Significance
Regulatory T cell therapy is a promising approach to control auto or alloimmunity. In cell therapy with conventional T cells, chronic activation can lead to a phenomenon called exhaustion, resulting in a blunted therapeutic effect. Whether or not Tregs are subject to exhaustion was unknown. We used a tonic signaling chimeric antigen receptor (CAR) system to induce chronic activation in human Tregs and showed that this resulted in the loss of suppressive function. Transcriptional and epigenetic comparisons between Tregs and conventional T cells revealed similarities and differences, indicating a Treg-specific signature. These data show that exhaustion-driven dysfunction has the potential to limit the effectiveness of CAR Treg therapy and provide a “benchmark” signature to assess states of Treg exhaustion in future studies.
Keywords: regulatory T cells, exhaustion, tolerance, chimeric antigen receptor, epigenetics
Abstract
Regulatory T cell (Treg) therapy is a promising approach to improve outcomes in transplantation and autoimmunity. In conventional T cell therapy, chronic stimulation can result in poor in vivo function, a phenomenon termed exhaustion. Whether or not Tregs are also susceptible to exhaustion, and if so, if this would limit their therapeutic effect, was unknown. To “benchmark” exhaustion in human Tregs, we used a method known to induce exhaustion in conventional T cells: expression of a tonic-signaling chimeric antigen receptor (TS-CAR). We found that TS-CAR-expressing Tregs rapidly acquired a phenotype that resembled exhaustion and had major changes in their transcriptome, metabolism, and epigenome. Similar to conventional T cells, TS-CAR Tregs upregulated expression of inhibitory receptors and transcription factors such as PD-1, TIM3, TOX and BLIMP1, and displayed a global increase in chromatin accessibility-enriched AP-1 family transcription factor binding sites. However, they also displayed Treg-specific changes such as high expression of 4-1BB, LAP, and GARP. DNA methylation analysis and comparison to a CD8+ T cell-based multipotency index showed that Tregs naturally exist in a relatively differentiated state, with further TS-CAR-induced changes. Functionally, TS-CAR Tregs remained stable and suppressive in vitro but were nonfunctional in vivo, as tested in a model of xenogeneic graft-versus-host disease. These data are the first comprehensive investigation of exhaustion in Tregs and reveal key similarities and differences with exhausted conventional T cells. The finding that human Tregs are susceptible to chronic stimulation–driven dysfunction has important implications for the design of CAR Treg adoptive immunotherapy strategies.
Regulatory T cells (Tregs) control immune homeostasis and their adoptive transfer is a promising therapeutic approach with multiple trials completed, ongoing, or planned to test their efficiency in autoimmunity, solid organ transplantation, or hematopoietic stem cell transplantation (reviewed in ref. 1). As Tregs are relatively rare, the majority of cell therapy protocols involve in vitro stimulation and expansion, in some cases culturing cells for up to 36 d to achieve > 2,000-fold expansion (2). Although these in vitro-expanded cells typically remain FOXP3+ and suppressive in vitro, the progressive DNA methylation changes that occur with prolonged culture (3) suggest that repeated Treg stimulation may affect their in vivo function.
In contrast to the paucity of knowledge about how chronic stimulation affects Treg function, the effects of chronic stimulation on conventional T cells (Tconvs) have been extensively studied. In the context of infection, cancer, or transplantation, chronic antigen exposure leads to a phenomenon termed exhaustion, characterized by high expression of inhibitory receptors such as programmed cell death protein 1 (PD-1), T-cell immunoglobulin and mucin-domain containing-3 (TIM-3), and lymphocyte-activation gene 3 (LAG-3), diminished proliferative and cytokine-producing capacity, and high apoptosis (4–7). Most research in this area examined effects on CD8+ T cells or unfractionated CD3+ T cells, but there is evidence that CD4+ T cells are similarly susceptible to exhaustion (8–10).
In addition to repeated TCR stimulation, T cell exhaustion can be induced by the expression of engineered receptors, including chimeric antigen receptors (CARs), which can mediate tonic signaling via spontaneous aggregation in the absence of antigen (8, 11, 12). CAR-mediated T cell exhaustion induces the expression of a variety of inhibitory receptors, causes heritable epigenetic changes, and diminishes their antitumor therapeutic efficacy (4, 13, 14). This state of functional exhaustion can be manipulated by limiting the activity of signaling pathways and altering the epigenetic program and/or metabolic pathways (8, 12, 15–17).
Whether or not CAR Tregs are also susceptible to exhaustion has not been explored. Here, we used a tonic-signaling (TS)-CAR to chronically stimulate human Tregs and investigated the phenotypic, metabolic, epigenomic, and functional consequences.
Results
Tonic Signaling CARs Induce a Phenotype Consistent with Exhaustion in Tregs.
To study the existence and biological relevance of Treg exhaustion, we investigated the effect of expressing a TS-CAR, which incorporates a high-affinity scFv specific for disialoganglioside (GD2) fused to a CD28 costimulatory endodomain and CD3ζ (8, 11, 12). Expression of this TS-CAR induces functional, phenotypic, transcriptional, and epigenetic features of exhaustion in Tconvs in vitro and abrogates antitumor effects in vivo (8, 11). Naive Tregs, or as a control naive Tconvs (SI Appendix, Fig. S1A), were untransduced (UT), or transduced with the TS-CAR or a CD19-specific CAR (non-TS-CAR) which neither mediates tonic signaling nor induces exhaustion (8, 11). After 7 d, CAR-expressing cells were sorted and then cultured for an additional 5 d before analysis. Phenotypic analysis revealed that in comparison to UT or non-TS-CAR Tregs, a significantly higher proportion of TS-CAR Tregs expressed inhibitory receptors associated with exhaustion, namely LAG-3, TIM-3, PD-1 (Fig. 1A and SI Appendix, Fig. S1B) as well as GITR and 4-1BB (Fig. 1B and SI Appendix, Fig. S1B). Similarly, a higher proportion of TS-CAR Tregs expressed the Treg-function-associated proteins CTLA-4, LAP, and GARP (Fig. 1C and SI Appendix, Fig. S1B). High expression of all these proteins was also observed in TS-CAR-expressing CD4+ Tconvs and CD8+ T cells (SI Appendix, Fig. S2A), except for 4-1BB, LAP, and GARP which were Treg-specific. Also, a higher proportion of TS-CAR Tregs and CD4+ Tconvs were Ki67+ (Fig. 1D and SI Appendix, Fig. S2B), which could be indicative of a high proportion of terminally exhausted cells as opposed to progenitor exhausted cells (18). Although viability was similar between the three groups, in accordance with exhausted T cells (19), TS-CAR Tregs were more apoptotic than controls (Fig. 1E and SI Appendix, Fig. S3).
Fig. 1.
Expression of a tonic signaling CAR in Tregs induces phenotypic changes without lineage destabilization. Naive Tregs were left transduced (UT) or transduced with retrovirus encoding a non-tonic signaling (non-TS) or a tonic signaling (TS-) CAR. After 11 to 12 d of culture, cells were analyzed for expression of (A) LAG-3, TIM-3, PD-1, (B) GITR, 41BB, (C) CTLA4, LAP, GARP, and (D) Ki67. (E) Viability was assessed using an automated cell counter and apoptosis by flow cytometry and (F) FOXP3 expression by intracellular staining. (G) DNA was isolated, and pyrosequencing was used to measure methylation status of 7 CpGs in the Treg-specific demethylation region (TSDR) (male samples). Data are the average methylation of the 7 CpGs. For reference, the average amount of methylation in Tconv is indicated by the dotted line. (H) Intracellular cytokine production was assessed by flow cytometry 4 h after a PMA/Ionomycin stimulation. Each dot represents a unique donor. Error bars represent median ± interquartile range. (A–H) One-way Anova with Turkey’s comparisons test. For A–E (Left), n = 11 to 14 from 5 to 6 independent experiments, except for 41BB n = 3. For the apoptosis assay (E), Right, n = 5 to 6 from 3 to 4 independent experiments. For F, n = 11 to 17 from 5 to 7 independent experiments. For G, n = 3 to 6 from 3 to 4 independent experiments. For H, n = 3 to 6 from three independent experiments. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
These phenotypic changes were not associated with loss of Treg stability since TS-CAR Tregs had no change in TSDR methylation and actually increased FOXP3 expression (Fig. 1 F and G). Nevertheless, compared to controls, a significantly higher proportion of TS-CAR Tregs spontaneously produced IL-2 (Fig. 1H), suggesting that at least upon stimulation with phorbol esters and ionomycin, they overcame FOXP3-mediated repression of IL2 transcription (20). Overall, these data show that Tregs expressing a TS-CAR acquire a phenotype which mirrors that acquired by exhausted CD8+ T cells and CD4+ Tconvs (SI Appendix, Fig. S2) (8).
Transcriptome Analysis of Tonic Signaling CAR-Tregs Reveals Changes in Metabolic Pathways.
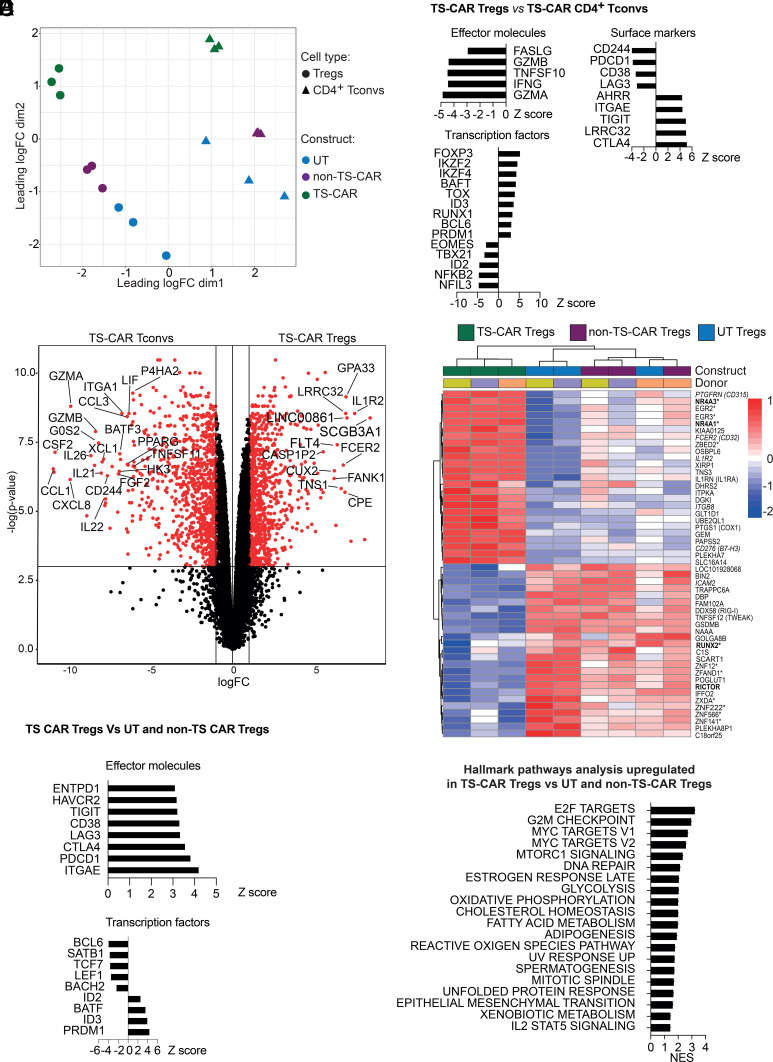
A limitation of studying exhaustion in Tregs is that several of the canonical exhaustion makers are also markers of Tregs or simply of activated T cells (4, 21). Moreover, since Tregs do not produce inflammatory cytokines, loss of cytokine production cannot be monitored as a surrogate marker of dysfunction. Therefore, we next took an unbiased approach and used RNA sequencing to determine how tonic signaling affected the Treg transcriptome in comparison to UT or non-TS-CAR Tregs, or CD4+ Tconvs. Multidimensional scaling plots revealed that Tregs and Tconvs clustered separately and that for both Tregs and Tconvs, the TS-CARs were further away from untransduced cells than cells expressing the non-TS-CAR cells (Fig. 2A). Furthermore, TS-CAR Tregs maintained their expected gene signature (22) and, compared to untransduced and non-TS-Car Tregs, had significantly increased expression of FOXP3, CTLA4, ZBRB38, IL1R2, and IL1RN and decreased DACT1, ID2, and LPIN2, the higher and lower expression of which define Tregs, respectively (SI Appendix, Fig. S4A). Preservation of gene expression characteristic of Tregs was further confirmed by analysis of subsets of genes known to be targets of FOXP3 (23) (SI Appendix, Fig. S4B).
Fig. 2.
Expression of a tonic signaling CAR results in differential effects on the transcriptome of Tregs and Tconvs. After 12 d of culture, RNA was extracted from Tregs or CD4+ Tconvs that were untransduced (UT) or transduced to express a non-tonic signaling (non-TS) or a tonic signaling (TS-) CAR, and subject to RNA sequencing. (A) Multidimensional scaling plots comparing the transcriptome of the indicated types of Tregs and Tconvs. (B) Z score of differentially expressed genes in TS-CAR Tregs compared to UT or non-TS-CAR Tregs. Threshold used was a logFC > 1 and adj. P value < 0.05 and logFC < −1 and adj. P value < 0.05. (C) Volcano plot comparing the gene expression profile of TS-CAR Tregs versus TS-CAR Tconvs after removal of genes differentially expressed (upregulated and downregulated) between UT Tconvs and UT Tregs (Fig. S5A). Genes in red are defined by a P value < 0.05. (D) Heatmap showing the top 50 differentially expressed genes between the different types of Tregs. Bold genes are associated with exhaustion in Tconvs, italicized genes are cell surface proteins and those marked with * are transcription factors. (E) Z score comparing genes that were significatively upregulated or downregulated in TS-CAR Tregs versus TS-CAR Tconvs. Threshold used was a logFC > 1 and adj. P value < 0.05, and logFC < −1 and adj. P value < 0.05. (F) Gene set enrichment analysis. Normalized enrichment score (NES) of hallmark pathways analysis overrepresented in TS-CAR Tregs compared to non-TS CAR or UT Tregs cells. Each dot represents a unique donor. n = 3 per group from two independent experiments.
To better understand the consequences of exhaustion in Tregs, we first sought to identify differences in gene expression between TS-CAR Tregs and TS-CAR CD4+ Tconvs. As expected, in comparison to TS-CAR CD4+ Tconvs, TS-CAR Tregs had low expression of effector molecules such as GZMA, GZMB (granzyme A &B), IFNG, TNFSF10 (TRAIL), and FASLG, and high expression of Treg-related genes such as FOXP3, IKZF2 (Helios), and LRRC32 (GARP), confirming their Treg identity (Fig. 2B). Interestingly, inhibitory surface receptors were differentially expressed in TS-CAR Tregs compared to TS-CAR CD4+ Tconvs, with lower expression of CD244, PDCD1 (PD-1), and LAG3 and higher expression of TIGIT and CTLA4. Similarly, there were differences in expression of exhaustion-related transcription factors (19), with higher expression of TOX, but not EOMES or TBX21 in TS-CAR Tregs compared to TS-CAR CD4+ T cells (Fig. 2B).
Since many of the genes differentially expressed between TS-CAR Tregs and Tconvs were simply characteristic of known transcriptional differences between these cell lineages, we next examined differential gene expression after removing genes that were differentially expressed between UT Tregs and UT Tconvs (Fig. 2C and SI Appendix, Fig. S5A). The resulting data revealed that compared to CD4+ TS-CAR Tconvs, TS-CAR Tregs expressed significantly less BATF3, a transcription factor expressed in exhausted TS-CAR T cells (8, 12), less CD244, an immunoregulatory receptor also associated with T cell exhaustion (24, 25), as well as a variety of chemokines (CCL1, CXCL1, and CXCL8) and cytokines (IL21, IL22, and LIF). On the other hand, TS-CAR Tregs overexpressed GPA33 and GARP (LLRC32), the expression of which is associated with stable Tregs (26), and other markers such as FCER2 (CD32), IL1R2 (IL1 decoy receptor), CD276 (B7-H3), and ITGB8 (integrin beta 8).
Within the 3 groups of Tregs, the transcriptome of the TS-CAR Tregs was clearly distinct from UT and non-TS-CAR Tregs, with a heat map of the 50 most differentially expressed genes between these groups shown in Fig. 2D. In comparison to both UT and non-TS CAR Tregs, TS-CAR Tregs had high expression of NR4A3 (NOR1) and NR4A1 (NUR77), two genes associated with exhaustion in CD8+ T cells (16, 27). TS-CAR Tregs also had lower RICTOR, which is part of the mammalian target of rapamycin (mTOR) complex 2 (mTORC2) and implicated in control of Treg suppressive function (28). There was also a decrease in RUNX2, the expression of which inversely correlates with TCR stimulation strength (29). Confirming the flow cytometry data, TS-CAR Tregs had high expression of exhaustion-related genes such as PDCD1 (PD-1), HAVCR2 (TIM-3), TIGIT, LAG3, and CTLA4 and low expression of several transcription factors known to be downregulated in terminally exhausted T cells, including BCL6, SATB1, TCF7 (encodes TCF-1), LEF1, and BACH2 (18, 30, 31) (Fig. 2E).
Hallmark pathway analysis, obtained by Gene Set Enrichment Analysis, revealed that TS-CAR Tregs upregulated pathways linked to metabolism, including MYC and MTORC1 signaling, glycolysis, oxidative phosphorylation, cholesterol homeostasis, fatty acid metabolism, and adipogenesis (Fig. 2F), suggesting that changes in cell metabolism may be key factors driving the observed phenotypic changes in TS-CAR Tregs. Of note, although these pathways were upregulated in TS-CAR Tregs compared to other Treg groups, they were still lower in comparison to TS-CAR CD4+ Tconvs (SI Appendix, Fig. S5B).
Tonic Signaling in Tregs Causes an Increase in Glycolysis.
A growing body of evidence suggests that differences in the metabolic pathway activity underlie functional differences between Tregs and Tconvs (32). Specifically, oxidative phosphorylation and mitochondrial activity, which are required for Treg suppressive function (33, 34), are promoted by FOXP3 via inhibition of c-Myc-driven glycolysis (35–38). In the context of exhaustion, metabolic dysfunction precedes its development in T cells (39) and is influenced by CAR expression (40). Transcriptomic analysis suggested that there could be TS-CAR-related changes in metabolic activity so we analyzed extracellular acidification rates (ECAR) and oxygen consumption rates (OCR) as surrogate measures of glycolysis and oxidative phosphorylation, respectively. In their basal state, TS-CAR Tregs had a tendency to have higher ECAR and OCR than UT or non-TS-CAR Tregs, suggesting both metabolic pathways were enhanced (Fig. 3 A and B). However, a significantly lower relative OCR/ECAR ratio showed that TS-CAR Tregs preferentially engaged glycolysis compared to their UT or non-TS-CAR counterparts (Fig. 3C).
Fig. 3.
Expression of a tonic signaling CAR signaling alters Treg metabolism. Tregs were untransduced (UT) or transduced to express a non-tonic signaling (non-TS) or a tonic signaling (TS-) CAR. After 11 to 12 d of culture, cells were plated in Cell-Tak coated wells and subject to the Agilent Seahorse XF Cell Mito Stress Test. (A) Extracellular acidification rate (ECAR) (Left) and normalized to baseline ECAR (Right) over time, (B) oxygen consumption rate (OCR) (Left) and normalized to baseline OCR (Right) over time. Oligomycin, FCCP, and a mix of Rotenone and antimycin A were added as indicated. (A&B) n = 3 to 5 donors from two independent experiments, using the average of technical replicates. Error bars represent mean ± SEM. (C) OCR/ECAR ratio at baseline. One-way Anova with Turkey’s comparisons test P = 0.0196. n = 4 to 6 from three independent experiments. (D) The spare respiratory capacity (SRC) was calculated by the average of the maximum OCR after FCCP injection minus the average basal respiration, divided by the average basal respiration, times 100 [(max-basal)/basal*100)]. One-way Anova with Turkey’s comparisons test P = 0.0054, n = 3 to 4 from two independent experiments. **P ≤ 0.01, *P ≤ 0.05.
To further interrogate these pathways, cells were exposed to oligomycin, a mitochondrial ATP synthase inhibitor which prevents oxidative phosphorylation and usually upregulates glycolysis. Since TS-CAR Tregs had high basal ECAR, the relative oligomycin-stimulated increase was modest compared to the other groups, suggesting limited spare glycolytic capacity (Fig. 3A). In contrast, there was comparable ATP-coupled mitochondrial respiration in all groups (Fig. 3B). Injection of carbonyl cyanide-4 (trigluoromethoxy) phenylhydrazone (FCCP) to maximize mitochondrial oxygen consumption revealed that TS-CAR Tregs also had lower spare respiratory capacity (Fig. 3D). Data in Tregs were mirrored with CD4+ Tconvs (SI Appendix, Fig. S5C). Overall, these data suggest that TS-CAR Tregs would poorly adapt to an increase in energy demand due to their high basal activity of both of these pathways (41, 42).
TS-CAR Tregs Have a Global Increase in Chromatin Accessibility.
The state of cellular exhaustion is known to be embedded in epigenomic changes so we next evaluated the epigenetic landscape of TS-CAR Tregs using an assay for transposase-accessible chromatin using sequencing (ATAC-seq) and whole genome DNA methylation analysis. For ATAC-seq, out of 73,762 regions, there were 3,373 differentially accessible regions (DAR)s, with a net increase in chromatin accessibility in TS-CAR Tregs, with almost twice the number of regions compared to non-TS-CAR Tregs (Fig. 4A). The proportions of DARs annotated in different genomic regions were similar between genes more accessible in TS versus non-TS-CAR Tregs (Fig. 4B).
Fig. 4.
Changes in chromatic accessibility in TS-CAR Tregs. Tonic signaling (TS) and non-TS CAR Tregs were subjected to transposase-accessible chromatin using sequencing (ATAC-seq) after 12 d of culture. (A) Differentially accessible regions (DAR)s between TS- and non-TS-CAR Tregs using a threshold of FC > 2 and adj. P value < 0 > 01. (B) Annotated genomic regions of DARs that are more accessible in TS-CAR Tregs (Left) or in non-TS-CAR Tregs (Right). (C) Heat map of 324 DAR between TS- and non-TS CAR Tregs using an absolute fold change > 3 and padj < 0.05, with known transcription factors indicated. (D) Transcription factor motifs enriched in TS-CAR Tregs were identified by HOMER. N = 3 to 4 donors tested in two independent experiments.
We then evaluated the most significant DARs between TS- and non-TS-CAR Tregs (padj < 0.05 and absolute fold change > 3) with a focus on transcription factor enrichment (Fig. 4C). Similar to exhausted Tconvs, we found increased accessibility of the PRDM1 (BLIMP-1) (43) and binding partner ZBTB32 loci (44). Interestingly, TS-CAR Tregs had decreased TET2 and increased NACC1 accessibility which could both affect Treg stability (45–47).
A functional deficiency in JUN, a basic region-leucine zipper (bZIP) transcription factor and AP-1 family member, mediates exhaustion in CAR Tconvs (8) so we used hypergeometric optimization of motif enrichment (HOMER) analysis to compare the accessibility of transcription factor binding regions (Fig. 4D). We found that accessibility of AP-1 bZIP and bZIP-IRF binding motifs was significantly increased in TS-CAR Tregs, with a similar enrichment in accessibility of NFkB, NFAT, and RUNX transcription factor motifs (48).
TS-CAR Tregs Have a Reprogrammed DNA Methylome.
In addition to changes in chromatin accessibility, T cell exhaustion is associated with altered DNA methylation (49). In CD8+ T cells, a methylation-based, multipotency index (MPI) which interrogates the methylation state of 245 CpG sites can be used to delineate their developmental hierarchy, with an MPI score of 1 representing a naive CD8+ T cell and a score of 0 representing exhausted/terminally differentiated cells (50, 51). To determine where Tregs fall on this MPI trajectory, we generated whole-genome bisulfite sequencing (WGBS) DNA methylation profiles from TS- and non-TS-CAR Tregs and Tconvs. A heat map of the WGBS data for the 245 MPI genes revealed segregation by cell type—i.e., Treg or Tconv—but not by TS-CAR expression (Fig. 5A). Moreover, analysis of the WGBS data in the MPI assay revealed that Tregs in general had significantly lower MPI score compared to Tconv cells; for both cell types, TS-CAR expression did not further decrease the MPI score (Fig. 5B). Nevertheless, we identified 1,458 differentially methylated regions (DMR) between TS and non-TS-CAR Tregs and 2,628 between TS and non-TS CAR Tconvs (SI Appendix, Fig. S6).
Fig. 5.
TS-CAR Tregs undergo DNA methylation reprogramming. Whole-genome bisulfite sequencing was performed in TS and non-TS-CAR Tregs and Tconvs. (A) Heat map of normalized methylation levels at 245 CpG methylation loci that are part of an established methylation-based multipotency index (MPI). (B) MPI values in relation to established values from naive (=1) and exhausted (=0) CD8+ T cells. (C) Gene Set Enrichment Analysis (GSEA) comparing the enrichment of an exhaustion program in Tregs vs Tconvs, comparing non-TS CARs on the left and TS-CARs on the right. (D) Representative differentially methylated regions (DMR) tracks for TCF7, TOX, and PRDM1 in TS and non-TS CAR Tregs and Tconv. Significant DMRs highlighted in the green boxes, with the significant comparison highlighted underneath in pink. TCF1/7, TOX and PRDM1 expression was assessed by flow cytometry, comparing TS or non-TS CAR to untransduced (UT) Tregs. N = 3 to 5 donors tested in two independent experiments. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
To further explore the effects on DNA methylation, we performed gene set enrichment analysis (GSEA) to assess enrichment for exhaustion programs (ranked from unmethylated to methylated) using an established gene expression signature characteristic of a progenitor subset of exhausted CD8+ T cells (51, 52). Compared to Tconvs, both non-TS CAR and TS-CAR Tregs had more demethylated regions in exhaustion-associated genes (Fig. 5C).
Since both RNAseq and ATACseq data revealed significant effects on transcription factor expression/potential function, we examined the methylation status of loci encoding three transcription factors associated with exhaustion: TCF-1, TOX1, and BLIMP1. For TCF7 (TCF1), TS-CAR Tregs were significantly more methylated compared to TS-CAR Tconvs, consistent with the fact that TCF-1 negatively correlates with inhibitory receptor expression (31). Although non-TS and TS-Tregs were not differentially methylated at the TCF7 locus, at the protein level, TS-CAR Tregs had significantly reduced TCF1 expression (Fig. 5D). TOX, which promotes an exhausted phenotype in CD8+ T cells (53–55), was more demethylated in Tregs compared to Tconvs, but only in cells with non-TS-CARs. However, TS-CAR Tregs expressed more TOX protein compared to non-TS CAR Tregs. Finally, BLIMP1 (PRDM1), associated with exhaustion (56), particularly in CD4+ T cells (57), was significantly less methylated in TS-CAR Tregs compared to all other cell types, a finding which correlated with similar differences in protein expression.
TS-CAR Tregs Are Suppressive In Vitro but Dysfunctional In Vivo.
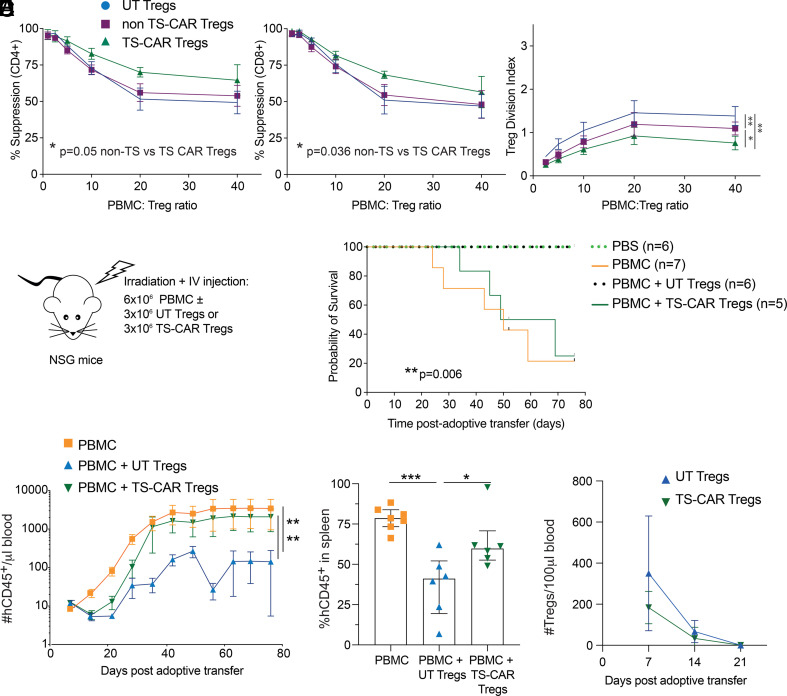
As a final approach, we assessed how TS-CAR expression affected Treg function. We first tested suppressive function in vitro by coculturing increasing numbers of UT, non-TS-, or TS-CAR Tregs with anti-CD3/28-stimulated PBMCs and measuring proliferation after 4 d. Unexpectedly, TS-CAR Tregs were significantly more potent than UT or non-TS Tregs at suppressing CD4+ and CD8+ T cell proliferation (Fig. 6A), but within the Treg population, the TS-CAR Tregs proliferated significantly less than UT or non-TS-CAR Tregs (Fig. 6B).
Fig. 6.
Tonic signaling CAR Tregs are functional in vitro but not in vivo. (A) Suppression of CPD-ef450-labeled PBMCs by untransduced (UT), non-tonic signaling (non-TS), or tonic signaling (TS-) CAR CPD-ef-670-labeled Tregs was determined after 4 d of coculture. n = 8 to 10 from four independent experiments (B) The Treg division index from cultures in (A) was determined. n = 6 to 7 from three independent experiments (A and B) Analysis were done using one-way ANOVA with Turkey’s multiple comparisons test. (C–F) Irradiated NSG mice were injected with PBS or 6 × 106 PBMCs without or with 3 × 106 UT or TS-CAR Tregs. Data from two independent experiments (C) Schematic diagram of the experiment set up. (D) Survival curve, log-rank (Mantel-Cox) test. (E) Absolute number of human CD45+ cells/µL of blood assessed weekly after adoptive transfer. One-way ANOVA with Dunnett’s multiple comparisons test (Left) and % Human CD45+ engraftment in the spleen upon experimental or humane endpoint (of live singlets) in the different groups (Right). (F) Absolute number of human Tregs/100 µL of blood assessed weekly after adoptive transfer. Tregs were gated as live human CD45+CD4+HLA-A2neg. Two-way ANOVA showed no differences between groups, mean ± 95% CIs are shown. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
Studies of TS-CARs in Tconvs showed that despite normal in vitro effector activity, functional defects were revealed in vivo (11) so we next tested Treg function in vivo using a humanized mouse model of xenogeneic graft-versus-host disease. NSG mice were injected with 6 × 106 HLA-A2 positive PBMCs without or with 3 × 106 UT or TS-CAR HLA-A2 negative Tregs (n.b. non-TS Tregs not examined as they would be stimulated by CD19+ B cells in the PBMCs), monitored and bled weekly (Fig. 6C). Surprisingly, TS-CAR Tregs completely lost their in vivo protective effect, with no improved survival in comparison to mice which only received PBMC. In contrast, mice which received UT Tregs were protected from mortality over this time period (Fig. 6D). Human cell engraftment mirrored the clinical effect, with only the UT Tregs able to reduce the number of circulating and splenic human CD45+ cells in comparison to mice that only received PBMCs (Fig. 6E). The injected Tregs were detectable in the peripheral blood for up to 14 d after adoptive transfer, with no significant differences in the absolute number of UT and TS-CAR Tregs (Fig. 6F).
Discussion
Continuous TCR signaling is essential for maintenance of immune tolerance by Tregs (58, 59), but whether or not Tregs become susceptible to exhaustion with prolonged, high levels of stimulation was an outstanding question. We used expression of a model TS-CAR to ask whether exhaustion has the potential to limit the therapeutic efficacy of Tregs. We found clear evidence for development of a functional deficit consistent with the concept of exhaustion. Expression of a TS-CAR caused rapid phenotypic, functional, and epigenetic changes and complete loss of suppression function in vivo. Importantly, loss of suppression function was not correlated with loss of Treg lineage stability. These findings indicate that Treg therapies should mitigate the potential for exhaustion-driven dysfunction.
Although the existence of Treg exhaustion has been speculated for many years, it was difficult to study since exhaustion-related inhibitory receptors are often associated with enhanced Treg suppressive potential (60–62). For example, CTLA-4 is essential for Treg function (63) and TIM-3+ Tregs are more potent than their TIM-3− counterparts in vitro (61, 64). Moreover, as Tregs do not normally secrete inflammatory cytokines or exhibit cytolytic activity, in vitro analysis of these activities cannot be used to gauge this phenomenon. We thus took advantage of the well-established TS-CAR model of exhaustion (8, 11, 12, 15, 65), reasoning that this extreme state would first reveal if Tregs were susceptible and then allow us to define Treg-specific exhaustion phenotypes.
Recently, Lamarthée et al. also studied the impact of a TS-CAR in human Tregs using a 4-1BB signaling domain to induce tonic signaling (66). Our results mirror some of their findings as TS-CAR expression induced changes in the phenotype, metabolic profile, and transcriptome of Tregs, without destabilizing the core Treg signature. Consistent with our findings, they also reported an increase in ENTPD1 (CD39) in TS-CAR compared to non-TS CARs. In contrast, they did not find higher LAG3 or HAVCR2 (TIM3) in TS-CAR Tregs as we found here. Differences could be related to specific CARs, the longer cell culture period, and/or progressive increase in FOXP3-HELIOS- cells observed by Lamarthée et al. (66).
Our data show that exhaustion-associated Treg dysfunction was only revealed upon analysis of Treg suppression in vivo, consistent with the finding that a TS-CAR specific for HLA-A2 reduced the in vivo suppressive capacity of Tregs (66). This finding is also consistent with the effects of TS CARs in Tconvs, where in vitro cytotoxicity is normal (11), but major dysfunction is revealed in vivo. Moreover, as with exhausted T cells (40, 42, 67), exhausted Tregs displayed increased apoptosis and underwent metabolic remodeling that reduced metabolic flexibility. This poor metabolic flexibility in exhausted Tregs could underlie the difference in suppressive function in vitro versus in vivo. In comparison to in vitro cultures with saturated metabolite availability, limited availability of metabolic substrates in vivo could contribute to the profound loss of TS-CAR Treg function in vivo. However, other mechanisms could also contribute to the in vivo findings that may not be reflected in vitro. Although not significant, there was a trend toward lower in vitro engraftment of TS-CAR Tregs. A similar reduction in engraftment has been reported in exhausted Tconvs (8, 15), suggesting this effect—which could be a consequence of increased death and/or decreased cell proliferation—is not Tregs specific.
We noted phenotypic similarities and differences between Treg and Tconv exhaustion. The TS-CAR induced expression of typical exhaustion-related surface markers (e.g., PD-1, TIM-3, LAG-3), but transcriptional analysis also revealed upregulation of several Treg-specific markers, such as FCER2 (CD32), IL1R2 (IL1 decoy receptor), CD276 (B7-H3), and ITGB8 (integrin beta 8), that could be used to monitor this state. Gene expression analysis revealed high expression of exhaustion-related transcription factors, including NR4A1 and 3, BATF, and PRDM1, suggesting there is at least some overlap with the transcriptional program that induces exhaustion in Tregs and in T cells.
Beyond phenotypic changes, exhaustion is associated with stable epigenetic changes such as changes in chromatin accessibility and DNA methylation (51, 68, 69). As with the transcriptomic data, ATAC-seq revealed similarities between exhausted Tregs and Tconvs, such as the increase accessibility of the PRDM1 and ZBTB32 loci. ATACseq also revealed that as for CD8+ and CD4+ Tconvs (8, 65), TS-CAR Tregs have increased chromatin accessibility in AP-1, JunB, BATF, RUNX, and ETS-1 motifs. In TS-CAR Tconvs, changes in chromatin accessibility of AP-1, and RUNX and ETS family transcription factor motifs are involved in exhaustion establishment and maintenance, respectively (65). Moreover, inhibitory AP-1 family transcription factors promote exhaustion by creating an imbalance between activating and immunoregulatory AP-1/IRF complexes (8).
To study the effects on DNA methylation, we took advantage of a “progenitor score”, finding that both TS and non-TS CARs had lower scores compared to Tconvs, suggesting that Tregs generally have a more differentiated/exhausted methylome. This conclusion was supported by GSEA analysis for an exhaustion program previously described in CD8+ T cells. Interestingly, expression of the TS-CAR in Tregs did not significantly alter the MPI or the GSEA score. A consideration is that most data on T cell exhaustion used bulk populations of CD8+ and/or CD4+ T cells, whereas we started from naive Tregs (CD4+CD127loCD25hi CD45RA+CD62Lhi) and Tconvs (CD4+CD45RA+). Thus, it is possible that more than 12 d of culture would be required to see additional changes in methylation (51). Moreover, Tregs may be generally resistant to DNA remodeling as they only showed limited changes in DNA methylation even after FOXP3 ablation (70).
Nevertheless, TS-CAR expression did impact methylation of loci encoding some exhaustion-related transcription factors, with effects substantiated by significant differences in protein expression. The manufacturing process of human Tregs for adoptive immunotherapy can result in similar epigenetic changes. Ou et al. performed genome-wide DNA methylation profiling of CD25+CD127− (i.e., not enriched for naive) Tregs subjected to repetitive TCR-mediated stimulation (3), finding progressive promotor hypomethylation of loci encoding inhibitory receptors implicated in Tconv exhaustion, such as PDCD1, HAVCR2, TIGIT, LGALS1, and CD5, which were associated a corresponding increase in protein expression (3).
An outstanding question is whether or not exhaustion will also be relevant to polyclonal, expanded Tregs and/or those genetically engineered with non-CAR receptors. Hippen et al reported that naive, blood Tregs expanded for up to 50 d with artificial antigen-presenting cells expressed high levels of CTLA-4, TIGIT, and LAG-3, but did not take on a gene expression signature that aligned with that induced by chronic viral infection (71). On the other hand, Ou et al. showed that repeated Treg stimulation with anti-CD3/CD28 beads resulted in significant hypomethylation in a number of promoters controlling expression of exhaustion-related genes [e.g., HAVCR2 (encoding for TIM-3), TIGIT, and CD160] (3). Our comprehensive set of data provide an important reference set which can be used for further study of exhaustion in other types of therapeutic Treg products and also highlight the need for in vivo functional studies to fully understand the biological relevance of phenotypic and epigenetic changes.
Finding ways to mitigate Treg exhaustion is critical for the continued development of successful CAR Treg adoptive therapy. Our data highlight that for optimal function, engineered receptors should be designed to limit antigen-independent activity. They also show the need to assess suppressive function in vivo, as similar to exhausted T cells, in vitro assays may not fully reveal functional defects. An open question is how repeated exposure of a Treg to its target in vivo would affect long-term function. For example, even in the absence of tonic signaling, how the continuous presence of an auto or alloantigen would affect a CAR- or TCR-engineered Tregs will be an important future direction. In this first formal study of Treg exhaustion, we report several similarities and differences with Tconv exhaustion and provide comprehensive expression and epigenetic data that can be used to “benchmark” states of Treg exhaustion in future studies.
Materials and Methods
Study Approval.
This study was approved by the University of British Columbia Clinical Research Ethics Board (H18-02553) and Canadian Blood Services Research Ethics Board (REB 2015.028 and REB_2020.038). Healthy volunteers gave written informed consent. Animal protocols were approved by the UBC Animal Care Committee (A18-0180).
T cell Sorting.
CD4+ T cells were isolated from healthy adults using the RosetteSep human CD4+ T cell enrichment cocktail (STEMCELL Technologies, 15062) followed by a density gradient isolation (STEMCELL Technologies, 07861). Naive Tregs were sorted from a CD25 enriched fraction ((Miltenyi, 130-092-983) CD4+CD127loCD25hi CD45RA+CD62Lhi cells and naive CD4+ Tconvs were sorted from the CD25 negative fraction as live CD4+ CD45RA+CD62Lhi cells using a MoFlo® Astrios (Beckman Coulter). The cells were resuspended at 1 × 106 cells/ml in XH media [X-Vivo 15 (Lonza) with 5% human serum, 1% penicillin/streptavidin, 1% glutamine, and phenol red] containing 300U IL2/mL (Proleukin).
Viral Vectors and Transduction.
MSGV retroviral vectors and retroviral supernatant encoding the CD19-28ζ and GD2-28ζ CARs were produced as previously described (8). For the retroviral transduction, sorted cells were stimulated with a three bead:1 cell ratio of Dynabeads in XH media containing 300U IL2/mL (Proleukin) as above. On the following day, nontissue culture-treated 96-well plates were coated overnight at 4 °C with 1 mL retronectin (Takara) at 25 μg/mL in PBS. On days 2 and 3, plates containing retronectin were washed with PBS and blocked with 2% BSA for 15 min. Then, 100 mL of thawed retroviral supernatant was then added to the well and centrifuged for 2h at 32 °C at 3,200 rpm before the addition of the same volume of media containing the activated cells. Media was refreshed on day 5. At day 7, the cells were collected and stained with a fixable viability dye (FVD, Thermo Fisher Scientific), CD4 (clone RPA-T4, BD), and an anti-14g2a idiotype antibody (clone 1A7, NIH) or Protein L (genscript) to sort cells expressing the GD2- or CD19-specific CARs, respectively. Live CD4+CAR+ cells or live CD4+ cells (for the untransduced controls) were sorted (MoFlo® Astrios) and resuspended at 0.5 × 106 cells/ml in XH media containing 300U IL2/mL for an addition 4 to 5 d. Throughout cell culture, cell number and viability were assessed using the Cellometer auto2000 and an AOPI Staining solution (Nexelcom Bioscience). Only cells that were > viable and, for Tregs, >75% FOXP3, were used.
Flow Cytometry.
Cells were stained with fixable viability dye (FVD, Thermo Fisher Scientific, 65-0865-14) and surface markers in PBS for 30 min at 4 °C before fixation and permeabilization using eBioscience FOXP3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific, 00-5523-00) and staining for intracellular proteins. Samples were analyzed on a BD LSR II (BD Bioscience) or a Cytoflex (Beckman Coulter), and the results were analyzed using FlowJo Software version 10.5.3. We used an FVD dye (Thermo Fisher Scientific, 65-0865-14), and the following antibodies from BD Biosciences: CD4 (RPA-T4), GARP (7B11), Ki67 (B56), CTLA4 (BNI3), CD8 (HIT8a), CD3 (UCHT1), IL-2 (MQ1-17H12), Blimp-1 (6D3), and an APC-Streptavidin conjugated antibody (554067). We also used CD25 (4E3) and TOX (REA473) antibodies from Miltenyi and the following antibodies from eBioscience: CD127 (eBioRDR5), CD45RA (HI100), CD62L (DREG-56), FOXP3 (236A/E7), LAG-3 (3DS223H), CD4 (RPA-T4) and TNF-a (MAb11), and IL-17A (eBio64DEC17). CD8 (RPA-T8), TIM-3 (F38-2E2), PD-1 (EH12.2H7), CD69 (FN50), IFN-γ (4S.B3), and Helios (22F6) were from Biolegend and GITR (110416) and LAP (27232) were from R&D, and TCF1/7 (C63D9) was from Cell Signaling.
Apoptosis was assessed using the apoptosis/necrosis detection kit (Abcam, ab176749) according to the manufacturer’s instructions. For intracellular cytokine staining, 0.1 × 106 cells were activated with PMA (10 ng/mL) and ionomycin (500 ng/mL) in the presence of Brefeldin A (10 μg/mL) for 4 h at 37 °C. Surface and intracellular staining were done as described above.
To monitor human cell engraftment in mice, 50 μL of blood was collected weekly and at the endpoint. The spleen was also collected at endpoint and weighed, and 3 to 6 μg was used for staining. Red blood cells were lysed with ammonium chloride, and remaining cells were resuspended in PBS containing anti-mouse CD16/32 (Thermo Fisher Scientific, 14-0161-82) for 10 min and then stained for extracellular markers using fixable viability dye (FVD; Thermo Fisher Scientific, 65-0865-14), anti-mouse CD45 (Thermo Fisher Scientific, 25-0451-82), anti-human CD45 (BD Biosciences, HI30), CD4 (Biolegend, RPA-T4), and CD8 (eBioscience, HIT8a) as previously described (72) and anti HLA-A2 (Biolegend, 343306). Ten thousand counting beads were added to every sample (Thermo Fisher Scientific, 01-1234-42).
TSDR.
DNA was isolated and bisulfite-converted using the EZ DNA Methylation-Direct Kit (Cedarlane labs, D5021). Methylation of the TSDR was calculated as previously described (70).
Seahorse Extracellular Flux Assay.
Seahorse XF Cell mito stress test assays were done using a Seahorse XFe96 Extracellular Flux Analyzer (Agilent) according to the manufacturer’s instructions. The plate was coated with Cell-Tak solution (BD) and both the plate and 25 mL of XF calibrant (Agilent) were placed overnight at 37 °C in a non-CO2 incubator. On the day of the assay, Cell-Tak was removed, washed with PBS, and cells were counted and resuspended in Seahorse XF RPMI media, pH 7.4 (Agilent), supplemented with 1% FBS, 1mM glutamax, and 10 mM glucose. Then, 2 × 105 live cells/well were plated in triplicate when possible. Oligomycin (1 μM), FCCP (1.5 μM), and Rotenone/Antimycin A (1 μM) (all Sigma) were added as indicated in ports A, B, and C.
Suppression Assay.
Allogeneic PBMCs were labeled with ef450 and Tregs with ef670 (ThermoFischer Scientific, 65-0842 and 65-0840, respectively). Then, 0.1 × 106 PBMCs were then plated in a 96-well plate with CD3/28 Dynabeads at a 1:16 cell:bead ratio. Labeled Tregs were added at the indicated ratios. After 4 d, cells were stained extracellularly and CD4+, CD8+, and Treg proliferation was assess by flow cytometry. Suppression was determined as 100-(Division index with Tregs/Division index without Tregs *100).
Xenogeneic Graft-versus-Host Disease.
Eight- to 12-wk-old male and female NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, The Jackson Laboratory, bred in house) received whole-body irradiation (150 cGy, RS-2000 Pro Biological System), and 1 d later, i.v. injected with PBS or 6 × 106 PBMCs in the absence or presence of 3 × 106 allogeneic UT or TS-CAR Tregs. Graft versus host disease was scored on the basis of weight, fur texture, posture, activity level, and skin integrity, with 0 to 3 points per category as described (72). Mice were killed during the course of the experiment if they had a total score of 6 or a score of 3 in any category. Flow cytometric analysis to measure human immune cell engraftment is described above.
RNA Sequencing.
RNA was extracted from 0.25 × 106 cells at day 12 of cell culture using the Monarch total RNA miniprep kit (New England Biolabs, T2010S) and quantified using the Qubit RNA HS Assay kit (Thermo Fisher Scientific, Q32855) according to the manufacturer’s instructions. RNA purity (RNA Integrity Number) was assessed using the RNA 6000 Pico assay (Agilent, 5067-1513), and samples were prepared using the TruSeq stranded mRNA library kit (Illumina) on the Illumina Neoprep automated nanofluidic prep instrument. Illumina NextSeq 500 with Paired End 42 bp × 42 bp reads was used for sequencing and demultiplexed read sequences and sequences were then aligned to the Homo sapiens (PAR-masked)/hg19 reference using STAR aligner (version 2.5.0a) and the RNA-Seq Alignment App (version 1.1.0) on Illumina Basespace. Demultiplexed read sequences and RnaReadCounter [part of the internal analysis tool IsisRNA (version 2.6.25.18)] were used for counting the number of aligned reads, as described previously (73). In R, raw count matrices were generated using HTSeq (v0.11.2), then scale factors were calculated to take into account differences in library sizes using edgeR (v3.24.3), and normalization was performed using limma (v3.38.3) as in (Law et al., 2016). Log (CPM) and visualization were performed using ggplot2 (3.2.1), RColorBrewer (v1.1.2), tibble (2.1.3), pheatmap (v1.0.12), stats (v3.5.1), and gplots (v3.0.1.2). The Gene Set Enrichment Analysis was performed with fgsea (v1.8.0) and Hallmark gene sets from MSigDB (v6.2). The code used for data analysis is available on GitHub: https://github.com/fransilvion/RNA_seq_Lamarche. RNAsequencing data are deposited on NCBI GEO, GSE153384.
Cell Preparation for ATAC-Seq and WGBS.
For ATAC-seq, 50,000 cells were taken from day 12 cultures and resuspended in Immunocult (STEMCELL) and processed immediately. For WGBS, 50,000–100,000 cells were taken from day 12 cultures and frozen as a dry pellet at -80 until transport for WBGS.
ATAC-Seq.
ATAC-Seq libraries from sorted T-cells were prepared using an adapted Omni-ATAC protocol as previously described, with minor modifications (74). Briefly, 50,000 cells were pelleted then lysed to isolate nuclei. For tagmentation, supernatants were removed and nuclei were resuspended in 50 μL of Tn5 transposition mix (100 nM Tn5 (Illumina, 20034197), 0.01% digitonin, 0.1% Tween-20 in 1× TD Buffer (Illumina, 20034197), and incubated at 37 °C for 30 min in a thermomixer while shaking at 700 rpm. Tagmented DNA was purified using a Qiagen Min-Elute PCR clean-up Kit (Qiagen, 28004) and PCR amplified for 8 to 13 cycles using Q5 High-Fidelity DNA polymerase (NEB, M0491S) with primers containing Illumina Nextera adaptors for library generation. The libraries were then size selected (described in ref. 75) with SPRISelect Beads (Beckman-Coulter, Cat# B23318), and concentrations were measured by a Qubit fluorometer using the Qubit dsDNA HS Assay kit (Life Technologies, Q32851) followed by bioanalyzer verification. Libraries were sequenced on the NextSeq 2000 platform (paired-end, 50 bp read lengths).
WGBS Data Processing and MPI Calculation.
For WGBS, 50,000–100,000 cells were taken from day 12 cultures and frozen as a dry pellet at −80 until transport. Sequencing data were aligned to the hg19 reference genome using the BSMAP 2.9.0 software (76). Differential methylation analysis was done by R package DSS 2.34 (75). Basic two-group comparisons were run, and P value threshold of 0.01 was used. The multipotency index calculation was based on 245 CpG sites listed in ref. 50. One naive and one HIV-exhausted sample were used to normalize the MPI scores. Heatmaps were generated using methylation levels from the 245 CpG sites, and z score normalized values were plotted. Pre-ranked Gene Set Enrichment Analysis (77) analysis was performed using DMR tables. Ranks for all genes were generated from areaStats column of DMR tables. Only one DMR with the largest absolute rank was used to represent a gene. The progenitor exhausted gene signature was derived from supplemental table 4 of ref. 52.
Statistical Analysis.
All statistics were done using Prism version 8.4.2 and are described in the figure legends. To compare Tregs, one-way Anova with Turkey’s comparisons test was done. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) FDN-154304. C.L. was supported by a CIHR fellowship and a KRESCENT award and is now supported by a Fonds de Recherche en Santé du Québec junior 1 and a Cole foundation award. M.K.L. receives a salary award from the BC Children's Hospital Research Institute and is a Canada Research Chair in Engineered Immune Tolerance. We thank Dr. Ramon Klein-Geltink for advice and critical reading of the manuscript and Dr. Lixin Xu for expert flow cytometry support.
Author contributions
C.L., K.W.-H., T.M., D.T.S.L., M.S.K., C.C.Z., and M.K.L. designed research; C.L., K.W.-H., T.M., D.T.S.L., Q.H., A.B., K.E., and C.N.Q. performed research; E.W.W. and C.L.M. contributed new reagents/analytic tools; C.L., K.W.-H., T.M., D.T.S.L., Q.H., A.B., K.E., G.E.N., and C.C.Z. analyzed data; and C.L., T.M., C.C.Z., E.W.W., C.L.M., and M.K.L. wrote the paper.
Competing interests
The authors have organizational affiliations to disclose. E.W.W. consults for Lyell Immunopharma, C.L.M. consults for Lyell Immunopharma, E.W.W. holds equity in Lyell Immunopharma, C.L.M. holds equity in Lyell Immunopharma, M.K.L. has patent applications related to CAR based immunotherapy, C.L.M. holds several patent applications in the area of CAR based immunotherapy.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Requests for the protocols, resources, and reagents should be directed to and will be fulfilled by author M.K.L. (mlevings@bcchr.ca). The data supporting the results in this study are available within the paper and its SI Appendix. For RNA sequencing, the code used for data analysis is available on GitHub: https://github.com/fransilvion/RNA_seq_Lamarche. RNAsequencing data are deposited on NCBI GEO, GSE153384 (78). ATAC-seq data are deposited on NCBI GEO, GSE209835 (79). DNA methylation data are deposited on NCBI GEO Gene Expression Omnibus, GSE209671 (80).
Supporting Information
References
- 1.Boardman D. A., Levings M. K., Emerging strategies for treating autoimmune disorders with genetically modified Treg cells. J. Allergy Clin. Immunol. 149, 1–11 (2022). [DOI] [PubMed] [Google Scholar]
- 2.MacDonald K. N., Piret J. M., Levings M. K., Methods to manufacture regulatory T cells for cell therapy. Clin. Exp. Immunol. 197, 52–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ou K., et al. , Strong expansion of human regulatory T cells for adoptive cell therapy results in epigenetic changes which may impact their survival and function. Front. Cell Dev. Biol. 9, 751590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fribourg M., et al. , T-cell exhaustion correlates with improved outcomes in kidney transplant recipients. Kidney Int. 96, 436–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorp E. B., Stehlik C., Ansari M. J., T-cell exhaustion in allograft rejection and tolerance. Curr. Opin Organ Transplant. 20, 37–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saeidi A., et al. , T-cell exhaustion in chronic infections: Reversing the state of exhaustion and reinvigorating optimal protective immune responses. Front. Immunol. 9, 2569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynn R. C., et al. , c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahbaz S., et al. , Galectin-9 and VISTA Expression Define Terminally Exhausted T Cells in HIV-1 Infection. J. Immunol. 204, 2474–2491 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Zou D., et al. , T cell exhaustion is associated with antigen abundance and promotes transplant acceptance. Am. J. Transplant 20, 2540–2550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long A. H., et al. , 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber E. W., Maus M. V., Mackall C. L., The emerging landscape of immune cell therapies. Cell 181, 46–62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finney O. C., et al. , CD19 CAR T cell product and disease attributes predict leukemia remission durability. J. Clin. Invest. 129, 2123–2132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zebley C. C., Gottschalk S., Youngblood B., Rewriting history: Epigenetic reprogramming of CD8(+) T cell differentiation to enhance immunotherapy. Trends Immunol. 41, 665–675 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber E. W., et al. , Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science 372, eaba1786 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., et al. , NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraietta J. A., et al. , Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller B. C., et al. , Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., et al. , T Cell Dysfunction and Exhaustion in Cancer. Front. Cell Dev. Biol. 8, 17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bettelli E., Dastrange M., Oukka M., Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. U.S.A. 102, 5138–5143 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A., Sakaguchi S., Targeting Treg cells in cancer immunotherapy. Eur. J. Immunol. 49, 1140–1146 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Pesenacker A. M., et al. , A regulatory T-cell gene signature is a specific and sensitive biomarker to identify children with new-onset type 1 diabetes. Diabetes 65, 1031–1039 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Sadlon T. J., et al. , Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J. Immunol. 185, 1071–1081 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Schlaphoff V., et al. , Dual function of the NK cell receptor 2B4 (CD244) in the regulation of HCV-specific CD8+ T cells. PLoS Pathog. 7, e1002045 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agresta L., Hoebe K. H. N., Janssen E. M., The emerging role of CD244 signaling in immune cells of the tumor microenvironment. Front. Immunol. 9, 2809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opstelten R., et al. , GPA33: A marker to identify stable human regulatory T cells. J. Immunol. 204, 3139–3148 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Liu X., et al. , Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charbonnier L. M., et al. , Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat. Immunol. 20, 1208–1219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olesin E., Nayar R., Saikumar-Lakshmi P., Berg L. J., The transcription factor Runx2 is required for long-term persistence of antiviral CD8(+) memory T cells. Immunohorizons 2, 251–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blank C. U., et al. , Defining “T cell exhaustion”. Nat. Rev. Immunol. 19, 665–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., et al. , The transcription factor TCF1 preserves the effector function of exhausted CD8 T cells during chronic viral infection. Front. Immunol. 10, 169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beier U. H., et al. , Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. FASEB J. 29, 2315–2326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saravia J., et al. , Homeostasis and transitional activation of regulatory T cells require c-Myc. Sci. Adv. 6, eaaw6443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pacella I., Piconese S., Immunometabolic checkpoints of Treg dynamics: Adaptation to microenvironmental opportunities and challenges. Front. Immunol. 10, 1889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howie D., et al. , Foxp3 drives oxidative phosphorylation and protection from lipotoxicity. JCI Insight 2, e89160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angelin A., et al. , Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293.e1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerriets V. A., et al. , Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 17, 1459–1466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang R., et al. , The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKinney E. F., Smith K. G. C., Metabolic exhaustion in infection, cancer and autoimmunity. Nat. Immunol. 19, 213–221 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Kawalekar O. U., et al. , Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016). [DOI] [PubMed] [Google Scholar]
- 41.van der Windt G. J., et al. , Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schurich A., et al. , Distinct Metabolic Requirements of Exhausted and Functional Virus-Specific CD8 T Cells in the Same Host. Cell Rep. 16, 1243–1252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu S. H., Yeh L. T., Chu C. C., Yen B. L., Sytwu H. K., New insights into Blimp-1 in T lymphocytes: A divergent regulator of cell destiny and effector function. J. Biomed. Sci. 24, 49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin H. M., et al. , Transient expression of ZBTB32 in anti-viral CD8+ T cells limits the magnitude of the effector response and the generation of memory. PLoS Pathog. 13, e1006544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue X., Lio C. J., Samaniego-Castruita D., Li X., Rao A., Loss of TET2 and TET3 in regulatory T cells unleashes effector function. Nat. Commun. 10, 2011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan L., et al. , Down-regulation of Tet2 is associated with Foxp3 TSDR hypermethylation in regulatory T cell of allergic rhinitis. Life Sci. 241, 117101 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Yang J. M., et al. , NAC1 modulates autoimmunity by suppressing regulatory T cell-mediated tolerance. Sci. Adv. 8, eabo0183 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauken K. E., et al. , Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnett K. R., et al. , ATAC-Me captures prolonged DNA methylation of dynamic chromatin accessibility loci during cell fate transitions. Mol. Cell 77, 1350–1364.e1356 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdelsamed H. A., et al. , Beta cell-specific CD8(+) T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol. 21, 578–587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zebley C. C., et al. , CD19-CAR T cells undergo exhaustion DNA methylation programming in patients with acute lymphoblastic leukemia. Cell Rep. 37, 110079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galletti G., et al. , Two subsets of stem-like CD8(+) memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 21, 1552–1562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bordon Y., TOX for tired T cells. Nat. Rev. Immunol. 19, 476 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Scott A. C., et al. , TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan O., et al. , TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin H., et al. , A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang S., Cobb D. A., Bhadra R., Youngblood B., Khan I. A., Blimp-1-mediated CD4 T cell exhaustion causes CD8 T cell dysfunction during chronic toxoplasmosis. J. Exp. Med. 213, 1799–1818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine A. G., Arvey A., Jin W., Rudensky A. Y., Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vahl J. C., et al. , Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity 41, 722–736 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Anderson A. C., Joller N., Kuchroo V. K., Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 44, 989–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gautron A. S., Dominguez-Villar M., de Marcken M., Hafler D. A., Enhanced suppressor function of TIM-3+ FoxP3+ regulatory T cells. Eur. J. Immunol. 44, 2703–2711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gianchecchi E., Fierabracci A., Inhibitory receptors and pathways of lymphocytes: The role of PD-1 in Treg development and their involvement in autoimmunity onset and cancer progression. Front. Immunol. 9, 2374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wing K., et al. , CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Gupta S., et al. , Allograft rejection is restrained by short-lived TIM-3+PD-1+Foxp3+ Tregs. J. Clin. Invest. 122, 2395–2404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gennert D. G., et al. , Dynamic chromatin regulatory landscape of human CAR T cell exhaustion. Proc. Natl. Acad. Sci. U.S.A. 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamarthée B., et al. , Transient mTOR inhibition rescues 4–1BB CAR-Tregs from tonic signal-induced dysfunction. Nat. Commun. 12, 6446 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wherry E. J., T cell exhaustion. Nat. Immunol. 12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Prinzing B., et al. , Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci. Transl. Med. 13, eabh0272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sen D. R., et al. , The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lam A. J., et al. , Optimized CRISPR-mediated gene knockin reveals FOXP3-independent maintenance of human Treg identity. Cell Rep. 36, 109494 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Hippen K. L., et al. , Multiply restimulated human thymic regulatory T cells express distinct signature regulatory T-cell transcription factors without evidence of exhaustion. Cytotherapy 23, 704–714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dawson N. A., et al. , Systematic testing and specificity mapping of alloantigen-specific chimeric antigen receptors in regulatory T cells. JCI Insight 4, e123672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoeppli R. E., et al. , Tailoring the homing capacity of human Tregs for directed migration to sites of Th1-inflammation or intestinal regions. Am. J. Transplant 19, 62–76 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Corces M. R., et al. , An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng H., Conneely K. N., Wu H., A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 42, e69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xi Y., Li W., BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinform. 10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamarche C., Levings M. K., RNA-seq profiling of human Tregs and conventional CD4+ cells that were left untransduced or transduced with a non-tonic signaling (CD19) or a tonic signaling (HA) CAR. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153384. Deposited 26 June 2020.
- 79.Lamarche C., Levings M. K., Brown A., Ward-Hartstonge K., ATAC-seq profiling of human Tregs and conventional CD4+ cells that were transduced with a non-tonic signaling (CD19) or a tonic signaling (HA) CAR. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE209835. Deposited 26 July 2022.
- 80.Lamarche C., Ward-Hartstonge K., Levings M. K., Tonic-signaling chimeric antigen receptors drive human regulatory T cell exhaustion. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE209671. Deposited 25 July 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Requests for the protocols, resources, and reagents should be directed to and will be fulfilled by author M.K.L. (mlevings@bcchr.ca). The data supporting the results in this study are available within the paper and its SI Appendix. For RNA sequencing, the code used for data analysis is available on GitHub: https://github.com/fransilvion/RNA_seq_Lamarche. RNAsequencing data are deposited on NCBI GEO, GSE153384 (78). ATAC-seq data are deposited on NCBI GEO, GSE209835 (79). DNA methylation data are deposited on NCBI GEO Gene Expression Omnibus, GSE209671 (80).