Significance
Crossing-over is a process of mixing chromosomes from mother and father to create new combinations of traits in the progeny. Plant breeders rely on it to produce new crop varieties with improved yields, resistances to new pests, and tolerance to extreme environmental conditions. Unfortunately, crossing-over events in crops are rare and do not always occur in places where breeders need them most. Thus, several methods have been developed to increase the number of crossing-over events and ensure that they occur at desired genome locations. We review these technologies and use simulations to show that they indeed can be helpful in plant breeding programs. We also discuss what needs to be done to implement these methods in breeding practice.
Keywords: recombination, crops, diversity, plant breeding
Abstract
Plant breeding relies on crossing-over to create novel combinations of alleles needed to confer increased productivity and other desired traits in new varieties. However, crossover (CO) events are rare, as usually only one or two of them occur per chromosome in each generation. In addition, COs are not distributed evenly along chromosomes. In plants with large genomes, which includes most crops, COs are predominantly formed close to chromosome ends, and there are few COs in the large chromosome swaths around centromeres. This situation has created interest in engineering CO landscape to improve breeding efficiency. Methods have been developed to boost COs globally by altering expression of anti-recombination genes and increase CO rates in certain chromosome parts by changing DNA methylation patterns. In addition, progress is being made to devise methods to target COs to specific chromosome sites. We review these approaches and examine using simulations whether they indeed have the capacity to improve efficiency of breeding programs. We found that the current methods to alter CO landscape can produce enough benefits for breeding programs to be attractive. They can increase genetic gain in recurrent selection and significantly decrease linkage drag around donor loci in schemes to introgress a trait from unimproved germplasm to an elite line. Methods to target COs to specific genome sites were also found to provide advantage when introgressing a chromosome segment harboring a desirable quantitative trait loci. We recommend avenues for future research to facilitate implementation of these methods in breeding programs.
The key aspect of a successful breeding program is the amount of genetic diversity (or genetic variance) in the founder population (1). High levels of genetic diversity are particularly important to attain new breeding targets, such as those needed to respond to climate change or new pest pressure. The main process governing the amount and distribution of genetic variation is meiotic recombination. In addition to generating new combinations of alleles underlying favorable traits, meiotic recombination is a driver of increased fitness, where it acts by purging deleterious mutations from populations (2). The rapid progress in understanding molecular mechanisms controlling the distribution of recombination events along chromosomes has brought interest in harnessing recombination to make breeding more efficient (3). In this paper, we review the current state of technology in this area, examine whether the existing technologies are indeed useful in breeding programs, and discuss which new developments are needed to make the latter possible.
Why is Recombination Important in Plant Breeding?
Breeders utilize recombination to increase variation in breeding populations in two main ways. Frequently, “wild” (i.e., unimproved) germplasm or germplasm not adapted to the intended growing environment is used as a source to introduce new desired traits into elite lines. These traits are often qualitative or influenced by only a few quantitative trait loci (QTL) exerting major effects (4). However, linkage drag can bring along with the target loci into the elite line undesirable characteristics of the donor germplasm, such as decreased agronomic performance (4, 5). It often takes several generations of backcrossing to limit the amount of donor genome “hitchhiking” with the target trait loci.
Another way to increase genetic variation of a population, if the desired variants already exist in elite breeding germplasm, are crosses among elite lines. In this case, there is no danger of bringing in agronomically undesirable traits. However, even here, it takes several generations of crossing and selection to obtain the most desired combination of traits.
Recombination Mechanisms
Meiotic recombination is initiated by the formation of double-strand breaks (DSBs) in chromosomal DNA in early meiotic prophase I (6) by a protein complex containing the topoisomerase-like protein SPO11 (7). In plants, the number of DSBs per meiosis is largely proportional to the genome size, with Arabidopsis exhibiting about 200 DSBs (8), maize ~500 (6), and wheat ~2,100 (9). Following their formation, DSBs are resected, and the single-ended DNA overhangs created in this way invade double-stranded DNA in search of homology (10). Eventually, cross-strand intermediates called double Holliday junction are formed, whose resolution leads to CO formation (11). However, this fate awaits only very few DSBs, ~4% in maize (12), as plants exhibit only from one to two COs per chromosome (13).
The majority of DSBs are repaired as non-COs (NCOs) by DNA synthesis utilizing the sister chromatid or the homologous chromosome as template (11). This process may result in a non-reciprocal replacement of the original DNA segment by the invading strand (11). If the replacing segment differs in sequence from the original segment, the result of the replacement is a gene conversion (GC). Considering the number of NCOs, the average length of DSB resection, which in maize has been found to be ~1,100 bp (14), and the level of intraspecies DNA sequence polymorphism, GCs should be a significant contributor to genetic variation in plants. However, detailed analyses of recombination products performed in Arabidopsis failed to produce evidence of substantial GC numbers (15, 16). It might be that presence of inter-parental DNA sequence polymorphism in DSB vicinity causes the DSB to be repaired as a CO rather than an NCO (17). Alternatively, the forming GCs might be repaired back to the state of the original DNA segment (18). Regardless of the mechanism, these observations imply that most of the meiotic recombination-generated genetic variation in plants comes from COs.
Within the CO pathway, there are two possibilities. Class I COs are the most common CO products constituting roughly 85% of all CO events in plants (19). These COs are formed by a group of proteins, which includes homolog of enhancer of cell invasion no.10 (HEI10) and MutL homolog (MLH) 1 and 3. Class I COs are sensitive to other COs forming nearby due to a phenomenon called CO interference. Interference prevents COs from forming close to each other (20). Therefore, class I COs on the same chromosome are limited in frequency. The second pathway produces class II COs, which are rarer than class I COs and not affected by CO interference (21). Proteins involved in class II CO formation include MMS4 and UV sensitive81 (MUS81). These proteins are not meiosis-specific and are also involved in the repair of spontaneous DNA breaks in somatic cells (22), suggesting that the class II CO pathway may be primarily utilized to resolve defective recombination intermediates. Several proteins affecting class I as well as II COs have been labeled anti-CO factors, including RecQ helicase 4 (RECQ4) (23), Fanconi Anemia of Complementation group M (FANCM) (24–26), and AAA-ATPase Fidgetin-like-1 (FIGL1) (27). RECQ4 and FANCM act by dissolving CO intermediates, while FIGL1 is thought to regulate DSB resection and limit strand invasion during DSB repair in class II CO formation. Mutating genes that encode these proteins leads to substantial CO number increases (28, 29).
CO Landscape in Crops
COs are not uniformly distributed along chromosomes but form distinct hotspots, which are usually defined as regions of 5 kb to 10 kb exhibiting recombination rates fivefold or more than the genome average (30). Centromeres have been found to be depleted of COs in all species studied (31). In addition, in plants with large and complex genomes, which includes most crops, such as wheat, barley, and maize, there are substantial pericentromeric regions that also exhibit strong CO suppression (32, 33). In contrast, in the small-genome plant Arabidopsis thaliana, CO suppression affects a relatively small centromeric/pericentromeric segment, and there is no CO rate increase along chromosome arms toward chromosome ends (34). The extent of the chromosome-end CO bias becomes larger with genome size increase. In maize, roughly one-quarter of the genome and one-fifth of the genes are in the CO-depleted pericentromeric areas (35). To put the size in perspective, the pericentromeric region of each of the largest maize chromosomes is approximately equal to the size of the entire Arabidopsis genome (36).
CO Landscape and Gene Distribution
At the fine scale, most COs are located relatively close to genes. In maize, about 90% of COs are located within 10 kb from genes, with peaks in gene promoters and terminators (37). Recombination occurring close to genes may be most beneficial from the point of view of creating genetic diversity. Recombination in promoter regions has the capacity to dramatically change the way a gene is expressed, and gene evolution frequently involves adaptive changes in regulatory sequences (38, 39). The close association of COs with genes implies that there should be a strong positive correlation between recombination rates and gene density. As gene density tends to be lower in pericentromeric regions in large-genome plants (33), such correlation could explain the chromosome-level bias of CO distribution. Indeed, studies using an early version of the maize genome sequence assembly found strong correlations between recombination rates and gene density (r = 0.767 to 0.960) (40). We re-examined this issue using the more accurate and complete recent maize genome assembly and annotation (36) utilizing recombination data from the maize Nested Association Mapping (NAM) recombinant inbred line population generated by crossing 25 diverse maize inbreds to a common parent, the B73 inbred. This population captures about ~100,000 CO events (41). Gene density was calculated in 200 kb-long intervals and computed by dividing the total number of unique genes by the interval length. Because of the limited CO number in the dataset relative to the number of genes, intervals containing genes but lacking COs were omitted. We found a positive Spearmen’s correlation coefficient of 0.7400122 (P < 2.2e−16). The same analysis conducted in rice, a species with a genome roughly one-sixth the size of maize (42), produced a correlation coefficient of −0.07860649 (P = 0.9991) indicating no significant correlation. Taken together, these results indicate that the gene density bias can only partially modulate the genetic effect of the biased CO distribution in large-genome plants. Thus, the biased CO distribution has direct impact on genetic diversity in crops.
Genetic Consequences of Biased CO Landscape
Suppression of CO events exposes genes in pericentromeric regions of chromosomes to a large deleterious mutation load over evolutionary history (43). The larger the size of pericentromeric regions, the larger this hypothetical mutation load. Deleterious mutations have the potential to impact plant fitness as well as agronomic productivity but can be purged by recombination (2). When these mutations first arise, they are at a very low allelic frequency within the population, and if recombination is efficient enough, they can be segregated out quickly.
In addition to harboring deleterious mutations, genetic linkage between loci influencing agronomic traits is important to consider. Loci harboring alleles that confer positive effects are less advantageous when they are linked to loci with alleles exerting negative effects. This phenomenon was described by Hill and Robertson in 1966 (44), who showed that loci under selection may have reduced fitness effects when they are linked to loci harboring deleterious alleles. In contrast to these repulsion phase linkages (i.e., a locus with a positive-effect allele next to a locus with a negative-effect allele), coupling linkages (i.e., a locus with a positive-effect allele next to a locus with a positive-effect allele or a locus with a negative-effect allele next to a locus with a negative-effect allele) are less affected by increased recombination. Coupling linkages may, however, affect heterosis, as absence of recombination can lead to pseudo-overdominance, which is one of the proposed mechanisms of heterosis (45). Altogether, these linkage-related phenomena indicate that recombination suppression in the pericentromeric region may have larger consequences than just accumulation of deleterious mutations.
Controlling CO Landscape by Changing DNA Methylation
Patterns of CO distribution have been associated with numerous chromatin, DNA sequence, and chromosome location characteristics in studies conducted in several plant species (46–49). A computational analysis using machine learning showed that by far the most important factor affecting CO landscape in plants is DNA methylation and chromatin openness measured by nucleosome occupancy (50). DNA methylation is of high importance in plants with large genomes, as in addition to controlling gene expression it is responsible for silencing of transposable element activity (51). Transposable elements make up substantial portions of large genomes; e.g., they constitute over 80% of the genome of maize (36). While removing DNA methylation in the small and compact Arabidopsis genome results in few immediate consequences (52), mutations in many DNA methylation-controlling genes in maize often result in embryo lethality (53).
Methylation of cytosine residues (5mC) can occur in plants at CG, CHG, and CHH sites, where H = A, C, or T (54, 55). Detailed studies in Arabidopsis have revealed that the different methylation types are conveyed by different DNA methyltransferases and depend on distinct molecular mechanisms (56, 57). Methylation at CG sites is maintained by the methylase MET1 and controlled by a DNA-replication-guided mechanism. CHG methylation is maintained by chromomethyltransferases and is heterochromatin guided. CHH methylation is maintained by DRM proteins and guided by an RNA-directed (RdDM) mechanism involving small interfering RNAs (58). The three DNA methylation types have distinct functions, although their exact roles have not been completely elucidated and may differ among species (57, 59). There are dramatic differences in DNA methylation levels among species. In maize, the average level of cytosine methylation is ~90% at CG sites, ~70% at CHG sites, and ~3% at CHH sites (60). In comparison, genome-wide DNA methylation levels in Arabidopsis at CG, CHG, and CHH sites are 24%, 6.7%, and 1.7%, respectively (56). In addition to the core DNA methylation pathway components, other proteins, such as nucleosome remodelers, also affect DNA methylation. One of the best-studied of them is decrease in DNA methylation 1 (DDM1), which is a SNF1/SNF2 protein family member and affects DNA methylation in all three sequence contexts in Arabidopsis and maize (46, 61, 62).
Mutant studies show that changing DNA methylation patterns alters CO distribution. Loss of CG methylation in the Arabidopsis met1 mutant results in CO landscape remodeling, although, interestingly, the increases are seen predominantly in the already hypomethylated chromosome arm regions (63). A similar pattern was observed in the Arabidopsis ddm1 mutant (47). The maize mop1 mutant, which shows reduced CHH methylation, exhibits elevated COs on chromosome arms (64). Maize plants defective in ddm1 exhibit similar but an even more pronounced recombination landscape alteration with steep CO increases at chromosome ends (65). In contrast, mutants affecting largely CHG methylation, cmt3 in Arabidopsis (66), and zmet2 in maize (65) exhibit increased pericentromeric COs with little to no effect anywhere else along chromosome arms. Altogether, these studies indicate that CO landscape could be manipulated in intricate ways by altering specific DNA methylation types.
Altering CO Landscape by Mutating CO Regulators
Studies in both Arabidopsis and large-genome plants: rice, tomato, and pea, have examined the possibility of increasing CO rates by altering expression of CO regulators. These studies used HEI10 overexpression as well as mutations in anti-CO genes RECQ4, FANCM, and FIGL1 (28, 29). These modifications exhibit effects on CO landscapes distinct from the effects of most DNA methylation changes, as they tend to boost COs along chromosome arms, where COs already exist and do not facilitate CO increase in pericentromeric regions (67). Plants combining several of these genetic changes have been shown to exhibit substantial elevation of CO numbers (28).
Will Altering CO Landscape Genome-Wide Make Breeding Programs More Effective?
Since it has been established that recombination is a limiting factor in creating genetic variation, it is interesting to explore whether increasing recombination rates would improve genetic gain in breeding programs. Genetic gain is the key characteristic of success in breeding programs and measures the improvement of the average value of the trait(s) of interest in response to selection (68). Genetic gain expected from selection is typically calculated as ΔG = i r σa, where ΔG is the genetic gain, i is selection intensity, r is selection accuracy (i.e., the ability to accurately identify the most beneficial individuals), and σa is the standard deviation of the additive genetic variance of the target trait within the breeding population.
Several past analyses have shown that increasing CO rates both globally and at specific chromosome sites could result in genetic gain increase and linkage drag reduction in plants (69–72) as well as animals (73). However, it is only now that detailed studies of CO landscape in large-genome crops (37, 67, 74) allow these assessments to be based on fully realistic assumptions. In addition, recent crop studies have identified new avenues for modulating DNA methylation to alter CO landscape in more elaborate ways (64, 65, 75). Given these advances, we sought to examine whether the currently available methods of CO landscape engineering are sufficient to produce meaningful benefits on the scale that makes them attractive in breeding programs.
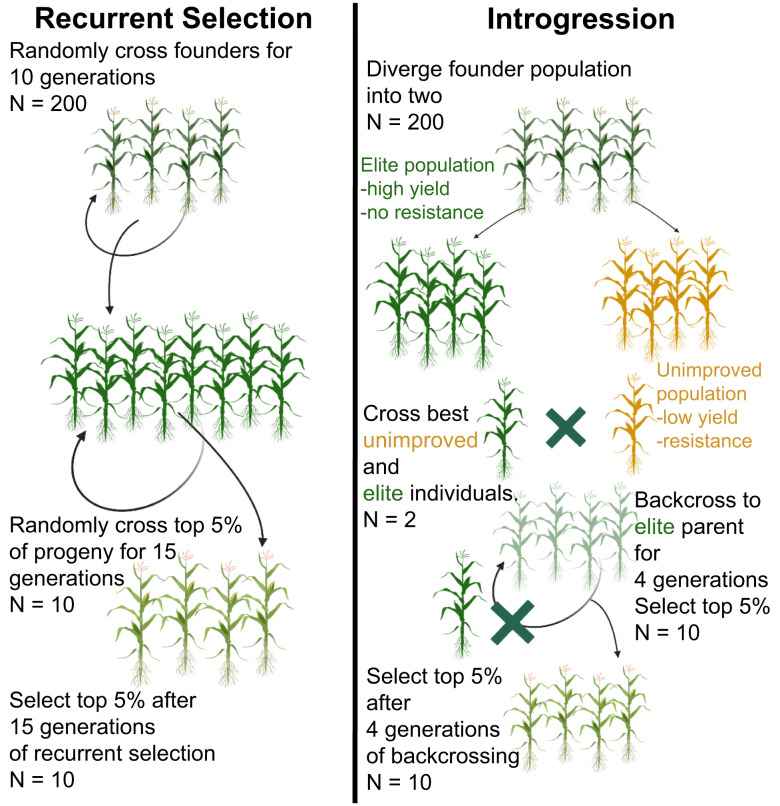
To conduct a comprehensive analysis of the impact of manipulating CO landscapes on genetic gain, we used simulations to examine two common breeding scenarios, i) intermating of two elite breeding lines followed by recurrent selection and ii) backcrossing to introgress desirable QTL from an unimproved parent into an elite breeding line (Fig. 1). We explored two crops with different genome sizes, rice (0.39 Gb) (42) and maize (2.4 Gb) (36), and used actual recombination landscapes in the two species that considered the pericentromeric suppression of COs and their elevation near chromosome ends.
Fig. 1.
Schematics of the recurrent selection and introgression scenarios used in simulations.
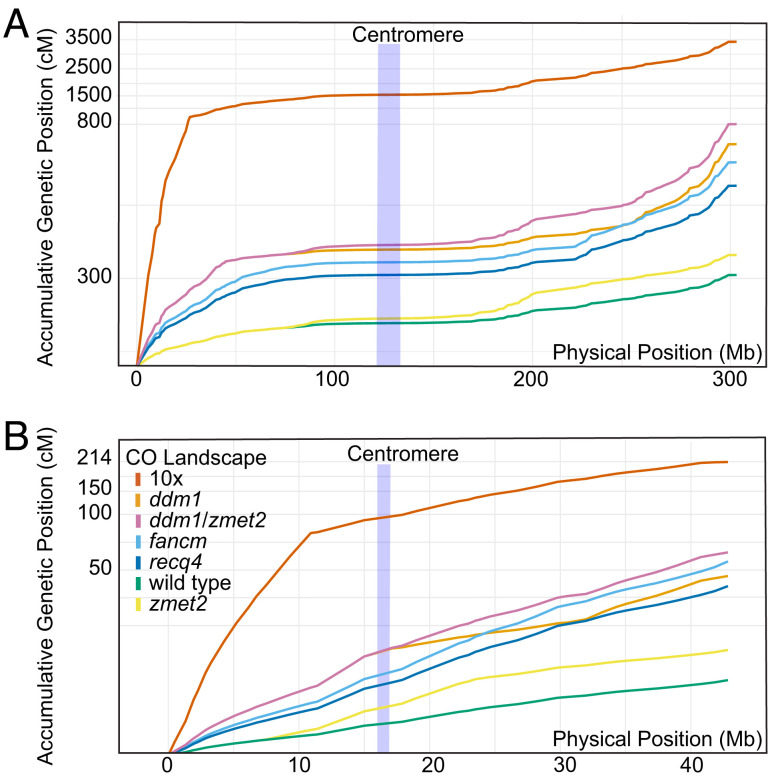
The goal of our simulation was to examine several alternative recombination landscapes (Fig. 2 and SI Appendix, Figs. S1 and S2). To do this, we modeled CO patterns from maize DNA methylation mutants showing CO increases in the pericentromeric regions (zmet2) and in chromosome arms (ddm1) as well as CO distribution of the rice fancm and recq4 mutants (59, 62). Since fancm and recq4 have not been studied in maize, their effects on recombination landscapes were extrapolated from rice to maize considering the size of the pericentromeric CO suppression region (see Materials and Methods). Similarly, the effects of maize DNA methylation mutants were extrapolated to rice, where such mutants have not been studied.
Fig. 2.
Recombination landscapes of wild type and mutants. (A) Maize chromosome 1. (B). Rice chromosome 1. Y-axis is compressed to fit the map for 10-fold global CO increase. Centromere positions are in blue.
Recurrent Selection Scenario.
The recurrent selection scenario modeled a polygenic trait underlain by 300 QTL with nearly equal additive effects, which is a genetic architecture resembling that of yield. Yield is the key target in most plant breeding programs. It is considered to be a highly polygenic trait and is most accurately predicted using an infinitesimal model, in which all loci are infinitely small and have equal effects on the trait (76). The 300 loci were placed genome-wide using gene density as a factor to guide their distribution (see Material and Methods). The simulated populations were tracked for 15 generations. Although most breeding programs perform recurrent selection for fewer than five generations, analyzing selection effects over a longer period should provide information on the limit of the potential impact of altered recombination landscapes.
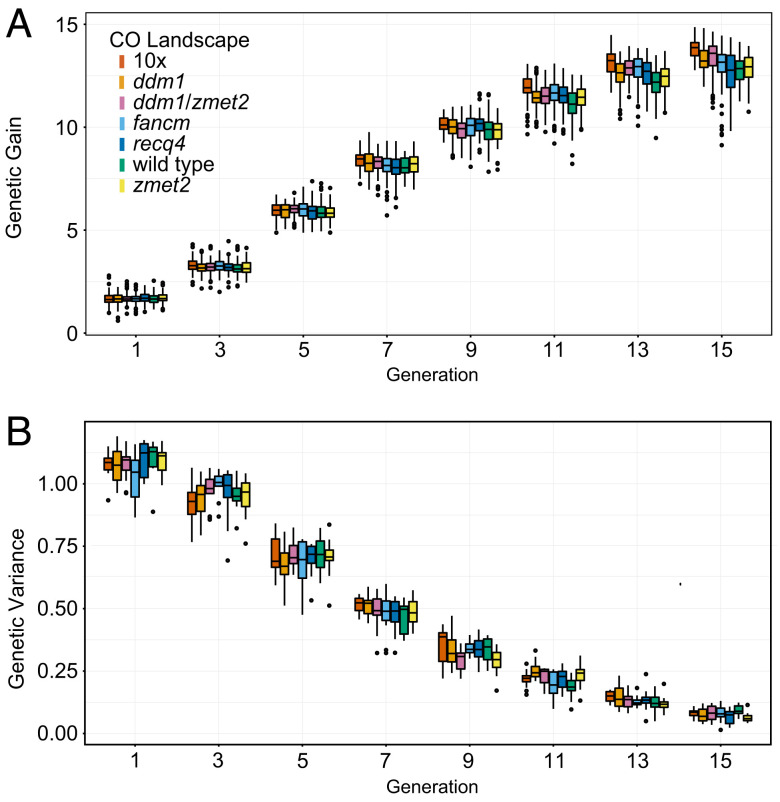
In maize, all existing mutant CO landscapes (i.e., ddm1, fancm, recq4, and zmet2) showed positive effects on genetic gain (Fig. 3A), exhibiting in generation 6 (i.e., after five generations of selection) an average 1.6% increase over the amount of genetic gain in wild type at the same generation. In generation 15, the increase was 3.1%. As these increases were modest, we created two idealistic scenarios, a global 10-fold CO rate increase and a hypothetical double mutant of ddm1 and zmet2 (Fig. 2 and SI Appendix, Figs. S1 and S2). Although the two latter CO landscapes have not yet been demonstrated in crops, they could be conceivably established by extending the currently existing CO landscape manipulation methods. We found that the 10-fold global increase outperformed the existing CO landscapes, yielding a 3.1% increase over the amount of genetic gain in wild type in generation 6 and a 11.0% increase in generation 15 (Fig. 3A). A similar conclusion was reached in rice: All existing CO landscapes showed positive effects on genetic gain with an average of 3.3% increase over the amount of genetic gain in wild type at generation 6 and 9.7% at generation 15 (SI Appendix, Fig. S3A). The 10-fold global CO rate increase in rice yielded a genetic gain increase of 9.7% over the amount of genetic gain in wild type at generation 6 and 27.5% at generation 15 (SI Appendix, Fig. S3A).
Fig. 3.
Simulating a maize breeding population over 15 generations of recurrent selection. (A) Genetic gain. (B) Genetic variance.
We also tracked genetic variance over the 15 generations of recurrent selection to determine how the various recombination landscapes affected the dynamics of genetic diversity (Fig. 3B). In generation 6, wild type had a similar amount of genetic variance as most mutant populations. However, in generations 7 to 13, the populations with mutant CO landscapes had retained more genetic variance than wild type. The increase in genetic variance was followed by an increase in genetic gain a couple generations later. By generation 15, most genetic variance was depleted in both mutants and wild type, and all populations again exhibited similar variance amounts. Similar trends were observed in rice (SI Appendix, Fig. S3B).
In addition to the polygenic trait, we examined whether an oligogenic trait could benefit from a recombination landscape change in a recurrent selection program. Pest and pathogen resistance are often oligogenic traits, exhibiting several contributing loci with known genomic positions (77). However, when we simulated a trait conditioned by 15 loci distributed genome-wide, we found that none of the mutant CO landscapes had detectable impact on genetic gain (SI Appendix, Fig. S4A). This outcome mirrors the results of Tourette et al. (69), who discovered that the more QTL a trait had, the more impact increased recombination had on genetic gain.
Altogether, our simulations show potential for genetic gain improvements through utilization of mutant recombination landscapes in breeding highly polygenic traits, such as yield. CO landscapes based on the existing mutants resulted in modest genetic gain increases. However, a more substantial improvement of ~10% over the amount of genetic gain in wild type in maize and nearly 30% in rice was achievable when using the idealistic recombination landscapes. Additionally, genetic variance was maintained over more generations in the populations with mutant recombination landscapes, which would improve the effectiveness of selection of the most desirable trait combinations.
Introgression Scenario.
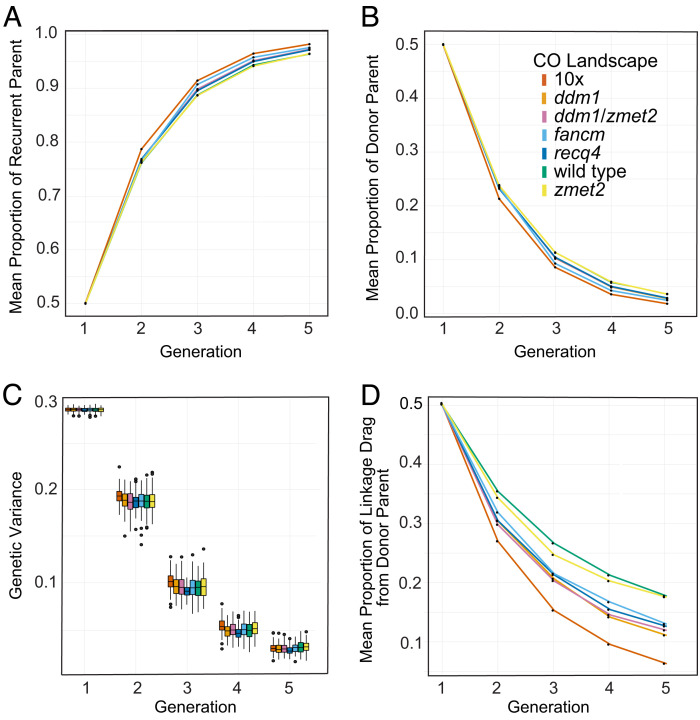
In the introgression scenario, we also modeled a yield-like polygenic trait with 300 QTL but added disease resistance loci. Introgressing disease resistance from unimproved germplasm to elite lines is a common practice in plant breeding programs. We examined a situation in which we believed increasing recombination rates could be most beneficial: three linked disease resistance loci placed about ~20 to 30 centiMorgans (cM) apart and interspersed with QTL for the yield-like trait. The three disease resistance loci were positioned either on a chromosome arm and away from the pericentromeric region or, alternatively, within the pericentromeric region. In maize, in both cases, the simulations showed that only the 10-fold global recombination rate increase accelerated the return to the recurrent parent genome and loss of the donor parent genome over four generations of backcrossing (Fig. 4 A and B). Genetic variance was maintained in similar ways in all populations, both mutant and wild type (Fig. 4C). In rice, the proportions of the recurrent and donor parent genomes (SI Appendix, Fig. S5 A and B) and the residual genetic variance recombination (SI Appendix, Fig. S4C) did not change with the use of any of the mutant recombination landscapes.
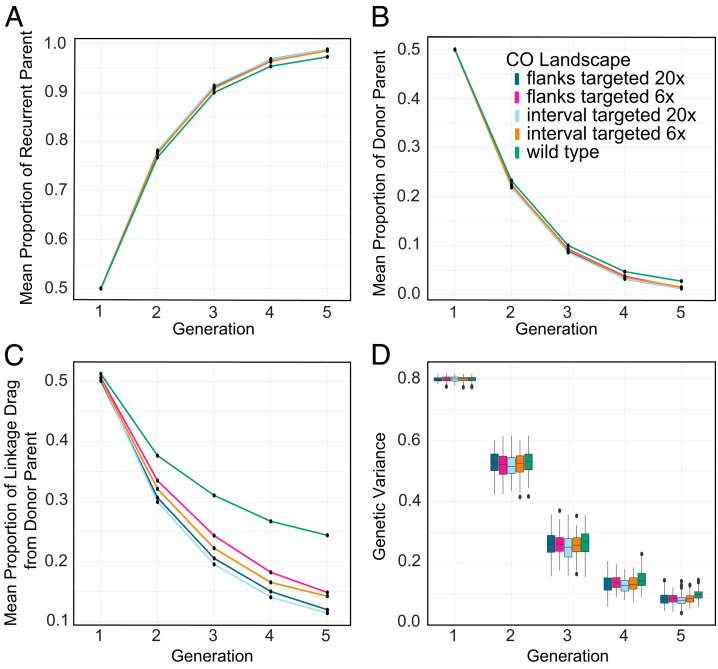
Fig. 4.
Simulating introgression of disease resistance loci placed on chromosome 1 in maize. Generation 1 represents F1 progeny from the initial elite parent x wild parent cross. Generations 2 to 4 represent progenies from backcrossing to the elite parent. (A) Proportion of recurrent (elite) parent. (B) Proportion of donor (wild) parent. (C) Genetic variance. (D) Linkage drag around the introgression target (disease resistance) loci.
To measure the effectiveness of the backcrossing process in eliminating the potentially undesirable donor parent characteristics linked to the introgressed disease resistance loci, we calculated the amount of linkage drag. To accomplish this goal, only the chromosome with the donor loci was considered and progeny genotypes for this chromosome were tracked in each generation. In both maize and rice when the disease resistance loci were placed on chromosome arms, most mutant CO landscapes, except for zmet2, resulted in significant reductions of linkage drag compared to wild type (Fig. 5D and SI Appendix, Fig. S5D). However, when the resistance loci were positioned in the pericentromeric region, only zmet2, ddm1/zmet2, and the 10-fold global CO increase landscape showed linkage drag reductions compared to wild type, and ddm1 even exhibited a linkage drag increase (SI Appendix, Fig. S6D). The latter could have occurred because of ddm1’s small ratio of pericentromeric to subtelomeric CO numbers.
Fig. 5.
Simulating the effect of targeted recombination at resistance loci placed on maize chromosome 1. Generation 1 represents F1 progeny from the initial elite parent x wild parent cross. Generations 2 to 4 represent progeny from backcrossing to the elite parent. (A) Proportion of recurrent (elite) parent. (B) Proportion of donor (wild) parent. (C) Linkage drag around the introgression target (disease resistance) loci. (D) Genetic variance.
Overall, the introgression scenario simulations showed that mutant recombination landscapes somewhat helped with the recovery of the recurrent parent genome and removal of the donor parent genome. Additionally, when QTL were at chromosome ends, most mutant CO landscapes decreased linkage drag around the donor loci, which is often a major goal in breeding programs. In contrast, only some mutant CO landscapes reduced linkage drag around pericentromerically located loci. The introgression scenario simulation results also demonstrate that CO landscape engineering still holds the promise shown in earlier studies (70), even when allowing for the restraints coming with using more realistic assumptions.
Selection Intensity.
As selection intensity is a key contributor to genetic gain, we explored the effect of altering recombination landscape at different selection intensity levels using the maize ddm1 mutant as an example. In the recurrent selection scenario, increasing selection intensity resulted, as expected, in stronger genetic gains and decreased genetic variance in both ddm1 and in wild type. However, the impact on genetic gain and genetic variance was proportionally greater in the ddm1 population (SI Appendix, Fig. S7 A and B). In the introgression scenario, higher selection intensities, which resulted in faster return to the recurrent parent genome and faster loss of the donor parent genome, affected ddm1 and wild type in similar ways, but with ddm1 allowing a faster return to the recurrent parent and loss of the donor parent (SI Appendix, Fig. S8 A and B). Higher selection intensities also reduced linkage drag, although the difference was small compared to the overall difference between ddm1 and wild type (SI Appendix, Fig. S8C). Genetic variance was substantially greater with the use of the lower selection intensities of 5% and 10% but was maintained at similar levels in ddm1 and wild type (SI Appendix, Fig. S7D). This result is consistent with the Bulmer effect (78), which is a reduction of genetic variance as a result of selection. Taken together, increasing selection intensities increased the impact of the ddm1 CO landscape, although the benefit was relatively small compared to the overall advantage of using ddm1.
Species Considerations.
Although the same breeding scenarios were used for maize and rice, there was a large difference in the genetic gain increase between the two species. The reason behind this phenomenon could be that the pericentromeric CO suppression is much smaller in rice than maize, resulting in the overall CO rate increase in the mutants being greater in rice than maize. Additionally, rice has more chromosomes (12) than maize (10), which results in more COs per genome.
Beyond the factors taken into account in our simulations, it is also important to consider that rice, in contrast to maize, is a natural self-pollinator. As a result, the genomes of rice lines are made up of large linkage blocks—the extent of linkage disequilibrium (LD) in maize is a fraction of that in rice (79). As these blocks can be shared among elite lines, the increase in genetic gain due to increased recombination may be more limited in rice (80).
Feasibility of Using Mutant CO Landscapes in Breeding Programs.
The existing approaches to modulate CO landscape genome-wide may not be without unintended consequences. Combining multiple anti-CO gene mutations has in some situations resulted in chromosome instabilities (81). These issues, possibly arising from defective CO intermediates being allowed to resolve as COs, may preclude application of anti-CO gene mutants to generate extreme CO number increases in breeding programs.
Similarly, although the maize DNA methylation mutants did not show obvious plant growth and development defects in the recombination studies (64, 82), full effects of genome demethylation in these mutants over multiple generations are not completely known. These effects may include changes in gene expression patterns as well as mobilization of transposable elements (83–86), both with potentially detrimental consequences. An alternative approach to avoid these problems would be to limit demethylation to reproductive tissues by transient treatment with DNA methylation-reducing drugs or transient silencing of DNA methylation machinery components (75). Demethylation could also be confined by targeting DNA demethylating enzymes to specific genome sites using the CRISPR/Cas system (87–89).
Targeted Recombination
There is significant interest in generating tools to target recombination to specific chromosome loci rather than increasing it genome-wide (3). Previous modeling studies have shown that by finding the most optimal breakpoints for targeted recombination, genetic gains could be increased for polygenic traits, such as yield, by as much as 15% (72, 80). Although this analysis assumed exact knowledge of the effects of each of the many QTL contributing to yield, which is not yet feasible for complex traits, it illustrates the high utility of CO targeting.
Studies in tomato, Arabidopsis, and maize (90–92) have shown that COs can be created by targeting DSBs in somatic tissues using CRISPR/Cas9. These DSBs are then repaired by non-homologous end joining (NHEJ) or, much less frequently, by homologous recombination (HR) resulting in COs. Since recombination can take place at any point during germline development, the efficiency of this method could be very high. Alternatively, instead of inducing DSBs with active Cas9, a catalytically inactive (dead) Cas9, or another site-specific DNA binding protein, can be fused to active SPO11 to target it to a specific locus. When expressed during meiosis, the recombinant SPO11 protein would initiate meiotic recombination at the targeted site. So far, this approach has been successfully tried in yeast Saccharomyces cerevisiae, where it increased CO rates at several loci up to sixfold compared to wild type (93).
An alternative to targeting COs to specific sites could be preventing their occurrence in all other regions on the chromosome. A recent study has shown that a CRISPR-Cas-generated chromosomal inversion resulted not only in a suppression of COs in the inverted region but also a redirection of COs toward the telomeres (94). This approach can be used to change CO landscape in more complex and elaborate ways.
Modeling the Effect of Targeted Recombination on Linkage Drag.
The most compelling case for using targeted recombination in a plant breeding program is to facilitate introgression of a beneficial locus or chromosome segment. Thus, to test the efficiency of CO targeting, we set up a scenario in maize, in which COs were increased at the flanks of an interval containing the loci to be introgressed from the donor parent. The goal of this setup was to mimic a frequent situation, in which exact coordinates of the donor locus are not known and, as a result, a relatively large donor chromosome chunk needs to be introgressed. We tested the effect of a sixfold CO increase at the donor segment flanks, based on the S. cerevisiae CO targeting efficiencies (93). Additionally, we tested an idealistic 20-fold increase in COs at the same sites. Modeling both scenarios led to a substantial decrease of linkage drag around the donor segment, as well as faster return to the recurrent parent genome and faster removal of the donor parent genome (Fig. 5 A–C). Genetic variance was reduced compared to wild type (Fig. 5D), likely because of the progeny recovering more of the recurrent parent genome.
To further explore the potential of CO targeting, we also examined an interval targeting approach, in which COs were increased in a wider chromosome region. In this scenario, we modeled uniform sixfold and 20-fold CO increases in a 1 Mb interval containing the donor locus. Technologies to create this type of recombination landscape do not exist yet. However, we hypothesized that interval targeting could be more advantageous than a site-specific CO increase, given that the 1 Mb introgression region may contain QTL with negative effect on the yield-like polygenic trait, resulting in linkage drag with the introgression target loci.
As in the case of CO targeting to interval ends, we observed faster return to the recurrent parent genome, faster removal of the donor parent genome, substantial decrease in linkage drag, and reduced genetic variance compared to wild type (Fig. 5). However, the interval targeting approach was, overall, not more efficient than targeting only the interval flanks. We believe that the reason could be that even if the introgressed region carried a few unfavorable yield QTL, their overall effect on the trait was minimal, given the large overall number of yield QTL in the genome. The magnitude of CO rate increase at the targeted region had a greater influence on linkage drag than the impact of the specific targeting approach that was used.
Taken together, targeting COs to specific genomic sites can be highly advantageous for improving introgression outcomes, with its overall efficiency matching even the idealistic 10-fold global CO increase. However, the effectiveness of CO targeting may be limited to the specific scenarios of introducing a small number of well-defined loci. Targeted recombination could be multiplexed to simultaneously pursue several genome sites, but it may not be easily scalable for a very large locus number.
Concluding Thoughts and Outlook
Altogether, our simulations have shown CO landscape engineering to be a promising tool in breeding programs. Even the currently existing DNA methylation and anti-CO gene mutants were able to produce significant decreases in linkage drag in a backcrossing scheme. However, achieving substantial genetic gain increases in recurrent selection required idealistic approaches for which methods do not yet exist. Furthermore, the specific outcomes were strongly dependent on the genetic architecture of the trait and QTL location. Mutant CO landscapes were able to increase genetic gain in the case of the highly polygenic yield-like trait but not for an oligogenic trait. More of the mutant CO landscapes showed significant linkage drag reduction when the introgressed QTL were positioned at chromosome arms versus the pericentromeric region. In summary, these results point to the need for further tool development. In particular, more effective methods are needed to boost pericentromeric COs.
Even though the current methods may be promising, they may not be ready for immediate deployment. Implementing these approaches will require developing novel technologies to avoid off-target effects, which might include genome instability and pleiotropic influence of DNA methylation changes on plant growth and development. For example, studies that used combinations of several Arabidopsis mutants to produce extreme CO increases (28) suggest that CO landscapes most effective for increasing genetic gain may be associated with genomic instability. Even a potential for genetic instability, or any other off-target effect, persisting in the final variety would make recombination landscape engineering unattractive to breeders. The technologies to-be-developed could alter recombination rates only in transient ways or allow for easy elimination of mutant gene copies, along with any lingering effects that may have, from the final varieties. In addition, implementing methods to increase CO rates may require adjustments in other technologies used to aid breeding. For example, genomic selection (GS) models used to help predict QTL effects for highly polygenic traits rely heavily on LD between the QTL and nearby markers. Increasing CO rates would necessitate increasing GS marker densities. Otherwise, improvements in breeding efficiency coming from increased recombination would be diminished by declines in GS prediction accuracy (95).
We found targeting COs very effective for improving introgression of a defined donor region. For a trait with a few underlying loci, such as most disease resistance traits, this approach could be very beneficial, if methods to induce targeted recombination become highly efficient. Using targeted recombination for polygenic traits could become a possibility only if every contributing locus is known and its effects could be predicted accurately. Future studies could focus on improving efficiency of CO targeting by increasing the odds of engineered DSBs to be repaired as COs. The route to achieve this goal is to better understand the genetic mechanisms controlling the efficiency of HR-type repair of somatic DSBs and the CO/NCO decision in meiosis. The efficiency of CO formation varies among genome sites (96, 97), and elucidating how the local genome context affects this process will help selecting more effective sites for CO targeting.
Another area of helpful future development will be improving the understanding of genomic features and architecture of the species of interest. Knowing the genomic characteristics, such as DNA methylation patterns, within-species DNA sequence diversity, marker density, and presence of whole or partial genome duplications, would be extremely useful in determining the specific approach for CO landscape engineering most productive for each specific situation.
Materials and Methods
Creation of Genetic Maps.
All data analyses were done using R Studio. To construct the wild-type genetic map in maize, raw CO data from the NAM population were downloaded from Cyverse (https://cyverse.org) and transformed to the v4 version of the maize genome assembly (98). To generate a set of genetic markers, single nucleotide polymorphism (SNP) data for the B73 × Mo17 hybrid were downloaded from maizeGDB (www.maizeGDB.org) and 2,000 of these SNPs were chosen at random but ensuring even coverage of each chromosome. Then, COs were binned into 1 Mb intervals. Recombination rates for each chromosome interval were calculated (as frequency of COs/population size *100)/interval length in Mb). SNPs that fell into an interval were assigned the recombination rate of the respective interval. To construct the wild-type genetic map of rice, high-resolution recombination rate data for O. japonica were taken from Marand et al. (74). Marand et al. (74) estimated recombination rates by treating phased haplotypes as haploid individuals within subsets of 2,000 consecutive SNP windows overlapping by 250 SNPs using the program interval from the software package LDhat v2.2 (74). Since recombination rates for rice were calculated for specific intervals in previous studies, the recombination rates were assigned to SNPs that fell into each rate interval.
To find the genetic position of each SNP marker, cumulative genetic distances between markers were calculated. To calculate the cumulative genetic position, the previous marker’s genetic position in cM was added to the difference in physical distance in Mb between the current marker and the previous marker multiplied by the recombination rate of the interval of the marker location (genetic position [current] = genetic position [previous] + (physical position [current] – physical position [previous]) *recombination rate). The first marker on each chromosome’s genetic map was assigned the physical position of 0 Mb and the genetic position of 0 cM.
Because CO studies in recombination mutants were done using different marker sets from the one we used to construct the wild-type maps and/or using relatively small numbers of genetic markers, we devised a procedure to convert our higher-resolution wild-type genetic maps into mutant genetic maps using the difference in CO rate between mutant and wild type. CO data for wild type and mutants were first normalized by multiplying each interval by 2 and dividing by the population size. Once normalized, mean differences in CO rates between the mutant and wild-type intervals were calculated. The wild-type CO rate was multiplied by the difference between the mutant and wild-type rate and then added to the wild-type recombination rate: mutant rate = wild-type rate + (wild-type rate × (mutant rate - wild-type rate)).
CO landscapes of the maize ddm1 and zmet2 mutants were extrapolated to rice taking into account the differences in the size of pericentromeric regions between the two species as well as the uneven effect of the mutations on CO rates in different parts of maize chromosomes. Each maize chromosome was split into six intervals of equal physical (Mb) size, with the first and last intervals constituting chromosome ends and the third and fourth intervals covering the pericentromeric region. Rice chromosomes were split into five intervals to account for the smaller size of pericentromeric regions in this species. Since the mutants influence CO rates in specific chromosome regions rather than chromosome-wide, the mean CO rate difference was only applied to those specific regions. For example, if ddm1 showed a mean 2.35× CO rate increase at chromosome ends compared to wild type, the increase was only applied to the first and fifth interval of rice chromosomes. A similar approach was employed to extrapolate to maize the CO landscape of the recq4 and fancm mutants, which were found to increase CO rates in all regions of rice chromosomes, except pericentromeres (67).
Simulation Parameters.
All simulations were done in AlphaSimR (99). The number of individuals in the founder population was 200. CO interference was modeled using the Kosambi mapping function assuming 15% of COs coming from the non-interfering pathway. The h2 heritability value was 0.8, chosen to increase the selection accuracy and therefore genetic gain outcomes. Previous studies have found that genetic gain outcomes did not change drastically when heritability was altered (69). To distribute the 300 polygenic trait QTL, gene densities were calculated in 200 kb windows genome-wide and used to guide the polygenic trait QTL placement, i.e., regions with higher gene density had higher probability of being selected as QTL locations (SI Appendix, Figs. S9 and S10). This approach was used to ensure that QTL were placed in accurate gene space because it has been shown in maize that QTL locations for highly polygenic traits like yield strongly correlate with gene density (100). For maize, the B73 v4 reference genome annotation was downloaded from maizeGDB (101), and for rice, the Nipponbare reference genome was downloaded from the Rice Annotation Project database (102). Gene densities in each of the 200 kb intervals were determined by dividing the number of genes in the interval by the actual length of the interval.
To create as realistic scenarios as possible, genetic markers represented a mixture of repulsion and coupling linkages. Marker effects were established using the rnorm() function in R. To generate a mixture of repulsion and coupling linkages, a mean of 0 with a standard deviation of 1 was used. This approach was used to follow the infinitesimal model. The absolute value of marker effects was summed to estimate the limit of genetic gain in individuals.
For both breeding scenarios, the same yield-like additive polygenic trait with 300 QTL was used. In the recurrent selection scenario, a founder population was created and randomly inter-mated. The polygenic trait was used as the basis for selection for 10 generations relying on the wild-type genetic map to create LD among the markers. The initial “burn-in” period was replicated 100 times, and all replicates were combined into one base population. The combined base population was then used to conduct the recurrent selection scheme. To simulate each recombination mutant, the genetic map of the base population was changed from wild type to the respective mutant genetic map. The base population was then randomly inter-mated, the top 5% individuals were selected, maintaining the population size of 200, in every generation for 15 generations.
To set up the introgression scenario, two burn-in populations were used to first diverge the founder population into an elite population and a wild population. In the elite population, individuals best-performing for the yield-like trait were selected every generation for a total of 10 generations. For the wild population, selection was simultaneously performed for two traits, the same yield-like trait as in the elite population, and a disease resistance-like trait with additive and dominance effects. In the wild population, the lowest performing individuals for the yield-like trait were selected first, and out of those, the best-performing individuals for the resistance-like trait were selected. This approach was used to replicate wild populations with low yield but with strong disease resistance. To create the F1 population, the best individuals from the elite and the wild populations were crossed. The F1 progeny was backcrossed to the same best individual from the elite population, and top 5% individuals were selected. Backcrossing was conducted for four generations.
The simulation scripts written in R are available at https://github.com/ruthkepstein/Recombination_sims. CO data for ddm1 and zmet2 and all genetic maps can be downloaded from Cyverse: https://data.cyverse.org/dav-anon/iplant/home/rke27/RecombinationData (65). Please e-mail rke27@cornell.edu with questions.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
The authors would like to thank Dr. Ellie Taagen for many concept discussions, Dr. Minghui Wang for advice and help with the development of genetic maps, Dr. Chris Gaynor for answering AlphaSimR questions, and Dr. Susan McCouch for advice on the rice simulations. This work was made possible by NSF grants IOS-1844588 and IOS-1546792 to W.P.P. R.E. was supported by NSF NRT training grant IOS-1922551. W.P.P. is a Cornell Institute for Food Systems Faculty Fellow.
Author contributions
R.E., N.S., K.R.R, and W.P.P. designed research; R.E. and N.S. performed research; M.Z. and A.Z. contributed new reagents/analytic tools; R.E., N.S., and K.R.R. analyzed data; and R.E. and W.P.P. wrote the paper.
Competing interests
The authors declare a competing interest. The authors have organizational affiliations to disclose, W.P.P. is a scientific advisor to Meiogenix Inc., a startup company located at the Cornell Center for Life Science Ventures, which works to develop tools for targeted recombination in crops. Meiogenix Inc. had no role in designing or performing the research or writing the paper.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Swarup S., et al. , Genetic diversity is indispensable for plant breeding to improve crops. Crop. Sci. 61, 839–852 (2021). [Google Scholar]
- 2.Muller H. J., The relation of recombination to mutational advance. Mutat. Res. 1, 2–9 (1964). [DOI] [PubMed] [Google Scholar]
- 3.Taagen E., Bogdanove A. J., Sorrells M. E., Counting on crossovers: Controlled recombination for plant breeding. Trends Plant Sci. 25, 455–465 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Young N. D., QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol. 34, 479–501 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Voss-Fels K. P., et al. , Linkage drag constrains the roots of modern wheat. Plant Cell Environ. 40, 717–725 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Pawlowski W. P., Golubovskaya I. N., Cande W. Z., Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15, 1807–1816 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grelon M., Vezon D., Gendrot G., Pelletier G., AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Moran E., Santos J. L., Jones G. H., Franklin F. C. H., ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21, 2220–2233 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardiner L. J., et al. , Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome. Biol. 20, 69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale M. J., Keeney S., Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442, 153–158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allers T., Lichten M., Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Sidhu G. K., et al. , Recombination patterns in maize reveal limits to crossover homeostasis. Proc. Natl. Acad. Sci. U.S.A. 112, 15982–15987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones G. H., Franklin F. C. H., Meiotic crossing-over: Obligation and interference. Cell 126, 246–248 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Zelkowski M., Crossing-over decision landscape in maize. bioXriv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.09.21.508771v1.full.pdf. (accessed 22 September 2022).
- 15.Sun Y., et al. , Deep genome-wide measurement of meiotic gene conversion using tetrad analysis in Arabidopsis thaliana. PLoS Genet. 8, 1002968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu P., et al. , Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22, 508–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dooner H. K., Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell 14, 1173–1183 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stahl F. W., Rehan M. B. M., Foss H. M., Borts R. H., Apparent epigenetic meiotic double-strand-break disparity in Saccharomyces cerevisiae: A meta-analysis. Genetics 204, 129–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falque M., Anderson L. K., Stack S. M., Gauthier F., Martin O. C., Two types of meiotic crossovers coexist in maize. Plant Cell 21, 3915–3925 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otto S. P., Payseur B. A., Crossover interference: Shedding light on the evolution of recombination. Annu. Rev. Genet. 53, 19–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3, 1355–1364 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J., Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730–2740 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartung F., Suer S., Puchta H., Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 18836–18841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crismani W., et al. , FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Knoll A., et al. , The fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24, 1448–1464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjardins S. D., et al. , FANCM promotes class I interfering crossovers and suppresses class II non-interfering crossovers in wheat meiosis. Nat. Commun. 13, 3644 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girard C., et al. , AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11, e1005448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serra H., et al. , Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl. Acad. Sci. U.S.A. 115, 2437–2442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes J. B., Séguéla-Arnaud M., Larchevêque C., Lloyd A. H., Mercier R., Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. U.S.A. 115, 2431–2436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mézard C., Tagliaro Jahns M., Grelon M., Where to cross? New insights into the location of meiotic crossovers. Trends Genet. 31, 393–401 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Fernandes J. B., Wlodzimierz P., Henderson I. R., Meiotic recombination within plant centromeres. Curr. Opin. Plant Biol. 48, 26–35 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Mayer K. F. X., et al. , A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Buerstmayr M., et al. , High-resolution mapping of the pericentromeric region on wheat chromosome arm 5AS harbouring the Fusarium head blight resistance QTL Qfhs.ifa-5A. Plant Biotechnol. J. 16, 1046–1056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomé P. A., et al. , The recombination landscape in Arabidopsis thaliana F2 populations. Heredity 108, 447–455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodgers-Melnick E., et al. , Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. U.S.A. 112, 3823–3828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao Y., et al. , Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kianian P. M. A., et al. , High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9, 2370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gage J. L., et al. , Variation in upstream open reading frames contributes to allelic diversity in maize protein abundance. Proc. Natl. Acad. Sci. U.S.A. 119, e2112516119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crisp P. A., et al. , Stable unmethylated DNA demarcates expressed genes and their cis-regulatory space in plant genomes. Proc. Natl. Acad. Sci. U.S.A. 117, 23991–24000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fengler K., Allen S. M., Li B., Rafalski A., Distribution of genes, recombination, and repetitive elements in the maize genome. Crop. Sci. 47, S83–S95 (2007). [Google Scholar]
- 41.McMullen M. D., et al. , Genetic properties of the maize nested association mapping population. Science 325, 737–740 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Du H., et al. , Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 8, 15324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., et al. , Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. 13, e1007019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill W. G., Robertson A., The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (1966). [PubMed] [Google Scholar]
- 45.Bingham E. T., Groose R. W., Woodfield D. R., Kidwell K. K., Complementary gene interactions in alfalfa are greater in autotetraploids than diploids. Crop Sci. 34, 823–829 (1994). [Google Scholar]
- 46.Fu F. F., Dawe R. K., Gent J. I., Loss of RNA-directed DNA methylation in maize chromomethylase and DDM1-type nucleosome remodeler mutants. Plant Cell 30, 1617–1627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melamed-Bessudo C., Levy A. A., Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, E981–E988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziolkowski P. A., et al. , Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodeling via interference during Arabidopsis meiosis. eLife 4, e03708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devos K. M., Dubcovsky J., Dvořák J., Chinoy C. N., Gale M. D., Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor. Appl. Genet. 91, 282–288 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Wang M., Machine learning reveals conserved chromatin patterns determining meiotic recombination sites in plants. bioXriv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.07.11.499557v1. (accessed 22 July 2022).
- 51.Slotkin R. K., Martienssen R., Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Jeddeloh J. A., Bender J., Richards E. J., The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12, 1714–1725 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao W., et al. , DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18, 805–814 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henderson I. R., Jacobsen S. E., Epigenetic inheritance in plants. Nature 447, 418–424 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Feng S., et al. , Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawashima T., Berger F., Epigenetic reprogramming in plant sexual reproduction. Nat. Rev. Genet. 15, 613–624 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Bewick A. J., et al. , The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bewick A. J., Schmitz R. J., Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 36, 103–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q., Eichten S. R., Hermanson P. J., Springer N. M., Inheritance patterns and stability of DNA methylation variation in maize near-isogenic lines. Genetics 196, 667–676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Long J., Liu J., Xia A., Springer N. M., He Y., Maize decrease in DNA methylation 1 targets RNA-directed DNA methylation on active chromatin. Plant Cell 33, 2183–2196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zemach A., et al. , The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirouze M., et al. , Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 5880–5885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao M., et al. , The mop1 mutation affects the recombination landscape in maize. Proc. Natl. Acad. Sci. U.S.A. 118, e2009475118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zelkowski M., CyVerse data commons. https://data.cyverse.org/dav-anon/iplant/home/rke27/RecombinationData. (accessed 03 September 2022).
- 66.Underwood C. J., et al. , Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28, 519–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mieulet D., et al. , Unleashing meiotic crossovers in crops. Nat. Plants 4, 1010–1016 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Lush J., Animal breeding plans. Q. Rev. Biol. 21, 292–293 (1946). [Google Scholar]
- 69.Tourrette E., Bernardo R., Falque M., Martin O. C., Assessing by modeling the consequences of increased recombination in recurrent selection of Oryza sativa and Brassica rapa. G3 (Bethesda; ) 9, 4169–4181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tourrette E., Falque M., Martin O. C., Enhancing backcross programs through increased recombination. Genet. Sel. Evol. 53, 25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernardo R., Prospective targeted recombination and genetic gains for quantitative traits in maize. Plant Genome 10, 2 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Ru S., Bernardo R., Targeted recombination to increase genetic gain in self-pollinated species. Theor. Appl. Genet. 132, 289–300 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Battagin M., Gorjanc G., Faux A. M., Johnston S. E., Hickey J. M., Effect of manipulating recombination rates on response to selection in livestock breeding programs. Genet. Sel. Evolut. 48, 44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marand A. P., et al. , Historical meiotic crossover hotspots fueled patterns of evolutionary divergence in rice. Plant Cell 31, 645–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raz A., Dahan-Meir T., Melamed-Bessudo C., Leshkowitz D., Levy A. A., Redistribution of meiotic crossovers along wheat chromosomes by virus-induced gene silencing. Front. Plant Sci. 11, 635139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fisher R. A., XV.—The correlation between relatives on the supposition of mendelian inheritance. T. R. Soc. Edinb. 52, 399–433 (1919). [Google Scholar]
- 77.Melchinger A. E., Use of molecular markers in breeding for oligogenic disease resistance. Plant Breed. 104, 1–19 (1990). [Google Scholar]
- 78.Bulmer M. G., The effect of selection on genetic variability. Am. Nat. 105, 201–211 (1971). [Google Scholar]
- 79.Mather K. A., et al. , The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 177, 2223–2232 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brandariz S. P., Bernardo R., Predicted genetic gains from targeted recombination in elite biparental maize populations. Plant Genome 12, 180062 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Hartung F., Puchta H., The RecQ gene family in plants. J. Plant. Physiol. 163, 287–296 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Anderson S. N., et al. , Subtle perturbations of the maize methylome reveal genes and transposons silenced by chromomethylase or RNA-directed DNA methylation pathways. G3 (Bethesda) 8, 1921–1932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reinders J., et al. , Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23, 939–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Osakabe A., et al. , The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W. Nat. Cell Biol. 23, 391–400 (2021). [DOI] [PubMed] [Google Scholar]
- 85.Griffin P. T., Niederhuth C. E., Schmitz R. J., A comparative analysis of 5-azacytidine-and zebularine-induced DNA demethylation. G3 (Bethesda) 6, 2773–2780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moritoh S., et al. , Targeted distribution of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs growth of rice plants by abnormal DNA methylation. Plant J. 1, 85–98 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Ji L., et al. , TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat. Commun. 9, 895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gallego-Bartolomé J., et al. , Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 115, E2125–E2134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shilo S., Melamed-Bessudo C., Dorone Y., Barkai N., Levy A. A., DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell 27, 2427–2436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ben Shlush I., et al. , CRISPR/Cas9 induced somatic recombination at the CRTISO locus in tomato. Genes 12, 59 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Filler-Hayut S., Kniazev K., Melamed-Bessudo C., Levy A. A., Targeted inter-homologs recombination in Arabidopsis euchromatin and heterochromatin. Int. J. Mol. Sci. 22, 12096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kouranov A., et al. , Demonstration of targeted crossovers in hybrid maize using CRISPR technology. Commun. Biol. 5, 53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sarno R., et al. , Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 45, e164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ronspies M., et al. , Massive crossover suppression by CRISPR-Cas-mediated plant chromosome engineering. Nat. Plants 8, 1153–1159 (2022). [DOI] [PubMed] [Google Scholar]
- 95.Taagen E., et al. , If it ain't broke, don't fix it: evaluating the effect of increased recombination on response to selection for wheat breeding. G3 jkac291; (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee P. S., et al. , A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5, e1000410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pazhayam N. M., Turcotte C. A., Sekelsky J., Meiotic crossover patterning. Front. Cell Dev. Biol. 9, 681123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiao Y., et al. , Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chris Gaynor R., Gorjanc G., Hickey J. M., AlphaSimR: An R package for breeding program simulations. G3 (Bethesda) 11, jkaa017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinez A. K., et al. , Yield QTLome distribution correlates with gene density in maize. Plant Sci. 242, 300–309 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Woodhouse M. R., et al. , A pan-genomic approach to genome databases using maize as a model system. BMC Plant Biol. 21, 385 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kawahara Y., et al. , Improvement of the Oryz a sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.