Significance
Human papillomavirus (HPV) causes 5% of cancers worldwide and is now the most common cause of oropharyngeal cancer. HPV has been implicated in inducing chromosomal instability (CIN), a rate of continuous chromosome missegregation over multiple cell divisions, but mechanistic insight is lacking and this has not been studied in head and neck cancer (HNC). Here, we report that the HPV oncoprotein E6 causes a specific type of CIN in which chromosomes remain chronically misaligned at the spindle pole during mitosis. HPV16 E6 induces this phenotype by causing the degradation of the mitotic kinesin CENP-E in an E6AP and proteasome-dependent manner. This work provides the first mechanistic insight into how HPV induces CIN in HNC, which likely contributes to carcinogenesis.
Keywords: mitosis, papillomavirus, CIN
Abstract
Chromosome segregation during mitosis is highly regulated to ensure production of genetically identical progeny. Recurrent mitotic errors cause chromosomal instability (CIN), a hallmark of tumors. The E6 and E7 oncoproteins of high-risk human papillomavirus (HPV), which causes cervical, anal, and head and neck cancers (HNC), cause mitotic defects consistent with CIN in models of anogenital cancers, but this has not been studied in the context of HNC. Here, we show that HPV16 induces a specific type of CIN in patient HNC tumors, patient-derived xenografts, and cell lines, which is due to defects in chromosome congression. These defects are specifically induced by the HPV16 oncogene E6 rather than E7. We show that HPV16 E6 expression causes degradation of the mitotic kinesin CENP-E, whose depletion produces chromosomes that are chronically misaligned near spindle poles (polar chromosomes) and fail to congress. Though the canonical oncogenic role of E6 is the degradation of the tumor suppressor p53, CENP-E degradation and polar chromosomes occur independently of p53. Instead, E6 directs CENP-E degradation in a proteasome-dependent manner via the E6-associated ubiquitin protein ligase E6AP/UBE3A. This study reveals a mechanism by which HPV induces CIN, which may impact HPV-mediated tumor initiation, progression, and therapeutic response.
Errors during mitosis have been recognized in cancer for over a century (1). Mitotic errors result in aneuploid progeny, which contain an atypical chromosome number that differs from a multiple of the haploid. Mitotic errors that recur over successive cell divisions cause chromosomal instability (CIN). Both aneuploidy and CIN are common in human cancers, which led to the hypothesis that aneuploid cells form the primordial cells of tumors (2, 3). More recent evidence in animal models has shown that the situation is more complex. Aneuploidy can promote tumors, suppress them, or do neither (reviewed in refs. 4–8). The impact of aneuploidy is determined by the specific aneuploidy-inducing insult, any additional interphase phenotypes caused, and the tissue context, as well as the rate of CIN. Low rates of CIN can be tumor-promoting through the gain of oncogenes or loss of tumor suppressors (9–13). Higher rates of CIN cause cell death, likely through loss of essential chromosomes, and tumor suppression (14–22). A preexisting rate of CIN can sensitize cells to another CIN-inducing insult, and cells with CIN are more sensitive to paclitaxel, which causes CIN due to multipolar spindles in patient tumors and at clinically relevant doses in cell culture (23–28). Thus, CIN is a common characteristic of cancer that, depending on the rate, can have differential impacts on tumor initiation, progression, and response to therapy.
High-risk human papillomavirus (HPV) causes ~5% of all cancers worldwide, including >95% of cervical and >85% of anal cancers (29–32). High-risk HPV also represents a significant and growing cause of oropharyngeal head and neck cancers (HNC), currently accounting for ~50% of oropharyngeal squamous cell carcinomas worldwide and >70% in North America (33, 34). Though HPV genotypes 16, 18, 31, 33, 45, 52, and 58 are recognized causes of cervical and anal cancers, HPV16 is the primary etiological agent in HNC, occurring in >90% of HPV+ HNC (33). HPV+ HNC are more sensitive to radiation than HPV− HNC and have improved outcomes to standard of care (chemo)radiation, with a 3-y survival rate of 82% as compared to 57% for HPV− HNC patients (35, 36). The basis for the differential sensitivity is an area of active investigation. An understanding of the relevant biological differences between HPV+ and HPV− HNC could be leveraged to improve outcomes in HPV− HNC.
HPV encodes two main oncoproteins, E6 and E7, which are necessary for host cellular transformation and have been implicated in numerous pathogenic pathways (reviewed in refs. 37–39). E6 binds p53 and the host ubiquitin ligase, E6AP (also known as UBE3A), which allows E6AP to ubiquitinate p53 leading to its degradation (40–42). E7 is known to bind Rb, which promotes S phase entry by releasing inhibition of E2F transcription factors (43–45). Thus together, viral E6 and E7 expression in the host cell leads to increased cell cycle progression and proliferation and reduced apoptosis, which are factors contributing to carcinogenesis. Additionally, HPV16 E6 and E7 have both been implicated in causing various types of mitotic defects that drive CIN. Ectopic expression of either E6 or E7 drives centrosome amplification in human foreskin keratinocytes, mouse embryonic fibroblasts, and the U2OS osteosarcoma cell line (46–50). Normally, one centrosome is found at each pole of a bipolar mitotic spindle, and centrosome amplification can cause multipolar spindles. Consistent with this, multipolar spindles have been observed due to expression of high levels of E6 or E7 in keratinocytes and U2OS cells (50–53). Multipolar spindles often result in CIN either because the two sets of replicated chromosomes segregate into three or more daughter cells or, if the multipolar spindle focuses into a bipolar spindle prior to chromosome segregation in anaphase, through an increase in chromosomes that lag behind the segregating masses of DNA (54, 55). E6 and E7, alone or in combination, also induce chromosomes that bridge the segregating DNA masses in anaphase and telophase (chromosome bridges) in keratinocytes (50, 52, 56, 57). Additionally, E6 expression causes a chromosome alignment defect, producing chromosomes that remain chronically misaligned near spindle poles (polar chromosomes) in nontransformed retinal pigment epithelial (RPE1) cells (58). Thus, high-level expression of high-risk HPV oncogenes has been reported to give rise to a variety of mitotic defects that can cause CIN and aneuploidy, yet mechanistic insight is lacking. Additionally, all prior work has occurred in the context of cervical cancer or utilized cell lines that are not physiologically relevant to HNC. We therefore aimed to determine the mitotic defects and associated mechanism in HPV+ HNC, which have not been well described, using primary tumors and physiologically relevant models.
Here, we use HNC patient tumor biopsies, patient-derived xenografts, clinically and physiologically relevant cell lines, and genetic engineering to demonstrate that HPV16 E6 oncogene expression causes a specific type of CIN in HNC that is due to chromosome congression defects that result in polar chromosomes. This HPV-induced mitotic error is specifically due to expression of E6 rather than E7 and is dependent on E6AP-mediated proteasomal degradation of the mitotic motor CENP-E. CENP-E is one of several proteins that link chromosomes to the microtubules of the mitotic spindle to facilitate congression to the metaphase plate during mitosis (reviewed in refs. 59 and 60). Reduction of CENP-E or inhibition of its ATPase activity results in a specific increase in polar chromosomes (61–64). These results support the conclusion that expression of the HPV16 E6 oncogene causes a specific type of CIN in HNC characterized by polar chromosomes that are due to E6AP-mediated CENP-E degradation.
Results
HPV16 Causes Chromosome Congression Defects in HNC.
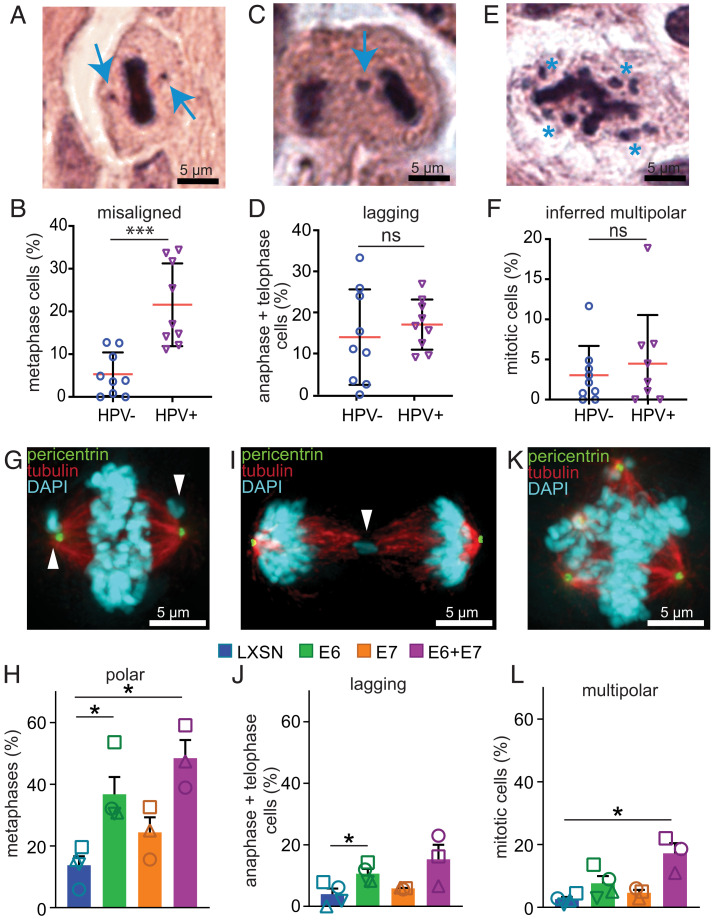
To define the types of CIN caused by HPV in HNC, we quantified mitotic defects in 9 HPV+ and 9 HPV− oropharyngeal HNC patient biopsies or surgical specimens prior to (chemo)radiation therapy. HPV status was determined based on p16 positivity, which is a clinically accepted surrogate for HPV positivity and has been shown to be 97% sensitive for HPV gene transcription in HNC (65). H&E-stained tumor sections were examined for mitotic defects that cause CIN, including misaligned (Fig. 1 A and B) and lagging (Fig. 1 C and D) chromosomes in mitotic cells. Multipolar spindles were inferred based on the position of the DNA since microtubules of the mitotic spindle are not visualized by H&E (Fig. 1 E and F). The most prominent mitotic defect observed in HPV+ tissues was misaligned chromosomes. (We note that since spindle poles cannot be visualized on H&E-stained slides, the term “misaligned chromosomes” is used to describe chromosomes that are not congressed to the metaphase plate, including those at or near the inferred spindle poles.) HPV+ patient HNC exhibited a significantly higher incidence of misaligned chromosomes than HPV− cancers (21.5% vs. 5%, Fig. 1B). In most cases, the misaligned chromosomes appeared to be adjacent to inferred spindle poles (Fig. 1A and SI Appendix, Fig. S1A). The frequency of lagging chromosomes (Fig. 1D) and inferred multipolar spindles (Fig. 1F) was similar in both HPV+ and HPV− patient samples. Analysis of HPV+ and HPV− HNC patient-derived xenograft (PDX) tumors (66–68) revealed similar results, with HPV+ tumors having increased misaligned chromosomes but not lagging chromosomes or multipolar spindles (SI Appendix, Fig. S1 A–F). We further investigated the presence of misaligned chromosomes in a panel of 4 HPV+ and 4 HPV− HNC cell lines and again found a significant increase in the incidence of misaligned chromosomes in HPV+ cells, with much more subtle effects on other types of mitotic defects (SI Appendix, Fig. S1 G–L). In these cells, visualization of the mitotic spindle confirmed that misaligned chromosomes were at or near spindle poles (termed “polar chromosomes”; SI Appendix, Fig. S1G). Thus, HPV causes a specific increase in misaligned chromosomes in HNC tumors, and this mitotic phenotype is faithfully recapitulated by HPV+ HNC cell and xenograft models.
Fig. 1.
HPV16 E6 induces polar chromosomes in HNC. (A–F) HPV causes a specific increase in misaligned chromosomes in HNC patient biopsies. (A) HPV+ mitotic cell with two misaligned chromosomes (arrows). (B) Quantitation of misaligned chromosomes from HNC biopsies. n = average of 82 metaphases from each of 9 HPV− and 9 HPV+ patients. (C and D) HPV+ HNC do not have a higher incidence of lagging chromosomes than HPV− HNC. (C) HPV− cell with a lagging chromosome (arrow). (D) Quantitation of lagging chromosomes in HNC biopsies. n = average of 25 anaphase + telophase cells from each of 9 HPV− and 9 HPV+ patients. (E and F) Both HPV+ and HPV− HNC have a low incidence of multipolar spindles. (E) HPV+ cell with an inferred multipolar spindle based on the location of the chromosomes. The expected location of the spindle poles is indicated with asterisks. (F) Quantitation of inferred multipolar spindles in HNC patient biopsies. n = average of 132 mitotic cells from each of 9 HPV− and 9 HPV+ patients. (G–L) Expression of HPV16 oncogenes in NOKs demonstrates that E6 causes a specific increase in misaligned chromosomes near spindle poles (polar chromosomes). (G) E6 expressing NOK with polar chromosomes (arrowheads). (H) Quantitation of polar chromosomes. n > 45 metaphases from each of ≥3 independent experiments. (I) E6 expressing NOK with a lagging chromosome (arrowhead). (J) Quantitation of lagging chromosomes. n > 100 anaphase + telophase cells from each of ≥3 independent experiments. (K) E6 expressing NOK with a multipolar spindle. Centrosomes at spindle poles are indicated by pericentrin staining (green). (L) Quantitation of multipolar mitoses. n > 250 mitotic cells from ≥3 independent experiments. (H, J, and L) Shapes indicate values obtained from a given independent experiment. Error bars indicate SD (B, D, and F ) or SEM (H, J, and L). ***P < 0.0001, *P < 0.05, ns = not significant.
To determine which HPV oncogene was responsible for inducing polar chromosomes in HNC, we tested the impact of stable expression of HPV16 E6 and/or E7 in a physiologically relevant cell line, normal oral keratinocytes (NOKs): gingival cells with a near diploid karyotype that have been immortalized with hTERT (69). We observed that E6 expression, with or without coexpression of E7, caused a substantial increase in polar chromosomes (Fig. 1 G and H), consistent with prior findings in E6-expressing RPE1 cells (58). E7 expression alone did not cause a significant increase in any mitotic phenotype (Fig. 1 G–L). Coexpression of E6 and E7 caused a significant increase in multipolar spindles as compared to pLXSN vector control, though the overall incidence of multipolar spindles remained substantially lower than the incidence of polar chromosomes. Thus, E6 expression is sufficient to cause the increase in misaligned chromosomes near spindle poles observed in HPV+ HNC.
Lagging chromosomes and chromosome bridges often result in micronuclei which have been implicated in the formation of structural aneuploidy (70, 71). In contrast, polar chromosomes are more likely to join with the main nucleus after chromosome segregation to spindle poles in early anaphase, leading to whole chromosome aneuploidy (72). Since HPV16 E6 specifically increases polar chromosomes, we wanted to determine whether HPV+ HNCs have higher levels of whole chromosome aneuploidy than HPV− HNCs. In The Cancer Genome Atlas (TCGA) dataset, 75 HNCs are HPV+ and 427 are HPV− (73). HPV+ tumors have fewer total aneuploidy events, with an average of seven chromosome arms affected compared to 12 in HPV− tumors (Student’s t test P-value = 1.29e-8). However, the fraction of aneuploidy due to whole chromosome changes is higher in HPV+ than HPV− cancers with a median of 31.6% compared to 28.9%, respectively (SI Appendix, Fig. S2A). It was previously shown that when clustered based on average aneuploidy alteration for each arm, HPV+ head and neck carcinomas (HNSCCs)cluster with cervical squamous cell cancers (CESCs), 95% of which are HPV+ (74). We therefore expanded this analysis to 1,352 squamous cell carcinoma (SCC) samples from the TCGA, spanning lung, head and neck, esophagus, cervix, and bladder with 318 HPV+ tumors. Although again HPV+ tumors have fewer aneuploidy events, the fraction of aneuploidy due to whole chromosome changes is significantly higher in HPV+ than HPV− cancers with a median of 33.1% compared to 28.7%, respectively (Student’s t test P value = 0.004, SI Appendix, Fig. S2B). Thus, the polar chromosome phenotype we observed in HNC tumors, PDXs, cell lines, and HPV16 E6-expressing NOKs correlates with whole chromosome aneuploidy observed in a large cohort of HPV-positive cancer patients.
HPV16 E6 Causes Polar Chromosomes through Reduced Protein Levels of CENP-E.
There are two known causes of the polar chromosome phenotype observed after E6 expression. First, polar chromosomes can occur after focusing of multipolar spindles. Chromosomes that were previously aligned between two separated poles become chronically pole-associated if those two poles subsequently focus into a single pole (54). Since pole focusing is typically incomplete, focused multipolar spindles generally remain pseudo-bipolar as opposed to monopolar. To assess the extent to which spindle pole focusing contributed to polar chromosomes in E6 expressing cells, we included the centrosome marker pericentrin to assess whether misaligned chromosomes primarily occurred in a) bipolar spindles (SI Appendix, Fig. S3A), b) pseudo-bipolar spindles in which multiple centrosomes had been focused into a single spindle pole (SI Appendix, Fig. S3B), or c) multipolar spindles (SI Appendix, Fig. S3C). A large majority of misaligned chromosomes in cells expressing E6 occurred on normal bipolar spindles, with <20% appearing on pseudobipolar spindles (SI Appendix, Fig. S3D). From this data, we conclude that focusing of multipolar spindles makes only a minor contribution to E6-induced polar chromosomes.
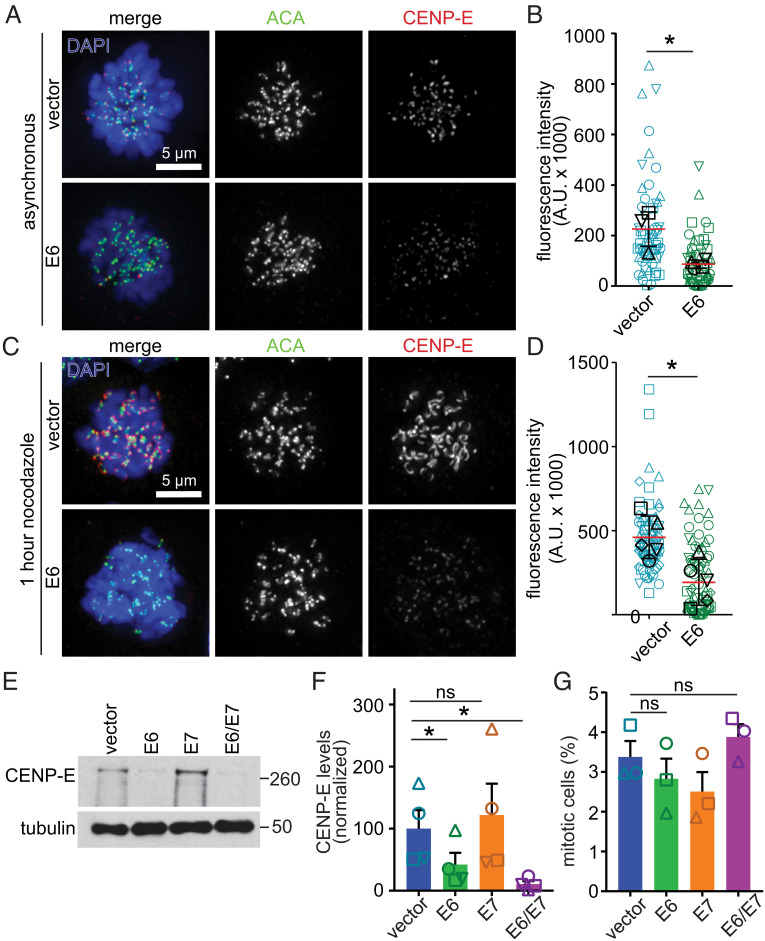
The other well-established mechanism of polar chromosome formation is reduced localization, expression, and/or activity of the mitotic kinesin CENP-E (61–64). To determine whether E6 expression affects CENP-E localization, we performed quantitative immunofluorescence. Mitotic NOKs expressing empty pLXSN vector (control) exhibited typical accumulation of CENP-E on kinetochores, the microtubule attachment sites of chromosomes (Fig. 2 A, Top and Fig. 2B). E6 expression substantially reduced kinetochore recruitment of CENP-E (Fig. 2 A, Bottom and Fig. 2B). As expected, 1 h of treatment with the microtubule destabilizer nocodazole enhanced CENP-E recruitment to kinetochores in control cells, which accentuated the deficiency in CENP-E localization in the presence of E6 (Fig. 2 C and D). Thus, HPV16 E6 leads to significantly decreased levels of CENP-E at kinetochores during mitosis.
Fig. 2.
HPV16 E6 reduces protein levels of CENP-E. (A–D) HPV16 E6 expression reduces CENP-E localization to kinetochores during mitosis. (A) Representative images of DMSO-treated NOKs expressing empty vector (Top) or E6 (Bottom). ACA, anti-centromere antibody. (B) Quantitation of CENP-E fluorescence intensity in asynchronous NOKs. n ≥ 12 prometaphase cells in each of four independent experiments. Shapes indicate values obtained from a given independent experiment. Black symbols represent average value for a given replicate. (C) Representative images of NOKs transduced with vector alone (Top) or E6 (Bottom) treated for 1 h with 200 ng/mL nocodazole. (D) Quantitation of CENP-E fluorescence intensity in nocodazole-treated NOKs. n ≥ 12 prometaphase cells from each of 5 independent experiments. Shapes indicate values obtained from independent biological replicates, as in B. (E–G) E6 expression reduces CENP-E protein level without decreasing the fraction of cells in mitosis. (E) NOKs expressing E6 +/− E7 exhibit reduced CENP-E protein expression. Tubulin, loading control. Numbers next to blots indicate molecular weight markers in kD. (F) Quantitation of CENP-E protein from four independent immunoblots. (G) Quantitation of mitotic index in NOKs showing that reduced CENP-E levels are not due to a decrease in mitotic cells. n > 375 cells from each of three independent experiments. Shapes indicate values obtained from a given independent experiment. Error bars indicate SD (B and D) or SEM (F and G). *P < 0.05. ns = not significant.
To determine whether reduced localization of CENP-E was due to a decrease in available protein, NOKs expressing vector control, E6, E7, or both were subjected to immunoblotting. Indeed, expression of E6, alone or in combination with E7, resulted in a significant reduction of CENP-E protein (Fig. 2 E and F). CENP-E is tightly cell cycle regulated, with protein levels peaking in late G2 and mitosis followed by quantitative proteasome-mediated degradation at the end of mitosis resulting in low levels in G1 and S phases (75). Thus, reduced protein expression of CENP-E could be specific or could be an indirect consequence of a decreased mitotic index in E6 expressing cells. However, expression of E6 did not affect the mitotic index (Fig. 2G). Moreover, among a panel of mitotic regulators (including Bub1, BubR1, Mad1, Mad2, and CENP-E), several of which are cell cycle regulated, only CENP-E was affected by E6 expression (SI Appendix, Fig. S3 E and F). Bub1 and BubR1 have both previously been implicated in correctly localizing CENP-E to kinetochores (76, 77). However, since expression of these proteins is unaffected, the impact of E6 is unlikely to be indirectly mediated via effects on Bub1 or BubR1. To determine if other proteins that link chromosomes to spindle microtubules are altered by E6, we quantified the levels of Hec1/Ndc80, a member of the protein complex that forms the core attachment sites for spindle microtubules (78, 79). Hec1 levels were unaffected by E6 expression (SI Appendix, Fig. S3 G and H). Thus, the polar chromosomes and reduced kinetochore localization of CENP-E in E6-expressing NOKs are likely due to a specific decrease in CENP-E protein levels.
To confirm that reduction of CENP-E induces polar chromosomes in the absence of E6 in NOKs, as it does in other cell types (61–64), we depleted CENP-E using siRNA in pLXSN control vector-expressing NOKs. This substantially decreased CENP-E protein expression and increased polar chromosomes (SI Appendix, Fig. S4 A–D). As further validation of the role of CENP-E in inducing polar chromosomes in NOKs, we inhibited CENP-E ATPase activity with GSK923295, which significantly increased the incidence of polar chromosomes (SI Appendix, Fig. S4E). These data demonstrate that a decrease in CENP-E levels or activity is sufficient to induce polar chromosomes in NOKs.
To determine if the polar chromosomes we observed in HPV+ patient tumors are also associated with a decrease in CENP-E protein, we analyzed tumor protein lysates of a second cohort of 4 HPV+ and 4 HPV− head and neck cancers. The HPV+ HNC samples typically expressed HPV16 E6, had reduced levels of CENP-E, and increased polar chromosomes compared to the HPV− samples (SI Appendix, Fig. S5 A–C). We then extended this analysis to cervical cancer. Interestingly, HPV16+ cervical cancer cells (SiHa and CaSki) also have significantly higher levels of polar chromosomes and lower levels of CENP-E than the HPV− cervical cancer cell line C33A (SI Appendix, Fig. S5 D–G). Thus, HPV16 infection causes CENP-E depletion and polar chromosomes in multiple contexts.
Because E6 is also known to cause decreased levels of p53, we tested whether decreased CENP-E was associated with p53 loss. We have previously shown in primary murine embryonic fibroblasts (MEFs) and intestine that p53 loss does not reduce CENP-E protein expression or cause polar chromosomes (22). To confirm this was also true in NOKs, we examined TP53−/− NOKs generated using CRISRP/Cas9 (80). Consistent with results in murine cells, loss of p53 did not impact CENP-E protein levels (SI Appendix, Fig. S6 A and B) or result in polar chromosomes (SI Appendix, Fig. S6C). We then transduced the TP53−/− NOKs with either pLXSN vector or E6. E6-expressing TP53−/− NOKs showed lower levels of E6 expression than TP53 wild-type NOKs (SI Appendix, Fig. S6 D–F). However, this low level of E6 expression was still sufficient to decrease CENP-E protein levels and increase polar chromosomes (SI Appendix, Fig. S6 D–G). Thus, E6-induced CENP-E depletion and chromosome congression defects are independent of p53.
High-risk HPV E6 oncoproteins, including HPV16 E6, contain a PDZ binding motif which regulates interaction among target proteins, often leading to their degradation, among other functions (81). In order to determine if the PDZ binding motif is necessary for E6-induced CENP-E degradation, we transfected NOKs with a mutant of E6 (E6Δ146−151) that lacks six amino acids at the C-terminus, which renders it unable to bind PDZ domain containing proteins, though it retains the ability to degrade p53 (82). Both WT E6 and E6Δ146−151 expression led to a decrease in CENP-E protein and a concomitant increase in polar chromosomes (SI Appendix, Fig. S6 H–J). Both were also able to degrade p53 (SI Appendix, Fig. S6H), consistent with prior reports (82, 83). We therefore conclude that E6-mediated degradation of CENP-E is independent of its interaction with PDZ domains.
E6 Regulates CENP-E Levels Posttranslationally.
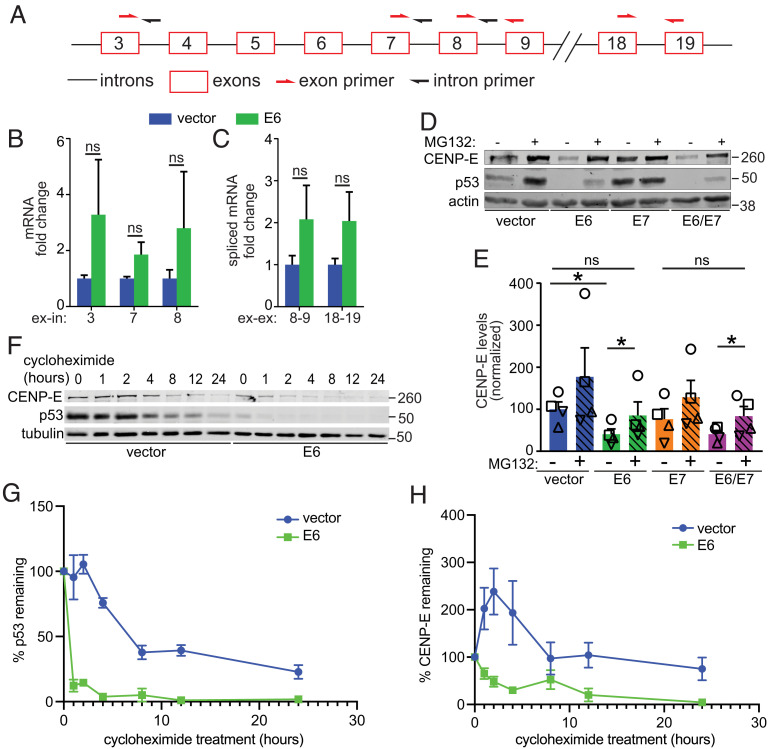
It was unclear whether reduced CENP-E protein levels after E6 expression were due to decreased transcription or to posttranscriptional events. CENP-E splicing is controlled by ERH (enhancer of rudimentary splicing) and reduced expression of ERH decreases CENP-E protein expression resulting in polar chromosomes (84). To determine whether E6 decreases CENP-E transcription or splicing, we performed qRT-PCR with RNA extracted from E6 or vector control expressing NOKs. We used three sets of exon-intron primers to quantify total RNA and two sets of exon–exon primers to measure mRNA (Fig. 3A). E6 expression did not decrease either CENP-E total RNA or mRNA (Fig. 3 B and C), suggesting that the reduction in CENP-E protein levels occurs due to decreased translation or increased protein turnover. As an initial test of whether an increase in CENP-E degradation accounted for the reduced levels of CENP-E in E6 expressing cells, NOKs expressing vector alone, E6, E7, or both, were treated with the proteasome inhibitor MG132. p53 was used as a positive control since its E6-mediated proteasomal degradation is well-established (40–42). MG132 treatment increased expression levels of both CENP-E and p53, consistent with E6 decreasing the posttranslational stability of CENP-E (Fig. 3 D and E). Pulse-chase experiments with the translation inhibitor cycloheximide revealed that, as expected, E6 expression sharply increased the rate at which p53 levels decreased (Fig. 3 F and G). CENP-E levels also declined more rapidly in E6-expressing cells (Fig. 3 F and H). We conclude that E6 regulates CENP-E posttranslationally through increasing its rate of proteasomal degradation.
Fig. 3.
HPV16 E6 regulates CENP-E levels posttranslationally. (A–C) E6 expression does not downregulate CENP-E transcription. (A) Schematic of CENP-E locus showing introns (black lines) and exons (red boxes). Primer locations are indicated with half arrows. (B and C) qRT-PCR quantitation of (B) total CENP-E RNA transcripts using exon–intron primers and (C) spliced mRNA transcripts using exon–exon primers. n ≥ 3 independent experiments with technical triplicates per experiment. (D–H) E6 expression decreases CENP-E protein levels through proteasome-mediated degradation. (D) Treatment with the proteasome inhibitor MG132 for 3 h rescues protein levels of CENP-E and p53 in E6-expressing NOKs. Actin, loading control. (E) Quantitation of CENP-E protein as in D. n = 4 independent experiments. Shapes indicate values obtained from a given independent experiment. (F) Addition of the translation inhibitor cycloheximide reveals more rapid protein degradation of both p53 and CENP-E in E6 expressing NOKs as compared to vector only controls. (G and H) Quantitation of the rate of degradation of p53 (G) and CENP-E (H) in E6-expressing and control (vector alone) NOKs. n = 3 independent experiments. Numbers next to blots indicate molecular weight markers (kD). Error bars indicate SD (B and C) or SEM (E, G, and H). *P < 0.05. ns = not significant.
Restoration of CENP-E Expression Rescues E6-Induced Polar Chromosomes.
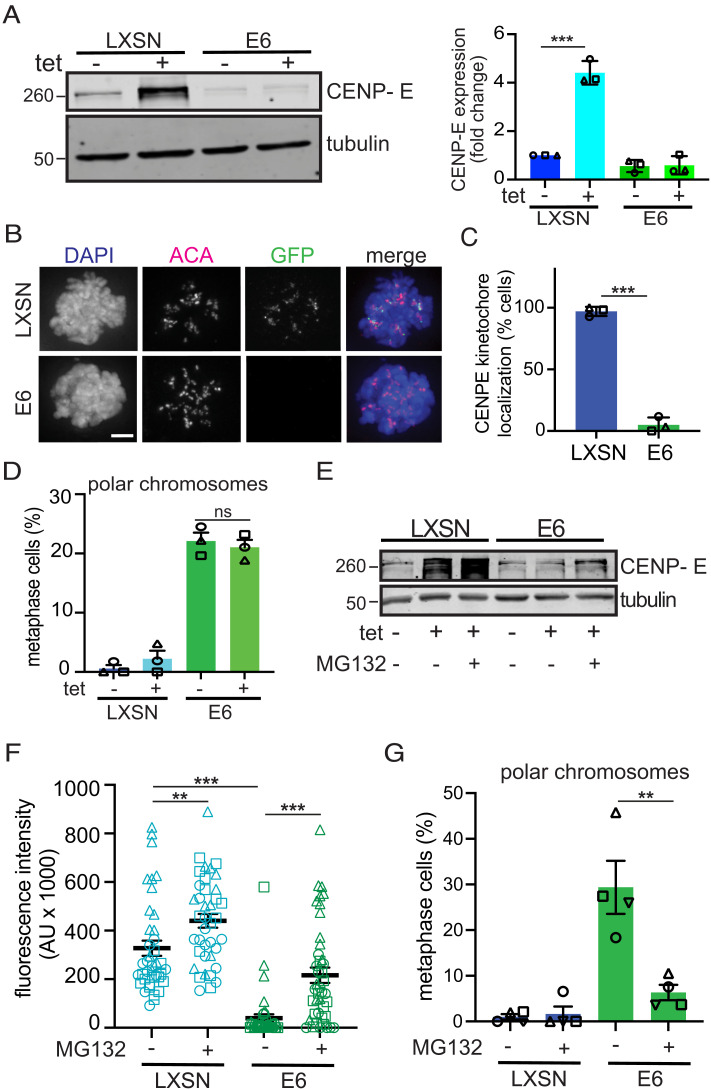
These results suggested that E6 induces CIN by increasing turnover of CENP-E. To test this directly, we restored CENP-E expression in E6 expressing NOKs to determine whether it could rescue E6-induced congression defects. Tetracycline (tet) inducible GFP-CENP-E (85) was introduced into NOKs expressing the Tet repressor using the PiggyBac transposon system. CENP-E protein expression was increased in subclones of these inducible GFP-CENP-E expressing NOKs upon tet induction (SI Appendix, Fig. S7A), which did not affect the mitotic index (SI Appendix, Fig. S7B). Time course experiments in cells transduced with either pLXSN control vector or HPV16 E6 (SI Appendix, Fig. S7C) revealed that tet induction led to a substantial increase in CENP-E expression in pLXSN control cells compared to the level of endogenous CENP-E (SI Appendix, Fig. S7D). We therefore tested whether inducible expression of GFP-CENP-E restored CENP-E levels in E6 expressing NOKs and rescued chromosome congression. While CENP-E protein levels increased approximately fourfold in pLXSN vector control expressing cells, they did not increase in E6-expressing cells (Fig. 4A and SI Appendix, Fig. S7D), despite equivalent upregulation of CENP-E and GFP mRNA after tet induction in the pLXSN and E6 expressing GFP-CENP-E NOKS (SI Appendix, Fig. S7E). This demonstrates a functional gene expression system and further supports the conclusion that E6 mediates robust CENP-E protein degradation, even when CENP-E is overexpressed. Importantly, GFP-CENP-E appropriately localized to kinetochores in vector control cells during mitosis but was almost completely absent in E6-expressing cells following tet induction (Fig. 4 B and C). While 97% of control cells expressed GFP-CENP-E at kinetochores, only 5% of E6 cells did (Fig. 4C). Thus, tet-inducible expression of GFP-CENP-E was insufficient to restore CENP-E levels at kinetochores or prevent polar chromosomes in the presence of E6 (Fig. 4D). However, proteasome inhibition with MG132 increased CENP-E protein levels to approximately the levels of pLXSN control cells (Fig. 4E) and restored levels of CENP-E at kinetochores to 66% of those in controls (Fig. 4F). This increase substantially reduced the incidence of polar chromosomes from 29% to 6% (Fig. 4G). These results support the conclusion that the highly efficient proteasome-mediated degradation of CENP-E mediated by E6 leads to polar chromosomes.
Fig. 4.
Restoration of CENP-E levels rescues polar chromosomes in E6 expressing cells. (A) CENP-E protein levels upon tetracycline induction in GFP-CENP-E NOKs expressing the control pLXSN vector or HPV16 E6 with tubulin as loading control. Immunoblot quantification is shown to the right with all samples normalized to pLXSN - tet. n = 3 independent experiments. (B) Immunofluorescence reveals appropriately localized GFP-CENP-E at kinetochores in mitotically arrested cells expressing the control vector, but not in E6-expressing cells. ACA, anti-centromere antibody. Scale bar, 5 μm. (C) Quantification of the percent of mitotic cells with GFP-CENP-E at kinetochores in pLXSN or E6-expressing cells. n ≥ 60 mitotic cells in each of three independent experiments. (D) Quantification of polar chromosomes in pLXSN and E6-expressing GFP-CENP-E NOKs with and without tet induction of CENP-E. n ≥ 50 metaphase cells from each of three independent experiments. (E) Protein levels of CENP-E in pLXSN and E6-expressing GFP-CENP-E NOKs after proteasome inhibition with MG132. Numbers next to blots indicate molecular weight markers (kD). (F) Quantitation of CENP-E fluorescence intensity at kinetochores in pLXSN and E6-expressing GFP-CENP-E NOKs +/−3 h of proteasome inhibition followed by mitotic arrest with nocodazole for 1 h. n ≥ 12 prometaphase cells from each of three independent experiments. (G) Quantification of polar chromosomes in pLXSN and E6-expressing WT NOKs following proteasome inhibition. n ≥ 50 metaphase cells from each of four independent experiments. In all panels above, shapes of data points indicate values obtained from a given independent experiment. Data indicate mean +/− SD. **P < 0.01, ***P < 0.0001, ns = not significant.
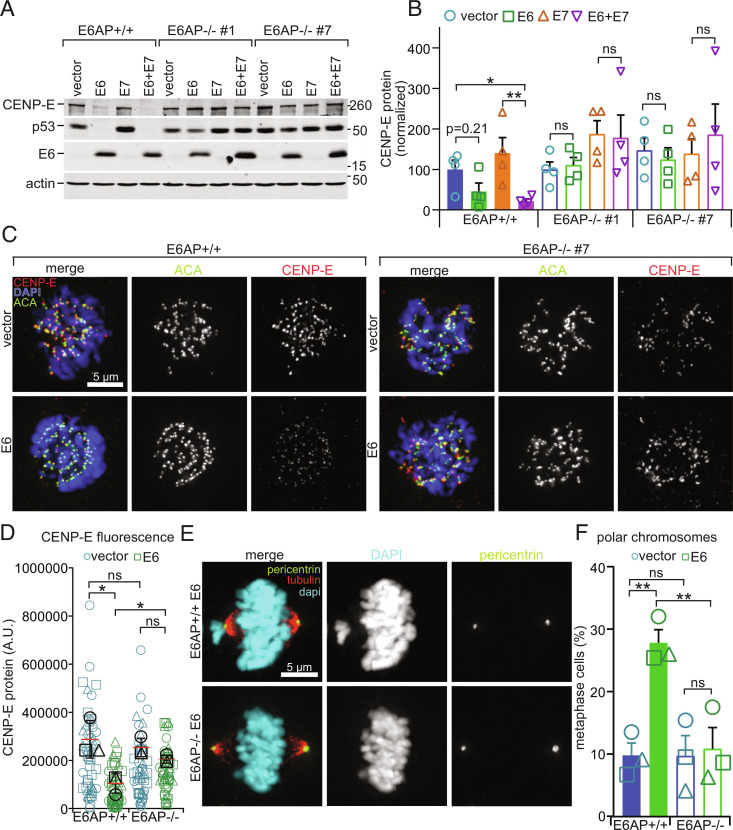
E6AP Is Necessary for E6-Mediated CENP-E Degradation and Formation of Polar Chromosomes.
E6 causes p53 degradation by driving the formation of a stable ternary complex between itself, the E3 ubiquitin ligase E6AP, and p53 (42, 86). E6AP is therefore necessary for E6 to induce proteasome-mediated p53 degradation. To determine whether E6AP is also responsible for ubiquitination and proteasomal degradation of CENP-E in the presence of E6, we used CRISPR Cas9 editing to generate E6AP null NOKs (SI Appendix, Fig. S8). Two clones lacking E6AP expression were selected for further analysis. Each clone was transduced with pLXSN control vector, E6, E7, or both and subclones with E6 expression levels equivalent to those in E6AP wild-type NOKs were evaluated (Fig. 5 A and B). In contrast to wild-type NOKs, both CENP-E and p53 levels remained stable upon expression of E6 or E6+E7 in E6AP null NOKs (Fig. 5 A and B). Consistent with this, while CENP-E kinetochore localization was substantially reduced in wild-type NOKs expressing E6, CENP-E kinetochore levels remained stable in E6AP null NOKs (Fig. 5 C and D). Importantly, E6 expression did not cause chromosome congression defects in the absence of E6AP (Fig. 5 E and F). Thus, the cellular ubiquitin ligase E6AP is necessary for E6-mediated CENP-E degradation and subsequent induction of polar chromosomes.
Fig. 5.
E6AP is necessary for E6-mediated degradation of CENP-E. (A and B) E6AP knockout restores CENP-E protein levels in E6-expressing NOKs. (A) E6 expression decreases CENP-E and p53 protein expression in wildtype but not in two different clones of E6AP null NOKs. Actin, loading control. Numbers next to blots indicate molecular weight markers (kD). (B) Quantitation of CENP-E protein levels as in A. n = 4 independent experiments. (C and D) E6AP knockout rescues kinetochore localization of CENP-E. (C) Representative images of CENP-E localization in wild-type and E6AP−/− NOKs expressing E6 or vector alone. ACA is used as a marker of kinetochores. (D) Quantitation of CENP-E fluorescence intensity in asynchronous NOKs, as in C. n ≥ 14 prometaphase cells from each of three independent experiments. Values from independent experiments are represented by different shapes. Black symbols represent average values for a given experiment. (E and F) E6AP knockout rescues the chromosome congression phenotype caused by E6 expression in NOKs. Data from E6AP−/− clone #7 are shown. (E) E6 expression in NOKs results in misaligned chromosomes at spindle poles in E6AP+/+ (Top) but not E6AP−/− (Bottom) NOKs. (F) Quantitation of polar chromosomes. n ≥ 35 metaphase cells from each of three independent experiments. Symbol shape indicates replicate. Error bars indicate SEM (B and F) or SD (D). *P < 0.05, **P < 0.01. ns = not significant.
Discussion
Here, we quantified mitotic defects in two independent cohorts of human HNC, HNC PDXs, HNC cell lines, and physiologically relevant cellular models. We demonstrate that HPV16 causes a specific type of CIN-inducing mitotic defect, which is characterized by chromosomes that fail to congress and remain misaligned at or near spindle poles. Mechanistically, we show that this is due to increased proteasomal degradation of the mitotic kinesin CENP-E. This is caused by the HPV16 E6 oncogene, is independent of HPV16 E7, p53, and the E6 PDZ binding domain, and requires the cellular E3 ubiquitin ligase E6AP.
HPV and HPV oncogenes have been reported to induce a variety of mitotic defects in anogenital cancers and associated cellular models. Images of mitotic defects in HPV+ cervical cancer typically depict what appear to be polar chromosomes, though these defects are described using variable terminology including HPV− associated atypical mitoses, ectopic chromosome around centrosome, three group metaphases, and lag-type mitosis (87–90). Though the terminology is unique, examination of the images of the abnormal mitotic figures reveals, in most cases, cells in which the majority of chromosomes are aligned at the metaphase plate and one or a few chromosomes are misaligned. The misaligned chromosomes are completely separate from the metaphase plate, in positions expected to be occupied by spindle poles based on the orientation and width of the metaphase plate, though this cannot be verified since the mitotic spindle is not visible in these H&E-stained tumor sections. This suggests that polar chromosomes may represent the dominant mechanism of HPV-induced CIN irrespective of cancer type. Indeed, we show that HPV16 infection also induces CENP-E degradation and polar chromosomes in cervical cancer cell models.
Though infrequent in models of HNC, HPV oncogenes can cause centrosome amplification in a variety of cell types, and centrosome amplification has been observed in HPV+ cervical (91) and anal (92) cancers. Both E6 and E7 could contribute to this. E6 has been proposed to cause centrosome amplification indirectly since decreased levels of p53 permit continued proliferation of tetraploid cells after cytokinesis failure, which doubles centrosome number. E7 can directly cause centriole overduplication during a prolonged S phase (53, 92–95). Over time, primary p53 null MEFs often fail cytokinesis and proliferate as tetraploid or near-tetraploid cells, suggesting that the minority of polar chromosomes in centrosome amplified NOK spindles that focused into bipolar spindles occur as a downstream consequence of p53 degradation. Though we did not assess centrosome amplification in our HNC patient samples and PDX tumors, multipolar spindles were relatively infrequent suggesting that, when present, supernumary centrosomes clustered during mitosis.
CENP-E protein is normally degraded rapidly as cells exit mitosis (75). This timing suggests that CENP-E is ubiquitinated by the Anaphase Promoting Complex/Cyclosome [APC/C; (75, 96)]. However, a screen for APC/C substrates did not identify CENP-E (97), and Skp1 of the SCF (Skp1-cullin-F box) E3 ubiquitin ligase interacts with CENP-E (98). Here, we demonstrate that the E3 ubiquitin ligase E6AP is responsible for degradation of CENP-E in the presence of HPV16 E6. We also show that E6-mediated CENP-E degradation occurs independently of the PDZ binding domain, which mediates many of E6’s protein interactions. It remains to be determined whether E6, E6AP, and CENP-E form a ternary complex and whether E6AP or another E3 ubiquitin ligase is responsible for the cell cycle–regulated degradation of CENP-E at the end of mitosis. These represent interesting questions for future studies.
Decreased levels of CENP-E have several implications for HPV-induced tumorigenesis. Low rates of CIN can promote tumor initiation by allowing cells to sample a variety of karyotypes to identify and select those that provide a proliferative advantage in their microenvironment. Heterozygous reduction of CENP-E increases the incidence of polar chromosomes, leading to aneuploidy and a low rate of CIN, which promotes the development of splenic lymphomas and lung adenomas (21, 61, 62). Aneuploidy, including gain of single, whole chromosomes caused by reduction of CENP-E, can result in genomic instability due to replication defects, DNA damage, and chromosomal rearrangements (99–101). Missegregated chromosomes or chromosome fragments that ultimately form individual micronuclei that are separated from the main nucleus can activate the proinflammatory cGAS/STING pathway and are often missegregated in subsequent divisions (102, 103). Thus, HPV16 E6-induced degradation of CENP-E may contribute to tumorigenesis by causing a low rate of CIN, thereby increasing aneuploidy, inflammation, and DNA damage.
Patients with HPV+ HNC have substantially improved overall and progression-free survival than patients with HPV− HNC following treatment with definitive chemoradiation (36, 104, 105). The mechanistic underpinning of this difference has been an area of intense investigation but remains unclear. However, it is generally accepted that HPV+ tumors are more sensitive to radiation and exhibit a delay in repairing damaged DNA when compared to HPV− HNC (35, 106, 107). Understanding the relevant differences between HPV+ and HPV− HNC is of crucial importance for improving treatment response in patients with HPV− HNC who continue to have a poor prognosis. Here, we have identified one significant difference between the mechanisms of CIN in HPV+ and HPV− HNC. We and others have found that CIN sensitizes cells to therapy that further increases CIN, such as taxane or radiation, in other cancer types (16, 17, 23–28, 108, 109). Thus, determining how distinct rates and mechanisms of CIN, such as polar chromosomes, influence radiation response is of substantial interest, and our group is actively studying how utilizing CIN may improve treatment response in HNC.
Materials and Methods
Cell Line Maintenance and Treatments.
All HNC and cervical cell line identities were confirmed via short tandem repeat testing. Cells were grown in DMEM with 4.5 g/dL glucose, 10% Fetal Bovine Serum (FBS), 1% penicillin/streptomycin with the exception of SCC-152, which was grown in EMEM, and CaSki, which was grown in RPMI 1640 with the same supplements as above. NOKs were cultured in keratinocyte serum-free medium (KSFM, Gibco) supplemented with 0.16 ng/mL epidermal growth factor, 25 µg/mL bovine pituitary extract plus 1% penicillin/streptomycin and cultured in 5% CO2 at 37 °C. Following cell detachment, trypsin (0.05% trypsin; Gibco) was inhibited with an equal volume of 2% FBS in PBS. Cells were then pelleted and resuspended in KSFM. All cell lines were routinely tested for mycoplasma.
Stable expression of pLXSN-based vector, E6, E7, or E6/E7 (transduced with a single retrovirus vector that carries both E6 and E7 genes) was achieved by transducing NOKs with Ɣ-retrovirus and 8 µg/mL polybrene. NOKs were selected with neomycin for >2 wk to achieve stable cell lines and were maintained under neomycin selection for all experiments. Following selection, subcloning was performed to select clones which had similar expression of E6. E6AP knockout cells were created in the same manner as in ref. 80 using CRISPR-Cas9 mutagenesis but the oligonucleotide guide RNAs were (5′-AAGGATAGGTGATAGCTCAC-3′ and 5′-GTGAGCTATCACCTATCCTT-3’. Tet-inducible GFP-CENP-E cells were created using the PiggyBac transposon system according to the manufacturer’s instructions. Briefly, the GFP-CENP-E gene was cloned into the PiggyBac vector backbone (System Biosciences #PB510B-1), and then cotransfected into NOKs with a transposase expression vector (System Biosciences #PB200PA-1) with a ratio of 3:1. Cells were then selected 48 h later for > 2 wk using zeocin and then subcloned.
NOKs were treated with 200 ng/mL nocodazole for 1 h. Three hours prior to protein harvest or fixation on coverslips, 10 µM MG132 was added to cells. Then, 30 µg/mL cycloheximide was used for the pulse-chase experiments, and protein was collected prior to treatment and at 1, 2, 4, 8, 12, and 24 h. CENP-E was knocked down with a SMARTpool of siRNA (four individual siRNAs targeting CENP-E) and used per the manufacturer’s instructions (Dharmacon). Cells were harvested or fixed 48 h after transfection. NOKs were treated with 0.01 nM of the CENP-E inhibitor GSK923295 for 24 h prior to fixation.
Patient Samples.
All patient samples were deidentified. Deidentified H&E-stained slides were obtained from pathology as part of the Head and Neck Cancer database at the University of Wisconsin. All patients provided informed consent to participate in the database, which is approved by the University of Wisconsin Institutional Review Board (IRB #2015-1216). Frozen head and neck tumor tissue was obtained from the University of Wisconsin Biobank and was used for Western blotting. The Biobank has an IRB-approved protocol (IRB #2016-0934) for acquiring specimens, though these samples were deidentified and are therefore not considered human subjects research. Polar chromosomes were quantified on an H&E-stained slide using formalin-fixed paraffin-embedded tissue, which was also provided from another portion of the tissue sample. All HPV+ tissue samples were p16+, and originated in the tonsil, or were taken from lymph node metastasis from a tonsillar primary cancer. All HPV− tissue samples were p16− and were from outside the oropharynx and included the larynx and oral cavity.
Immunofluorescence.
Cells were washed with microtubule stabilizing buffer (MTSB:100 mM 1,4-piperazinediethanesulfonic acid, pH 6.9, 30% glycerol, 1 mM ethylene glycol tetraacetic acid, and 1 mM MgSO4) then either fixed in MTSB plus 4% formaldehyde and 0.1% glutaraldehyde for 10 min at room temperature and permeabilized for 5 min in MTSB plus 0.05% Triton X-100 at room temperature or permeabilized in MTSB plus 0.05% Triton X-100 for 5 min at room temperature and fixed in MTSB + 4% formaldehyde for 10 min at room temperature. Coverslips were blocked for 1 h or overnight in Triton Block (0.2 M glycine, 2.5% fetal bovine serum [FBS], and 0.1% Triton X-100 in 1× phosphate buffered saline (PBS)). Primary antibody incubation (α-tubulin YL1/2, 1:1,000, Bio-Rad; pericentrin, 1:500, Abcam; CENP-E Hpx, 1:500 (110); anti-centromere antibody 1:1,000, ImmunoVision; anti-GFP (3E6) 1:500, ThermoFisher) was performed for 1 h at room temperature in Triton Block. Coverslips were washed 3× in PBS plus 0.1% Triton X-100 and incubated in Alexa Fluor–conjugated secondary antibodies diluted 1:200 in Triton Block for 45 min at room temperature. Coverslips were washed 3× in PBS plus 0.1% Triton X-100, incubated for 3 min in 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) in PBS, rinsed 1× in PBS, and mounted in Vectashield (Vector Labs, Burlingame, CA) mounting medium.
Images were acquired on a Nikon Eclipse Ti-E inverted fluorescence microscope using a CoolSNAP HQ2 or Hamamatsu Orca Flash 4.0 camera and a 100×/1.4 numerical aperture (NA) oil objective. Images are maximum projections of 0.2-μm z-stacks deconvolved using Autoquant unless otherwise indicated. Quantification was performed using Nikon Elements software by measuring the volume of signal over a uniform intensity threshold. The average intensity was then multiplied by the volume, and the resulting values were averaged and normalized to control conditions. CIN was quantified in mitotic cells as follows: Metaphase defects such as polar/misaligned chromosomes were quantified as the number of cells in metaphase with the defect of interest divided by the total number of cells in metaphase. Anaphase and telophase defects such as lagging and bridge chromosomes were quantified as the number of cells in anaphase or telophase with the defect of interest divided by the total number of cells in anaphase and telophase.
Paraffin-embedded sections of PDX tissue of 5-μm thickness were first deparaffinized in xylenes 3 × 10 min, rinsed in 100% ethanol, and hydrated in a series of 100, 95, and 70% ethanol for 5 min each, followed by 5 min in double-distilled H2O. Antigen retrieval was performed for 30 min in a beaker at 100 °C in citrate buffer (10 mM citric acid plus 0.05% Tween-20, pH 6.7). Slides were then washed in H2O and blocked overnight at 4 °C in a humidified chamber in Tris-buffered saline + 0.05% Tween 20 (TBST) plus 10% bovine serum albumin and 1% goat serum. Primary antibody incubation (α-tubulin YL1/2, 1:1,000; pericentrin, 1:200, Abcam; pHH3 S10 [6G3] Cell Signaling Technologies 1:200) was performed overnight at 4 °C in a humidified chamber in blocking buffer. Slides were washed 3 × 5 min in TBST and incubated in secondary antibodies (Alexa Fluor, 1:200 in TBST) for 1 h at room temperature. After three subsequent washes, slides were treated for 5 min with 0.05% Sudan Black B in 70% ethanol. Slides were rinsed with H2O and then incubated in 5 μg/mL DAPI in PBS for 10 min, washed 2× in PBS, and mounted using Vectashield (Vector Labs).
Images were acquired using a Nikon Eclipse Ti-E inverted microscope with a Hamamatsu ORCA Flash 4.0 camera using 40× (0.75 NA), 60× (1.4 NA), or 100× (1.4 NA) objectives. Image acquisition, analysis, and processing were performed using Nikon Elements AR. Autoquant was used for deconvolution.
Immunoblotting.
Cells from ~70% confluent 6-well plates were washed with PBS, harvested in 100 µL of PBS and 100 μL of 2× sample buffer, boiled for 10 min, and stored at −80 °C. Samples were sonicated prior to loading. Tumor tissue samples were homogenized in NP-40 lysis buffer + PMSF and protease inhibitors using dounce homogenizers. Samples were then centrifuged, and a DC protein assay (BioRad) was performed on the supernatant. Samples were run on 10% (CENP-E, actin, and Mad1) or 15% (p53, E6, E6AP, Bub1, BubR1, and Mad2) acrylamide gels, transferred to nitrocellulose overnight, and blocked in 5% milk in TBST for 1 h at room temperature before incubation with primary antibodies, which were diluted in 2% bovine serum albumin + 0.02% sodium azide in PBS. Secondary antibodies were diluted in 5% milk in TBS + 0.1% Tween 20. Primary antibody dilutions: CENP-E 1:500 (110), p53 1:500 (DO-1; Santa Cruz Biotechnology), actin 1:500 (JLA20; Developmental Studies Hybridoma Bank), E6 1:500 (GeneTex), E6AP 1:1,000 (7526; Cell Signaling Technologies), Mad1 1:1,000 (111), Mad2 1:2,000 (112), Bub1 1:500 (113), and BubR1 1:300 (113). Secondary antibodies were diluted at 1:10,000 (Licor IRDye).
qRT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA was reverse transcribed (iScript, Bio-Rad Laboratories), and the resulting cDNA was used for real-time PCR analysis. Intron–exon primer sets were used to detect total RNA and exon–exon primer sets were used to detect spliced mRNA (84). Reactions were performed in IQ SYBR Green Supermix (Bio-Rad). Thermocycling was performed using an annealing temperature of 60 °C. Relative RNA levels were calculated using the ΔΔCt method. Each sample was run in triplicate, and cycle threshold (Ct) values were averaged. GAPDH was used as a housekeeping gene for qRT-PCR reactions. A minimum of three biological replicates were analyzed. Primer sequences can be found in SI Appendix, Table S1.
TCGA Analysis.
First, 1,352 TCGA squamous cell carcinoma samples (73) from lung (n = 479), head and neck (n = 502), bladder (n = 43), esophagus (n = 82), and cervix (n = 246) were analyzed for this study. HPV status for each tumor is publicly available (73) as well as aneuploidy information (74). Then, to calculate fraction of aneuploidy due to whole chromosome changes, we divided the number of chromosome arms affected by whole chromosome events by the total number of chromosome arms affected by aneuploidy (aneuploidy score, 74). We excluded acrocentric chromosomes for this analysis, as for these chromosomes we only have copy number information for one chromosome arm.
Statistical Analysis.
Significant differences were determined using a two-tailed Student’s t test (mitotic defects, quantitative immunofluorescence, mitotic index, qRT-PCR, and protein quantitation). Slopes of protein decay after cycloheximide treatment were determined using linear regression analysis.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Dr. Andrew Holland and Dr. Karl Munger for CENP-E and HPV cDNA constructs, respectively, and members of the Weaver, Burkard, Lambert, and Suzuki laboratories for insightful discussions. Portions of this work were developed as part of the doctoral dissertation of co-first author Laura C. Funk, who has since changed her name to L.C.F.H. We thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory, supported by the UW Department of Pathology and Laboratory Medicine and NIH S10 OD023526 for use of its facilities and services. This project was supported in part by NIH P50DE026787 (DRP to B.A.W.) and P50CA278595 (pathology core), R01CA234904 (B.A.W.), T32CA009135 (L.F.H.), K08CA256166 (P.F.C.), P01CA022443 (D.L.L. and P.F.L.), R35CA210807 (D.L.L. and P.F.L.), the University of Wisconsin Carbone Cancer Center Support Grant P30CA014520, which supports the University of Wisconsin Carbone Cancer Center BioBank, the Radiological Society of North America (Research Fellow Grant RF1904 to P.F.C.), and the American Society of Clinical Oncology (Young Investigator Award to P.F.C.).
Author contributions
P.F.C., L.C.F.H., and B.A.W. designed research; P.F.C., L.C.F.H., J.W., K.J., S.A.B., K.N., S.M., and A.M.T. performed research; D.L.L., A.M.T., R.J.K., and P.F.L. contributed new reagents/analytic tools; P.F.C., L.C.F.H., and M.P. analyzed data; and P.F.C., L.C.F.H., and B.A.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.von Hansemann D., Ueber asymmetrische zelltheilung in Epithelkrebsen und deren biologische bedeutung. Virchow’s Arch. Path. Anat. 119, 299–326 (1890). [Google Scholar]
- 2.Boveri T., Ueber mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Vehr. d. phys. med. Ges. zu Wurzburg, N.F. (available in English translation at: http://8e.devbio.com/article.php?ch=4&id=24) Bd. 35 (1902). [Google Scholar]
- 3.Boveri T., The Origin of Malignant Tumors by Theodor Boveri Baltimore, Translated by Marcella Boveri (Williams and Wilkins, 1929, 1914). [Google Scholar]
- 4.Funk L. C., Zasadil L. M., Weaver B. A., Living in CIN: Mitotic infidelity and its consequences for tumor promotion and suppression. Dev. Cell 39, 638–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zasadil L. M., Britigan E. M., Weaver B. A., 2n or not 2n: Aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Semin Cell Dev. Biol. 24, 370–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon J. E., Bakker B., Foijer F., CINcere modelling: What have mouse models for chromosome instability taught us? Recent Results Cancer Res. 200, 39–60 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Ricke R. M., van Ree J. H., van Deursen J. M., Whole chromosome instability and cancer: A complex relationship. Trends Genet. 24, 457–466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosper P. F., Copeland S. E., Tucker J. B., Weaver B. A., Chromosome missegregation as a modulator of radiation sensitivity. Semin. Radiat. Oncol. 32, 54–63 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel L. S., et al. , MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409, 355–359 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga Y., et al. , Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 67, 160–166 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Baker D. J., van Deursen J. M., Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle 9, 1711–1716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker D. J., Jin F., Jeganathan K. B., van Deursen J. M., Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16, 475–486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schvartzman J. M., Duijf P. H., Sotillo R., Coker C., Benezra R., Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 19, 701–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kops G. J., Foltz D. R., Cleveland D. W., Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. U.S.A. 101, 8699–8704 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel L., et al. , Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 101, 4459–4464 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zasadil L. M., et al. , High rates of chromosome missegregation suppress tumor progression, but do not inhibit tumor initiation. Mol. Biol. Cell 27, 1981–1989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silk A. D., et al. , Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. U.S.A. 110, E4134–4141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowald K., et al. , Negative selection and chromosome instability induced by Mad2 overexpression delay breast cancer but facilitate oncogene-independent outgrowth. Cell Rep. 15, 2679–2691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laucius C. D., Orr B., Compton D. A., Chromosomal instability suppresses the growth of K-Ras-induced lung adenomas. Cell Cycle 18, 1702–1713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godek K. M., et al. , Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 6, 532–545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver B. A., Silk A. D., Montagna C., Verdier-Pinard P., Cleveland D. W., Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Funk L. C., et al. , p53 Is not required for high CIN to induce tumor suppression. Mol. Cancer Res. 19, 112–123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zasadil L. M., et al. , Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 6, 229ra243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maia A. R. R., et al. , Mps1 inhibitors synergise with low doses of taxanes in promoting tumour cell death by enhancement of errors in cell division. Br J. Cancer 118, 1586–1595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maia A. R., et al. , Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann. Oncol. 26, 2180–2192 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Janssen A., Kops G. J., Medema R. H., Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. U.S.A. 106, 19108–19113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schukken K. M., et al. , Altering microtubule dynamics is synergistically toxic with spindle assembly checkpoint inhibition. Life Sci. Alliance 3, e201900499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scribano C. M., et al. , Chromosomal instability sensitizes patient breast tumors to multipolar divisions induced by paclitaxel. Sci. Transl. Med. 13, eabd4811 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walboomers J. M., et al. , Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Alemany L., et al. , Human papillomavirus DNA prevalence and type distribution in anal carcinomas worldwide. Int. J. Cancer 136, 98–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miralles-Guri C., et al. , Human papillomavirus prevalence and type distribution in penile carcinoma. J. Clin. Pathol. 62, 870–878 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Roden R. B. S., Stern P. L., Opportunities and challenges for human papillomavirus vaccination in cancer. Nat. Rev. Cancer 18, 240–254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein A. P., et al. , Prevalence of human papillomavirus in oropharyngeal cancer: A systematic review. Cancer J. 21, 138–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturvedi A. K., et al. , Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 29, 4294–4301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimple R. J., et al. , Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 73, 4791–4800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ang K. K., et al. , Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 363, 24–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. z. Hausen, Papillomaviruses in the causation of human cancers - a brief historical account. Virology 384, 260–265 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Moody C. A., Laimins L. A., Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 10, 550–560 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Cosper P. F., Bradley S., Luo L., Kimple R. J., Biology of HPV mediated carcinogenesis and tumor progression. Semin. Radiat. Oncol. 31, 265–273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huibregtse J. M., Scheffner M., Howley P. M., A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. Embo J. 10, 4129–4135 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huibregtse J. M., Scheffner M., Howley P. M., Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell Biol. 13, 4918–4927 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M., The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Chellappan S., et al. , Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. U.S.A. 89, 4549–4553 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagano M., Durst M., Joswig S., Draetta G., Jansen-Durr P., Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene 7, 1681–1686 (1992). [PubMed] [Google Scholar]
- 45.Chellappan S. P., Hiebert S., Mudryj M., Horowitz J. M., Nevins J. R., The E2F transcription factor is a cellular target for the RB protein. Cell 65, 1053–1061 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Duensing S., Munger K., Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J. Virol. 77, 12331–12335 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duensing S., et al. , Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J. Virol. 75, 7712–7716 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen C. L., Eichwald C., Nibert M. L., Munger K., Human papillomavirus type 16 E7 oncoprotein associates with the centrosomal component gamma-tubulin. J. Virol. 81, 13533–13543 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace N. A., Robinson K., Galloway D. A., Beta human papillomavirus E6 expression inhibits stabilization of p53 and increases tolerance of genomic instability. J. Virol. 88, 6112–6127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marsh E. K., et al. , Mitotic control of human papillomavirus genome-containing cells is regulated by the function of the PDZ-binding motif of the E6 oncoprotein. Oncotarget 8, 19491–19506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plug-Demaggio A. W., McDougall J. K., The human papillomavirus type 16 E6 oncogene induces premature mitotic chromosome segregation. Oncogene 21, 7507–7513 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Duensing S., Munger K., The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 62, 7075–7082 (2002). [PubMed] [Google Scholar]
- 53.Duensing S., et al. , The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. U.S.A. 97, 10002–10007 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganem N. J., Godinho S. A., Pellman D., A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silkworth W. T., Nardi I. K., Scholl L. M., Cimini D., Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One 4, e6564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabet A. S., et al. , Impairment of the telomere/telomerase system and genomic instability are associated with keratinocyte immortalization induced by the skin human papillomavirus type 38. FASEB J. 22, 622–632 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Plug-DeMaggio A. W., et al. , Telomere erosion and chromosomal instability in cells expressing the HPV oncogene 16E6. Oncogene 23, 3561–3571 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Shirnekhi H. K., Kelley E. P., DeLuca J. G., Herman J. A., Spindle assembly checkpoint signaling and sister chromatid cohesion are disrupted by HPV E6-mediated transformation. Mol. Biol. Cell 28, 2035–2041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monda J. K., Cheeseman I. M., The kinetochore-microtubule interface at a glance. J. Cell Sci. 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maiato H., Gomes A. M., Sousa F., Barisic M., Mechanisms of chromosome congression during mitosis. Biology (Basel) 6, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Putkey F. R., et al. , Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell 3, 351–365 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Weaver B. A., et al. , Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551–563 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim Y., Heuser J. E., Waterman C. M., Cleveland D. W., CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J. Cell Biol. 181, 411–419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood K. W., et al. , Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. U.S.A. 107, 5839–5844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao G., et al. , A novel RT-PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int. J. Cancer 132, 882–890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimple R. J., et al. , Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clin. Cancer Res. 19, 855–864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stein A. P., et al. , Influence of handling conditions on the establishment and propagation of head and neck cancer patient derived xenografts. PLoS One 9, e100995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swick A. D., et al. , Defining the boundaries and expanding the utility of head and neck cancer patient derived xenografts. Oral. Oncol. 64, 65–72 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piboonniyom S. O., et al. , Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 63, 476–483 (2003). [PubMed] [Google Scholar]
- 70.Zhang C. Z., et al. , Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maciejowski J., Li Y., Bosco N., Campbell P. J., de Lange T., Chromothripsis and kataegis induced by telomere crisis. Cell 163, 1641–1654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tucker J. B., et al. , Misaligned chromosomes are a major source of chromosomal instability in breast cancer. Cancer Res. Commun. 3, 54–65 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell J. D., et al. , Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep 23, 194–212.e196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor A. M., et al. , Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown K. D., Coulson R. M., Yen T. J., Cleveland D. W., Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J. Cell Biol. 125, 1303–1312 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Legal T., et al. , The C-terminal helix of BubR1 is essential for CENP-E-dependent chromosome alignment. J. Cell Sci. 133, jcs246025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson V. L., Scott M. I., Holt S. V., Hussein D., Taylor S. S., Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117, 1577–1589 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A., The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 (2006). [DOI] [PubMed] [Google Scholar]
- 79.DeLuca J. G., et al. , Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Kraus R. J., et al. , Reactivation of epstein-barr virus by HIF-1α requires p53. J. Virol. 94, e00722-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ganti K., et al. , The human papillomavirus E6 PDZ binding motif: From life cycle to malignancy. Viruses 7, 3530–3551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen M. L., Nguyen M. M., Lee D., Griep A. E., Lambert P. F., The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6’s induction of epithelial hyperplasia in vivo. J. Virol. 77, 6957–6964 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simonson S. J., Difilippantonio M. J., Lambert P. F., Two distinct activities contribute to human papillomavirus 16 E6’s oncogenic potential. Cancer Res. 65, 8266–8273 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Weng M. T., et al. , Evolutionarily conserved protein ERH controls CENP-E mRNA splicing and is required for the survival of KRAS mutant cancer cells. Proc. Natl. Acad. Sci. U.S.A. 109, E3659–3667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim Y., Holland A. J., Lan W., Cleveland D. W., Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142, 444–455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Masuda Y., et al. , Stepwise multipolyubiquitination of p53 by the E6AP-E6 ubiquitin ligase complex. J. Biol. Chem. 294, 14860–14875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burger M. P., van Leeuwen A. M., Hollema H., Quint W. G., Pieters W. J., Human papillomavirus type influences the extent of chromosomal lag during mitosis in cervical intraepithelial neoplasia grade III. Int. J. Gynecol. Pathol. 16, 10–14 (1997). [DOI] [PubMed] [Google Scholar]
- 88.Scurry J., Baskota S. U., HPV-associated atypical mitotic figures in squamous intraepithelial lesions of the lower female genital tract. J. Low. Genit. Tract Dis. 20, 165–168 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Claas E. C., et al. , Human papillomavirus and the three group metaphase figure as markers of an increased risk for the development of cervical carcinoma. Am. J. Pathol. 140, 497–502 (1992). [PMC free article] [PubMed] [Google Scholar]
- 90.Furuta R., et al. , Ectopic chromosome around centrosome in metaphase cells as a marker of high-risk human papillomavirus-associated cervical intraepithelial neoplasias. Int. J. Cancer 106, 167–171 (2003). [DOI] [PubMed] [Google Scholar]
- 91.Skyldberg B., et al. , Human papillomavirus infection, centrosome aberration, and genetic stability in cervical lesions. Mod. Pathol. 14, 279–284 (2001). [DOI] [PubMed] [Google Scholar]
- 92.Duensing A., Chin A., Wang L., Kuan S. F., Duensing S., Analysis of centrosome overduplication in correlation to cell division errors in high-risk human papillomavirus (HPV)-associated anal neoplasms. Virology 372, 157–164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korzeniewski N., Treat B., Duensing S., The HPV-16 E7 oncoprotein induces centriole multiplication through deregulation of polo-like kinase 4 expression. Mol. Cancer 10, 61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guarguaglini G., et al. , The forkhead-associated domain protein Cep170 interacts with polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell 16, 1095–1107 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duensing A., et al. , Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene 26, 6280–6288 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holland A. J., et al. , Preventing farnesylation of the dynein adaptor spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol. Biol. Cell 26, 1845–1856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh S. A., et al. , Co-regulation proteomics reveals substrates and mechanisms of APC/C-dependent degradation. EMBO J. 33, 385–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu D., et al. , Interaction of Skp1 with CENP-E at the midbody is essential for cytokinesis. Biochem. Biophys. Res. Commun. 345, 394–402 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Passerini V., et al. , The presence of extra chromosomes leads to genomic instability. Nat. Commun. 7, 10754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamm N., et al. , Genomic instability in human pluripotent stem cells arises from replicative stress and chromosome condensation defects. Cell Stem Cell 18, 253–261 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Santaguida S., et al. , Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651.e635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mackenzie K. J., et al. , cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He B., et al. , Chromosomes missegregated into micronuclei contribute to chromosomal instability by missegregating at the next division. Oncotarget 10, 2660–2674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fakhry C., et al. , Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J. Natl. Cancer Inst. 100, 261–269 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Ko H. C., et al. , Prognostic implications of human papillomavirus status for patients with non-oropharyngeal head and neck squamous cell carcinomas. J Cancer Res. Clin Oncol. 143, 2341–2350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park J. W., et al. , Human papillomavirus type 16 E7 oncoprotein causes a delay in repair of DNA damage. Radiother Oncol. 113, 337–344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rieckmann T., et al. , HNSCC cell lines positive for HPV and p16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiother Oncol. 107, 242–246 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Bakhoum S. F., et al. , Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat. Commun. 6, 5990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bakhoum S. F., Kabeche L., Murnane J. P., Zaki B. I., Compton D. A., DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 4, 1281–1289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brown K. D., Wood K. W., Cleveland D. W., The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J. Cell Sci. 109, 961–969 (1996). [DOI] [PubMed] [Google Scholar]
- 111.Ryan S. D., et al. , Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc. Natl. Acad. Sci. U.S.A. 109, E2205–2214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kops G. J., et al. , ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169, 49–60 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor S. S., Hussein D., Wang Y., Elderkin S., Morrow C. J., Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114, 4385–4395 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.