purified RNA from samples of COVID‐19 patients. Also, RT‐LAMP was susceptible to producing false‐negative results in samples with low viral loads. 55 This assay is rapid and cost‐effective. According to a study result, when all related costs are included, the average per‐test cost for the real‐time RT‐PCR was $14.75 and for RT‐LAMP was $8.45. 56 The sensitivity and specificity of CRISPR‐based assays are high. 53 SHERLOCK for SARS‐CoV‐2 detection has demonstrated 93.1% sensitivity and is significantly faster than qRT‐PCR. 57 It was reported that depending on the assay technique, CRISPR‐COVID assay costs less than $3.50 for a single reaction. 58 In the near future, CRISPR‐based assays have the potential to become less complicated, more reliable, faster, and more affordable. 59 The detection time is 2–8 h for ELISA. The sensitivity is low and specificity is intermediate. 53 Many ELISA kits have been developed during the COVID‐19 pandemic. The sensitivity range can be from 84% to 94% and the specificity can be up to 100%. 60 , 61 , 62 Costs of these tests range from $445 to $785 for 96 tests. 63 Rapid antigen tests are relatively inexpensive and give results within 10–15 min. 64 But the accuracy of rapid antigen tests is lower than standard RT‐PCR tests. 65 According to a meta‐analysis including 17,171 suspected COVID‐19 patients, sensitivity and specificity of rapid antigen test kits were detected as 68.4% and 99.4%, respectively. 66 For the early detection of patients suspected of having COVID‐19, the use of rapid antigen test kits is often advised, especially in remote, densely populated areas with few resources and laboratory equipment.
7. VACCINE STUDIES FOR COVID‐19 DISEASE
7.1. Vaccine studies against viral infections
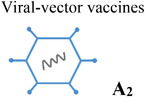
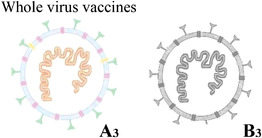
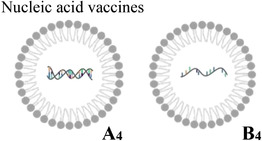
Four major platforms have been investigated for the SARS‐CoV‐2 vaccines: protein‐based, viral‐vector, whole virus, and nucleic acid vaccines. 103 , 104 The contents, advantages, disadvantages, storage conditions, and side effects of these vaccine platforms are summarized in Table 3. 103 , 105 , 106 , 107
TABLE 3.
Comparison of four vaccine types.
| Vaccine platform | Contains | Advantages | Disadvantages | Storage | Side effects |
|---|---|---|---|---|---|
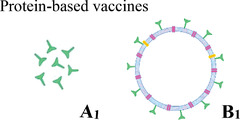
|
Isolated and purified viral proteins (A1: subunit) or viral proteins, which mimic the structure of the virus, but no genetic material (B1: virus‐like particle) | Well‐established technology, no risk of triggering the disease, broad antigenic profile | Complex to manufacture, poorly immunogenic | 2–8°C | Low |
|
Viral genetic material packaged inside another harmless virus, which can copy itself (replicating viral vector) or cannot copy itself (nonreplicating viral vector) | Well‐established technology, strong immune response | Complex to manufacture, pre‐existing immunity against the vector could reduce the effectiveness | 2–8°C | Pain at the injection site, joint pain, headaches, fatigue, chills, muscle pain, fever |
|
Copies of the virus that have been killed (A3: attenuated) or weakened (B3: inactivated) | Well established technology, simple to manufacture, broad antigenic profile | Live attenuated vaccine may trigger disease in very rare cases | 2–8°C | Elevated blood pressure, injection site pain, headache, rash, dizziness |
|
Viral genetic material (A4: deoxyribonucleic acid [DNA]‐based or B4: ribonucleic acid [RNA]‐based) that provides the instructions for making viral proteins | Strong immune response, easy production, no risk of the vaccine triggering disease | Need ultra‐cold storage and special delivery systems | −70°C | Tiredness, headache, muscle pain, chills, fever, nausea |
7.1.1. Protein‐based vaccines
Protein‐based vaccines could be divided into two groups that are subunit vaccines and virus‐like particle (VLP) vaccines. 103 Protein subunit vaccines, which are produced through recombinant protein strategies, are generated easily. Also, they are relatively well‐tolerated and safe, compared to whole virus vaccines. Adjuvants are generally used with these vaccines because of their low immunogenicity. Moreover, the adjuvant could be optimized, and booster doses could be increased to provide a more robust and consistent level of immunity. 103 , 108 The chances of unfavorable reactions to the vaccine are lower because subunit vaccines comprise only the essential part of the antigens and not the whole virus. 109 Thus, concerns such as virulence recovery, pre‐existing antivector immunity, and incomplete viral inactivation could be eliminated. 110 They may include variable antigens ranging from 1 to 20 antigens (Table 3, A1). The subunit vaccines have been produced for hepatitis B and Haemophilus influenza A and B. 111
On the other hand, VLP vaccines use empty virus shells, which mimic the structure of the virus (Table 3, B1), and induce a robust immune response against the antigen presented on its surface. These vaccines are not infectious because they lack genetic material. 103 , 108 VLPs mimic virions morphologically and immunologically, so they could induce high titers of neutralizing antibodies (nAbs) to conformational epitopes. They can induce an immune response as if the immune system has been exposed to an actual virus. 109 VLPs, which fall in the size range of viruses (generally 22–200 nm), are unable to replicate in the recipient; however, they can stimulate the immune system via recognition of repetitive subunits, generating high humoral and cellular immune responses. 112 , 113
Moreover, their repetitive, high‐density display of epitopes is potentially highly effective in enhanced immunogenicity responses. 112 VLP vaccines can be made by recombinant expression and allow for the incorporation of ligands, targeting moieties, and immunomodulators. 114 They are excellent prophylactics and eliminate virus‐based vaccine limitations. 109 VLPs offer a promising approach to produce vaccines against many diseases, including Ebola, influenza A, Zika, and respiratory syncytial virus infections. 115
7.1.2. Viral‐vector vaccines
Viral‐vector vaccines, whose concept is different from that of subunit vaccines, could combine numerous properties of DNA vaccines with live attenuated vaccines. 116 , 117 The main advantage of this vaccine is that the vector alone can achieve the goal of eliciting a protective immune response, so it may not need an adjuvant. 118 The viral vector acts as a delivery system that invades the cell, inserts the code for various viral antigens, and finally transduces cells to create the desired immunized result. 119 The virus used as a vector, including the adenovirus, vaccinia virus, and measles virus, is chemically weakened. That is why it cannot cause the disease. 103 The efficacy and safety of viral‐vector vaccines need to be evaluated using different assays involving genetic stability, immunogenicity, genotoxicity, replication deficiency, or attenuation. As infectious diseases are one of the significant problems in developing countries, it is crucial to enable large‐scale manufacturing of viral vectors. 116 Viral‐vector vaccines are divided into two groups that are replicative and nonreplicative vector‐based vaccines where key genes have been disabled. These vaccines are produced by integrating exogenous protective antigen‐encoding genes into the viral genome whose deleterious genes have been removed (Table 3, A2). 10 , 103 These vaccines appear to be safe and provoke a robust immune response.
On the other hand, if a person has been exposed to the viral vector and has an improved immune response, this situation could blunt the vaccine's effectiveness. 103 , 120 Although different types of viral vectors have been developed, only one vaccine had been approved in humans against Ebola virus disease, a fatal illness, before the onset of SARS‐CoV‐2. 103 , 121 The Ebola vaccine uses the vesicular stomatitis virus as a replicative viral vector, which has little or no effect on humans. 103 , 122
7.1.3. Whole virus vaccines
Whole virus vaccines use an attenuated or inactivated form of the virus to induce protective immunity. 103 An essential advantage of these vaccines is their inherent immunogenicity and ability to stimulate toll‐like receptors (TLRs), including TLR3, TLR7/8, and TLR9. 123 Live attenuated vaccines utilize a weakened form of the virus that can still replicate and grow; however, they are almost or entirely devoid of pathogenicity (Table 3, A3). They do not cause disease but can induce a protective immune response against various virus antigens. The other advantage of these vaccines is the possibility for scale‐up for mass production. 103 , 110 , 111 These vaccines are like natural infections and can have a long‐lasting effect. Most live vaccines only require one or two doses to achieve the desired immune response. However, live vaccines require extended safety and extra caution for people with long‐term health problems, weakened immune systems, or organ transplants.
Moreover, they should be well preserved in cold storage. 109 Another disadvantage of these vaccines is that secondary mutations may cause the reversion of virulence and finally lead to disease. 111 Live attenuated vaccines against different viruses, including poliovirus, influenza, measles, rotavirus, and yellow fever virus, have been commercialized. 110
Inactivated vaccines comprise viruses whose genetic material has been destroyed using chemicals such as formaldehyde or β‐propiolactane and/or physical methods such as heat or radiation (Table 3, B3). Therefore, they cannot infect the cells and replicate; however, they can trigger an immune response. 103 , 111 Inactivated vaccines generally do not provide immunity as strong as live vaccines, so multiple doses are required. On the other hand, inactivated vaccines are safer and more stable than live vaccines because dead viruses cannot mutate back to their pathogenic state. In addition, they are easy to store and transport because they do not require cold storage. 109 Also, they do not involve genetic manipulation and have low production costs. 124 Many approved vaccines are inactivated, including influenza, polio, cholera, typhoid, pertussis, and plague. 110
7.1.4. Nucleic acid vaccines
Nucleic acid vaccines utilize genetic instructions, in the form of DNA or RNA (ribonucleic acid), as depicted in Table 3 A4, for a protein that prompts an immune response. 103 DNA or RNA vaccines indicate CD8+ cytotoxic T‐cell responses that play an essential role in eradicating the virus, in addition to antibody and CD4+ T‐cell responses. 114 DNA vaccines, which have emerged as a promising alternative to traditional protein‐based vaccines, include selected gene/genes of the virus in the form of DNA. 109 , 110 They are based on a recombinant eukaryotic expression vector that encodes a particular protein antigen and is directly injected into the organism. Thus, the foreign gene is expressed in vivo, and the antigen induces the immune system.
Furthermore finally, specific cellular and humoral immune responses are stimulated. Adjuvants can also enhance the prophylactic and treatment effects. 10 These vaccines could not cause disease because they contain only copies of a few virus genes. DNA vaccines offer many advantages, including safety, increased stability, scalability for mass production, rapid and inexpensive production, long‐term persistence of immunogens, and flexibility to generate vaccines for various infectious diseases. 10 , 109 , 110 DNA vaccines can be produced within weeks as no culture or fermentation is required. 124
On the other hand, these vaccines have some disadvantages. DNA injected into the body is rapidly degraded and may risk autoimmunity. 10 In addition, there is limited positive clinical data on DNA vaccines in the literature. Unfortunately, no DNA vaccine has been licensed. 110
RNA vaccines include selected virus genes in the form of mRNA (messenger RNA) as depicted in Table 3, B4, and, after cytosolic delivery, these genes are translated into viral proteins. 110 DNA vaccines need to get into the nucleus, whereas mRNA vaccines only require to get into the cytoplasm to express the target antigen. Therefore, mRNA vaccines are safer than DNA vaccines theoretically. On the other hand, DNA vaccines are considerably more temparature‐stable compared to mRNA vaccines. The immunogenicity, stability, and half‐life of mRNA could be modulated by established modifications. 10 , 114 mRNA vaccines can be produced quickly, making the process comparatively easy to monitor. 10 The RNA could be encapsulated within nanoparticles, injected by itself, or driven into cells through some techniques. When RNA is inside the cell and begins generating antigens, they are displayed on its surface, and the immune response is triggered. 103 The prophylactic mRNA vaccines could be divided into two main groups: nonreplicating and self‐amplifying RNA vaccines. The nonreplicating mRNA vaccines, also called conventional mRNA vaccines, contain 5′ untranslated region (5′UTR) and 3′UTR that flank the antigenic or immunomodulatory sequence. 10 , 125 A poly(A) tail could be added enzymatically after in vitro transcription or incorporated from the 3′ end of the plasmid DNA (pDNA) template.
On the other hand, self‐amplifying RNA vaccine pDNA templates include additional alphavirus replicon genes and conserved sequence elements. Conventional mRNA and synthetic self‐amplifying RNA vaccines are produced in the same way. An mRNA expression plasmid encoding a DNA‐dependent RNA polymerase promoter and the RNA vaccine candidate is designed as a template for in vitro transcription, which is cell‐free. Compared to conventional mRNA, self‐amplifying RNAs have enhanced antigen expression at lower doses. On the other hand, conventional and self‐amplifying mRNA vaccines have demonstrated a protective effect in preclinical trials against various infectious diseases, including Ebola, influenza, Human Immunodeficiency Virus‐1 (HIV‐1), Rabies, and Respiratory Syncytial Virus (RSV). 125 However, nonreplicating mRNA vaccines have a short RNA sequence and simple structure. Except for the antigen, no additional proteins are required. 10 One of the disadvantages of mRNA vaccines is that they can cause an anaphylactic reaction. 126 Also, the storage and transportation of mRNA vaccines need ultra‐low temperatures. 127
7.2. Steps in vaccine development
The first step in the vaccine development process is identifying the vaccine candidate. This stage is called preclinical development, which determines whether the vaccine is effective and safe. First, antigen and suitable technology are selected, and in vitro and in vivo tests are applied. 128 The second step is the clinical stage, which aims to demonstrate efficacy, immunogenicity, and safety, including phase I, phase II, and phase III vaccine trials. These three phases may overlap, and multiple phase I or phase II trials could be performed. In phase I, the candidate vaccine is evaluated by its application on a limited number of volunteers (usually between 20 and 80) to evaluate its effectiveness, decide the dosage, and classify the side effects. These results are collected by comparing the vaccine with a control or a placebo (e.g., saline), an inactive product. Data on the dosage and the period between vaccinations needed to ensure the optimum response of the immune system are determined herein. Vaccines that pass the first phase are applied to a larger group of volunteers (usually between 100 and 300) in the second phase to determine their safety and immunogenicity. In phase II, which takes at least 2 years, the vaccine's appropriate dosage and administration schedule are investigated in detail. In addition, this phase is well‐controlled and frequently randomized. 128 , 129 , 130 Phase III and phase II designs are similar but in different sizes. 131 Phase III, where 3000–50,000 volunteers are enrolled, is usually performed with the most promising candidate vaccines. This process, which may take 3–5 years, aims to assess the efficacy of the vaccines in a large‐scale population. In addition, concomitant administration with other vaccines is checked herein. In the next step, these vaccines are approved by regulatory agencies, including the European Medicines Agency in the European Union and Food and Drug Administration (FDA) in the United States. After this stage, the vaccine can be produced on a large scale. The vaccines reached phase IV, also defined as pharmacovigilance, which aims to detect adverse events early and enter the pharmaceutical industry. This phase includes strict monitoring of vaccines in order to identify, evaluate, recognize, prevent, and communicate any adverse effects following immunization or any other aspects relevant to the immunization. Summarily, the development of a vaccine takes more than a decade as depicted in Figure 3A since (i) it is necessary to test on tens of thousands of volunteers, (ii) the safety and efficacy of vaccines have to be regulated, not only by suppliers but also by competent authorities, (iii) requirement to determine whether the vaccine's safety is long‐lasting. However, in order to overcome the pandemic in a short time, scientists around the world have managed to develop COVID‐19 vaccines against the SARS‐CoV‐2 virus bypassing all these stages quickly 128 , 130 , 132 as seen in Figure 3B. So, the development of COVID‐19 vaccines include new concepts such as large production capacity, parallel and adaptive development phases, and innovative regulatory processes. It should not be forgotten that the principles of the 3Rs, which are replacement, reduction, and refinement, must be considered in the vaccine development process. 130
FIGURE 3.
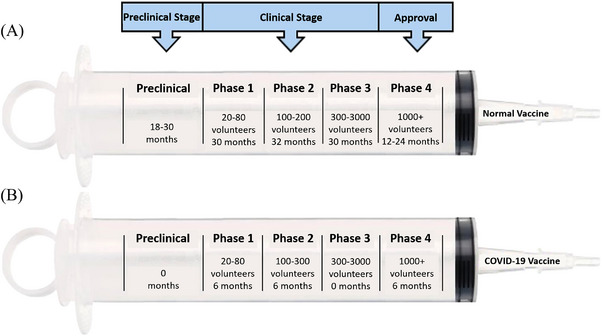
A scheme to compare (A) regular and (B) potential COVID‐19 vaccines.
7.3. COVID‐19 vaccine studies
To control the COVID‐19 pandemic, producing effective vaccines is very important. Vaccines could reduce person‐to‐person transmission, disease severity, and viral shedding. There are many promising targets, including the S protein and RBD subunit for SARS‐CoV‐2. 124 Especially the composition of the S protein, which is a trimeric class I fusion protein, has become significant for COVID‐19 vaccine development because of its essential role in membrane fusion and receptor binding. S protein typically exists in a metastable and prefusion conformation. When the virus interacts with the host cell, the S protein undergoes structural changes, and the viral membrane fuses with the host cell membrane. Characterizing the prefusion S structure at the atomic level guides the design and development of the vaccine for COVID‐19. 110 , 133 Nowadays, there are many different vaccine studies in the production process to protect from COVID‐19. As of January 21, 2023, 168,47 vaccine doses have been administered per 100 population. 9
As of April 8, 2022, WHO has evaluated nine vaccines: Pfizer/BioNTech, Moderna, Johnson and Johnson, AstraZeneca/Oxford, Sinovac, Sinopharm, Covaxin, Covovax, and Nuvaxovid against COVID‐19 that have met the necessary criteria for safety and efficacy. 134 The immunogenicity of these vaccines characterizes their clinical efficacy. However, these vaccines have common adverse events such as erythema, pain, swelling at the injection site, myalgia, chills, and weakness. 135
Pfizer/BioNTech (BNT162b2) vaccine is based on mRNA technology, which is delivered in a lipid nanoparticle to express a full‐length SARS‐CoV‐2 S protein. 136 This vaccine induces both nAbs and cellular immune responses. The administration of this vaccine in two doses 21 days apart gave 95% efficacy in preventing COVID‐19 in subjects. 137 As requires storage at −80 to −60°C, logistic problems may occur. 138 It could be stored for 1 month at 2 to 8°C. 139 Moderna (mRNA‐1273) utilizes the same mRNA technology as Pfizer/BioNTech and uses mRNA delivered in a lipid nanoparticle to express a full‐length SARS‐CoV‐2 S protein. This vaccine has a similarly high efficacy (∼94%) and storage temperature is −25 to −15°C, 136 , 138 , 140 , 141 Johnson and Johnson's vaccine is based on a replication‐incompetent adenovirus 26 vector that encodes a stabilized S protein. 136 This vaccine requires refrigeration (2 to 8°C). 138 Only one dose of this vaccine is 66% effective in preventing moderate to severe COVID‐19 and 100% effective in preventing COVID‐19‐related hospitalization and death. 142 AstraZeneca/Oxford vaccine, which requires refrigeration (2 to 8°C), is based on a replication‐incompetent chimpanzee adenovirus vector that expresses the S protein. 136 , 138 It has 76% efficacy against symptomatic COVID‐19, while 100% efficacy against severe or critical disease in addition to hospitalizations. 143 Sinovac is an inactivated vaccine with an aluminum hydroxide adjuvant. 136 The storage temperature is 2 to 8°C. 138 This vaccine showed 50.7% efficacy in Brazil, 144 and 83.5% efficacy in Turkey. 145 Sinopharm is inactivated, whole‐virus vaccine 136 with 79% efficacy against symptomatic SARS‐CoV‐2 infection 14 or more days after the second dose. 143 Covaxin (BBV152), a whole‐virion inactivated Vero cell‐derived platform, has an aluminum hydroxide and a TLR agonist adjuvant. This vaccine, which is stable at 2–8°C, results in an efficacy of 80.6%. 136 , 146 , 147 Covovax is a recombinant S protein nanoparticle vaccine with 89.3% efficacy. 147 Nuvaxovid (NVX‐CoV2373), another recombinant S protein nanoparticle‐based vaccine, includes the full‐length S protein and a saponin‐based Matrix‐M adjuvant. 148 The efficacy of Nuvaxovid to prevent the onset of COVID‐19 from 7 days after the second dose is reported as 90.4%. 149 Besides these, as of December 2, 2022, 50 vaccines have been approved by at least one country (Table 4). 106
TABLE 4.
COVID‐19 vaccines approved by at least one country. 106
| Vaccine name | Vaccine platform | Manufacturer | Number of countries approving | Number of trials |
|---|---|---|---|---|
| Zifivax (ZF2001) | Protein subunit | Anhui Zhifei Longcom, Beijing, People's Republic of China | 4 countries | 21 trials in 5 countries |
| Noora vaccine | Protein subunit | Bagheiat‐Allah University of Medical Sciences, Tehran, Iran | 1 country | 3 trials in 1 country |
| Covaxin (BBV152) | Inactivated | Bharat Biotech, Hyderabad, India | 14 countries | 16 trials in 2 countries |
| iNVOVACC | Nonreplicating viral vector | Bharat Biotech, Hyderabad, India | 1 country | 4 trials in 1 country |
| Corbevax (BECOV2A) | Protein subunit | Biological E Limited, Hyderabad, India | 2 countries | 7 trials in 1 country |
| Convidecia Air (Ad5‐nCoV‐IH) | Nonreplicating viral vector | CanSino, Tianjin, People's Republic of China | 2 countries | 5 trials in 4 countries |
| Convidecia (Ad5‐nCoV) | Nonreplicating viral vector | CanSino, Tianjin, People's Republic of China | 10 countries | 14 trials in 6 countries |
| Abdala (CIGB‐66) | Protein subunit | Cuban Center for Genetic Engineering and Biotechnology, Havana, Republic of Cuba | 6 countries | 5 trials in 1 country |
| KoviVac | Inactivated | Chumakov Federal Scientific Center for Research and Development of Immune‐and‐Biological Products of Russian Academy of Sciences, Moscow, Russian Federation | 3 countries | 3 trials in 1 country |
| Gam‐COVID‐Vac (Sputnik, rAd5) | Nonreplicating viral vector | Gamaleya National Center of Epidemiology and Microbiology, Moscow, Russian Federation | 1 country | 2 trials in 0 countries |
| Sputnik Light | Nonreplicating viral vector | Gamaleya National Center of Epidemiology and Microbiology, Moscow, Russian Federation | 26 countries | 7 trials in 3 countries |
| Sputnik V (Gam‐COVID‐Vac) | Nonreplicating viral vector | Gamaleya National Center of Epidemiology and Microbiology, Moscow, Russian Federation | 74 countries | 25 trials in 8 countries |
| GEMCOVAC‐19 | RNA | Gennova Biopharmaceuticals Limited, Maharashtra, India | 1 country | 2 trials in 1 country |
| Turkovac (ERUCOV‐VAC) | Inactivated | Health Institutes of Turkey, Istanbul, Turkey | 1 country | 8 trials in 1 country |
| Soberana 02 (FINLAY‐FR‐2, Pastu Covac) | Protein subunit | Instituto Finlay de Vacunas Cuba, Havana, Republic of Cuba | 4 countries | 7 trials in 2 countries |
| Soberana Plus (FINLAY‐FR‐1A) | Protein subunit | Instituto Finlay de Vacunas Cuba, Havana, Republic of Cuba | 2 countries | 5 trials in 1 country |
| Jcovden (Ad26.COV2.S) | Nonreplicating viral vector | Janssen (Johnson & Johnson), Beerse, Belgium | 113 countries | 26 trials in 25 countries |
| V‐01 | Protein subunit | Livzon Mabpharm Inc., Guanghong, China | 1 country | 7 trials in 3 countries |
| Covifenz (CoVLP, MT‐2766, Plant‐based virus‐like particle [VLP]) | VLP | Medicago, Quebec City, Canada | 1 country | 6 trials in 6 countries |
| MVC‐COV1901 | Protein subunit | Medigen, Taipei, Taiwan, People's Republic of China | 4 countries | 15 trials in 4 countries |
| Spikevax (mRNA‐1273, Elasomeran) | RNA | Moderna, Cambridge, MA, USA | 88 countries | 70 trials in 24 countries |
| Spikevax Bivalent Original/Omicron BA.1 | RNA | Moderna, Cambridge, MA, USA | 38 countries | 5 trials in 4 countries |
| Spikevax Bivalent Original/Omicron BA.4/BA.5 | RNA | Moderna, Cambridge, MA, USA | 33 countries | 2 trials in 1 country |
| Recombinant SARS‐CoV‐2 Vaccine (CHO Cell, NVSI‐06‐08) | Protein subunit | National Vaccine and Serum Institute, Beijing, People's Republic of China | 1 country | 3 trials in 2 countries |
| Nuvaxovid (NVX‐CoV2373) | Protein subunit | Novavax, Gaithersburg, MD, USA | 40 countries | 22 trials in 14 countries |
| FAKHRAVAC (MIVAC) | Inactivated | Organization of Defensive Innovation and Research, Tehran, Iran | 1 country | 3 trials in 1 country |
| Vaxzevria (AZD1222, ChAdOx1 nCoV‐19) | Nonreplicating viral vector | Oxford University/AstraZeneca, Södertälje, Sweden | 149 countries | 73 trials in 34 countries |
| Comirnaty (BNT162b2, Tozinameran) | RNA | Pfizer/Biontech, Mainz, Germany | 149 countries | 100 trials in 31 countries |
| Comirnaty Bivalent Original/Omicron BA.1 | RNA | Pfizer/Biontech, Mainz, Germany | 35 countries | 3 trials in 5 countries |
| Comirnaty Bivalent Original/Omicron BA.4/BA.5 | RNA | Pfizer/Biontech, Mainz, Germany | 33 countries | 4 trials in 1 country |
| IndoVac | Protein subunit | PT Bio Farma, Jawa Barat Indonesia | 1 country | 4 trials in 1 country |
| Razi Cov Pars | Protein subunit | Razi Vaccine and Serum Research Institute, Karaj, Iran | 1 country | 5 trials in 1 country |
| QazVac (QazCovid‐in) | Inactivated | Research Institute for Biological Safety Problems, Guardeyskiy, Republic of Kazakhstan | 2 countries | 3 trials in 1 country |
| VidPrevtyn Beta | Protein subunit | Sanofi/GSK | 30 countries | 3 trials in 2 countries |
| Covishield (Oxford/ AstraZeneca formulation) | Nonreplicating viral vector | Serum Institute of India, Pune, India (based on AstraZeneca technology) | 49 countries | 6 trials in 1 country |
| COVOVAX (Novavax formulation) | Protein subunit | Serum Institute of India, Pune, India (based on Novavax technology) | 6 countries | 7 trials in 3 countries |
| KCONVAC (SARS‐CoV‐2 Vaccine (Vero Cells), KconecaVac | Inactivated | Shenzhen Kangtai Biological Products Co., Shenzhen, China | 2 countries | 7 trials in 1 country |
| COVIran Barekat (COVID‐19 Inactivated Vaccine) | Inactivated | Shifa Pharmed Industrial Co, Karaj, Iran | 1 country | 6 trials in 1 country |
| Covilo (BBIBP‐CorV (Vero Cells)) | Inactivated | Sinopharm (Beijing), People's Republic of China | 93 countries | 39 trials in 18 countries |
| Inactivated (Vero Cells) | Inactivated | Sinopharm (Wuhan), People's Republic of China | 2 countries | 9 trials in 7 countries |
| CoronaVac | Inactivated | Sinovac, Beijing, People's Republic of China | 56 countries | 42 trials in 10 countries |
| SKYCovione | Protein subunit | SK Bioscience Co. Ltd., Republic of Korea | 1 country | 7 trials in 6 countries |
| TAK‐019 (NovaVax formulation) | Protein subunit | Takeda, Tokyo, Japan (based on Moderna technology) | 1 country | 3 trials in 1 country |
| TAK‐919 (Moderna formulation) | RNA | Takeda, Tokyo, Japan (based on Moderna technology) | 1 country | 2 trials in 1 country |
| VLA2001 | Inactivated | Valneva, Saint‐Herblain, France | 33 countries | 9 trials in 4 countries |
| SpikoGen (COVAX‐19) | Protein subunit | Vaxine/CinnaGen Co., Tehran, Iran | 1 country | 8 trials in 2 countries |
| Aurora‐CoV (EpiVacCorona‐N) | Protein subunit | “Vector,” National Research Center for Virology and Biotechnology, Novosibirsk, Russian Federation | 1 country | 2 trials in 1 country |
| EpiVacCorona | Protein subunit | “Vector,” National Research Center for Virology and Biotechnology, Novosibirsk, Russian Federation | 4 countries | 4 trials in 1 country |
| AWcorna | RNA | Walvax, Yunnan, China | 1 country | 4 trials in 3 countries |
| ZyCoV‐D | DNA | Zydus Cadila, Ahmedabad, India | 1 country | 6 trials in 1 country |
7.3.1. Efficacy and effectiveness of vaccines against SARS‐CoV‐2 variants
According to a trial of Novavax (NVX‐CoV2373) vaccine, a post hoc analysis demonstrated that it has an efficacy of 86.3% against Alpha variant (Lineage B.1.1.7). 150 When Alpha variant (Lineage B.1.1.7) was dominant in Israel, Pfizer‐BioNTech (BNT162b2) vaccine demonstrated 92% protection against asymptomatic infection, 97% against severe disease, and 97% against symptomatic. Pfizer‐BioNTech (BNT162b2) vaccine and AstraZeneca vaccine showed 85% and 94% reduction in hospitalizations, respectively, against Alpha variant (Lineage B.1.1.7) in Scotland. 151 The effectiveness of the Pfizer‐BioNTech (BNT162b2) vaccine against the severe disease was 97.4%, while against any documented infection with the Alpha variant (Lineage B.1.1.7) was 89.5% in Qatar. 152 Vaxzevria (AZD1222) was 70% effective against Alpha variant (Lineage B.1.1.7) infection as determined by nucleic acid amplification tests, while the effectiveness of the vaccine was 77% against lineages other than Alpha variant (Lineage B.1.1.7). 153
The protection after the first dose of the Pfizer‐BioNTech (BNT162b2) vaccine was decreased in Qatar. In addition, after the second dose, it showed 97.4% protection against severe disease and 75% effectiveness against any documented infection with Beta variant (Lineage B.1.351). 152 J&J/Janssen's vaccine (Ad26.COV2.S) clinical trial demonstrated 65%–66% protection against hospitalization, while 91%–95% protection against mortality in South Africa. These efficacy results are similar to studies conducted in the United States. 154 Also, another clinical trial showed that Novavax provided 60% protection against the Beta variant (Lineage B.1.351). 155 Additionally, a significant decrease in the neutralization of Beta variant (Lineage B.1.351) by sera from humans vaccinated with Moderna (mRNA‐1273) was detected; however, there was no significant impact against Alpha variant (Lineage B.1.1.7). 156
In Brazil, where the Gamma variant (Lineage P1) was predominant, comparable efficacy to the J&J/Janssen vaccination was reported. Also, comparable real‐world effectiveness was demonstrated by the Sinovac vaccination (CoronaVac) and the AstraZeneca vaccination against symptomatic COVID‐19 during wide Gamma variant (Lineage P1) circulation. 154
In England, vaccine effectiveness of 67% and 88% against symptomatic disease following infection with Delta variant (Lineage B.1.617.2) has been noted after two doses of Vaxzevria (AZD1222) and Pfizer‐BioNTech (BNT162b2), respectively. 157 In addition, in Scotland, Vaxzevria (AZD1222) and Pfizer‐BioNTech (BNT162b2) vaccines were effective to decrease the risk of infection and also hospitalization against Delta variant (Lineage B.1.617.2). But the level of protection was not as high as against Alpha variant (Lineage B.1.1.7). 158 In Qatar, the effectiveness against the Delta variant (Lineage B.1.617.2) was 86% with Moderna (mRNA‐1273) and 60% with BioNTech (BNT162b2) vaccines. 151
Two shots of the Pfizer‐BioNTech (BNT162b2) vaccines have been reported to be 70% effective in preventing hospitalization from infection with the Omicron variant (Lineage B.1.1.529). 159 On the other hand, a third dose of the Pfizer‐BioNTech (BNT162b2) vaccine has been shown to neutralize the Omicron variant (Lineage B.1.1.529). Different studies have shown that CoronaVac and Moderna (mRNA‐1273) and Pfizer/BioNTech (BNT162b2) mRNA vaccines have significantly decreased neutralizing activity against the Omicron variant (Lineage B.1.1.529). 160
8. DRUG DEVELOPMENT STUDIES FOR COVID‐19
Anticoronavirus therapy is divided into two categories according to the target. One of this therapy acts on the coronavirus itself, and the other is the human immune system. The human immune system plays a significant role in controlling the infection and replication of coronavirus. Therefore, blocking the human signaling pathways required for virus replication might show a particular antiviral effect. 161 The therapies that act on the coronavirus itself contain blocking the binding of the virus to the human cell receptors, preventing the synthesis of viruses’ RNA, inhibiting processing of the viral assembly through acting on some structural proteins, or inhibiting viral replication via acting on crucial enzymes of the virus. 162
Researchers have enhanced three strategies for developing the drug against coronavirus. 27 One of them is to test existing broad‐spectrum antiviral agents. In this strategy, interferons, cyclophilin inhibitors, and ribavirin are used to treat coronavirus pneumonia. Their advantages are that their potential efficacy, metabolic characteristics, side effects, and applied dose are clearly known because they have been approved for treating viral infections. However, they cannot kill coronavirus in a targeted manner. 162 In the second strategy, the existing molecular databases are used for high‐throughput screening of specific molecules, which may have a therapeutic effect on coronavirus. Thus, new functions of drug molecules could be identified. In the third strategy, pathological characteristics and genomic information of different CoVs are determined to develop new targeted drugs, which would show better therapeutic effects. 163 It should not be forgotten that the research procedure for new drugs may take several years. 162
As of January 21, 2023, 8842 COVID‐19 studies, completed or still in progress, are listed on ClinicalTrials.gov. 164 Different target drugs have been identified to date, and various immunomodulators and antiviral molecules were applied to patients with severe COVID‐19. These antiviral molecules include nucleoside analogs such as remdesivir and molnupiravir, protease inhibitors such as lopinavir/ritonavir, antibiotics such as azithromycin, antimalarial such as hydroxychloroquine, nAbs such as sotrovimab, antiparasitic such as ivermectin. On the other hand, immunomodulators contain antibodies such as convalescent plasma and tocilizumab anti‐IL‐6 (Interleukin‐6) receptor mAb, interferon such as IFNb‐1a (Interferon beta‐1a) and peg‐IFNλ‐1 (Interferon lambda‐1), corticosteroids such as dexamethasone and inhaled budesonide, anticoagulants such as heparin, angiotensin receptor blocker such as telmisartan. 165
In addition to viral polymerase and the Mpro, the PLpro was considered to have significant potential as therapeutic targets. 166 The SARS‐CoV‐2 PLpro mediates the cleavage of viral polyprotein and also modulates the innate immune response of the host upon viral infection. PLpro has deISGylating and deubiquitinating activities, which are implicated in suppressing innate immune responses and promoting viral replication. It can remove ISG15 (interferon‐stimulated gene 15) and ubiquitin modifications from host proteins. 167 The active site of PLpro is now the subject of the most research because of its significance, yet due to its structural characteristics, the active site itself has proven to be quite a difficult target for rational drug design. 166
Antiviral drugs with the capacity to prevent virus attachment and entry by specifically targeting the S protein and cell surface protease could block the virus replication cycle and may be used as treatment approaches for COVID‐19 patients 168 , 169 (Figure 4). The FDA has now issued an emergency use authorization for three antiviral drugs. 170 Remdesivir, which can block the activity of RNA‐dependent RNA polymerase with in vitro inhibitory activity against SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2, was reported early as a promising therapeutic candidate for COVID‐19 infection. 171 Therefore, in May 2020, the FDA approved remdesivir as the first drug to treat COVID‐19 in adults and children requiring hospitalization. 172 However, the clinical application of remdesivir has been highly restricted because of the requirement of intravenous administration, variable antiviral activity in different organelles, and unstable concentrations in plasma. Moreover, it did not reduce the mortality caused by COVID‐19. 173 , 174 On the other hand, molnupiravir, which is an oral antiviral drug with β‐D‐N4‐hydroxycytidine as the active ingredient, has been used since December 2021. Molnupiravir shows antiviral activity by causing catastrophic mutations during viral RNA replication using RNA‐dependent RNA polymerase. 175 , 176 The other antiviral drug is nirmatrelvir/ritonavir (Paxlovid), which inhibits the Mpro, 3CL protease, of SARS‐CoV‐2 via reversible covalent bonding between the catalytic cysteine (Cys145) of Mpro and the nitrile warhead of nirmatrelvir. This drug has been used since December 2021 and has extensive antiviral activity and good off‐target selectivity. 176 , 177 , 178
FIGURE 4.
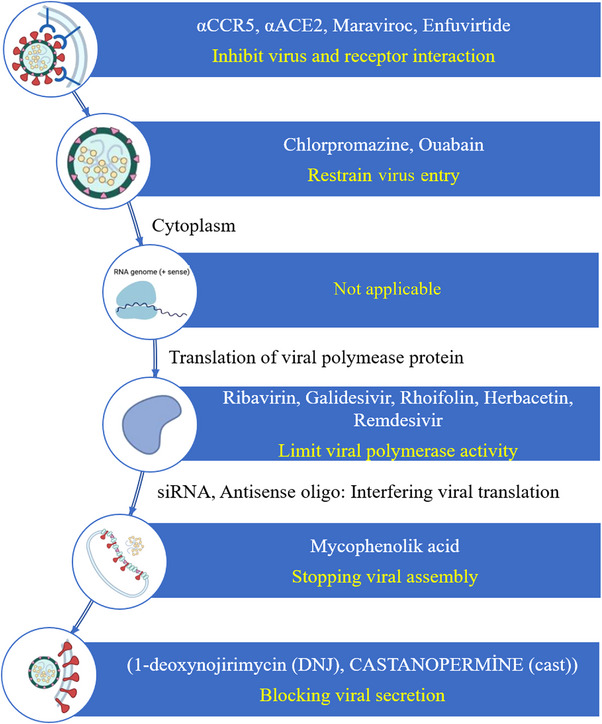
Virus replication cycle and the steps that are targeted by antiviral drugs. PCR, polymerase chain reaction; RNA, ribonucleic acid.
In order to accelerate the development of antiviral drugs to combat the pandemic, it is essential to predict drug resistance before it becomes prevalent in clinical settings. 179 Drug resistance against remdesivir and nirmatrelvir has been reported in some studies. RdRp mutations that confer resistance to remdesivir are well known through in vitro studies. Numerous SARS‐CoV‐2 sequencing efforts have allowed to assess emergence and fitness of antiviral resistance in vivo. 180 The de novo emergence of remdesivir resistance mutation, E802D, has been reported in an immunocompromised patient with acquired B‐cell deficiency, who had persistent SARS‐CoV‐2 infection. In vitro tests revealed that this mutation conferred an approximately six‐fold increase in remdesivir IC50; however, resulted in a fitness cost in the absence of remdesivir. 181 In another study, 100 naturally occurring Mpro mutations, which are located at the nirmatrelvir binding site, have been studied. Twenty of these mutants demonstrated comparable enzymatic activity to the wild type (10‐fold change) and resistance to nirmatrelvir (> 10‐fold increase). 179
Since viruses need host factors to translate their transcripts, targeting the host factor(s) are important for developing novel antiviral drugs. 182 Host‐targeting antivirals (HTAs) consist reagents which target the host proteins that are involved in the life cycle of virus, regulating the function of the immune system or other cellular processes in host cells. 183 Additionally, a genetic barrier to resistance is produced via the minimal diversity of host components that HTAs target. 182 Even before comprehensive information about a virus becomes available, host‐targeting drugs can be used to combat newly developing infections. 184
Also, cases of Paxlovid rebound have been reported where mild COVID‐19 symptoms recur after a period of initial recovery post‐treatment with Paxlovid. According to a preliminary study, this occurrence is likely caused by insufficient drug exposure because infected cells that are not exposed to the drug would allow the virus to continue replicating and symptoms to reappear. 185 , 186 , 187 These rebound cases are still rare, though, as in the Paxlovid clinical study, < 1% of 5287 patients required hospitalization or care in the emergency department for COVID‐19 symptoms 5–15 days following Paxlovid treatment. 185 , 188
To date, convalescent plasma, classic adaptive immunotherapy, has been exerted to prevent and treat various infectious diseases. The FDA has approved using blood plasma from patients that have recovered from COVID‐19 infection with a high nAb titer because of being an essential source of convalescent plasma. 189 Besides the optimal dose, the treatment time point is also significant. Studies have shown that convalescent plasma therapy has low risk and a potential therapeutic effect for severe COVID‐19 patients. 190 However, there was a lack of procedure control and standardization regarding the level of antibodies in convalescent plasma units and the donor selection process. These issues may lead to various therapeutic effects in patients with the same disease. 191
Antibody therapies for COVID‐19 also contain hyperimmune gamma globulin as well as mAbs preparations. 192 Production of hyperimmune globulin has been pursued in parallel with research into convalescent plasma during COVID‐19. Hyperimmune globulin is a licensed product made from convalescent plasma from thousands of donors. Hyperimmune globulin includes concentrated polyclonal immune globulin fraction and is subject to pathogen reduction techniques. 193
On the other hand, several mAb, which recognize a single antigen, have been discovered to bind the SARS‐CoV‐2 S protein and block this virus from invading human cells. An mAb may be administered intravenously to COVID‐19 patients, usually at an emergency department, an infusion center, or another outpatient settings—such as a nursing home or patient's home. Some potential risks of mAbs including allergic or nonallergic infusion‐related reaction have been reported. mAbs are intended for patients just been diagnosed with COVID‐19 who have some risk factors for severe infection but who are not sick enough to be hospitalized. 194 Thus, results from current trials do not show a benefit of mAbs in the majority of hospitalized patients. 195
Antibody‐based COVID‐19 therapies have demonstrated promising clinical efficacy and safety. To date, different nAbs have been approved by the FDA for urgent treatment of mild to moderate COVID‐19. Some nAbs such as imdevimab and casirivimab target viral RBD and have demonstrated strong neutralizing activity against wild‐type SARS‐CoV‐2 infection. On the other hand, Omicron variants exhibit notable resistance to the majority of approved nAbs such as bamlanivimab, casirivimab, imdevimab, and regdanvimab. 196
9. CONCLUSION AND PERSPECTIVES
In the 21st century, due to human activities and rapid globalization, the risk of viral transmission has increased across continents, and various pandemics have been observed. The latest pandemic, COVID‐19, has affected nearly every aspect of our lives. We have been forced to adapt to the new normal such as quarantine, working from home, having to homeschool, wearing face masks, and so forth. In addition, the global prevalence of depression and anxiety has increased. 3 years have passed since this pandemic, which claimed more than six million lives and affected billions. COVID‐19 continues to threaten us for reasons such as new variants of SARS‐CoV‐2, high transmissibility, some people not being vaccinated, and not using personal protective equipment. Early and accurate diagnoses are essential to control infectious diseases. Various diagnoses, vaccines, and drug studies have been carried out to support the fight against the COVID‐19 pandemic. The presence of SARS‐CoV‐2 has been determined by molecular techniques such as RT‐PCR, RT‐LAMP, CRISPR, amplicon‐based metagenomic sequencing, and nucleic acid hybridization using microarray and serological techniques such as lateral flow test and ELISA. The fact that this disease shows symptoms similar to other viral infections leads people to think they do not carry the SARS‐CoV‐2 virus, even if they have it. In addition, those with the suspected disease do not prefer to go for testing because they are afraid of the risk of contamination in the hospital. Therefore, people have begun to use COVID‐19 self‐test kits, which give rapid results and are easy to use. However, these serological tests have low accuracy and may give false positive or false negative results. In such cases, the need to develop nucleic acid‐based rapid diagnostic kits with high specificity and sensitivity, which are not on the market, has emerged.
On the other hand, numerous scientists and pharmaceutical companies have tried to develop safe and effective vaccines against COVID‐19 infection, and 50 vaccines have been approved by at least one country. The primary concern in vaccine studies is the continuous mutation in SARS‐CoV‐2. The Omicron variant, detected recently, is less virulent but more infectious than previous SARS‐CoV‐2 variants. Unfortunately, COVID‐19 continues to run its course. Omicron will most likely not be the last COVID‐19 variant. In the future, the currently available vaccines may be less effective in fighting new emerging variants. Although several clinical trials have been carried out until now, it is still urgent to develop highly effective and low‐toxic antiviral drugs that can control the SARS‐CoV‐2 infection. Moreover, developing rapid diagnostic kits for different variants including the Omicron sublineages with which patients are infected is important to guide the selection of antibody drugs. To assess the therapeutic potential of combining PLpro inhibitors with RdRp or Mpro inhibitors, combination experiments may be planned. On the other hand, due to COVID‐19 and the threat of bacterial coinfections, antibiotic use has increased, perhaps contributing to the development of drug resistance. This pandemic has taught that new strategies must be developed to combat a possible pandemic, such as (i) quickly obtaining and sharing the genetic sequences of viruses as they emerge, (ii) production of vaccines, therapeutics, and diagnostic kits in regional center that will enable them to be easily distributed everywhere, and (iii) building plug‐and‐play technical platforms such as rapidly modifying mRNA technology or adenovirus vectors.
AUTHOR CONTRIBUTIONS
İ.P. performed writing and editing original draft, conceptualization and investigation. T.O.O. performed writing and editing original draft, conceptualization, and investigation. I.D. performed writing original draft. S.O. performed writing original draft. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
No financial support was received from any institution or organization in the writing of this review article.
ETHICS STATEMENT
This review does not require an ethical statement.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
The authors have nothing to report.
Polatoğlu I, Oncu‐Oner T, Dalman I, Ozdogan S. COVID‐19 in early 2023: Structure, replication mechanism, variants of SARS‐CoV‐2, diagnostic tests, and vaccine & drug development studies. MedComm. 2023;4:e228. 10.1002/mco2.228
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Kang S, Peng W, Zhu Y, et al. Recent progress in understanding 2019 novel coronavirus (SARS‐CoV‐2) associated with human respiratory disease: detection, mechanisms and treatment. Int J Antimicrob Agents. 2020;55:105950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shereen MA, Khan S, Kazmi A, et al. COVID‐19 infection: emergence, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gorbalenya AE, Enjuanes L, Ziebuhr J, et al. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sevajol M, Subissi L, Decroly E, et al. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS‐coronavirus. Virus Res. 2014;194:90‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ather A, Patel B, Ruparel NB, et al. Coronavirus disease 19 (COVID‐19): implications for clinical dental care. J Endod. 2020;46:584‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. National Center for Immunization and Respiratory Diseases (NCIRD). Division of viral diseases. 2020. Accessed October 26, 2020. https://www.cdc.gov/coronavirus/types.html
- 7. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Health Organization . WHO target product profiles for COVID‐19 vaccines. 2020. Accessed December 26, 2020. https://www.who.int/blueprint/priority‐diseases/key‐action/WHO_Target_Product_Profiles_for_COVID‐19_web.pdf
- 9. World Health Organization . WHO coronavirus disease (COVID‐19) dashboard. 2023. Accessed February 4, 2021 and January 21, 2023. https://covid19.who.int/table
- 10. Peng XL, Cheng JSY, Gong HL, et al. Advances in the design and development of SARS‐CoV‐2 vaccines. Mil Med Res. 2021;8:1‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parra‐Lucares A, Segura P, Rojas V, et al. Emergence of SARS‐CoV‐2 variants in the world: how could this happen? Life. 2022;12:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. World Health Organization . WHO tracking SARS‐CoV‐2 variants. 2022. Accessed October 12, 2022 and January 13, 2023. https://www.who.int/en/activities/tracking‐SARS‐CoV‐2‐variants/
- 13. Puhach O, Adea K, Hulo N, et al. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS‐CoV‐2. Nat Med. 2022;28:1491‐1500. [DOI] [PubMed] [Google Scholar]
- 14. Suzuki R, Yamasoba D, Kimura I, et al. Attenuated fusogenicity and pathogenicity of SARS‐CoV‐2 Omicron variant. Nature. 2022;603:700‐705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abu‐Raddad LJ, Chemaitelly H, Ayoub HH, et al. Severity, criticality, and fatality of the SARS‐CoV‐2 Beta variant. Clin Infect Dis. 2022;75(1):e1188‐e1191. doi: 10.1093/cid/ciab909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID‐19: their roles in pathogenesis. J Microbiol Immunol Infect. 2021;54:159‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Boheemen S, De Graaf M, Lauber C, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao YC, Deng QX, Dai SX. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID‐19: an evaluation of the evidence. Travel Med Infect Dis. 2020;35:101647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID‐19) outbreak–an update on the status. Mil Med Res. 2020;7:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang N, Shi X, Jiang L, et al. Structure of MERS‐CoV spike receptor‐binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986‐993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581:215‐220. [DOI] [PubMed] [Google Scholar]
- 23. World Health Organization . Advice for the public: coronavirus disease (COVID‐19). 2021. Accessed March 3, 2021. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/advice‐for‐public
- 24. Chen L, Xiong J, Bao L, et al. Convalescent plasma as a potential therapy for COVID‐19. Lancet Infect Dis. 2020;20:398‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma DK. Pathogenicity and prevention of COVID‐19. In: Sharma DK, ed. Understanding COVID‐19. SpotWrite Publications; 2022:1‐16. [Google Scholar]
- 26. Zhang Q, Xiang R, Huo S, et al. Molecular mechanism of interaction between SARS‐CoV‐2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021;6:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kubina R, Dziedzic A. Molecular and serological tests for COVID‐19. A comparative review of SARS‐CoV‐2 coronavirus laboratory and point‐of‐care diagnostics. Diagnostics. 2020;10:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Liu SM, Yu XH, et al. Coronavirus disease 2019 (COVID‐19): current status and future perspectives. Int J Antimicrob Agents. 2020;55:105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carter LJ, Garner LV, Smoot JW, et al. Assay techniques and test development for COVID‐19 diagnosis. ACS Cent Sci. 2020;6:591‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Machado BAS, Hodel KVS, Barbosa‐Júnior VG, et al. The main molecular and serological methods for diagnosing COVID‐19: an overview based on the literature. Viruses. 2021;13:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Izzo MM, Kirkland PD, Gu X, et al. Comparison of three diagnostic techniques for detection of rotavirus and coronavirus in calf faeces in Australia. Aust Vet J. 2012;90:122‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alpdagtas S, Ilhan E, Uysal E, et al. Evaluation of current diagnostic methods for COVID‐19. APL Bioeng. 2020;4:041506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mlcochova P, Collier D, Ritchie A, et al. Combined point‐of‐care nucleic acid and antibody testing for SARS‐CoV‐2 following emergence of D614G spike variant. Cell Rep Med. 2020;1:100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Woo PC, Lau SK, Wong BHL, et al. False‐positive results in a recombinant severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV) nucleocapsid enzyme‐linked immunosorbent assay due to HCoV‐OC43 and HCoV‐229E rectified by Western blotting with recombinant SARS‐CoV spike polypeptide. J Clin Microbiol. 2004;42:5885‐5888. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Mohamadian M, Chiti H, Shoghli A, et al. COVID‐19: virology, biology and novel laboratory diagnosis. J Gene Med. 2021;23:e3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones H. The importance of diagnostic testing for COVID‐19. Infectious Diseases Hub. 2020. Accessed December 30, 2020. https://www.id‐hub.com/2020/04/02/the‐importance‐of‐diagnostic‐testing‐for‐covid‐19/
- 37. Ishige T, Murata S, Taniguchi T, et al. Highly sensitive detection of SARS‐CoV‐2 RNA by multiplex rRT‐PCR for molecular diagnosis of COVID‐19 by clinical laboratories. Clin Chim Acta. 2020;507:139‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hong TCT, Mai QL, Cuong DV, et al. Development and evaluation of a novel loop‐mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1956‐1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Santiago I. Trends and innovations in biosensors for COVID‐19 mass testing. Chembiochem. 2020;21:2880‐2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alhassan A, Li Z, Poole CB, et al. Expanding the MDx toolbox for filarial diagnosis and surveillance. Trends Parasitol. 2015;31:391‐400. [DOI] [PubMed] [Google Scholar]
- 41. Cecchetelli A. Finding nucleic acids with SHERLOCK and DETECTR. Addgene Blog. 2020. Accessed March 3, 2021. https://blog.addgene.org/finding‐nucleic‐acids‐with‐sherlock‐and‐detectr [Google Scholar]
- 42. Illumina . Amplicon sequencing solutions. 2020. Accessed April 23, 2020. https://www.illumina.com/techniques/sequencing/dna‐sequencing/targeted‐resequencing/amplicon‐sequencing.html
- 43. Moore SC, Penrice‐Randal R, Alruwaili M, et al. Amplicon based MinION sequencing of SARS‐CoV‐2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID‐19. MedRxiv. Preprint posted online March 8, 2020. doi: 10.1101/2020.03.05.20032011 [DOI] [Google Scholar]
- 44. Kafetzopoulou LE, Pullan ST, Lemey P, et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science. 2019;363:74‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen M, Zhou Y, Ye J, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020;10:97‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo X, Geng P, Wang Q, et al. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J Microbiol Biotechnol. 2014;24:1445‐1454. [DOI] [PubMed] [Google Scholar]
- 47. Perera RA, Mok CK, Tsang OT, et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Euro Surveill. 2020;25:2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hou H, Wang T, Zhang B, et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology. 2020;9:e1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams ER, Ainsworth M, Anand R, et al. Antibody testing for COVID‐19: a report from the National COVID Scientific Advisory Panel. Wellcome Open Res. 2020;5:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brandstetter S, Roth S, Harner S, et al. Symptoms and immunoglobulin development in hospital staff exposed to a SARS‐CoV‐2 outbreak. Pediatr Allergy Immunol. 2020;31:841‐847. [DOI] [PubMed] [Google Scholar]
- 51. BioMedomics . COVID‐19 IgM/IgG rapid test. 2020. Accessed March 3, 2022. https://www.biomedomics.com/products/infectious‐disease/covid‐19‐rt/
- 52. Giri B, Pandey S, Shrestha R, et al. (2021). Review of analytical performance of COVID‐19 detection methods. Anal Bioanal Chem. 2021;413:35‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhu N, Wong PK. Advances in viral diagnostic technologies for combating COVID‐19 and future pandemics. Slas Technol. 2020;25:513‐521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Böger B, Fachi MM, Vilhena RO, et al. Systematic review with meta‐analysis of the accuracy of diagnostic tests for COVID‐19. Am J Infect Control. 2021;49:21‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Subsoontorn P, Lohitnavy M, Kongkaew C. The diagnostic accuracy of isothermal nucleic acid point‐of‐care tests for human coronaviruses: a systematic review and meta‐analysis. Sci Rep. 2020;10:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Iqbal BN, Arunasalam S, Divarathna MV, et al. Diagnostic utility and validation of a newly developed real time loop mediated isothermal amplification method for the detection of SARS CoV‐2 infection. J Clin Virology Plus. 2022;2:100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sharma A, Balda S, Apreja M, et al. COVID‐19 diagnosis: current and future techniques. Int J Biol Macromol. 2021;193:1835‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shademan B, Nourazarian A, Hajazimian S, et al. CRISPR technology in gene‐editing‐based detection and treatment of SARS‐CoV‐2. Front Mol Biosci. 2021:8:772788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ganbaatar U, Liu C. CRISPR‐based COVID‐19 testing: toward next‐generation point‐of‐care diagnostics. Front Cell Infect Microbiol. 2021;11:663949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. MacMullan MA, Ibrayeva A, Trettner K, et al. ELISA detection of SARS‐CoV‐2 antibodies in saliva. Sci Rep. 2020;10:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sapkal G, Shete‐Aich A, Jain R, et al. Development of indigenous IgG ELISA for the detection of anti‐SARS‐CoV‐2 IgG. Indian J Med Res. 2020;151:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sharma S, Shrivastava S, Kausley SB, et al. Coronavirus: a comparative analysis of detection technologies in the wake of emerging variants. Infection. 2022;51(1):1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID‐19 International Summit, 23 March 2020: value of diagnostic testing for SARS–CoV‐2/COVID‐19. MBio. 2020;11:e00722‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smithgall MC, Dowlatshahi M, Spitalnik SL, et al. Types of assays for SARS‐CoV‐2 testing: a review. Lab Med. 2020;51:e59‐e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Olearo F, Nörz D, Heinrich F, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS‐CoV‐2 compared to RT‐qPCR. J Clin Virol. 2021;137:104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Centers for Disease Control and Prevention . CDC 2019‐novel coronavirus (2019‐nCoV) real‐time RT‐PCR diagnostic panel. 2020. Accessed March 3, 2022. https://www.fda.gov/media/134922/download
- 68. DiaSorin Molecular . Simplexa COVID‐19 direct kit—enabling swift action against the COVID‐19 pandemic. 2021. Accessed March 3, 2022. https://molecular.diasorin.com/us/wp‐content/uploads/2020/03/USC19BR0320‐C19‐Direct‐Brochure‐8.5x11‐FINAL_3‐27‐20.pdf
- 69. U.S. Food and Drug Administration . Emergency use authorization (EUA) summary COVID‐19 RT‐PCR test (Laboratory Corporation of America). 2021. Accessed March 3, 2022. https://www.fda.gov/media/136151/download
- 70. Luminex Corporation . Aries ARIES® SARS‐CoV‐2 assay package insert. 2020. Accessed March 3, 2022. https://www.fda.gov/media/136693/download
- 71. Sansure Biotech . Novel coronavirus (2019‐nCoV) nucleic acid diagnostic kit (PCR‐fluorescence probing). 2022. Accessed May 3, 2022. https://www.fda.gov/media/137651/download
- 72. Eagle Biosciences . COVID‐19 coronavirus real time PCR kit. 2022. Accessed May 3, 2022. https://eaglebio.com/product/covid‐19‐coronavirus‐real‐time‐pcr‐kit/
- 73. BioTNS . COVID‐19 RT‐PCR PNA kit. 2022. Accessed May 3, 2022. https://www.fda.gov/media/139680/download
- 74. SD Biosensor . STANDARD M nCoV Real‐Time detection kit. 2020. Accessed May 3, 2022. https://www.fda.gov/media/137302/download
- 75. Bioeksen R&D Technologies . Bio‐speedy direct RT‐qPCR SARS‐CoV‐2. 2021. Accessed May 3, 2022. https://www.fda.gov/media/141823/download
- 76. Abbott Diagnostics . ID NOWTM COVID‐19. 2021. Accessed May 3, 2022. https://www.fda.gov/media/136525/download
- 77. M Monitor . Isopollo COVID‐19 detection kit. 2021. Accessed May 3, 2021. https://www.medica.de/vis‐content/event‐medcom2020.MEDICA/exh‐medcom2020.2676180/MEDICA‐2020‐M‐MONITOR‐INC‐Product‐medcom2020.2676180‐ZIJnmNYZQy2xJpEOhenpvw.pdf
- 78. Eiken Chemical . Loopamp™ SARS‐CoV‐2 detection kit. 2021. Accessed May 3, 2021. https://www.eiken.co.jp/en/products/lamp/covid19test/
- 79. Detectachem . MobileDetect Bio BCC19 test kit. 2020. Accessed May 3, 2022. https://www.fda.gov/media/141791/download
- 80. Detect . Detect COVID‐19 test. 2021. Accessed May 3, 2022. https://www.fda.gov/media/153661/download
- 81. Lucira Health . Lucira™ COVID‐19 All‐In‐One test kit. 2022. Accessed May 3, 2022. https://www.fda.gov/media/143808/download
- 82. Mammoth Biosciences . SARS‐CoV‐2 DETECTR. 2020. Accessed May 3, 2022. https://mammoth.bio/wp‐content/uploads/2020/03/Mammoth‐Biosciences‐A‐protocol‐for‐rapid‐detection‐of‐SARS‐CoV‐2‐using‐CRISPR‐diagnostics‐DETECTR.pdf
- 83. Sherlock Biosciences . Sherlock CRISPR SARS‐CoV‐2 kit. 2022. Accessed May 3, 2022. https://www.fda.gov/media/137746/download
- 84. Tata Medical and Diagnostics . TATA MD check CRISPR SARS‐COV‐2 kit 1.0. 2022. Accessed May 3, 2022. https://www.finddx.org/product/tata‐md‐check‐crispr‐sars‐cov‐2‐kit‐1‐0/
- 85. PathogenDx, DetectX‐Rv . 2021. Accessed May 3, 2022. https://www.fda.gov/media/147803/download
- 86. Damin F, Galbiati S, Gagliardi S, et al. CovidArray: a microarray‐based assay with high sensitivity for the detection of Sars‐Cov‐2 in nasopharyngeal swabs. Sensors. 2021;21:2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Creative Diagnostics . SARS‐CoV‐2 IgG ELISA kit. 2022. Accessed May 3, 2022. https://www.creative‐diagnostics.com/pdf/DEIASL019.pdf
- 88. Epitope Diagnostics . EDI novel coronavirus COVID‐19 IgM ELISA kit. 2020. Accessed May 3, 2022. https://static1.squarespace.com/static/52545951e4b021818110f9cf/t/5fca9ffd453475472aaf6e6d/1607114754760/KT‐1033+IVD%2C+CE+V10.pdf
- 89. AMEDA Labordiagnostik GmbH . AMP ELISA test SARS‐COV‐2 AB. 2022. Accessed May 3, 2022. https://www.amp‐med.com/news‐elisa‐sars‐cov‐2
- 90. EUROIMMUN AG . Anti‐SARS‐CoV‐2 QuantiVac ELISA (IgG). 2021. Accessed May 3, 2022. https://www.coronavirus‐diagnostics.com/documents/Indications/Infections/Coronavirus/EI_2606_D_UK_E.pdf
- 91. General Biologicals Corporation . GB SARS‐CoV‐2 Ab ELISA. 2020. Accessed May 3, 2022. https://3aw3qz1wxc523mqoui3jyc615c‐wpengine.netdna‐ssl.com/wp‐content/uploads/2020/10/IFUIVD‐GB‐CoV‐2‐ELISA‐Ab‐v2.1en_20200703.pdf
- 92. Coris Bioconcept . COVID‐19 Ag Respi‐Strip. 2022. Accessed May 3, 2022. https://www.corisbio.com/products/covid19/ [DOI] [PMC free article] [PubMed]
- 93. Oncosem Onkolojik Sistemler . CRT COVID‐19 rapid test. 2022. Accessed May 3, 2022. https://oncosem.com/en/crt_kit_10_eng/
- 94. Megna Health . Rapid COVID‐19 IgM/IgG combo test kit. 2021. Accessed May 3, 2022. https://www.fda.gov/media/140297/download
- 95. SD Biosensor . PilotTM COVID‐19 at‐home test. 2022. Accessed May 3, 2022. https://www.fda.gov/media/155125/download
- 96. PHASE Scientific International . INDICAID COVID‐19 rapid antigen at‐home test. 2022. Accessed May 3, 2022. https://www.fda.gov/media/156954/download
- 97. VivaChek Biotech . VivaDiag™ SARS‐CoV‐2 IgM/IgG rapid test. 2022. Accessed May 3, 2022. https://www.vivachek.com/en/prods/prod‐rapidtest.html
- 98. InBios International . ScoV‐2 Ag detect rapid self‐test. 2022. Accessed May 3, 2022. https://www.fda.gov/media/154336/download
- 99. Abbott Diagnostics . BinaxNOW COVID‐19 Ag card home test. 2022. Accessed May 3, 2022. https://www.fda.gov/media/144574/download
- 100. iHealth Labs . iHealth COVID‐19 antigen rapid test pro. 2022. Accessed May 3, 2022. https://www.fda.gov/media/155490/download
- 101. Xtrava Health . SPERA COVID‐19 Ag test. 2021. Accessed May 3, 2022. https://www.fda.gov/media/153096/download
- 102. ANP Technologies . NIDS COVID‐19 antigen rapid test kit. 2021. Accessed May 3, 2022. https://www.fda.gov/media/152464/download
- 103. Ndwandwe D, Wiysonge CS. COVID‐19 vaccines. Curr Opin Immunol. 2021;71:111‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li Y, Tenchov R, Smoot J, et al. A comprehensive review of the global efforts on COVID‐19 vaccine development. ACS Cent Sci. 2021;7:512‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. CNN Philippines Staff . Five most common side effects of AstraZeneca, Sinovac vaccines. 2021. Accessed May 12, 2021. https://www.cnnphilippines.com/news/2021/4/20/Five‐most‐common‐side‐effects‐AstraZeneca‐Sinovac‐vaccines.html
- 106. VIPER Group COVID19 Vaccine Tracker Team . COVID‐19 vaccine tracker. 2022. Accessed January 21, 2023. https://covid19.trackvaccines.org/
- 107. Centers for Disease Control and Prevention . Moderna COVID‐19 vaccine (also known as Spikevax): overview and safety. 2022. Accessed April 12, 2022. https://www.cdc.gov/coronavirus/2019‐ncov/vaccines/different‐vaccines/Moderna.html
- 108. Kyriakidis NC, López‐Cortés A, González EV, et al. SARS‐CoV‐2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Uddin MN, Allon A, Roni MA, et al. Overview and future potential of fast dissolving buccal films as drug delivery system for vaccines. J Pharm Pharm Sci. 2019;22:388‐406. [DOI] [PubMed] [Google Scholar]
- 110. Frederiksen LSF, Zhang Y, Foged C, et al. The long road toward COVID‐19 herd immunity: vaccine platform technologies and mass immunization strategies. Front Immunol. 2020;11:1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yadav DK, Yadav N, Khurana SMP. Vaccines: present status and applications. In: Verma AS, Singh A, eds. Animal Biotechnology., Academic Press; 2020:523‐542. [Google Scholar]
- 112. Vicente T, Roldão A, Peixoto C, et al. Large‐scale production and purification of VLP‐based vaccines. J Invertebr Pathol. 2011;107:S42‐S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Fuenmayor J, Gòdia F, Cervera L. Production of virus‐like particles for vaccines. N Biotechnol. 2017;39:174‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shin MD, Shukla S, Chung YH, et al. COVID‐19 vaccine development and a potential nanomaterial path forward. Nat Nanotechnol. 2020;15:646‐655. [DOI] [PubMed] [Google Scholar]
- 115. Queiroz AMV, Oliveira JWDF, Moreno CJ, et al. VLP‐based vaccines as a suitable technology to target trypanosomatid diseases. Vaccines. 2021;9:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ura T, Okuda K, Shimada M. Developments in viral vector‐based vaccines. Vaccines. 2014;2:624‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mohammed S, Bakshi N, Chaudri N, et al. Cancer vaccines: past, present, and future. Adv Anat Pathol. 2016;23:180‐191. [DOI] [PubMed] [Google Scholar]
- 118. Mahony TJ. The role of adjuvants in the application of viral vector vaccines. In: Samal SK, Tikoo SK, Vanniasinkam T, eds. Viral vectors in veterinary vaccine development. Springer; Cham; 2021:37‐50. [Google Scholar]
- 119. Soriano R. Viral vector vaccines: a borrowed technology from gene therapy advancements. Malvern Panalytical. 2021. Accessed February 5, 2022. https://www.materials‐talks.com/viral‐vector‐vaccines‐a‐borrowed‐technology‐from‐gene‐therapy‐advancements/
- 120. Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580:576‐577. [DOI] [PubMed] [Google Scholar]
- 121. Wirsiy FS, Boock AU, Akoachere JFTK. Assessing the determinants of Ebola virus disease transmission in Baka community of the tropical rainforest of Cameroon. BMC Infect Dis. 2021;21:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. European Medicine Agency . Ervebo: Ebola Zaire Vaccine (rVSV∆G‐ZEBOV‐GP, live). 2019. Accessed February 5, 2022. https://www.ema.europa.eu/en/medicines/human/EPAR/ervebo
- 123. Chen WH, Strych U, Hotez PJ, et al. The SARS‐CoV‐2 vaccine pipeline: an overview. Curr Trop Med Rep. 2020;7:61‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Belete TM. A review on promising vaccine development progress for COVID‐19 disease. Vacunas. 2020;21:121‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bloom K, van den Berg F, Arbuthnot P. Self‐amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28:117‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rijkers GT, Weterings N, Obregon‐Henao A, et al. Antigen presentation of mRNA based and viral vector corona vaccines. Preprints. Preprint posted online on June 30, 2021. doi: 10.20944/preprints202106.0725.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Park JW, Lagniton PN, Liu Y, et al. mRNA vaccines for COVID‐19: what, why and how. Int J Biol Sci. 2021;17:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. International Federation of Pharmaceutical Manufacturers & Associations (IFPMA) . The complex journey of a vaccine: the steps behind developing a new vaccine. 2019. Accessed February 5, 2022. https://www.ifpma.org/wp‐content/uploads/2019/07/IFPMA‐ComplexJourney‐2019_FINAL.pdf
- 129. Artaud C, Kara L, Launay O. Vaccine development: from preclinical studies to phase 1/2 clinical trials. Malar Control Elimin. 2019;2013:165‐176. [DOI] [PubMed] [Google Scholar]
- 130. Calina D, Sarkar C, Arsene AL, et al. Recent advances, approaches and challenges in targeting pathways for potential COVID‐19 vaccines development. Immunol Res. 2020;68:315‐324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Han S. Clinical vaccine development. Clin Exp Vaccine Res. 2015;4:46‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. De Mattia F, Chapsal JM, Descamps J, et al. The consistency approach for quality control of vaccines—a strategy to improve quality control and implement 3Rs. Biologicals. 2011;39:59‐65. [DOI] [PubMed] [Google Scholar]
- 133. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367:1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. World Health Organization . WHO COVID‐19 advice for the public: getting vaccinated. 2022. Accessed August 9, 2022. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/covid‐19‐vaccines/advice
- 135. Kudlay D, Svistunov A. COVID‐19 vaccines: an overview of different platforms. Bioengineering. 2022;9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Edwards KM, Orenstein WA. COVID‐19: vaccines, up to date. 2022. Accessed May 5, 2022. https://www.uptodate.com/contents/covid‐19‐vaccines
- 137. Polack FP, Thomas SJ, Kitchin N, et al. C4591001 Clinical Trial Group, Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383:2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kashte S, Gulbake A, El‐Amin SF III, et al. COVID‐19 vaccines: rapid development, implications, challenges and future prospects. Hum Cell. 2021;34:711‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Sharma G. An overview of recent developments in COVID‐19 vaccine. In: Sharma DK, ed. Understanding COVID‐19. SpotWrite Publications; 2022:50. [Google Scholar]
- 140. Wu K, Werner AP, Moliva JI, et al. mRNA‐1273 vaccine induces neutralizing antibodies against spike mutants from global SARS‐CoV‐2 variants. BioRvix. Preprint posted online January 25, 2021. doi: 10.1101/2021.01.25.427948 [DOI] [Google Scholar]
- 141. Katella K. Comparing the COVID‐19 vaccines: how are they different? Yale Medicine: Family Health. 2021. Accessed February 5, 2022. https://palmtreewellness.com/wp‐content/uploads/2021/05/Comparing‐the‐COVID‐19‐Vaccines‐How‐Are‐They‐Different‐News‐Yale‐Medicine.pdf
- 142. Luchsinger LL, Hillyer CD. Vaccine efficacy probable against COVID‐19 variants. Science. 2021;371:1116. [DOI] [PubMed] [Google Scholar]
- 143. Ghiasi N, Valizadeh R, Arabsorkhi M, et al. Efficacy and side effects of Sputnik V, Sinopharm and AstraZeneca vaccines to stop COVID‐19; a review and discussion. Immunopathol Persa. 2021;7:31. [Google Scholar]
- 144. Costa AG. Clinical study proving greater effectiveness of Coronavac is sent to Lancet, CNN Brasil. 2021. Accessed February 5, 2022. www.cnnbrasil.com.br/saude/2021/04/11/estudo‐clinico‐que‐comprova‐maior‐eficacia‐da‐coronavac‐e‐enviado‐para‐lancet
- 145. Reuters Staff . Turkish study revises down Sinovac COVID‐19 vaccine efficacy to 83.5%, Reuters. 2021. Accessed February 5, 2022. https://www.reuters.com/article/health‐coronavirus‐turkey‐sinovac‐int‐idUSKBN2AV18P
- 146. Bharat Biotech . COVAXIN ®‐India's first indigenous COVID‐19 vaccine. 2020. Accessed May 5, 2022. https://www.bharatbiotech.com/covaxin.html
- 147. Sharun K, Dhama K. India's role in COVID‐19 vaccine diplomacy. J Travel Med. 2021;28:taab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. World Health Organization . WHO Background document on the Novavax (NVX‐CoV2373) vaccine against COVID‐19. 2021. Accessed May 5, 2022. https://www.who.int/publications/i/item/WHO‐2019‐nCoV‐vaccines‐SAGE‐recommendation‐Novavax‐NVX‐CoV2373‐background
- 149. European Medicines Agency . 2022. Accessed May 5, 2022. https://www.ema.europa.eu/en/documents/product‐information/nuvaxovid‐epar‐product‐information_en.pdf
- 150. Heath PT, Galiza EP, Baxter DN, et al. Safety and efficacy of NVX‐CoV2373 Covid‐19 vaccine. N Engl J Med. 2021;385:1172‐1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Cevik M, Grubaugh ND, Iwasaki A, et al. COVID‐19 vaccines: keeping pace with SARS‐CoV‐2 variants. Cell. 2021;184:5077‐5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Abu‐Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid‐19 vaccine against the B. 1.1. 7 and B. 1.351 variants. N Engl J Med. 2021;385:187‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Emary KR, Golubchik T, Aley PK, et al. Efficacy of ChAdOx1 nCoV‐19 (AZD1222) vaccine against SARS‐CoV‐2 variant of concern 202012/01 (B. 1.1. 7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351‐1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tregoning JS, Flight KE, Higham SL, et al. Progress of the COVID‐19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol. 2021;21:626‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mahase E. Covid‐19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021:372:n296. [DOI] [PubMed] [Google Scholar]
- 156. Wu K, Werner AP, Moliva JI, et al. mRNA‐1273 vaccine induces neutralizing antibodies against spike mutants from global SARS‐CoV‐2 variants. BioRxiv. Preprint posted online January 25, 2021. doi: 10.1101/2021.01.25.427948 [DOI] [Google Scholar]
- 157. Wall EC, Wu M, Harvey R, et al. Neutralising antibody activity against SARS‐CoV‐2 VOCs B. 1.617. 2 and B. 1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331‐2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Sheikh A, McMenamin J, Taylor B, et al. SARS‐CoV‐2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461‐2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Sharma V, Rai H, Gautam DN, et al. Emerging evidence on Omicron (B. 1.1. 529) SARS‐CoV‐2 variant. J Med Virol. 2022;94:1876‐1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. de Oliveira Campos DM, da Silva MK, Silva de Oliveira CB, et al. Effectiveness of COVID‐19 vaccines against Omicron variant. Immunotherapy. 2022;14:903‐904. [DOI] [PubMed] [Google Scholar]
- 161. Erlina L, Paramita RI, Kusuma WA, et al. Virtual screening on Indonesian herbal compounds as COVID‐19 supportive therapy: machine learning and pharmacophore modeling approaches. BMC Complement Med The. 2022;22:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS‐CoV‐2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Noor R. Antiviral drugs against severe acute respiratory syndrome coronavirus 2 infection triggering the coronavirus disease‐19 pandemic. Tzu‐Chi Med J. 2021;33:7‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. U.S. National Library of Medicine . Views of COVID‐19 studies listed on ClinicalTrials.gov (Beta). 2023. Accessed January 21, 2023. https://clinicaltrials.gov/ct2/covid_view
- 165. Salasc F, Lahlali T, Laurent E, et al. Treatments for COVID‐19: lessons from 2020 and new therapeutic options. Curr Opin Pharmacol. 2022;62:43‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Ton AT, Pandey M, Smith JR, et al. Targeting SARS‐CoV‐2 papain‐like protease in the post‐vaccine era. Trends Pharmacol Sci. 2022;43:906‐919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Tan H, Hu Y, Jadhav P, et al. Progress and challenges in targeting the SARS‐CoV‐2 papain‐like protease. J Med Chem. 2022;65: 7561–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Ghanbari R, Teimoori A, Sadeghi A, et al. Existing antiviral options against SARS‐CoV‐2 replication in COVID‐19 patients. Future Microbiol. 2020;15:1747‐1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Yang KC, Lin JC, Tsai HH, et al. Nanotechnology advances in pathogen‐and host‐targeted antiviral delivery: multipronged therapeutic intervention for pandemic control. Drug Deliv Transl Res. 2021;11:1420‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kumar P, Ratia KM, Richner JM, et al. Dual inhibition of Cathepsin L and 3CL‐Pro by GC‐376 constrains SARS Cov2 infection including Omicron variant. BioRxiv. Preprint posted online February 10, 2022. doi: 10.1101/2022.02.09.479835 [DOI] [Google Scholar]
- 171. John H, Beigel MD, Kay M, Tomashek MD, et al. ACTT‐1 Study Group. Remdesivir for the treatment of Covid‐19: final report. N Engl J Med. 2020;383:1813‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. U.S. Food and Drug Administration . FDA's approval of Veklury (remdesivir) for the treatment of COVID‐19—the science of safety and effectiveness. 2020. Accessed May 21, 2022. https://www.fda.gov/drugs/news‐events‐human‐drugs/fdas‐approval‐veklury‐remdesivir‐treatment‐covid‐19‐science‐safety‐and‐effectiveness
- 173. Fan H, Lou F, Fan J, et al. The emergence of powerful oral anti‐COVID‐19 drugs in the post‐vaccine era. Lancet Microbe. 2022;3:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ni Y, Liao J, Qian Z, et al. Synthesis and evaluation of enantiomers of hydroxychloroquine against SARS‐CoV‐2 in vitro. Bioorg Med Chem. 2022;53:116523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Negi M, Chawla PA, Teli G, et al. Molnupiravir–A prospective silver bullet to mitigate severe acute respiratory syndrome corona virus‐2. 2021. Accessed May 10, 2022. http://isfcppharmaspire.com/abstract.aspx?Article=PHARMASPIRE_14_2021
- 176. Infectious Diseases Society of America . Antivirals. 2022. Accessed May 10, 2022. https://www.idsociety.org/covid‐19‐real‐time‐learning‐network/therapeutics‐and‐interventions/antivirals/
- 177. Hung YP, Lee JC, Chiu CW, et al. Oral Nirmatrelvir/Ritonavir therapy for COVID‐19: the dawn in the dark? Antibiotics. 2022;11:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Brady DK, Gurijala AR, Huang L, et al. A guide to COVID‐19 antiviral therapeutics: a summary and perspective of the antiviral weapons against SARS‐CoV‐2 infection. FEBS J. Preprint posted online October 20, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Hu Y, Lewandowski EM, Tan H, et al. Naturally occurring mutations of SARS‐CoV‐2 main protease confer drug resistance to nirmatrelvir. BioRxiv. Preprint posted online September 6, 2022. doi: 10.1101/2022.06.28.497978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Focosi D, Maggi F, McConnell S, et al. Very low levels of remdesivir resistance in SARS‐COV‐2 genomes after 18 months of massive usage during the COVID19 pandemic: a GISAID exploratory analysis. Antivir Res. 2022;198:105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Gandhi S, Klein J, Robertson AJ, et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS‐CoV‐2 infection in an immunocompromised patient: a case report. Nat Commun. 2022;13:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ullah H, Hou W, Dakshanamurthy S, et al. Host targeted antiviral (HTA): functional inhibitor compounds of scaffold protein RACK1 inhibit herpes simplex virus proliferation. Oncotarget. 2019;10:3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Lou Z, Sun Y, Rao Z. Current progress in antiviral strategies. Trends Pharmacol Sci. 2014;35:86‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Dwek RA, Bell JI, Feldmann M, et al. Host‐targeting oral antiviral drugs to prevent pandemics. Lancet. 2022;399:1381‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Center for Disease Control and Prevention. COVID‐19 Rebound after Paxlovid Treatment . CDC; 2022. [Google Scholar]
- 186. Alshanqeeti S, Bhargava A. COVID‐19 rebound after Paxlovid treatment: a case series and review of literature. Cureus. 2022;14(6):e26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Carlin AF, Clark AE, Chaillon A, et al. Virologic and immunologic characterization of COVID‐19 recrudescence after nirmatrelvir/ritonavir treatment. Res Sq. Preprint posted online May 18, 2022. doi: 10.21203/rs.3.rs-1662783/v1 [DOI] [Google Scholar]
- 188. Kuehn BM. Rehospitalization, emergency visits after Paxlovid treatment are rare. Jama. 2022;328:323‐323. [DOI] [PubMed] [Google Scholar]
- 189. Abd El‐Aziz TM, Stockand JD. Recent progress and challenges in drug development against COVID‐19 coronavirus (SARS‐CoV‐2)‐an update on the status. Infect Genet Evol. 2020;83:104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Duan K, Liu B, Li C, et al. Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proc Natl Acad Sci USA. 2020;117:9490‐9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Li L, Zhang W, Hu Y, et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life‐threatening COVID‐19: a randomized clinical trial. Jama. 2020;324:460‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Casadevall A, Pirofski LA, Joyner MJ. The principles of antibody therapy for infectious diseases with relevance for COVID‐19. Mbio. 2021;12:e03372‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Bloch EM, Feldweg AM. COVID‐19: convalescent plasma and hyperimmune globulin. UpToDate. Waltham, MA: UpToDate Inc. 2021. Accessed December 21, 2022. https://www.uptodate.com/contents/covid‐19‐convalescent‐plasma‐and‐hyperimmune‐globulin [Google Scholar]
- 194. Lloyd EC, Gandhi TN, Petty LA. Monoclonal antibodies for COVID‐19. Jama. 2021;325:1015‐1015. [DOI] [PubMed] [Google Scholar]
- 195. Manis JP, Feldweg AM. Overview of therapeutic monoclonal antibodies. UpToDate. Waltham, MA: UpToDate Inc. Accessed December 21, 2022. https://www.uptodate.com/contents/overview‐of‐therapeutic‐monoclonal‐antibodies/print [Google Scholar]
- 196. Xia S, Wang L, Zhu Y, et al. Origin, virological features, immune evasion and intervention of SARS‐CoV‐2 Omicron sublineages. Signal Transduct Target Ther. 2022;7:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Not applicable.


