Significance
Life is a product of many genes that interact to enhance fitness. Genetics has made much progress in mapping quantitative trait loci (QTLs) that affect complex traits while the probing of epistasis is still rudimentary. To characterize epistasis—the nonlinear interaction of genes—we developed interspecific backcross inbred lines of unparalleled size and mapping resolution. Of the 80 cases of QTL epistasis, we selected validation of QTLs on chromosomes 1 and 7 that independently had no effect on yield, but surprisingly, together they increased yield by 20 to 50% over a period of 4 y in irrigated and dry fields. Our work demonstrates the power of a very large interspecific population to highlight rare epistatic QTLs that improve productivity via heterosis.
Keywords: epistasis, heterosis, breeding
Abstract
Controlled population development and genome-wide association studies have proven powerful in uncovering genes and alleles underlying complex traits. An underexplored dimension of such studies is the phenotypic contribution of nonadditive interactions between quantitative trait loci (QTLs). Capturing of such epistasis in a genome-wide manner requires very large populations to represent replicated combinations of loci whose interactions determine phenotypic outcomes. Here, we dissect epistasis using a densely genotyped population of 1,400 backcross inbred lines (BILs) between a modern processing tomato inbred (Solanum lycopersicum) and the Lost Accession (LA5240) of a distant, green-fruited, drought-tolerant wild species, Solanum pennellii. The homozygous BILs, each harboring an average of 11 introgressions and their hybrids with the recurrent parents, were phenotyped for tomato yield components. Population-wide mean yield of the BILs was less than 50% of that of their hybrids (BILHs). All the homozygous introgressions across the genome reduced yield relative to recurrent parent, while several QTLs of the BILHs independently improved productivity. Analysis of two QTL scans showed 61 cases of less-than-additive interactions and 19 cases of more-than-additive interactions. Strikingly, a single epistatic interaction involving S. pennellii QTLs on chromosomes 1 and 7, that independently did not affect yield, increased fruit yield by 20 to 50% in the double introgression hybrid grown in irrigated and dry fields over a period of 4 y. Our work demonstrates the power of large, interspecific controlled population development to uncover hidden QTL phenotypes and how rare epistatic interactions can improve crop productivity via heterosis.
In mapping quantitative trait loci (QTLs), we compare the mean values for a trait in the different genotypic groups in segregating populations or genome-wide association studies (GWAS) “one marker at a time” to generate plots of the effects on a trait and its significance along the linkage groups. This approach made marker-assisted selection a prerequisite for the success of breeding programs for resistances to diseases and for improved crop quality. Currently, we can score thousands of single nucleotide polymorphisms (SNPs) but still we analyze them mostly “one at a time” along the genome. A deeper view of the analysis of complex traits is to conduct genome-wide scans for epistatic QTLs that have small or no effect on a trait but a surprising outcome when combined with other QTL positions. According to Weinreich et al. (1), epistasis can be regarded as our surprise at the phenotype when QTLs are combined, given the constituent QTL individual effects.
Crop plants are often used to study complex traits since large populations of sessile individuals can be assayed in relatively uniform agriculture environments. Tomato (Solanum lycopersicum; 2n = 2x = 24) is one of the models for studying the genetic basis of yield associated traits including the deployment of wild species variation for the breeding of cutting-edge commercial hybrids (2). One of the populations used for tomato genetics and breeding has been the Solanum pennellii (LA716) introgression lines (ILs) that provide a complete coverage of the genome in a set of 76 lines each containing a single genomic segment from the wild species (3). The ILs are nearly isogenic to an inbred processing tomato variety, and this greatly reduced the variation associated with QTL–QTL interactions compared to populations that segregate for the entire genome. Soon after the development of the S. pennellii ILs, we tested for epistasis of yield-associated traits by crossing between ten different homozygous ILs and evaluating in the field the 45 resulting hybrids (4). For the fruit weight and fruit sugar content (% Brix), the epistasis was predominantly of the less-than-additive mode, i.e., the phenotypic value of the double heterozygotes was lower than the sum of the effects of the single heterozygotes. For total fruit yield however, five of the cases of IL epistasis followed the mode of more than additive, suggesting that it is possible to identify epistatic QTLs that improve productivity beyond the sum of the effects of the individual QTL.
There are two main factors, which limit our ability to detect significant epistasis in segregating populations (5): 1) Usually such populations number 100–200 individuals, which is much too low to identify significant QTL–QTL interactions since the number of individuals that carry specific combinations of two (or more) genomic regions is low. 2) A second limitation results from the “multiple testing penalty” of all pairs of genomic regions (in the case of digenic interactions), which causes a strong downward adjustment of the statistical significance threshold. To overcome these limitations, we embarked in 2007 on the development of a very large backcross inbred line (BIL) population where each BC2S6 line can be traced to a different BC1 plant. The wild donor for the BILs was an unexplored accession of S. pennellii that was rediscovered in the Gatersleben collection (Lost Accession, LA5240) (6). This accession is completely self-compatible (a rare trait in this species) and importantly does not carry the necrotic dwarf trait that is characteristic of S. pennellii LA716 and which often interferes with the phenotypic evaluations (3). The Lost Accession was introgressed into a parental line suited for the processing tomato industry (determinate inbred, LEA). Here, we describe the genotype and the yield components of 1,400 homozygous BILs and their hybrids back to LEA (BILHs). We identified and validated two epistatic QTLs that by themselves have no effect on yield but whose hybrids showed 20–50% heterosis in a consistent manner over a period of 4 y.
Results
Genomic Composition of the Biparental BILs.
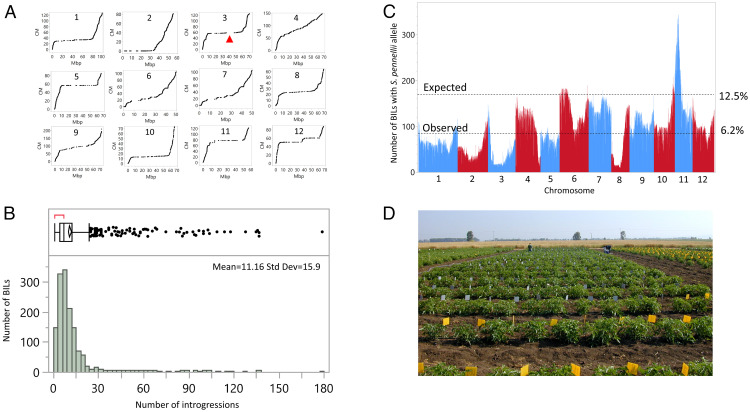
In 2018, we genotyped the BC2S6 BILs using the single primer enrichment technology (SPET) (7). Of the 173,000 SNPs that were scored, we used, after filtering, 7,699 SNPs. We calculated the recombination frequencies between the markers and plotted the position of each marker on the genetic (centi-Morgan) and physical maps (Mbp) of the 12 tomato chromosomes (Fig. 1A). The chromosome plots show a classic distribution that can be seen in many organisms in which the euchromatic regions show high recombination rates for a physical distance (an average of one recombinant every 18 Kb), whereas in the heterochromatic region, there are few recombination events (one recombinant every 700 Kb). It is noteworthy that along the different chromosomes in the heterochromatic regions, there are multiple gaps that represent deletions in the S. pennellii genome. The markers that map the gap regions all showed homozygosity for the LEA alleles. Similarly, the interspecific F1 hybrid also shows only the LEA alleles in the gap regions, the largest of which is on chromosome 3 (11 Mbp; Fig. 1A). Since the SPET marker design was based on SNPs of the red-fruited species, we assume that had we used S. pennellii–based SPET design instead, we would have found many more S. lycopersicum deletions relative to the wild species since the genome size of S. pennellii is between 1 and 1.2 Gb compared to 0.9 Gb for the cultivated tomato (6). The genotyping analysis of the BILs indicated that an average line carried 11.2 wild species introgressions with a few lines that harbor more than 100 S. pennellii genome segments (Fig. 1B).
Fig. 1.
Genomic composition of the biparental BILs. (A) Plots of the physical (Mbp) and genetic (cM) distances of the 7,699 SPET markers of the 12 tomato chromosomes. The red arrowhead points to the largest gap detected in chromosome 3 (11 Mpb). (B) Frequency distribution of number of wild species introgressions per BIL. (C) The prevalence of the S. pennellii alleles in BC2S6 relative to the expected value of 12.5%. (D) A picture of the experimental plots in Akko showing the replicated single young plants each covering an area of 1 m2.
The BIL population was designed to allow whole-genome two-dimensional QTL scans. The original F1 interspecific hybrid carried the entire genome of the wild species and of the cultivated tomato LEA. In the BC1 and BC2, the average proportion of the wild species genome was reduced by half each generation such that BC2 carried 25% of the wild genome in a heterozygous state. Upon each of the six selfing generations, heterozygosity was expected to be reduced by 50%, and thus, in the sixth generation of selfing, we would expect every introgression to be present in a homozygous state in 12.5% of the BILs. Consequently, of the 1,400 BILs, we would expect 21 BILs to carry two random independent introgressions, which is more than necessary for a comparison with the plants that carry a single introgression. However, due to segregation distortion against the wild species alleles, which are common in interspecific crosses (8), we observed that in most of the chromosomes, the average prevalence of the S. pennellii introgressions was 6% (Fig. 1C).
Identification of Single and Epistatic QTLs.
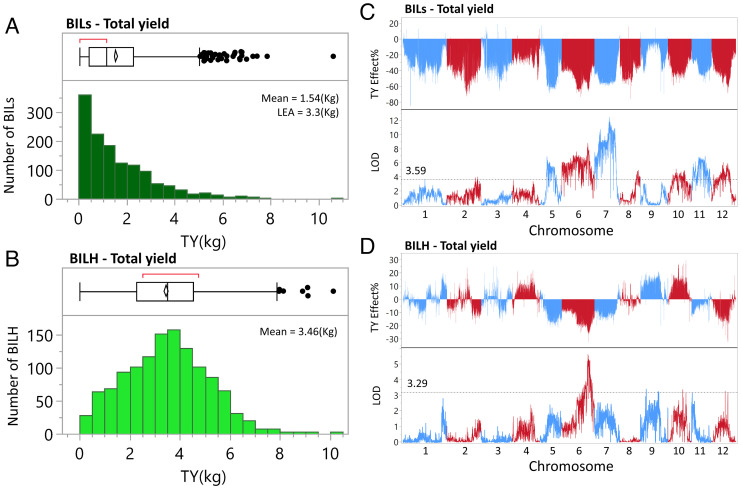
Plants of the BILs and the BILHs were transplanted in heavy soil in field capacity and irrigated with ~20% of the amount of water usually used in tomato cultivation (Fig. 1D). Each plant at harvest time was phenotyped for the following traits: plant weight (PW) (only the vegetative part), average fruit weight, Brix (%, total soluble solids, mainly sugars), and total yield (TY). As we expected based on previous studies of tomato interspecific crosses, the homozygous BILs produced 50% of the yield relative to the BILHs and LEA (Fig. 2 A and B). Since our objective in this research was to improve the productivity of the processing tomato, we focused on breeding F1 hybrids that carry the wild species introgressions in a heterozygous state. This observation is also supported by the single QTL analysis that showed that all the homozygous BIL introgressions reduced yield relative to the common control LEA by as much as 50%, while in the BILHs, we detected introgressions that significantly increased yield (Fig. 2 C and D). The frequency distributions of the other yield-associated traits were close to normal both in the BILs and in the BILHs. The mean fruit weight of the BILHs was more than double that of the BILs; the single QTL that affected the measured traits and their significance are indicated (SI Appendix, Figs. S1 and S2). The S. pennellii introgression on chromosome 4, that did not suffer from negative linkage drag, increased yield by 20% relative to the BILH without the introgression (LOD 2.36). This introgression showed in a validation study a 20% yield increase in the LEA background as well as in some additional genetic backgrounds (SI Appendix, Fig. S3 A–C). This analysis validated the reproducibility of the QTLs detected in 2018 and gave us the confidence to explore epistasis using the BILs.
Fig. 2.
Identification of single yield QTLs in the BILs and BILHs. (A) Frequency distribution of total yield (TY, kg) in the 1,389 homozygote BILs. (B) Frequency distribution of TY (kg) in the 1,233 heterozygous BILHs. (C) Single marker analysis of TY (kg) in the BILs relative to the TY of LEA. LOD scores for each of the marker effects were calculated by the Haley–Knott regression, and the LOD threshold was determined by 1,000 permutation tests. (D) Single marker analysis of TY (kg) in the BILHs relative to the TY of LEA. LOD scores for each of the marker effects were calculated by the Haley–Knott regression, and the LOD threshold was determined by 1,000 permutation tests.
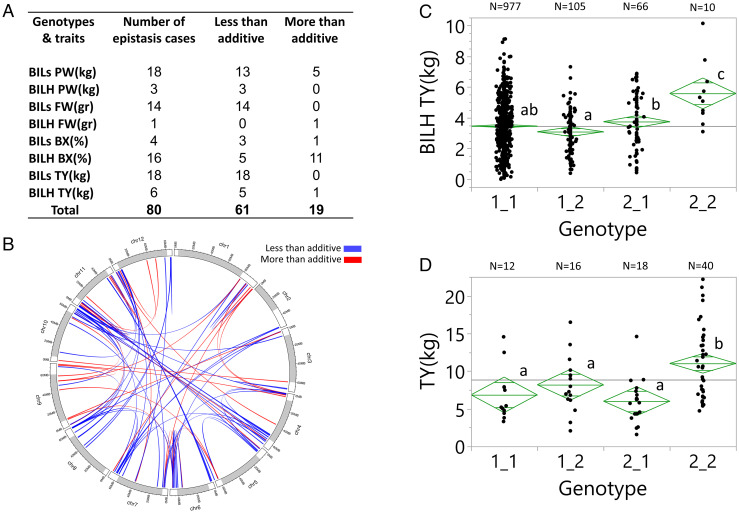
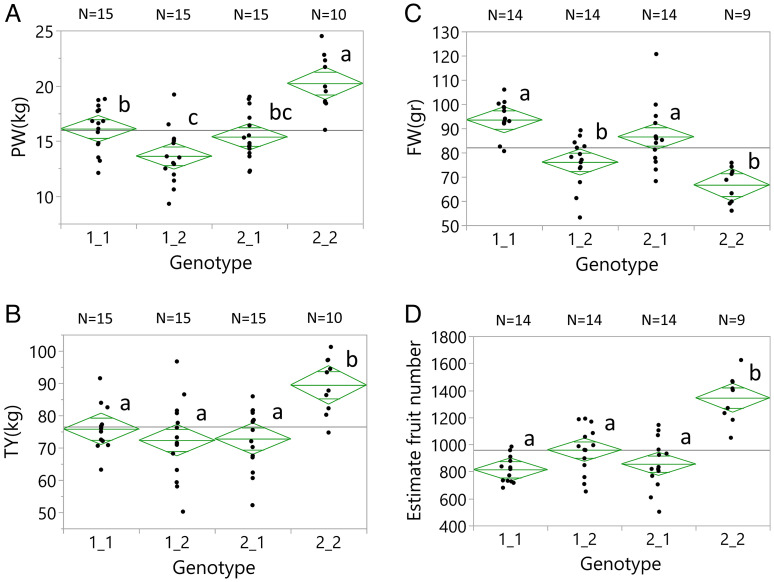
Our ability to identify epistasis between two QTLs depends on the number of plants that carry the pair of introgressions whose phenotype is compared to that of single QTL hybrids. Due to the deviations from the Mendelian segregations (Fig. 1C), we included in the epistasis analysis only digenic scans with at least ten plants that carried both QTL alleles from the wild species. A total of 80 epistasis cases were found in the BILs and BILHs, with 61 being less than additive and 19 more than additive (Fig. 3A and SI Appendix, Table S1). In the BILs, we detected the majority of the less-than-additive interactions (total 48). For example, for fruit weight and TY, all the homozygous QTLs reduced the phenotype relative to the lines that did not carry the introgressions. Based on additivity, we would expect the phenotype of the BILs that carries both introgressions to be the sum of two QTL effects. However, in all cases, the effect measured on yield and fruit weight in the double introgression BILs was less than the sum of the two QTLs. It is important to note that homozygous alleles in the BILs and heterozygous alleles in the BILHs can show epistasis but not necessarily through heterosis. The cases of digenic epistasis were also diagrammed on the circus physical map showing that an overwhelming majority of loci involved mapped the gene-rich regions (Fig. 3B). Strikingly, a single epistatic interaction in the BILHs involving introgressions on chromosomes 1 and 7 of S. pennellii, that independently had no effect on yield, when put together the mean of the ten double introgression hybrids increased TY by 58% (Fig. 3C). To validate this surprising result, we planted selfed seed from the double introgression hybrid in the field and ran the markers for chromosome 1 (CHR-1, SSL2.50CH01_95261222) and chromosome 7 (CHR-7, SSL2.50CH07_65737800) to select plants from the following four genotypic groups: 1) homozygous for the cultivated tomato alleles in chromosomes 1 and 7 (1_1), 2) heterozygous for the chromosome 1 introgression (2_1), 3) heterozygous for the chromosome 7 introgression (1_2), and 4) plants that were heterozygous for both introgressions (2_2). All the selected seedlings were planted randomly in an irrigated field where the double heterozygous group (the largest group obtained from the F2) had 37% higher yield than the mean of the three other genotypic groups (Fig. 3D).
Fig. 3.
Identification and validation of epistatic QTLs. (A) Two-dimensional QTL scan of the BILs and BILHs for plant weight (PW), fruit weight (FW), Brix (BX), and total yield (TY) in dry conditions (Akko 2018). We used LOD fv1>316 to indicate significant epistasis. (B) Whole-genome circus epistasis plot. White segments on each of the chromosomes signify the euchromatic regions, and gray segments signify the heterochromatin. (C) Yield epistasis (Akko 2018) detected for markers on chromosomes 1 (SSL2.50CH01_95261222) and 7 (SSL2.50CH07_65737800) showing the number (N) of plants in each of the genotypic groups: 1) homozygous for the cultivated tomato alleles in chromosomes 1 and 7 (1_1), 2) heterozygous for the chromosome 1 introgression (2_1), 3) heterozygous for the chromosome 7 introgression (1_2), and 4) heterozygous for both introgressions (2_2). Genotypic group means showing the same letters are not significantly different at the 5% level based on the Tukey–Kramer test. (D) A validation test of the chromosomes 1 and 7 epistasis in F2 progenies of the double heterozygous BILHs (Akko 2020). Markers, genotypic groups, and statistical tests are the same as in Fig. 3C.
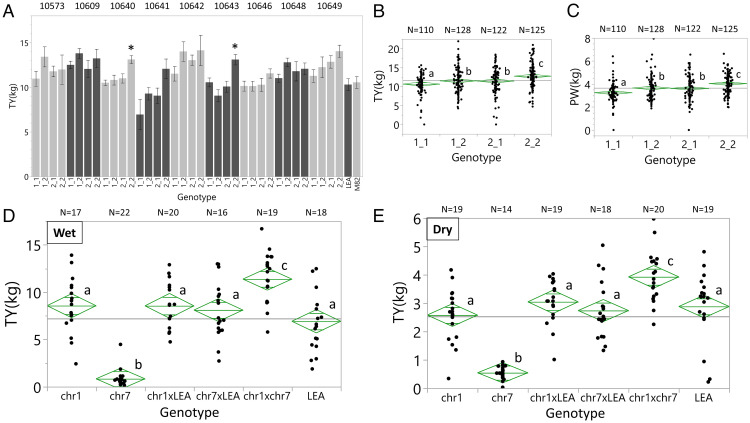
The next question we addressed related to the effect of the genetic background on the epistatic interaction. For this, we crossed the double heterozygous LEA hybrid to nine different processing tomato inbred lines and planted 100 plants from each of the crosses in the field. Genotyping of the plants was done after planting, and for each of the families, there were between 10 and 15 plants for each of the four genotypic groups (1_1, 2_1, 1_2, and 2_2). The results in Fig. 4A show that in two of the crosses (10,640 and 10,643), the 2_2 group had a statistically significant higher yield. The average effect of the four genotypic groups over all genetic backgrounds indicates that the double heterozygous group had 13% higher yield (Fig. 4B). The only yield-associated trait that correlated with TY was plant weight (r = 0.54), where plants of the 2_2 group had larger vines than those of the other groups (Fig. 4C). These results indicate that the two introgressions do not function the same way in all genetic backgrounds, and their use in variety development would require the breeder to identify the genetic backgrounds that maximize the effect of the wild species genes.
Fig. 4.
Epistatic QTLs for yield heterosis. (A) A LEA background hybrid heterozygous for the QTLs on chromosomes 1 and 7 was crossed to nine different processing tomato inbreds to produce progenies of four genotypic groups, which were assayed for TY in Akko 2021. TY of the double heterozygous hybrids (2_2) with inbreds 10,640 and 10,643 was significantly higher than that of the other genotypes (5% level based on the Tukey–Kramer test). (B) A test of the pooled TY from all the nine genetic backgrounds (Fig. 4A) shows that the double introgression hybrids had the highest yield of the four genotypes. Means with the same letters are not significantly different at the 5% level based on the Tukey–Kramer test. (C) A test of the pooled PW data from all the nine genetic backgrounds shows that the double introgression hybrids had the largest PW. Means with the same letters are not significantly different at the 5% level based on the Tukey–Kramer test. (D and E) To test whether the epistatic QTLs on chromosomes 1 and 7 drive yield heterosis, we selected two homozygous BILs, each carrying a single QTL, and crossed the two BILs to LEA and to each other. The yield of the two BILs, their hybrids with LEA, and the double introgression hybrid was tested in Akko 2020 in well-irrigated conditions (D) and in drought (E). Means with the same letters are not significantly different at the 5% level based on the Tukey–Kramer test.
Epistatic QTLs for Yield Heterosis.
To validate the observation that yield heterosis is driven by digenic epistasis, we used a homozygous BIL for chromosome 1 (p-427) and a BIL for chromosome 7 (p-1573) both covering the QTL locations. The TY of p-427 was high, but p-1573 was partially sterile likely due to deleterious recessive alleles derived from the wild species. For this reason, we crossed both BILs to LEA to produce single introgression BILH that could be compared phenotypically to the double introgression heterozygotes. The derived genotypes and the LEA control were planted in both wet and dry plots since in the initial experiment in 2018, this epistasis improved yield under drought. In both treatments, the yield of the double introgression hybrid was significantly higher than that of the inbred LEA and the best heterozygous parent (chromosome 1 × LEA; 29% yield improvement in the dry and 33% in the wet; Fig. 4 D and E). These results support the conclusion that the observed heterosis is driven by more-than-additive digenic interaction.
In the experiments described above, each replication consisted of a single plant grown on 1 m2. This planting density is a good way to generate, in an economical way, the many replications that are needed for analysis of complex traits. However, the commercial stand of processing tomatoes is 2.5 plants per m2. To evaluate the relevance of the observed heterosis to the tomato crop, we conducted in 2022 an experiment in which each replication consisted of a plot of 5 m2 with 12 plants. A control experiment of the same genotypes was planted with single plants on 1 m2 and yielded results like the previously described experiments (SI Appendix, Fig. S4). The double heterozygous (2_2) had the largest PW, the smallest fruit weight, and significantly higher yield (13.1 kg) than the mean of the other three genotypes (1_1, 1_2, and 2_1; 10 kg per plant). Dividing the TY by the average fruit weight showed that the double introgression hybrid produced 58% higher number of fruits than the three control genotypes. The dense spacing plots experiment followed the same trends that were observed in the single plants’ experiments. PW for the plots of the double introgression hybrid plots was the highest, and their fruit weight was the lowest (Fig. 5 A and B). TY of the 2_2 plots was 22% higher than that of the control genotypes, and fruit number was 53% higher (Fig. 5 C and D). In summary, the reduction of 30% in fruit weight in the double heterozygous genotype was more than compensated for by a large increase in fruit number, which resulted in 22% yield increase in the plots. Both in the single plant wide spacing and in the dense spacing plot experiments, the TY did not correlate to fruit weight but correlated strongly to PW (r2 0.59 and 0.60, respectively; SI Appendix, Table S2). To understand the cause of the yield heterosis, we planted in the greenhouse plants of the F2 population of the double heterozygote in the LEA background and the progeny of its crosses to 10,640 and 10,643 and counted the number of inflorescences along the main stem (SI Appendix, Fig. S5). In both cases, the double heterozygotes produced significantly more inflorescences along the main shoot, suggesting that an increase in inflorescence number is responsible, at least in part, to the observed yield heterosis.
Fig. 5.
Validation of the epistatic QTLs in plots of commercial plant density. Yield components (Akko 2022) measured in 5-m2 plots with 12 plants each. We present the plots of the four genotypic groups with respect to the markers on chromosomes 1 (SSL2.50CH01_95261222) and 7 (SSL2.50CH07_65737800): 1) homozygous for the cultivated tomato alleles in chromosomes 1 and 7 (1_1), 2) heterozygous for the chromosome 1 introgression (2_1), 3) heterozygous for the chromosome 7 introgression (1_2), and 4) heterozygous for both introgressions (2_2). Genotypic group means showing the same letters are not significantly different at the 5% level based on the Tukey–Kramer test. The traits measured were PW (A), TY (B), fruit weight (C), and the estimated fruit number (D).
Discussion
Heterosis is an agricultural phenomenon that describes the superior productivity of the F1 hybrid over its best parent where the yield of the hybrids is in the range of the leading varieties in the market. The genetic basis of heterosis includes diverse mechanisms such as dominance, overdominance, epistasis, and perhaps additional contributors (9–13). In a recent tomato study, epistasis between two independently segregating homologous MADS box mutations was investigated (14). One of the mutations promotes enlargement of a leaf-like organ on fruit sepals and was favored during domestication due to its effect of increasing fruit size. The second recessive mutation eliminates the flower abscission zone leading to “jointless” fruit stems that improve the efficiency of the mechanical harvest. Lines that combine both mutations in a homozygous state produce an extremely overbranched inflorescence with partial sterility, and thus, the yield is low. However, when the highly branched inflorescence lines were crossed with a wild-type line, heterosis for yield was observed due to slightly branched inflorescences in the heterozygous line that increase fruit set in the hybrid compared to its parents. A similar case of epistasis in tomato was discovered in relation to the heterotic mutation single flower truss (sft) and its interaction with the self-pruning gene that belongs to the same family of CET factors (15). Plants heterozygous for loss-of-function mutations in the sft, the florigen gene, produce 50% higher yield compared to the nonmutant isogenic cultivar M82 and the homozygous sft mutant. The heterotic effect on yield was only detected in the determinate genetic background, which is homozygous for the self-pruning (sp) mutation (sft/+; sp−/sp−), while in indeterminate tomatoes (Sp+/−), there was no effect of sft/+ on yield, indicating that Sp+ nullifies the heterotic effect of its family member sft. These two epistasis examples were discovered by chance, whereas in this paper, we present an attempt to systematically discover favorable epistatic interactions in a genome-wide manner.
The first factor that limits the mapping of epistatic interactions in segregating populations is the number of individuals. For this, we developed a very large, highly polymorphic, permanent mapping population of BILs from a cross of a divergent wild species (S. pennellii; the Lost Accession) and a processing tomato inbred (LEA). The population was composed of 1,400 plants in BC2S6 generation, and we expected that it would be large enough to analyze whole-genome digenic epistasis for yield-associated traits. Unfortunately, many of the wild species’ alleles deviated from the expected Mendelian segregations, (8) causing the proportion of plants that carry the S. pennellii alleles to be lower than expected (6% compared to 12.5%; Fig. 1D). For this reason, we conducted the analysis of epistasis only for digenic combinations where the double introgression genotype included at least ten individuals. When we analyzed the phenotypes of the homozygous BILs, containing an average of 11 introgressions per BIL, their yield was 50% lower than that of the LEA control. However in the BILHs (BIL × LEA), the mean yield was doubled, indicating that the best use of this resource is for breeding of hybrids. A second limitation for the mapping of epistatic interactions results from the multiple testing penalty of all pairs of genomic regions (in the case of digenic interactions), which causes a strong downward adjustment of the statistical significance threshold. The BILs dataset included 7,699 SPET markers and thus close to 60 × 106 digenic combinations whose testing could lead to many false positives. An important advantage of the BILs is that they provide the means for quick and simple validation of candidate cases of epistasis and thus eliminate the need to use very stringent statistical thresholds in the epistasis discovery phase. The validation is simply done by crossing the double introgression hybrid to LEA or other tomato inbreds and planting ~100 plants whose genotypes are expected to be 1_1, 2_1, 1_2, and 2_2 in roughly equal numbers. Genotyping and phenotyping of these plants provide a quick and easy validation protocol that does not suffer from the multiple testing penalty, which can eliminate favorable combinations that can be of value for the breeder.
The BILs were found to include a recombinant every 18 Kb in the euchromatin, which is equivalent to a recombinant between every tomato gene. Since the majority of the 80 cases of epistasis that we detected for the yield-related traits involved genomic regions in the euchromatin, the BILs also provide the means for fine mapping of the QTLs involved by virtue of the high number of mapped recombinants. For fine mapping of the epistatic QTLs on chromosomes 1 and 7, we can use the two BILs that create the epistasis and the mapped recombinant BILs for each of the chromosomes (SI Appendix, Fig. S6). The BILH for the chromosome 1 QTL (green chromosome) is crossed to homozygous BILs that are recombinant in chromosome 7. The progeny of such crosses would produce nearly isogenic hybrids of two genotypes: with or without the S. pennellii QTL on chromosome 1. The yield of the isogenic hybrids would be compared, and if the hybrids with the chromosome 1 QTL would have a higher yield, then the assumption is that the recombined segment of the BIL on chromosome 7 carries the second QTL needed to generate heterosis. A comparison of the values of the two genotypic groups using multiple recombinant BILs would indicate the location of the chromosome 7 QTL. To map the QTL on chromosome 1, which is involved in the interaction, we need to cross the BILH of the chromosome 7 QTL (orange chromosome) to recombinant BILs of chromosome 1 and follow the scheme described above to fine map the genomic region of the QTL on chromosome 1.
The BIL resource is also valuable for the mapping of other traits that are of interest to biologists. We are currently distributing to the tomato community seed of the parents (LEA and the Lost Accession), their F1, and a set of 60 BILs that provide complete coverage of the S. pennellii genome. These BILs can be used to map traits of interest, and once the mapping is done, it will be possible to screen the recombinants in the defined interval and identify the genes involved—provided they are in the euchromatin. Beyond the academic value of the BILs, they can be used to breed new processing tomato hybrids including varieties that are tolerant to drought stress (Fig. 4E). However, translation of this know-how to competitive processing hybrid requires several important steps: 1) As shown in Fig. 4A, not all genetic backgrounds benefit from the epistatic interaction of the QTLs on chromosomes 1 and 7, and thus, the breeder would need to identify the genetic backgrounds that maximize the heterotic effect. 2) The two introgressions reduce fruit weight compared to LEA (SI Appendix, Fig. S2D), and this apparent linkage drag needs to be recombined out of the genetic background in a similar manner as described in the fine mapping scheme (SI Appendix, Fig. S6).
It is well recognized in genetics that when a gene or QTL is introduced into different genetic backgrounds, the phenotypic outcome may vary, indicating epistatic interactions with unknown factor(s) in the receptor genomes. The role of epistasis is also well known in hybrid breeding where often hundreds of homozygous inbreds are crossed to create many experimental F1 hybrids, which are evaluated for yield heterosis and other traits. In processing tomatoes in California, ~10,000 new hybrids are tested every year, and roughly only ten of them make it to the commercial market. Thus, only few F1 hybrids show competitive heterosis, indicating that very specific interactions between the parents’ genomes are needed to produce a top-yielding variety. Development of very large, controlled populations can provide the means to identify the genomic components that amplify heterosis, and using such information, it will be possible to be more scientific in establishing crossing blocks of new experimental hybrids.
Materials and Methods
Plant Material.
In 2007, we started to develop a new BIL population for genetic analysis of epistasis. The donor wild species for this resource was an unexplored accession of S. pennellii that was discovered in the Gatersleben collection (Lost Accession, LA5240). This accession which is self-compatible was crossed to the modern determinate processing tomato inbred called LEA. A single F1 hybrid was crossed as a male to LEA to produce a backcross1 (BC1) of 2,000 plants, which were crossed again to LEA to produce 2,000 BC2 plants. Selfing of the BC2 was carried out until backcross-2-self-6 (BC2S6). Throughout the BIL selfing, we planted two to six plants from each BIL and extracted seed from a random fertile plant. In BC2S6 and previous generations, we observed close phenotypic resemblance between the siblings of a particular BIL, indicating that the semiindustrial project of pollination, fruit harvest, seed extraction, and plantings was carried out accurately. In the more advanced selfing generation, we encountered higher sterility, which resulted in the production of 1,400 BILs.
In fall 2017, all the 1,400 BILs were grown in a greenhouse and were crossed to LEA to create a set of BILHs. In spring 2018, seedlings were grown in a greenhouse for 35–40 d, and then, 1,389 BILs, 1,233 BILHs, and the controls LEA and M82 were transplanted in a drip-irrigated field in Akko. The planting density was one plant per m2, where the distance between the plants was 0.5 m, and the width of the bed was 2 m. Both the dry and wet field plots started the growing season at field capacity, which represents the maximum amount of water that the soil could hold. For the dry treatments, only 60 m3 of water was applied per 1,000 m2 of field immediately after transplanting. In the irrigated treatment, 320 m3 of water was applied per 1,000 m2 of field throughout the growing season according to the irrigation protocols in the area. 3 wk after planting, the irrigation of the dry treatment was stopped, and the plants grew under drought condition with ~20% of the amount of water that is given to the normal crop. In spring 2022, we established an experiment where the yield epistasis combinations were tested in a commercial stand in replicated plots of 5 m2 with 12 plants per plot.
Genotyping.
Leaflets of each of the 1,389 BILs were collected from the field-grown plants in Akko. DNA was extracted using the CTAB protocol and was diluted to a final concentration of 40–60 ng in a volume of 40 μl. DNA quantity and quality were determined using a Nanodrop ND-1000 spectrophotometer followed by electrophoresis on a 1% agarose gel. DNA quantity was validated using the Qubit dsDNA BR Assay Kit (Life Technologies, Eugene, OR, USA). The BIL DNA and that of the controls LA5240, LEA, and M82 were genotyped by SPET7 using the HiSeq2000 platform by IGA Technology Services, Udine, Italy. A total of 173,000 SNPs were called and subjected to filtering using TASSEL v5.2.43: Sites with a depth of less than three reads or >50% missing data were filtered out as well as heterozygous markers, nonpolymorphic markers, and markers with a minor allele frequency of <1%. The final SNP set included 7,699 markers across the 1,389 homozygous BILs. Introgression bins and genetic distances were calculated using ASMap package in R software (https://cran.rproject.org/web/packages/ASMap/index.html).
For the experimental validation of the epistatic interactions of chromosomes 1 and 7, we designed primers for plant genotyping using the site https://www.snapgene.com/software for alignment of the S. pennellii genome sequence and for the detection the polymorphisms in the target regions.
Primer sequences for the chromosome 1 QTL:
F – CTATCACTGAAGCAACTAGTGAGG and R – CGTTGTTGGTGAATATGAGCTTCAC.
The PCR product was 729 bp that was digested with EcoRI: The S. pennellii allele was not cut, while the LEA allele generated two fragments: 503 bp and 226 bp.
Primer sequences for the chromosome 7 QTL:
F – ATGGATCGATCGGCTCTGATAC and R – GGTAGTCAAAGTTTGACCGACCTT.
The PCR product was 528 bp that was digested with HphI: The S. pennellii allele was not cut, while the LEA allele generated two fragments: 354 bp and 174 bp.
Phenotyping.
Fruits of all the genotypes were harvested when 95–100% of the tomatoes were red (105–115 d after transplanting). The following measurements were taken for each of the plants: PW kg of the vegetative part, average fruit weight (FW g) calculated from a random sample of 10 red fruits per plant, total soluble solids concentration (Brix) of the fruit (% Bx) assayed on the same 10 red fruits, and total fresh yield per plant (TY) (including both red and green fruits if there were any).
Data Analysis.
Single-QTL mapping analysis and two-dimensional genome scans for the detection for epistasis were performed using R/qtl (16) by the Haley–Knott regression. In specific cases, single marker effects were reanalyzed by ANOVA using JMP Pro 16 software package (SAS Institute, Cary, NC, USA). Multiple comparison corrections to significance thresholds were performed for each of the measured phenotypes using 1,000 permutation tests that generated the maximum LOD. The thresholds were set to the 95th percentile (P < 0.05) from the obtained distributions.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Author contributions
D.Z. designed research; S.T. analyzed data; and S.T. and D.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The complete raw data (17) of the genotypes and phenotypes is deposited in DRYAD (https://datadryad.org/stash/share/J9Fxf54OEq975m4K_4PiB97BcrdGdjdR7QJZw_t_QzU).
Supporting Information
References
- 1.Weinreich D. M., Lan Y., Wylie C. S., Heckendorn R. B., Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin. Genet. Dev. 23, 700–707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dempewolf H., et al. , Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082 (2017). [Google Scholar]
- 3.Eshed Y., Zamir D., An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eshed Y., Zamir D., Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143, 1807–1817 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackay T. F. C., Epistasis and quantitative traits: Using model organisms to study gene–gene interactions. Nat. Rev. Genet. 15, 22–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt M. H. W., et al. , De novo assembly of a new Solanum pennellii accession using nanopore sequencing. Plant Cell 29, 2336–2348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barchi L., et al. , Single primer enrichment technology (SPET) for high-throughput genotyping in tomato and eggplant germplasm. Front. Plant Sci. 10, 1005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamir D., Tadmor Y., Unequal segregation of nuclear genes in plants. Bot. Gaz. 147, 355–358 (1986). [Google Scholar]
- 9.Birchler J. A., Yao H., Chudalayandi S., Unraveling the genetic basis of hybrid vigor. Proc. Natl. Acad. Sci. U.S.A. 103, 12957–12958 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lippman Z. B., Zamir D., Heterosis: Revisiting the magic. Trends Genet. 23, 60–66 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Schnable P. S., Springer N. M., Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 64, 71–88 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Li Z. K., et al. , Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158, 1737–1753 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., et al. , Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics 180, 1725–1742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soyk S., et al. , Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169, 1142–1155 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Krieger U., Lippman Z. B., Zamir D., The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–463 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Broman K. W., Sen S., A Guide to QTL Mapping with R/qtl (Springer, New York, 2009), vol. 46. [Google Scholar]
- 17.S. Torgeman, D.Zamir, Genotypes and Phenotypes. Dryad. https://datadryad.org/stash/share/J9Fxf54OEq975m4K_4PiB97BcrdGdjdR7QJZw_t_QzU. Deposited 8 November2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The complete raw data (17) of the genotypes and phenotypes is deposited in DRYAD (https://datadryad.org/stash/share/J9Fxf54OEq975m4K_4PiB97BcrdGdjdR7QJZw_t_QzU).