Summary
Background
The SARS-CoV-2 delta (B.1.617.2 lineage) variant was first identified at the end of 2020 and possessed two unique amino acid substitutions in its spike protein: S-P681R, at the S1/S2 cleavage site, and S-D950N, in the HR1 of the S2 subunit. However, the roles of these substitutions in virus phenotypes have not been fully characterized.
Methods
We used reverse genetics to generate Wuhan-D614G viruses with these substitutions and delta viruses lacking these substitutions and explored how these changes affected their viral characteristics in vitro and in vivo.
Findings
S-P681R enhanced spike cleavage and membrane fusion, whereas S-D950N slightly promoted membrane fusion. Although S-681R reduced the virus replicative ability especially in VeroE6 cells, neither substitution affected virus replication in Calu-3 cells and hamsters. The pathogenicity of all recombinant viruses tested in hamsters was slightly but not significantly affected.
Interpretation
Our observations suggest that the S-P681R and S-D950N substitutions alone do not increase virus pathogenicity, despite of their enhancement of spike cleavage or fusogenicity.
Funding
A full list of funding bodies that contributed to this study can be found under Acknowledgments.
Keywords: SARS-CoV-2, COVID-19, Reverse genetics, Hamster
Research in context.
Evidence before this study
The SARS-CoV-2 delta variant (lineage B.1.617.2) identified at the end of 2020 obtained multiple substitutions. Although the delta variant rapidly became globally dominant, how these substitutions contributed to its rapid spread remained unclear. Published reports state that the introduction of S-P681R into the ancestral Wuhan-D614G virus enhances virus fusogenicity and pathogenicity. A PubMed search up to 2022 using the keywords “SARS-CoV-2” and “P681R” or “D950N” found that the P681R and D950N substitutions are characteristic substitutions for the delta variant; however, the roles of S-P681R and S-D950N have not been fully characterized.
Added value of this study
We found that S-P681R enhances spike cleavage efficiency and membrane fusion and S-D950N slightly promotes membrane fusion. However, these substitutions did not significantly change the virus pathogenicity in hamsters.
Implications of all the available evidence
Although previous papers suggested that the S-P681R substitution enhanced virus pathogenicity in animal models, our results indicate that the S-P681R and S-D950N substitutions alone do not increase virus pathogenicity significantly. However, these two substitutions in combination with other substitutions could enhance the pathogenicity. Therefore, we must continue to monitor the emergence of novel variants bearing these substitutions.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which was identified as a novel human pathogen in China at the end of 2019, is responsible for COVID-19.1,2 It spread rapidly around the world causing the World Health Organization (WHO) to declare a pandemic on 11 March 2020. Since SARS-CoV-2 is an RNA virus, mutations frequently occur in its genome, and many variants have emerged and caused waves of infection that have spread repeatedly. One such variant, the delta variant (lineage B.1.617.2), was identified at the end of 2020 in India3 and rapidly became globally dominant. Amino acid substitutions in its spike (S) protein enhanced spike cleavage and fusion activity relative to the ancestral Wuhan-D614G virus and alpha variant (lineage B.1.1.7)4,5 and increased pathogenicity compared with previous variants in animal models and humans.4,6,7 The L452R substitution in its spike protein increases the binding affinity to human ACE2, protein stability, and fusogenicity, resulting in superior replication.8 This substitution also contributed to immune evasion from some neutralizing antibodies.8 The P681R and D950N substitutions in the spike protein of delta virus (S-P681R and S-D950N) have not been detected in other major variants such as alpha (lineage B.1.1.7), beta (lineage B.1.351), gamma (lineage P.1), and omicron (lineage B.1.1.529). S-P681R is located at the cleavage site at the boundary between the S1 and S2 domains. The introduction of S-P681R into the ancestral Wuhan-D614G virus was found to enhance virus fusogenicity and pathogenicity,4 and reversion of S-681R to wild-type S-681P reduced the replication and spike cleavage of the delta variant5; however, the contribution of S-P681R to virus pathogenicity in the context of the delta variant per se has not been demonstrated. S-D950N faces the trimer boundary of the S protein and is located in the heptad repeat 1 (HR1) of the S2 subunit that is essential for the conformational change for fusion.9, 10, 11, 12 Therefore, it may affect virus fusogenicity. However, its effect has not been investigated.
In this study, we explored how S-P681R and S-D950N affect the viral characteristics of SARS-CoV-2. We generated recombinant SARS-CoV-2 with or without these mutations based on ancestral Wuhan-D614G and delta viruses. These viruses were then examined for spike cleavage efficiency, fusion activity, virus replication in vitro, and pathogenicity in hamsters.
Methods
Cells
VeroE6/TMPRSS2 (JCRB 1819) cells13,14 were propagated in the presence of 1 mg/ml geneticin (G418; Invivogen) and 5 μg/ml plasmocin prophylactic (Invivogen) in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). VeroE6 (ATCC CRL-1586) cells and Calu-3 (ATCC HTB-55) cells were maintained in Eagle's minimal essential media (EMEM) containing 10% FCS and antibiotics. HEK293T cells were cultured in DMEM supplemented with 10% FCS. BHK-21 (JCRB 9020) cells were cultured in EMEM with non-essential amino acids (Gibco) and 10% FCS. All cells were maintained at 37 °C with 5% CO2. The cells were regularly tested and confirmed to be negative for mycoplasma contamination by using PCR.
Bacterial Artificial Chromosome (BAC) construction
The full-genome nucleotide sequence of SARS-CoV-2 was assembled into the pBeloBAC11 vector to generate infectious cDNA clones under the control of a cytomegalovirus (CMV) promoter by using Gibson Assembly Master Mix (NEB) as described previously15,16 with some modifications. Five fragments (Fa to Fe) covering the full-length SARS-CoV-2 genome with a 30-bp overlap and restriction sites and linearized pBeloBAC11 (linker fragment) were amplified by PCR using PrimeSTAR GXL DNA Polymerase (TaKaRa) and six primer sets (Table 1).16 The pBeloBAC11 vector was used as a template for PCR to amplify the linker fragment. The artificially synthesized DNA of SARS-CoV-2 Wuhan/Hu-1/2019 or cDNA synthesized from hCoV-19/USA/WI-UW-5250/2021 was used as a template for PCR to amplify the five fragments, Fa to Fe. Mutations were introduced into the Fd fragments by using primer-guided mutagenesis. First, Fa and Fe were ligated with the linker by using Gibson Assembly Master Mix (NEB) resulting in a BAC containing Fa and Fe. Fb and Fc were subsequently ligated into the BAC digested with FseI and AscI. Finally, Fd was cloned into the BAC digested with FseI and AscI resulting in the BAC carrying the full-length SARS-CoV-2 genome. The constructed BACs were introduced into DH10B E. coli (NEB) by electroporation. The E. coli was amplified at 37 °C and BACs were extracted by using NucleoBond Xtra Maxi (TaKaRa).
Table 1.
Primers used in this study.
| Fragment | Direction | Sequence (5′→3′) |
|---|---|---|
| linker | Forward | TAGCTTCTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAAAAAAAGGGTCGGCATGGCAT |
| Reverse | CTGGGAAGGTATAAACCTTTAATACGGTTCACTAAACGAGCTC | |
| Fa | Forward | ATTAAAGGTTTATACCTTCCCAG |
| Reverse | AGATTCTCATAAACAAATCCATAAGTTCGTGGCGCGCCGATGGCCGGCCGATGAAATGGTAATTTGTATAGTTTCTAAA | |
| Fb | Forward | TTTAGAAACTATACAAATTACCATTTCATCTTTTAAATGGGATTTAACTG |
| Reverse | AAACATTAAAGTTTGCACAATGCAGAATGCATCTGTCATCCAAACAGTTA | |
| Fc | Forward | GCATTCTGCATTGTGCAAACTTTAATGTTTTATTCTCTACAGTGTTCCCA |
| Reverse | AGATTCTCATAAACAAATCCATAAGTTCGTGGCGCGCCGATGGCCGGCCTGTTCGTTTAGTTGTTAACAAGAACATCAC | |
| Fd | Forward | GTGATGTTCTTGTTAACAACTAAACGAACAATGTTTGTTTTTCTTGTTTT |
| Reverse | AGATTCTCATAAACAAATCCATAAGTTCGTTTATGTGTAATGTAATTTGA | |
| Fe | Forward | ACGAACTTATGGATTTGTTTATGAGAATCTTCACAATTGGAACTGTAACT |
| Reverse | TTTTTTTTTTTTTTTTTTTTTTTTTGTCATTCTCCTAAGAAGCTA |
SARS-CoV-2 rescue
To recover recombinant SARS-CoV-2, BACs encoding the full-length SARS-CoV-2 genome were transfected into HEK293T cells by using TransIT-293 (TaKaRa) according to the manufacturer's protocol. At 3 days post-transfection, the supernatant containing viruses was collected and inoculated onto VeroE6/TMPRSS2 at 37 °C to prepare the virus stock. The virus titers of the stock viruses were determined by using plaque assays in VeroE6/TMPRSS2. The stock viruses were subjected to NGS as described17,18 to confirm the absence of unwanted mutations. All experiments with SARS-CoV-2 were performed in enhanced biosafety level 3 (BSL3) containment laboratories at the University of Tokyo.
Growth kinetics in cultured cells
VeroE6, VeroE6/TMPRSS2, or Calu-3 cells grown on 24-well plates in triplicate were infected with the indicated virus at an MOI of 0.001. The inoculum was removed after 60 min of incubation at 37 °C, and the cells were further incubated at 37 °C. Cell culture supernatants were collected at 6, 24, 48, and 72 h post-infection. Virus titers were determined by use of a plaque assay in VeroE6/TMPRSS2 cells.
Western blotting
To determine the level of the S2 protein in the SARS-CoV-2-infected cells, VeroE6/TMPRSS2 cells were infected with each virus at an MOI of 1. Cells were lysed at 12 h post-infection with Tris-Glycine SDS sample buffer (Invitrogen). The cell lysates were sonicated, heated for 10 min at 95 °C, and then subjected to SDS-PAGE on Any kD Mini-PROTEAN TGM Precast Protein Gels (Bio-Rad). Proteins in the SDS-PAGE gels were transferred to a polyvinylidene fluoride membrane (Millipore), and detected by using the indicated primary antibodies [mouse anti-SARS-CoV-2 S monoclonal antibody (1A9, GeneTex, GTX632604, 1:10,000) or mouse anti-α-tubulin (TUBA) monoclonal antibody (clone DM1A, Sigma–Aldrich, T9026, 1:10,000)], followed by secondary antibodies [sheep horseradish peroxidase (HRP)-conjugated anti-mouse IgG (GE Healthcare)]. Signals were developed by using ECL Prime Western Blotting Detection Reagent (GE Healthcare). Images were captured with the ChemiDoc Touch Imaging System (Bio-Rad) and quantified by using Image Lab Software (Bio-Rad).
Fusion assay
To prepare spike-expressing cells, BHK-21 cells grown on 12-well plates were transfected with 500 ng of pCAGGS-spike, which encodes a codon-optimized spike gene, 600 ng of plasmid encoding T7 promoter-driven firefly luciferase, and 20 ng of pCAGGS-Renilla luciferase, by using TransIT-LT1 (TaKaRa). To prepare ACE2-expressing cells, VeroE6 or VeroE6/TMPRSS2 cells grown on 12-well plates were transfected with 500 ng of pCAGGS-human ACE2 and 800 ng of pCAGGS-T7 polymerase, by using TransIT-LT1 (TaKaRa). After 24 h, these cells were detached by using EDTA without trypsin, mixed 1:1, and reseeded into 24-well plates. After 24 h of co-culture, firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega). The firefly luciferase activity was normalized to the Renilla luciferase activity.
Animal experiments and approvals
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo (approval numbers: PA19-75). Virus inoculations were performed under anesthesia, and all efforts were made to minimize animal suffering. In vivo studies were not blinded, and animals were randomly assigned to infection groups. No sample-size calculations were performed to power each study. Instead, sample sizes were determined based on prior in vivo virus challenge experiments.
Experimental infection of Syrian hamsters
Five- to six-week-old male wild-type Syrian hamsters (Japan SLC Inc., Shizuoka, Japan) were used in this study. Baseline body weights were measured before infection. Under isoflurane anesthesia, five hamsters per group were intranasally inoculated with 105 PFU (in 30 μL) of the indicated virus. Body weight was monitored daily for 10 days. For virological and pathological examinations, ten hamsters per group were intranasally infected with 105 PFU (in 30 μL) of the indicated virus; 3 and 7 days post-infection, five animals per group were euthanized and nasal turbinates and lungs were collected and homogenized. The virus titers in the nasal turbinate and lung samples were determined by use of plaque assays in VeroE6/TMPRSS2 cells.
Measuring lung functions
Respiratory parameters were measured by using a whole-body plethysmography system (PrimeBioscience) according to the manufacturer's instructions. In brief, hamsters were placed in the unrestrained plethysmography chambers and allowed to acclimatize for 1 min before data were acquired over a 3-min period by using FinePointe software.
Pathology
Excised animal tissues were fixed in 4% paraformaldehyde in PBS and processed for paraffin embedding. The paraffin blocks were cut into 3-μm-thick sections and mounted on silane-coated glass slides for histopathological examination. To detect SARS-CoV-2 RNA, in situ hybridization was performed using an RNA scope 2.5 HD Red Detection kit (Advanced Cell Diagnostics, Newark, California) with an antisense probe targeting the nucleocapsid gene of SARS-CoV-2 (Advanced Cell Diagnostics) as previously described.19 Tissue sections were also processed for immunohistochemical staining with a rabbit polyclonal antibody for SARS-CoV nucleocapsid protein (ProSpec; ANT-180, Rehovot, Israel), which cross-reacts with SARS-CoV-2 nucleocapsid protein. Specific antigen–antibody reactions were visualized by means of 3,3′-diaminobenzidine tetrahydrochloride staining using the Dako Envision system (Dako Cytomation, Glostrup, Denmark).
Statistical analysis
GraphPad Prism software was used to analyze all data. Statistical analysis was conducted by use of a one-way or two-way ANOVA with multiple comparison post-hoc corrections. Differences among groups were considered significant for p values <0.05.
Role of funders
The funders had no role in the study design, data collection, analysis, or interpretation, or any aspect pertinent to this study.
Results
The spike substitution at position 681 is important for its cleavage
To elucidate the roles of S-P681R or S-D950N in the delta variant, we generated Wuhan/Hu-1/2019 (Wuhan) possessing the S-D614G substitution and mutants bearing one or both mutations by reverse genetics using the BAC system: Wuhan-D614G, Wuhan-D614G/P681R, Wuhan-D614G/D950N, and Wuhan-D614G/P681R/D950N. We also generated a delta variant (hCoV-19/USA/WI-UW-5250/2021) and its mutants lacking one or both substitutions: delta, delta-R681P, delta-N950D, and delta-R681P/N950D. The viral genome sequences of the stock viruses were confirmed to have no unintended mutations by deep sequencing. No unintended nucleotide substitutions with a frequency of greater than 10% were observed in all virus stocks, indicating that these stocks have a sufficiently low genetic mixture to use to explore the functions of a single amino acid substitution.
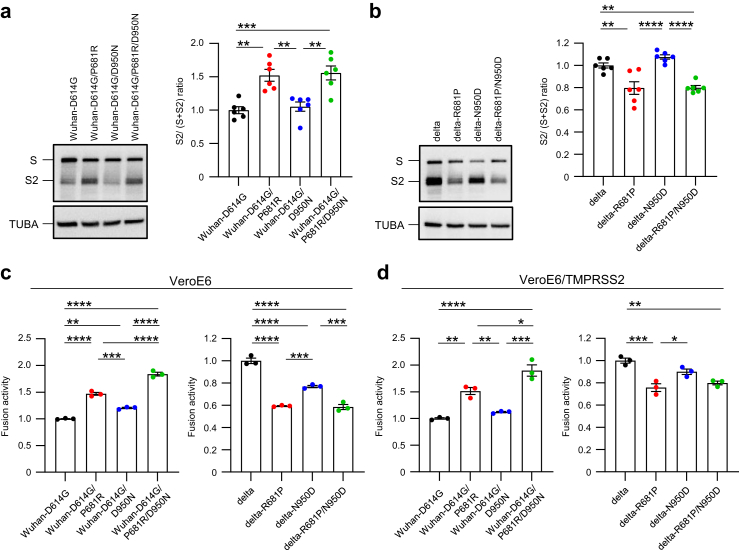
First, we compared the cleavage efficiency of the spike protein. Newly synthesized spike protein is cleaved at the S1/S2 site by furin protease. Since the cleavage of the spike protein is required for virus entry20 and S-681 is located at the S1/S2 site, S-681 could influence spike cleavage. To assess the efficiency of spike cleavage for each virus, VeroE6/TMPRSS2 cells were infected at a multiplicity of infection (MOI) of 1 and lysed at 12 hours post-infection (h.p.i). The levels of the cleaved S2 protein in the infected cells were determined by western blotting and were quantified based on band intensity (Fig. 1a and b). The ratio of cleaved S2 to full-length S protein was significantly increased by S-P681R in Wuhan-D614G (Fig. 1a) and the loss of S-P681R in delta variant reduced the cleavage efficiency (Fig. 1b). S-D950N in Wuhan-D614G and its loss in delta variant did not affect spike cleavage. These results suggest that S-P681R but not S-D950N promotes spike cleavage.
Fig. 1.
Characterization of amino acid substitutions at positions 681 and 950 in vitro. a and b, VeroE6/TMPRSS2 cells were infected with each virus at an MOI of 1. (Left) At 12 h post-infection, cells were lysed, and spike and TUBA (internal control) expression was analyzed by western blotting. (Right) Ratios of S2 to full-length S were determined based on the band intensity of the western blots (n = 6, mean ± s.e.m). c and d, spike-expressing BHK cells and human ACE2-expressing VeroE6 (c) or VeroE6/TMPRSS2 (d) cells were co-cultured and fusion activity was determined by using the dual-luciferase assay (n = 3, mean ± s.e.m.). a–d, Data were analyzed by using a one-way ANOVA with Tukey's multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Spike substitutions at positions 681 and 950 change fusion activity
Next, we assessed the fusion activity of each spike protein because the level of spike protein cleavage affects the fusion of the cell membrane with the viral envelope.20 We measured the fusion activity between spike-expressing effector cells and human ACE2-expressing target cells as surrogates. The effector cells were prepared by transfecting BHK cells with a plasmid encoding each spike protein together with a plasmid encoding the firefly luciferase gene under the control of the T7 promoter. The target cells were prepared by transfecting VeroE6 or VeroE6/TMPRSS2 cells with a human ACE2-expressing plasmid and a T7 polymerase-expressing plasmid. Co-culture of the effector cells and target cells induces cell fusion and then translation of firefly luciferase is driven by T7 polymerase. Therefore, the fusion activity of the spike protein could be estimated by measuring the firefly luciferase activity. As previously described,4 S-P681R in Wuhan-D614G significantly increased fusion activity and its loss in the delta variant significantly decreased fusion activity with or without overexpression of TMPRSS2 (Fig. 1c and d). S-D950N in Wuhan-D614G slightly increased the fusion activity under both conditions (Fig. 1c and d). Loss of S-D950N in the delta variant significantly or slightly decreased fusion activity in VeroE6 cells or VeroE6/TMPRSS2 cells, respectively. Moreover, the fusion activity of Wuhan-D614G bearing both P681R and D950N was greater than that of Wuhan-D614G and Wuhan-D614G bearing P681R. These results demonstrate that S-P681R enhances membrane fusion whereas S-D950N may only slightly enhance membrane fusion.
Roles of amino acid substitutions at positions 681 and 950 in the spike protein in virus replication in cultured cells
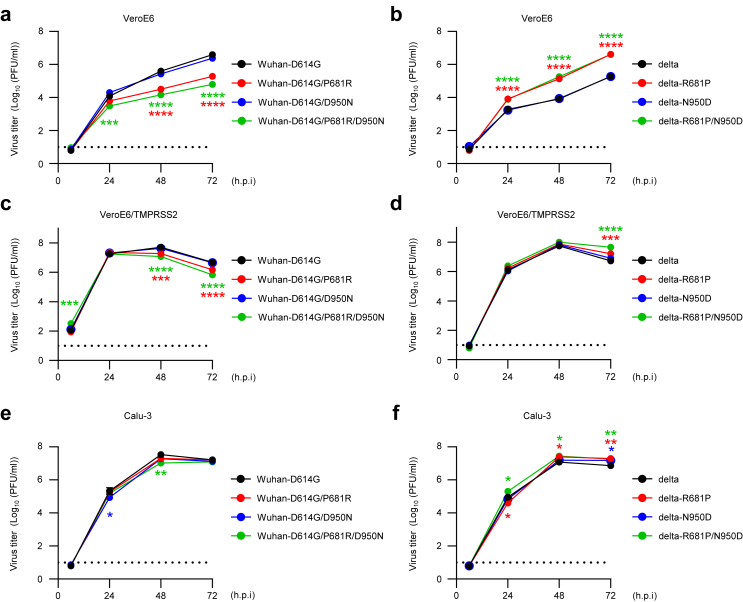
To compare the growth kinetics of the viruses in vitro, VeroE6, VeroE6/TMPRSS2, or Calu-3 cells were infected with each virus at an MOI of 0.001. The supernatants were collected at the indicated timepoints, and virus titers were determined by use of plaque assays. The growth kinetics of Wuhan-D614G-backbone viruses with S-P681R in VeroE6 cells were significantly lower than those of Wuhan-D614G-backbone viruses without S-P681R, and vice versa for delta-backbone viruses (Fig. 2a and b). In VeroE6/TMPRSS2 cells, replication of all tested viruses was improved and reached approximately 108 PFU/ml (Fig. 2c and d). In particular, viruses possessing R at position 681 in the spike protein propagated at a similar efficiency as viruses possessing P at position 681. In Calu-3 cells, the virus titers of each virus were similar at each timepoint (Fig. 2e and f). These results show that the S-P681R substitution reduces virus growth in VeroE6 cells and overexpression of TMPRSS2 broadly enhances the growth of the viruses, especially S-P681R viruses. Taken together with the data from the spike cleavage and fusion assays, our findings show that efficient spike cleavage and high fusogenicity do not enhance viral fitness in vitro.
Fig. 2.
Growth kinetics of mutant viruses. VereE6 (a and b), VeroE6/TMPRSS2 (c and d), or Calu-3 cells (e and f) were infected with each virus at an MOI of 0.001. Virus titers at the indicated timepoints were determined by using plaque assays (n = 3, mean ± s.e.m.). Data were analyzed by using a two-way ANOVA with Dunnet's multiple comparisons test. Statistical significance was calculated against the values in Wuhan-D614G- or delta-infected cells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The lower limit of detection is indicated by the horizontal dashed line.
The spike substitutions P681R and/or D950N alone do not significantly increase virus pathogenicity in Syrian hamsters
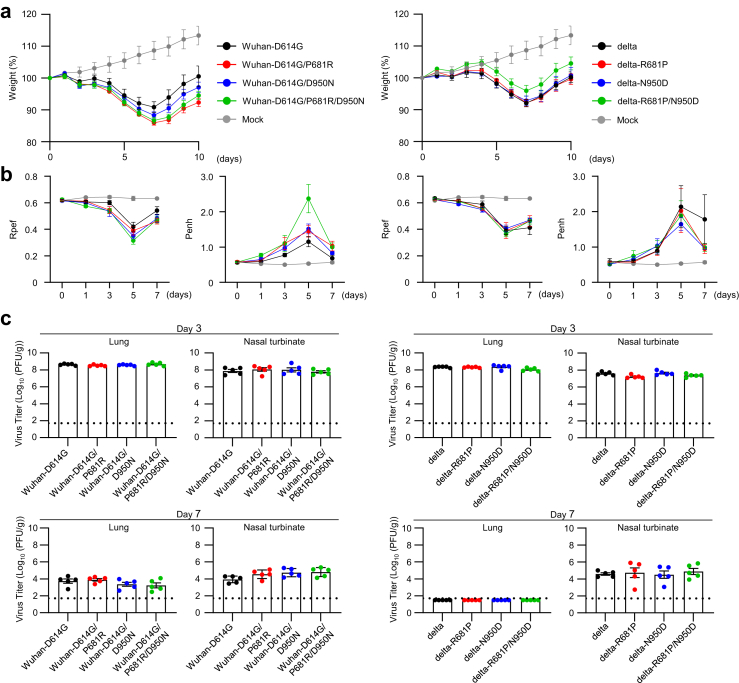
We next evaluated the replication and pathogenicity of each virus in Syrian hamsters, a well-established small animal model for SARS-CoV-2.21 Hamsters were intranasally inoculated with 105 PFU of each virus and their body weights were measured for 10 days (Fig. 3a). There was no statistically significant difference between any of the infected hamsters. However, Wuhan-D614G bearing P681R and/or D950N caused slightly greater weight loss than Wuhan-D614G, and delta bearing both R681P and N950D caused slightly less weight loss compared with delta variant.
Fig. 3.
The effect of amino acid substitutions at 681 and/or 950 on viral in vivo characteristics. Syrian hamsters were intranasally inoculated with 105 PFU (in 30 μL) of the indicated virus. a, Body weights of virus-infected and mock-infected hamsters (n = 5) were monitored daily for 10 days. Data are presented as the mean percentages of the starting weight (±s.e.m.). b, Pulmonary function analyses in infected hamsters. Penh and Rpef were measured by using whole-body plethysmography. (n = 5, mean ± s.e.m.). c, Virus titers in infected Syrian hamsters. Hamsters (n = 5) were euthanized at 3 and 7 days post-infection for virus titration. Virus titers in the nasal turbinates and lungs were determined by using plaque assays. Vertical bars show the mean ± s.e.m. Points indicate data from individual hamsters. The lower limit of detection is indicated by the horizontal dashed line.
We then evaluated the pulmonary functions Penh and Rpef in the infected hamsters by using the whole-body plethysmography system (Fig. 3b). No statistically significant differences in either Penh or Rpef were observed between all groups of infected hamsters. However, P681R and/or D950N in Wuhan-D614G caused a slight exacerbation of Rpef and Penh on days 3, 5, and 7 post-infection. Penh recovered slightly on day 7 post-infection in delta-R681P and/or -N950D group. These results suggest that these substitutions may have a limited effect on pathogenicity in hamsters.
On days 3 and 7 post-infection, virus titers in the nasal turbinates and lungs of hamsters infected with each virus were measured by use of plaque assays. Virus titers in the lungs and nasal turbinates were not affected by the substitutions at positions 681 and 950 (Fig. 3c). This result indicates that substitutions at positions 681 and 950 do not affect virus replication in hamsters. Whole genome sequencing of viruses in the nasal turbinates and lungs at days 3 and 7 post-infection, respectively, revealed that no revertant mutants had emerged. Of note, delta-backbone viruses were eliminated from the lungs of infected hamsters earlier than from the nasal turbinates, as previously reported,22 and delta-backbone viruses were eliminated earlier than Wuhan-backbone viruses from the lungs, as previously reported.23
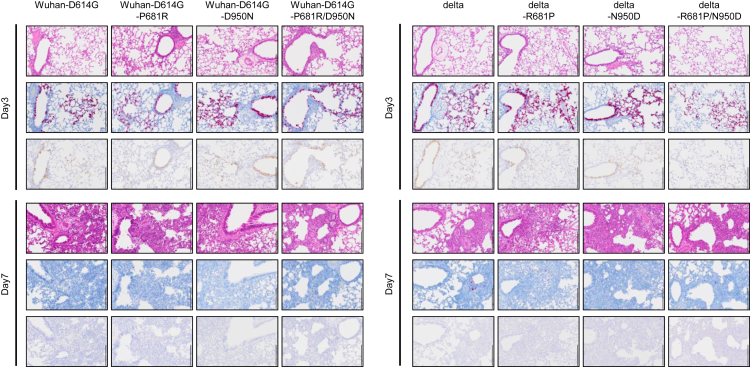
Histopathological analysis of the lungs of the infected hamsters was also conducted. There was no obvious difference in the magnitude of inflammation in the bronchi, bronchioles and alveoli or the distribution of viral RNA and antigen with or without S-P681R and S-D950N (Fig. 4). These observations suggest that S-P681R and S-D950N do not have obvious pathological effects in lungs of hamsters.
Fig. 4.
Pathological findings in the lungs of infected Syrian hamsters. Representative histopathological images of the lungs of hamsters infected with the indicated virus on days 3 and 7 post-infection. (Top) Hematoxylin/eosin (H&E) staining. (Middle) in situ hybridization for SARS-CoV-2 viral RNA. (Bottom) Immunohistochemistry (IHC) for SARS-CoV-2 nucleocapsid protein. Scale bars, 200 μm.
Discussion
Here we characterized Wuhan-D614G-backbone viruses with S-P681R and/or S-D950N and delta-backbone viruses that lacked these substitutions. The pathogenicity analysis of these recombinant viruses in the hamster model indicates that these two amino acid substitutions alone do not significantly increase the pathogenicity of the virus, and Wuhan-D614G-backbone viruses show similar or slightly higher pathogenicity compared with delta-backbone viruses. In a previous report,4 we and our collaborators concluded from data similar to those presented in this study that the pathogenicity of the delta variant was higher than that of the ancestral strains and that the S-P681R substitution was responsible for this high pathogenicity. Therefore, two differences exist between the previous4 and current studies: high pathogenicity of the delta variant and the contribution of S-P681R to the high pathogenicity. With regard to the first point, the differences in body weight change after infection and in the inflammatory response between the ancestral strain and delta variant in the previous report were marginal.4 However, another group showed that Wuhan-like viruses caused greater body weight loss in hamsters than the delta variant.24 Therefore, the data are inconsistent as to whether the delta variant is more pathogenic than Wuhan in the hamster infection model. In fact, some papers have suggested that the delta variant was more pathogenic in humans,6,7 but it is hard to evaluate virus pathogenicity based on human clinical data as it is highly dependent on a variety of factors, such as individual immunological status and medical treatments. With regard to the second point, the differences may be caused by the experimental design: this study assessed both Wuhan-D614G-backbone and delta-backbone mutants, whereas the previous study assessed Wuhan-D614G-backbone only. In addition, the method to generate recombinant viruses differed between this study and the previous study. In the previous study, the recombinant viruses were prepared by using a PCR-based method, CPER, which has the risk of PCR errors16 In fact, the recombinant viruses produced by Saito et al.4 contained unintended mutations (their Wuhan-D614G virus stock has nonsynonymous mutations at Nsp13 and ORF3a), and the sequence of their parental Wuhan virus differs at position ORF8-84 from our parental Wuhan virus (Supplementary Table S1), which could affect the pathogenicity of the virus. By contrast, we produced recombinant viruses by using cloned and sequence-verified BACs, which can yield recombinant viruses with a very low genetic mixture.16 However, without a direct comparison of the virus stocks, we cannot definitively state that the difference in recombinant virus production between the two studies caused the discrepancy. Nevertheless, since the differences in pathogenicity of the parental viruses and mutant viruses in both studies were similar, that is, in both studies the differences were small, we can conclude that the S-P681R substitution alone does not significantly enhance pathogenicity. The roles of S-P681R and S-D950N in virus transmission efficiency remains unclear; further studies are needed to clarify this feature.
S-P681R significantly enhanced spike cleavage and membrane fusion, but reduced virus replication significantly in VeroE6 cells and slightly in VeroE6/TMPRSS2 cells. This could be explained by the low stability of cleaved spike proteins. Cleaved spikes may cause the premature shedding of the S1 subunit, leading to loss of receptor binding function.25, 26, 27 The stability of spikes is an important factor for virus entry especially in the endosome-mediated pathway, since the spikes must retain their conformation under acidic conditions.27 S-P681R enhances spike cleavage, resulting in a reduction in protein stability, and it increases the ratio of abortive infection, leading to a reduction in virus fitness. The advantage for viruses with low spike cleavage efficiency in VeroE6 cells has been reported in several papers.28, 29, 30, 31, 32, 33, 34
Overexpression of TMPRSS2 in VeroE6 cells improved the replication efficiency of viruses possessing R at position 681 in the spike protein. SARS-CoV-2 that attaches to the ACE2 receptor on host cell surfaces enters the host cells via two distinct pathways: TMPRSS2-mediated and endosome-mediated pathways.20 In the former pathway, at the cell surface, spike proteins cleaved at the S1/S2 cleavage site are further cleaved by TMPRSS2 at the S2’ cleavage site, which induces membrane fusion. In contrast, viruses with un-cleaved spikes enter host cells through endocytosis independent of TMPRSS2 expression. In TMPRSS2-negative cells, viruses with spikes cleaved at the S1/S2 site are also taken up through endocytosis. Both types of viruses are delivered to the endosome where the spikes can be cleaved by another host protease cathepsin at both the S1/S2 and S2’ cleavage sites or the S2’ cleavage site, respectively, and induce membrane fusion. The expression pattern of the cathepsins and TMPRSS2 determines which pathway is primarily selected. In VeroE6 cells, viruses enter through endosome-mediated pathways because VeroE6 cells express cathepsins but not TMPRSS2.25,27,35 In VeroE6/TMPRSS2 cells, which express both cathepsins and TMPRSS2, viruses enter via the TMPRSS2-mediated pathway rather than the endosome-mediated pathway.25,27,35 Therefore, the overexpression of TMPRSS2 changes the entry pathway of viruses to the TMPRSS2-mediated pathway through which viruses with unstable cleaved spikes (especially viruses bearing S-P681R) can be processed by TMPRSS2 and enter cells efficiently. This change restores the inefficient virus replication in VeroE6 cells. S-P681R did not affect virus replication in Calu-3. In Calu-3 cells, which express TMPRSS2 and very low levels of cathepsins, viruses enter mainly via the TMPRSS2-mediated pathway.27 Taken together, our findings suggest that the disadvantages of spike destabilization by S-P681R may be offset by the facilitation of the TMPRSS2-mediated pathway due to the increased cleavage at the S2’ site by TMPRSS2 in target cells. Further studies are needed to test this hypothesis because spike destabilization by S-P681R has not been evaluated experimentally.
We found that S-P681R contributes to efficient spike cleavage and fusion. Although the S-P681R substitution is unique to the delta variant, alpha and omicron variants have an amino acid substitution at this position: S-P681H. S-P681H increases cleavage efficiency but does not enhance fusion activity,36 suggesting that the amino acid residue R at this position is superior for both spike cleavage and membrane fusion. The S-D950N substitution, which lies in the HR1 of the S2 site, was involved in fusion activity but did not affect spike cleavage. Although the contribution of this substitution to fusion activity was limited compared with that of the substitution at position 681, S-D950N enhanced the fusion activity of S-P681R when both substitutions were present.
Reverse genetics enabled us to engineer these specific virus genomes. Since the emergence of SARS-CoV-2, several reverse genetics systems for SARS-CoV-2 have been established.37, 38, 39, 40, 41, 42 Reverse genetics systems using BAC are classical methods to generate viruses with a large genome such as coronaviruses and herpes viruses.41,43,44 Although this method is inferior in terms of rapidity to some other methods, such as PCR-based methods, we can efficiently generate recombinant viruses with a very low genetic mixture by using cloned and sequence-verified BACs. The homogeneity of the virus stocks generated by BACs is highly reproducible and higher than that generated by PCR-based methods such as CPER.16 This is why we utilized the BAC system to generate the viruses in this study. Indeed, we confirmed by NGS analysis that the frequency of unwanted mutations in the stock viruses was sufficiently low.
In conclusion, by using reverse genetics, we demonstrated that of the amino acid substitutions that are unique to the delta variant, namely, S-P681R and S-D950N, S-P681R enhances spike cleavage efficiency and membrane fusion and S-D950N slightly promotes membrane fusion alone or in combination with S-P681R. Furthermore, S-P681R reduces viral fitness under the condition of low TMPRSS2, in which the virus uses the endosome-mediated pathway. Although these substitutions did not drastically change virus pathogenicity in hamsters, these two substitutions in combination with other substitutions might contribute to high pathogenicity. Comparison of the pathogenicity of recombinant Wuhan-backbone virus with a delta spike with that of a Delta-backbone virus with a Wuhan-D614G spike would clarify this point. Regardless, we need to monitor the emergence of novel variants bearing these substitutions.
Contributors
Y.F., S.Y., and Y.K. designed the study. Y.F., M.K., S.I., R.U., Y.H., M.I., T.S., and S.Y. performed the experiments. Y.F. and S.Y. analyzed the data. Y.F., S.Y., and Y.K. wrote the manuscript. Y.F., S.Y., and Y.K. verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
The data needed to support the conclusions of this study have been included in the paper. All other data supporting the conclusions of this study are available from the corresponding authors upon reasonable request.
Declaration of interests
Y.K. has received collaborative research funds from the following companies. For antiviral studies; FUJIFILM Toyama Chemical Co. LTD (various viruses), Shionogi &; Co. LTD (influenza viruses). For vaccine and other studies; Daiichi Sankyo Pharmaceutical, Otsuka Pharmaceutical, KM Biologics, Kyoritsu Seiyaku, Fuji Rebio, Tauns Laboratories, Inc., Matsubara Co. LTD. As the Flagship Center of the Japan Initiative for World-leading Vaccine Research and Development Centers, we plan to initiate collaborative research related to vaccine development with the following companies: Shionogi &Co. LTD, Daiichi Sankyo Pharmaceutical, KM Biologics, Cytiva, Sysmex Corporation, NEC.
Acknowledgments
We would like to thank Dr. Susan Watson for editing the manuscript, and Drs. Wataru Kamitani (Gunma university) and Yasushi Kawaguchi for their assistance with the BAC construction. We also thank Yuko Sato and Seiya Ozono for their technical assistance.
This work was supported by grants from the Japan Agency for Medical Research and Development (JP22fk0108637, JP22wm0125001, JP22wm0125002, and JP223fa627001), from the Center for Research on Influenza Pathogenesis (HHSN272201400008C), and from the Center for Research on Influenza Pathogenesis and Transmission (75N93021C00014) of the National Institute of Allergy and Infectious Diseases.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104561.
Contributor Information
Seiya Yamayoshi, Email: yamayo@ims.u-tokyo.ac.jp.
Yoshihiro Kawaoka, Email: yoshihiro.kawaoka@wisc.edu.
Appendix A. Supplementary data
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 4.Saito A., Irie T., Suzuki R., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Liu J., Johnson B.A., et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022;39:110829. doi: 10.1016/j.celrep.2022.110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheikh A., McMenamin J., Taylor B., Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twohig K.A., Nyberg T., Zaidi A., et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22:35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motozono C., Toyoda M., Zahradnik J., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X., He C., Hong W., Zhang K., Wei X. The challenges of COVID-19 delta variant: prevention and vaccine development. MedComm (2020) 2021;2:846–854. doi: 10.1002/mco2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrov D.A., Knox G.W. Emerging mutation patterns in SARS-CoV-2 variants. Biochem Biophys Res Commun. 2022;586:87–92. doi: 10.1016/j.bbrc.2021.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Xiao T., Cai Y., et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuyama S., Nao N., Shirato K., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai M., Halfmann P.J., Yamayoshi S., et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2106535118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X., Quan S., Xu M., et al. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2025866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furusawa Y., Yamayoshi S., Kawaoka Y. The accuracy of reverse genetics systems for SARS-CoV-2: circular polymerase extension reaction versus bacterial artificial chromosome. Influenza Other Respir Viruses. 2023;17 doi: 10.1111/irv.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takashita E., Kinoshita N., Yamayoshi S., et al. Efficacy of antibodies and antiviral drugs against Covid-19 omicron variant. N Engl J Med. 2022;386:995–998. doi: 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett K.S., Werner A.P., Connell S.O., et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat Immunol. 2021;22:1306–1315. doi: 10.1038/s41590-021-01021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., et al. SARS-CoV-2 omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang J., Wan Y., Luo C., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai M., Iwatsuki-Horimoto K., Hatta M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uraki R., Halfmann P.J., Iida S., et al. Characterization of SARS-CoV-2 omicron BA.4 and BA.5 isolates in rodents. Nature. 2022;612:540–545. doi: 10.1038/s41586-022-05482-7. [DOI] [PubMed] [Google Scholar]
- 23.Suryawanshi R.K., Chen I.P., Ma T., et al. Limited cross-variant immunity from SARS-CoV-2 omicron without vaccination. Nature. 2022;607:351–355. doi: 10.1038/s41586-022-04865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahan K., Giffin V., Tostanoski L.H., et al. Reduced pathogenicity of the SARS-CoV-2 omicron variant in hamsters. Med. 2022;3:262–268.e4. doi: 10.1016/j.medj.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peacock T.P., Goldhill D.H., Zhou J., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L., Jackson C.B., Mou H., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laporte M., Raeymaekers V., Berwaer R.V., et al. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau S.-Y., Wang P., Mok B.W.-Y., et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z., Zheng H., Lin H., et al. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94(17) doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong Y.C., Lau S.Y., Wang To K.K., et al. Natural transmission of bat-like severe acute respiratory syndrome coronavirus 2 without proline-arginine-arginine-alanine variants in coronavirus disease 2019 patients. Clin Infect Dis. 2021;73:e437–e444. doi: 10.1093/cid/ciaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klimstra W.B., Tilston-Lunel N.L., Nambulli S., et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J Gen Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson A.D., Williamson M.K., Lewis S., et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki M., Uemura K., Sato A., et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubinski B., Fernandes M.H.V., Frazier L., et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (alpha) spike. iScience. 2021;25:103589. doi: 10.1016/j.isci.2021.103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torii S., Ono C., Suzuki R., et al. Establishment of a reverse genetics system for SARS-CoV-2 using circular polymerase extension reaction. Cell Rep. 2021;35:109014. doi: 10.1016/j.celrep.2021.109014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amarilla A.A., Sng J.D.J., Parry R., et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat Commun. 2021;12:3431. doi: 10.1038/s41467-021-23779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X., Lokugamage K.G., Zhang X., et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat Protoc. 2021;16:1761–1784. doi: 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiem K., Ye C., Martinez-Sobrido L. Generation of recombinant SARS-CoV-2 using a bacterial artificial chromosome. Curr Protoc Microbiol. 2020;59:e126. doi: 10.1002/cpmc.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye C., Chiem K., Park J.-G., et al. Rescue of SARS-CoV-2 from a single bacterial artificial chromosome. mBio. 2020;11(5) doi: 10.1128/mBio.02168-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Y.J., Okuda K., Edwards C.E., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St-Jean J.R., Desforges M., Almazán F., Jacomy H., Enjuanes L., Talbot P.J. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J Virol. 2006;80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messerle M., Crnkovic I., Hammerschmidt W., Ziegler H., Koszinowski U.H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci U S A. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.