Abstract
In recent years, the development and implementation of animal-free approaches to chemical and pharmaceutical hazard and risk assessment has taken off. Alternative approaches are being developed starting from the perspective of human biology and physiology.
Neural tube closure is a vital step that occurs early in human development. Correct closure of the neural tube depends on a complex interplay between proteins along a number of protein concentration gradients. The sensitivity of neural tube closure to chemical disturbance of signalling pathways such as the retinoid pathway, is well known. To map the pathways underlying neural tube closure, literature data on the molecular regulation of neural tube closure were collected. As the process of neural tube closure is highly conserved in vertebrates, the extensive literature available for the mouse was used whilst considering its relevance for humans. Thus, important cell compartments, regulatory pathways, and protein interactions essential for neural tube closure under physiological circumstances were identified and mapped. An understanding of aberrant processes leading to neural tube defects (NTDs) requires detailed maps of neural tube embryology, including the complex genetic signals and responses underlying critical cellular dynamical and biomechanical processes. The retinoid signaling pathway serves as a case study for this ontology because of well-defined crosstalk with the genetic control of neural tube patterning and morphogenesis. It is a known target for mechanistically-diverse chemical structures that disrupt neural tube closure
The data presented in this manuscript will set the stage for constructing mathematical models and computer simulation of neural tube closure for human-relevant AOPs and predictive toxicology.
Keywords: Developmental neurotoxicology, Retinoic acid, Neural tube defect, Systems biology
1. Introduction
The development and implementation of animal-free approaches to chemical and pharmaceutical hazard and risk assessment has reached a critical crossroad. The realization grows that the approach of implementing individual animal-free alternative methods is limited by the complexity of toxicities at the level of the intact organism [1,2]. A novel paradigm emerges that takes a fundamentally different starting point in contrast to the approach that replaces individual animal studies with reductionistic in vitro assays [3]. Alternatively, an approach from the perspective of human biology, physiology and toxicology takes an open view towards what knowledge is needed to sufficiently cover all aspects necessary for an inclusive human hazard and risk assessment [4,5]. Briefly, the general idea is that a map of human biology will allow one to identify comprehensive networks of quantitative Adverse Outcome Pathways (qAOP) in the future. The human biology map, when captured in an in silico model, has been referred to as the virtual human [6]. The molecular network underlying human biology responding to toxic insults has been named the toxicological ontology [7,8]. The quantitative aspect of this ontology will allow the selection of a limited number of steps in the network that need to be monitored to reliably calculate the response of the network as a whole and hence to predict the adverse outcome. Based on these selected steps, which are comparable to key events in an AOP, dedicated animal-free, preferably human-based, assays can be selected with which quantitative concentration-responses to chemical exposures can be measured. The integration of individual quantitative key event responses requires an intelligent computational tool that calculates dose-dependent compound-induced changes in the ontology leading to the adverse outcome prediction [9]. For application in integrated risk assessments, this dynamic model calculating quantitative concentration-dependent adverse outcomes needs to be appended with kinetic models and exposure estimates [4].
This open view approach allows a fresh perspective on what toxicities and diseases need to be considered, which can be significantly broader than currently required under existing legislation. Given that the integral human biology is the starting point, this approach includes all possible adverse outcomes, and therefore is in principle more inclusive than current practice which is limited by the spectrum of end points addressed in current regulatory guideline animal studies.
The current paradigm is considered to be adequately health protective, but it does not scale to the problem of testing 80 K chemicals in the human exposure landscape. The computational models can provide a tier 1 screen to inform targeted testing for, in this case, developmental toxicity. Virtual embryo simulations such as those developed in US EPA’s ‘Virtual Embryo’ program can work to translate data-driven machine learning models into mechanistic simulations for critical developmental transitions. These models can feed into Integrated Approaches to Testing and Assessment (IATA) in which the dynamics of those key events that represent tipping points in the qAOP network are combined with exposure and dosimetry to predict adversity and to achieve an integrated risk assessment.
The human biology map, when captured in an in silico model, has been referred to as the virtual human [10]. This concept provides an integral model of human physiology, which increasingly finds applications in clinical medicine as well as toxicological approaches [11]. In toxicology, it facilitates the integration of a wide variety of data types relevant for toxicity assessment, including kinetics and dynamics of chemicals in biological systems such as the wealth in vitro assays compiled in the Tox Cast library [12,13]. Thus, the virtual human concept aims at data integration towards computational modelling of the causation of adverse health effects, and consequently of chemical hazard and risk assessment [14]. Although it should be acknowledged that building the virtual human and the toxicological ontology require significant effort and time, its principal point of departure, together with the slow progress of implementing animal-free alternatives in current human safety assessment, merit strong investments in this innovative approach. Ongoing efforts in the realm of computational models for human physiology and disease, diagnostics and therapy, coupled with big data analysis through artificial intelligence and machine learning, indicate that in other areas of human health these virtual approaches are rapidly becoming mainstream [15–19]. Toxicological risk assessment need such innovations to move away from the scientifically and ethically challenged animal experimentation.
This manuscript follows the concept of the virtual human, focusing on one specific area in developmental toxicology that is highly relevant to human risk [20,21]. It describes the biology of neural tube closure from a molecular and cellular perspective. Neural tube defects are among the most prevalent human congenital malformations, which warrants specific attention in chemical safety assessment [22]. We took advantage of the highly conserved nature of the molecular mechanisms underlying neural tube closure throughout vertebrate biology, which leads to the pharyngula stage embryo that all vertebrates pass through during development before species-specific developmental differences become morphologically apparent [23]. It allowed us to mine the extensive literature of the molecular regulation of vertebrate neural tube closure, considering the relevance for humans where possible, starting from the literature available for the mouse. This approach has of course been performed with a critical eye towards species specificity. This description of the molecular and cellular developmental biology underlying neural tube closure follows up on our earlier studies focused on the morphogenetic role of mesoderm-derived all-trans-retinoic acid (ATRA) in neural tube development [24,25].
ATRA provides a small but well-known and important fraction of the essential molecular regulators of neural tube formation. The current manuscript explores in more detail the developmental biology of neural tube closure, focusing on ATRA-related molecular pathways linked to the various cell types in which they occur, and their role in driving intercellular interaction and its morphogenetic consequences, ultimately leading to closure of the neural tube. ATRA gradients play critical roles in early embryonic cell differentiation, and are regulated in time and space throughout embryo development. Retinoic acid response has also emerged from an extensive ToxCast library multi-assay response analysis as the most prominent developmental toxicant response [26]. It is the local balance between ATRA-producing retinol dehydrogenase families and ATRA-metabolizing cytochrome P450 family 26 (CYP26) enzyme families that determines local ATRA concentrations. In the neural tube ATRA as a differentiation inducer counteracts the activity of fibroblast growth factor (FGF) which stimulates cell proliferation. Opposite gradients of ATRA and FGF direct development along the rostro-caudal axis of the vertebrate embryo. In the ventro-dorsal direction a host of different factors such as chorda-derived sonic hedgehog (SHH) and neuroectoderm-derived WNT3 co-determine specific morphogenetic differentiation avenues. The resulting molecular neural tube closure map was collected in CellDesigner® software [27]. In follow-up research, based on existing data on the perturbation of gene expression by chemicals in cellular assays, from this map a qAOP network representing a toxicological ontology can be derived and represented in an in silico model. This ontology can inform the assays that need to be applied and combined to build the in silico model to calculate the adverse outcome at the level of the intact embryo.
The present manuscript compiles and integrates existing information on the molecular, cellular and spatial regulation of mammalian (mouse and/or human) neural tube closure in a systems biology network, representing the first step towards the generation of an in silico model for spinal/caudal neural tube closure. The construction of the biological regulation map underlying the in silico model is dependent on existing knowledge of the molecular regulation on embryogenesis. Although our systems model is presented as a two-dimensional map, morphogenesis is critically three-dimensional. This allows anterior-posterior, and dorsoventral gradients to interact in driving morphogenesis and generate left-right symmetry. While mammalian development at the early stages of embryogenesis up to the pharyngula stage is highly conserved [28], it should be kept in mind that building a virtual human embryo based on animal (mouse) data comes with unknown limitations.
2. Methods
The morphology of neural tube closure was used to define cell compartments playing a role in neural tube closure. Data were collected on changes in these cell compartments required for normal neural tube closure. Underlying genetic processes and interactions, and establishment of gradients of key molecular factors were identified based on literature search using the Abstract Sifter tool [29]. Publications from the PubMed database (until 2016) were selected if they were annotated with the following terms: Cauda Equina, Meningocele, Meningomyelocele, Neural Crest, Neural Plate, Neural Stem Cells, Neural Tube, Neural Tube Defects, Neuroepithelial Cells, Neuroglia, Neurons, Primitive Streak, Sacral defect and anterior sacral meningocele, Sacrococcygeal Region, Spina Bifida Cystica, Spina Bifida Occulta, Spinal Dysraphism, Spine. The database was manually curated for relevance to development. This set of publications was searched for the role of ATRA in development based on the schematic representation by Tonk and coworkers [30] which resulted in selection of the spinal section of the neural tube for further elucidation of molecular processes playing a role. Specific searches on genes and cell types were conducted from the selected publications or, for more recent publications, in PubMed to elucidate direct or indirect interactions between genes or between cells and genes or to confirm interactions with other research. When available, data from genetic mouse models (including (conditional) knock-out models) was used to substantiate relationships between regulatory molecules and pathways. As the data should be applicable for human toxicological responses, processes obtained from human data were preferred. However, since human data are limited, the abundant mouse data were also used as the biological process of neural tube closure is highly conserved between vertebrates. Interactions that play a role in other species or results from in vitro assays were only used as an indicator and required confirmation by human or mouse data.
Cell designer software was used to qualitatively visualize the molecular processes of neural tube closure. All required signaling pathways, regulatory loops and underlying gene networks leading to phenotypical changes necessary for normal development were described integrally using a dedicated graphical software package (CellDesigner v4.4.2) (www.celldesigner.org). This software package is designed to capture systems biology networks while using a unified systems biology markup language (SBML). An SBML-compliant language is needed to enable the incorporation of signaling approaches as it relates to the novel contribution of the Cell Designer. This will ultimately enable incorporation of signaling networks in modelling approaches. Kinetic and dynamic parameters were therefore not included since this will be part of future computational modelling of the neural tube closure process.
3. Results
To develop a systems biology network that is ready for integration in a computer model, we focused on five tissue compartments or cell populations for their roles in autonomous signalling in the developing neural tube. These are the (non-neural or surface) ectoderm, the (future) neural crest cells (ectodermal of origin), the neuroectoderm, the paraxial mesoderm and the notochord. Within the neuroectoderm, two populations of cells were operationally discriminated based on their behavior at the extreme ventral and dorsal ends of the neural folds, induced by dorsoventral gradients of inducing factors. These contain the cell populations that during the development will form the median hinge point and the dorsolateral hinge points. The sequence of events that starts with the formation of the neural groove and ultimately leads to a fully closed neural tube has been captured in 2D in a series of six diagrams (Fig. 1) to visualize the whole process and thus to facilitate understanding of the chronology of the different processes. Below we describe the biomechanical processes, their origin in regulatory networks and their dependence on the different molecular gradients of the spinal part of the neural tube (summarized in Tables 1 and 2).
Fig. 1.
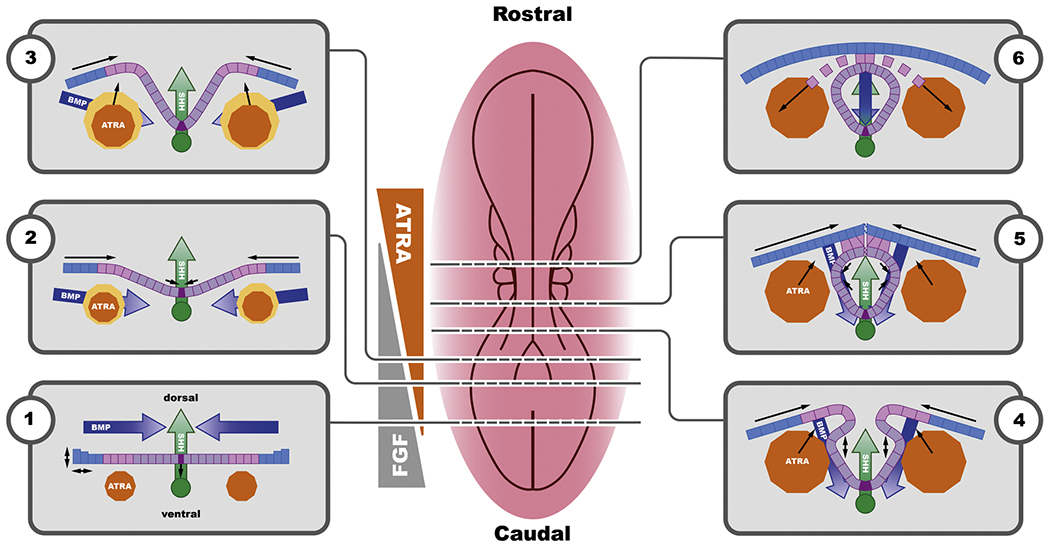
Two-dimensional description of the series of events leading to neural tube closure starting with the formation of the neural groove and ultimately leading to a fully closed neural tube (1-6). The centrally placed overview of a developing embryo can be considered a snapshot of the progressive closing neural tube. Color coding in the transverse sections: orange: paraxial mesoderm, primary source of ATRA; dark blue: BMP gradient originating from the surface ectoderm; pink: neural crest; green: notochord and the related SHH gradient; purple: neuroectoderm; dark purple: median hinge point cells. Colored arrows indicate directionality of the respective gradients; line arrows indicate physical movement of tissue or cell layer. The schematic representation of the (static) mutually antagonistic ATRA (orange) - FGF8 (grey) gradient illustrates a general concept and it should be noted that in the developing embryo this is a dynamic gradient that changes as the embryo grows and elongates.
Table 1.
Summary of important protein concentration gradients and their origin.
| Gradient | maintained by (cell type) |
|---|---|
| BMP | Surface Ectoderm |
| SHH | Notochord and MHP |
| ATRA | Paraxial Mesoderm |
| FGF | Paraxial Mesoderm |
| WNT | Surface Ectoderm |
Table 2.
Summary table of key molecules in mammalian neural tube closure.
| Cell Type | Behavior | Signal molecule | Reference |
|---|---|---|---|
| Surface Ectoderm | Differentiation and migration | BMP, FGF, NOG, CHRD, FST | 31, 32, 43–45 |
| Fusion of SE | WNT-PCP signalling, RAC-1, CDC42, RHOA, CDH1, GHRL2 | 56, 70–74 | |
| Neural Crest | Inhibition of differentiation | FGF8, PAX3 | 77, 78 |
| EMT | BMP, MSX1, TGFbeta, SNAI1, SNAI2, CDH7, CDH11 | 65, 76–79 | |
| Neuro Ectoderm (DLHP) | Cytoskeletal reorganization | ATRA, FGF, WNT, RHOA, ROCK | 52, 67, 68 |
| Bending | NOG, ZIC, BMP, ATRA, GLI2, FGF, MSX1 | 31, 33, 38, 41, 62, 63 | |
| Neuro Ectoderm (MHP) | Cytoskeletal reorganization | ZIC, GLI2 | 31, 33, 34, 56–58 |
| Apical constriction/bending | WNT-PCP signalling, ATRA, BMP, SHH | 40, 41, 59 | |
| Fusion of NE | FGF8, GHRL2, CDH2 | 74, 75 | |
| Notochord | SHH gradient shift | SHH | 61 |
| Stable SHH gradient | FGF | 36, 38, 39, 40, 41, 42 | |
| Paraxial Mesoderm | Growth and body axis extension | FGF, ZIC1, ZIC3 | 35, 36, 38, 52–55, 60 |
| Differentiation | ATRA, RHD10, RALDH2, CYP26, DHRS3 | 30, 36, 47–52 |
Neural tube closure starts with a flat ectoderm with a bone morphogenetic protein (BMP) gradient originating from the non-neural or surface ectoderm [31,32], an SHH gradient originating from the notochord [33,34] and all-trans retinoic acid (ATRA) being produced by the paraxial mesoderm (future somites) [35,36] (Fig. 1.1). At this stage, ATRA levels are relatively low and FGF levels are high, resulting in cell division and caudal body axis extension while at the same time inhibiting neural crest specification as well as neuronal differentiation [36]. At the level of the mesoderm, ATRA regulates somitogenesis through direct transcriptional repression of FGF8. With body axis elongation, the boundary between the area under control of the FGF8 gradient coming from the caudal epiblast and the area under control of the ATRA gradient from the presomitic mesoderm shifts caudally with ATRA restricting the anterior extent of FGF8 expression [35,37].
SHH is induced by the relatively high levels of FGF [36,38,39] and is produced in the notochord, which causes a ventro-dorsal gradient of SHH [40,41]. Following stimulation of SHH production by FGF10, SHH expression is self-sustained via binding of SHH to its own receptor [40, 42]. BMP is produced in the cells of the surface ectoderm and is stimulated by FGF [43,44]. BMP function can be inhibited through expression of one of the many BMP inhibitors like noggin (NOG), chordin (CHRD) and follistatin (FST), which block the binding of BMP to its receptor [31,32,44,45]. Since BMP plays an important role in late processes such as differentiation and migration, the effect of BMP is repressed at this early stage, to avoid premature differentiation [46].
Like FGF, ATRA is also produced in the paraxial mesoderm. The first step of ATRA formation is uptake of Vitamin A by the cells via the signaling receptor and transporter of retinol (STRA6 receptor) and subsequent metabolization to retinaldehyde by retinol dehydrogenase 10 (RHD10) [30,47,48]. Retinaldehyde is further metabolized to ATRA by aldehyde dehydrogenase 1 family member A2 (RALDH2) [30,49,50]. RA inhibits the expression of both RDH10 and RALDH2, and upregulates the expression of dehydrogenase/reductase 3 (DHRS3) that converts all-trans retinal back to Vitamin A thus ensuring a tight feedback loop on its formation [30,47]. RA is metabolized to inactive metabolites by CYP26-enzymes, specifically cytochrome P450, family 26, subfamily a, polypeptide 1 (CYP26A1) [51]. Comparable to RDH10 and RALDH, expression of CYP26-enzymes for RA inactivation is regulated by intracellular RA levels [50]. Both the inhibition of RA forming enzymes and the induction of RA metabolizing enzymes are crucial in keeping the RA levels well controlled in the developing embryo. The balance between FGF and RA determines whether cells proliferate or differentiate [35,52]. This interaction is controlled in the somites through direct transcriptional repression of FGF by ATRA and FGF-induced expression of zinc finger proteins 1 and 3 (ZIC1 and ZIC3 respectively), which induce the expression of CYP26 enzymes [35,52–54]. In addition, ZIC1 and ZIC3 induce the expression of RALDH2, but FGF inhibits RALDH2 expression [52,55]. Both routes are important in controlling the balance between FGF and RA [38].
SHH, originating from the notochord is relatively high at the site of the neuroectoderm closest to the notochord. In these cells, which will become the floor plate, SHH binds to its receptor and inhibits the expression of ZIC [33,34,56], either directly or by inhibiting glioma-associated oncogene family zinc finger 2 (GLI2) which also inhibits ZIC [31]. This results in cytoskeletal reorganization and the formation of the Median Hinge Point (MHP; Fig. 2) [57,58]. Cells of the MHP start to contract on the apical side (inside of the developing tube) in response to Wnt signaling pathway / planar cell polarity pathway (WNT-PCP signaling) [59]. The resulting bend of the MHP is the start of the process of invagination of the neural groove. This process is facilitated by the growth of the paraxial mesoderm providing upward forces [60]. ATRA plays a role in this process by inhibiting the expression of GLI2 [38]. Median hinge point cells will eventually start producing SHH themselves [61] strengthening the sustained SHH gradient along the ventro-dorsal axis.
Fig. 2.
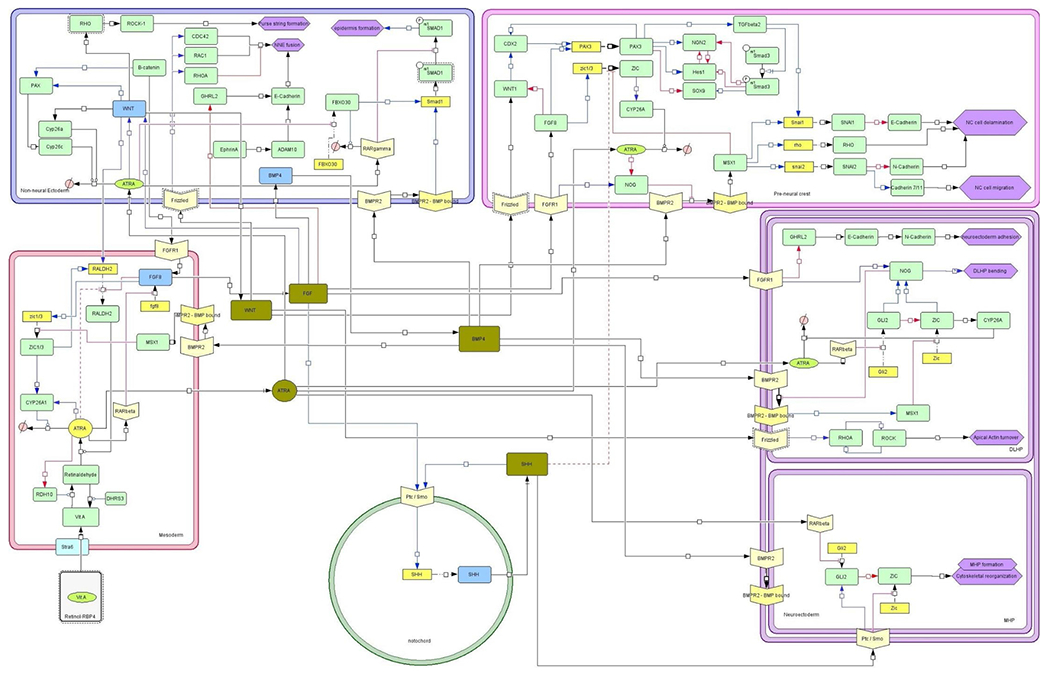
Systems biology network (CellDesigner® format) describing the cellular and molecular interactions in the most important cell populations/compartments underlying neural tube closure. Compartments: Green circle: notochord; Red rectangle: paraxial mesoderm; Blue rectangle: surface ectoderm; Pink rectangle: Neural crest; Purple rectangle: Neuroectoderm divided in DLHP (top) and MHP (bottom). Blue lines and arrows are stimulating, red lines and arrows and blocked lines are inhibitory, black lines and arrows indicate an interaction. Interactions indicated with a dashed line (irrespective of color) are subject to uncertainty which is defined as the interaction being presented in or deductible from literature but without experimental substantiation. Light-yellow v-shaped boxes are receptors. Yellow boxes are genes if expressed resulting in proteins in green boxes. Blue boxes indicate the main source of that protein. Olive green boxes indicate the gradients excreted from different cell types and interacting with other compartments. Purple hexagons indicate morphological changes. Pink circle with a diagonal line represent ‘inactive metabolites’.
Upon further growth of the mesoderm and the paraxial mesoderm, and facilitated by the bending of the MHP, two ridges will start to form on either side of the neural groove (Fig. 1.3). With the upward movement of the neuroectoderm forming the neural folds, the BMP gradient changes direction relative to the SHH gradient which becomes increasingly dorsoventral [40,41]. This change in the BMP gradient implies a reduced repression of BMP on ATRA formation resulting in increasing ATRA levels. At this stage the surface ectoderm starts to provide an additional driving force via cell division, stiffening and flattening of the cells pushing the ridges of the neural folds to the central line [60]. This results in further bending of the MHP and formation of the neural groove and a further shift in the BMP-SHH gradient.
In the neuroectoderm ZIC induces NOG expression [41]. NOG inhibits BMP binding to its receptor, which results in a positive feedback loop on the expression of NOG [62,63]. ATRA plays a role in the inhibition of GLI2 in the neuroectoderm [38]. Ensuring relatively low levels of RA at this stage, and high levels of FGF is required to maintain high NOG levels. This upregulated NOG expression is required for the subsequent formation and bending of the dorsal lateral hinge points (DLHPs) [31,33,41,63]. This further facilitates bending of the neural tube on the lateral sides and will eventually bring the ridges of the neural folds in juxtaposition in the dorsal midline [59,60] (Fig. 1.4). DLHP formation comprises cell proliferation, a dorsal movement of cells in the neuroepithelium and cytoskeletal rearrangement via apical movement of nuclei creating wedge-shaped cells, effectively bending the neural folds [64]. The anatomical localization of the DLHPs is related to the balance between various signal molecules/gradients including opposing BMP and SHH gradients and the local (FGF-mediated) expression of the BMP inhibitors such as NOG [41,65,66]. Conditional knock-out data show that ZIC plays a crucial role in this process [41]. Driving the cytoskeletal reorganization involved in DLHP formation is ATRA-and FGF-mediated expression of WNT [52]. WNT binds to the Frizzled receptor on the neuroectoderm and induces the expression of ras homolog family member A (RHOA), which induces the expression of Rho associated coiled-coil (ROCK). ROCK expression on its turn, results in a positive feedback loop by induction of RHOA [67,68]. Increased expression of ROCK then results in the apical actin turnover required for bending of the apical side of the DLHP cells [67].
With progressing neural tube closure come increasing levels of RA and decreasing levels of FGF leading to an inhibition of NOG [69]. This inhibition of the BMP inhibitor effectively stimulates BMP signalling and enhances further development and differentiation of the neuroectoderm and preparation for closure.
Once the neural ridges are in juxtaposition, the final act of closure of the neural tube involves the fusion of the respective surface ectoderm and neuroectodermal layers and remodeling of the tissue layers (Fig. 1.5). Two types of cellular protrusions play a role in the fusion act, Rac family small GTPase 1 (RAC-1) mediated ruffles (lamellopodia-like) and cell division cycle 42 (CDC42)-mediated protrusions (filopodia) [70]. CDC42 and RAC1 are both upregulated by the non-canonical WNT-PCP pathway and balanced through RHOA-mediated stabilization of the actin network [71,72]. In parallel, WNT-signalling activating Rho GTPase (RHO) signalling results in the assembly of an actin cable that will form a purse string narrowing the dorsal opening of the NT as closure progresses [73].
Expression of E-cadherin (CDH1), required for fusion of the surface ectoderm, is induced by grainyhead-like transcription factor 2 (GHRL2) [56]. GHRL2 expression is negatively regulated by FGF leading to increased expression of GHRL2 and thus E-cadherin once closure progresses and the FGF gradient fades [71,74]. In the neuroectoderm, a switch is made from E-cadherin to N-cadherin (CDH2) which is absent in the surface ectoderm [74,75].
Upon closure of the neural tube and fusion of the ectodermal layers, the former neural crest cells undergo epithelial-to-mesenchymal transition (EMT), delaminate from the layers of neural and non-neural ectoderm and migrate to their final destination (Fig. 1.6). To delaminate, the cells need to loosen intercellular connections, including the intercellular E- and N-cadherin connections between the neural crest cells and the non-neural ectoderm and neuroectoderm respectively [76]. On the border with the non-neural ectoderm a transforming growth factor beta (TGFbeta) regulated increase in SNAIL results in the disruption of E-Cadherin [77,78]. This process is supported by an increased BMP-mediated stimulation of snail family transcriptional repressor 1 (SNAI1) expression through MSX1 [65,79]. On the border with the neuroectoderm, increased expression of snail family transcriptional repressor 1 (SNAI2) downstream of MSX1 results in a suppression of N-cadherin in the neural crest cells, and an increase in Cadherin 7 and 11 (CDH7 and CDH11 respectively) expression required for migration [76,79]. Premature differentiation of neural crest cells prior to migration is inhibited through an FGF8-induced stimulation of paired box 3 (PAX3) expression [77,78].
4. Conclusion/ discussion
The molecular network underlying neural tube closure presented here is a work in progress and does not visualize all genes and intermediate steps that may play a role in this process. Specifically, those genes that have an intermediate role are not always included for reasons of simplification. Relations between genes are suggested to be direct in the network, but may actually be missing intermediate steps. Whether these steps serve a functional “gate-keeper” role remains to be elucidated. However, the network presented visualizes all important biological processes involved in neural tube closure based on current knowledge. Quantitative aspects will need to be included, either based on scarce literature information or as relative gradients in the computational model. As indicated in general terms above, from this network the major rate-limiting regulating aspects need to be identified that can be used as biomarkers to monitor compound-induced effects in in vitro assays. The next step in this process therefore entails an extensive analysis of the abundant and still growing literature on compound-induced gene expression changes and their consequences for cell behavior in in vitro cell models. In developmental neurotoxicity a wealth of assays have been developed for monitoring cell proliferation, migration, differentiation, neural network formation, and development of electrical activity [80,81]. Molecular pathways that respond to compound exposure in these assays, as well as data from genetic models such as (conditional) knock-outs, may provide candidates for biomarkers of developmental toxicity to be prioritized for monitoring to feed the in silico model. As an example, the enzyme system regulating retinoic acid homeostasis has been shown to provide a sensitive biomarker set for the (neuro) developmental toxicity of triazole antifungal compounds [82]. Retinoic acid being a morphogen suggests that its dysregulation may provide a more broadly applicable biomarker set for developmental toxicity. Its prominent role in neural tube closure as shown in this review underpins this notion.
Building the in silico model can be done using dedicated software packages. As an example, the Virtual Embryo® program of US-EPA published in silico 2D models for other elements of embryogenesis, such as for blood vessel formation, palatal closure, and urogenital development in recent years [42,83,84]. The current model development effort for NTD is focusing on a 3D model. Although building the model in 3D implicates an increase in complexity, 2D models are not able to capture the crucial dynamics of protein gradients and their interplay throughout development. This will not be a mere reconstruction of anatomy but provide a model that captures the dynamics of development and can be perturbed for sensitivity analysis of imputing data while looking for quantitative relationships between genetic and toxicological effects on key cellular processes. The resulting in silico model will be instrumental in predicting the developmental effects in the intact embryo of changes in gene expression and cell behavior that are observed in cell-based assays as a consequence of compound exposure. Thus, results from a battery of underlying cell assays can be fed into the in silico model for prediction of toxicity in the intact embryo. In order to define the test battery, the rate-limiting components in the gene- and cell-interactions need to be identified and represented in relevant assays. At a higher level of integration, the neural tube closure model should ultimately be combined with other models representing additional morphogenetic pathways. Given the abundance of rodent data, a substantial fraction of these models will still be (partially) based on rodent data. However, with the extensive knowledge of human homologs of rodent genes and proteins, it will be possible to draw a human overlay of the network and followed by targeted verification using e.g. genomics and proteomics technologies in well-defined (human) in vitro assays. These efforts provide proof of principle for the concept of the virtual embryo, which may in the future replace animal testing and allow fine-tuning the developmental toxicity assessment of chemicals and pharmaceuticals in humans.
It should be noted that in vitro assays, the main anticipated data providers for feeding the in silico model, will not be able to cover every aspect of embryogenesis. Particularly when it comes to pattern formation and morphogenesis at the level of the intact embryo, the virtual embryo is needed to provide the necessary level of integration. Therefore, the in silico model needs to be sufficiently reliable, as evidenced by sufficient coverage of the biology and by in depth case studies on data-rich compounds. The ToxCast and ToxRefDB databases provide ample examples of extensive in vitro and in vivo information available for such case studies [85,86]. This is a significant challenge that requires a critical appraisal throughout [87,88]. The ultimate aim of replacing animal studies and the high need for extending the rapidly progressing innovation in biomedical and clinical research into human chemical and pharmaceutical safety assessment makes this effort worthwhile.
Acknowledgements
We gratefully acknowledge the financial support from CEFIC-LRI under project codes AIMT-5 and AIMT5.2. The authors would like to thank Eric Gremmer for valuable contribution to the artwork.
Footnotes
Declaration of Competing Interest
The authors reported no declarations of interest.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- [1].Piersma AH, Burgdorf T, Louekari K, Desprez B, Taalman R, Landsiedel R, Barroso J, Rogiers V, Eskes C, Oelgeschläger M, Whelan M, Braeuning A, Vinggaard AM, Kienhuis A, van Benthem J, Ezendam J, Workshop on acceleration of the validation and regulatory acceptance of alternative methods and implementation of testing strategies, Toxicol. Vitr 50 (2018) 62–74. [DOI] [PubMed] [Google Scholar]
- [2].Burgdorf T, Piersma AH, Landsiedel R, Clewell R, Kleinstreuer N, Oelgeschläger M, Desprez B, Kienhuis A, Bos P, de Vries R, de Wit L, Seidle T, Scheel J, Schönfelder G, van Benthem J, Vinggaard AM, Eskes C, Ezendam J, Workshop on the validation and regulatory acceptance of innovative 3R approaches in regulatory toxicology – evolution versus revolution, Toxicol. Vitr 59 (2019) 1–11. [DOI] [PubMed] [Google Scholar]
- [3].Piersma AH, Ezendam J, Luijten M, Muller JJA, Rorije E, van der Ven LTM, van Benthem J, A critical appraisal of the process of regulatory implementation of novel in vivo and in vitro methods for chemical hazard and risk assessment, Crit. Rev. Toxicol 24 (0) (2014) 1–19. [DOI] [PubMed] [Google Scholar]
- [4].Staal YCM, Pennings JLA, Hessel EVS, Piersma AH, Advanced toxicological risk assessment by implementation of ontologies operationalized in computational models, Appl. In Vitro Toxicol 3 (4) (2017) 325–332. [Google Scholar]
- [5].Scialli AR, Daston G, Chen C, Coder PS, Euling SY, Foreman J, Hoberman AM, Hui J, Knudsen T, Makris SL, Morford L, Piersma AH,Stanislaus D, Thompson KE, Rethinking developmental toxicity testing: Evolution or revolution? Birth Defects Res 110 (10) (2018) 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Piersma A, van Benthem J, Ezendam J, Staal YCM, Kienhuis AS, The virtual human in chemical safety assessment, Curr. Opin. Toxicol 15 (2019) 26–32. [Google Scholar]
- [7].Baker N, Boobis A, Burgoon L, Carney E, Currie R, Fritsche E, Knudsen T, Laffont M, Piersma AH, Poole A, Schneider S, Daston G, Building a developmental toxicity ontology, Birth Defects Res. 110 (6) (2018) 502–518. [DOI] [PubMed] [Google Scholar]
- [8].Desprez B, Birk B, Blaauboer B, Boobis A, Carmichael P, Cronin MTD, Curie R, Daston G, Hubesch B, Jennings P, Klaric M, Kroese D, Mahony C, Ouédraogo G, Piersma A, Richarz A-N, Schwarz M, van Benthem J, van de Water B, Vinken M, A mode-of-action ontology model for safety evaluation of chemicals: outcome of a series of workshops on repeated dose toxicity, Toxicol. Vitr 59 (2019) 44–50. [DOI] [PubMed] [Google Scholar]
- [9].Thomas RS, Bahadori T, Buckley TJ, Cowden J, Deisenroth C, Dionisio KL, Frithsen JB, Grulke CM, Gwinn MR, Harrill JA, Higuchi M, Houck KA, Hughes MF, Hunter ES, Isaacs KK, Judson RS, Knudsen TB, Lambert JC, Linnenbrink M, Martin TM, Newton SR, Padilla S, Patlewicz G, Paul-Friedman K, Phillips KA, Richard AM, Sams R, Shafer TJ, Setzer RW, Shah I, Simmons JE, Simmons SO, Singh A, Sobus JR, Strynar M, Swank A, Tornero-Valez R, Ulrich EM, Villeneuve DL, Wambaugh JF, Wetmore BA, Williams AJ, The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency, Toxicol. Sci 169 (2) (2019) 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Piersma AH, van Benthem J, Ezendam J, Staal YCM, Kienhuis AS, The virtual human in chemical safety assessment, Curr. Opin. Toxicol 15 (2019) 26–32. [Google Scholar]
- [11].Van Sint Jan S, Geris L, Modelling towards a more holistic medicine: the Virtual Physiological Human (VPH), Morphologic 103 (343) (2019) 127–130. [DOI] [PubMed] [Google Scholar]
- [12].Saili KS, Franzosa JA, Baker NC, Ellis-Hutchings RG, Settivari RS, Carney EW, Spencer R, Zurlinden TJ, Kleinstreuer NC, Li S, Xia M, Knudsen TB, Systems modeling of developmental vascular toxicity, Curr. Opin. Toxicol 15 (1) (2019) 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shah I, Wambaugh J, Virtual tissues in toxicology, J. Toxicol. Environ. Health B Crit. Rev 13 (2-4) (2010) 314–328. [DOI] [PubMed] [Google Scholar]
- [14].Kavlock R, Dix D, Computational toxicology as implemented by the U.S. EPA: providing high throughput decision support tools for screening and assessing chemical exposure, hazard and risk, J. Toxicol. Environ. Health B Crit. Rev 13 (2-4) (2010) 197–217. [DOI] [PubMed] [Google Scholar]
- [15].Viceconti M, Hunter P, The virtual physiological human: ten years after, Annu. Rev. Biomed. Eng 18 (1) (2016) 103–123. [DOI] [PubMed] [Google Scholar]
- [16].Carpenter KA, Huang X, Machine learning-based virtual screening and its applications to alzheimer’s drug discovery: a review, Curr. Pharm. Des 24 (28) (2018) 3347–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yuan Y, Bai X, Luo C, Wang K, Zhang H, The virtual heart as a platform for screening drug cardiotoxicity, Br. J. Pharmacol 172 (23) (2015) 5531–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shinbane JS, Saxon LA, Digital monitoring and care: virtual medicine, Trends Cardiovasc. Med 26 (8) (2016) 722–730. [DOI] [PubMed] [Google Scholar]
- [19].Du P, Paskaranandavadivel N, Angeli TR, Cheng LK, O’Grady G, The virtual intestine: in silico modeling of small intestinal electrophysiology and motility and the applications, WIRES Syst. Biol. Med 8 (1) (2016) 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hutson MS, Leung MCK, Baker NC, Spencer RM, Knudsen TB, Computational model of secondary palate fusion and disruption, Chem. Res. Toxicol 30 (4) (2017) 965–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Leung MCK, Hutson MS, Seifert AW, Spencer RM, Knudsen TB, Computational modeling and simulation of genital tubercle development, Reprod. Toxicol 64 (2016) 151–161. [DOI] [PubMed] [Google Scholar]
- [22].Zaganjor I, Sekkarie A, Tsang BL, Williams J, Razzaghi H, Mulinare J, Sniezek JE, Cannon MJ, Rosenthal J, Describing the prevalence of neural tube defects worldwide: a systematic literature review, PLoS One 11 (4) (2016) e0151586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Irie N, Kuratani S, The developmental hourglass model: a predictor of the basic body plan? Development 141 (24) (2014) 4649. [DOI] [PubMed] [Google Scholar]
- [24].Tonk ECM, Pennings JLA, Piersma AH, An adverse outcome pathway framework for neural tube and axial defects mediated by modulation of retinoic acid homeostasis, Reprod. Toxicol 55 (2015) 104–113. [DOI] [PubMed] [Google Scholar]
- [25].Piersma AH, Hessel EV, Staal YC, Retinoic acid in developmental toxicology: teratogen, morphogen and biomarker, Reprod. Toxicol 72 (2017) 53–61. [DOI] [PubMed] [Google Scholar]
- [26].Sipes NS, Martin MT, Reif DM, Kleinstreuer NC, Judson RS, Singh AV, Chandler KJ, Dix DJ, Kavlock RJ, Knudsen TB, Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data, Toxicol. Sci 124 (1) (2011) 109–127. [DOI] [PubMed] [Google Scholar]
- [27].Matsuoka Y, Funahashi A, Ghosh S, Kitano H, Modeling and simulation using Cell Designer, in: Miyamoto-Sato E, Ohashi H, Sasaki H, Nishikawa J.-i., Yanagawa H(Eds.), Eds.), Transcription Factor Regulatory Networks: Methods and Protocols, Springer, New York, 2014, pp. 121–145. [Google Scholar]
- [28].Irie N, Kuratani S, The developmental hourglass model: a predictor of the basic body plan? Development 141 (24) (2014) 4649–4655. [DOI] [PubMed] [Google Scholar]
- [29].Baker N, Knudsen T, Williams A, Abstract Sifter: a comprehensive front-end system to PubMed, F1000Research 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tonk EC, Pennings JL, Piersma AH, An adverse outcome pathway framework for neural tube and axial defects mediated by modulation of retinoic acid homeostasis, Reprod. Toxicol 55 (2015) 104–113. [DOI] [PubMed] [Google Scholar]
- [31].Copp AJ, Greene ND, Genetics and development of neural tube defects, J. Pathol 220 (2) (2010) 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu A, Niswander LA, Bone morphogenetic protein signalling and vertebrate nervous system development, nature reviews, Neuroscience 6 (12) (2005) 945–954. [DOI] [PubMed] [Google Scholar]
- [33].Aruga J, The role of Zic genes in neural development, Mol. Cell. Neurosci 26 (2) (2004) 205–221. [DOI] [PubMed] [Google Scholar]
- [34].Caspary T, Anderson KV, Patterning cell types in the dorsal spinal cord: what the mouse mutants say, nature reviews, Neuroscience 4 (4) (2003) 289–297. [DOI] [PubMed] [Google Scholar]
- [35].Cunningham TJ, Brade T, Sandell LL, Lewandoski M, Trainor PA, Colas A, Mercola M, Duester G, Retinoic acid activity in undifferentiated neural progenitors is sufficient to fulfill its role in restricting Fgf8 expression for somitogenesis, PLoS One 10 (9) (2015), e0137894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Diez Del Corral R, Morales AV, The multiple roles of FGF signaling in the developing spinal cord, Front. Cell Dev. Biol 5 (2017) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kumar S, Duester G, Retinoic acid controls body axis extension by directly repressing Fgf8 transcription, Development (Cambridge, England) 141 (15) (2014) 2972–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ribes V, Le Roux I, Rhinn M, Schuhbaur B, Dolle P, Early mouse caudal development relies on crosstalk between retinoic acid, Shh and Fgf signalling pathways, Development (Cambridge, England) 136 (4) (2009) 665–676. [DOI] [PubMed] [Google Scholar]
- [39].Tsurubuchi T, Allender EV, Siddiqui MR, Shim KW, Ichi S, Boshnjaku V, Mania-Farnell B, Xi G, Finnell RH, McLone DG, Tomita T, Mayanil CS, A critical role of noggin in developing folate-nonresponsive NTD in Fkbp8 −/− embryos, Childs Nerv. Syst 30 (8) (2014) 1343–1353. [DOI] [PubMed] [Google Scholar]
- [40].Liem KF, Jessell TM, Briscoe J, Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites, Development (Cambridge, England) 127 (22) (2000) 4855–4866. [DOI] [PubMed] [Google Scholar]
- [41].Ybot-Gonzalez P, Gaston-Massuet C, Girdler G, Klingensmith J, Arkell R, Greene ND, Copp AJ, Neural plate morphogenesis during mouse neurulation is regulated by antagonism of Bmp signalling, Development (Cambridge, England) 134 (17) (2007) 3203–3211. [DOI] [PubMed] [Google Scholar]
- [42].Hutson MS, Leung MC, Baker NC, Spencer RM, Knudsen TB, Computational model of secondary palate fusion and disruption, Chem. Res. Toxicol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sheng N, Xie Z, Wang C, Bai G, Zhang K, Zhu Q, Song J, Guillemot F, Chen Y, Lin A, Jing N, Retinoic acid regulates bone morphogenic protein signal duration by promoting the degradation of phosphorylated Smad1, Proc. Natl. Acad. Sci. U.S.A 107 (44) (2010) 18886–18891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Anderson MJ, Schimmang T, Lewandoski M, An FGF3-BMP signaling Axis Regulates caudal neural tube closure, neural crest specification and anterior-posterior Axis extension, PLoS Genet. 12 (5) (2016), e1006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tuazon FB, Mullins MC, Temporally coordinated signals progressively pattern the anteroposterior and dorsoventral body axes, Semin. Cell Dev. Biol 42 (2015) 118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Delaune E, Lemaire P, Kodjabachian L, Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition, Development (Cambridge, England) 132 (2) (2005) 299–310. [DOI] [PubMed] [Google Scholar]
- [47].Schilling TF, Nie Q, Lander AD, Dynamics and precision in retinoic acid morphogen gradients, Curr. Opin. Genet. Dev 22 (6) (2012) 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhong M, Kawaguchi R, Costabile B, Tang Y, Hu J, Cheng G, Kassai M, Ribalet B, Mancia F, Bok D, Sun H, Regulatory mechanism for the transmembrane receptor that mediates bidirectional vitamin A transport, Proc. Natl. Acad. Sci. U.S.A 117 (18) (2020) 9857–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu L, Suzuki K, Nakagata N, Mihara K, Matsumaru D, Ogino Y, Yashiro K, Hamada H, Liu Z, Evans SM, Mendelsohn C, Yamada G, Retinoic acid signaling regulates sonic hedgehog and bone morphogenetic protein signalings during genital tubercle development, Birth Defects Res. B Dev. Reprod. Toxicol 95 (1) (2012) 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M, The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis, Birth defects research. Part A, Clin. Mol. Teratol 88 (10) (2010) 883–894. [DOI] [PubMed] [Google Scholar]
- [51].Janesick A, Nguyen TT, Aisaki K, Igarashi K, Kitajima S, Chandraratna RA, Kanno J, Blumberg B, Active repression by RARgamma signaling is required for vertebrate axial elongation, Development (Cambridge, England) 141 (11) (2014) 2260–2270. [DOI] [PubMed] [Google Scholar]
- [52].Pera EM, Acosta H, Gouignard N, Climent M, Arregi I, Active signals, gradient formation and regional specificity in neural induction, Exp. Cell Res 321 (1) (2014) 25–31. [DOI] [PubMed] [Google Scholar]
- [53].Marchal L, Luxardi G, Thome V, Kodjabachian L, BMP inhibition initiates neural induction via FGF signaling and Zic genes, Proc. Natl. Acad. Sci. U.S.A 106 (41) (2009) 17437–17442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Plouhinec JL, Roche DD, Pegoraro C, Figueiredo AL, Maczkowiak F, Brunet LJ, Milet C, Vert JP, Pollet N, Harland RM, Monsoro-Burq AH, Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers, Dev. Biol 386 (2) (2014) 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Drummond DL, Cheng CS, Selland LG, Hocking JC, Prichard LB, Waskiewicz AJ, The role of Zic transcription factors in regulating hindbrain retinoic acid signaling, BMC Dev. Biol 13 (2013) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Greene ND, Copp AJ, Neural tube defects, Annu. Rev. Neurosci 37 (2014) 221–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Amarnath S, Agarwala S, Cell-cycle-dependent TGFbeta-BMP antagonism regulates neural tube closure by modulating tight junctions, J. Cell. Sci 130 (1) (2017) 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Eom DS, Amarnath S, Fogel JL, Agarwala S, Bone morphogenetic proteins regulate hinge point formation during neural tube closure by dynamic modulation of apicobasal polarity, Birth defects research. Part A, Clin. Mol. Teratol 94 (10) (2012) 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nikolopoulou E, Galea GL, Rolo A, Greene ND, Copp AJ, Neural tube closure: cellular, molecular and biomechanical mechanisms, Development (Cambridge, England) 144 (4) (2017) 552–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhang J, Raghunathan R, Rippy J, Wu C, Finnell RH, Larin KV, Scarcelli G, Tissue biomechanics during cranial neural tube closure measured by Brillouin microscopy and optical coherence tomography, Birth Defects Res. 111 (14) (2019) 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ybot-Gonzalez P, Cogram P, Gerrelli D, Copp AJ, Sonic hedgehog and the molecular regulation of mouse neural tube closure, Development (Cambridge, England) 129 (10) (2002) 2507–2517. [DOI] [PubMed] [Google Scholar]
- [62].Sela-Donenfeld D, Kalcheim C, Inhibition of noggin expression in the dorsal neural tube by somitogenesis: a mechanism for coordinating the timing of neural crest emigration, Development (Cambridge, England) 127 (22) (2000) 4845–4854. [DOI] [PubMed] [Google Scholar]
- [63].Stottmann RW, Berrong M, Matta K, Choi M, Klingensmith J, The BMP antagonist Noggin promotes cranial and spinal neurulation by distinct mechanisms, Dev. Biol 295 (2) (2006) 647–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Juriloff DM, Harris MJ, Insights into the etiology of mammalian neural tube closure defects from developmental, genetic and evolutionary studies, J. Dev. Biol 6 (3) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Martinez-Morales PL, Diez del Corral R, Olivera-Martinez I, Quiroga AC, Das RM, Barbas JA, Storey KG, Morales AV, FGF and retinoic acid activity gradients control the timing of neural crest cell emigration in the trunk, Cell Biol. 194 (3) (2011) 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yamaguchi Y, Miura M, How to form and close the brain: insight into the mechanism of cranial neural tube closure in mammals, Cell. Mol. Life Sci 70 (17) (2013)3171–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gray JD, Kholmanskikh S, Castaldo BS, Hansler A, Chung H, Klotz B, Singh S, Brown AM, Ross ME, LRP6 exerts non-canonical effects on Wnt signaling during neural tube closure, Hum. Mol. Genet 22 (21) (2013) 4267–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rolo A, Escuin S, Greene NDE, Copp AJ, Rho GTPases in mammalian spinal neural tube closure, Small GTPases 9 (4) (2018) 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kalcheim C, Epithelial-mesenchymal transitions during neural crest and somite development, J. Clin. Med 5 (1) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Rolo A, Savery D, Escuin S, de Castro SC, Armer HE, Munro PM, Mole MA, Greene ND, Copp AJ, Regulation of cell protrusions by small GTPases during fusion of the neural folds, Elife 5 (2016), e13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pai YJ, Abdullah NL, Mohd-Zin SW, Mohammed RS, Rolo A, Greene ND, Abdul-Aziz NM, Copp AJ, Epithelial fusion during neural tube morphogenesis, Birth defects research, Part A, Clin. Mol. Teratol 94 (10) (2012) 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schlessinger K, Hall A, Tolwinski N, Wnt signaling pathways meet rho GTPases, Genes Dev. 23 (3) (2009) 265–277. [DOI] [PubMed] [Google Scholar]
- [73].Jacinto A, Wood W, Woolner S, Hiley C, Turner L, Wilson C, Martinez-Arias A, Martin P, Dynamic analysis of actin cable function during Drosophila dorsal closure, Curr. Biol 12 (14) (2002) 1245–1250. [DOI] [PubMed] [Google Scholar]
- [74].Pyrgaki C, Liu A, Niswander L, Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion, Dev. Biol 353 (1) (2011) 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Taneyhill LA, To adhere or not to adhere: the role of Cadherins in neural crest development, Cell Adh. Migr 2 (4) (2008) 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Nieto MA, The early steps of neural crest development, Mech. Dev 105 (1-2) (2001) 27–35. [DOI] [PubMed] [Google Scholar]
- [77].Ichi S, Costa FF, Bischof JM, Nakazaki H, Shen YW, Boshnjaku V, Sharma S, Mania-Farnell B, McLone DG, Tomita T, Soares MB, Mayanil CS, Folic acid remodels chromatin on Hes1 and Neurog2 promoters during caudal neural tube development, J. Biol. Chem 285 (47) (2010) 36922–36932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nakazaki H, Shen YW, Yun B, Reddy A, Khanna V, Mania-Farnell B, Ichi S, McLone DG, Tomita T, Mayanil CS, Transcriptional regulation by Pax3 and TGFbeta2 signaling: a potential gene regulatory network in neural crest development, Int. J. Dev. Biol 53 (1) (2009) 69–79. [DOI] [PubMed] [Google Scholar]
- [79].Meulemans D, Bronner-Fraser M, Gene-regulatory interactions in neural crest evolution and development, Dev. Cell 7 (3) (2004) 291–299. [DOI] [PubMed] [Google Scholar]
- [80].Fritsche E, Barenys M, Klose J, Masjosthusmann S, Nimtz L, Schmuck M, Wuttke S, Tigges J, Current availability of stem cell-based in vitro methods for developmental neurotoxicity (DNT) testing, Toxicol. Sci 165 (1) (2018) 21–30. [DOI] [PubMed] [Google Scholar]
- [81].Hessel EVS, Staal YCM, Piersma AH, Design and validation of an ontology-driven animal-free testing strategy for developmental neurotoxicity testing, Toxicol. Appl. Pharmacol 354 (2018) 136–152. [DOI] [PubMed] [Google Scholar]
- [82].Theunissen PT, Robinson JF, Pennings JL, de Jong E, Claessen SM, Kleinjans J, Piersma AH, Transcriptomic concentration-response evaluation of valproic acid, cyproconazole, and hexaconazole in the neural embryonic stem cell test (ESTn), Toxicol. Sci 125 (2) (2012) 430–438. [DOI] [PubMed] [Google Scholar]
- [83].Kleinstreuer N, Dix D, Rountree M, Baker N, Sipes N, Reif D, Spencer R,Knudsen T, A computational model predicting disruption of blood vessel development, PLoS Comput. Biol 9 (4) (2013), e1002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Leung MC, Hutson MS, Seifert AW, Spencer RM, Knudsen TB, Computational modeling and simulation of genital tubercle development, Reprod. Toxicol 64 (2016) 151–161. [DOI] [PubMed] [Google Scholar]
- [85].Knudsen T, Martin M, Chandler K, Kleinstreuer N, Judson R, Sipes N, Predictive models and computational toxicology, Methods Mol. Biol 947 (2013) 343–374. [DOI] [PubMed] [Google Scholar]
- [86].Silva M, Pham N, Lewis C, Iyer S, Kwok E, Solomon G, Zeise L, A comparison of ToxCast test results with in vivo and other in vitro endpoints for neuro, endocrine, and developmental toxicities: a case study using endosulfan and methidathion, Birth Defects Res. B Dev. Reprod. Toxicol 104 (2) (2015) 71–89. [DOI] [PubMed] [Google Scholar]
- [87].Daston GP, Naciff JM, Predicting developmental toxicity through toxicogenomics, Birth Defects Res. C Embryo Today 90 (2) (2010) 110–117. [DOI] [PubMed] [Google Scholar]
- [88].Laufersweiler MC, Gadagbui B, Baskerville-Abraham IM, Maier A, Willis A, Scialli AR, Carr GJ, Felter SP, Blackburn K, Daston G, Correlation of chemical structure with reproductive and developmental toxicity as it relates to the use of the threshold of toxicological concern, Regul. Toxicol. Pharmacol 62 (1) (2012) 160–182. [DOI] [PubMed] [Google Scholar]


