Abstract
Since its first demonstration, stimulated Raman scattering (SRS) microscopy has become a powerful chemical imaging tool that shows promise in numerous biological and biomedical applications. The spectroscopic capability of SRS enables identification and tracking of specific molecules or classes of molecules, often without labeling. SRS microscopy also has the hallmark advantage of signal strength that is directly proportional to molecular concentration, allowing for in situ quantitative analysis of chemical composition of heterogeneous samples with submicron spatial resolution and subminute temporal resolution. However, it is important to recognize that quantification through SRS microscopy requires assumptions regarding both system and sample. Such assumptions are often taken axiomatically, which may lead to erroneous conclusions without proper validation. In this review, we focus on the tacitly accepted, yet complex, quantitative aspect of SRS microscopy. We discuss the various approaches to quantitative analysis, examples of such approaches, challenges in different systems, and potential solutions. Through our examination of published literature, we conclude that a scrupulous approach to experimental design can further expand the powerful and incisive quantitative capabilities of SRS microscopy.
Keywords: stimulated Raman scattering, Raman microscopy, quantitative microscopy, chemical imaging
INTRODUCTION
Stimulated Raman scattering (SRS) microscopy was first demonstrated in 2008 by Freudiger et al. (1) as an alternative coherent Raman microscopy technique to the then-more-popular coherent anti-Stokes Raman scattering (CARS) microscopy. Both SRS and CARS microscopy use synchronized pulsed lasers to coherently excite the inherent chemical vibrations of molecules. This contrasts with so-called spontaneous Raman scattering, where a single (typically continuous wave) laser source focused on a sample will cause spontaneous emission of photons of different wavelengths (i.e., energies) corresponding to vibrational energy differences with respect to the original laser source. While spontaneous Raman scattering affords broad and readily interpretable spectral information about a sample, the probability of spontaneously emitted Raman photons is typically quite low with differential scattering cross-sections in the ~10−30 cm2 regime (2–4). Thus, if microscopy is desired via spontaneous Raman, acquisition times are often egregiously slow given the typical excitation wavelengths and maximum laser powers appropriate for biological samples. Coherent Raman scattering microscopies offer significantly faster and more sensitive Raman imaging via coherent nonlinear optical interactions, typically at the expense of broad spectral coverage and some spectral specificity (5, 6).
CARS microscopy has seen broad use in biophotonics research since its first demonstration but has largely fallen to the wayside of SRS microscopy (7–13). This is due largely to the shortcomings of CARS microscopy with respect to SRS microscopy. Specifically, CARS microscopy, though technically easier to implement and detect signal, suffers from a significant nonresonant background effect and spectral distortion (with respect to the spontaneous Raman spectrum). This nonresonant background and resultant spectral distortions (e.g., wavenumber shifts, peak broadening, peak intensity changes) make quantitative chemical analysis of CARS microscopy images difficult (14, 15).
SRS microcopy, in contrast, does not suffer from the nonresonant background issue. SRS signal arises from energy transfer between the two laser sources and the molecular vibration, with spectral features nearly identical to spontaneous Raman except for slight peak broadening related to the pulse duration and bandwidth of the lasers (16). Additionally, SRS microscopy offers the distinct advantage of signal strength that is linearly dependent on sample concentration. This affords SRS microscopy with fast, sensitive, label-free, and quantitatively intuitive detection of chemically specific imaging contrast of samples. Moreover, SRS microscopy is well suited for biological samples because Raman scattering of water vibrations is relatively weak and the typically used near infrared picosecond laser sources cause minimal photodamage due to heating and multiphoton ionization. Indeed, SRS microscopy has found its largest use in biophotonics as a label-free yet chemically specific alternative to more ubiquitously adopted fluorescence microscopy techniques. While fluorescence microscopy is a valuable tool with a relatively low barrier to utilize, the use of exogenous fluorophores has some commonly cited shortcomings (6, 12, 17–20), including label specificity, cytotoxicity, photobleaching, and color cross talk. SRS microscopy has found utility in a wide variety of biological applications, such as imaging pharmacokinetics (21–25), cellular and physiological metabolism (26–30), antibiotic susceptibility and resistance (31, 32), and histopathological diagnosis (33–36). Although such experiments could also be performed using fluorescent labels, the value of a label-free method that also provides quantitative insight is highly desirable for modern biological studies.
The growing popularity of SRS microscopy has led to an explosive growth of publications in recent years. There are abundant literature reviews dedicated to the discussion of coherent Raman scattering microscopy (9–12, 37–44). Instead of summarizing recent advances in SRS instrumentation and applications, we take a different approach and focus on an underappreciated aspect: quantification in SRS microscopy. With the combination of spatial information and vibrational chemical information acquired in SRS, even complex and temporally dynamic samples can be observed with quantitative insight. However, there are many pitfalls associated with quantification owing to complications such as sample scattering and non-Raman background. The problem is analogous to one in fluorescence microscopy: while it is simple to compare the intensity of two fluorescent specimens, quantitation of concentration or other molecular properties is error prone and requires a thorough understanding of limitations and the careful use of proper methods (45–47). A detailed discussion of SRS-based quantitative analysis is missing in the literature. In this article, we focus on the common quantitative metrics afforded by SRS microscopy. The review is organized based on the five main categories of quantitative measurements in SRS microscopy based on a Web of Science search for “quantitative/quantify/quantification” + “SRS”:
Morphological measurements such as size, shape, or count of objects in a field of view;
Relative concentration measurements, where SRS signal strength is taken as a proxy for molecular concentration;
Absolute concentration measurements, where SRS signal is calibrated with respect to an analyte such that exact spatially resolved concentrations can be calculated;
Ratiometric measurements, where SRS signals from different vibrational peaks are proportionally compared to provide compositional insight; and
Spectrally dynamic measurements, where a vibrational peak (typically of a sensor molecule) shifts in wavenumber in response to changes in the microenvironment.
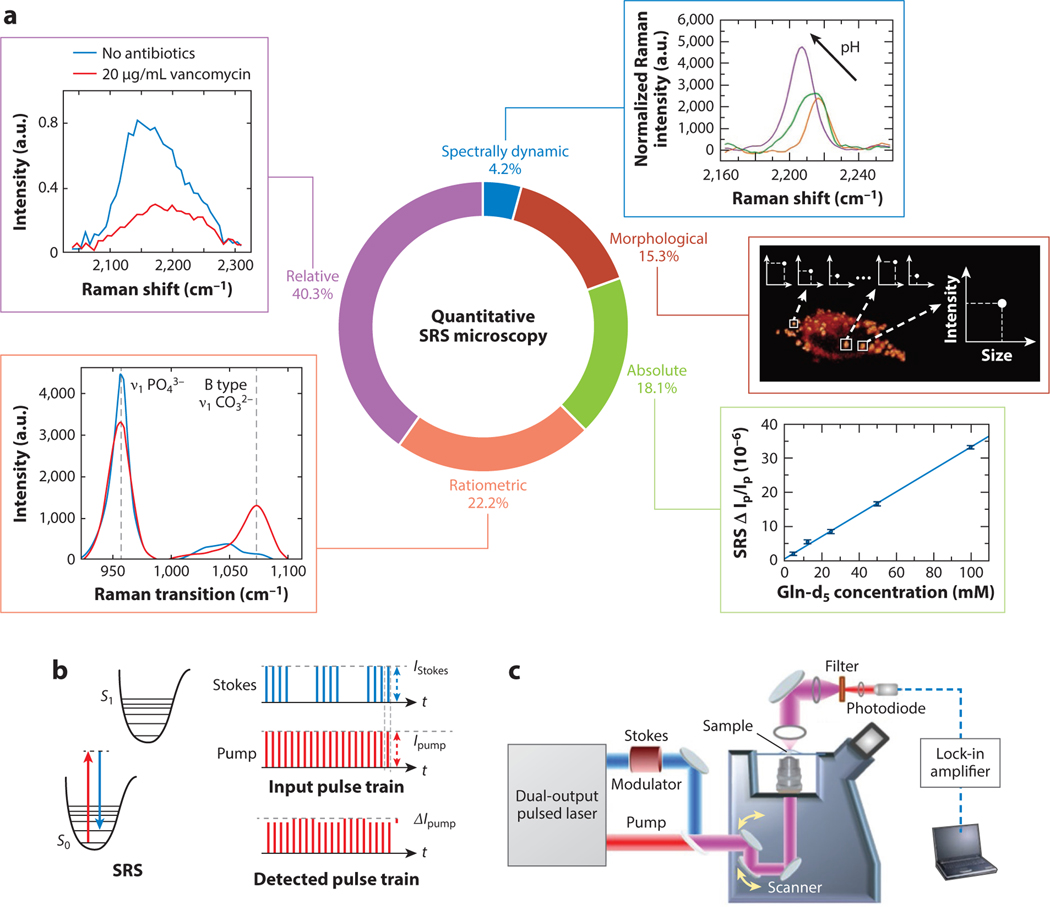
The relative prevalence of these five categories based on the search is shown in Figure 1a. We explore these five categories of quantitative SRS microscopy and discuss their respective prevalence, utility, and potential pitfalls or shortcomings. For each category, we highlight only a few example applications to demonstrate the relevant method for quantitative analysis.
Figure 1.
Types of quantitative stimulated Raman scattering (SRS) measurements and principles of SRS microscopy. (a) The five types of quantitative SRS measurements discussed in this review, representative figures for each, and their relative proportions in the literature based on a Web of Science search for “quantitative/quantify/quantification” + “SRS.” Percentages represent 73 publications in total. (b) Energy level diagram for the SRS process (left) and the pulse train modulation detection scheme commonly used for SRS microscopy (right). (c) A representative schematic diagram for a basic SRS microscope. In panel a, the representative spectrally dynamic subpanel adapted with permission from Reference 105; copyright 2021 American Chemical Society. Morphological subpanel adapted with permission from Reference 66; copyright 2016 American Chemical Society. Absolute subpanel adapted with permission from Reference 82; copyright 2020 American Chemical Society. Ratiometric subpanel adapted with permission from Reference 103; copyright 2020 Nature Publishing Group. Relative subpanel adapted with permission from Reference 31; copyright 2018 American Chemical Society.
PRINCIPLES OF QUANTITATIVE STIMULATED RAMAN SCATTERING MICROSCOPY AND POTENTIAL CHALLENGES
Specific implementations of SRS microscopy have been extensively discussed in previous articles and reviews (1, 6, 41, 48). Briefly, pulsed laser sources produce synchronized pump and Stokes beams. The Stokes pulse train is amplitude modulated at >1 MHz frequency, as shown in Figure 1b. The pulse trains are spatially and temporally overlapped and aligned through a microscope, as depicted in Figure 1c. The detected beam is isolated and collected by a photodiode. The voltage output from the photodiode is sent into a lock-in amplifier to demodulate the SRS signal . Finally, the signal is fed to computer software that produces an image where pixel values correspond to SRS signal strength. The signal intensity detected is proportional to the concentration of the target molecule as shown in Equation 1:
| 1. |
where C is the concentration of the target molecule, is the focal volume, is the differential Raman cross section of the molecule, and and are the intensities of the pump and Stokes beams, respectively. In principle, given the Raman cross-section, the concentration can be directly calculated. In practice, a calibration curve of SRS intensity versus concentration is typically obtained for quantitative analysis. The procedure is similar to the classical spectrophotometric analysis using Beer-Lambert’s law, except that the cuvette length is a hidden parameter (typically ~1 μm) that depends on the imaging condition (including beam size, objective numerical aperture, and condenser numerical aperture). When the sample and the calibration solutions are measured under the same imaging condition, the concentration of the unknown sample can be simply calculated as
| 2. |
where is the signal strength of the unknown signal, is the signal strength of the calibration sample, and is the concentration of the calibration sample.
Although this seems straightforward, many pitfalls can obfuscate quantitative interpretation in imaging complex samples. The most notable challenge is light scattering. Light scattering is a ubiquitous process in optical imaging of biological tissues and other heterogeneous materials. Scattering decreases the amount of light reaching the objective focus, thus decreasing SRS signal intensity. It also reduces the amount of light collected by the photodiode, which further decreases the measured signal. Because the amount of light scattering cannot be directly measured, it is challenging to quantify concentration based on measured SRS intensity. This is due to calibration samples likely not having the same light scattering–induced signal intensity loss. To circumvent this challenge, either relative comparisons are made on similar samples, or an internal standard is necessary to correct for light scattering.
In imaging monolayer cell cultures, light scattering is typically negligible. However, there are other important considerations when absolute concentration is to be quantified. The first one is the non-Raman background. Non-Raman-induced pump intensity change is detected by the lock-in amplifier. This extraneous background may be due to transient absorption, photothermal process, and cross-phase modulation (49–51). These parasitic processes can contribute interfering background with respect to the SRS signal. This is particularly relevant when imaging molecules at low concentrations. In practice, any SRS imaging that is not targeting dominant species such as proteins, lipids, water, and nucleic acids should be scrutinized for background contribution. Various background removal methods have been developed to overcome this challenge (37, 50, 52, 53). The second consideration is cross talk. Like fluorescence, Raman peaks of different molecules may overlap with one another. Thus, separation of their contributions is necessary for quantitative analysis. Cross talk is particularly poignant for SRS signal from carbon-hydrogen (C-H) or carbon-deuterium (C-D) stretching, which is commonly used in SRS imaging. The third consideration is the nonuniform imaging intensity across the field of view. Due to chromatic and spatial aberration of the pump and Stokes beams at the objective focus, it is common that the intensity at the edge of the field of view is weaker than that at the center. Such field variation must be corrected before applying any calibration curve obtained from solutions (54). A related but more subtle point is sample-dependent optical aberration that may influence quantification accuracy. Calibration is typically done in solutions, which introduce different optical aberrations than biological cells and tissue. This difference in optical aberration changes the effective laser intensity at focus and focal volume. Consequently, applying calibration of solutions to cells or tissues may bias the result. This effect is rarely considered but could lead to significant difference in signal intensity with high numerical aperture (NA) objectives and a large refractive index mismatch (55, 56).
Lastly, we must acknowledge the distinction between effective concentration and genuine concentration. In SRS imaging where absolute concentration measurements are made (by creating appropriate calibrations and accounting for the above pitfalls), it is ultimately a measurement of average concentration within the focal volume (i.e., effective concentration). When the measured molecule is contained within an area smaller than the focal spot size, the actual local concentration can be much larger than the effective concentration. This is an important consideration when measuring molecules in lipid droplets, vesicles, or other organelles (57, 58).
To ensure robust quantitative analysis, it is necessary to consider the aforementioned challenges and design proper calibration or other quantitative metrics that are not solely intensity dependent. In the following sections, we discuss a few major types of SRS-based quantitative analysis and provide some example applications.
MORPHOLOGICAL ANALYSIS
The first quantitative metric, morphological measurement, is not strictly unique to SRS microscopy and is, perhaps, the most straightforward and familiar. These measurements include qualities such as shape, size, number, orientation, spatial relation to other observed objects, or dynamics. Indeed, any microscopy with an appropriate contrast will provide morphological or spatial information about a sample. More unique to SRS, however, is the origin, quality, and temporal resolution of such acquired spatial information.
Specifically, the origin of the spatial information is still inherently chemical. Because the observed signal is ideally related only to the SRS process, any images formed can be thought of as a chemical map showing the presence of molecules vibrationally resonant with the chosen laser frequencies. Further, the relative intensity of pixels (i.e., the spatial contrast) is directly related to the concentration of resonant molecules at a given location within a field of view.
Regarding the quality of spatial information, there are a few important factors for SRS microscopy. First, the spatial resolution of images acquired with SRS microscopy is limited by the laser wavelengths in accordance with Abbe’s limit of diffraction. The lateral resolution (with a high NA objective) can be approximated as
| 3. |
where and are the two wavelengths used for SRS, and NA is the numerical aperture of the objective (59, 60). As an example, a system using a pump wavelength at 800 nm and Stokes wavelength at 1,040 nm for C-H SRS imaging using a 1.0 NA objective should have a spatial resolution of ~343 nm.
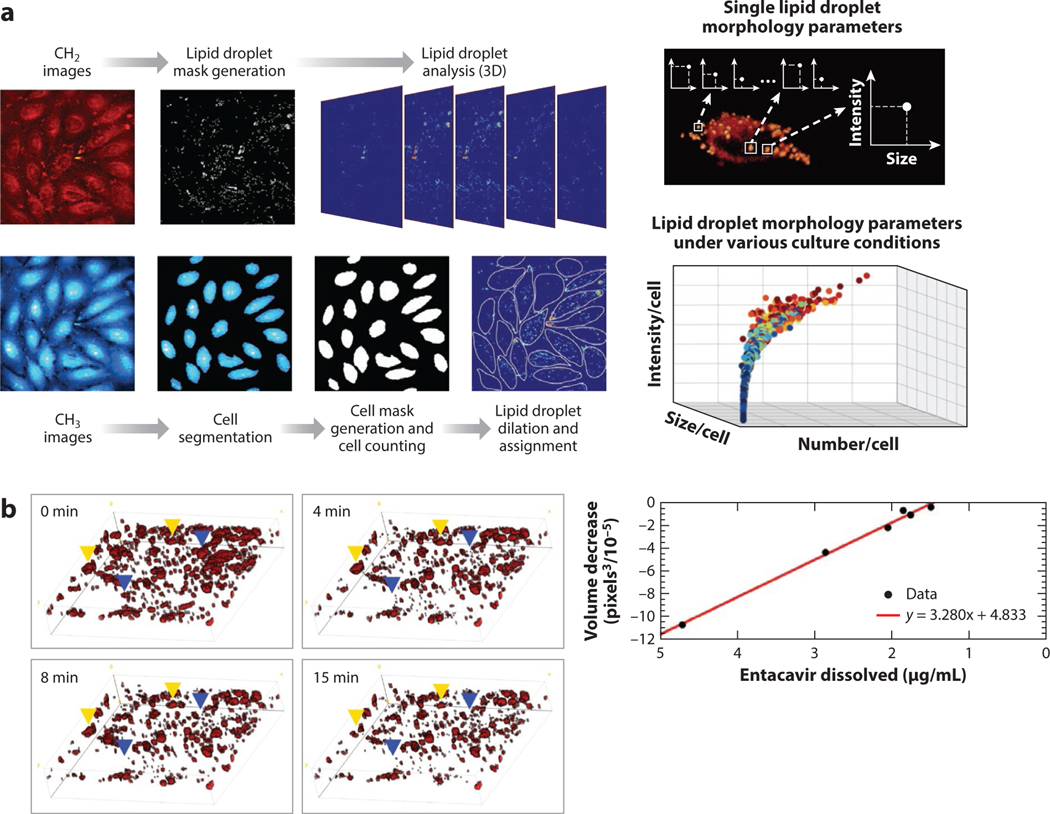
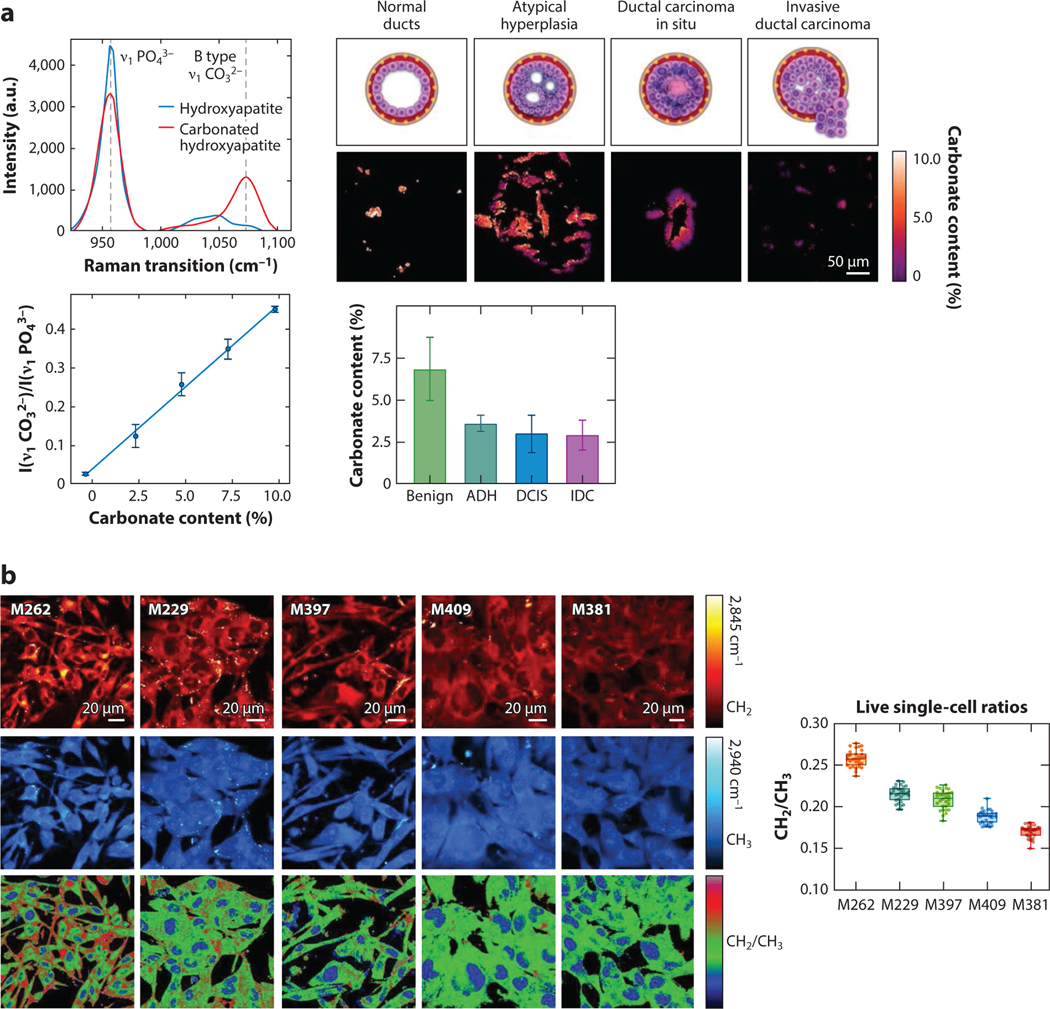
Quantitative metrics based solely on morphological characteristics have found many uses, particularly in lipid droplet analysis (24, 61–65). Cao et al. (66) demonstrate an excellent use of morphological measurements in their work quantifying lipid droplets within cells in a microfluidic platform. They measure the number, size, distribution, and intensity of lipid droplets within thousands of cells. They manage the vast numbers of spatially segmented lipid droplets for analysis in a workflow depicted in Figure 2a. By leveraging the chemical difference between lipids (where CH2 signal is strong) and the rest of the cell (mostly CH3 signal), they create segmentation maps for the lipids within cells in a field of view and then parameterize the lipid droplets based on an intensity threshold. The quantitative parameters they derive demonstrate the capabilities of SRS microscopy to produce relevant phenotypic markers in a label-free manner. In addition to size, shape, and number, SRS microscopy can also monitor lipid droplets over time within live cells (28). Time-lapse imaging provides dynamic information about the lipids and can be used to characterize lipid droplet function and potential disease states (67).
Figure 2.
Quantitative morphological analysis through SRS microscopy. (a) Imaging scheme and workflow of lipid droplet imaging and analysis based on lipid droplet size, number, and intensity. (Left) Representative images of lipid signal (CH2), protein signal (CH3), the respective lipid droplet masks, and cell masks created are shown left. (Right) The distribution of measured lipid droplet parameters across all cell images are shown right. Panel adapted with permission from Reference 66; copyright 2016 American Chemical Society. (b) Representative time-lapse images of entecavir within a polymer matrix dissolving into solution (left) and the correlation between measured volume decrease and dissolved entecavir (right). Panel adapted with permission from Reference 68; copyright 2018 American Chemical Society. Abbreviations: 3D, three-dimensional; SRS, stimulated Raman scattering.
Aside from phenotyping biological samples, spatial metrics can also offer insight into other chemical and physical properties (63). As another example, Francis et al. (68) utilize the precise spatial metrics of SRS microscopy to track the dissolution of drug particles within a polymer matrix designed for slow release. By targeting the vibrational signature of entecavir versus the polymer matrix, they visualize drug particles in three dimensions (3D) using SRS, as shown in Figure 2b. The drug particles and their sizes are then monitored over time as the drug dissolves into the solution. The authors also validate that the change in size is proportional to the dissolved drug amount. This example shows that morphological measurements from SRS microscopy can have a variety of quantitative ends based on the experiment.
Because morphological metrics are independent of concentration, they are typically not affected by the challenges mentioned in the previous section. However, one potential pitfall is the segmentation method. As the metrics are strongly based on the effective pixel area designated as a lipid droplet or drug particle, it is important to distinguish signal from background. Improper thresholding in this regard would lead to potentially erroneous observations in size, location, or effective intensity of the area. To mitigate this risk, it is important to choose molecular targets that are strong and distinct from the background or other signal sources for the sample. In the above examples, this is achieved by targeting CH2 bonds (strong and abundant, chiefly in lipid droplets) and carbon–carbon double bonds in the drug (strong and highly specific to the drug). However, for other samples, features may be less distinct within a heterogeneous mixture or weak in Raman signal. Even for lipid droplets, smaller droplets may be obscured by the limited spatial resolution of SRS, leading to erroneous results if the thresholding was done improperly (28). We note that contrast can be improved with deuterium labeling or exogenous vibrational labels with distinct spectral features and minimal Raman background (27, 69–72) or by utilizing machine learning or deep learning to enhance visual contrast (73–77).
RELATIVE CONCENTRATION MEASUREMENTS
Relative concentration measurements comprise the largest proportion of studies that utilize SRS microscopy for quantitative measurements (Figure 1a). This is perhaps not surprising given the relative ease by which relative measurements can provide quantitative information without the complications and validations of more precise concentration measurements. Here we take relative concentration to mean any measurement or visualization of molecular presence via SRS microscopy that does not report an exact concentration for the molecule. This is often discussed in publications as SRS intensity. SRS intensity can be depicted as either normalized or non-normalized and is typically reported as arbitrary units (a.u.). These relative measurements are then used to draw quantitative comparisons between different samples or samples at different conditions. Such comparisons still rely on the linear dependence between SRS signal and molecular concentration. However, the conclusions being drawn do not require exact concentration knowledge.
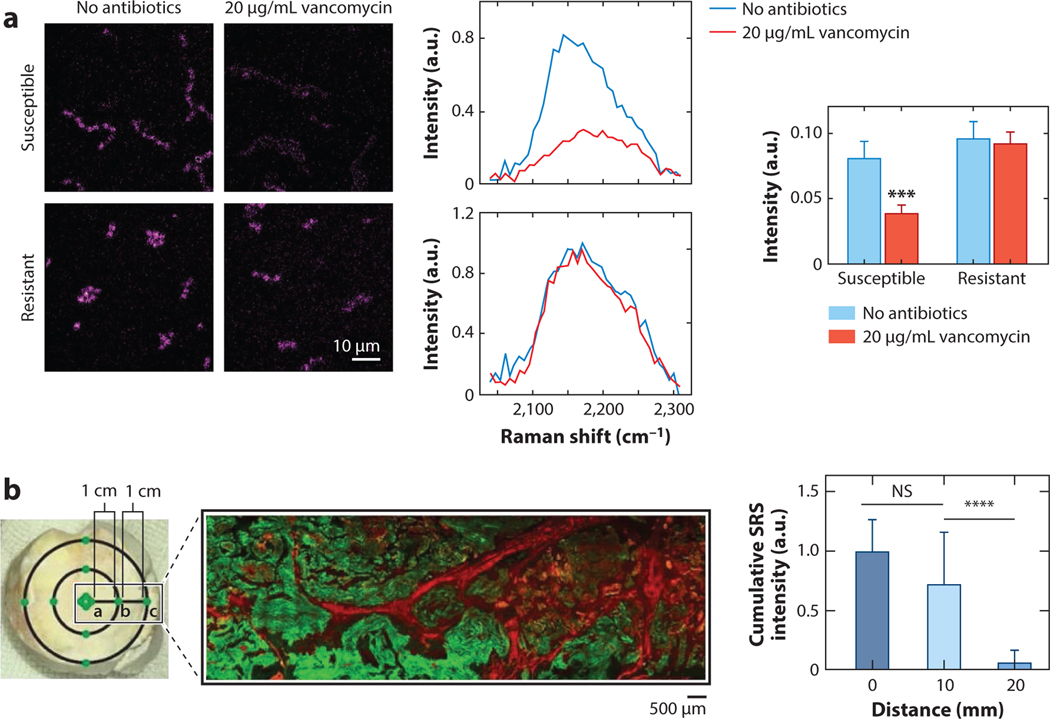
Relative measurements can take on a variety of tasks in SRS imaging. One example is time-lapse measurements that provide chemical insight into the dynamics of biological systems. For example, Hong et al. (31) demonstrate antibiotic susceptibility testing via SRS metabolic imaging. They show that antibiotic susceptibility can be predicted by monitoring glucose uptake within a single cell cycle. By targeting the C-D vibrational peak of deuterated glucose, they visualize uptake within live cells over the course of hours, as seen in Figure 3a. As the bacteria cells take up deuterated glucose, SRS signal at the corresponding 2,178 cm−1 peak increases. When the strains are additionally treated with vancomycin, the susceptible line shows a marked decrease in deuterated glucose uptake while the resistant line remains the same. Although exact concentrations are not calculated here, the relative SRS intensity provides insight into uptake dynamics and thus antibiotic susceptibility.
Figure 3.
Relative concentration measurements through SRS microscopy. (a) Representative SRS images (left) of deuterated glucose uptake in vancomycin-susceptible and vancomycin-resistant enterococci with and without 20 μg/mL vancomycin dosing. Raman intensity spectra (middle) of the carbon-deuterium signal in the two lines with and without vancomycin. Comparison of average Raman intensity for the two conditions for each enterococci line (right). *** indicates p-value <0.001 Panel adapted with permission from Reference 31; copyright 2018 American Chemical Society. (b) Brightfield (left) and SRS/second harmonic generation (green/red, respectively) of human gout tissue. Distances from the center of the tophus are shown. The cumulative SRS intensity of the monosodium urate crystals at 0, 10, and 20 mm away from the tophus center. N = 40 for each distance group using Kruskal-Wallis test followed by Dunn’s multiple comparisons test. NS indicates “No Significance” in the difference. **** indicates p-value <0.0001 Panel adapted with permission from Reference 33; copyright 2021 Ivyspring International Publisher. Abbreviations: NS, no significance in the difference; SRS, stimulated Raman scattering.
Another example of how relative concentration metrics from SRS microscopy can provide valuable insight is Zhang et al.’s (33) label-free SRS imaging of monosodium urate (MSU) crystals in human gout tissue. In this work, Zhang and colleagues measured the relative intensity of the MSU as a function of distance from the center of tophi (the relevant gout tissue). A clear trend can be observed with respect to the MSU crystal presence in a patient’s tissue. Specifically, the cumulative SRS intensity, and thus, the total MSU crystal amount, increases towards the center of a tophus. This diagnostic information is potentially useful in understanding and characterizing the progression of gout.
These examples of relative metrics highlight the value of SRS microscopy’s inherent quantitative nature while avoiding the litany of problems associated with measurements of exact concentrations. Where scattering and background signal will obfuscate direct quantification, relative measurements are valid so long as comparison among sufficiently similar samples is performed under uniform conditions. Ultimately, relative metrics trade the precision of quantitative information for an ease of understanding and avoidance of particularly difficult, if not impossible, experimental calibrations. This can be particularly appropriate for methods involving machine learning for further mining data (78–80). It is worth noting, however, that relative measurements make it difficult to compare results between labs, or even results from the same lab but obtained at different times where imaging conditions may be different. Reported results may be relevant more within the paradigm of one study rather than broadly applicable.
ABSOLUTE CONCENTRATION MEASUREMENTS
In contrast to relative measurements, absolute measurements provide exact numbers for concentration of observed molecular species in SRS imaging. They are important for comparison with other analytical techniques or deeper mechanistic understanding. Applications of absolute measurements are often shown in imaging of cultured monolayer cells, where scattering is negligible.
As discussed earlier, attaining absolute concentrations numbers from SRS images is, unfortunately, rarely as easy as making a calibration curve and then imaging the sample. In spatially and spectrally complex samples, interfering species, background, aberration, absorption, and scattering can all distort SRS signal with respect to a calibration curve based on solution images. Thus, significant care must be taken toward verifying the concentration numbers reported are accurate.
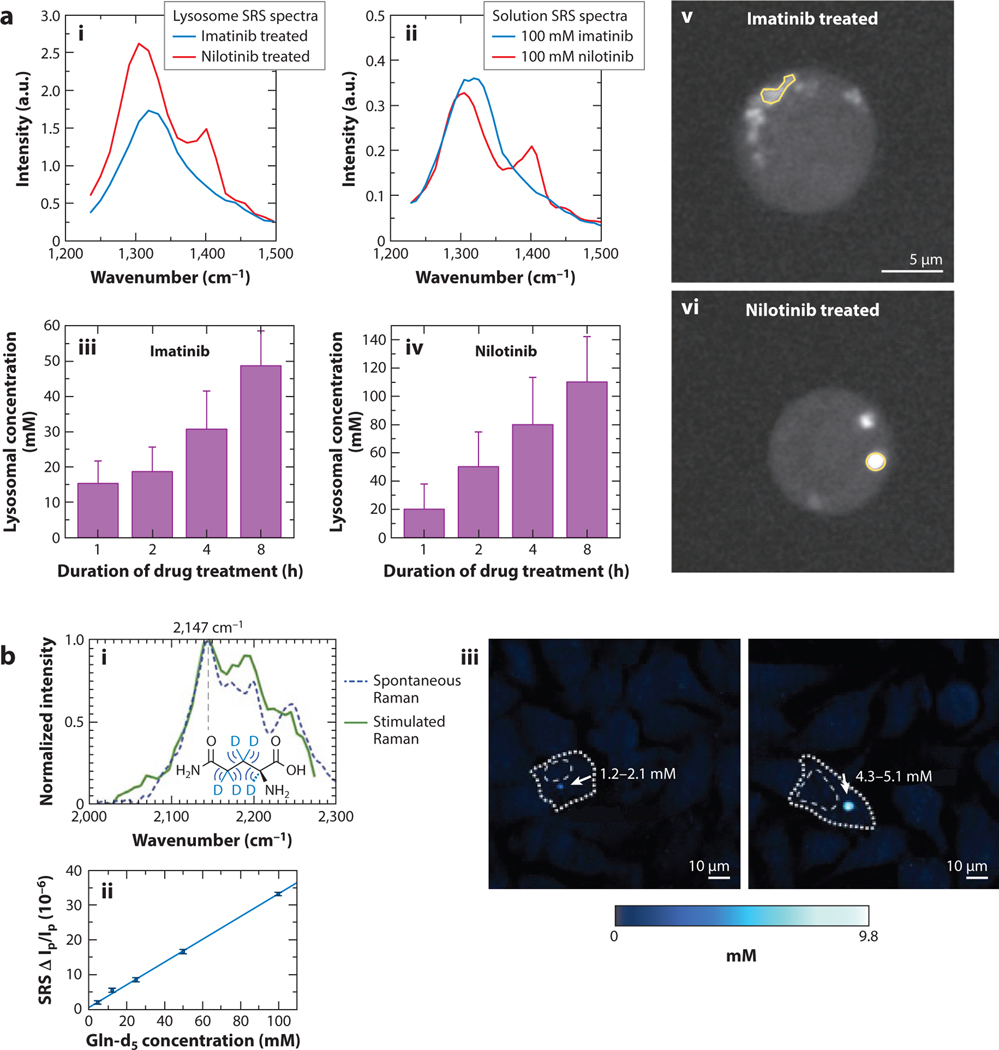
An example of absolute concentration measurement is Fu et al.’s (81) demonstration of intracellular quantification of tyrosine kinase inhibitors (TKIs). TKIs are drugs used in the treatment of specific types of cancers to inhibit cancer cell growth and proliferation. In the study, solution spectra of two TKIs, imatinib and nilotinib, were first acquired and then used to calculate intracellular drug concentrations of treated cells. The concentrations are calculated after spectral unmixing and background subtraction (Figure 4a). Specifically, compared to extracellular drug concentration, the drugs were shown to exhibit over 1,000-fold enrichment in the lysosomes of the cells while cytosolic drug presence was below the sensitivity for the system. The enrichment of imatinib agrees with the expected results from the lysosomotropic effect. Disagreement in nilotinib enrichment can be attributed to its precipitation in lysosomes. Recent reports have also shown similar results, but without the same absolute concentration measurements (21). Here, the impact and potential of measured concentrations over simple relative uptake over time are clear: The drug uptake for given dosage concentrations can be tracked quantitatively to better understand drug disposition across many drugs and cell types.
Figure 4.
Absolute concentration measurements with stimulated Raman scattering (SRS) microscopy. (a) Spectra of lysosome-sequestered (i) imatinib and (ii) nilotinib and solution spectra of the same drugs, time-lapse observed intracellular concentration of (iii) imatinib and nilotinib (iv), and representative images of BaF3 cells treated with (v) imatinib and nilotinib (vi). Panel adapted permission from Reference 81; copyright 2014 Nature Publishing Group. (b) Spontaneous and stimulated Raman spectra of the deuterated glutamine (i) and the linear response of its SRS signal with respect to concentration (ii). (iii) Representative carbon-deuterium SRS images with noted deuterated glutamine-tagged mHtt-97Q protein in intracellular protein aggregates. Panel adapted with permission from Reference 82; copyright 2020 American Chemical Society.
One important note regarding label-free imaging for concentration measurements such as Fu et al.’s (81) demonstration is that hyperspectral imaging is imperative for proper calibration. This is due to background signal that interferes with the target molecule’s spectrum. Various spectral unmixing techniques have been developed and reported to this end (40, 79, 80). Unmixing techniques allow the background to be accounted for, thus enabling more precise concentration measurements.
Another approach to mitigate background contribution is to target deuterated signal molecules or other exogenous Raman vibrational reporters by working in the so-called cell silent region (~1,800–2,800 cm−1), where endogenous biomolecules have no vibrational contributions. For example, Miao & Wei (82) demonstrate the ability to characterize polyglutamine protein aggregates within cells and provide concentration measurements of the different constituent proteins within aggregates (Figure 4b). They show significant aggregation of the proteins with deuterated glutamine levels in the millimolar range. In combination with size and ratiometric measurements of the aggregates, they also show the composition and steady concentration of nontoxic proteins within aggregates, suggesting a toxic protein scavenging functionality for the aggregates. It is worth noting that in deuterated samples, non-Raman background contribution may still contribute significantly, and either hyperspectral SRS or frequency modulation SRS may be needed to provide quantitative measurements of concentrations (52).
Absolute concentration measurement from SRS imaging is a somewhat underdeveloped branch of quantitative SRS measurements, likely due to the difficulty of obtaining controlled and verifiable quantitative information. However, subcellular concentration measurements within live and dynamic systems are significant and can provide unique mechanistic insight into various small molecules within cells or tissue. The demonstrations so far have been promising but ultimately remain limited by the challenges associated with attaining precise chemical information in complex samples. For example, Fu et al. (58) demonstrate measurements of acetylcholine concentrations at neuromuscular junctions in frog pectoral muscle using frequency modulation SRS. While concentrations were obtained, the small size of acetylcholine vesicles likely leads to underestimation of its concentration. Further, the tissue samples, though thin, still have a small amount of scattering, which may degrade the signal. Another example is Li et al.’s (84) work demonstrating the ability to measure concentrations of deuterated fatty acids within Caenorhabditis elegans tissue. It is challenging to determine the degradation of measurement accuracy due to scattering in experiments such as these. When unaccounted for, concentration may be underestimated. The problem is exacerbated in thick tissue. Indeed, to the best of our knowledge, absolute quantitative concentration measurements in thick tissue via SRS imaging have not yet been reported.
RATIOMETRIC ANALYSIS
Ratiometric measurements are a category of quantitative measurements based on the ratio of two vibrational peaks that provide quantitative information about the chemical composition of one molecular species or the relative abundance of two species. Ratiometric measurements can provide information about sample composition (62, 82–86), diagnostic histology (34, 87–90), chemical reaction dynamics (91), or biological metabolism and homeostasis (62, 92–95). The main advantage of ratiometric measurements is that the quantitative information is not susceptible to distortion due to absorption and scattering because they affect both peaks equally and thus cancel out in the ratio. However, background signals often have a different ratio that affects quantification when they have a non-negligible contribution to the overall signal. In this case, background subtraction is necessary before ratiometric analysis. To this end, spectral coverage and pulse duration play a significant role in the relative strength of SRS to background signal when broadband pulses are used. Spectral coverage refers to the observable vibrational bandwidth for a given SRS setup based on the laser pulse wavelengths, duration, and chirp (in the case of spectral-focusing SRS). If the peaks to be measured are far apart, yet are to be imaged either simultaneously (74, 96, 97) or quickly in sequence (23, 48, 98), large spectral coverage is necessary to cover both peaks. Methods to broaden spectral coverage have also demonstrated significant background reduction (52, 99–101).
Shin et al. (102) demonstrate the ability to quantitatively measure breast cancer calcification composition by ratiometric SRS imaging, as shown in Figure 5a. Specifically, a calibration of carbonate content in hydroxyapatite based on Raman transitions in the fingerprint region is first acquired. The ratio between the carbonated and noncarbonated hydroxyapatite changes as a function of calcification composition. The calibration is then used to create compositional maps of calcifications in breast tissue ducts from human patients. As different samples are imaged, relationships between carbonate content and breast cancer pathology are elucidated. The significant change in carbonate percentage between benign calcification and neoplastic conditions provides a reliable quantitative metric for diagnosis. This example shows how ratiometric measurements of an endogenous species can give quantitative insight (here, compositional information) that augments regular diagnostic pathways for breast cancer.
Figure 5.
Ratiometric measurements through SRS microscopy. (a) SRS spectra (upper left) of hydroxyapatite (blue) and carbonated hydroxyapatite (red) and the relevant Raman transitions for ratiometric comparison (960 cm−1 and 1,070 cm−1). The ratiometric calibration curve of carbonate content (lower left). Representative ratiometric images of carbonate content in breast calcifications at various levels of neoplastic progression and measured carbonate content for each pathological category. Panel adapted with permission from Reference 102; copyright 2020 Ivyspring International Publisher. (b) Representative SRS images (left) of various melanoma cell lines of varying differentiation at the lipid peak (2,845 cm−1; red), protein peak (2,940 cm−1; blue), and the ratio of lipid/protein (bottom). The average lipid/protein signal ratios across the different cell lines (right). Panel adapted with permission from Reference 103; copyright 2020 Nature Publishing Group. Abbreviations: ADH, atypical ductal hyperplasia; DCIS, ductal carcinoma in situ; IDC, invasive ductal carcinoma; SRS, stimulated Raman scattering.
Ratiometric imaging can also be used for metabolic imaging, where dynamic compositional information relates to cellular uptake and growth. For example, Du et al. (103) show that transcriptome data–mined phenotypic metabolic susceptibilities could be visualized with SRS by measuring the ratio between lipid and protein synthesis for different metastatic melanoma cell lines, as illustrated in Figure 5b. They note that the hypothesized correlation between differentiation and metabolism is treatment-targetable through the elevated fatty acid synthesis as visualized in the elevated CH2/CH3 (lipid/protein) signal ratio. This demonstrates the ability of ratiometric imaging to quantitatively measure metabolism among different cancer cell lines and elucidate potential cancer treatment targets. It is worth nothing that deuterium labeling SRS imaging has also be widely used in ratiometric measurements to study metabolism (27, 97, 104).
In ratiometric imaging, when the excitation wavelengths remain unchanged, we can assume that the scattering-induced signal loss for both SRS peaks is the same, thus obviating the significant challenges in acquiring absolute chemical measurements in tissue. This is likely why it is highly favored in SRS imaging of tissue. Ratiometric SRS imaging offers the practical quantitative information necessary to study and understand relevant chemical compositions of systems. Ultimately, ratiometric imaging provides a wealth of quantitative information with similar levels of ease as with relative concentration but with the added benefit of providing absolute metrics that can be compared across experiments.
SPECTRALLY DYNAMIC MEASUREMENTS
Spectrally dynamic measurements are dependent on the shift of a vibrational peak of a molecule due to the molecule undergoing some chemical or physical change. This, in effect, utilizes the molecule as a sensor of the local environment through SRS microscopy. The spectral shift can be measured through either hyperspectral SRS imaging or ratiometric imaging. However, in these measurements, the information sought is not about the peak intensity or the concentration, but the implied changes in the local environment of the molecule. The sensor molecule can be either endogenous or exogenous. Compared to fluorescent or phosphorescent sensors, SRS sensors have the advantage of being less susceptible to unrelated environmental changes.
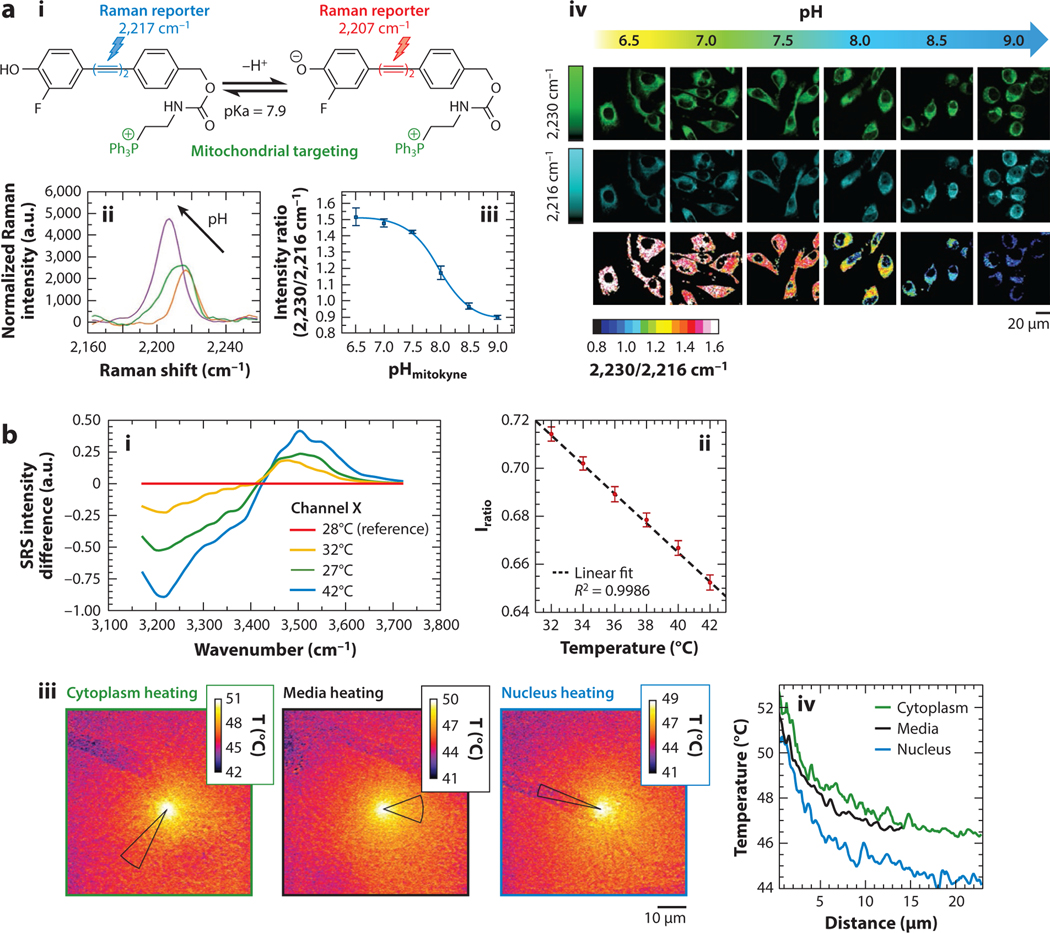
One example of spectrally dynamic measurements using a reporter is Wilson et al.’s (105) demonstration of a pH-sensitive mitochondria Raman probe. The reporter molecule shifts the center wavenumber of its alkyne peak in response to pH (Figure 6a). Further, this alkyne peak acts as a strong signal molecule in the cell silent region. The molecule is specific to the mitochondria within the cell, where pH can indicate mitochondrial health. As the pH environment of the probe changes, the ratio between the molecule’s SRS peaks at 2,230 cm−1 and 2,216 cm−1 changes accordingly, with the highest sensitivity corresponding to the pKa of the probe. Ratiometric imaging of the cells allows for the exact calculation of pH maps of the mitochondria within the cells. Further, because the reporting molecules are based on Raman signal rather than fluorescence, background fluorescence and photobleaching of the molecule are not concerns.
Figure 6.
Spectrally dynamic measurements with SRS microscopy. (a) The Raman dye molecule (i), mitokyne, and its SRS spectra (ii) showing the shifting peak in response to different pH. (iii) The SRS intensity ratio of 2,230/2,216 cm−1 with respect to pH (iv). Representative SRS images of HeLa cells treated with mitokyne at different controlled pH values at 2,216 cm−1, 2,230 cm−1, and their ratio. Panel adapted with permission from Reference 105; copyright 2021 American Chemical Society. (b) SRS difference spectra of liquid water at various temperatures (i). The calibration of the spectral intensity ratios and temperatures (ii). Temperature map images (iii) based on SRS signal of an A549 cell being heated with an infrared laser at different positions. Thermal decay curves (iv) with respect to distance for each field of view. Panel adapted with permission from Reference 106; copyright 2020 American Chemical Society. Abbreviations: a.u., arbitrary unit; SRS, stimulated Raman scattering.
Aside from imaging the microenvironment using reporter molecules, more exotic measurements may also be attained through spectrally dynamic SRS measurements. For example, Figueroa et al. (106) demonstrate the ability to measure microscale temperatures by ratiometrically imaging water’s temperature-dependent vibrational peaks. Using a simultaneous imaging scheme, the authors ratiometrically calibrated two points in water’s O-H vibrational band to known temperatures. The same transitions were then imaged for cells being heated by an infrared laser. In this case, the intracellular water acts as an endogenous reporter to create temperature maps of cells. These temperature maps show that SRS imaging can provide quantitative insight into biological heating, as shown in Figure 6b. The temperature maps shown by Figueroa et al. provide direct evidence of significantly lower mitochondria temperature than previously measured with fluorescence. Compared to commonly used fluorescent probes, the water-based Raman probe is less influenced by other environmental changes and may provide more reliable results. The use of simultaneous dual-band SRS imaging is critical to remove artifacts due to motion or perturbation of the system.
Besides small molecules for pH and temperature sensing, there have also been recent reports of using Raman reporters for gas molecule sensing (107) and mapping water solvation within cells (108, 109), showcasing the potential diversity of quantitative measurements possible through spectral shifts. The unique advantage of this type of measurement is that the spectral shape or peak position can be determined much more reliably and accurately than intensity. Though the examples are few so far for this type of quantitative SRS imaging, we expect significant growth in spectrally dynamic sensor molecule–based SRS imaging.
CONCLUSIONS AND OUTLOOK
SRS microscopy has established itself as a useful tool for quantitative chemical imaging. Through the wide variety of quantitative metrics, we expect SRS microscopy to continue growing in use across a broad range of biological and biomedical applications. While the challenges associated with quantitative measurements can be onerous, we have discussed potential strategies and solutions to maximize the capabilities of SRS microscopy. We have also pointed out types of quantitative SRS imaging approaches that obviate the challenges of light scattering, such as ratiometric SRS imaging and imaging of Raman reporter molecules that are sensitive to their local environment. Although relative concentration metrics are still most common owing to their ease of use, we point out that the more accurate and robust ratiometric measurements offer desirable quantitative information that can be compared across experiments or even across different labs. Sample preparation and system configuration are much less demanding because scattering and aberration contributions are largely removed. However, for ratiometric measurements to work, it is necessary to have large and reproducible SRS spectral changes due to either the compositional variation of one molecular species or relative abundance change of two molecular species. Moreover, many of the ratiometric examples shown here used two distinct Raman peaks. It is common to have more subtle spectral changes of a group of Raman peaks (often with overlapping features). In those cases, chemometric approaches are necessary to quantitatively disentangle the spectral changes (84).
Determining the absolute, spatially resolved concentration of molecular species in living cells and tissues with SRS is highly desired but largely unfulfilled owing to many challenges. In a few limited cases (mostly cells in monolayer culture), absolute concentration is shown to offer unique insights into the molecular processes that are largely unattainable with relative concentration measurements. However, these measurements cannot be directly translated to tissue due to light scattering. A universal approach to scattering correction using internal standards is needed to enable quantitative concentration measurements in tissue. Toward that end, a recent approach using water as that internal standard offers hope for quantitative SRS imaging in tissue (110).
Regardless of the quantitative metrics used, it is important to stress that rigorous consideration of background and proper control or calibration are required for any attempt to generate accurate and reproducible SRS measurements of concentration, composition, or microenvironment. Non-Raman background is ubiquitous in SRS imaging. Its contribution to SRS imaging of minority species (including deuterated compounds) can be significant. Thus, it is recommended that hyperspectral SRS imaging is performed for quantitative measurements to better assess background contribution. In the literature, off-resonance images are often used for background assessment or removal. While it is a widely accepted practice, we need to exercise caution in choosing an appropriate off-resonance peak. Typically, a peak that is close to the on-resonance peak (e.g., <50 cm−1) should be used to avoid laser intensity or alignment changes.
Overall, the utility of SRS microscopy as a quantitative technique is thus far well demonstrated with many applications. With awareness of quantitative SRS microscopy’s challenges and limitations, we expect that the continuing advances in optical systems, technical implementations, and computer-aided data interpretation will only serve to further quantitative SRS microscopy as a powerful tool for the chemical imaging of biological and nonbiological systems.
ACKNOWLEDGMENTS
The work is supported by an NSF CAREER grant 1846503 (D.F.) and the US National Institutes of Health (NIH) grant R35 GM133435 (D.F.).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, et al. 2008. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322(5909):1857–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colles MJ, Griffiths JE. 1972. Relative and absolute Raman scattering cross sections in liquids. J. Chem. Phys 56(7):3384–91 [Google Scholar]
- 3.McAnally MO, Phelan BT, Young RM, Wasielewski MR, Schatz GC, Van Duyne RP. 2017. Quantitative determination of the differential Raman scattering cross sections of glucose by femtosecond stimulated Raman scattering. Anal. Chem 89(13):6931–35 [DOI] [PubMed] [Google Scholar]
- 4.Xiong H, Qian N, Miao Y, Zhao Z, Min W. 2019. Stimulated Raman excited fluorescence spectroscopy of visible dyes. J. Phys. Chem. Lett 10(13):3563–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng J-X, Xie XS. 2015. Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science 350(6264):aaa8870 [DOI] [PubMed] [Google Scholar]
- 6.Hill AH, Fu D. 2019. Cellular imaging using stimulated Raman scattering microscopy. Anal. Chem 91(15):9333–42 [DOI] [PubMed] [Google Scholar]
- 7.Duncan MD, Reintjes J, Manuccia TJ. 1982. Scanning coherent anti-Stokes Raman microscope. Opt. Lett 7(8):350–52 [DOI] [PubMed] [Google Scholar]
- 8.Cheng J-X, Xie XS. 2004. Coherent anti-Stokes Raman scattering microscopy: instrumentation, theory, and applications. J. Phys. Chem. B 108(3):827–40 [Google Scholar]
- 9.Fu D 2017. Quantitative chemical imaging with stimulated Raman scattering microscopy. Curr. Opin. Chem. Biol 39:24–31 [DOI] [PubMed] [Google Scholar]
- 10.Stender A, Marchuk K, Liu C, Sander S, Meyer M, et al. 2013. Single cell optical imaging and spectroscopy. Chem. Rev. 113(4):2469–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schie I, Krafft C, Popp J. 2015. Applications of coherent Raman scattering microscopies to clinical and biological studies. Analyst 140(12):3897–909 [DOI] [PubMed] [Google Scholar]
- 12.Min W, Freudiger CW, Lu S, Xie XS. 2011. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annu. Rev. Phys. Chem 62:507–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camp CH Jr, Cicerone MT. 2015. Chemically sensitive bioimaging with coherent Raman scattering. Nat. Photon 9(5):295–305 [Google Scholar]
- 14.Masia F, Karuna A, Borri P, Langbein W. 2015. Hyperspectral image analysis for CARS, SRS, and Raman data. J. Ram. Spectrosc 46(8):727–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandakumar P, Kovalev A, Volkmer A. 2009. Vibrational imaging based on stimulated Raman scattering microscopy. New J. Phys 11:033026 [Google Scholar]
- 16.Cheng J-X, Xie XS. 2013. Coherent Raman Scattering Microscopy. Boca Raton, FL: CRC Press [Google Scholar]
- 17.Sanderson MJ, Smith I, Parker I, Bootman MD. 2014. Fluorescence microscopy. Cold Spring Harb. Protoc 2014(10): 10.1101/pdb.top071795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santi PA. 2011. Light sheet fluorescence microscopy: a review. J. Histochem. Cytochem 59(2):129–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjit S, Lanzanò L, Libby AE, Gratton E, Levi M. 2021. Advances in fluorescence microscopy techniques to study kidney function. Nat. Rev. Nephrol 17(2):128–44 [DOI] [PubMed] [Google Scholar]
- 20.Icha J, Weber M, Waters JC, Norden C. 2017. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays 39(8):1700003. [DOI] [PubMed] [Google Scholar]
- 21.Sepp K, Lee M, Bluntzer MTJ, Helgason GV, Hulme AN, Brunton VG. 2020. Utilizing stimulated Raman scattering microscopy to study intracellular distribution of label-free ponatinib in live cells. J. Med. Chem 63(5):2028–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feizpour A, Marstrand T, Bastholm L, Eirefelt S, Evans CL. 2021. Label-free quantification of pharmacokinetics in skin with stimulated Raman scattering microscopy and deep learning. J. Investig. Dermatol 141(2):395–403 [DOI] [PubMed] [Google Scholar]
- 23.Liao C-S, Slipchenko MN, Wang P, Li J, Lee S-Y, et al. 2015. Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light Sci. Appl 4(3):e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae K, Zheng W, Ma Y, Huang Z. 2020. Real-time monitoring of pharmacokinetics of mitochondria-targeting molecules in live cells with bioorthogonal hyperspectral stimulated Raman scattering microscopy. Anal. Chem 92(1):740–48 [DOI] [PubMed] [Google Scholar]
- 25.Belsey N, Garrett N, Contreras-Rojas L, Pickup-Gerlaugh A, Price G, et al. 2014. Evaluation of drug delivery to intact and porated skin by coherent Raman scattering and fluorescence microscopies. J. Control. Release 174:37–42 [DOI] [PubMed] [Google Scholar]
- 26.Stiebing C, Meyer T, Rimke I, Matthäus C, Schmitt M, et al. 2017. Real-time Raman and SRS imaging of living human macrophages reveals cell-to-cell heterogeneity and dynamics of lipid uptake. J. Biophoton 10(9):1217–26 [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Shi L, Shen Y, Miao Y, Wei M, et al. 2019. Spectral tracing of deuterium for imaging glucose metabolism. Nat. Biomed. Eng 3(5):402–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Li J, Lan L, Cheng J-X. 2017. Quantification of lipid metabolism in living cells through the dynamics of lipid droplets measured by stimulated Raman scattering imaging. Anal. Chem 89(8):4502–7 [DOI] [PubMed] [Google Scholar]
- 29.Huang K-C, Li J, Zhang C, Tan Y, Cheng J-X. 2020. Multiplex stimulated Raman scattering imaging cytometry reveals lipid-rich protrusions in cancer cells under stress condition. iScience 23(3):100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Min W, Freudiger C, Ruvkun G, Xie X. 2011. RNAi screening for fat regulatory genes with SRS microscopy. Nat. Methods 8(2):135–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong W, Karanja CW, Abutaleb NS, Younis W, Zhang X, et al. 2018. Antibiotic susceptibility determination within one cell cycle at single-bacterium level by stimulated Raman metabolic imaging. Anal. Chem 90(6):3737–43 [DOI] [PubMed] [Google Scholar]
- 32.Bae K, Zheng W, Ma Y, Huang Z. 2019. Real-time monitoring of pharmacokinetics of antibiotics in biofilms with Raman-tagged hyperspectral stimulated Raman scattering microscopy. Theranostics 9(5):1348–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Xu H, Chen J, Zhu X, Xue Y, et al. 2021. Highly specific and label-free histological identification of microcrystals in fresh human gout tissues with stimulated Raman scattering. Theranostics 11(7):3074–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Wu Y, Zheng B, Su L, Chen Y, et al. 2019. Rapid histology of laryngeal squamous cell carcinoma with deep-learning based stimulated Raman scattering microscopy. Theranostics 9(9):2541–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji M, Arbel M, Zhang L, Freudiger CW, Hou SS, et al. 2018. Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Sci. Adv 4(11):eaat7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, Yang Y, Liu Z, Guo L, Li S, et al. 2021. Microcalcification-based tumor malignancy evaluation in fresh breast biopsies with hyperspectral stimulated Raman scattering. Anal. Chem 93(15):6223–31 [DOI] [PubMed] [Google Scholar]
- 37.Fu D, Lu F, Zhang X, Freudiger C, Pernik D, et al. 2012. Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J. Am. Chem. Soc 134(8):3623–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Fung A, Zhou A. 2021. Advances in stimulated Raman scattering imaging for tissues and animals. Quant. Imaging Med. Surg 11(3):1078–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Streets A, Li A, Chen T, Huang Y. 2014. Imaging without fluorescence: nonlinear optical microscopy for quantitative cellular imaging. Anal. Chem 86(17):8506–13 [DOI] [PubMed] [Google Scholar]
- 40.Zhang D, Wang P, Slipchenko M, Cheng J. 2014. Fast vibrational imaging of single cells and tissues by stimulated Raman scattering microscopy. Acc. Chem. Res 47(8):2282–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HJ, Cheng J-X. 2017. Imaging chemistry inside living cells by stimulated Raman scattering microscopy. Methods 128:119–28 [DOI] [PubMed] [Google Scholar]
- 42.Lee M, Simon Herrington C, Ravindra M, Sepp K, Davies A, et al. 2021. Recent advances in the use of stimulated Raman scattering in histopathology. Analyst 146(3):789–802 [DOI] [PubMed] [Google Scholar]
- 43.Shen Y, Hu F, Min W. 2019. Raman imaging of small biomolecules. Annu. Rev. Biophys 48:347–69 [DOI] [PubMed] [Google Scholar]
- 44.Vanden-Hehir S, Tipping WJ, Lee M, Brunton VG, Williams A, Hulme AN. 2019. Raman imaging of nanocarriers for drug delivery. Nanomaterials 9(3):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawley J 2000. The 39 steps: a cautionary tale of quantitative 3-D fluorescence microscopy. Bio Techniques 28(5):884–87 [DOI] [PubMed] [Google Scholar]
- 46.Waters JC, Wittmann T. 2014. Concepts in quantitative fluorescence microscopy. Methods Cell Biol 123:1–18 [DOI] [PubMed] [Google Scholar]
- 47.Lee J-Y, Kitaoka M 2018. A beginner’s guide to rigor and reproducibility in fluorescence imaging experiments. Mol. Biol. Cell 29(13):1519–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu D, Holtom G, Freudiger C, Zhang X, Xie XS. 2013. Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J. Phys. Chem. B 117(16):4634–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fimpel P, Choorakuttil A, Pruccoli A, Ebner L, Tanaka S, et al. 2020. Double modulation SRS and SREF microscopy: signal contributions under pre-resonance conditions. Phys. Chem. Chem. Phys 22(37):21421–27 [DOI] [PubMed] [Google Scholar]
- 50.Zhang D, Slipchenko MN, Leaird DE, Weiner AM, Cheng J-X. 2013. Spectrally modulated stimulated Raman scattering imaging with an angle-to-wavelength pulse shaper. Opt. Express 21(11):13864–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu D, Ye T, Matthews TE, Yurtsever G, Warren SW. 2007. Two-color, two-photon, and excited-state absorption microscopy. J. Biomed. Optics 12(5):054004. [DOI] [PubMed] [Google Scholar]
- 52.Hill AH, Munger E, Francis AT, Manifold B, Fu D. 2019. Frequency modulation stimulated Raman scattering microscopy through polarization encoding. J. Phys. Chem. B 123(40):8397–404 [DOI] [PubMed] [Google Scholar]
- 53.Berto P, Andresen ER, Rigneault H. 2014. Background-free stimulated Raman spectroscopy and microscopy. Phys. Rev. Lett 112(5):053905. [DOI] [PubMed] [Google Scholar]
- 54.Nahmad-Rohen A, Regan D, Masia F, McPhee C, Pope I, et al. 2020. Quantitative label-free imaging of lipid domains in single bilayers by hyperspectral coherent Raman scattering. Anal. Chem 92(21):14657–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pillai RS, Brakenhoff GJ, Müller M. 2006. Analysis of the influence of spherical aberration from focusing through a dielectric slab in quantitative nonlinear optical susceptibility measurements using third-harmonic generation. Opt. Express 14(1):260–69 [DOI] [PubMed] [Google Scholar]
- 56.Gelber MK, Kole MR, Kim N, Aluru NR, Bhargava R. 2017. Quantitative chemical imaging of nonplanar microfluidics. Anal. Chem 89(3):1716–23 [DOI] [PubMed] [Google Scholar]
- 57.Li X, Jiang M, Lam JWY, Tang BZ, Qu JY. 2017. Mitochondrial imaging with combined fluorescence and stimulated Raman scattering microscopy using a probe of the aggregation-induced emission characteristic. J. Am. Chem. Soc 139(47):17022–30 [DOI] [PubMed] [Google Scholar]
- 58.Fu D, Yang W, Xie XS. 2017. Label-free imaging of neurotransmitter acetylcholine at neuromuscular junctions with stimulated Raman scattering. J. Am. Chem. Soc 139(2):583–86 [DOI] [PubMed] [Google Scholar]
- 59.Bi Y, Yang C, Chen Y, Yan S, Yang G, et al. 2018. Near-resonance enhanced label-free stimulated Raman scattering microscopy with spatial resolution near 130 nm. Light Sci. Appl 7(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zipfel WR, Williams RM, Webb WW. 2003. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol 21(11):1369–77 [DOI] [PubMed] [Google Scholar]
- 61.Ji M, Lewis S, Camelo-Piragua S, Ramkissoon SH, Snuderl M, et al. 2015. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci. Transl. Med 7(309):309ra163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee D, Du J, Yu R, Su Y, Heath JR, Wei L. 2020. Visualizing subcellular enrichment of glycogen in live cancer cells by stimulated Raman scattering. Anal. Chem 92(19):13182–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nixdorf J, Di Florio G, Brockers L, Borbeck C, Hermes H, et al. 2019. Uptake of methanol by poly(methyl methacrylate): an old problem addressed by a novel Raman technique. Macromolecules 52(13):4997–5005 [Google Scholar]
- 64.Tian F, Yang W, Mordes D, Wang J, Salameh J, et al. 2016. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat. Commun 7:13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta A, Dorlhiac G, Streets A. 2019. Quantitative imaging of lipid droplets in single cells. Analyst 144(3):753–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao C, Zhou D, Chen T, Streets AM, Huang Y. 2016. Label-free digital quantification of lipid droplets in single cells by stimulated Raman microscopy on a microfluidic platform. Anal. Chem 88(9):4931–39 [DOI] [PubMed] [Google Scholar]
- 67.Urasaki Y, Zhang C, Cheng J-X, Le TT. 2018. Quantitative assessment of liver steatosis and affected pathways with molecular imaging and proteomic profiling. Sci. Rep 8(1):3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Francis AT, Nguyen TT, Lamm MS, Teller R, Forster SP, et al. 2018. In situ stimulated Raman scattering (SRS) microscopy study of the dissolution of sustained-release implant formulation. Mol. Pharm 15(12):5793–801 [DOI] [PubMed] [Google Scholar]
- 69.Wei L, Chen Z, Shi L, Long R, Anzalone AV, et al. 2017. Super-multiplex vibrational imaging. Nature 544(7651):465–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C, Zhao Z, Qian N, Wei S, Hu F, Min W. 2021. Multiplexed live-cell profiling with Raman probes. Nat. Commun 12(1):3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao Y, Qian N, Shi L, Hu F, Min W. 2021. 9-Cyanopyronin probe palette for super-multiplexed vibrational imaging. Nat. Commun 12(1):4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J, Yan S, He Z, Ding C, Zhai T, et al. 2018. Small unnatural amino acid carried Raman tag for molecular imaging of genetically targeted proteins. J. Phys. Chem. Lett 9(16):4679–85 [DOI] [PubMed] [Google Scholar]
- 73.Manifold B, Thomas E, Francis AT, Hill AH, Fu D. 2019. Denoising of stimulated Raman scattering microscopy images via deep learning. Biomed. Opt. Express 10(8):3860–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill AH, Hill AH, Manifold B, Manifold B, Fu D. 2020. Tissue imaging depth limit of stimulated Raman scattering microscopy. Biomed. Opt. Express 11(2):762–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manifold B, Men S, Hu R, Fu D. 2021. A versatile deep learning architecture for classification and label-free prediction of hyperspectral images. Nat. Mach. Intell 3(4):306–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin H, Lee HJ, Tague N, Lugagne J-B, Zong C, et al. 2021. Microsecond fingerprint stimulated Raman spectroscopic imaging by ultrafast tuning and spatial-spectral learning. Nat. Commun 12(1):3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, Zhao J, Lin H, Tan Y, Cheng J-X. 2020. High-speed chemical imaging by dense-net learning of femtosecond stimulated Raman scattering. J. Phys. Chem. Lett 11(20):8573–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alfonso-Garcia A, Paugh J, Farid M, Garg S, Jester J, Potma E. 2017. A machine learning framework to analyze hyperspectral stimulated Raman scattering microscopy images of expressed human meibum. J. Raman Spectrosc 48(6):803–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ragupathy I, Schweikhard V, Zumbusch A. 2021. Multivariate analysis of hyperspectral stimulated Raman scattering microscopy images. J. Raman Spectrosc 52:1630–42 [Google Scholar]
- 80.Zhang D, Wang P, Slipchenko MN, Ben-Amotz D, Weiner AM, Cheng J-X. 2013. Quantitative vibrational imaging by hyperspectral stimulated Raman scattering microscopy and multivariate curve resolution analysis. Anal. Chem 85(1):98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu D, Zhou J, Zhu WS, Manley PW, Wang YK, et al. 2014. Imaging the intracellular distribution of tyrosine kinase inhibitors in living cells with quantitative hyperspectral stimulated Raman scattering. Nat. Chem 6(7):614–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miao K, Wei L. 2020. Live-cell imaging and quantification of PolyQ aggregates by stimulated Raman scattering of selective deuterium labeling. ACS Cent. Sci 6(4):478–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Li Y, Jiang M, Wu W, He S, et al. 2019. Quantitative imaging of lipid synthesis and lipolysis dynamics in Caenorhabditis elegans by stimulated Raman scattering microscopy. Anal. Chem 91(3):2279–87 [DOI] [PubMed] [Google Scholar]
- 84.Figueroa B, Nguyen T, Sotthivirat S, Xu W, Rhodes T, et al. 2019. Detecting and quantifying microscale chemical reactions in pharmaceutical tablets by stimulated Raman scattering microscopy. Anal. Chem 91(10):6894–6901 [DOI] [PubMed] [Google Scholar]
- 85.Wei L, Yu Y, Shen Y, Wang MC, Min W. 2013. Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. PNAS 110(28):11226–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang P, Li J, Wang P, Hu C, Zhang D, et al. 2013. Label-free quantitative imaging of cholesterol in intact tissues by hyperspectral stimulated Raman scattering microscopy. Angew. Chem. Int. Ed 52(49):13042–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orringer D, Pandian B, Niknafs Y, Hollon T, Boyle J, et al. 2017. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng 1(2): 0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pekmezci M, Morshed RA, Chunduru P, Pandian B, Young J, et al. 2021. Detection of glioma infiltration at the tumor margin using quantitative stimulated Raman scattering histology. Sci. Rep 11(1):12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sarri B, Poizat F, Heuke S, Wojak J, Franchi F, et al. 2019. Stimulated Raman histology: one to one comparison with standard hematoxylin and eosin staining. Biomed. Opt. Express 10(10):5378–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin KS, Francis AT, Hill AH, Laohajaratsang M, Cimino PJ, et al. 2019. Intraoperative assessment of skull base tumors using stimulated Raman scattering microscopy. Sci. Rep 9(1):20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H, Cheng Y, Tang H, Bi Y, Chen Y, et al. 2020. Imaging chemical kinetics of radical polymerization with an ultrafast coherent Raman microscope. Adv. Sci 7(10):1903644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu B, Lee H, Zhang D, Liao C, Ji N, et al. 2015. Label-free spectroscopic detection of membrane potential using stimulated Raman scattering. Appl. Phys. Lett 106(17):173704 [Google Scholar]
- 93.Li J, Cheng J-X. 2014. Direct visualization of de novo lipogenesis in single living cells. Sci. Rep 4(1):6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen Y, Xu F, Wei L, Hu F, Min W. 2014. Live-Cell quantitative imaging of proteome degradation by stimulated Raman scattering. Angew. Chem. Int. Ed 53(22):5596–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shen Y, Zhao Z, Zhang L, Shi L, Shahriar S, et al. 2017. Metabolic activity induces membrane phase separation in endoplasmic reticulum. PNAS 114(51):13394–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.He R, Xu Y, Zhang L, Ma S, Wang X, et al. 2017. Dual-phase stimulated Raman scattering microscopy for real-time two-color imaging. Optica 4(1):44–47 [Google Scholar]
- 97.Long R, Zhang L, Shi L, Shen Y, Hu F, et al. 2018. Two-color vibrational imaging of glucose metabolism using stimulated Raman scattering. Chem. Commun 54(2):152–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Laptenok S, Rajamanickam V, Genchi L, Monfort T, Lee Y, et al. 2019. Fingerprint-to-CH stretch continuously tunable high spectral resolution stimulated Raman scattering microscope. J. Biophoton 12(9): e201900028. [DOI] [PubMed] [Google Scholar]
- 99.Figueroa B, Fu W, Nguyen T, Shin K, Manifold B, et al. 2018. Broadband hyperspectral stimulated Raman scattering microscopy with a parabolic fiber amplifier source. Biomed. Opt. Express 9(12):6116–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shin S, Kim D, Kim K, Park Y. 2018. Super-resolution three-dimensional fluorescence and optical diffraction tomography of live cells using structured illumination generated by a digital micromirror device. Sci. Rep 8(1):9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karpf S, Eibl M, Wieser W, Klein T, Huber R. 2015. A time-encoded technique for fibre-based hyperspectral broadband stimulated Raman microscopy. Nat. Commun 6:6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shin KS, Laohajaratsang M, Men S, Figueroa B, Dintzis SM, Fu D. 2020. Quantitative chemical imaging of breast calcifications in association with neoplastic processes. Theranostics 10(13):5865–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Du J, Su Y, Qian C, Yuan D, Miao K, et al. 2020. Raman-guided subcellular pharmaco-metabolomics for metastatic melanoma cells. Nat. Commun 11(1):4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shi L, Shen Y, Min W. 2018. Invited article: visualizing protein synthesis in mice with in vivo labeling of deuterated amino acids using vibrational imaging. APL Photon. 3(9):092401 [Google Scholar]
- 105.Wilson LT, Tipping WJ, Wetherill C, Henley Z, Faulds K, et al. 2021. Mitokyne: A ratiometric Raman probe for mitochondrial pH. Anal. Chem 93(37):12786–12792 [DOI] [PubMed] [Google Scholar]
- 106.Figueroa B, Hu R, Rayner SG, Zheng Y, Fu D. 2020. Real-time microscale temperature imaging by stimulated Raman scattering. J. Phys. Chem. Lett 11(17):7083–89 [DOI] [PubMed] [Google Scholar]
- 107.Zeng C, Hu F, Long R, Min W. 2018. A ratiometric Raman probe for live-cell imaging of hydrogen sulfide in mitochondria by stimulated Raman scattering. Analyst 143(20):4844–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi L, Hu F, Min W. 2019. Optical mapping of biological water in single live cells by stimulated Raman excited fluorescence microscopy. Nat. Commun 10(1):4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lang X, Welsher K. 2020. Mapping solvation heterogeneity in live cells by hyperspectral stimulated Raman scattering microscopy. J. Chem. Phys 152(17):174201. [DOI] [PubMed] [Google Scholar]
- 110.Oh S, Lee C, Fu D, Yang W, Li A, et al. 2019. In situ measurement of absolute concentrations by Normalized Raman Imaging. bioRxiv 629543 [Google Scholar]