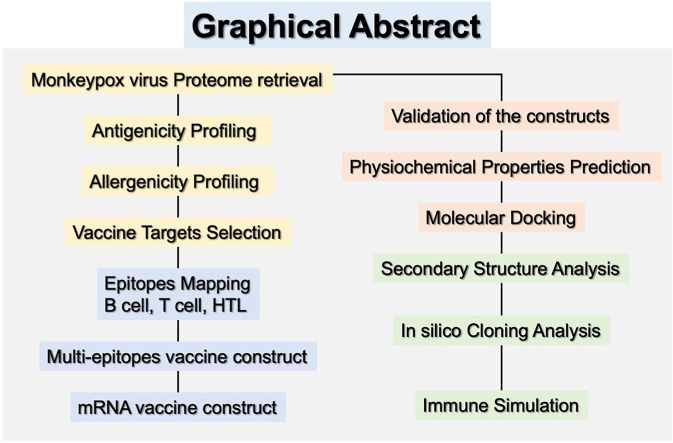
Abstract
Monkeypox Virus (MPXV) is a growing public health threat with increasing cases and fatalities globally. To date, no specific vaccine or small molecule therapeutic choices are available for the treatment of MPXV disease. In this work, we employed proteomics and structural vaccinology approaches to design mRNA and multi-epitopes-based vaccines (MVC) against MPXV. We first identified ten proteins from the whole proteome of MPXV as potential vaccine targets. We then employed structural vaccinology approaches to map potential epitopes of these proteins for B cell, cytotoxic T lymphocytes (CTL), and Helper T lymphocytes (HTL). Finally, 9 CTL, 6 B cell, and 5 HTL epitopes were joined together through suitable linkers to construct MVC (multi-epitope vaccine) and mRNA-based vaccines. Molecular docking, binding free energy calculation, and in silico cloning revealed robust interaction of the designed MVC with toll-like receptor 2 (TLR2) and efficient expression in E. Coli K12 strain. The immune simulation results revealed that the antigen titer after the injection reached to the maximum level on the 5th day and an abrupt decline in the antigen titer was observed upon the production of IgM, IgG and IgM + IgG, dendritic cells, IFN-gamma, and IL (interleukins), which suggested the potential of our designed vaccine candidate for inducing an immune response against MPXV.
Keywords: Monkeypox, Vaccines, Multi-epitopes, mRNA, Immune simulation
Graphical abstract
1. Introduction
The Monkeypox virus, which was isolated from cynomolgus monkeys in 1958, can cause human Monkeypox, which somewhat looks like smallpox [[1], [2], [3]]. There were several local outbreaks in the past few decades, mostly within African countries and a few in other continents [[4], [5], [6], [7], [8]]. However, in 2022, a new outbreak of this virus spreads much more rapidly, and it has become a global threat [9,10]. Once infected, patients not only get a rash but may experience other symptoms like fever, headache, cough, and fatigue. In extreme cases, this disease may be fatal [11]. As of March 16, 2023, the current active cases are 86,494 in 110 localities while 111 deaths have been reported [12]. Since the virus can be transmitted from human to human through multiple pathways, especially body fluid, the number of people infected is expected to continue to rise [10,13]. As a consequence, it becomes increasingly emergent to develop innovative strategies to treat or prevent this disease.
Vaccination is one of the most effective methods to prevent infectious diseases [14]. Regarding the Monkeypox virus, it has been identified that individuals vaccinated against smallpox are likely to have the capability to protect against it [[15], [16], [17]]. However, there are limited smallpox vaccines that are currently available, and they are not accessible to all countries [14]. Moreover, since they are genetically distinct, vaccines against the smallpox virus may lose their potency when dealing with the Monkeypox virus. For instance, a patient vaccinated with the ACAM2000 smallpox vaccine in the United States was still infected by the Monkeypox virus [18]. Furthermore, globally, the number of patients has already become enormous, and there is significant diversity in geographical regions, gender, race, age, and occupation. Therefore, the effectiveness of smallpox vaccines against the Monkeypox virus may be more questionable.
Since the goal of the vaccine is to protect against the Monkeypox virus, one of the most straightforward approaches is to design it from this virus directly. One of the traditional techniques is based on killed, attenuated, or purified pathogens [19]. Although they can be highly productive when properly designed, however large-scale manufacturing may be challenging [20]. Another pathway that gradually became popular in the past decade is the creation of nucleic acids based vaccines such as DNA, viral vectors, or RNA [21]. Nuclei-acid-based vaccines can emulate the immunization of inactivated vaccines safely and efficiently. In addition the industrial production of such vaccines is cost-effective and easy [22]. Among these mRNA vaccines have been more and more widely used in recent years, especially when dealing with the COVID-19 pandemic virus [23,24].
As a virus belonging to the orthopoxvirus genus, the Monkeypox virus has been found to exist in two different clades [25,26]. The one that spreads rapidly in 2022 belongs to the West African clade, whose sequence overlaps mostly with another clade found in Central Africa but has its unique genomic regions [26,27]. As its complete genome sequence is available, antigens can be identified with bioinformatics tools, and eventually, a multi-epitope mRNA vaccine can be constructed.
As two types of white blood cells, B and T cells can generate antibodies and eliminate pathogens, respectively, endowing their predominant position for anything related to vaccines [[28], [29], [30]]. The primary goal of this work is to design such a Monkeypox vaccine by joining several B cells and T cell epitopes. These epitopes are obtained by filtering out all the proteins in the Monkeypox virus proteome based on the immunization potential. Next, they were joined by using linkers to generate the sequence of the MVC and mRNA vaccines, and the corresponding structure was predicted. Once the structure is generated, its effectiveness is further verified through various tools. This work will provide a valuable guideline for producing a commercial vaccine against the current Monkeypox virus.
2. Methodology
2.1. Vaccinomics targets mining in Monkeypox virus proteome
The entire proteome of the Monkeypox virus strain responsible for the recent epidemic in 2022 contains 198 proteins, which were downloaded from UniProt [31] (https://www.ncbi.nlm.nih.gov/genbank/) under the accession number ON563414. For each protein in the proteome, whether it can potentially be a protective viral antigen was predicted by the server VirVACPRED [32] (https://virvacpred.herokuapp.com/). The result for each protein was either viral or non-viral antigens with a probability score. Furthermore, proteins that are labeled as viral antigens were submitted to the AllerTOP v.2 server [33] (https://www.ddg-pharmfac.net/AllerTOP/) to predict the allergenicity. Antigenic proteins that were also predicted as non-allergic were filtered out for the subsequent designing of mRNA and MVC.
2.2. Prediction of immunogenic B cell and T cell epitopes
Among the proteins that are both antigenic and not allergenic in the Monkeypox virus proteome, only ten with the highest antigenic probability score were selected. In the next step, the appropriate B and T cell epitopes were mapped. The B cell epitopes on each protein, which can potentially recognize the B cell receptors, were predicted by the server ABCpred [34,35] (https://webs.iiitd.edu.in/raghava/abcpred/index.html). A threshold of 0.51 was used. The length of each epitope was kept at 16aa during the prediction. Equally important is the identification of T cell epitopes that are recognized by the major histocompatibility complex-I (MHC-I), which is also called as the human leukocyte antigen (HLA) system for human beings [36,37]. The cytotoxic T lymphocyte (CTL) epitopes, fragments of intracellular antigens binding to class I MHC molecules (MHC-I), were predicted by NetCTL-1.2 server [38] (https://services.healthtech.dtu.dk/service.php?NetCTL-1.2). A cutoff value of 0.75 for the prediction of potential CTL epitope was applied.
2.3. Mapping of helper T lymphocyte (HTL) epitopes
The helper T lymphocyte (HTL) epitopes, extracellular antigenic peptides linking to class II MHC (MHC-II), were evaluated by the IEDB web server [39] (http://tools.iedb.org/mhcii/). The prediction was made against the human HLAs considering 7-alleles as the reference set (HLA-DRB1*03:01; HLA-DRB1*07:01; HLA-DRB1*15:01; HLA-DRB3*01:01; HLA-DRB3*02:02; HLA-DRB4*01:01; HLA-DRB5*01:01). The length of each epitope was kept as 15aa. The server gives a list of HTL epitopes against the specific HLAs, which can be ranked by using the percentile rank parameter. The lower the percentile rank, the better the HTL epitopes.
2.4. Prediction of interferon-γ (IFN-γ) inducing epitopes
A family of signaling proteins known as interferons (IFNs) is essential for the immune system's response instigation against viruses and other pathogens. In response to viral infections and other forms of cellular stress, immune system cells produce IFNs and released which then send a signal to the nearby cells to enter an antiviral state, which consequently helps to stop the virus from spreading. The overexpression of several immune response-related genes, such as those involved in antigen presentation, cytokine signaling, and apoptosis, is a signature of this antiviral state. IFNs are also essential for the activation of immune cells such as T cells, macrophages, and natural killer cells. They can augment macrophage phagocytic activity, induce natural killer cell cytotoxicity, and increase T cell proliferation and differentiation [40]. Since the cytokine interferon-γ (IFN-γ) can lead to MHC II expression, it is worthwhile to evaluate whether an epitope is IFN positive or negative. Considering the HTL epitopes prediction results in the previous step, ensuring minimal sequence overlap, five HTL epitopes with the lower percentile rank from each protein were submitted to IFN-epitope predicting server [41] (http://crdd.osdd.net/raghava/ifnepitope/). For the subsequent process, we only included IFN-positive HTL epitopes for vaccine construct designing.
2.5. Construction of mRNA vaccine
For the construction of an mRNA vaccine, five critical elements are required in the Open Reading Frame for effective antigenic mRNA vaccine construction.
-
a.
Kozak sequence;
-
b.
Linkers;
-
c.
B and T cell Epitopes;
-
d.
Stop codon; and
-
e.
Signal Peptide.
A start codon (AUG) must be a part of the Kozak sequence, whereas the stop codon can be optimized. Rigid, cleavable, and flexible linkers are critical in the development of a multi-epitope vaccine (MVC) because it permits different parts of MVC to interact freely while preventing self-contact. To enhance the epitopes/antigen presentation, tissue Plasminogen Activator (tPA) (UniProt ID: P00750) secretory signal was chosen as a signal peptide that was added to the ORF region [50]. The stability of the mRNA-based vaccine was enhanced by adding poly(A) tail, 5′cap, and 5′-3′ untranslated regions. A long poly (A) tail of 115–150 nucleotides was added for stable transmission and efficient translation which is also used by various earlier studies. Poly (A) tails have been discovered to work in tandem with 5′ m7G cap sequences. We included the NCA-7d at the 5′ UTR and S27a + R3U as the 3′ UTR regions in our vaccine sequence because they are known to stabilize the mRNAs [42]. For the secondary structure of mRNA, the RNAFold webserver (http://rna.tbi.univie.ac.at/) was used [43].
2.6. Designing of multi-epitopes vaccine (MVC)
In addition to the B and T cell and HTL epitopes, few other components are indispensable for an efficient MVC, including an adjuvant and a certain number of linkers. The candidate of the Monkeypox vaccine designed in this work is composed of 20 epitopes. The sequence of the adjuvant at the N-terminal is taken from human beta-defensin-2 (hDB-2) which can stabilize the structure and improve its ability for innate immunity [44]. Starting from this adjuvant and an EAAAK linker, there are nine CTL epitopes followed which are connected by eight AAY linkers. Right after this region are five HTL epitopes coupled by GPGPG linkers. At the end the vaccine construct is composed of six B cell epitopes, which are joined together through KK linkers. These linkers attenuate the immunogenicity of the vaccine, avoid self-folding of the epitopes and finally improve the attachment of one epitope with another. These building blocks collectively generate the sequence of the Monkeypox vaccine.
2.7. Secondary structure (SS) and physiochemical properties of MVC
Protparam, a web-based physiochemical properties estimation resource, was used to compute the estimated half-life, molecular weight, pI, and other features of the MVC [45]. Moreover, for the secondary structural component's determination, PSIPRED webserver was used [46].
2.8. Modeling the 3D-structure of MVC and evaluation
The Robetta server [47] (https://robetta.bakerlab.org/) and the deep learning-based method RoseTTAFold were used to predict the 3D structure of the MVC. The server-generated five predicted models, and the first one which had the smallest estimated error was taken as the final structure.
2.9. Molecular docking of vaccine-TLR2
The toll-like receptor (TLR) plays an essential role in recognizing pathogens and is crucial for the appropriate development of immune responses within the cells [48]. To analyze the binding of the designed vaccine to immune cell receptors, it is docked to TLR2 (PDB ID: 6NIU) [49]. The docking is completed on the HawkDock server [50], and the default MM/GBSA scoring function was applied when finding the docking positions. The MM/GBSA is also responsible for the free energy decomposition analysis.
2.10. In silico cloning
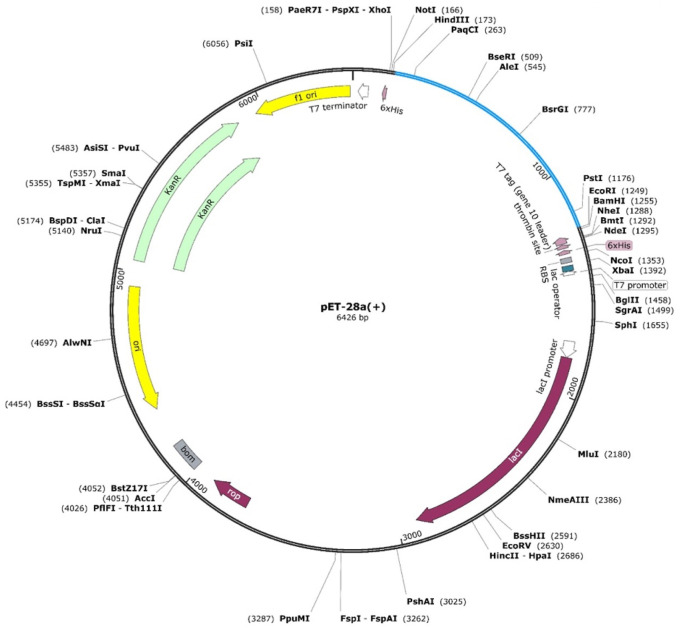
To make sure that host-based expression is effective, the vaccine created was cloned in Escherichia coli (strain K12). To evaluate whether the Monkeypox virus can respond specifically in such a system, codon optimization via reverse translation was performed on JCat (Java Codon Adaptation tool) [51]. To ensure the expected results can be obtained, three additional alternatives are selected: the prokaryote ribosome binding site, rho-independent transcript termination, and cleavage limit enzymes. Those exhibited codon adaptation index (CAI) and GC (Guanine-Cytosine) content by the JCat program were considered to guarantee the possibility of high levels of protein expression. The generated final vaccine is then cloned in protein expression specific E. coli pET-28a vector with certain restriction sites including NdeI and XhoI located at C and N terminals. Also, the pET-28a (β) plasmid maps with adapted vaccine sequences were constructed for optimized vaccine expression having futility in further experimental designs.
2.11. Immune simulation
To simulate the impact of foreign peptides as an antigen on the immune system, modeling based on agents (discrete entities) was performed on the C-ImmSim server (https://kraken.iac.rm.cnr.it/C-IMMSIM/index.php?page=1) [52,53]. It utilizes the PSSM (Position-specific score matrices) method to assess the antigen-specific immune response, including cytokines, interferon, and the production of antibodies for the injected Monkeypox vaccine. Since it is recommended that there is a gap of four weeks between the first and second doses for most of the vaccines nowadays, the default parameters of the simulation are maintained. It also predicts the Th1 and Th2 (T Helper Cell Type 1 & 2) responses. Also, the Simpson Index D is plotted with the default parameters.
3. Results and discussion
3.1. Vaccinomics targets annotation and antigenic profiling
There are 202 genes in total for the entire genome of the Monkeypox virus. Among the translated proteins, 61 are predicted to be potentially eligible to serve as antigens, in which 46 of which are not likely to have allergic side effects. Ten of them that have the highest antigenicity scores are selected for extracting epitopes to build up the vaccine. The basic information about these ten proteins is summarized in Table 1 .
Table 1.
Components of the monkeypox virus that are most antigenic and are non-allergic.
| UniProt ID | Protein Name | Length | Antigenicity Score |
|---|---|---|---|
| Q3I8I4 | B13R | 149 | 0.98 |
| Q3I8W2 | Abundant component of virosome | 67 | 0.97 |
| Q5QC90 | Bifunctional zinc finger-like protein/E | 242 | 0.97 |
| Q3I911 | Ankyrin | 56 | 0.94 |
| Q5IXT7 | DNA-directed RNA polymerase subunit | 185 | 0.93 |
| Q3I8N5 | A18L | 204 | 0.93 |
| Q8AZ47 | Bifunctional hemagglutinin/type-I membrane | 313 | 0.93 |
| Q5IXU5 | IMV membrane protein | 250 | 0.90 |
| Q3I907 | Interleukin-1 receptor antagonist-like protein | 83 | 0.89 |
| Q6YLY9 | CC-type chemokine binding protein | 246 | 0.89 |
3.2. Prediction of B cell epitopes
As an effective antigen, one of the fundamental requirements is that it can be recognized by the binding region of at least one type of B cell receptors. This receptor, which is a tester antibody, recognizes the antigen of the Monkeypox virus with its Fab region and makes the B cell with this specific antibody multiply. Since in reality, the B cell receptor binds only to a small region of the antigen, the vaccine with a number of these epitopes is more likely to make B cells produce antibodies that take the virus to the phagocytes [54,55]. This was the primary motivation to find such epitopes on the protein sequences of the Monkeypox virus. Within those ten protein sequences, due to the variation of the lengths, the number of epitopes predicted to be recognizable by B cell receptors also varies. Keeping the length of to the B cell epitopes as 16, there were a total of 157 epitopes that are qualified. Since it is difficult for an antigen to accommodate a large number of epitopes, we selected the one with the highest prediction score from each protein, and the relevant information is summarized in Table 2 .
Table 2.
Predicted B cell epitopes with their respective prediction scores.
| UniProt ID | Start position | Epitope sequence | Score |
|---|---|---|---|
| Q3I8I4 | 114 | YWGGEDHPTSNSLNAL | 0.90 |
| Q3I8W2 | 52 | ICLEMTIDSFVIECIQ | 0.80 |
| Q5QC90 | 78 | TGIQQSKLTETIRNCQ | 0.94 |
| Q3I911 | 2 | YSQIIKDITTELINNE | 0.82 |
| Q5IXT7 | 79 | GEPIIITSYLQKGHNK | 0.92 |
| Q3I8N5 | 45 | YGGIMNDYLGIFKNND | 0.93 |
| Q8AZ47 | 237 | KSTTDDADLYDTYNDN | 0.95 |
| Q5IXU5 | 15 | SERISSKLEQEANASA | 0.90 |
| Q3I907 | 66 | CVDIDNIITFMKYDPN | 0.93 |
| Q6YLY9 | 107 | ISESSDGNTVNTRLSS | 0.92 |
3.3. Prediction of CTL epitopes
Antibodies produced by B cells can label the viruses to make them be devoured by phagocytes such as macrophages, but this process cannot take place if the viruses have already entered the cells. In such a case, the killer T cell, that is, the cytotoxic T lymphocyte (CTL) is required to destroy the infected cells. To make this process happen, a fragment of antigen needs to be loaded onto the surface of the infected cell so that it can be recognized by CTL. In the human body, it is the major histocompatibility complex (MHC) that presents the antigen to the T cells, and class I MHC proteins are responsible specifically for sending the signal to the CTL. It loads this fragment of antigen, that is, the CTL epitope, to the surface of the infected cells [54]. To make the vaccine more effective, it is necessary to include a certain number of this type of epitope on the antigen. NetCTL-1.2 server makes such a prediction. Since the groove of class I MHC proteins is closed and can fit a peptide with almost exactly nine amino acid residues, the predicted CTL epitopes are restricted to this length. There are overall 51 peptide chains eligible to match the groove of class I MHC for the ten proteins. However, no qualified epitope is found on the protein Q3I911 (Ankyrin). Among the rest of the nine protein sequences, only one epitope from each protein with the highest prediction score was selected, which represents its binding affinity with class I MHC. The results are listed in Table 3 .
Table 3.
Predicted CTL epitopes with their respective prediction score.
3.4. Prediction of HTL epitopes
Besides the killer T cells (CTLs), it is equally essential to pass the signal of infection to the helper T cells (helper T lymphocytes, HTLs), who can then secrete cytokines during an immune response. The epitopes which can be recognized by HTLs are displayed on the surface of antigen-presenting cells, and it is the class II MHC molecules, encoded by genes in the HLA-D region, that present these epitopes. Since there are alleles of HLA-D on a chromosome, the IEDB server considers various possibilities and predicts whether an epitope can fit into the groove of the class II MHC molecule concerning a specific allele. As the groove of the class II MHC molecule is open, it can accommodate longer peptide chains. By keeping the length of each epitope to 15, the server predicts its affinity to class II MCH and compares it to other sequences with equal size in the database, and the final result is printed as a percentile rank. Similar to the previous two predictions, the epitope with the smallest rank is selected, and the relevant information is summarized in Table 4 .
Table 4.
Predicted HTL epitopes with their respective allele information and percentile ranks.
| UniProt ID | Alleles | Start position | HTL epitope sequence | Percentile rank |
|---|---|---|---|---|
| Q3I8I4 | HLA-DRB1*03:01 | 61 | AESVFIDVRELVKNM | 0.71 |
| Q3I8W2 | HLA-DRB3*02:02 | 20 | KTYKLFSDISVIGKA | 5.70 |
| Q5QC90 | HLA-DRB1*07:01 | 226 | RTRFRKITMSKFYKL | 0.22 |
| Q3I911 | HLA-DRB4*01:01 | 35 | DNNIRMMRTRIDRKK | 0.77 |
| Q5IXT7 | HLA-DRB3*02:02 | 161 | IVRFFRNNMVTGVEI | 0.01 |
| Q3I8N5 | HLA-DRB1*15:01 | 68 | ILFVLALYSPPLISI | 0.08 |
| Q8AZ47 | HLA-DRB5*01:01 | 37 | NNTNYYVVMSAWYKE | 0.06 |
| Q5IXU5 | HLA-DRB1*07:01 | 230 | TYMDTFFRTSPMIIA | 0.27 |
| Q3I907 | HLA-DRB1*03:01 | 21 | CIRNDIRELFKHVTD | 0.12 |
| Q6YLY9 | HLA-DRB3*01:01 | 74 | EEVVKGDPTTYYNIV | 0.91 |
3.5. Construction of multi-epitope vaccine
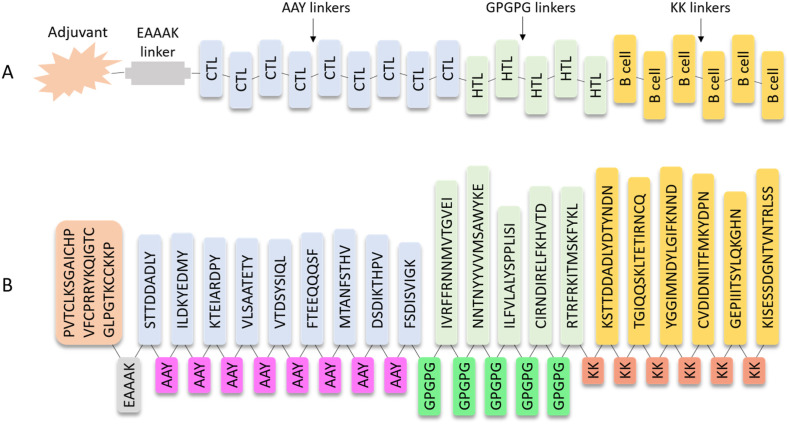
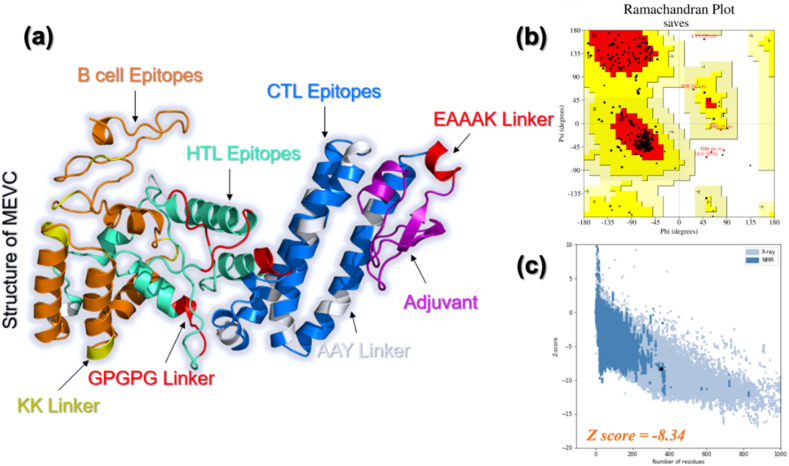
Based on the information above, we constructed a peptide by joining a certain number of epitopes that can be recognized by B cells or fit into class I and II MHC molecules together. This polypeptide is expected to be powerful to prevent the Monkeypox virus. In other words, it should be able to activate the adaptive immune system in various pathways so that the virus invaded can either be consumed by phagocytes or be destroyed by killer T cells. An adjuvant, which was constituted by a fragment of hDB-2 with 37 amino acid residues, was connected with nine CTL epitopes, five HTL epitopes, and six B cell epitopes reported in Table 2, Table 3, Table 4 using specific linkers (Fig. 1 A). The selection of the epitopes was based on their scores or percentile ranks in Table 2, Table 3, Table 4. The specific choice also obeys the defined molecular weight and experimental feasibility rules. The length of the designed vaccine is 355, and the entire sequence is displayed in Fig. 1B.
Fig. 1.
Construction of the multi-epitope vaccine. (A) General representation of designed multi-epitope vaccine against the Monkeypox virus and (B) Detailed sequence of the vaccine.
3.6. Construction of mRNA vaccine for MPXV
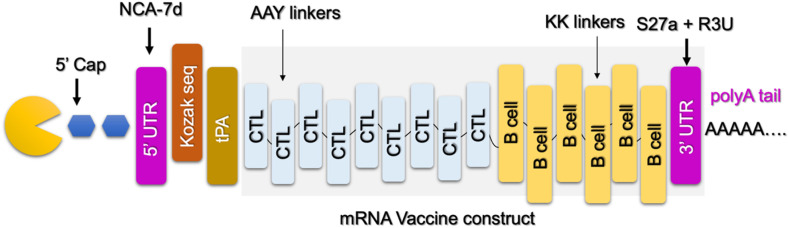
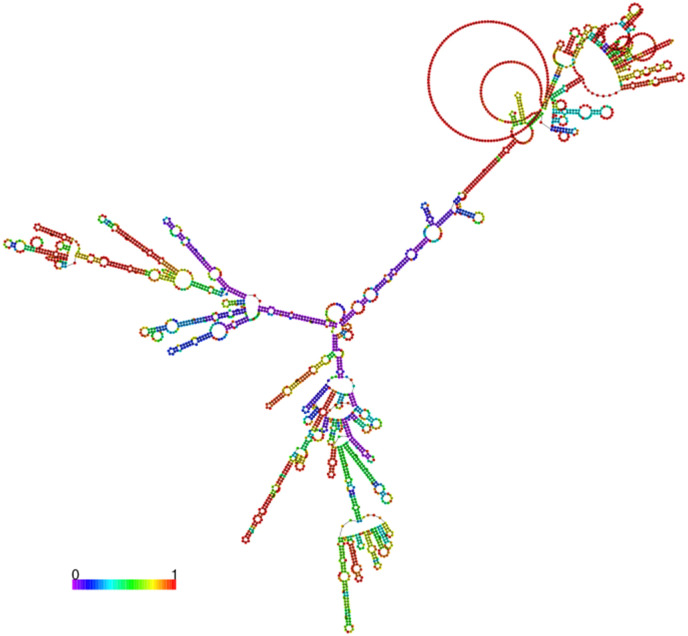
mRNA vaccines function by delivering a portion of mRNA that matches to a viral protein, often a short fragment of a protein present on the virus's outer membrane. Cells can manufacture the viral protein by utilizing this mRNA. The immune system identifies that the protein is foreign and creates specific proteins known as antibodies as part of a normal immunological response. To design the mRNA vaccine from these selected proteins and mapped epitopes, we only considered CTL and B cell epitopes joined by specific linkers. Nine CTL epitopes and 6 B cell epitopes were joined together to construct the mRNA vaccine. Moreover, essential elements required for the stability of mRNA, protection from degradation, and transportation were added. The designed mRNA vaccine for MPXV is shown in Fig. 2 . The secondary structure of mRNA was predicted which revealed the free energy of the thermodynamic ensemble as −1047.81 kcal/mol. The frequency of the MFE structure in the ensemble was calculated to be 0.00% while the ensemble diversity was calculated to be 667.12. The secondary structure of the mRNA is given in Fig. 3 .
Fig. 2.
Topographical organization of mRNA vaccine constructed for MPXV. The different elements of the mRNA vaccine such as 5′ capping, poly-A tail, and Kozak sequences are tagged at a particular point.
Fig. 3.
Secondary structure of the designed mRNA vaccine construct using RNAFold webserver.
3.7. Structural modeling and validation of MVC
The complete sequence of the designed MVC was then submitted to the Robetta server to predict the three-dimensional structure by RoseTTAFold algorithm. The generated geometry successfully adapted most of the secondary structural elements by the MVC, especially the fragment composed of CTL epitopes. The structure contains 2 beta-strands, 17 helices, and loops which connect these elements with each other. This shows that the structure modeled by Robetta contains enough secondary structural elements thus attaining proper folding and consequently causing proper functionality. The geometry of this vaccine plotted using the PyMOL program is demonstrated in Fig. 4 a. Various structure-based validation tools revealed that the 3D structure of MPXV-MVC has excellent structural folding with no deformities. For instance, ERRAT predicted an overall quality factor of 94%, which shows the correct folding, and bonding of atoms in the structure. VERIFY-3D demonstrated that 84.51% of the residues have averaged 3D-1D score ≥ 0.2 which further validates the accurate folding of the structure. Similarly, WHATCHECK PROVES and PROCHECK validated the 3D structure of the vaccine candidate. Ramachandran plot analysis as given in Fig. 4b, revealed that only 1% of the residues are in the disallowed region while the rest are in the favoured, additionally allowed, and generously allowed regions. On the other hand, the ProSA-web server predicted a Z-score of −8.34 which further validates the overall accurate folding of MVC. The ProSA-web results are shown in Fig. 4c.
Fig. 4.
3D-structural modeling and evaluation of MVC. (a) The predicted three-dimensional structure of the Monkeypox vaccine with the distribution of different epitopes and linkers. (b) The Ramachandran Plot validation results for MVC. (c) The ProSA-web results for MVC (the dark point, the deep and light blue points represent the Z-scores for available NMR and X-ray structures respectively).
3.8. Secondary structure (SS) analysis and physiochemical properties calculation
Analysis of the secondary structural elements i.e., alpha helices, beta-sheets, and loops regions demonstrate the functionality of a protein and determine the accurate folding. Determination of key secondary structure elements could be helpful in predicting the function of a protein. We herein predicted different SS elements in the structure of MVC. It was observed that 60% of the residues lie in the helices, 2.54% are beta-strands and the rest 37.46% are loops that connect different helices and beta-sheets. The SS distribution is shown in Fig. 5 . This shows that the designed MVC has proper SS and folding and can be used for further processing. Physiochemical properties of the MVC revealed an optimum molecular weight of 43.70 kDa, enough expression for downstream processing and good GRAVY (−0.410), and pI value of 9.10. Moreover, the estimated half-life was more than 20 h which is appropriate for downstream recovery.
Fig. 5.
Distribution of the secondary structural elements i.e., Alpha-helices (pink), beta-strand (yellow), and loops (gray) in the MVC.
3.9. Molecular docking and binding free energy calculation for TLR2-MVC complex
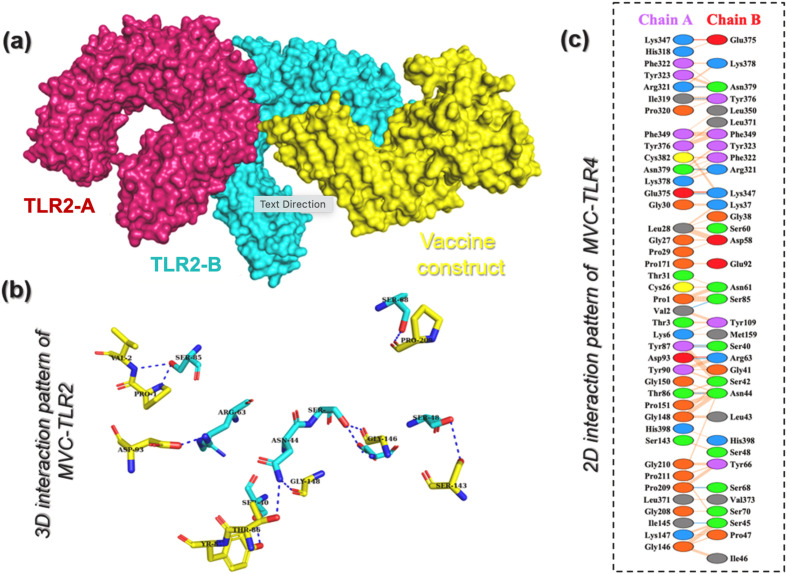
After invading the host, MPXV attacks dendritic cells (DCs) and macrophages and inhibits the stimulation of the host's innate immunity. TLRs are essential inflammatory pathway regulators that play critical roles in modulating immunogenicity against infections. TLRs identify pathogen-associated molecular patterns (PAMPs), causing gene expression to alter the intracellular cascades. The host's innate immune system recognizes the invaders and responds appropriately via TLR recognition. TLRs have a vital role in identifying different components of Monkeypox viruses, such as nucleic acids and envelope glycoproteins, which trigger several cascades that include the creation of IFN-I, inflammatory cytokines, and chemokines. Furthermore, when TLRs stimulate dendritic cell maturation, adaptive immune responses are triggered (DCs) [48]. Thus, considering the interaction between TLRs and the designed vaccine is essential to forecast the immune response. Docking of the MVC designed in this work to TLR2 revealed a docking score of −274.68 kcal/mol which shows that the designed MVC in this work can robustly instigate the immune response more than our previously designed MVC [48]. The interaction pattern of MVC with TLR2 in 3D is shown in Fig. 6 a and b. Analysis of the interaction pattern revealed that the complex established 11 hydrogen bonds including Val2-Ser85, Thr86-Asn44, Tyr87-Ser40, Tyr87-Ser40, Asp93-Arg63, Ile145-Ser45, Pro209-Ser68, Ile319-Tyr376, Arg321-Asn379, Glu375-Lys347, and Asn379-Arg321, and three salt bridges including Asp93-Arg63, Lys347-Glu375, and Lys347-Glu375. The MVC designed in this work showed more interactions with TLR2 than our prevised designed MVC, implying its higher efficiency in instigating immune response. The 2D interaction pattern of TLR2-MVC is presented in Fig. 6c. Furthermore, the HAWKDOCK server also used MMPBSA.py module of AMBER package to calculate the binding free energy, which revealed a vdW of −250.56 kcal/mol, electrostatic energy of −180.34 kcal/mol, and total binding energy of −77.0 kcal/mol respectively. This shows robust stabilized binding of MVC with TLR2 and thus may produce a stronger immune response upon the interaction.
Fig. 6.
Interaction analysis of TLR2 and MVC. (a) Represent the surface view of the TLR2 in complex with MVC. (b) Show the 3D interaction pattern of TLR2 and MVC while (c) demonstrate the 2D interaction pattern of TLR2 and MVC. The blue lines represent hydrogen bonds, the red lines represent salt bridges while orange lines show non-bonded contacts.
3.10. In silico cloning and expression
The maximum expression of the vaccine construct was achieved through codon optimization using in silico JCat codon optimization tool. Our results revealed that the vaccine construct sequence optimized for E. Coli K12 strain had a GC content of 52% (optimum range = 30–70%) and a CAI value of 0.96, which indicated reasonable stability and high expression of our designed MVC. Moreover, these values are better than our previous in silico designed vaccine candidates for MPXV [48]. Furthermore, Xho1 and EcoR1 restriction sites were added to the optimized MVC sequence and cloned into the pET-28a (+) expression vector (Fig. 7 ). These results demonstrate stable and high expression in the experimental model.
Fig. 7.
Codon optimization and in silico cloning of MVC. The designed vaccine is shown in sky blue colour in the expression vector with their respective restriction sites.
3.11. In silico immune simulation
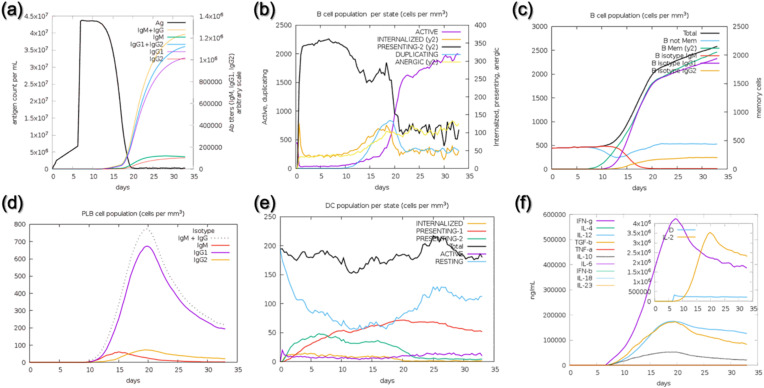
Investigation of whether or not the designed vaccine will instigate the immune response is a critical step in defining the immune potential of such agents against the pathogen. In order to validate our MVC, we employed an agent-based modeling approach to determine the immune potential and see which immune components are released immediately. Specifically, we used C-ImmSim to simulate the immune reaction as a consequence of a single injection of our designed MVC. As given in Fig. 8 a, the antigen titer after the injection reached to a maximum level on the 5th day however, after the antigen recognition, primary and secondary immune response factors were released as a response to the invader. After the instigation of immune response abrupt decline in the antigen titer was observed. On the 20th day, a complete clearance of the antigen can be seen with an increase of the primary and secondary antibodies. The B cell population count can also be seen in Fig. 8b and c, where time neutralization of the Ag was observed. The production of primary immune factors i.e., IgM, IgG, and IgM + IgG together can be seen to have a maximum level after the 10th day of injection. The results of IgM, IgG, IgM + IgG, and dendritic cell production are shown in Fig. 8d and e. We also investigated the level of secondary immune response factors i.e., IFN, IL and other types of IL (s) to further evaluate the potential of MVC. IFN-gamma and IL (interleukins) levels reached to a maximum after the 10th day. (Fig. 8f). The levels of the aforementioned immune factors were significantly higher, which shows the vigorous and steady immune triggering response upon injection. The above data showed that the constructed vaccines can robustly induce immune responses against the invading Monkeypox virus.
Fig. 8.
Immune simulation-based validation of the designed MVC. (a) Injection of Ag and production of an immune response, (b–c) levels of B cell population per mm3, (d) production of primary and secondary antibodies, (e) Dendritic cell population after the Ag injection while (f) shows the other immune response factors required for Ag neutralization.
4. Conclusions
When compared to traditional vaccine development, computationally developed vaccines are thermodynamically stable, efficacious, specific, and affordable in cost. We employed immunoinformatics techniques to design highly immunogenic, thermostable, and non-allergenic MVC and mRNA-based vaccines against the Monkeypox virus in this study. The whole proteome of MPXV was analyzed and potential vaccine targets were obtained which were then used for epitope mapping. We generated MVC and mRNA-based vaccines that possess a highly stable nature, efficient expression properties, and immunogenic. The designed vaccines were validated through in silico immune simulation which instigated vigorous immune response by releasing higher titers of primary and secondary antibodies. The current study lacks experimental validation and optimization of the vaccine construct through different linkers and adjuvant attachment may further increase the immune response against MPXV. In conclusion, the current work developed dynamic and effective vaccines against the Monkeypox virus that are built of highly antigenic and non-allergenic peptides, prompting additional experimental testing.
Funding
This work was supported by the National Key R&D Program of China (Grant No. 2021YFA0911500) and Shanghai Pujiang Program (Grant No. 21PJ1405100). Dong-Qing Wei was supported by the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (No. 19430750600), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2021ZD02). Yanjing Wang was supported by the grants from the National Natural Science Foundation of China (No.32200531), the Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2022QN114 and YG2022QN082), and Startup Fund for Young Faculty at SJTU (SFYF at SJTU). Yifan Jin was supported by China Postdoctoral Science Foundation (2022M722100).
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgments
The computations were partially performed at the Center for High-Performance Computing of Shanghai Jiao Tong University and Pengcheng Lab. Authors are also thankful to the Researchers Supporting Project number (RSP2023R491), King Saud University, Riyadh, Saudi Arabia.
References
- 1.Magnus P.v., Andersen E.K., Petersen K.B., Birch-Andersen A. A POX-like disease in cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 1959;46:156–176. [Google Scholar]
- 2.Dou Y.M., Yuan H., Tian H.W. Monkeypox virus: past and present. World J. Pediatr. 2022;19 doi: 10.1007/s12519-022-00618-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gessain A., Nakoune E., Yazdanpanah Y. Monkeypox. New Engl. J. Med. 2022;10 doi: 10.1056/NEJMra2208860. [DOI] [PubMed] [Google Scholar]
- 4.Hutin Y.J.F., Williams R.J., Malfait P., Pebody R., Loparev V.N., Ropp S.L., Rodriguez M., Knight J.C., Tshioko F.K., Khan A.S., Szczeniowski M.V., Esposito J.J. Outbreak of human monkeypox, democratic republic of Congo, 1996-1997. Emerg. Infect. Dis. 2001;7:434–438. doi: 10.3201/eid0703.010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Giulio D.B., Eckburg P.B. Human monkeypox: an emerging zoonosis. Lancet Infect. Dis. 2004;4:15–25. doi: 10.1016/S1473-3099(03)00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nalca A., Rimoin A.W., Bavari S., Whitehouse C.A. Reemergence of monkeypox: prevalence, diagnostics, and countermeasures. Clin. Infect. Dis. 2005;41:1765–1771. doi: 10.1086/498155. [DOI] [PubMed] [Google Scholar]
- 7.Rimoin A.W., Mulembakani P.M., Johnston S.C., Lloyd-Smith J.O., Kisalu N.K., Kinkela T.L., Blumberg S., Thomassen H.A., Pike B.L., Fair J.N., Wolfe N.D., Shongo R.L., Graham B.S., Formenty P., Okitolonda E., Hensley L.E., Meyer H., Wright L.L., Muyembe J.J. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16262–16267. doi: 10.1073/pnas.1005769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yinka-Ogunleye A., Aruna O., Dalhat M., Ogoina D., McCollum A., Disu Y., Mamadu I., Akinpelu A., Ahmad A., Burga J., Ndoreraho A., Nkunzimana E., Manneh L., Mohammed A., Adeoye O., Tom-Aba D., Silenou B., Ipadeola O., Saleh M., Adeyemo A., Nwadiutor I., Aworabhi N., Uke P., John D., Wakama P., Reynolds M., Mauldin M.R., Doty J., Wilkins K., Musa J., Khalakdina A., Adedeji A., Mba N., Ojo O., Krause G., Ihekweazu C., Mandra A., Davidson W., Reynolds M.G., Olson V., McCollum A.M., Doty J., Wilkins K., Li Y., Radford K., Mauldin M.R., Zhao H., Townsend M., Burgado J., Satheshkumar P.S., Team C.D.C.M.O. Outbreak of human monkeypox in Nigeria in 2017-18: a clinical and epidemiological report. Lancet Infect. Dis. 2019;19:872–879. doi: 10.1016/S1473-3099(19)30294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shafaati M., Zandi M. Human monkeypox (hMPXV) re-emergence: host immunity status and current vaccines landscape. J. Med. Virol. 2022;n/a doi: 10.1002/jmv.28251. [DOI] [PubMed] [Google Scholar]
- 10.Lai C.-C., Hsu C.-K., Yen M.-Y., Lee P.-I., Ko W.-C., Hsueh P.-R. Monkeypox: an emerging global threat during the COVID-19 pandemic. J. Microbiol. Immunol. Infect. 2022;55:787–794. doi: 10.1016/j.jmii.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farasani A. Monkeypox virus: future role in Human population. J. Infect. Public Health. 2022;15:1270–1275. doi: 10.1016/j.jiph.2022.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vivancos R., Anderson C., Blomquist P., Balasegaram S., Bell A., Bishop L., Brown C.S., Chow Y., Edeghere O., Florence I., Logan S., Manley P., Crowe W., McAuley A., Shankar A.G., Mora-Peris B., Paranthaman K., Prochazka M., Ryan C., Simons D., Vipond R., Byers C., Watkins N.A., Welfare W., Whittaker E., Dewsnap C., Wilson A., Young Y., Chand M., Riley S., Hopkins S., Managemen U.M.I. Community transmission of monkeypox in the United Kingdom, april to may 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poland G.A., Kennedy R.B., Tosh P.K. Prevention of monkeypox with vaccines: a rapid review. Lancet Infect. Dis. 2022;22 doi: 10.1016/S1473-3099(22)00574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earl P.L., Americo J.L., Wyatt L.S., Eller L.A., Whitbeck J.C., Cohen G.H., Eisenberg R.J., Hartmann C.J., Jackson D.L., Kulesh D.A., Martinez M.J., Miller D.M., Mucker E.M., Shamblin J.D., Zwiers S.H., Huggins J.W., Jahrling P.B., Moss B. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature. 2004;428:182–185. doi: 10.1038/nature02331. [DOI] [PubMed] [Google Scholar]
- 16.Edghill-Smith Y., Golding H., Manischewitz J., King L.R., Scott D., Bray M., Nalca A., Hooper J.W., Whitehouse C.A., Schmitz J.E., Reimann K.A., Franchini G. Smallpox vaccine–induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 2005;11:740–747. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 17.Hirao L.A., Draghia-Akli R., Prigge J.T., Yang M., Satishchandran A., Wu L., Hammarlund E., Khan A.S., Babas T., Rhodes L., Silvera P., Slifka M., Sardesai N.Y., Weiner D.B. Multivalent smallpox DNA vaccine delivered by intradermal electroporation drives protective immunity in nonhuman primates against lethal monkeypox challenge. J. Infect. Dis. 2011;203:95–102. doi: 10.1093/infdis/jiq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner M., Mandia J., Keltner C., Haynes R., Faestel P., Mease L. Monkeypox in patient immunized with ACAM2000 smallpox vaccine during 2022 outbreak. Emerg. Infect. Dis. 2022;28:2336. doi: 10.3201/eid2811.221215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdelaal A., Reda A., Lashin B.I., Katamesh B.E., Brakat A.M., Al-Manaseer B.M., Kaur S., Asija A., Patel N.K., Basnyat S., Rabaan A.A., Alhumaid S., Albayat H., Aljeldah M., Al Shammari B.R., Al-Najjar A.H., Al-Jassem A.K., AlShurbaji S.T., Alshahrani F.S., Alynbiawi A., Alfaraj Z.H., Alfaraj D.H., Aldawood A.H., Sedhai Y.R., Mumbo V., Rodriguez-Morales A.J., Sah R. Preventing the next pandemic: is live vaccine efficacious against monkeypox, or is there a need for killed virus and mRNA vaccines? Vaccines. 2022;10 doi: 10.3390/vaccines10091419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappuoli R. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 2007;25:1361–1366. doi: 10.1038/nbt1207-1361. [DOI] [PubMed] [Google Scholar]
- 21.Ulmer J.B., Mason P.W., Geall A., Mandl C.W. RNA-based vaccines. Vaccine. 2012;30:4414–4418. doi: 10.1016/j.vaccine.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 22.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K.J., Southern J., Swerdlow D.L., Jodar L., Levy Y., Alroy-Preis S. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397:1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., Feldman J., Roederer A.L., Gregory D.J., Poznansky M.C., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–+. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Likos A.M., Sammons S.A., Olson V.A., Frace A.M., Li Y., Olsen-Rasmussen M., Davidson W., Galloway R., Khristova M.L., Reynolds M.G., Zhao H., Carroll D.S., Curns A., Formenty P., Esposito J.J., Regnery R.L., Damon I.K. A tale of two clades: monkeypox viruses. J. Gen. Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- 26.Chen N., Li G., Liszewski M.K., Atkinson J.P., Jahrling P.B., Feng Z., Schriewer J., Buck C., Wang C., Lefkowitz E.J., Esposito J.J., Harms T., Damon I.K., Roper R.L., Upton C., Buller R.M.L. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopera J.G., Falendysz E.A., Rocke T.E., Osorio J.E. Attenuation of monkeypox virus by deletion of genomic regions. Virology. 2015;475:129–138. doi: 10.1016/j.virol.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caoili S.E.C. Benchmarking B-cell epitope prediction for the design of peptide-based vaccines: problems and prospects. J. Biomed. Biotechnol. 2010;2010 doi: 10.1155/2010/910524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patronov A., Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3 doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaech S.M., Wherry E.J., Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 31.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W., GenBank Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera-Bravo J., Farías J.G., Contreras F.P., Herrera-Belén L., Norambuena J.-A., Beltrán J.F. VirVACPRED: a web server for prediction of protective viral antigens. Int. J. Pept. Res. Therapeut. 2021;28:35. doi: 10.1007/s10989-021-10345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP v.2—a server for in silico prediction of allergens. J. Mol. Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 34.Saha S., Raghava G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: Struct., Funct., Bioinf. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 35.Saha S., Raghava G.P.S. In: Immunoinformatics: Predicting Immunogenicity in Silico. Flower D.R., editor. Humana Press; Totowa, NJ: 2007. Prediction methods for B-cell epitopes; pp. 387–394. [Google Scholar]
- 36.Crux N.B., Elahi S. Human leukocyte antigen (HLA) and immune regulation: how do classical and non-classical HLA alleles modulate immune response to human immunodeficiency virus and hepatitis C virus infections? Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wieczorek M., Abualrous E.T., Sticht J., Alvaro-Benito M., Stolzenberg S., Noe F., Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P., Sidney J., Dow C., Mothe B., Sette A., Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delves P.J., Roitt I.M. The immune system. N. Engl. J. Med. 2000;343:108–117. doi: 10.1056/NEJM200007133430207. [DOI] [PubMed] [Google Scholar]
- 41.Rubinstein N.D., Mayrose I., Martz E., Pupko T. Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinf. 2009;10:287. doi: 10.1186/1471-2105-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA website. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoover D.M., Rajashankar K.R., Blumenthal R., Puri A., Oppenheim J.J., Chertov O., Lubkowski J. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 2000;275:32911–32918. doi: 10.1074/jbc.M006098200. [DOI] [PubMed] [Google Scholar]
- 45.Garg V.K., Avashthi H., Tiwari A., Jain P.A., Ramkete P.W., Kayastha A.M., Singh V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation. 2016;12:74. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchan D.W., Jones D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., Wang J., Cong Q., Kinch L.N., Schaeffer R.D., Millán C., Park H., Adams C., Glassman C.R., DeGiovanni A., Pereira J.H., Rodrigues A.V., van Dijk A.A., Ebrecht A.C., Opperman D.J., Sagmeister T., Buhlheller C., Pavkov-Keller T., Rathinaswamy M.K., Dalwadi U., Yip C.K., Burke J.E., Garcia K.C., Grishin N.V., Adams P.D., Read R.J., Baker D. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suleman M., Rashid F., Ali S., Sher H., Luo S., Xie L., Xie Z. Immunoinformatic-based design of immune-boosting multiepitope subunit vaccines against monkeypox virus and validation through molecular dynamics and immune simulation. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1042997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishida H., Asami J., Zhang Z., Nishizawa T., Shigematsu H., Ohto U., Shimizu T. Cryo-EM structures of Toll-like receptors in complex with UNC93B1. Nat. Struct. Mol. Biol. 2021;28:173–180. doi: 10.1038/s41594-020-00542-w. [DOI] [PubMed] [Google Scholar]
- 50.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rapin N., Lund O., Bernaschi M., Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castiglione F., Deb D., Srivastava A.P., Liò P., Liso A. From infection to immunity: understanding the response to SARS-CoV2 through in-silico modeling. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.646972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan A., Khan S., Saleem S., Nizam-Uddin N., Mohammad A., Khan T., Ahmad S., Arshad M., Ali S.S., Suleman M. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai D.-Y., Hung K.-H., Chang C.-W., Lin K.-I. Regulatory mechanisms of B cell responses and the implication in B cell-related diseases. J. Biomed. Sci. 2019;26:1–13. doi: 10.1186/s12929-019-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]