Abstract
Due to the difficulty in accurately identifying structural variants (SVs) across genomes, their impact on cis-regulatory divergence of closely related species, especially fish, remains to be explored. Recently identified broad H3K4me3 domains are essential for the regulation of genes involved in several biological processes. However, the role of broad H3K4me3 domains in phenotypic divergence remains poorly understood. Siniperca chuatsi and S. scherzeri are closely related but divergent in several phenotypic traits, making them an ideal model to study cis-regulatory evolution in sister species. Here, we generated chromosome-level genomes of S. chuatsi and S. scherzeri, with assembled genome sizes of 716.35 and 740.54 Mb, respectively. The evolutionary histories of S. chuatsi and S. scherzeri were studied by inferring dynamic changes in ancestral population sizes. To explore the genetic basis of adaptation in S. chuatsi and S. scherzeri, we performed gene family expansion and contraction analysis and identified positively selected genes (PSGs). To investigate the role of SVs in cis-regulatory divergence of closely related fish species, we identified high-quality SVs as well as divergent H3K27ac and H3K4me3 domains in the genomes of S. chuatsi and S. scherzeri. Integrated analysis revealed that cis-regulatory divergence caused by SVs played an essential role in phenotypic divergence between S. chuatsi and S. scherzeri. Additionally, divergent broad H3K4me3 domains were mostly associated with cancer-related genes in S. chuatsi and S. scherzeri and contributed to their phenotypic divergence.
Keywords: cis-regulatory divergence, Structural variants, H3K27ac, Broad H3K4me3, Siniperca chuatsi, Siniperca scherzeri
INTRODUCTION
Evolutionary biologists have long sought to elucidate which genetic variants contribute to the evolution of morphological diversity (Carroll, 2008). Accurate and robust regulation of gene expression is critical for the development of organisms and is a common source of evolutionary change (Long et al., 2016). Accumulating empirical evidence suggests that variation in the regulation of gene expression contributes to morphological variation among species, populations, and individuals, which may, in turn, lead to adaptation and speciation of various species (Brawand et al., 2014; Verta & Jones, 2019). Thus, dissecting the genetic basis of variation in the regulation of gene expression is essential for understanding phenotypic evolution (Wittkopp & Kalay, 2012).
Variation in gene expression can result from mutations in cis-regulatory elements (CREs), which are collections of transcription binding sites and other non-coding DNA required for transcriptional activation (Carroll, 2013; Ong & Corces, 2011). Several types of mutations responsible for cis-regulatory divergence between species have been identified. Nucleotide substitutions due to point mutations can cause divergence in cis-regulatory activity (Wittkopp & Kalay, 2012). In addition, structural variants (SVs) intersecting with CREs of several developmental regulatory genes can cause morphological divergence between species (Chan et al., 2010; Livraghi et al., 2021). Recently, the availability of high-quality reference genomes for several species has enabled genome-wide analysis of SVs in cis-regulatory divergence, revealing that gene expression is widely impacted by SVs affecting CREs (Alonge et al., 2020; Chiang et al., 2017). However, comprehensive studies of the role of SVs in cis-regulatory divergence between closely related species, especially fish, are still scarce.
Histones at different CREs, including promoters, enhancers, silencers, and insulators, exhibit specific post-translational modifications (Blakey & Litt, 2015). Genome-wide profiling of CREs using chromatin immunoprecipitation sequencing (ChIP-seq) or the recently developed Cleavage Under Targets and Tagmentation (CUT&Tag) strategy revealed the evolution of these regulatory elements (Kaya-Okur et al., 2019; Park, 2009). Histone H3 lysine 27 acetylation (H3K27ac) is an important epigenetic mark associated with active enhancers and promoters (Creyghton et al., 2010). Study of H3K27ac in the genome found that enhancers are variable among species and responsible for cis-regulatory divergence (Levine, 2010). In addition to H3K27ac, typical histone H3 lysine 4 trimethylation (H3K4me3), which is restricted to narrow regions (1–2 kb) at the 5’ end of genes, is one of the most well-recognized epigenetic marks of active transcription (Dong et al., 2012). Recent studies have revealed that enrichment site breadth plays a key role in determining the functions of H3K4me3 (Lv & Chen, 2016). Broad H3K4me3 domains are essential for the regulation of genes involved in cell identity specification, embryonic development, and tumor suppression (Benayoun et al., 2014; Chen et al., 2015; Dahl et al., 2016). Nevertheless, the role of broad H3K4me3 domains in phenotypic divergence remains poorly understood.
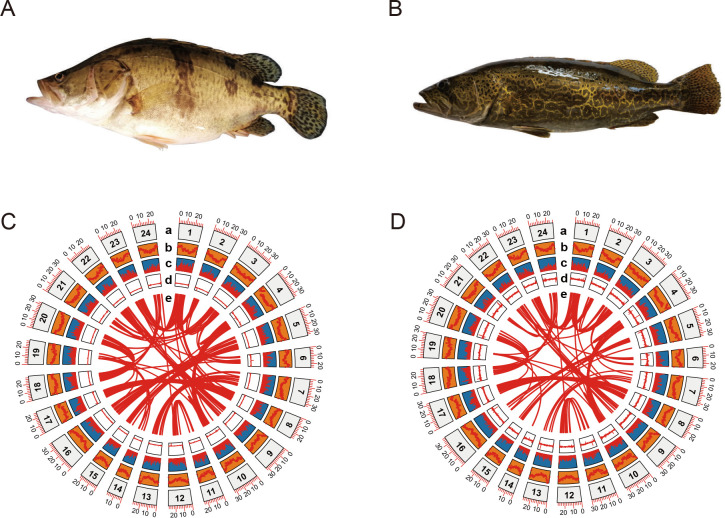
Sinipercidae (Perciforms) is a subfamily of freshwater fish comprised of three genera, including Siniperca, Coreoperca, and Coreosiniperca. Sinipercids are endemic to East Asia (China, Korea, Japan, and Vietnam) and most species are distributed in China. Mandarin fish (Siniperca chuatsi) and leopard mandarin fish (Siniperca scherzeri) are two closely related Siniperca species (Song et al., 2017). Unlike other sinipercids distributed in the river systems of South China, S. chuatsi and S. scherzeri are widely dispersed from south to north in China and are well-adapted to diverse environments (Li, 1991). Siniperca chuatsi and S. scherzeri differ in several phenotypic traits. First, the two species are divergent in body length, body width, and skin pigmentation (Figure 1A, B) (Guan et al., 2022). Second, S. chuatsi exhibits a substantially higher growth rate than S. scherzeri but is also more susceptible to disease (Ding et al., 2022). Third, although sinipercids are typical innate and obligate piscivores that feed solely on the live fry of other fish species, S. scherzeri will accept dead prey fish or artificial diets more easily than S. chuatsi (He et al., 2013). Their strong phenotypic differences make S. chuatsi and S. scherzeri ideal models for studying cis-regulatory divergence in closely related species.
Figure 1.
Genome assemblies of S. chuatsi and S. scherzeri
A, B: Large morphological divergence exists between S. chuatsi (A) and S. scherzeri (B). C, D: Concentric circles show structural, functional, and evolutionary aspects of S. chuatsi (C) and S. scherzeri (D) genomes. a: Chromosome number; b: Repeat density; c: Gene density; d: GC content; e: Paralogous relationships within genome.
Here, we used Nanopore sequencing and Hi-C data to assemble high-quality genomes of S. chuatsi and S. scherzeri. The evolutionary histories of S. chuatsi and S. scherzeri were studied by inferring dynamic changes in ancestral population sizes. The genetic basis of adaptation in S. chuatsi and S. scherzeri was dissected by performing gene family expansion and contraction analysis and identifying positively selected genes (PSGs). To investigate the role of SVs in cis-regulatory divergence of closely related fish species, we identified high-quality SVs between S. chuatsi and S. scherzeri, as well as the H3K27ac and H3K4me3 domains. Integrated analysis revealed that gene expression variation caused by SVs affecting cis-regulatory regions played an essential role in the phenotypic divergence between S. chuatsi and S. scherzeri. Additionally, the broad H3K4me3 domains contributed to phenotypic divergence between S. chuatsi and S. scherzeri.
MATERIALS AND METHODS
Ethics approval
All fish operations were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (Protocol No. SYSU-IACUC-2020-B0975). All efforts were made to minimize animal suffering.
Genome sequencing
One male S. chuatsi and one male S. scherzeri collected from Fujian Province, China, were used for genome sequencing. High-quality DNA was extracted from the muscle cells of S. chuatsi and S. scherzeri using a DNeasy Blood & Tissue Kit (Qiagen, Germany) in accordance with the manufacturer’s protocols. DNA quality and quantity were measured via standard agarose-gel electrophoresis and a Qubit 3.0 fluorometer (Invitrogen, USA), respectively. Nanopore sequencing libraries of S. chuatsi and S. scherzeri were constructed and sequenced using the Nanopore PromethION platform (Oxford Nanopore Technologies, UK) (140X raw-read coverage for S. chuatsi; 127X raw-read coverage for S. scherzeri). For Illumina sequencing, short-insert paired-end (PE) (150 bp) DNA libraries of S. chuatsi and S. scherzeri were constructed in accordance with the manufacturer’s instructions. Sequencing of PE libraries was performed (2×150 bp) on the Illumina NovaSeq 6000 platform (Illumina, USA).
Transcriptome sequencing for genome annotation
Eye, gill, heart, intestine, kidney, stomach, testis, liver, and spleen samples were collected from the S. chuatsi and S. scherzeri specimens to construct sequencing libraries for strand-specific RNA-sequencing (RNA-seq). Total RNA was extracted with TRIzol reagent (Invitrogen, USA). Purity and integrity were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and Bioanalyzer 2100 (Agilent, USA), respectively. The mRNA was enriched from total RNA using poly-T oligo-attached magnetic beads. rRNA was removed using a TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA). A PE library was constructed using a VAHTSTM mRNA-seq V2 Library Prep Kit for Illumina (Vazyme, China) and sequenced (2×150 bp) using the Illumina HiSeq NovaSeq 6000 platform (Illumina, USA).
To construct full-length RNA-seq libraries, total RNA was extracted from muscle cells of S. chuatsi and S. scherzeri with TRNzol Universal reagent (TIANGEN, China). RNA integrity was determined using a NanoDrop One Microvolume UV-Vis spectrophotometer (Thermo Fisher Scientific, USA) and Bioanalyzer 2100 (Agilent Technologies, USA). RNA concentration was determined using a Qubit 3.0 fluorometer (Invitrogen, USA). A SMARTer PCR cDNA Synthesis Kit (Takara, Japan) was used for first-strand cDNA synthesis. Polymerase chain reaction (PCR) was performed to generate double-stranded cDNA using PrimeSTAR GXL DNA polymerase (Takara, Japan). Sequencing libraries of both species were then constructed using a SMRTbell Express Template Prep Kit 2.0 and Sequel Binding Kit 3.0 (Pacific Biosciences, USA) in accordance with the manufacturer’s protocols. Sequencing was performed using the PacBio Sequel II platform (Pacific Bioscience, USA).
Genome size estimation and genome assembly
Low-quality reads (≥10% unidentified nucleotides and/or ≥50% nucleotides with a Phred score <5) and sequencing adapter-contaminated Illumina reads were filtered and trimmed with Fastp (v0.21.0) (Chen et al., 2018a). The resulting high-quality Illumina reads were used in the following analyses. Siniperca chuatsi and S. scherzeri genome size and heterozygosity were estimated using high-quality Illumina reads based on k-mer frequency distribution. The number of k-mers and peak depth of k-mer size at k=21 were obtained using Jellyfish (v2.3.0) (Marçais & Kingsford, 2011) with the -C setting. The Jellyfish results were then inputted into GenomeScope2 (v1.0.0) (Ranallo-Benavidez et al., 2020) to estimate genome size and heterozygosity rate. The size of the S. chuatsi genome was also estimated based on previous flow cytometry results (Cui et al., 1991).
Low-quality Nanopore reads were filtered using a previously published Python script (Zhang et al., 2022). The filtered reads were then corrected using NextDenovo (v1.0) (https://github.com/Nextomics/NextDenovo). Draft genome assemblies for S. chuatsi and S. scherzeri were generated using filtered and corrected reads with WTDBG (v1.2.8) (Ruan & Li, 2020). The contigs of the two draft assemblies were subjected to error correction using high-quality Illumina reads with Pilon (v1.23) three times (Walker et al., 2014).
We used Hi-C to correct mis-joins, order and orient contigs, and merge overlaps. Liver samples from S. chuatsi and S. scherzeri were collected and used to construct Hi-C libraries based on a previously published approach (Belton et al., 2012). The Hi-C libraries of S. chuatsi and S. scherzeri were sequenced (2×150 bp) on the Illumina MiSeq platform (Illumina, USA). Low-quality sequencing reads were filtered using fastp (v0.21.0) (Chen et al., 2018a) with default parameters. Filtered Illumina reads were aligned to the respective assembled contigs of each species using Juicer (v1.5.7) (Durand et al., 2016). Scaffolding was accomplished using the 3D-DNA pipeline (v180419) (Dudchenko et al., 2017). Juicebox (v1.9.9) was used to modify the order and direction of certain scaffolds in a Hi-C contact map and to help determine chromosome boundaries (Robinson et al., 2018).
The completeness and quality of the final S. chuatsi and S. scherzeri genome assemblies were first evaluated using Benchmarking Universal Single-Copy Orthologs (BUSCO) (v4.0.5) (Simão et al., 2015) against the conserved Actinopterygii dataset (odb10). Second, RNA-seq reads of S. chuatsi brain, intestine, liver, and muscle were downloaded from the NCBI database (He et al., 2020). Previously published reads and our RNA-seq reads of S. chuatsi were aligned to the previously published assembly and our assembly of the S. chuatsi genome using HISAT2 (v2-2.1) (Kim et al., 2019). RNA-seq reads of S. scherzeri generated in this and previous studies were aligned to the previously published assembly and our assembly of the S. scherzeri genome using HISAT2 (v2-2.1). Third, Merqury (v1.3) (Rhie et al., 2020) was used to assess the completeness and quality of the four assemblies with k-mer set to 20.
Genome annotation
Repetitive elements in the assembly were identified by de novo predictions using RepeatMasker (v4.1.0) (https://www.repeatmasker.org/). RepeatModeler (v2.0.1) (Flynn et al., 2020) was used to build the de novo repeat libraries of S. chuatsi and S. scherzeri. To identify repetitive elements, sequences from the assemblies were aligned to the de novo repeat library using RepeatMasker (v4.1.0). Additionally, repetitive elements in the S. chuatsi and S. scherzeri genome assemblies were identified by homology searches against known repeat databases using RepeatMasker (v4.1.0).
Protein-coding genes in the S. chuatsi and S. scherzeri genomes were predicted using homology-based, ab initio, and RNA-seq-based prediction. For homology-based prediction, protein-coding sequences of Danio rerio, Gasterosteus aculeatus, Takifugu rubripes, Oryzias latipes, and Tetraodon nigroviridis were downloaded from Ensembl (v96) (Yates et al., 2020), and aligned to both the S. chuatsi and S. scherzeri genomes using tBLASTn. GenomeThreader (v1.7.0) (Gremme et al., 2005) was employed to predict gene models based on alignment with an E-value cut-off of 10−5. For ab initio gene prediction, short RNA-seq reads of nine samples (eye, gill, heart, intestine, kidney, stomach, testis, liver, spleen) of S. chuatsi and S. scherzeri were aligned to the respective assembled genome sequences using STAR (v2.7.0) (Dobin et al., 2013). Additionally, full-length RNA-seq reads of S. chuatsi and S. scherzeri were aligned to the respective assembled genome sequences using GMAP (2018-07-04) (Wu & Watanabe, 2005). Gene models were predicted based on the alignment results of short and full-length RNA-seq reads using BRAKER2 (v2.1.5) (Brůna et al., 2021).
For RNA-seq-based prediction, short RNA-seq reads of S. chuatsi and S. scherzeri were first aligned to the respective reference sequences using HISAT2 (v2-2.1). Gene models were predicted based on the alignment results of HISAT2 using StringTie (v2.1.4) (Pertea et al., 2016), and coding regions were identified using TransDecoder (v5.5.0). Second, short RNA-seq reads of S. chuatsi and S. scherzeri were assembled using Trinity (v2.8.5) (Grabherr et al., 2011). Third, full-length RNA-seq reads of S. chuatsi and S. scherzeri were assembled using Iso-Seq3 (v3.1) (https://github.com/PacificBiosciences/IsoSeq). The Iso-Seq3-assembled full-length RNA-seq reads were subjected to error correction using high-quality Illumina reads with LoRDEC (v0.5.3) (Salmela & Rivals, 2014). Finally, PASA (v2.5.0) was used to predict gene models of the genomes of both species based on the Trinity and Iso-Seq3 assembly results, with StringTie-predicted gene models as a reference. Coding regions of PASA-predicted gene models were then identified using TransDecoder (v5.5.0) (Grabherr et al., 2011).
The BRAKER2-, GenomeThreader-, and PASA-predicted gene models of S. chuatsi and S. scherzeri were integrated into a nonredundant consensus gene set using EvidenceModeler (v1.1.1) (Haas et al., 2008). The EvidenceModeler-integrated gene models were updated with short and full-length RNA-seq reads using PASA (v2.5.0) three times. Completeness of the predicted gene models of the two species was evaluated using BUSCO (v4.0.5) (Simão et al., 2015) against the conserved Actinopterygii dataset (odb10).
To assign functions to the predicted proteins, we aligned the S. chuatsi and S. scherzeri protein models against the NCBI nonredundant (NR) amino acid sequences, UniProt, Translated EMBL-Bank (TrEMBL), Cluster of Orthologous Groups for Eukaryotic Complete Genomes (KOG), and SwissProt databases using BLASTP with an E-value cutoff of 10−5. Protein models were also aligned against the eggNOG database (Huerta-Cepas et al., 2019) using eggNOG-Mapper (Huerta-Cepas et al., 2017). Finally, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation of the protein models was performed using BlastKOALA (Kanehisa et al., 2016).
Genome resequencing and single nucleotide polymorphism (SNP) calling
To investigate the evolutionary history and adaptation of S. chuatsi and S. scherzeri, we collected six wild S. chuatsi individuals from Hunan Province, China, six wild S. scherzeri individuals from Guangdong Province, China, and six wild S. scherzeri individuals from Jilin Province, China. Sequencing libraries were constructed using an Illumina TruSeq Nano DNA HT Sample Preparation Kit (Illumina, USA) in accordance with the manufacturer’s protocols. All individuals were whole-genome re-sequenced to an average coverage of 12.5× with 2×150 bp chemistry using the NovaSeq 6000 platform (Illumina, USA).
Low-quality and sequencing adapter-contaminated Illumina reads were filtered and trimmed with Trimmomatic (v0.36) (Bolger et al., 2014). High-quality PE reads of S. chuatsi and S. scherzeri were aligned to the respective reference sequences using BWA (v0.7.17) with the “mem” function (Li & Durbin, 2009). PCR duplicates were removed using the MarkDuplicate program in Picard (v2.18.27) (https://broadinstitute.github.io/picard/). SNP variants were identified using the HaplotypeCaller program in the Genome Analysis Toolkit (GATK) (v4.1.0.0). Raw SNP calling datasets were filtered using the VariantFiltration program in GATK (v4.1.0.0) with the parameters “QD<2.0 || QUAL<30.0 || FS>60.0 || MQ<0.0 || MQRankSum<−12.5 || ReadPosRankSum<−8.0 || SOR>3.0”.
Demographic inference of S. chuatsi and S. scherzeri
The historical effective population sizes of S. chuatsi and S. scherzeri were first estimated based on SNP variants of six wild-caught S. chuatsi individuals and six wild-caught S. scherzeri individuals using SMC++ (v1.15.3). The substitution mutation rate and generation time of S. chuatsi and S. scherzeri were set to 2.22×10-9 and 2 years, respectively, according to a previous study on Siniperca knerii (big-eye mandarin fish) (Lu et al., 2020).
To infer historical effective population sizes using PSMC (Li & Durbin, 2011), high-quality Illumina reads of S. chuatsi and S. scherzeri generated for genome assembly were aligned to the respective reference sequences using BWA (v0.7.17) with the “mem” function. Genetic variants were identified using SAMtools (v1.9-52) (Li et al., 2009). Whole-genome consensus sequences were generated using the genetic variants with SAMtools (v1.9-52) (Li et al., 2009). PSMC (v0.6.5) was applied to infer population size history of S. chuatsi and S. scherzeri using the whole-genome consensus sequences. The substitution mutation rate and generation time of S. chuatsi and S. scherzeri were set to 2.22×10-9 and 2 years, respectively.
Phylogenetic reconstruction
Protein sequences of eight species (D. rerio, Oreochromis niloticus, Maylandia zebra, Amphiprion percula, Lates calcarifer, Larimichthys crocea, Dicentrarchus labrax, and Sander lucioperca) were downloaded from Ensembl (v96) (Yates et al., 2020). Protein sequences of Epinephelus lanceolatus were downloaded from NCBI (Zhou et al., 2019). Protein sequences shorter than 50 amino acids were removed. OrthoFinder (v2.5.4) (Emms & Kelly, 2019) was applied to identify and cluster gene families among the nine species as well as S. chuatsi and S. scherzeri. Gene clusters with >100 gene copies in one or more species were removed. Single-copy orthologs in each gene cluster were aligned using MAFFT (v7.490) (Katoh et al., 2002). Alignments were then trimmed using Gblocks (v0.91b) (Talavera & Castresana, 2007). The phylogenetic tree was reconstructed with trimmed alignments using maximum-likelihood implemented in IQ-TREE2 (v2.2.0) with D. rerio as the outgroup. The best-fit substitution model was selected using the ModelFinder algorithm (Kalyaanamoorthy et al., 2017). Branch supports were assessed using the ultrafast bootstrap (UFBoot) approach with 1 000 replicates (Hoang et al., 2018).
Divergence time was estimated using the MCMCTree module in the PAML package (v4.9) (Yang, 2007). MCMCTree analysis was performed using the maximum-likelihood tree reconstructed by IQ-TREE2 as a guide tree and calibrated with the divergence times obtained from the TimeTree database (minimum=206 million years and soft maximum=252 million years between D. rerio and O. niloticus; minimum=18.08 million years and soft maximum=35.16 million years between M. zebra and O. niloticus; minimum=82 million years and soft maximum=131 million years between A. percula and O. niloticus; minimum=104 million years and soft maximum=145 million years between A. percula and L. calcarifer; minimum=94 million years and soft maximum=115 million years between S. lucioperca and L. calcarifer; minimum=69 million years and soft maximum=88 million years between S. lucioperca and E. lanceolatus; minimum=99 million years and soft maximum=127 million years between S. lucioperca and L. crocea; and minimum=87 million years and soft maximum=105 million years between D. labrax and L. crocea) (retrieved June 2021) (Kumar et al., 2017).
Gene family expansion and contraction analysis
CAFÉ (v5) (De Bie et al., 2006) was applied to determine the significance of gene family expansion and contraction among the 11 teleost species based on the MCMCTree-generated ultrametric tree and OrthoMCL-determined gene clusters used for species tree reconstruction.
The p300 protein family was significantly expanded in the S. scherzeri genome compared with all other perciform fish. Phylogenetic tree reconstruction was performed to investigate the evolutionary relationships of p300 proteins from S. chuatsi, S. scherzeri, and other teleost species. The p300 protein sequences from D. rerio, O. niloticus, M. zebra, L. calcarifer, L. crocea, D. labrax, S. lucioperca, E. lanceolatus, and G. aculeatus were downloaded from Ensembl (v96) (Yates et al., 2020) and aligned using MAFFT (v7.490) (Katoh et al., 2002). The phylogenetic tree was reconstructed using maximum-likelihood alignment implemented in IQ-TREE2 (v2.2.0), with a p300 protein sequence (XP_040032248.1) from G. aculeatus as the outgroup. The best-fit substitution model was selected using the ModelFinder algorithm (Kalyaanamoorthy et al., 2017). Branch supports were assessed using UFBoot with 1 000 replicates (Hoang et al., 2018).
Analysis of olfactory receptor (OR) genes
Sinipercids are extreme piscivores and only accept live prey once fry start feeding. Studies have shown that S. chuatsi fish use vision and mechanoreception but not olfaction for predation (Liang et al., 1998). Thus, investigating OR genes can provide insight into the adaptive evolution of reduced olfaction in S. chuatsi and S. scherzeri. Here, we identified OR genes in the genomes of D. rerio, O. niloticus, M. zebra, A. ocellaris, L. calcarifer, L. crocea, D. labrax, S. lucioperca, E. lanceolatus, S. chuatsi, and S. scherzeri using a previously described approach (Niimura et al., 2014). In brief, a tBLASTn search was performed with a E-value cut-off of 10-10 using a set of known functional OR genes from D. rerio, L. crocea, L. calcarifer, O. niloticus, O. latipes, and T. rubripes as queries (Liu et al., 2021). The best-hit regions were extracted and extended 5 000 bp in both the 3’ and 5’ directions along the genome sequences using SAMtools (v1.9-52) (Li et al., 2009). Gene structure was predicted using the extended best-hit regions with Exonerate (v2.4.0) (Slater & Birney, 2005). The resulting protein-coding sequences were aligned against the UniProt database using BLASTP (v2.8.1) and those with the highest alignment scores to known ORs were retained. We used CD-HIT (v4.6.2) (Fu et al., 2012) to remove redundant sequences and cluster filtered OR sequences. We classified OR genes with intact sequences and without loss-of-function mutations as functional genes. The functional OR protein sequences were aligned using MAFFT (v7.490) (Katoh et al., 2002). Phylogenetic tree reconstruction was performed with the alignments using maximum-likelihood in IQ-TREE2 (v2.2.0). The best-fit substitution model was selected using the ModelFinder algorithm (Kalyaanamoorthy et al., 2017). Branch supports were assessed using UFBoot with 1 000 replicates (Hoang et al., 2018).
Identification of PSGs
PSGs in the S. chuatsi and S. scherzeri genomes were identified using PosiGene (v0.1) (Sahm et al., 2017) with parameters “-as=D.rerio, -ts=S.chuatsi and S.scherzeri -rs=D.rerio, -nhsbr”. Genes with P<0.05 were determined to have undergone positive selection.
SV identification
SVs between the S. chuatsi and S. scherzeri genomes were identified using two methods. First, SVs were identified by aligning the genome of S. scherzeri against the genome of S. chuatsi using BLASR (Chaisson & Tesler, 2012). SVs between the two genomes were identified by combining the results of smartie-sv (Kronenberg et al., 2018) and SyRI (v4.1) (Goel et al., 2019). Second, high-quality Nanopore reads of S. scherzeri were aligned against the genome of S. chuatsi, and S. chuatsi reads were aligned against the genome of S. scherzeri using NGMLR (v0.2.7) (Sedlazeck et al., 2018). SVs were identified based on both alignment results using Sniffles (v1.0.11) (Sedlazeck et al., 2018). SVs shorter than 50 bp were removed. Samplot (v1.3.0) was used to validate the candidate SVs with high-quality Nanopore and Illumina read alignment.
Histone modification analysis
The CUT&Tag assay was performed as described previously with some modifications (Kaya-Okur et al., 2019). Briefly, liver cells of S. chuatsi and S. scherzeri were harvested and gently washed twice in 300 μL of wash buffer (20 mmol/L HEPES pH 7.5; 150 mmol/L NaCl; 0.5 mmol/L spermidine; 1×protease inhibitor cocktail). A 1:50 dilution of H3K27ac rabbit pAb (ab4729, Abcam), H3K4me3 rabbit pAb (ab8580, Abcam), or H3K4me1 rabbit pAb (ab8895, Abcam) was used as the primary antibody for incubation. A 1:50 dilution of goat anti-rabbit IgG (ab8580, Abcam) was used as the secondary antibody. To construct a negative control library (IgG), we added the secondary antibody without the primary antibody. Cells were washed with Dig-Wash buffer to remove unbound antibodies. A 1:200 dilution of pG-Tn5 adapter complex was added to the cells and incubated with pG-Tn5 protein at 37 °C for 1 h. Cells were then washed twice in Dig-300 buffer to remove unbound pG-Tn5 protein. Next, the cells were resuspended in tagmentation buffer (10 mmol/L MgCl2 in Dig-300 buffer) and incubated at 37 °C for 1 h. To stop tagmentation, 10 μL of 0.5 mol/L EDTA, 3 μL of 10% sodium dodecyl sulfate (SDS), and 2.5 μL of 20 mg/mL proteinase K were added to the sample, followed by incubation at 55 °C for 1 h. DNA was purified using phenol-chloroform-isoamyl alcohol and ethanol, washed with 100% ethanol, and suspended in water. The libraries were amplified by mixing DNA with 2 μL of a universal i5 and uniquely barcoded i7 primer. After DNA quantification and qualification, all libraries were sequenced (2×150 bp) on the Illumina Nova-seq 6000 platform (Illumina, USA).
The CUT&Tag sequencing reads were processed using a previously described pipeline (Kaya-Okur et al., 2019). Low-quality and sequencing adapter-contaminated Illumina reads were filtered and trimmed with Trimmomatic (v0.39) (Bolger et al., 2014). The filtered reads of S. chuatsi and S. scherzeri were aligned to the respective genomes using Bowtie2 (v2.3.2) (Langmead & Salzberg, 2012) with parameters “--local --very-sensitive --no-mixed --no-discordant -I 10 -X 700”. Duplicate reads and reads with mapping quality scores (MAPQ) less than 30 were removed using SAMtools (v1.9-52) (Li et al., 2009). MACS2 (v2.1.1) (Zhang et al., 2008) was used to identify broad and narrow peaks of H3K27ac and H3K4me3 histone modifications with parameters “--keep-dup all”, and only broad peaks for H3K4me1 histone modifications with parameters “--keep-dup all --broad”. Significantly differentiated peaks between the S. chuatsi and S. scherzeri genomes (|log2FC|>=1, P<=0.05) were identified using MAnorm (v1.1.4) (Shao et al., 2012). Genes potentially regulated by the differentiated enhancers and promoters were identified using ChIPseeker (Yu et al., 2015). Profiles of CUT&Tag signals of H3K27ac, H3K4me1, and H3K4me3 were visualized using the plotProfile function in deepTools (v3.5.1) (Ramírez et al., 2016). Clustering of the CUT&Tag signals of the three histone marks around transcription start sites (TSSs) of genes were visualized using the computeMatrix and plotHeatmap functions in deepTools (v3.5.1) (Ramírez et al., 2016).
Open chromatin region (OCR) identification
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) libraries were constructed based on a previously described approach (Corces et al., 2017). Liver cells of S. chuatsi and S. scherzeri were harvested, and cell suspensions were prepared. Cell membranes were lysed to obtain the nucleus. Then, Tn5 transposase was added to cut open the DNA. The DNA fragments were amplified by PCR and sequenced (2×150 bp) on the Illumina HiSeq X Ten platform (Illumina, USA).
Low-quality and sequencing adapter-contaminated ATAC-seq reads were filtered and trimmed with Trimmomatic (v0.39) (Bolger et al., 2014). The filtered reads of S. chuatsi and S. scherzeri were aligned to the respective genomes using Bowtie2 (v2.3.2) (Langmead & Salzberg, 2012) with parameters “--local --very-sensitive --no-mixed --no-discordant -I 10 -X 700”. Duplicate reads and reads with MAPQ scores less than 30 were removed using SAMtools (v1.9-52) (Li et al., 2009). MACS2 (v2.1.1) (Zhang et al., 2008) was used to identify peaks with parameters “--shift -75 --extsize 150 --nomodel -B --SPMR --keep-dup all”.
Transcriptome sequencing for epigenomic analyses
Liver samples from S. chuatsi and S. scherzeri were collected to construct RNA-seq libraries for epigenomic analyses. Total RNA was extracted using TRIzol reagent (Invitrogen). RNA purity and integrity were monitored using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and Bioanalyzer 2100 (Agilent, USA), respectively. The mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. PE sequencing libraries were generated from the purified mRNA using a VAHTS Universal V6 RNA-seq Library Kit for MGI (Vazyme, China) with unique index codes. Sequencing was performed (2×150 bp) using the MGISEQ 2000 platform (MGI Tech, China).
Low-quality and sequencing adapter-contaminated RNA-seq reads were filtered using SOAPnuke (v2.1.6) (Chen et al., 2018b) with parameters “-n 0.05 -q 0.5 -l 20”. Filtered reads of S. chuatsi and S. scherzeri were aligned to the respective reference genomes using STAR (v2.7.0) (Dobin et al., 2013). RSEM (v1.3.3) (Li & Dewey, 2011) was used to map and calculate gene expression levels represented as fragments per kilobase of exon per million mapped fragments (FPKM). Differential expression analysis was performed using DESeq2 (R4.1.2) (Love et al., 2014).
RESULTS
Chromosome-level genome assemblies of S. chuatsi and S. scherzeri
The S. chuatsi and S. scherzeri genomes were sequenced using a combination of Nanopore and Illumina shotgun sequencing. A total of 100.7 Gb of Nanopore reads and 61 Gb of Illumina reads were obtained for S. chuatsi, and 94.3 Gb of Nanopore reads and 46 Gb of Illumina reads were obtained for S. scherzeri (Supplementary Tables S1, S2). Based on the k-mer distribution of Illumina reads, the genome sizes of S. chuatsi and S. scherzeri were estimated to be 695.9 Mb and 708.4 Mb, respectively (Supplementary Figure S1). Additionally, based on previous flow cytometry analysis (Cui et al., 1991), the genome size of S. chuatsi was estimated to be 731 Mb. The S. chuatsi and S. scherzeri genomes were first assembled into contigs with Nanopore reads using the WTDBG assembler (Ruan & Li, 2020). The contigs were then subjected to error correction with Illumina reads. Contigs of S. chuatsi and S. scherzeri were scaffolded using proximity ligation data from the respective Hi-C libraries to yield genome assemblies (Supplementary Figures S2, S3 and Tables S3–S5). The final genome assembly of S. chuatsi was composed of 191 scaffolds (contig N50: 21.55 Mb, scaffold N50: 29.96 Mb) assembled into 24 pseudomolecules, resulting in a total assembly size of 716.35 Mb. The final genome assembly of S. scherzeri consisted of 252 scaffolds (contig N50: 16.04 Mb, scaffold N50: 30.49 Mb) assembled into 24 pseudomolecules with a total assembly size of 740.54 Mb (Figure 1C, D; Table 1). BUSCO analysis indicated that 97.9% and 98.7% of conserved single-copy ray-fin fish (Actinopterygii) genes (odb10) were captured in the S. chuatsi and S. scherzeri genomes, respectively (Supplementary Table S6).
Table 1. Genome assembly statistics of S. chuatsi and S. scherzeri.
|
S. chuatsi (This study) |
S. chuatsi(sinChu7) (He et al., 2020) |
S. scherzeri
(This study) |
S. scherzeri(sinSch6b) (He et al., 2020) |
|
| Total length (Mb) | 716.34 | 755.06 | 740.54 | 736.22 |
| Number of scaffolds | 191 | 1 156 | 252 | 2 826 |
| Scaffold N50 (bp) | 29 956 575 | 30 508 166 | 30 486 584 | 30 166 107 |
| Scaffold N90 (bp) | 24 860 938 | 23 880 225 | 27 297 733 | 23 838 788 |
| Number of scaffolds (>N90 length) | 21 | 22 | 21 | 22 |
| Number of contigs | 328 | 1 464 | 334 | 23 070 |
| Contig N50 (bp) | 21 551 097 | 12 191 788 | 16 036 871 | 83 589 |
| Contig N90 (bp) | 3 004 397 | 1 214 209 | 3 383 819 | 16 890 |
| Number of contigs (>N90 length) | 42 | 89 | 53 | 9 466 |
We compared the sequence consistency and integrity of our assemblies to previously published genome assemblies of S. chuatsi (sinChu7, GCA_011952085.1) and S. scherzeri (sinSch6b, GCA_011952095.1) (He et al., 2020). First, the number of contigs and scaffolds was greatly reduced, and contig N50 was substantially increased in our genome assemblies compared to previously published assemblies (Table 1), suggesting better contiguity. Second, RNA-seq reads of different tissues were aligned to our and previously published genome assemblies. The mapping rates of the RNA-seq reads of nine tissues to the S. chuatsi and S. scherzeri assemblies were 92.24% and 96.29%, while the mapping rates to previously published assemblies were 83.37% and 94.28%, respectively (Supplementary Tables S7, S8). Third, based on Merqury evaluation, the consensus quality values (QVs) of our S. chuatsi and S. scherzeri assemblies were 36.91 and 35.73, respectively, compared to 32.37 and 30.96 for previously published assemblies, thus suggesting high quality (Supplementary Table S9) (Rhie et al., 2020).
The S. chuatsi and S. scherzeri genomes carried 196.35 Mb (27.41%) and 209.40 Mb (28.28%) of repetitive sequences, respectively (Supplementary Tables S10, S11). DNA transposons were the largest class of annotated transposable elements (TEs), accounting for 7.51% and 7.70% of the S. chuatsi and S. scherzeri genomes, respectively. Protein-coding genes in the genomes were identified through a combination of ab initio, homology-based, and RNA-seq-based prediction approaches. In total, 29 278 and 29 543 protein-coding genes were identified in the S. chuatsi and S. scherzeri genomes, respectively (Supplementary Tables S12, S13). In the predicted gene models of S. chuatsi and S. scherzeri, BUSCO analysis identified 3 353 (92.1%) and 3 337 (91.7%) complete conserved single-copy ray-fin fish (Actinopterygii) genes (odb10), respectively (Supplementary Figure S4 and Table S14). In total, 26 623 (90.93%) gene models in the S. chuatsi genome and 27 024 (91.47%) gene models in the S. scherzeri genome were annotated in at least one database (InterPro, eggNOG, KEGG, Swiss-Prot, and TrEMBL) (Supplementary Table S15).
Demographic history of S. chuatsi and S. scherzeri
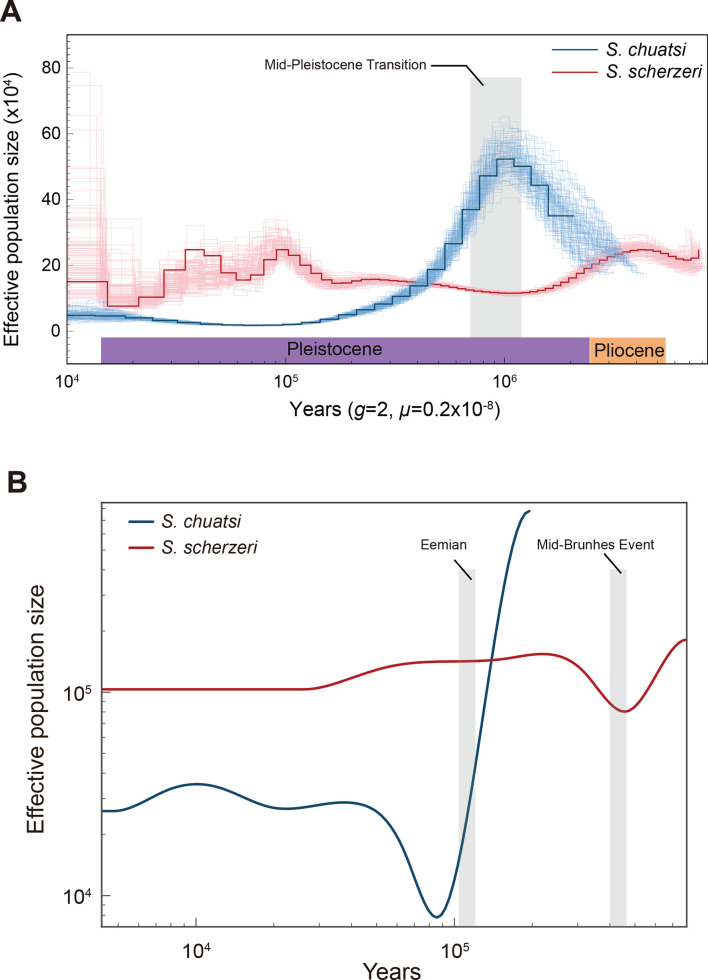
To investigate the evolutionary history of S. chuatsi and S. scherzeri, we inferred their ancestral population sizes using the PSMC program (Li & Durbin, 2011) (Figure 2A). The ancestral population size of S. scherzeri was relatively stable. In contrast, S. chuatsi populations expanded in the early Pleistocene (~2 million years ago, Ma) and declined during the early phase of the Mid-Pleistocene Transition (~0.9 Ma), when the duration of the Pleistocene glacial cycles increased from 41 to 100 thousand years (Berends et al., 2021). These results suggest that S. chuatsi began to colonize new habitats after glacial cycle prolongation. Ancestral population size dynamics of S. chuatsi and S. scherzeri were also inferred using the SMC++ program based on genetic variants of six wild S. chuatsi individuals and six wild S. scherzeri individuals from South China (Guangdong Province) and six wild S. scherzeri individuals from North China (Jilin Province) (Supplementary Table S16) (Terhorst et al., 2017). SMC++ analysis showed that S. chuatsi experienced a population bottleneck at the beginning of the last glacial period (~90 ka), suggesting that the warm temperature during the Eemian interglacial period (129~116 ka) may have facilitated diversification of the species (Hoffman et al., 2017). After the Mid-Brunhes Event (MBE, ~430 ka), the effective population size of S. scherzeri in southern China experienced a decline at ~400 ka, with several species also undergoing dramatic changes in ancestral population size (Figure 2B) (Bowen et al., 2006; Kozma et al., 2016; Yin & Berger, 2010). The ancestral population size of S. scherzeri in northern China declined at ~300 ka, approximately 100 ka after the population bottleneck of their counterparts in southern China (Supplementary Figure S5).
Figure 2.
Demographic history of S. chuatsi and S. scherzeri
A: Ancestral population sizes of S. chuatsi (blue) and S. scherzeri (red) inferred using PSMC. B: Dynamic changes in ancestral population size of S. chuatsi (blue) and S. scherzeri from South China (red) using SMC++.
Genetic basis of adaptation in S. chuatsi and S. scherzeri
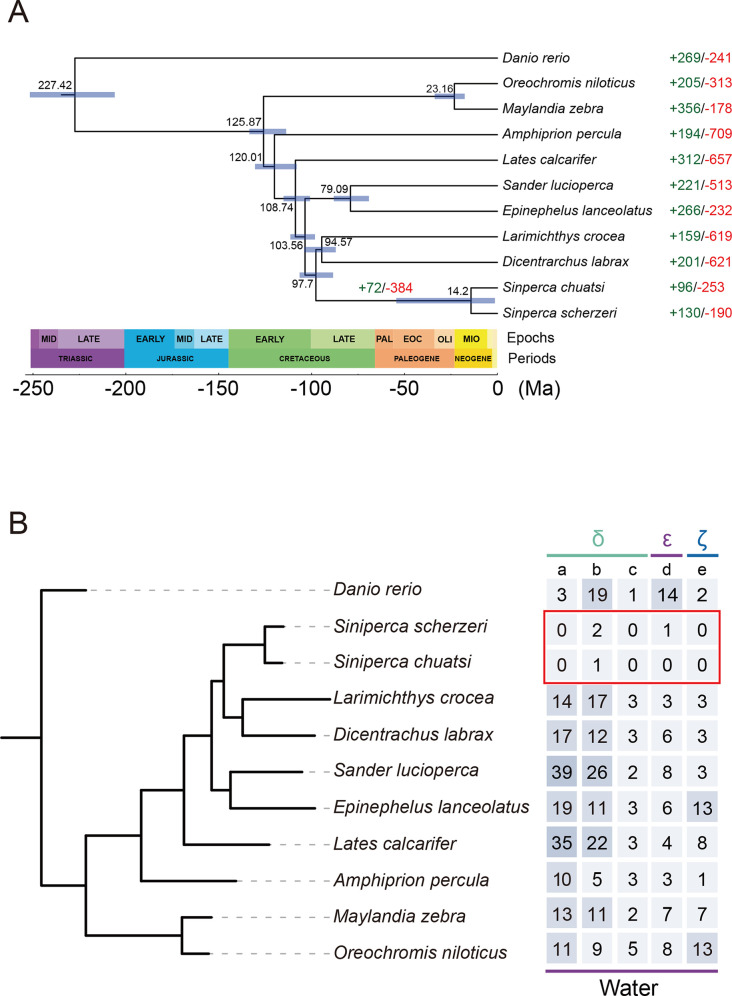
Gene family expansion and contraction analyses were performed to dissect the genetic basis of adaptation in S. chuatsi and S. scherzeri. First, a phylogenetic tree of 10 perciform fish was reconstructed with Danio rerio as the outgroup (Figure 3A). Divergence times among the 10 fish species were determined. Divergence between S. chuatsi and S. scherzeri was estimated at 14.2 Ma (CI: 1.26–54.45 Ma), consistent with previous studies (He et al., 2020; Song et al., 2017). Second, gene family analysis was performed based on the phylogenetic tree (Figure 3A). Compared with other perciform fish, 72 gene families were expanded and 384 gene families were contracted in the Siniperca clade (P<0.05).
Figure 3.
Genetic basis of adaptation in S. chuatsi and S. scherzeri
A: Species tree of 10 perciform species with Danio rerio as the outgroup. Divergence time between species pairs is listed above each node, and 95% confidence interval of estimated divergence time is denoted as a blue bar. Numbers of protein families significantly expanded (green) and contracted (red) (P<0.05) in each species are denoted beside species names. Numbers of expanded and contracted protein families in Siniperca genus are denoted above the node. B: OrthoFinder-identified ortholog groups of OR genes contracted in S. chuatsi and S. scherzeri. a: OG0000155; b: OG0000197; c: OG0000029; d: OG0000041; e: OG0000713.
The olfactory system in fish plays a critical role in feeding, reproduction, predator avoidance, and migration (Hara, 1975). The number and diversity of OR genes are positively correlated with olfactory epithelium complexity and contribute to olfactory specialization and ecological adaptation in fish (Policarpo et al., 2021). Sinipercids are extreme piscivores and will only accept live prey once fry start feeding. Previous research has shown that S. chuatsi uses vision and mechanoreception but not olfaction for predation (Liang et al., 1998). Thus, investigating OR genes can provide insight into the adaptive evolution of olfaction in S. chuatsi and S. scherzeri. The nine subfamilies of OR genes are classified into two types in vertebrates (Type 1: α, β, γ, δ, ε, and ζ; Type 2: η, θ, and κ), in which α and γ detect airborne molecules, δ, ε, ζ, and η detect water-soluble molecules, and β senses both airborne and soluble odorants (Hara & Zhang, 1996; Niimura, 2009; Niimura & Nei, 2003). Interestingly, three OrthoFinder-identified ortholog groups of OR genes in subfamily δ (OG0000029, OG0000041, and OG0000713), one ortholog group in subfamily ε (OG0000155), and one ortholog group in subfamily ζ (OG0000197) were significantly contracted in the Siniperca clade (Figure 3B). In addition, comprehensive genomic screening of functional OR genes showed that S. chuatsi and S. scherzeri had fewer OR genes than seven other perciform fish (Supplementary Figure S6). These results suggest that the loss of OR genes, especially those in the four ortholog groups, could be attributed to the special feeding habits of S. chuatsi and S. scherzeri.
Compared with other perciform fish, 96 and 130 gene families were expanded in S. chuatsi and S. scherzeri, respectively (P<0.05) (Figure 3A). The CREB-binding protein and p300 (CBP/p300) protein family is a group of transcriptional coactivators that acetylate several histones and non-histone targets (Iyer et al., 2004). CBP and p300 act as non-DNA-binding co-factors for proteins involved in multiple biological processes, including melanoblast specification, DNA damage response, and circadian rhythm (Curtis et al., 2004; Goding & Arnheiter, 2019; Xu et al., 2008). Interestingly, p300 proteins were significantly expanded in the S. scherzeri genome (five copies) compared with all other perciform fish (two copies) (P<0.05) (Supplementary Figure S7), which may facilitate the rapid responses of this species to internal and external stimuli.
Identification and analysis of PSGs can provide insight into how natural selection shapes individual traits during evolution (Smith & Eyre-Walker, 2002). Therefore, we identified PSGs in the S. chuatsi and S. scherzeri genomes. In total, 15 PSGs were identified in the S. chuatsi genome compared to S. scherzeri and eight teleosts (D. rerio, O. niloticus, M. zebra, L. calcarifer, L. crocea, D. labrax, S. lucioperca, and Epinephelus lanceolatus) (Supplementary Table S17). Interestingly, four genes related to growth and development (smpd3, mbtps1, fn1a, and uchl3) were positively selected in S. chuatsi, which may contribute to the higher growth rate of S. chuatsi compared with S. scherzeri (Achilleos et al., 2015; Gao et al., 2021; Semenova et al., 2003; Stoffel et al., 2005). Furthermore, 25 PSGs were identified in the S. scherzeri genome compared to S. chuatsi and eight teleosts (Supplementary Table S18). Among these genes, three are involved in skin pigmentation (cdc42, rbpjb, and atrn), and may contribute to darker skin in S. scherzeri than in S. chuatsi (Gunn et al., 2001; Moriyama et al., 2006; Woodham et al., 2017).
Role of SVs in cis-regulatory divergence between S. chuatsi and S. scherzeri
We investigated the role of SVs in cis-regulatory divergence between S. chuatsi and S. scherzeri. The genomic locations of H3K27ac and H3K4me3 were identified in the livers of S. chuatsi and S. scherzeri using CUT&Tag (Supplementary Table S19). Analysis identified 17 015 and 15 150 H3K27ac regions (hereafter referred to as peaks) and 16 955 and 16 239 H3K4me3 peaks in the S. chuatsi and S. scherzeri genomes, respectively (Supplementary Figure S8). Fraction of reads in peaks (FRiP) and TSS enrichment analyses showed that the CUT&Tag data were of high quality and sufficient for further analysis (Supplementary Figure S9 and Table S19). We also identified OCRs in the two Siniperca genomes using ATAC-seq (Supplementary Figures S10, S11 and Table S20). In the S. chuatsi genome, we identified 22 588 nonredundant CREs (17 623 putative promoters and 4 965 potential distal enhancers) using CUT&Tag data and 74 170 OCRs using ATAC-seq data. In addition, we identified 19 858 nonredundant CREs (16 726 putative promoters and 3 132 potential distal enhancers) and 75 972 OCRs in the S. scherzeri genome (Supplementary Table S21).
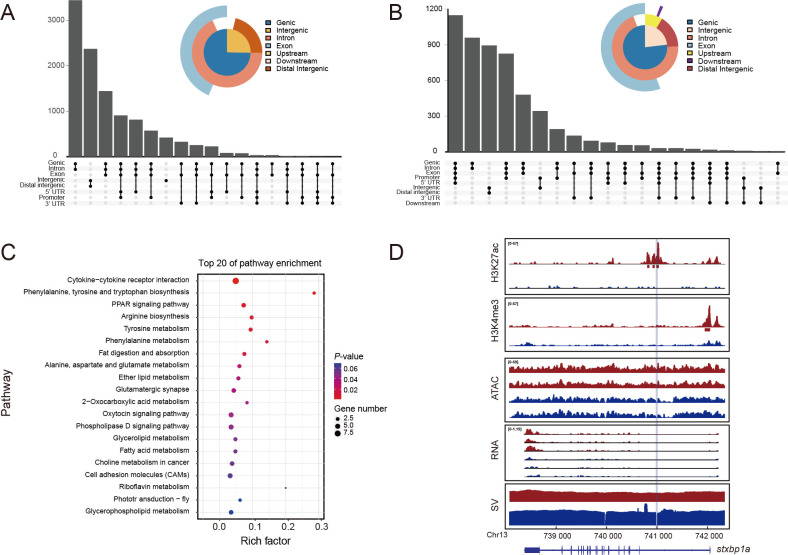
A total of 11 050 differential H3K27ac peaks (|log2FC|>=1, P<=0.05) were identified between S. chuatsi and S. scherzeri, including 6 632 up-regulated and 4 418 down-regulated in S. chuatsi. The distribution of differential H3K27ac peaks was associated with introns and intergenic regions (Figure 4A). We identified 5 437 differential H3K4me3 peaks (|log2FC|>=1, P<=0.05) between the two species, which were mostly located in introns and promoters (Figure 4B). In addition, 3 870 differentially expressed genes (DEGs) (|log2FC|>=1, FDR<=0.05) were identified between the S. chuatsi and S. scherzeri livers, including 2 099 up-regulated and 1 771 down-regulated in S. chuatsi compared with S. scherzeri (Supplementary Figure S12).
Figure 4.
Role of SVs in cis-regulatory divergence between S. chuatsi and S. scherzeri
A: Genomic distribution of differential H3K27ac peaks. B: Genomic distribution of differential H3K4me3 peaks. C: KEGG enrichment of genes associated with differential H3K27ac peaks intersecting high-confidence SVs. D: Cis-regulatory divergence between S. chuatsi (red) and S. scherzeri (blue) at stxbp1a gene. Intensities of H3K27ac, H3K4me3, ATAC-seq, and gene expression are shown. Mapping coverage of Nanopore reads is plotted to indicate genomic SVs. Deletion in S. scherzeri is denoted in light blue. MACS2-identified H3K27ac and H3K4me3 peaks are denoted with rectangles below tracks. Transcripts (with exons as boxes) are depicted.
Based on the high-quality assemblies, SVs were identified between the S. chuatsi and S. scherzeri genomes using three different tools (Supplementary Figure S13). Of note, SyRI and smartie-sv identify SVs by aligning two genome assemblies, while NGMLR-Sniffles calls SVs by aligning Nanopore reads to reference genome sequences (Goel et al., 2019; Kronenberg et al., 2018; Sedlazeck et al., 2018). By combining the results of these three tools, we identified 75 988 deletions, 84 189 insertions, 1 714 duplications, and 310 inversions between the S. chuatsi and S. scherzeri genomes.
In total, 1 358 SVs overlapped with differential H3K27ac peaks between the S. chuatsi and S. scherzeri genomes. We validated these SVs using Samplot and obtained 905 high-confidence SVs (Belyeu et al., 2021). In total, 1 205 genes were associated with the differential H3K27ac peaks that intersected with high-confidence SVs. Of these genes, 301 showed significantly differential expression (|log2FC|>=1, FDR<0.05) between S. chuatsi and S. scherzeri. KEGG enrichment analysis indicated that the 301 DEGs were mainly enriched in pathways related to lipid and amino acid metabolism, including arginine biosynthesis (ko00220), fat digestion and absorption (ko04975), and fatty acid metabolism (ko01212) (Figure 4C). Interestingly, three genes involved in fatty acid catabolism (acsl1a, acadl, and got2a) showed higher signal intensity of H3K27ac peaks and gene expression levels in S. chuatsi than in S. scherzeri (Supplementary Figures S14–S16). The expression of acsl1a, a key member of the long-chain acyl-CoA synthetase family responsible for fatty acid degradation and lipid synthesis, is positively correlated with lipid uptake (Soupene & Kuypers, 2008; Zhan et al., 2012). The acadl gene plays a critical role in lipid catabolism by catalyzing the initial step of β-oxidation of long-chain fatty acyl-CoAs (Kurtz et al., 1998). Furthermore, got2a promotes fatty acid metabolism by transporting long-chain fatty acids into cells (Chabowski et al., 2007). The elevated expression of these three genes suggests that S. chuatsi may utilize fatty acids more efficiently than S. scherzeri, which may contribute to its higher growth rate.
Three genes (stxbp1a, mkln1, and myca) involved in skin pigmentation showed differential expression and H3K27ac peak intensity between S. chuatsi and S. scherzeri. Deletion of the first intron of stxbp1a completely removed a putative enhancer in S. scherzeri, and expression of this gene was lower in S. scherzeri than in S. chuatsi (Figure 4D). Zebrafish with a stxbp1a mutation display darker pigmentation on their heads and backs (Grone et al., 2016). Therefore, reduced expression of this gene in S. scherzeri may contribute to its darker pigmentation. Deletion of the first intron of mkln1 reduced the intensity of the H3K27ac peaks in S. chuatsi, resulting in lower expression of this gene in S. chuatsi than in S. scherzeri (Supplementary Figure S17). Mice with mkln1 mutation develop brighter fur over time (Heisler et al., 2011). Thus, the darker pigmentation in S. scherzeri could be attributed to elevated expression of mkln1. These findings suggest that cis-regulatory variation caused by SVs results in differences in expression between the two genes, leading to pigmentation differences between S. chuatsi and S. scherzeri.
In addition to genes related to metabolism and skin pigmentation, we found that the intensities of H3K27ac peaks associated with four immune genes (prss16, lama4, cd22, and tecpr1b) were significantly decreased (|log2FC|>=1, P<=0.05) in S. chuatsi compared with S. scherzeri (Supplementary Table S22). Moreover, the expression levels of these genes in the spleen, a major immune organ in fish, were substantially higher in S. scherzeri than in S. chuatsi. Thymus-specific serine protease (TSSP), encoded by prss16, plays a critical role in T cell maturation (Brisson et al., 2015). The ability of immune cells to penetrate the vessel wall is reduced in lama4 mutant mice (Kenne et al., 2010). The cd22 gene is a regulator of innate and adaptive B cell responses (Clark & Giltiay, 2018). A tecpr1-dependent pathway is important in targeting bacterial pathogens for selective autophagy (Ogawa et al., 2011). Previous studies have shown that S. scherzeri is more resistant to disease than S. chuatsi (Ding et al., 2022). Therefore, cis-regulatory variation caused by SVs in these four genes may contribute to the divergence in disease resistance between S. scherzeri and S. chuatsi.
Divergence in broad H3K4me3 domains between S. chuatsi and S. scherzeri
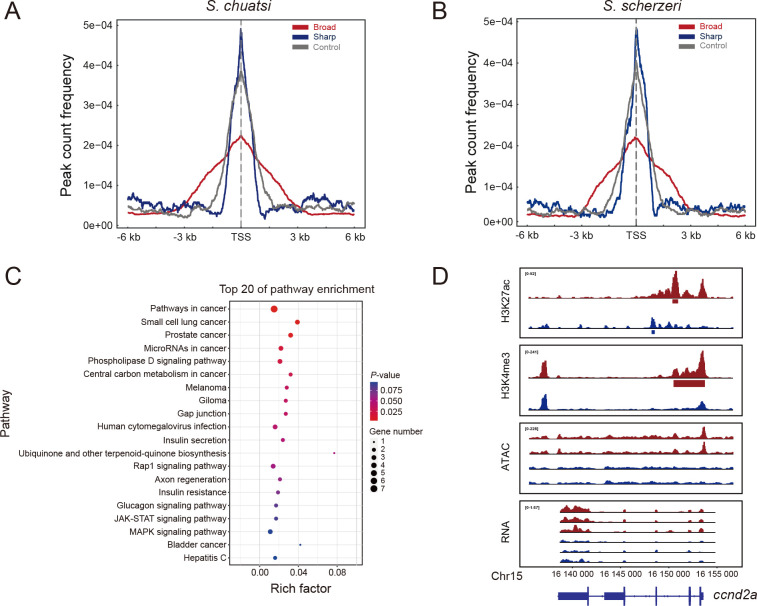
To determine whether broad H3K4me3 domains are involved in the phenotypic divergence between S. chuatsi and S. scherzeri, we identified and compared broad H3K4me3 peaks in the genomes of both species. Analysis revealed 491 and 481 broad H3K4me3 peaks (top 3% broadest H3K4me3 domains) in the S. chuatsi and S. scherzeri genomes, respectively. The broad H3K4me3 peaks were mostly found close to genes (Figure 5A, B). Consistent with the previous results, the signal intensity of broad H3K4me3 peaks was lower than that of narrow H3K4me3 peaks (Chen et al., 2015). In addition, 93.5% and 92.3% of the broad H3K4me3 peaks in S. chuatsi and S. scherzeri overlapped with H3K27ac peaks, respectively, confirming that broad H3K4me3 domains tend to overlap with H3K27ac domains (Supplementary Table S23) (Beacon et al., 2021). A total of 194 differential broad H3K4me3 peaks (|log2FC|>=1, P<=0.05) were identified between the S. chuatsi and S. scherzeri genomes, including 119 up-regulated and 75 down-regulated in S. chuatsi. KEGG pathway analysis of the function of genes associated with differential broad H3K4me3 peaks (Figure 5C; Supplementary Table S24) showed that the genes were enriched in several pathways related to cancer, including microRNA in cancer (ko05206), pathways in cancer (ko05200), and choline metabolism in cancer (ko05231). Among these enriched genes, the gene expression levels (log2FC>=1, FDR<=0.05) and broad H3K4me3 peak intensities (log2FC>=1, P<=0.05) of four genes (ccnd2a, egln2, kita, and f2r) were significantly up-regulated in S. chuatsi compared with S. scherzeri (Figure 5D; Supplementary Figures S18–20 and Table S25). Most of these genes are involved in developmental processes, including cardiovascular and melanocyte development (Ellertsdottir et al., 2012; Nicoli et al., 2012; O'Reilly-Pol & Johnson, 2013). These results suggest that broad H3K4me3 domain divergence contributes to the phenotypic divergence between S. chuatsi and S. scherzeri.
Figure 5.
Divergence of broad H3K4me3 domains between S. chuatsi and S. scherzeri
A, B: Signal intensities of 400 broad H3K4me3 peaks (red), 400 sharp H3K4me3 peaks (blue), and 400 randomly selected H3K4me3 peaks (control, gray) of S. chuatsi (A) and S. scherzeri (B). C: KEGG enrichment of genes associated with divergent broad H3K4me3 peaks. D: Cis-regulatory divergence between S. chuatsi (red) and S. scherzeri (blue) at ccnd2 gene. Intensities of H3K27ac, H3K4me3, ATAC-seq, and gene expression are shown. MACS2-identified H3K27ac and broad H3K4me3 peaks are denoted with rectangles below tracks. Transcripts (with exons as boxes) are depicted.
DISCUSSION
By integrating Nanopore and Hi-C sequencing strategies, we generated two high-quality chromosome-level assemblies of S. chuatsi and S. scherzeri. Based on the nearly complete genome assemblies, we first studied the evolutionary history of both species. Second, we dissected the role of SVs intersecting CREs in the phenotypic divergence between S. chuatsi and S. scherzeri. Finally, we investigated the role of broad H3K4me3 domains in the phenotypic differences between these closely related species.
Demographic history analysis of S. chuatsi and S. scherzeri provides insight into sinipercid evolution
Siniperca chuatsi and S. scherzeri are two of the most widely distributed sinipercids and are well adapted to diverse environments in East Asia, especially China (Song et al., 2017). Siniperca chuatsi is also a major aquaculture species in China due to its excellent flesh quality (FAO, 2006). Therefore, investigating the evolutionary history of these species can provide insight into the adaptation in this subfamily and facilitate potential breeding. The sinipercid ancestors repeatedly invaded freshwater systems from the ocean during the early Cenozoic Era (Li, 1991). Common ancestors of the subfamily Sinipercidae and genus Siniperca are estimated to have occurred 43.4 Ma and 12.2 Ma, respectively (Song et al., 2017). However, the demographic history of sinipercid fish is largely unknown. We estimated the ancestral population sizes of S. chuatsi and S. scherzeri using two approaches. PSMC inferred that the ancestral population of S. chuatsi declined during the early phase of the Mid-Pleistocene Transition (~0.9 Ma), when the duration of the Pleistocene glacial cycles increased from 41 to 100 ka (Berends et al., 2021). SMC++ analysis revealed a population bottleneck at the beginning of the last glacial period (~90 ka) after the Eemian interglacial period (129~116 ka). These results suggest that the colonization of new habitats and the diversification in S. chuatsi were largely affected by changes in temperature.
Sinipercids are dispersed over two major zoogeographic regions (Palearctic and Oriental) in China. Furthermore, two populations of S. scherzeri are identified in northern and southern China (Chen et al., 2014). We therefore inferred the dynamics of the ancestral population sizes of two S. scherzeri populations from northern (Jilin Province) and southern (Guangdong Province) China. SMC++ inferred that S. scherzeri from southern China experienced a decline in effective population size at ~400 ka, after the Mid-Brunhes Event (~430 ka), while the ancestral population of S. scherzeri from northern China declined at ~300 ka. These results imply that different populations of this widely distributed species have distinct local adaptation histories.
Cis-regulatory divergence by SVs plays an important role in phenotypic divergence between S. chuatsi and S. scherzeri
Gene expression plays a major role in the form, fitness, and function of organisms (Gordon & Ruvinsky, 2012). Differences in cis-regulatory activity contribute to gene expression divergence over a wide range of evolutionary scales and play a major role in phenotypic divergence (Coolon et al., 2014; Emerson et al., 2010; He et al., 2016). SVs are essential in the phenotypic evolution of both plants and animals (Mérot et al., 2020). Additionally, SVs alter the cis-regulatory activity of several developmental regulatory genes, resulting in morphological divergence between species (Wittkopp & Kalay, 2012). However, most SVs identified using short-read sequencing are unreliable, making comprehensive investigation of the impacts of SVs on cis-regulatory divergence largely impossible. However, with the advent of third-generation sequencing, researchers have revealed the role of SVs in phenotypic variation among diverse species. Here, we identified genome-wide high-quality SVs between S. chuatsi and S. scherzeri and investigated their role in the divergence of CREs marked with H3K27ac modification. We found that the signal intensity of H3K27ac peaks and expression levels of several genes involved in amino acid metabolism and fatty acid catabolism were elevated in S. chuatsi, indicating that lipid and amino acid metabolism efficiency is higher in S. chuatsi than in S. scherzeri. The growth rate of S. chuatsi is substantially higher than that of S. scherzeri (Ding et al., 2022). Organisms with high growth rates tend to have higher rates of lipid and amino acid metabolism (Møller & Jørgensen, 2009). These results suggest that cis-regulatory divergence induced by SVs causes expression divergence of genes involved in lipid and amino acid metabolism, leading to differences in the growth rates of S. chuatsi and S. scherzeri. In addition, we found that SVs caused cis-regulatory divergence in genes involved in skin pigmentation between S. chuatsi and S. scherzeri, resulting in darker pigmentation in S. scherzeri. Furthermore, the expression levels of four immune genes associated with SV-intersected divergent H3K27ac peaks were also substantially higher in S. scherzeri than in S. chuatsi. These four genes play critical roles in immunity, suggesting that SV-caused cis-regulatory variation contributes to the divergence in disease resistance between S. chuatsi and S. scherzeri. Taken together, our results suggest that cis-regulatory divergence caused by SVs plays an important role in the phenotypic divergence between these two closely related Siniperca species.
The relative contribution of coding and regulatory changes to speciation remains unclear. Studies in different organisms have shown that coding sequence variations in adaptive loci contribute to speciation and adaptation (Hoekstra & Coyne, 2007). In contrast, modification in gene expression by changes in regulatory regions is reported to play a prominent role in evolution and adaptation (Wray, 2007). Our analysis of PSGs using protein-coding sequences found that three genes associated to pigmentation were positively selected in S. scherzeri compared with S. chuatsi and eight teleost species. In addition, cis-regulatory divergence was found in two pigmentation-related genes between S. chuatsi and S. scherzeri. These findings indicate that both coding and regulatory changes contribute to speciation and adaptation in S. chuatsi and S. scherzeri.
Broad H3K4me3 domains are associated with cancer-related genes in S. chuatsi and S. scherzeri and contribute to their phenotypic divergence
There are two classes of H3K4me3 domains, including the well-studied narrow domains and newly identified broad domains. Most H3K4me3-enriched nucleosomes are detected as narrow peaks (1–2 kb), associated with the promoters of actively transcribed genes and acting as gene transcription switches (Dong et al., 2012). A small number of broad H3K4me3 domains, which can span up to 60 kb, have recently been identified in several species, including mammals, flies, worms, and plants (Beacon et al., 2021). Broad H3K4me3 domains are correlated with increased transcriptional elongation and enhancer activity, resulting in high expression levels of associated genes (Chen et al., 2015). This special histone modification preferentially marks genes associated with cell identity and cell-specific function (Benayoun et al., 2014). Broad H3K4me3 domains are reported to modulate maternal-to-zygotic transition in mouse oocytes (Dahl et al., 2016). Furthermore, broad H3K4me3 domains explicitly mark cancer-suppressor genes in normal human cells (Chen et al., 2015). This suggests that broad H3K4me3 domains may play a role in phenotypic divergence in animals by regulating developmental processes. However, comparative analysis of broad H3K4me3 domains between closely related species is still lacking. Previous studies of these domains have been largely restricted to a single species or distantly related species. In this study, we compared broad H3K4me3 domains in the genomes of two closely related fish species. Genes associated with divergent domains were mostly enriched in cancer-related pathways. Most of these genes are also involved in various developmental processes. These results suggest that divergence in broad H3K4me3 domains contributed to phenotypic divergence between two closely related species.
In conclusion, we generated high-quality chromosome-level genome assemblies of S. chuatsi and S. scherzeri, facilitating comparative genomic analysis of these two closely related species. First, demographic analysis indicated that S. chuatsi experienced two population bottlenecks in the early phase of the Mid-Pleistocene Transition (~0.9 Ma) and at the beginning of the last glacial period (~90 ka), respectively. This suggests that the colonization of new habitats and the diversification of S. chuatsi were largely affected by changes in temperature. Furthermore, the effective population size of S. scherzeri from northern China experienced a decline ~100 ka later than that of S. scherzeri from southern China, indicating that different populations of this widely distributed species have distinct local adaptation histories. Second, based on the chromosome-level assemblies, we identified high-quality SVs between the S. chuatsi and S. scherzeri genomes. Integrative analysis of SVs and H3K27ac domains indicated that cis-regulatory divergence caused by SVs played an essential role in the divergence of lipid and amino acid metabolism, skin pigmentation, and immunity between S. chuatsi and S. scherzeri. This suggests that cis-regulatory divergence caused by SVs played an important role in phenotypic divergence between these two closely related species. Finally, we found that divergence of broad H3K4me3 domains contributed to the phenotypic divergence between S. chuatsi and S. scherzeri.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
M.W. and J.G.H. conceived the project and designed the research; X.S.Z. and Li Z. sequenced the genomes; G.X.T. and R.R.J. assembled and annotated the genomes; G.X.T., Long Z., C.J.L, and Z.Y.Y. performed the evolutionary analyses; G.X.T., Y.R.L., and S.P.W. performed the epigenomic analyses; M.W., J.G.H., and G.X.T. wrote the paper with contribution from all authors. All authors read and approved the final version of the manuscript.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
ACKNOWLEDGEMENTS
We thank Dr. Jin-Long Wang for providing resequencing samples of S. chuatsi, Dr. Chuan-Fu Dong and Mr. Fei Li for providing resequencing samples of S. scherzeri from Jilin Province, Mr. Gen-Cheng Xian for providing resequencing samples of S. scherzeri from Guangdong Province. We gratefully acknowledge the National Supercomputing Center in Guangzhou for provision of computational resources.
Funding Statement
This study was supported by the National Natural Science Foundation of China (31900309), Guangdong Basic and Applied Basic Research Foundation (2019A1515011644), Key-Area Research and Development Program of Guangdong Province (2021B0202020001), Seed Industry Development Project of Agricultural and Rural Department of Guangdong Province (2022), and Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (311021006)
Contributor Information
Jian-Guo He, Email: lsshjg@mail.sysu.edu.cn.
Muhua Wang, Email: wangmuh@mail.sysu.edu.cn.
References
- Achilleos A, Huffman NT, Marcinkiewicyz E, et al MBTPS1/SKI-1/S1P proprotein convertase is required for ECM signaling and axial elongation during somitogenesis and vertebral development. Human Molecular Genetics. 2015;24(10):2884–2898. doi: 10.1093/hmg/ddv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Wang XG, Benoit M, et al major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182:145–161.e23. doi: 10.1016/j.cell.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacon TH, Delcuve GP, López C, et al The dynamic broad epigenetic (H3K4me3, H3K27ac) domain as a mark of essential genes. Clinical Epigenetics. 2021;13(1):138. doi: 10.1186/s13148-021-01126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, et al Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyeu JR, Chowdhury M, Brown J, et al Samplot: a platform for structural variant visual validation and automated filtering. Genome Biology. 2021;22(1):161. doi: 10.1186/s13059-021-02380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, et al H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158(3):673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends CJ, Köhler P, Lourens LJ, et al On the cause of the mid-Pleistocene transition. Reviews of Geophysics. 2021;59(2):e2020RG000727. [Google Scholar]
- Blakey CA, Litt MD. 2015. Histone modifications—models and mechanisms. In: Huang SM, Litt MD, Blakey CA. Epigenetic Gene Expression and Regulation. Amsterdam: Academic Press, 21–42.
- Bolger AM, Lohse M, Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen BW, Muss A, Rocha LA, et al Shallow mtDNA coalescence in Atlantic pygmy angelfishes (genus Centropyge) indicates a recent invasion from the Indian Ocean. Journal of Heredity. 2006;97(1):1–12. doi: 10.1093/jhered/esj006. [DOI] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, et al The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513(7518):375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson L, Pouyet L, N'guessan P, et al The thymus-specific serine protease TSSP/PRSS16 is crucial for the antitumoral role of CD4+ T cells. Cell Reports. 2015;10(1):39–46. doi: 10.1016/j.celrep.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, et al BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics and Bioinformatics. 2021;3(1):lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Carroll SB Evo-devo and an expanding evolutionary synthesis. The FASEB Journal. 2013;27(S1):194. [Google Scholar]
- Chabowski A, Górski J, Luiken JJFP, et al. 2007. Evidence for concerted action of FAT/CD36 and FABPpm to increase fatty acid transport across the plasma membrane. Prostaglandins, Leukotrienes and Essential Fatty Acids, 77(5–6): 345–353.
- Chaisson MJ, Tesler G Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, et al Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327(5963):302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Li Y, Li H, et al The genetic diversity of sinipercid fishes based on complete mitochondrial DNA of six sinipercid fishes from different drainages in China. Current Molecular Medicine. 2014;14(10):1279–1285. doi: 10.2174/1566524014666141202162058. [DOI] [PubMed] [Google Scholar]
- Chen KF, Chen Z, Wu DY, et al Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nature Genetics. 2015;47(10):1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SF, Zhou YQ, Chen YR, et al fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018a;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YX, Chen YS, Shi CM, et al SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018b;7(1):gix120. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Scott AJ, Davis JR, et al The impact of structural variation on human gene expression. Nature Genetics. 2017;49(5):692–699. doi: 10.1038/ng.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Giltiay NV CD22: A regulator of innate and adaptive B cell responses and autoimmunity. Frontiers in Immunology. 2018;9:2235. doi: 10.3389/fimmu.2018.02235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, McManus CJ, Stevenson KR, et al. 2014. Tempo and mode of regulatory evolution in Drosophila. Genome Research, 24(5): 797–808.
- Corces MR, Trevino AE, Hamilton EG, et al An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nature Methods. 2017;14(10):959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, et al Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JX, Ren XH, Yu QX Nuclear DNA content variation in fishes. Cytologia. 1991;56(3):425–429. doi: 10.1508/cytologia.56.425. [DOI] [Google Scholar]
- Curtis AM, Seo SB, Westgate EJ, et al Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. Journal of Biological Chemistry. 2004;279(8):7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, et al Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537(7621):548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, et al CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Ding SY, Shi YB, Hao C, et al Molecular mechanisms of growth and disease resistance in hybrid mandarin (Siniperca chuatsi ♀ × Siniperca scherzeri♂) revealed by combined miRNA‐mRNA transcriptome analysis. Aquaculture Research. 2022;53(6):2146–2158. doi: 10.1111/are.15734. [DOI] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, et al STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XJ, Greven MC, Kundaje A, et al Modeling gene expression using chromatin features in various cellular contexts. Genome Biology. 2012;13(9):R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, et al De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356(6333):92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, et al Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Systems. 2016;3(1):95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellertsdottir E, Berthold PR, Bouzaffour M, et al Developmental role of zebrafish protease-activated receptor 1 (PAR1) in the cardio-vascular system. PLoS One. 2012;7(7):e42131. doi: 10.1371/journal.pone.0042131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, et al Natural selection on cis and trans regulation in yeasts. Genome Research. 2010;20(6):826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. 2006(2006-05-30). Siniperca chuatsi. Cultured aquatic species information programme. Rome. https://www.fao.org/fishery/en/culturedspecies/siniperca_chuatsi/en.
- Flynn JM, Hubley R, Goubert C, et al RepeatModeler2 for automated genomic discovery of transposable element families. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu LM, Niu BF, Zhu ZW, et al CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FX, Lu WJ, Shi Y, et al. 2021. Transcriptome profiling revealed the growth superiority of hybrid pufferfish derived from Takifugu obscurus ♀ × Takifugu rubripes ♂. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 40: 100912.
- Goding CR, Arnheiter H. 2019. MITF-the first 25 years. Genes & Development, 33(15–16): 983–1007.
- Goel M, Sun HQ, Jiao WB, et al SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biology. 2019;20(1):277. doi: 10.1186/s13059-019-1911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I Tempo and mode in evolution of transcriptional regulation. PLoS Genetics. 2012;8(1):e1002432. doi: 10.1371/journal.pgen.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremme G, Brendel V, Sparks ME, et al Engineering a software tool for gene structure prediction in higher organisms. Information and Software Technology. 2005;47(15):965–978. doi: 10.1016/j.infsof.2005.09.005. [DOI] [Google Scholar]
- Grone BP, Marchese M, Hamling KR, et al Epilepsy, behavioral abnormalities, and physiological comorbidities in syntaxin-binding protein 1 (STXBP1) mutant zebrafish. PLoS One. 2016;11(3):e0151148. doi: 10.1371/journal.pone.0151148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WZF, Qiu G, Feng L Comparative analysis of the morphology, karyotypes and biochemical composition of muscle in Siniperca chuatsi, Siniperca scherzeri and the F1 hybrid (S. chuatsi ♀ × S. scherzeri ♂) Aquaculture and Fisheries. 2022;7(4):382–388. doi: 10.1016/j.aaf.2020.07.020. [DOI] [Google Scholar]
- Gunn TM, Inui T, Kitada K, et al Molecular and phenotypic analysis of Attractin mutant mice. Genetics. 2001;158(4):1683–1695. doi: 10.1093/genetics/158.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, et al Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biology. 2008;9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara TJ Olfaction in fish. Progress in Neurobiology. 1975;5(4):271–335. doi: 10.1016/0301-0082(75)90014-3. [DOI] [PubMed] [Google Scholar]
- Hara TJ, Zhang CB Spatial projections to the olfactory bulb of functionally distinct and randomly distributed primary neurons in salmonid fishes. Neuroscience Research. 1996;26(1):65–74. doi: 10.1016/0168-0102(96)01078-4. [DOI] [PubMed] [Google Scholar]
- He F, Arce AL, Schmitz G, et al. 2016. The footprint of polygenic adaptation on stress-responsive Cis-regulatory divergence in the Arabidopsis Genus. Molecular Biology and Evolution, 33(8): 2088–2101.
- He S, Li L, Lv LY, et al Mandarin fish (Sinipercidae) genomes provide insights into innate predatory feeding. Communications Biology. 2020;3(1):361. doi: 10.1038/s42003-020-1094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liang XF, Sun J, et al Insights into food preference in hybrid F1 of Siniperca chuatsi (♀) × Siniperca scherzeri (♂) mandarin fish through transcriptome analysis. BMC Genomics. 2013;14:601. doi: 10.1186/1471-2164-14-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler FF, Loebrich S, Pechmann Y, et al Muskelin regulates actin filament- and microtubule-based GABAA receptor transport in neurons. Neuron. 2011;70(1):66–81. doi: 10.1016/j.neuron.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, et al UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution. 2018;35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61(5):995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hoffman JS, Clark PU, Parnell AC, et al Regional and global sea-surface temperatures during the last interglaciation. Science. 2017;355(6322):276–279. doi: 10.1126/science.aai8464. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, et al Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Molecular Biology and Evolution. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, et al eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Research. 2019;47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Özdag H, Caldas C p300/CBP and cancer. Oncogene. 2004;23(24):4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of Molecular Biology. 2016;428(4):726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, et al MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, et al CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nature Communications. 2019;10(1):1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenne E, Soehnlein O, Genové G, et al Immune cell recruitment to inflammatory loci is impaired in mice deficient in basement membrane protein laminin α4. Journal of Leukocyte Biology. 2010;88(3):523–528. doi: 10.1189/jlb.0110043. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, et al Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma R, Melsted P, Magnússon KP, et al Looking into the past - the reaction of three grouse species to climate change over the last million years using whole genome sequences. Molecular Ecology. 2016;25(2):570–580. doi: 10.1111/mec.13496. [DOI] [PubMed] [Google Scholar]
- Kronenberg ZN, Fiddes IT, Gordon D, et al High-resolution comparative analysis of great ape genomes. Science. 2018;360(6393):eaar6343. doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, et al TimeTree: a resource for timelines, timetrees, and divergence times. Molecular Biology and Evolution. 2017;34(7):1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Kurtz DM, Tolwani RJ, Wood PA Structural characterization of the mouse long-chain acyl-CoA dehydrogenase gene and 5' regulatory region. Mammalian Genome. 1998;9(5):361–365. doi: 10.1007/s003359900770. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M Transcriptional enhancers in animal development and evolution. Current Biology. 2010;20(17):R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SZ Geographical distribution of the Sinipercine fishes. Chinese Journal of Zoology. 1991;26(4):40–44. [Google Scholar]
- Liang XF, Kiu JK, Huang BY The role of sense organs in the feeding behaviour of Chinese perch. Journal of Fish Biology. 1998;52(5):1058–1067. doi: 10.1111/j.1095-8649.1998.tb00603.x. [DOI] [Google Scholar]
- Liu H, Chen CH, Lv ML, et al A chromosome-level assembly of blunt snout bream (Megalobrama amblycephala) genome reveals an expansion of olfactory receptor genes in freshwater fish. Molecular Biology and Evolution. 2021;38(10):4238–4251. doi: 10.1093/molbev/msab152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livraghi L, Hanly JJ, Van Bellghem SM, et al. 2021. Cortex cis-regulatory switches establish scale colour identity and pattern diversity in Heliconius. eLife, 10: e68549.
- Long HK, Prescott SL, Wysocka J Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167(5):1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhao JL, Li CH High-quality genome assembly and annotation of the big-eye mandarin fish (Siniperca knerii) G3 Genes| Genomes| Genetics. 2020;10(3):877–880. doi: 10.1534/g3.119.400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Chen KF Broad H3K4me3 as a novel epigenetic signature for normal development and disease. Genomics, Proteomics & Bioinformatics. 2016;14(5):262–264. doi: 10.1016/j.gpb.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot C, Oomen RA, Tigano A, et al A roadmap for understanding the evolutionary significance of structural genomic variation. Trends in Ecology & Evolution. 2020;35(7):561–572. doi: 10.1016/j.tree.2020.03.002. [DOI] [PubMed] [Google Scholar]
- Møller N, Jørgensen JOL Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocrine Reviews. 2009;30(2):152–177. doi: 10.1210/er.2008-0027. [DOI] [PubMed] [Google Scholar]
- Moriyama M, Osawa M, Mak SS, et al Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. Journal of Cell Biology. 2006;173(3):333–339. doi: 10.1083/jcb.200509084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Knyphausen CP, Zhu LJ, et al miR-221 is required for endothelial tip cell behaviors during vascular development. Developmental Cell. 2012;22(2):418–429. doi: 10.1016/j.devcel.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biology and Evolution. 2009;1:34–44. doi: 10.1093/gbe/evp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Research. 2014;24(9):1485–1496. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M Evolution of olfactory receptor genes in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly-Pol T, Johnson SL Kit signaling is involved in melanocyte stem cell fate decisions in zebrafish embryos. Development. 2013;140(5):996–1002. doi: 10.1242/dev.088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshikawa Y, Kobayashi T, et al A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host & Microbe. 2011;9(5):376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Reviews Genetics. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ ChIP-seq: advantages and challenges of a maturing technology. Nature Reviews Genetics. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, et al Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature Protocols. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policarpo M, Bemis KE, Tyler JC, et al Evolutionary dynamics of the OR gene repertoire in teleost fishes: evidence of an association with changes in olfactory epithelium shape. Molecular Biology and Evolution. 2021;38(9):3742–3753. doi: 10.1093/molbev/msab145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, et al deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, et al Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biology. 2020;21(1):245. doi: 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Turner D, Durand NC, et al Juicebox. js provides a cloud-based visualization system for Hi-C data. Cell Systems. 2018;6(2):256–258.e1. doi: 10.1016/j.cels.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H Fast and accurate long-read assembly with wtdbg2. Nature Methods. 2020;17(2):155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm A, Bens M, Platzer M, et al PosiGene: automated and easy-to-use pipeline for genome-wide detection of positively selected genes. Nucleic Acids Research. 2017;45(11):e100. doi: 10.1093/nar/gkx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L, Rivals E LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30(24):3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, et al Accurate detection of complex structural variations using single-molecule sequencing. Nature Methods. 2018;15(6):461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Wang XF, Jablonski MM, et al An engineered 800 kilobase deletion of Uchl3 and Lmo7 on mouse chromosome 14 causes defects in viability, postnatal growth and degeneration of muscle and retina. Human Molecular Genetics. 2003;12(11):1301–1312. doi: 10.1093/hmg/ddg140. [DOI] [PubMed] [Google Scholar]
- Shao Z, Zhang YJ, Yuan GC, et al MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biology. 2012;13(3):R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. 2002. Adaptive protein evolution in Drosophila. Nature, 415(6875): 1022–1024.
- Song SL, Zhao JL, Li CH Species delimitation and phylogenetic reconstruction of the sinipercids (Perciformes: Sinipercidae) based on target enrichment of thousands of nuclear coding sequences. Molecular Phylogenetics and Evolution. 2017;111:44–55. doi: 10.1016/j.ympev.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Soupene E, Kuypers FA Mammalian long-chain acyl-CoA synthetases. Experimental Biology and Medicine. 2008;233(5):507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel W, Jenke B, Blöck B, et al Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(12):4554–4559. doi: 10.1073/pnas.0406380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Terhorst J, Kamm JA, Song YS Robust and scalable inference of population history from hundreds of unphased whole genomes. Nature Genetics. 2017;49(2):303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verta JP, Jones FC Predominance of cis-regulatory changes in parallel expression divergence of sticklebacks. eLife. 2019;8:e43785. doi: 10.7554/eLife.43785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, et al Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics. 2012;13(1):59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Woodham EF, Paul NR, Tyrrell B, et al Coordination by Cdc42 of actin, contractility, and adhesion for melanoblast movement in mouse skin. Current Biology. 2017;27(5):624–637. doi: 10.1016/j.cub.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8(3):206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Xu DM, Zalmas LP, La Thangue NB A transcription cofactor required for the heat-shock response. EMBO Reports. 2008;9(7):662–669. doi: 10.1038/embor.2008.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni W, et al Ensembl 2020. Nucleic Acids Research. 2020;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QZ, Berger A Insolation and CO2 contribution to the interglacial climate before and after the Mid-Brunhes Event. Nature Geoscience. 2010;3(4):243–246. doi: 10.1038/ngeo771. [DOI] [Google Scholar]
- Yu GC, Wang LG, He QY ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zhan TZ, Poppelreuther M, Ehehalt R, et al Overexpressed FATP1, ACSVL4/FATP4 and ACSL1 increase the cellular fatty acid uptake of 3T3-L1 adipocytes but are localized on intracellular membranes. PLoS One. 2012;7(9):e45087. doi: 10.1371/journal.pone.0045087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, He J, Tan PP, et al The genome of an apodid holothuroid (Chiridota heheva) provides insights into its adaptation to a deep-sea reducing environment. Communications Biology. 2022;5(1):224. doi: 10.1038/s42003-022-03176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, et al Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Gao HY, Zhang Y, et al A chromosome-level genome assembly of the giant grouper (Epinephelus lanceolatus) provides insights into its innate immunity and rapid growth. Molecular Ecology Resources. 2019;19(5):1322–1332. doi: 10.1111/1755-0998.13048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.