Abstract
Treatment of severe cases of coronavirus disease 2019 (COVID-19) is extremely important to minimize death and end-organ damage. Here we performed a proteomic analysis of plasma samples from mild, moderate and severe COVID-19 patients. Analysis revealed differentially expressed proteins and different therapeutic potential targets related to innate immune responses such as fetuin-A, tetranectin (TN) and paraoxonase-1 (PON1). Furthermore, protein changes in plasma showed dysregulation of complement and coagulation cascades in COVID-19 patients compared to healthy controls. In conclusion, our proteomics data suggested fetuin-A and TN as potential targets that might be used for diagnosis as well as signatures for a better understanding of the pathogenesis of COVID-19 disease.
Keywords: Innate immunity, Mass spectrometry, Proteomics, Fetuin-A, Tetranectin, COVID-19
Graphical abstract
Highlights
-
•
Proteomic analysis revealed differentially expressed proteins associated with COVID-19 disease severity.
-
•
Down regulation of Fetuin-A, Tetranectin (TN) and Paraoxonase-1 (PON1) are correlated with disease severity.
-
•
Dysregulation of complement and coagulation cascades in COVID-19 patients compared to healthy controls.
1. Introduction
Severe acute respiratory syndrome, coronavirus 2 (SARS-CoV-2), caused coronavirus disease 2019 (COVID-19) [1]. COVID-19 disease is characterized by different clinical phenotypes ranging from asymptomatic, mild, and moderate to severe or extremely severe ones, characterized by sever pneumonia and high risk of death in the elderly population especially in patients over 70 years old [2,3]. Vaccines were generated late in 2020 and most people worldwide were vaccinated with at least 2 doses. Despite the decrease in the number of infected subjects as well as the severe cases reported, the problem is so far from being resolved and the cause of the progression of the disease to severe condition remains unknown. Furthermore, SARS-CoV-2 like other viruses change over time and new variants may have a modification in original proprieties such as the spread and associated disease severity and become less (omicron variant) [4] or more (delta variant) [5] severe. The potential emergence of new variants is still possible and understanding the mechanism of pathways behind COVID-19 disease is very critical.
It is well known that the dysfunction of immune responses is the major issue against clearance of the virus and recovery from the illness. Additionally, innate immune responses are considered the key factors in modulating the progress of the disease towards asymptomatic, mild, moderate, severe or extremely severe clinical phenotypes [6,7]. Neutrophils and monocyte/macrophages are the crucial frontline defense elements driving the innate immune responses during infection [8,9]. SARS-CoV-2 spike protein might activate macrophages directly via the ACE2 receptor present in these cells [10]. Throughout infection, immune cells released several proteins, cytokines and mediators to regulate inflammation and maintain the physiological homeostasis of the body. Plasma-based proteomics is a practical approach to analyze protein profile and to sort out changes associated with pathophysiologic conditions or disease progression [11]. Different proteomics studies were performed to identify and characterize proteome profiles in different COVID-19 cohorts using tissues [12,13], cells [14] or plasma [15,16]. Hence, we applied plasma-based proteomics to identify potential innate immune system-associated proteins relevant to COVID-19 disease and severity progression. We aim to study the pattern of plasmatic protein expression in healthy and COVID-19-infected patients and to evaluate potential interaction with innate immune responses.
2. Results
2.1. Plasma proteomic profiles
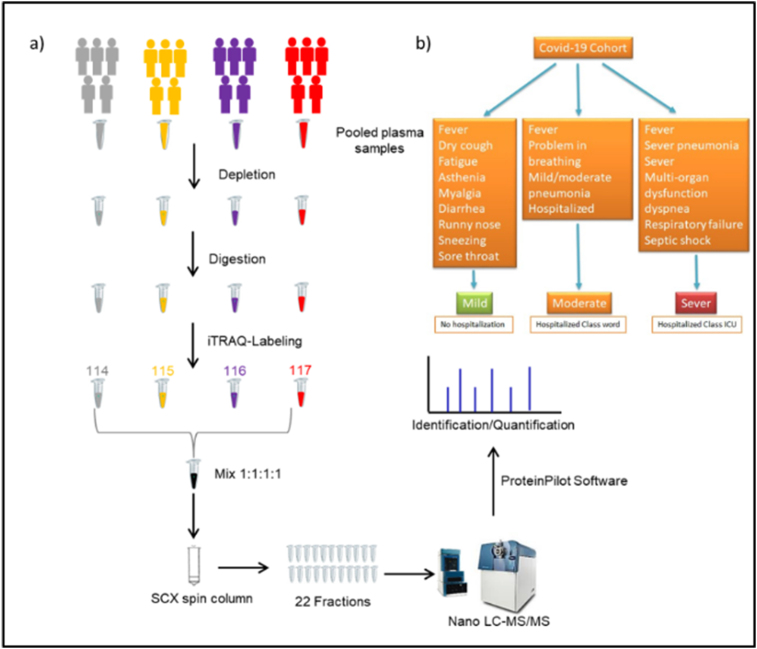
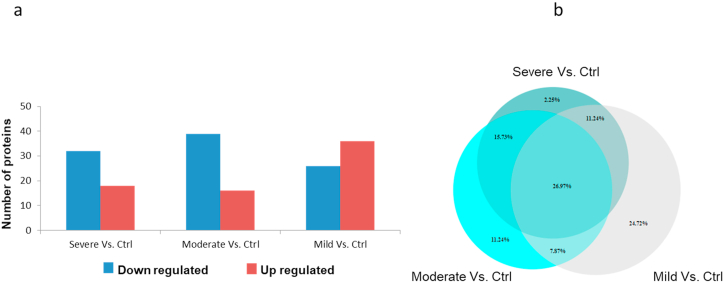
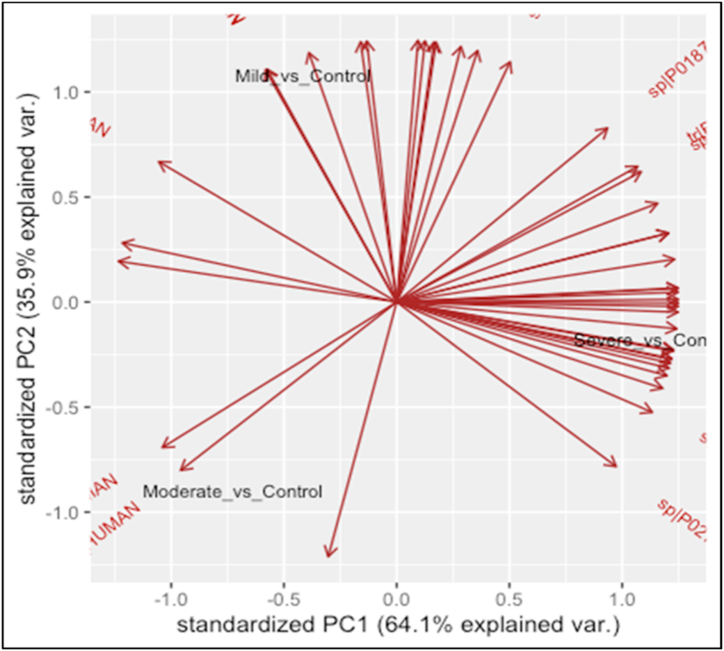
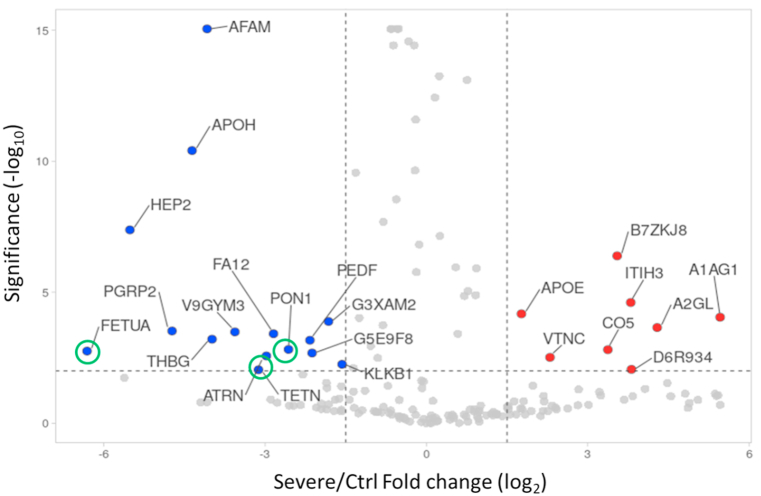
To assess the host responses of SARS-CoV-2, a plasma proteomics analysis of a cohort of COVID-19 patients and healthy controls was performed. In our study, iTRAQ 4plex method was carefully chosen over other proteomics labelled methods due to the high reproducibility and proteome coverage [17]. Moreover, for a better depth of total proteome coverage, we combined strong cation exchange peptide fractionation to iTRAQ. Illustration of the study workflow is presented in Fig. 1. A total of 176 proteins were identified in all COVID-19 and healthy control plasma samples with a false discovery rate of <1% at protein level. Nevertheless, our prime interest was to identify biomarkers and investigate the pathogenesis of COVID-19. Consequently, differentially expressed proteins analysis was performed for healthy control versus other COVID-19 groups (severe, moderate, and mild) by applying pairwise comparison. Supplementary Table S1 shows a list of differentially expressed proteins in all pairwise comparisons including ‘severe v. Control’, ‘moderate v. Control’, and ‘mild v. Control’. The statistical criteria for the quantitative analysis were set with a cutoff for fold change with >1.20 for up-regulation and <0.83 for down-regulation and a p-value of <0.05 (Supplementary Table S2). Results revealed that the number of proteins that altered among groups of comparisons varied as shown in Fig. 2a. Comparative results showed that 27% of the differentially expressed protein measurements overlap in all three groups (Fig. 2b). The Venn diagram also displayed that the number of unique proteins in the pairwise compassion of mild versus control has a higher number compared to other groups. This does not necessarily reflect the differences at the protein identification level but the quantitative level considering that the statistical criteria were selected for the fold change and p-value. To assess whether the differentially expressed protein measurements for all comparisons can be distinguished among COVID-19 groups or not, PCA analysis was applied. Interestingly, results showed that there was a clear distinction among COVID-19 groups as shown in Fig. 3. Furthermore, differentially expressed proteins have been presented by creating a volcano plot for all the pairwise comparisons of healthy control versus COVID-19 groups (Fig. 4). Several of the differentially expressed proteins in our dataset have been previously reported as biomarkers for COVID-19 [16,18].
Fig. 1.
Illustration of the plasma proteomics study design for healthy controls and COVID-19 patients. Clinical characterization of cohort study. a) Illustration of the plasma proteomics study design for healthy controls and COVID-19 patients. b) Flowchart of the clinical strategy for characterization of covid-19 symptoms and category.
Fig. 2.
Differentially expressed protein analysis. (a) Bar chart shows the number of up and down regulated proteins in different pairwise comparisons. Red color indicated the up regulated proteins and blue color for down regulation. (b) Venn diagram shows the unique and common percentage of differentially expressed protein in all three groups.
Fig. 3.
Principal Component Analysis (PCA) of the mild, moderate, and severe patient groups using the identified proteins. The data underlying the PCA analysis is the fold change values for the identified proteins in different patient groups compared to the normal control. PCA was done using R statistical environment. PC1 and PC2 provide the maximum separation in two-dimensional space among the mild, moderate, and severe covid-19 patients. The arrows show the projections of each variable (identified proteins) in two dimensional space.
Fig. 4.
Volcano plot shows differentially expressed protein. Red dot indicate the significant up-regulated proteins and blue for down regulated proteins. Gray dot indicate not significant and unregulated proteins. Green circle for target proteins (FETUA, TETN, and PON1) implicated in innate immune responses.
Gene ontology and pathway analysis using the online tool ‘STRING’ was performed on the differentially expressed protein measurements for healthy control and COVID-19 patients regardless of the group they are. For go enrichment analysis, the majority of differentially expressed proteins were found to be involved in biological regulation, extracellular region, and molecular function regulator for the biological process, cellular complement, and molecular function respectively (supplementary material, Fig. S1). Furthermore, we have studied the differentially expressed proteins and their involvement in pathway analysis using the KEGG database. The statistical criteria of the enrichment analysis were chosen to be p < 0.01 and the ranking of pathway terms was based on fold enrichment as shown in the supplementary material (Fig. S2). Throughout infection, damaged and dead cells initiate large immune responses and release different cell components and endotoxins that activate the blood coagulation cascade [19]. Activation of such pathways leads to endothelial dysfunction and inflammation as well as stimulation of leucocyte migration and tissue infiltration, thus triggering activation of innate immune local responses [20]. In our study, the complement system which is the central modulator of innate immunity and coagulation cascade pathways was found to be the most significant one among other pathway terms. Several proteomics studies on plasma from COVID-19 patients showed that the complement and coagulation cascades were one of the most enriched pathways [21,22]. However, the focus in our discussion is on three proteins fetuin-A, TN and PON-1 as potential new targets involved in innate immune responses to SARS-COV-2 infection and severity of COVID-19 disease.
2.2. Plasma levels of TNFα are associated with COVID-19 disease symptoms
Cytokine storm induced by Sars-CoV2 is one signature of severe cases in COVID-19 disease [23]. It was reported that over-release of TNFα is associated with the severity and poor prognosis of patients with COVID-19 disease [24,25]. In our study, measurement of plasma levels of TNFα showed an expected increase in COVID-19 groups compared to healthy controls corroborated with further increase in severe samples compared to moderate or mild samples.
3. Discussion
Several untargeted proteomics studies have been employed to investigate the change in COVID-19 serum/plasma samples. The number of identified proteins from these studies varies because of the different methods that were used. Label-free quantification approach using data-independent acquisition method displayed higher plasma proteome coverage [26]. For labeling-based proteomic approach, Orbitrap LC-MS/MS-based TMT quantitation method has shown higher plasma/serum proteome coverage with the number of identified proteins reaching 900 [27]. In our study, the total number of identified proteins was 176, which is very close to two published studies used the same mass analyzer (TripleTOF) where they identified 179 and 189 proteins [28,29]. Circulated proteins in plasma of COVID-19 patients present a window for detection of disease progress and severity, as well as signature of inflammation and innate and adaptive immune responses.
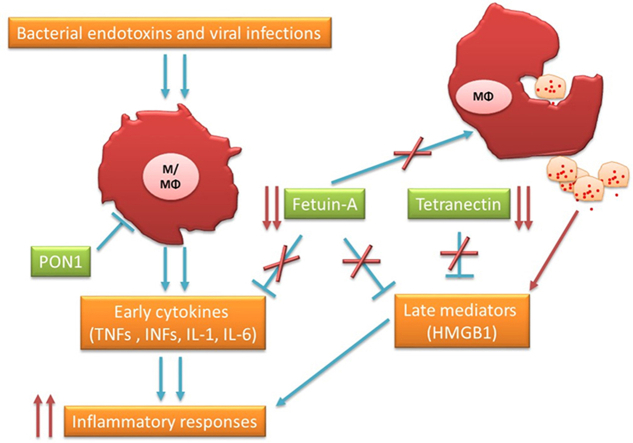
Production and over-release of proinflammatory cytokines are considered the key point in shifting the clinical phenotype from moderate to severe status in COVID-19 patients. In the course of infection, innate immune responses are modulated by damage-associated molecular patterns (DAMPs) or pathogen-associated molecular patterns (PAMPs) [30]. Acute phase proteins (APP) such as fetuin‐A are systematically released in plasma to support the anti-inflammatory process during inflammation. In our study, we observe a clear down-regulation of plasmatic fetuin‐A in moderate and severe cases compared to healthy controls (Fig. 4). It was demonstrated that fetuin‐A may inhibit early (TNF, IL-1, IL-6 and INF) and late (HMGB1) mediators released by innate immune cells such as macrophages [31]. Analysis of TNFα in our COVID-19 cohort indicates a trend of increase in the plasmatic concentration compared to healthy donors which are more potentiated in severe cases (supplementary material, Fig. S3). During inflammation, activated monocyte/macrophages released pro-inflammatory cytokines which are attenuated by a ubiquitous biogenic amine called spermine [32]. However, spermine anti-inflammatory effects are dependent on fetuin-A. Thus spermine could not inhibit the function of monocyte/macrophage fetuin-A deficient cells [33]. Furthermore, a decrease in the fetuin-A level might be related to vascular abnormalities and thrombosis detected in extremely severe cases due to an increase in vascular calcification and endothelial dysfunction [34,35] and oxidative stress [36] associated with fetuin-A levels.
After viral infection, cell apoptosis is one of the hosts' defence mechanisms. However, over-accumulation of apoptotic cells for the reason of defects in apoptotic cell clearance is associated with hyperinflammation and autoimmunity [37]. Decrease of fetuin-A plasmatic level could be at least in part involved in this process as it increases the phagocytosis capacity to clean apoptotic cells by macrophages [38] and vesicles by VSMC [39].
Recently it was reported that fetuin-A plays a crucial role not only as a calcification inhibitor but as a multi-protective component by reducing fibrosis and inflammation and modulating macrophage polarization [40].
Rudloff et al. demonstrated that fetuin-A is a HIF target gene that has a protective role in hypoxia-induced kidney dysfunction [40]. Fetuin-A attenuates macrophage calcium overload due to hypoxia. We recently demonstrated that SARS-CoV-2 coronavirus spike protein activates thp-1-like macrophages and increases intracellular calcium levels [41]; thus fetuin-A might play the role of calcium scavenger to regulate cell homeostasis and modulate adequate polarization for reasonable innate immune responses, thereby avoiding over-inflammation.
Multiple organ dysfunction syndromes (MODS) or infection-related MODS are considered one of the major causes of death in COVID-19 severe cases [42]. Hyperinflammation and immune dysregulation during infection lead to end organ damage due to failure of innate immune response and hypercytokinemia. Over systemic infection, bacterial endotoxins and viral RNAs are recognized by the innate immune system which triggers the expression and release of early cytokines (TNF and INFs). Furthermore, the front-line innate immune responses stimulate the production of late mediators such as high-mobility group box 1 (HMGB1) [43]. Intra or extracellular HMGB1 expressed in most cells is an alarmin released upon stimulation or after the death of the cell [44]. Hyper-release of HMGB1 triggers cytokine production such as IL-6 [45]. Increased IL-6 concentration was associated with severe outcomes in multisystem inflammatory syndrome in children (MIS-C) related to SARS-CoV-2 infection [46]. IL-6 is secreted mainly by monocytes/macrophages specifically when polarized to an M1-like phenotype. It was demonstrated also that HMGB1 may polarize macrophages via Toll-like receptor (TLR) 4 [47].
Tetranectin (CLEC3B) is one of the HMGB1-binding proteins and its interaction might modulate the biological function and inflammatory process during infection. Highly purified TN protein inhibited dose-dependently LPS-induced HMGB1 release in macrophages [48]. Dysregulation of the innate immune system leading to immune over-activation is a common future of COVID-19 severe cases and sepsis. It was demonstrated recently that tetranectin blood concentration is dramatically decreased in extremely severe sepsis patients [48]. Recently, Yuming Li et al. [49] reported the downregulation of tetranectin in severe cases of COVID-19 patients using the label-free quantitative proteomics method. We confirmed the significantly plasmatic decrease of this protein in our study (Fig. 4) with different groups and based on the label-quantitative method. Thus tetranectin can be considered a potential marker of innate immune dysregulation associated with the severity of COVID-19 symptoms.
Tetranectin is involved in the fibrinolysis system leading to control of abnormal coagulation; thus downregulation of TN is associated with cardiovascular diseases such as coronary artery disease (CAD) [50]. Noteworthy, many severe cases of SARS-COV-2 infected patients present vascular dysfunction possibly due to TN deficiency. The decline of TN serum level is accompanied by a reduction of paraoxonase 1 (PON1) in different cardiovascular diseases. In our study, down-regulation of PON1 (Fig. 4) could be a marker of disease progression to severe symptoms as it was reported that the presence of inflammation and oxidative stress is associated with a decrease of serum PON1 in cardiovascular diseases [51]. Furthermore, a low level of PON1 may exacerbate the innate proinflammatory reaction to virus infection as it was demonstrated that PON1 suppresses macrophage overreaction and sustains inflammation in atherosclerosis [52]. Dysregulation of TN exerts influence on AMPK downstream effectors such as HIF1α that regulates phenotype and function of innate immune cells and modulates macrophage polarization towards M1-like phenotype as well as DC maturation [53,54].
Collectively, down-regulation of tetranectin, fetuin-A and PON1 observed in moderate and severe cases might be associated with dysregulation of innate immune responses through monocyte/macrophage dysfunction, leading to hyper-release of cytokines and progression of COVID-19 disease to severe status.
Additionally, our proteomic analysis revealed an up-regulation of APOE (supplementary material, Fig. S4) in severe cases compared to healthy controls. APOE plasmatic levels are known to modulate lipoprotein function and cardio-metabolic diseases. Furthermore, APOE is involved in the inflammatory process by switching macrophage polarization and affecting the pro-/anti-inflammatory phenotypes [55]. In agreement with this, it was reported that an increase in APOE levels might be associated with COVID-19 severity [56].
In the course of infection, macrophage-complement system interaction is important for innate immune responses as well as modulation of complement functions and transition from innate to adaptive immune conditions [57]. Complement component interactivity with different receptors on macrophages drives the inflammatory process and cytokine production. It was reported in different proteomics studies the dysregulation of complement cascade [58,59]in COVID-19 patients and that activation of complement system intensify T cell cytotoxicity in severe cases [60]. We reported a significant increase of complements 1q, 5, 6, 7 (supplementary material, Fig. S5) in moderate and severe COVID-19 patients. Over the process of apoptotic cell clearance, C1q control the monocyte/macrophage polarization and inhibit inflammasome activity [61]. Enrichment analysis showed a significant link between complement components C1q, C5, C6, C7, and coagulation cascades, where we find also the involvement of kallikrein-kinin system through kininogen-1 which modulates innate immune reactions through the bradykinin pathway.
The present study has a limitation in term of sample size. However, our results have shown a good agreement with previous proteomics covid-19 studies where we identified biomarkers show similar expression. This is show that the proteomics approach followed in this study is valid, but maybe not optimal.
4. Conclusion
During SARS-COV-2 infection, a disorder in immune responses is the major issue driving the escalation of COVID-19 disease to severe cases. Mechanistic pathways leading to innate immune dysfunction are the keystones in resolving the complications associated with uncontrolled SARS‐CoV‐2 infection. To the best of our knowledge, this is the first study to employ the iTRAQ approach on COVID-19 plasma/serum samples. Serum levels of TN, fetuin-A and PON1 are potential markers implicated at least in part in the innate immune system, specifically in macrophage hyper-activation, and, consequently, could be considered potential targets for COVID-19 treatment.
5. Materials and methods
5.1. Study design and population
Our study cohorts include 25 subjects: Healthy control (n = 5) and 20 SARS-COV-2 infected patients grouped according to the disease severity (Fig. 1) into Mild (n = 5), Moderate (n = 5), Severe (n = 5), and Dead (n = 5). The dead group was excluded from the beginning of the study due to comorbidities of subjects that could affect the specificity of COVID-19 effects. All patients were recruited in King Abdallah Specialist Children Hospital (KASCH) and King Abdulaziz Medical City (KAMC). Informed consent was signed by all participants while they filled out a questionnaire about clinical symptoms and demographic data.
Blood collection: 3–5 ml of venous blood was collected from each patient. EDTA tubes were used to collect blood samples; then plasma was separated by centrifugation before storage at 80 °C for further analysis.
5.2. iTRAQ proteomics plasma sample preparation
Five biological samples from each group were pooled in one sample. All plasma samples were then subjected to the Agilent Human-14 Multiple Affinity Removal Spin cartridge to remove the most 14 abundant proteins from plasma following the manufacture protocol (Agilent, USA). Afterwards, protein digests were labelled using 4-plex iTRAQ reagents (Sciex, Canada) following the manufacture protocol with minor modifications. In short, 100 μg proteins were denatured, reduced and alkylated using 8 M urea/100 mM Triethylammonium bicarbonate (TEAB), 50 mM Tris (2-carboxyethyl) phosphine (TCEP), and methyl methanethiosulfonate (MMTS) respectively. Protein samples were then digested into peptides using trypsin with a ratio of 1:40 (trypsin toprotein). Subsequently, peptide samples were labelled with 114, 115, 116, and 117 for healthy control, severe, moderate, and mild respectively. All 4 labelled samples were then mixed and pooled in one sample.
5.3. Peptides fractionation and desalting
The iTRAQ pooled sample was fractionated offline based on cation exchange using Pierce Strong Cation Exchange (SCX) spin column (Thermo Scientific, USA) following the manufacture instruction. In brief, the spin column was first conditioned using 0.1% formic acid followed by loading the labelled peptides into the column. Two steps of washing were performed using 0.1% formic acid. Peptides were then eluted into 22 fractions by changing the ratio of 10% Acetonitrile and Ammonium formate concentration from 15 mM to 900 mM. Subsequently, the desalting and cleanup step was performed by polymeric reversed-phase sorbent using Oasis HLB Cartridge (Waters, USA). In short, the cartridge was conditioned and equilibrated with Acetonitrile and 0.1% formic acid respectively. Labelled peptides were then loaded and two steps of wash with 0.1% formic acid were applied. The elution of peptides was performed using 70% acetonitrile.
5.4. NanoLC-MS/MS
Each fraction (total of 22) was loaded on an Ekspert nanoLC 425 (Eksigent, Dublin, CA, USA), coupled to a TripleTOF 5600 (Sciex, Concord, Canada) in triplicate. The separation of peptides on NanoLC was performed as described previously [41]. The MS1 and MS/MS for iTRAQ analysis were set as follows: TOF mass range 250–1500 with accumulation time 0.25 s. IDA criteria was for ions greater than 400 m/z with charge state of 2–5. The abundance threshold exceeds 200 cps. The adjust CE when iTRAQ reagent parameter was checked. The ion source parameters were set as follow: ion source gas 1 (GS1), 25; Curtain Gas (CUR), 25; IonSpray Voltage Floating (ISVF), 2250; Interface Heater Temperature (HIT), 60.
5.5. iTRAQ quantitative proteomics analysis
The raw MS iTRAQ data were processed using ProteinPilot Software (v 4.5, Sciex) for identification and quantification. The parameters were set as follows: sample type, iTRAQ 4plex, Cys, Alkylation, MMTS, Digestion, Trypsin, Special factors, Urea denaturation, Species, Homo sapiens, Database, uniprot Proteome Humans database (downloaded on October 29, 2019); globalFDR was checked. Fold change and p-value were calculated based on the average of peptide intensity and Student’s t-test respectively. The iTRAQ MS data was deposited in ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with identifier PXD037400.
5.6. Bioinformatics analysis
PCA analysis was performed using ‘princop’ function in R statistical software. VolcaNoseR [62] was used to generate the volcano plots. STRING online tool [63] was used for Gene enrichment and pathway analysis with statistical criteria set at p < 0.05. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was applied for the pathway analysis.
5.7. Enzyme-linked immunosorbent assays (ELISA) measurements
Samples were prepared as previously described [64] using Tumour necrosis factor α (TNF-α) and ELISA assay kit (SEKH-0047, Solarbio life sciences). Briefly, 100 μl of samples or standards were added to each well and then incubated at 37 °C for 1 h 30 min. After washing four times with washing buffer, Biotin-Conjugated detection antibody (100 μl) was added before incubation for 1 h at 37 °C. A second washing step (four times) was performed and Streptavidin-HRP (100 μl) was added before incubation for 30 min. After washing, a Substrate solution (TMB) (100 μl) was added and incubated for 15 min in dark before adding stop solution, and measurement of absorbance was detected at 450 nm.
Funding
This article was supported by a grant from King Abdullah International Research Center no. RC20/153/R and RC20/205/R and BioBank KAIMRC NGHA.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (IRB) of King Abdullah International Medical Research Center (KAIMRC) Ethics Committee with IRB RC20/153/R and RC20/205/R. Written informed consent was obtained from all participants in the study.
Authors' contributions
B.A: Conceived and designed the experiments and analyzed and interpreted the data. F.A.M, H·S, K.A and H.A: Performed the experiments. F.A, H.S.A and M.R: Analyzed and interpreted the data. F.A, M.J., A.H and M.B: Contributed reagents, materials, analysis tools or data and wrote the paper. T.B: Conceived and designed the experiments and analyzed and interpreted the data.
Data availability statement
Data associated with this study has been deposited at The iTRAQ MS data was deposited in ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with identifier PXD037400.
Declaration of interest’s statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP-2023/17) at King Saud University, Riyadh, Saudi Arabia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e15224.
Contributor Information
Bandar Alghanem, Email: ghanemba@ngha.med.sa.
Tlili Barhoumi, Email: barhoumitl@ngha.med.sa.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
List of differentially expressed proteins in all pairwise comparisons ‘severe v. Control’, ‘moderate v. Control’, and ‘mild v. Control'.
Multimedia component 3Figure S1. The bar chart shows the go-enrichment analysis for pairwise comparison of healthy control against COVID-19 including biological process, cellular components, and molecular function. Figure S2. KEGG enrichment analysis of differentially expressed proteins. The statistical criteria of the enrichment analysis were set as p < 0.01.Figure S4 Apolipoprotein E expression. Severe (ICU), Moderate (Ward) and Mild (quarantine) p < 0.01. Figure S5 Complement component expression. Severe (ICU), Moderate (Ward) and Mild (quarantine) p < 0.01. Figure S3. Soluble TNF-α concentrations in the serum of healthy (Ctrl) subjects, Mild, hospitalized (Moderate) and ICU (Severe) COVID-19 patients. (n = 3 to 4).
References
- 1.Almutlaq M., et al. Classical and counter-regulatory renin-angiotensin system: potential key roles in COVID-19 pathophysiology. CJC Open. 2021;3(8):1060–1074. doi: 10.1016/j.cjco.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccarelli M., et al. Untargeted lipidomics reveals specific lipid profiles in COVID-19 patients with different severity from Campania region (Italy) J. Pharm. Biomed. Anal. 2022;217 doi: 10.1016/j.jpba.2022.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., et al. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603(7902):587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiello T.F., et al. Infection with the Omicron variant of SARS-CoV-2 is associated with less severe disease in hospitalized patients with COVID-19. J. Infect. 2022;85(5):e152–e154. doi: 10.1016/j.jinf.2022.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Wintersdorff C.J.H., et al. Infections with the SARS-CoV-2 Delta variant exhibit fourfold increased viral loads in the upper airways compared to Alpha or non-variants of concern. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-18279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Munoz A.D., et al. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Sci. Adv. 2022;8(31):eabp9770. doi: 10.1126/sciadv.abp9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso M.S., et al. Physical interactions with bacteria and Protozoan parasites establish the scavenger receptor SSC4D as a broad-spectrum pattern recognition receptor. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.760770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei X., et al. EDIL3 deficiency ameliorates adverse cardiac remodelling by neutrophil extracellular traps (NET)-mediated macrophage polarization. Cardiovasc. Res. 2022;118(9):2179–2195. doi: 10.1093/cvr/cvab269. [DOI] [PubMed] [Google Scholar]
- 10.Cao X., et al. 2020. Spike Protein of SARS-CoV-2 Activates Macrophages and Contributes to Induction of Acute Lung Inflammations in Mice. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angel T.E., et al. Mass spectrometry-based proteomics: existing capabilities and future directions. Chem. Soc. Rev. 2012;41(10):3912–3928. doi: 10.1039/c2cs15331a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T., et al. Proteomic and metabolomic characterization of SARS-CoV-2-infected cynomolgus macaque at early stage. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.954121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie X., et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184(3):775–791 e14. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.julian.knight@well.ox.ac.uk, C.O.-M.-o.B.A.C.E.a. and C.O.-M.-o.B.A. Consortium A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell. 2022;185(5):916–938 e58. doi: 10.1016/j.cell.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammed Y., et al. Longitudinal plasma proteomics analysis reveals novel candidate biomarkers in acute COVID-19. J. Proteome Res. 2022;21(4):975–992. doi: 10.1021/acs.jproteome.1c00863. [DOI] [PubMed] [Google Scholar]
- 17.Casey T.M., et al. Analysis of reproducibility of proteome coverage and quantitation using isobaric mass tags (iTRAQ and TMT) J. Proteome Res. 2017;16(2):384–392. doi: 10.1021/acs.jproteome.5b01154. [DOI] [PubMed] [Google Scholar]
- 18.Shu T., et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53(5):1108–1122 e5. doi: 10.1016/j.immuni.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen G.Y., Nunez G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 2010;10(12):826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esmon C.T., Xu J., Lupu F. Innate immunity and coagulation. J. Thromb. Haemostasis. 2011;9(Suppl 1):182–188. doi: 10.1111/j.1538-7836.2011.04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J., et al. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-80120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messner C.B., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11–24 e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Valle D.M., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danlos F.X., et al. High levels of TNFalpha in patients with COVID-19 refractory to tocilizumab. Eur. J. Cancer. 2021;149:102–104. doi: 10.1016/j.ejca.2021.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., et al. Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge. Cell Death Dis. 2022;13(3):235. doi: 10.1038/s41419-022-04674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen B., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72 e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarif J., et al. Plasma gradient of soluble Urokinase-type plasminogen activator receptor is linked to pathogenic plasma proteome and immune transcriptome and stratifies outcomes in severe COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.738093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villar M., et al. Characterization by quantitative serum proteomics of immune-related prognostic biomarkers for COVID-19 symptomatology. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.730710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Sama A.E. Anti-inflammatory role of fetuin-A in injury and infection. Curr. Mol. Med. 2012;12(5):625–633. doi: 10.2174/156652412800620039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., et al. A hepatic protein, fetuin-A, occupies a protective role in lethal systemic inflammation. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M., Wang H., Tracey K.J. Regulation of macrophage activation and inflammation by spermine: a new chapter in an old story. Crit. Care Med. 2000;28(4 Suppl):N60–N66. doi: 10.1097/00003246-200004001-00007. [DOI] [PubMed] [Google Scholar]
- 33.Wang H., et al. Fetuin protects the fetus from TNF. Lancet. 1997;350(9081):861–862. doi: 10.1016/S0140-6736(05)62030-2. [DOI] [PubMed] [Google Scholar]
- 34.Ketteler M., et al. Association of low fetuin-A (AHSG) concentrations in serum with cardiovascular mortality in patients on dialysis: a cross-sectional study. Lancet. 2003;361(9360):827–833. doi: 10.1016/S0140-6736(03)12710-9. [DOI] [PubMed] [Google Scholar]
- 35.Caglar K., et al. Serum fetuin-a concentration and endothelial dysfunction in chronic kidney disease. Nephron Clin. Pract. 2008;108(3):c233–c240. doi: 10.1159/000120209. [DOI] [PubMed] [Google Scholar]
- 36.El-Malkey N.F., et al. Fetuin-A exerts a protective effect against experimentally induced intestinal ischemia/reperfusion by suppressing autophagic cell death. Exp. Biol. Med. (Maywood) 2021;246(11):1307–1317. doi: 10.1177/1535370221995207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon I.K., et al. Apoptotic cell clearance: basic biology and therapeutic potential. Nat. Rev. Immunol. 2014;14(3):166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jersmann H.P., Dransfield I., Hart S.P. Fetuin/alpha2-HS glycoprotein enhances phagocytosis of apoptotic cells and macropinocytosis by human macrophages. Clin. Sci. (Lond.) 2003;105(3):273–278. doi: 10.1042/CS20030126. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds J.L., et al. Multifunctional roles for serum protein fetuin-a in inhibition of human vascular smooth muscle cell calcification. J. Am. Soc. Nephrol. 2005;16(10):2920–2930. doi: 10.1681/ASN.2004100895. [DOI] [PubMed] [Google Scholar]
- 40.Rudloff S., et al. Fetuin-A is a HIF target that safeguards tissue integrity during hypoxic stress. Nat. Commun. 2021;12(1):549. doi: 10.1038/s41467-020-20832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barhoumi T., et al. SARS-CoV-2 coronavirus spike protein-induced apoptosis, inflammatory, and oxidative stress responses in THP-1-like-macrophages: potential role of angiotensin-converting enzyme inhibitor (perindopril) Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.728896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Roquetaillade C., et al. Timing and causes of death in severe COVID-19 patients. Crit. Care. 2021;25(1):224. doi: 10.1186/s13054-021-03639-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu C.S., et al. Endogenous regulation and pharmacological modulation of sepsis-induced HMGB1 release and action: an updated Review. Cells. 2021;10(9) doi: 10.3390/cells10092220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H., Wang H., Andersson U. Targeting inflammation driven by HMGB1. Front. Immunol. 2020;11:484. doi: 10.3389/fimmu.2020.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamiya N., Kim H.K. Elevation of proinflammatory cytokine HMGB1 in the synovial fluid of patients with Legg-calve-perthes disease and correlation with IL-6. JBMR Plus. 2021;5(2) doi: 10.1002/jbm4.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abrams J.Y., et al. Factors linked to severe outcomes in multisystem inflammatory syndrome in children (MIS-C) in the USA: a retrospective surveillance study. Lancet Child Adolesc. Health. 2021;5(5):323–331. doi: 10.1016/S2352-4642(21)00050-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu H., et al. Transcriptomic profiling reveals that HMGB1 induces macrophage polarization different from classical M1. Biomolecules. 2022;12(6) doi: 10.3390/biom12060779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W., et al. Identification of tetranectin-targeting monoclonal antibodies to treat potentially lethal sepsis. Sci. Transl. Med. 2020;12(539) doi: 10.1126/scitranslmed.aaz3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., et al. Multi-platform omics analysis reveals molecular signature for COVID-19 pathogenesis, prognosis and drug target discovery. Signal Transduct. Targeted Ther. 2021;6(1):155. doi: 10.1038/s41392-021-00508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y., et al. Tetranectin as a potential biomarker for stable coronary artery disease. Sci. Rep. 2015;5 doi: 10.1038/srep17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saril A., et al. Serum proteomic changes in dogs with different stages of chronic heart failure. Animals (Basel) 2022;12(4) doi: 10.3390/ani12040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aharoni S., Aviram M., Fuhrman B. Paraoxonase 1 (PON1) reduces macrophage inflammatory responses. Atherosclerosis. 2013;228(2):353–361. doi: 10.1016/j.atherosclerosis.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 53.McGettrick A.F., O’Neill L.A.J. The role of HIF in immunity and inflammation. Cell Metabol. 2020;32(4):524–536. doi: 10.1016/j.cmet.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Dai W., et al. Downregulation of exosomal CLEC3B in hepatocellular carcinoma promotes metastasis and angiogenesis via AMPK and VEGF signals. Cell Commun. Signal. 2019;17(1):113. doi: 10.1186/s12964-019-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tudorache I.F., Trusca V.G., Gafencu A.V. Apolipoprotein E - a multifunctional protein with implications in various pathologies as a result of its structural features. Comput. Struct. Biotechnol. J. 2017;15:359–365. doi: 10.1016/j.csbj.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo C.L., et al. APOE e4 Genotype Predicts Severe COVID-19 in the UK Biobank Community Cohort. J. Gerontol. A Biol. Sci. Med. Sci. 2020;75(11):2231–2232. doi: 10.1093/gerona/glaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bohlson S.S., et al. Complement, c1q, and c1q-related molecules regulate macrophage polarization. Front. Immunol. 2014;5:402. doi: 10.3389/fimmu.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Alessandro A., et al. Serum proteomics in COVID-19 patients: altered coagulation and complement status as a function of IL-6 level. J. Proteome Res. 2020;19(11):4417–4427. doi: 10.1021/acs.jproteome.0c00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doykov I., et al. Quantitative, multiplexed, targeted proteomics for ascertaining variant specific SARS-CoV-2 antibody response. Cell Rep. Methods. 2022;2(9) doi: 10.1016/j.crmeth.2022.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Georg P., et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell. 2022;185(3):493–512 e25. doi: 10.1016/j.cell.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benoit M.E., et al. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J. Immunol. 2012;188(11):5682–5693. doi: 10.4049/jimmunol.1103760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goedhart J., Luijsterburg M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-76603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szklarczyk D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alghamdi J., et al. Interferon-induced transmembrane protein-3 genetic variant rs12252 is associated with COVID-19 mortality. Genomics. 2021;113(4):1733–1741. doi: 10.1016/j.ygeno.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of differentially expressed proteins in all pairwise comparisons ‘severe v. Control’, ‘moderate v. Control’, and ‘mild v. Control'.
Multimedia component 3Figure S1. The bar chart shows the go-enrichment analysis for pairwise comparison of healthy control against COVID-19 including biological process, cellular components, and molecular function. Figure S2. KEGG enrichment analysis of differentially expressed proteins. The statistical criteria of the enrichment analysis were set as p < 0.01.Figure S4 Apolipoprotein E expression. Severe (ICU), Moderate (Ward) and Mild (quarantine) p < 0.01. Figure S5 Complement component expression. Severe (ICU), Moderate (Ward) and Mild (quarantine) p < 0.01. Figure S3. Soluble TNF-α concentrations in the serum of healthy (Ctrl) subjects, Mild, hospitalized (Moderate) and ICU (Severe) COVID-19 patients. (n = 3 to 4).
Data Availability Statement
Data associated with this study has been deposited at The iTRAQ MS data was deposited in ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with identifier PXD037400.