Abstract
Background
The incorporation of co-stimulatory signaling domains into second-generation chimeric antigen receptors (CARs) significantly enhances the proliferation and persistence of CAR-T cells in vivo, leading to successful clinical outcomes.
Methods
To achieve such functional enhancement in transgenic T-cell receptor-engineered T-cell (TCR-T) therapy, we designed a second-generation TCR-T cell in which CD3ζ genes modified to contain the intracellular domain (ICD) of the 4-1BB receptor were selectively inserted into the CD247 locus.
Results
This modification enabled the simultaneous recruitment of key adaptor molecules for signals 1 and 2 on TCR engagement. However, the addition of full-length 4-1BB ICD unexpectedly impaired the expression and signaling of TCRs, leading to suboptimal antitumor activity of the resulting TCR-T cells in vivo. We found that the basic-rich motif (BRM) in the 4-1BB ICD was responsible for the undesirable outcomes, and that fusion of minimal tumor necrosis factor receptor-associated factor (TRAF)-binding motifs at the C-terminus of CD3ζ (zBBΔBRM) was sufficient to recruit TRAF2, the key adaptor molecule in 4-1BB signaling, while retaining the expression and proximal signaling of the transgenic TCR. Consequently, TCR-T cells expressing zBBΔBRM exhibited improved persistence and expansion in vitro and in vivo, resulting in superior antitumor activity in a mouse xenograft model.
Conclusions
Our findings offer a promising strategy for improving the intracellular signaling of TCR-T cells and their application in treating solid tumors.
Keywords: cell engineering, costimulatory and inhibitory T-cell receptors, immunotherapy, T-lymphocytes
WHAT IS ALREADY KNOWN ON THIS TOPIC
T cell receptor (TCR) signaling along with co-stimulatory signaling is crucial for proliferation and persistence of T cells. The clinical success of second-generation chimeric antigen receptor (CAR)-T therapy demonstrated the importance of including a co-stimulatory domain in their cytoplasmic domain. Unlike CAR T cells, however, few studies have modulated the signaling domains of the TCR complex to improve the efficacy of TCR-T cells. Therefore, there is an essential need to develop new strategies for enhancing the efficacy of TCR-T cells.
WHAT THIS STUDY ADDS
We designed a second-generation TCR-T cell that simultaneously delivers TCR and co-stimulatory signals on TCR stimulation by using a newly designed modified-CD3ζ that is fused with a truncated 4-1BB signaling domain containing the tumor receptor-associated factor-binding motif. Second-generation TCR-T cells exhibited enhanced persistence with a highly upregulated expression of anti-apoptotic molecules, leading to superior antitumor efficacy in a mouse xenograft model.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This finding offers a promising strategy for improving the intracellular signaling of TCR-T cells and provides a rationale to improve TCR-T therapies in patients with solid tumor.
Background
Adoptive T-cell therapy (ACT) has the potential to revolutionize drug development for cancer therapy.1 The effectiveness of ACT, which is closely linked to the number of tumor-reactive T cells, can be enhanced by uniformly redirecting the antigen specificity of polyclonal T cells, through the expression of either chimeric antigen receptors (CARs) or transgenic T-cell receptors (TCRs).2 3 The former traditionally combines an antigen-specific single-chain variable fragment with a hinge region, a transmembrane domain, and an intracellular domain (ICD).4 5 In first-generation CAR constructs, the ICD contained the signaling domain of either CD3ζ or FcRγ, which are crucial signaling components of the TCR and Fc receptor, respectively.6–8 Despite promising in vitro activity, first-generation CAR-T cells failed to exhibit long-term persistence, ultimately leading to treatment failure in vivo.9 10 In the second-generation CAR constructs, the addition of ICDs from the co-stimulatory molecules CD28 or 4-1BB resulted in robust in vivo proliferation and persistence.11–13 Indeed, US Food and Drug Administration-approved second-generation CAR-T cells have proven to be effective against various relapsed and refractory hematological malignancies.14 15
Another promising ACT involves the use of transgenic TCR-T cells, in which the genes encoding antigen-specific TCR α and β chains are introduced into T cells. Unlike CARs, which is typically a single polypeptide chain, the TCR complex is composed of eight subunits that serve distinct roles in antigen recognition (TCRα/β) or signal transduction (CD3 ε/δ, ε/γ, and ζ/ζ).16 Most studies on TCR-T cells have sought to identify TCRα/β sequences specific to tumor-specific antigens or tumor-associated antigens or to increase their affinity.17–21 Further, several groups have recently modified the extracellular domain of TCR complex subunits by incorporating or replacing them with variable domains of an antibody.22–25 However, few studies have explored modification of signaling domains of the TCR complex as a strategy for functionally improving TCR-T cells.26 Interaction of the TCR with the peptide-presenting major histocompatibility complex (MHC) of an antigen-presenting cell triggers TCR signaling (signal 1), which promotes the activation of T cells.27 28 However, binding of co-stimulatory ligands, which leads to co-stimulatory signaling (signal 2), is also essential for enhancing T-cell proliferation and survival.29 30 In the absence of co-stimulatory ligand binding, T cells become hyporesponsive and are ineffective in resolving diseases.31 32 Indeed, the progression from first-generation to second-generation CARs provided researchers the critical lesson that co-stimulatory signaling is required for the robust functionality of engineered T cells.
In the present study, we sought to generate second-generation TCR-T cells capable of simultaneously delivering signals 1 and 2 upon engagement of target cells via TCRα/β. To this end, we used the CRISPR/Cas9 system to selectively knock-in (KI) CD3ζ variants modified to contain the 4-1BB signaling domain within the CD247 (CD3ζ) locus. We found that the position of 4-1BB relative to CD3ζ ICD (N-terminus or C-terminus) and the presence of a positively charged basic-rich motif (BRM) of 4-1BB significantly affected surface expression of the TCR complex and the membrane-proximal signaling. Consequently, integration of the minimal tumor necrosis factor (TNF) receptor-associated factor (TRAF)-binding motifs of 4-1BB at the C-terminus of CD3ζ preserved the expression of the transgenic NY-ESO-1-specific TCR and its proximal signaling, while enhancing the in vitro and in vivo expansion of TCR-T cells, leading to superior antitumor efficacy in a mouse xenograft model.
Materials and methods
Animals
Female 6–8 weeks old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from the Jackson Laboratory (Bar Harbor, Maine, USA). All mice were housed in a specific pathogen-free facility. Animal care and experimental procedures were performed as per the protocols approved by the Animal Care Committee of the Korea Advanced Institute of Science and Technology (KAIST).
Cell lines and isolation of human primary T cells
Jurkat, K562, and A375 cells were purchased from the American Type Culture Collection (ATCC, USA) and cultured according to ATCC recommendations. A375 cells were transduced to express Zsgreen to generated A375-Zsgreen cells. The A375 cells with herpes simplex virus 1–thymidine kinase, green fluorescent protein (GFP), and firefly luciferase, referred to herein as A375-TGL, were kindly provided by Professor Mi-Young Kim (KAIST). Whole peripheral blood from healthy donors were isolated and provided by Asan Medical Hospital (Seoul, Korea), using an Institutional Review Board-approved protocol. Primary human peripheral blood mononuclear cells (PBMCs) were isolated from the blood of an anonymous healthy donors using Ficoll centrifugation with SepMate tubes (STEMCELL Technologies, cat# 86460). Pan T cells were isolated from PBMCs by means of magnetic negative selection using a Pan T Cell Isolation Kit (Miltenyi Biotec, cat# 130-096-535) according to the manufacturer’s instructions. T cells were cultured in human T-cell medium consisting of RPMI-1640 (Gibco, cat# 21 870–092), 10% fetal bovine serum (FBS; Thermo Fisher Scientific, cat# 26140079), 55 µM 2-mercaptoethanol (Thermo Fisher Scientific, cat# 31350010), and 1% GlutaMAX (Gibco, cat# 35 050–061), supplemented with 300 units/mL interleukin (IL)-2 (BMI, Korea). T cells were cryopreserved in a mixed solution of 80% heat-inactivated FBS, 10% dimethyl sulfoxide (Sigma, cat# 2650), and 10% T-cell media.
Construction of modified-CD3ζ-expressing genes
Modified-CD3ζ constructs were generated by inserting the cytoplasmic domain of 4-1BB between the transmembrane and cytoplasmic domains of CD3ζ (BBz) or next to the C-terminus of CD3ζ (zBB). To block the ubiquitination of the modified-CD3ζ, the zBBKtoR construct was designed by substituting all lysines with arginine within the 4-1BB ICD of the zBB construct. To remove the BRM of the 4-1BB ICD from the modified-CD3ζ construct, the zBBK/RtoS construct was generated by mutating four lysines and two arginines, located at the N-terminus of 4-1BB ICD from the zBB construct, to charge-neutral serine. For the same purpose, the zBBΔBRM construct was prepared in which the BRM moiety (amino acid sequence, KRGRKKLLYIFK) of the 4-1BB ICD was truncated from the zBB construct. All modified-CD3ζ-encoding genes were linked to the sequence encoding truncated delta low-affinity nerve growth factor receptor (dLNGFR) using a self-cleaving porcine teshovirus-1 2A (P2A) sequence and subcloned into a lentiviral pELPS vector,14 downstream of an EF1α promoter and upstream of a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE).
For immunoprecipitation of modified-CD3ζ, the modified-CD3ζ constructs were fused to a 3×FLAG, linked to the sequence encoding RQR833 using an internal ribosome entry site (IRES), and cloned into the lentiviral pELPS vector. pAAV-modified-CD3ζ (CD3z, zBB, zBBΔBRM) vector variants were constructed by substituting the modified-CD3ζ sequence into a pAAV-EF1a double-floxed-EYFP-WPRE-human growth hormone polyA signal (hGHpA) construct (Addgene, cat# 20296). The modified-CD3ζ construct was integrated at the CD247 locus by cloning the modified-CD3ζ sequence with 1.2 kb of genomic CD247 exon 1 elements (each ~0.6 kb in length) flanking the single-guide RNA (sgRNA) targeting sequences, resulting in expression of modified-CD3ζ variants under the control of the CD3ζ promoter. The modified-CD3ζ construct was followed by a WPRE and an hGHpA.
Recombinant lentivirus and AAV production
1G4 TCR lentiviral particles were created by subcloning the transgene of 1G4 TCR into a lentiviral pELPS vector. For lentivirus production, HEK293T cells (ATCC) were subcultured in Dulbecco’s Modified Eagle Medium supplemented with 10% heat-inactivated FBS, 1% GlutaMAX, 1 mM MEM sodium pyruvate, and 1% penicillin/streptomycin (all from Gibco). HEK293FT cells (3.6×106) were transferred to a poly-D-lysine-coated 100 mm cell culture plate in antibiotic-free medium. The cells were transfected with pMDLg/p.RRE (6 µg), pRSV.REV (6 µg), pMDG.1 (2 μg), and the modified-CD3ζ-containing lentiviral vector (7.5 µg) using the Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, invitrogen, cat# 11668019), according to the manufacturer’s instructions. After incubation of the cells with the DNA-Lipofectamine complex for 6 hours, the medium was replaced with fresh medium. Forty-two hours after transfection, the lentiviral supernatants were collected, filtered using a 0.45 µm polyethersulfone membrane filter to remove cell debris, and stored at −80°C.
For production of recombinant AAV6 viruses, cloned AAV donor plasmid and packaging plasmids (Takara, cat# 6665) were transfected into HEK293T cells using the polyethylenimine transfection method. Seventy-two hours after transfection, the resulting AAV viruses were purified, and the titer of the virus was determined as previously described.34
Lentiviral transduction
Pan T cells were activated by means of incubation with Human T-Activator CD3/CD28 Dynabeads (from Life Technologies; bead-to-PBMC ratio of 1:1) and IL-2 (300 IU/mL; BMI Korea) for 24 hours. Activated T cells were then transduced with the lentivirus, in a medium containing serum mixed with protamine sulfate (Sigma; 10 µg/mL) by means of spinfection (1000×g for 90 min). Cells were incubated with the lentivirus for 24 hours, after which the medium was replaced with T-cell medium. The percentage of transduced T cells was evaluated based on the dLNGFR-positive population on day 5 after transduction. dLNGFR-positive transduced T cells were isolated using the human CD271 MicroBead kit (Miltenyi Biotec, cat# 130-099-023), according to the manufacturer’s instructions. Modified-CD3ζ+ dLNGFR+ T cells were maintained at a concentration range of 0.5×106–2.0×106 cells/mL at all times during expansion.
Generation of modified-CD3ζ-KI NY-ESO-1-specific TCR-T cells
Twenty-four hours after initiating T-cell activation, lentiviruses containing the 1G4 TCR were transduced into activated T cells. Forty-eight hours later, CD3/CD28 Microbeads (Gibco) were magnetically removed from the transduced T cells, and electroporated, for use in genome editing. CD247 sgRNA, targeting exon 1 (5′- GTGGAAGGCGCTTTTCACCG −3′) was synthesized by means of in vitro transcription using T7 RNA polymerase according to a previously described method.35 Ribonucleoprotein (RNP) was prepared immediately before electroporation by mixing Cas9 protein (Enzynomics, cat# M058U) and CD247 sgRNA at a molar ratio of 1:5 and incubated at 37°C for 10 min. T cells (1×106) were mixed with RNP (sgRNA-Cas9) and electroporated using the Neon Transfection System (Invitrogen, Carlsbad, California, USA). For the Cas9-only treatment group, activated T cells were electroporated as above, without sgRNA. Following electroporation, the cells were diluted in culture medium and incubated at 37°C in a humidified 5% CO2-containing environment. Recombinant AAV6 viruses were added to the culture 15 min after electroporation, at an indicated multiplicity of infection (2–5×104). Edited T cells were subsequently cultured at 37°C under standard conditions, and expanded in T-cell growth medium, which was replenished as needed (~2–3 days) to maintain a density of ~1×106 cells/mL. KI T cells were obtained by isolating dLNGFR-positive T cells using allophycocyanin (APC)-tagged anti-CD271 antibodies (Miltenyi Biotec, cat# 130-113-980), anti-APC Microbeads (Miltenyi Biotec, cat# 130-090-855), and LS columns (Miltenyi Biotec, cat# 130-042-401).
Flow cytometry
All fluorescence-activated cell sorting (FACS) analyses were performed on an LSRFortessa X-20 cytometer (BD Biosciences, USA) and evaluated using FlowJo software (Tree Star, USA). For cell surface staining, 2×105 cells were stained with antibodies and resuspended in 100 µL FACS buffer (2% FBS and 2 mM EDTA in phosphate-buffered saline (PBS)) for 20 min at 4°C in the dark. Cells were washed with 2 mL FACS buffer, resuspended in the same buffer, and analyzed. For intracellular staining, T cells were fixed and permeabilized by incubating in 250 µL of Cytofix/Cytoperm Fixation/Permeabilization solution (BD Biosciences, cat# 554714) at 4°C for 20 min. T cells were washed with 2 mL of intracellular staining buffer and stained with antibodies. Viable cells were determined using Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific, Invitrogen, cat# 65-0865-14). dLNGFR was labeled with an APC-conjugated anti-human CD271 antibody (clone ME20.4–1 .H4; Miltenyi Biotec, cat# 130-113-980). FACS analyses were performed using the following antibodies against: BV421-anti-human CD3ε (clone OKT3; BioLegend, cat# 317344), PE-anti-human CD3ζ (clone 6B10.2; BioLegend, cat# 644106), PerCP/Cyanine5.5-anti human CD4 (clone A161A1; BioLegend, cat# 357414), BV605-anti-human CD8 (clone SK1; BioLegend, cat# 344742), AF488-anti-human TCRα/β (clone IP26; BioLegend, cat# 306718), PE-MHC Dextramer (Immudex, cat# VB3247-PE), APC-anti-human TCRVβ13.1 (clone H131; BioLegend, cat# 362408), AF488-anti-human pCD3ζ (BD Phosflow, cat# 558486), AF647-anti-human pZAP-70 (BD Phosflow, cat# 557817), Fluorescein isothiocyanate (FITC)-anti-human human leukocyte antigen (HLA)-A2 (BD Pharmingen, cat# 561107), BV650-anti-human CD45 (clone HI30; BioLegend, cat# 304044), FITC-anti-human CD69 (clone FN50; BioLegend, cat# 310904), APC-anti-human 4-1BBL (clone 5F4; BioLegend, cat# 311506), PE-anti-human CD80 (clone 2D10; BioLegend, cat# 305208), BV421-anti human CD86 (clone IT2.2, BioLegend, cat# 305426), PE-anti-human CD34 (R&D systems, cat# FAB7227P), and AF647-anti-mouse IgG, F(ab')2 (Jackson ImmunoResearch, cat# 109-606-097).
Stimulation of cells by cross-linked anti-CD3 antibodies
1×106 Jurkat cells or T cells were either unstimulated or incubated with 5 µg/mL of anti-human CD3 (OKT3; eBioscience, cat# 14-0037-82) antibodies on ice for 15 min and then washed by cold-Dulbecco's phosphate buffered saline (dPBS). Surface bound antibodies were subsequently cross-linked with 10 µg/mL goat anti-mouse IgG (Sigma, cat# M8642) for 15 min at 4°C and then washed by cold-dPBS. Following washing, cells were resuspended in pre-warmed RPMI1640 media and incubated at 37°C for 5 min.
For flow cytometric analysis of phosphorylated CD3ζ and zeta-associated protein-70 (ZAP-70), stimulated cells fixed and permeabilized by incubating in 250 µL of Cytofix/Cytoperm Fixation/Permeabilization solution (BD Biosciences, cat# 554714) at 4°C for 20 min. Fixed cells were washed with 2 mL of intracellular staining buffer, and was added PE-anti-human CD3ζ (clone 6B10.2; BioLegend, cat# 644106), AF488-anti-human pCD3ζ (BD Phosflow, cat# 558486), and AF647-anti-human pZAP-70 (BD Phosflow, cat# 557817) for 30 min on ice. Cells were washed and suspended in FACS buffer and analyzed by flow cytometry.
Immunoblot analysis
For immunoblot analyses, approximately 10 µL of whole-cell lysates, prepared by incubating cells with 1% Nonidet P (NP)−40 lysis buffer (ELPISBIO, cat# EBA-1049), was loaded on 4%–12% acrylamide gradient gels (Thermo Fisher Scientific, cat# NP0322BOX) and transferred to polyvinylidene difluoride (PVDF) membranes (Merck, cat# IPVF00010). Fusion proteins were detected using an anti-CD3 (human) monoclonal antibody (mAb) (clone 8D3; BD Biosciences, cat# 551034, 1:1000 dilution). Immunoreactive proteins were detected using an enhanced chemiluminescence detection system. Additional antibodies used for immunoblotting were as follows: Anti-β-actin (clone AC-15; Sigma, cat# A5441, 1:20,000 dilution), anti-Bcl-xL (clone H-5; Santa Cruz Biotechnology, cat# sc-8392, 1:1000 dilution), anti-mouse IgG-horseradish peroxidase (HRP) (Thermo Fisher Scientific, cat# 31437, 1:20,000 dilution), and anti-rabbit IgG-HRP (Thermo Fisher Scientific, cat# 31460, 1:20,000 dilution).
Co-immunoprecipitation
Activated Jurkat cells (1×107) were lysed in 350 µL lysis buffer containing 1% NP-40, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, protease inhibitors (cOmplete; Roche) and phosphatase inhibitor (PhosSTOP; Roche), at 4°C for 30 min, after which the cell lysates were pelleted by means of centrifugation at 13,000×g for 15 min. For anti-3×FLAG immunoprecipitation, 250 µL of the cleared cell lysate was incubated with 5 µL of a 50% protein G Sepharose slurry and 1 µg anti-3×FLAG tag antibody, for 2 hours at 4°C. The immunoprecipitated material was washed four times with 200 µL wash buffer and then resolved by means of a reducing SDS-PAGE, on 4%–12% gradient gels. Separated proteins were transferred to PVDF membranes using a semi-dry transfer procedure. Membranes were blocked in PBS with 0.1% Tween-20 (PBST) containing 5% bovine serum albumin, then incubated with primary antibodies against TRAF2 (Abclonal, cat#A19129, 1:1000 dilution), ZAP-70 (Novus, cat# NB110-60490, 1:100 dilution), and 3×FLAG (MBL International, cat# M185-3L, 1:1000 dilution) in PBST, followed by incubation with HRP-conjugated secondary antibodies (1:10,000 dilution). Immunoreactive proteins were detected using a ChemiDoc XRS+Imaging System (Bio-Rad).
For ubiquitination assays, 1×107 activated Jurkat cells per condition were harvested and washed with chilled PBS. The cells were lysed with lysis buffer consisting of 1% NP-40, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, protease inhibitors (cOmplete; Roche) and phosphatase inhibitor (PhosSTOP; Roche). The lysates were precleared by incubation for 4 hours at 4°C with anti-3×FLAG mAb-Magnetic Beads (clone FLA-1GS; MBL, cat# M185-11R). Immunoprecipitates were washed first with PBS and then with PBST containing 0.2% NP-40, after which they were resuspended in PBS containing 0.2% NP-40 and 0.5 M LiCl and boiled in sample buffer containing a reducing agent (Invitrogen, cat# NP0004). Antibodies against the following proteins were used: Ubiquitin (clone P4D1; BioLegend, cat# 646302), CD3ζ (clone 8D3; BD, cat# 550133), 3×FLAG (clone FLA; MBL, cat# M185-3L), and β-actin (clone AC-15; Sigma, cat# A5441, 1:20,000 dilution). Immunoreactive proteins were detected by means of chemiluminescence using a ChemiDoc XRS+Imaging System (Bio-Rad). Protein was quantified using ImageJ software and normalized the levels of the loading control.
Cytokine production assay
Interferon (IFN)-γ, IL-2, and TNF-γ levels were simultaneously measured in supernatant samples using a Cytometry Bead Array (BD Biosciences, cat# 551809) according to the manufacturer’s protocol. Data were acquired with a BD LSRFortessa X-20 and analyzed using FACSBD Diva software.
Real-time quantitative reverse transcription-PCR
Modified-CD3ζ-KI 1G4 T cells were restimulated with γ-irradiated A375 cells for 8 hours and then sorted using dLNGFR microbeads. Total RNA from the sorted cells was extracted using an RNeasy Mini Kit (Qiagen, cat# 74104) and reverse transcribed into complementary DNA using the ReverTra Ace qPCR RT Master Mix with gDNA remover (TOYOBO, cat# FSQ-301). Real-time quantitative reverse transcription-PCR was performed using the SYBR Green Real-time PCR Master Mix (TOYOBO, cat# QPK-201), with three parallel replicates on a CFX96 Real-Time System (Bio-Rad). The expression level of each indicated gene was normalized to that of the housekeeping gene, ribosomal 18S. The following quantitative PCR primer pairs were used: 18s rRNA, 5′-CAG CCA CCC GAG ATT GAG CA-3′ (forward) and 5′-TAG TAG CGA CGG GCG GTG TG-3′ (reverse); CD3ζ, 5′-GCC GAG AAG GAA GAA CCC TC-3′ (forward) and 5′-GTG GCT GTA CTG AGA CCC TG-3′ (reverse); Bcl-xL, 5′-GAG CTG GTG GTT GAC TTT CTC-3′ (forward) and 5′-TCC ATC TCC GAT TCA GTC CCT-3′ (reverse); Bcl-2, 5′-TCG CCC TGT GGA TGAC TGA-3′ (forward) and 5′-CAG AGA CAG CCA GGA GAA ATC A-3′ (reverse); and Bfl1, 5′-TAC AGG CTG GCT CAG GAC TA-3′ (forward) and 5′-CGC AAC ATT TTG TAG CAC TC-3′ (reverse).
In vitro cytotoxicity assay
A375-Zsgreen cells (1×104) were cultured with 1G4 TCR-T cells at effector-to-target ratios of 0.1:1 to 1:1 in 200 µL complete T-cell medium, in a 96-well flat-bottom plate, for up to 72 hours. Quintuplicate wells were plated for each 1G4 TCR-T group. The GFP fluorescence intensity of the target cells was measured every 2 hours, using the IncuCyte S3 Live-Cell Analysis System (Sartorius, Germany). Integrated total GFP intensity per well was used as a quantitative measure of the amount of viable target cells. The integrated total GFP intensity was normalized to the GFP intensity at the starting point.
T-cell long-term expansion assay
At the first stimulation, 1×106 modified-CD3ζ-KI 1G4 TCR-T cells were co-cultured with γ-irradiated (80 Gy) feeder cells at a 1:1 ratio together with IL-2 (100 IU/mL) in a 24-well plate. Single-chain trimer (SCT)-transduced K562 cells were prepared as a feeder cell layer, as described previously.36 T-cell number was counted every 5 days and the cells were restimulated with irradiated target cells at the same cell ratio. T-cell number counts and restimulation were repeated three times, for a total of 15 days.
Apoptosis assay
An FITC Annexin V Apoptosis Detection Kit Ⅰ (BD Pharmingen, cat# 556547) was used to stain apoptotic cells, according to the manufacturer’s instructions. After acquiring 1G4 T cells in the long-term expansion assay on days 5, 10, 15, the cells were labeled with APC-tagged anti-human dLNGFR, annexin V, and propidium iodide (PI), and detected using a BD LSRFortessa X-20 flow cytometer.
Cell proliferation assay
1G4 TCR-T cells were stained with CellTrace Violet (Thermo Fisher Scientific, cat# C34557) according to the manufacturer’s protocol. The stained 1G4 TCR-T cells (5×105 cells) were co-cultured with A375 cells (5×105 cells) in a 24-well tissue culture plate for 4 days. Cells were harvested, immunostained for T-cell surface markers, co-stained with Fixable Viability Dye eFluor 780, and analyzed using a flow cytometer.
Xenograft mouse models
All mouse experiments were performed in accordance with the protocol of Institutional Animal Care and Use Committee, KAIST. Xenograft models were prepared by subcutaneously injecting 1×106 A375 human melanoma cells (ATCC CRL-1619) into the shaved right flank of 8–12 weeks old male NSG mice (Jackson Laboratory). After implantation, the mice were randomly allocated to different experimental groups and 2.5×106 T cells that were purified based on dLNGFR expression (equivalent to 1×106 1G4+ TCR-T cells) were intravenously injected into each mouse on day 7. Tumor length and width were measured using a digital caliper, and tumor volume (V) was calculated as V=1/6 × π×length × width × (length+width)/2. Mice were euthanized when the major axis of the tumor reached ~20 mm. The tumor burden of each mouse was measured at 7-day intervals using a bioluminescent in vivo imaging system (IVIS) and quantified as the luminescence of the region of interest (generally the whole area of one mouse).
Isolation and analysis of tumor-infiltrating lymphocytes
Mice were euthanized on the indicated days, following which their tumors were resected and transferred into a gentleMACS C tube (Miltenyi Biotec, cat# 130-094-237) containing RPMI-1640. The resected A375 tumors were then chopped into small pieces of 2–4 mm with a pair of surgical scissors and dissociated into a single cell suspension using the human Tumor Dissociation Kit (Miltenyi Biotec, cat# 130-095-929) and gentleMACS Octo Dissociator with Heators (Miltenyi Biotec) following the standard protocol (program, 37C_h-TDK_1) for soft tumors. After enzymatic/mechanical dissociation, the tumor suspension was filtered using a 70 µm cell strainer. Red blood cells were lysed using ACK lysis buffer, followed by two washes in PBS containing 1% FBS and 0.5 mM EDTA. The cell mixture was stained with Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific, Invitrogen, cat# 65-0865-14) and fluorescence-conjugated antibodies as described above. Live CD3+dLNGFR+tumor-infiltrating lymphocytes (TILs) were quantified.
Statistical analysis
Statistically significant differences between two groups were assessed using a two-tailed unpaired student’s t-test. Comparisons among more than two groups were carried out using an one-way analysis of variance (ANOVA) with Tukey’s multiple-comparisons post hoc test. In the mouse experiments, comparisons of tumor burden were made using a two-way ANOVA with Holm-Sidak’s multiple comparisons post hoc test. The overall survival of mice was estimated using the Kaplan-Meier method, and the statistical significance of differences between groups was calculated using a log-rank test. All statistical analyses were performed using Prism V.8 (GraphPad Software), and a p value<0.05 was considered statistically significant.
Result
CRISPR/Cas9- and AAV-mediated site-specific modifications of CD3ζ in human primary T cells
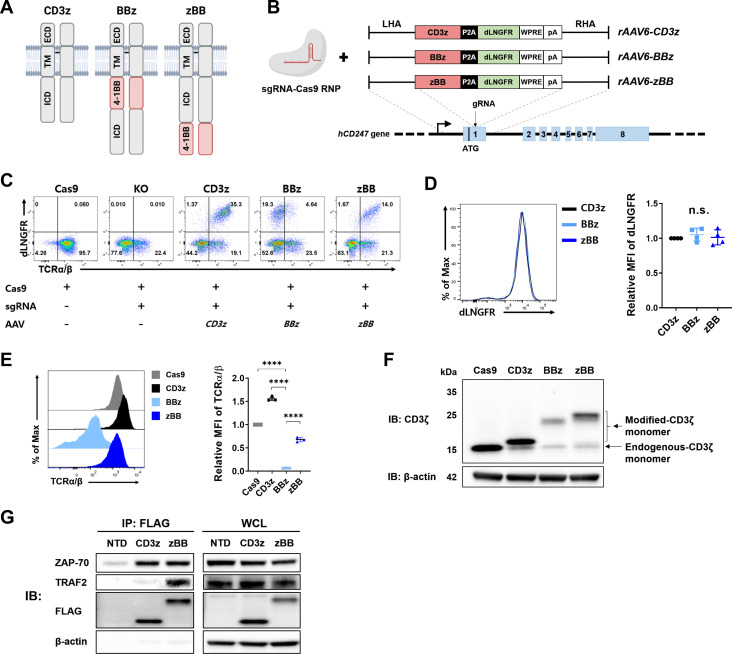
We opted for a CRISPR/Cas9 KI approach rather than the commonly used lentiviral method for site-specific integration of modified-CD3ζ genes into CD247 locus, as it allows to rule out the effects of endogenous-CD3ζ proteins and permits endogenous transcriptional control. In modified-CD3ζ genes, the ICD of the co-stimulatory receptor 4-1BB was placed at the N-terminus (termed BBz) or C-terminus (termed zBB) of the CD3ζ ICD (figure 1A, online supplemental figure 1A). The unmodified-CD3ζ construct (termed CD3z) was also prepared as a control. AAV6 donor vectors were designed to deliver a modified-CD3ζ cassette followed by self-cleaving P2A and the dLNGFR tag (figure 1B). To disrupt the CD247 locus, we designed four sgRNAs for electroporation of Cas9/sgRNA RNP complexes into activated human primary T cells (online supplemental table 1, online supplemental figure 1B). We chose sgRNA#3 for its high knockout (KO) efficiency (up to 80%). Subsequent AAV transduction used for KI of the modified-CD3ζ constructs showed robust efficiency (~35%), measured by monitoring expression of the dLNGFR tag (figure 1C).
Figure 1.
Design and cellular characteristics of CRISPR-engineered T cells expressing modified-CD3ζ. (A) Schematic diagram of the intact CD3ζ homodimer and two different modified-CD3ζ dimers containing the 4-1BB signaling domain. (B) CRISPR/Cas9-targeted integration of the modified-CD3ζ gene into the CD247 locus. Top, rAAV6 containing a modified-CD3ζ cassette flanked by homology arms; bottom, CD247 locus. (C) Representative flow cytometry plots of knock-in efficiency at 5 days after CD247 targeting. (D) Representative histogram (left) and mean fluorescence intensity (MFI; right) of dLNGFR in CD3ζ-targeted T cells, obtained using magnetic-activated cell sorting. (E) Surface expression (left) and MFI (right) of TCR complex (TCRα/β). (F) Expression of modified-CD3ζ protein. (G) Co-immunoprecipitation of zBB. Jurkat cells were stimulated by cross-linked anti-human CD3 antibody. CD3z and zBB were immunoprecipitated from the indicated Jurkat cell lysates, and then subsequently analyzed using SDS-PAGE. Western blots were probed with anti-TRAF2, anti-ZAP-70, anti-CD3ζ, and anti-β-actin antibodies. n.s., not significant, as assessed using one-way analysis of variance with Tukey’s multiple comparisons post hoc test (D and E). dLNGFR, delta low-affinity nerve growth factor receptor; ECD, extracellular domain; ICD, intracellular domain; MFI, mean fluorescence intensity; P2A, porcine teshovirus-1 2A; sg RNA, single-guide RNA; TCR, T cell receptor; TRAF2, tumor receptor-associated factor; TM, transmembrane; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; ZAP-70, zeta-associated protein-70.
jitc-2022-005519supp001.pdf (13.8MB, pdf)
dLNGFR-positive CD3z cells showed recovery of TCRα/β expression relative to cells treated only with the Cas9/sgRNA RNP, reflecting the fact that CD3ζ is indispensable for TCR complex formation and surface expression.37 Interestingly, despite successful KI of the BBz construct in T cells, as measured by dLNGFR expression, TCRα/β expression was markedly reduced compared with that in dLNGFR-positive CD3z and zBB T cells (figure 1C). In-out PCR analysis revealed seamless and precise targeting of modified-CD3ζ constructs into the CD247 locus (online supplemental figure 1C). There was also no significant difference in the messenger RNA (mRNA) expression between all the groups (online supplemental figure 1D). Next, we isolated dLNGFR-positive cells using magnetic separation (figure 1D). Consistent with our previous observation, TCRα/β expression was significantly reduced in BBz cells compared with CD3z or zBB cells (figure 1E), despite comparable mean fluorescence intensity (MFI) of dLNGFR (figure 1D). Western blot analysis also showed that expression of BBz was the lowest compared with other constructs (figure 1F). Since the same BBz sequence was used in the ICD of the approved second-generation CAR-T cells, we generated lentiviral vectors for two CAR constructs targeting CD19, 19BBz, and 19zBB (online supplemental figure 2A). Interestingly, in contrast to the case for TCR expression, 19zBB CAR was expressed poorly on human primary T cells compared with 19BBz (online supplemental figure 2B). These results indicate that the location of the 4-1BB ICD within CD3ζ affects protein expression and TCR complex formation, and that the preferred configuration for this particular modification is distinct from that of the CAR design. Given its inability to form TCR complexes, BBz was excluded in the subsequent experiments.
ZAP-70 is a key adaptor protein for the signaling cascade downstream of TCR stimulation through its association with CD3ζ.38 In addition, TRAF2 plays a key role in 4-1BB signaling by binding to the TRAF-binding motif (TBM) in the ICD of 4-1BB.39 40 To determine whether zBB simultaneously transmits TCR and 4-1BB signals, the association of ZAP-70 with the modified-CD3ζ and binding of TRAF2 to TBM were assessed using co-immunoprecipitation in zBB-transduced Jurkat cells (online supplemental figure 3A, B). On activation, we found that zBB was simultaneously associated with ZAP-70 and TRAF2, whereas CD3z was associated only with ZAP-70 (figure 1G). Collectively, these observations confirm that, on TCR stimulation, the zBB simultaneously recruits the adaptor molecules required for both signal 1 and 2.
Establishment and functional evaluation of zBB-KI NY-ESO-1-specific TCR-T cells
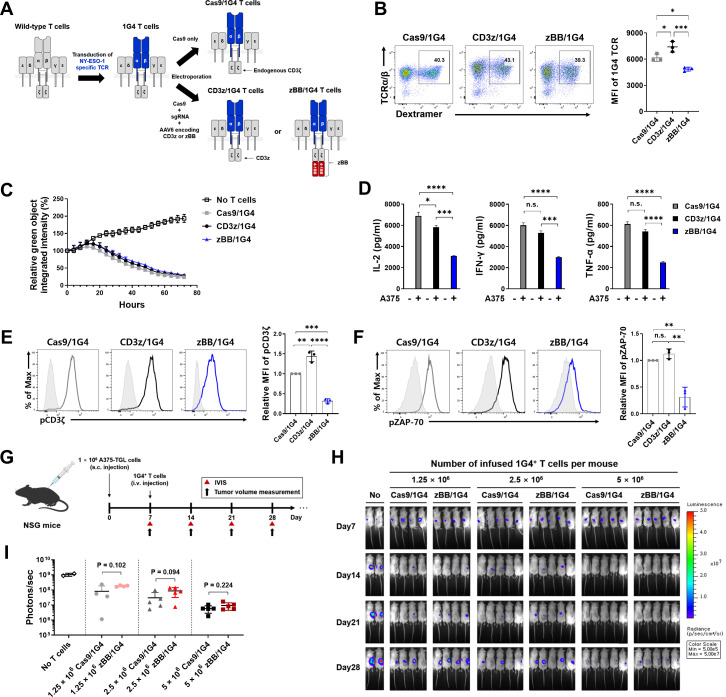
Next, to determine if our modified-CD3ζ constructs enhance the function of TCR-T cells, we transduced human primary T cells with lentiviral vectors expressing the 1G4-α95:LY TCR, an affinity-enhanced TCR that recognizes the NY-ESO-1-derived peptide, SLLMWITQC, presented by the HLA genotype HLA-A*02:01 (online supplemental figure 4A, B).18 This previously described TCR has been clinically tested in patients with metastatic synovial cell sarcoma and melanoma.41 42 We further knocked in the CD3z and zBB constructs into these 1G4-α95:LY TCR T cells (figure 2A), referred to hereafter as CD3z/1G4 and zBB/1G4 T cells, respectively. A Cas9/1G4 group with endogenous-CD3ζ was also generated as a control. In all groups, approximately 40% of cells were 1G4 TCR-positive (figure 2B, left), although the surface expression of the TCR complex was lower in zBB/1G4 T cells than that in the other T-cell groups (figure 2B, right).
Figure 2.
Establishment and functional evaluation of zBB-KI NY-ESO-1-specific TCR-T cells. (A) Schematic diagram showing the generation of modified-CD3ζ-KI NY-ESO-1-specific TCR-T cells. (B) Population (left) and MFI (right) of NY-ESO-1-specific TCR complexes in modified-CD3ζ-KI 1G4 T cells. (C) In vitro cytotoxicity of modified-CD3ζ-KI 1G4 T cells against NY-ESO-1-expressing A375 melanoma cells. Modified-CD3ζ-KI 1G4 T cells were incubated with Zsgreen-expressing A375 cells at an effector:target ratio of 1:1. Green fluorescence intensity was measured using the IncuCyte S3 live-cell imaging system. (D) Production of the cytokines, IL-2 (left), IFN-γ (middle), and TNF-α (right) by the indicated modified-CD3ζ-KI 1G4 T cells, after stimulation with A375 cells for 24 hours (n=3). (E and F) Phosphorylation of CD3ζ (E) and ZAP-70 (F) in modified-CD3ζ-KI 1G4 T cells. P values (*p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001) were determined using one-way analysis of variance, with Tukey’s multiple comparisons post hoc test (B, and D to F). Data have been presented as mean±SEM. n.s., not significant. (G) Experimental design showing subcutaneous inoculation of NSG mice with A375-TGL cells in the right flank, followed by random distribution to experimental groups and intravenous injection of the indicated modified-CD3ζ-KI 1G4 T cells on day 7 (n=4–5 mice/group). Tumor burden was monitored based on bioluminescence intensity, which was determined using an in vivo imaging system (IVIS). (H) Bioluminescence image of A375 tumor burden in NSG mice treated with the indicated doses of modified-CD3ζ-KI 1G4 T cells. (I) Quantification of total photon counts on day 28 after A375 tumor cell injection. The indicated p values, as determined using student’s t-test, correspond to comparisons of the Cas9/1G4 T-cell group with the zBB/1G4 T cell group. IFN, interferon; IL, interleukin; MFI, mean fluorescence intensity; TCR, T-cell receptor; TNF, tumor necrosis factor; ZAP-70, zeta-associated protein-70.
The cytolytic potential of the modified T cells was measured by co-culturing them with NY-ESO-1-expressing A375 melanoma cells (online supplemental figure 4C-E). All three modified T cells were specific for their target cells and showed nearly identical cytolytic potential (figure 2C), consistent with previous studies showing that first-generation and second-generation CAR constructs exhibit comparable in vitro cytotoxicity.11 43 There was also no significant difference in the composition of CD4-positive and CD8-positive T cells (online supplemental figure 4F). However, zBB/1G4 T cells produced significantly lower levels of the pro-inflammatory cytokines, IL-2, IFN-γ, and TNF-α, on antigen stimulation (figure 2D). Moreover, phosphorylation of both CD3ζ and ZAP-70 was significantly reduced in zBB/1G4 T cells (figure 2E, F).
To further compare the therapeutic potential of the modified-CD3ζ-KI 1G4 T cells, we used a xenograft mouse model of A375 cells labeled with GFP-luciferase (A375-TGL). Seven days after subcutaneous inoculation of the A375 cells, NOD.Cg-PrkdcscidIL2γgtm1Wjl/SzJ (NSG) mice were intravenously injected with Cas9/1G4 or zBB/1G4, at doses of 1.25×106, 2.5×106, or 5×106 T cells (figure 2G). Tumor burden diminished in a dose-dependent manner (figure 2H). Contrary to expectations, we found no significant difference in the antitumor effect of zBB/1G4 T cells compared with Cas9/1G4 T cells at all doses, although zBB/1G4 cells showed a tendency towards decreased efficacy (figure 2I). Collectively, these results suggest that our zBB/1G4 T cells lack superior functional and antitumor effects compared with Cas9/1G4 and CD3z/1G4 T cells.
The BRM in 4-1BB affects TCR complex formation and membrane-proximal TCR signaling
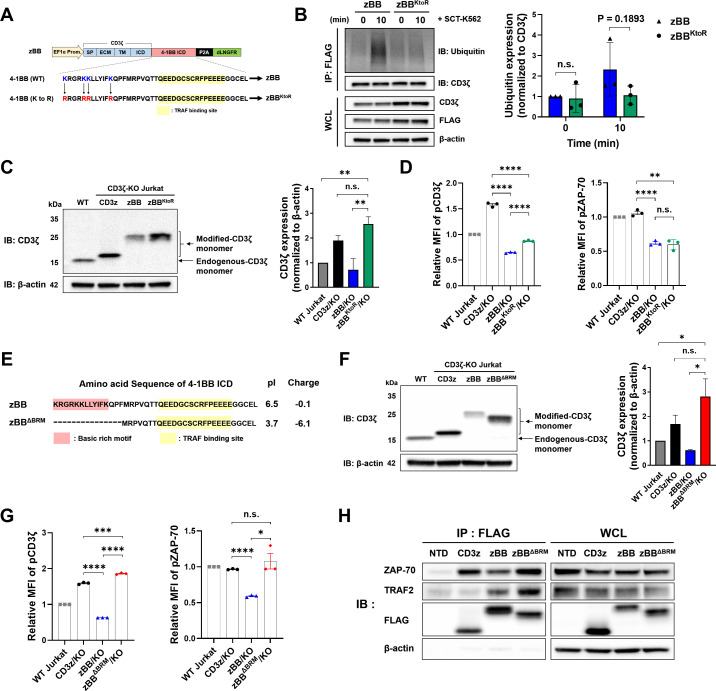
The insufficient antitumor effects observed in zBB/1G4 T cells, despite TRAF2 association with zBB, is possibly attributable to the low expression level of zBB (figure 1F, figure 2B), an observation confirmed by lentiviral transduction of modified-CD3ζ into CD3ζ-KO Jurkat cells (online supplemental figure 5C-G). Expression level of modified-CD3ζ mRNA (online supplemental figure 1D) and dLNGFR (figure 1D) among all the constructs were similar to each other, indicating that the low expression of zBB might be caused by degradation during a post-translational modification step. Therefore, we attempted to increase the expression of zBB by introducing mutations into the 4-1BB ICD that would prevent ubiquitination of zBB.44 To this end, we designed zBBKtoR, in which all lysines within the 4-1BB ICD were mutated to arginine (figure 3A). To study ubiquitination and degradation, we used lentiviruses to transduce zBBKtoR into CD3ζ-KO Jurkat cells (online supplemental figure 6A-C). As expected, ubiquitination of zBBKtoR was reduced compared with that of zBB (figure 3B). We also confirmed that expression of zBBKtoR protein was significantly increased compared with zBB (figure 3C), and that surface TCR expression of zBBKtoR was slightly higher as compared with that of zBB (online supplemental figure 6D). Despite this, TCR surface expression was not fully restored, and phosphorylation levels of CD3ζ and ZAP-70 were not substantially different from those of zBB (figure 3D). These results suggest that ubiquitination and post-translational degradation of zBB is not the main factor responsible for the failure of TCR complex formation and signaling.
Figure 3.
The BRM in the 4-1BB ICD affects membrane-proximal TCR signaling. (A) Schematic and amino acid sequences showing lysine-to-arginine mutations in zBB. (B) Immunoblot analysis of zBBKtoR ubiquitination. Jurkat cells were stimulated with SCT-K562 for 10 min. Modified-CD3ζ protein was immunoprecipitated with anti-3×FLAG beads, separated using SDS-PAGE, and probed using western blot, with anti-ubiquitin and anti-CD3ζ antibodies (left). Quantification of the relative abundances of ubiquitin, normalized to CD3ζ protein (right). (C) Expression of modified-CD3ζ protein in zBBKtoR-transduced CD3ζ-KO Jurkat cells (left). Quantification of the relative abundances of modified-CD3ζ, normalized to β-actin protein (right). (D) Phosphorylation of CD3ζ (left) and ZAP-70 (right) in zBBKtoR-transduced CD3ζ-KO Jurkat cells. (E) Amino acid sequences of zBB and zBBΔBRM. (F) Expression of modified-CD3ζ protein in zBBΔBRM-transduced CD3ζ-KO Jurkat cells (left). Quantification of the relative abundances of modified-CD3ζ, normalized to β-actin protein (right). (G) Phosphorylation of CD3ζ (left) and ZAP-70 (right) in zBBΔBRM-transduced CD3ζ-KO Jurkat cells. (H) Co-immunoprecipitation of zBBΔBRM. CD3z, zBB, and zBBΔBRM were immunoprecipitated from the indicated Jurkat cell lysates and separated by means of SDS-PAGE. Western blots were probed with anti-TRAF2, anti-ZAP-70, anti-CD3ζ, and anti-β-actin antibodies. P values (*p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001) were determined using multiple unpaired student’s t-test (B) and one-way analysis of variance, with Tukey’s multiple comparisons post hoc test (C, D, F, and G). Data have been presented as mean±SEM. BRM, basic-rich motif; dLNGFR, delta low-affinity nerve growth factor receptor; ICD, intracellular domain; KO, knockout; MFI, mean fluorescence intensity; n.s., not significant; P2A, porcine teshovirus-1 2A; TCR, T cell receptor; TM, transmembrane, TRAF2, tumor receptor-associated factor; ZAP-70, zeta-associated protein-70.
The BRMs within the cytoplasmic domains of CD3ε and CD3ζ are important for the association of the respective proteins with the plasma membrane and thus modulation of TCR signaling.45 46 We hypothesized that the BRM of 4-1BB within our zBB construct enhances the association of the modified CD3ζ with the plasma membrane, further attenuating TCR signaling. The BRM of 4-1BB is located at the N-terminal region of the ICD (online supplemental figure 7A). To test our hypothesis, we designed zBBK/RtoS, in which the positively charged lysine and arginine residues within the BRM were mutated to serine (online supplemental figure 7A), while preserving the TBM at the C-terminus of CD3ζ.47 The zBBK/RtoS construct was transduced into CD3ζ-KO Jurkat cells, and then dLNGFR-positive cells were isolated by magnetic separation (online supplemental figure 7B). There was also no significant difference in the mRNA expression (online supplemental figure 7C) and dLNGFR MFI (online supplemental figure 7D) between all groups. However, there was increased expression of zBBK/RtoS protein compared with zBB, as measured by western blot (online supplemental figure 7E). In addition, zBBK/RtoS induced greater TCR complex formation (online supplemental figure 7F) as well as CD3ζ and ZAP-70 phosphorylation (online supplemental figure 7G, H) compared with zBB and zBBKtoR, although to a lesser extent compared with unmodified CD3z.
In parallel with the mutagenesis approach, we also constructed zBBΔBRM in which the BRM residues within 4-1BB were truncated, leaving a minimal TBM for fusion with CD3ζ (figure 3E). On transduction of CD3ζ-KO Jurkat cells with zBBΔBRM lentiviral vectors (online supplemental figure 8A, B), we found that zBBΔBRM protein expression (figure 3F, online supplemental figure 8C), TCR complex formation (online supplemental figure 8D), and CD3ζ and ZAP-70 phosphorylation (figure 3G) were significantly higher compared with those of zBB and were comparable to those of CD3z cells. As shown in online supplemental figure 8E, mRNA expression levels of all constructs were similar. To determine whether the presence of BRM in BBz also contributes to the compromised expression of BBz, we generated BBΔBRMz by removing the BRM sequence from BBz (online supplemental figure 8F). We then transduced CD3ζ-deficient Jurkat cells with lentivirus encoding BBΔBRMz construct and measured the TCRα/β expression levels. We found that BBΔBRMz expressing cells had a significantly higher TCRα/β MFI than BBz expressing cells (online supplemental figure 8G). Therefore, BRM appears to have the same effect in BBz as in zBB with respect to TCRα/β expression. Additionally, these results indicate that the location of the 4-1BB sequence relative to the plasma membrane also has a profound effect on TCRα/β expression with or without BRM, as determined by the higher MFI of TCRα/β in zBB and zBBΔBRM as compared with BBz and BBΔBRMz. Interestingly, there was higher TCR complex expression in zBBΔBRM cells compared with zBBK/RtoS cells (online supplemental figure 8H). Next, using co-immunoprecipitation to determine whether deletion of the BRM in zBBΔBRM altered the association of ZAP-70 and TRAF2, we found that zBBΔBRM and CD3z showed comparable association with ZAP-70 and that this association was greater than that of zBB (figure 3H). Furthermore, zBBΔBRM retained the highest association with TRAF2 among the constructs tested. Collectively, these results indicate that basic residues in the BRM of the 4-1BB ICD negatively affect TCR complex formation and signaling, and that addition of the minimal 4-1BB TBM is sufficient for TRAF2 binding to the modified-CD3ζ, supporting further investigation of the function of zBBΔBRM.
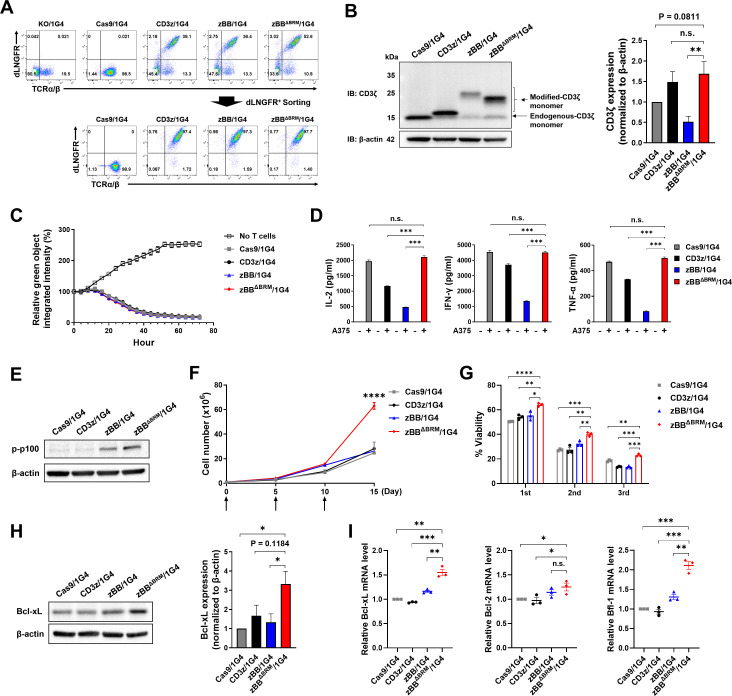
Primary human zBBΔBRM/1G4 T cells exhibit more robust in vitro expansion than conventional 1G4 T cells
To characterize the zBBΔBRM construct in primary human T cells, we generated zBBΔBRM/1G4 T cells with a KI efficiency of approximately 30%–50% (figure 4A, online supplemental figure 9A, B). Cells were subsequently sorted with dLNGFR. Western blot analysis revealed that CD3ζ protein expression in zBBΔBRM/1G4 T cells was comparable to that in Cas9/1G4 and CD3z/1G4 T cells, but higher than that in zBB/1G4 T cells. (figure 4B). We further found that TCR complex MFI was highest in zBBΔBRM/1G4 T cells, followed by CD3z/1G4, Cas9/1G4, and zBB/1G4 T cells (online supplemental figure 9C). Finally, phosphorylation of CD3ζ and ZAP-70 in Cas9/1G4, CD3z/1G4, and zBBΔBRM/1G4 T cells was comparable to each other but was much higher than that in zBB/1G4 T cells (online supplemental figure 9D, E), consistent with our analysis of zBBΔBRM in Jurkat cells (figure 3). Next, using in vitro cytotoxicity and cytokine production assays performed on co-cultures of T cells with A375 cells, we found that the Cas9/1G4, CD3z/1G4, zBB/1G4, and zBBΔBRM T cells were equally cytotoxic (figure 4C) and that all exhibited comparable CD4/CD8 ratios (online supplemental figure 9F). Interestingly, the production of IL-2, IFN-γ, and TNF-α in zBBΔBRM/1G4 T cells was restored to the levels observed in Cas9/1G4 T cells (figure 4D). A previous study found that 4-1BB-expressing second-generation CAR-T cells exhibited enhanced non-canonical nuclear factor kappa B (NF-κB) signaling as compared with the first generation or CD28-expressing CAR-T cells.48 To determine whether 4-1BB signaling can also activate non-canonical NF-κB signaling in zBBΔBRM/1G4 T cells, p100 phosphorylation status was measured by western blot after co-culturing with A375 cells for 24 hours. As shown in the figure 4E, we found that phosphorylated p100 was significantly increased only in zBB/1G4 and zBBΔBRM/1G4 T cells, providing further evidence of functional 4-1BB signaling activation.
Figure 4.
zBBΔBRM-KI 1G4 T cells exhibit more robust in vitro expansion than conventional 1G4 T cells. (A) KI (top) and sorting (bottom) efficiency of zBBΔBRM in primary human T cells. (B) Expression of modified-CD3ζ protein in zBBΔBRM/1G4 T cells (left). Quantification of the relative abundances of modified-CD3ζ, normalized to β-actin protein (right). (C) In vitro cytotoxicity of zBBΔBRM/1G4 T cells against Zsgreen-expressing A375 cells (effector-to-target ratio=1:1). (D) Production of the cytokines, IL-2 (left), IFN-γ (middle), and TNF-α (right) in the indicated zBBΔBRM/1G4 T cells, following stimulation with A375 cells for 24 hours (n=3). (E) Expression levels of phosphorylated p-100 protein in indicated TCR-T cells after co-culturing with target cells for 24 hours. (F) In vitro T-cell expansion assay of zBBΔBRM/1G4 T cells. Cumulative cell counts of TCR-T cells during serial stimulation by irradiated SCT-K562 cells. Arrows indicate time points of TCR-T cell stimulation with irradiated SCT-K562 cells. Results are representative of two donors. (G) Flow cytometric analysis of annexin V/PI-stained, apoptotic TCR-T cells (n=4) after 4-day co-culture with target cells. (H) Expression levels of Bcl-xL protein in indicated TCR-T cells after co-culturing with target cells for 24 hours (left). Quantification of the relative abundances of Bcl-xL, normalized to β-actin protein (right). (I) mRNA expression levels of Bcl-xL, Bcl-2, and Bfl-1 in indicated TCR-T cells after co-culturing with target cells for 16 hours. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001, as assessed using one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons post hoc test (B, D, and G-I) and two-way ANOVA with Tukey’s multiple comparisons post hoc test (F). Data have been presented as mean±SEM. BRM, basic-rich motif; dLNGFR, delta low-affinity nerve growth factor receptor; IFN, interferon; IL, interleukin; KI, knock-in; KO, knockout; mRNA, messenger RNA; n.s., not significant; PI, propidium iodide; TCR, T-cell receptor; TNF, tumor necrosis factor.
Multiple studies have shown that the addition of co-stimulatory signaling domains improves the long-term expansion of CAR-T cells both in vitro and in vivo, leading to superior antitumor effects.11–13 To evaluate whether zBBΔBRM affects TCR-T cell expansion in vitro, we quantified T cells on repeated antigen stimulation on days 0, 5, and 10 with irradiated K562 cells expressing an SCT of HLA-A2 linked to the NY-ESO-1 peptide (online supplemental figure 10A–C). There was no difference in cell number between groups on days 5 or 10, but zBBΔBRM/1G4 T cells were markedly more abundant as of day 15 (figure 4F). To determine whether the increase in the zBBΔBRM/1G4 T-cell number was attributable to reduced T-cell apoptosis or increased cell division, we performed annexin V and PI staining on days 5, 10, and 15. zBBΔBRM/1G4 T cells were more viable than CD3z/1G4 and zBB/1G4 T cells, on repeated stimulation (figure 4G). Consistent with this, zBBΔBRM/1G4 T cells showed increased expression of Bcl-xL, an anti-apoptotic protein, compared with the other groups after a 24-hour stimulation (figure 4H). In addition, mRNA expression levels of the anti-apoptotic proteins, Bcl-xL, Bcl-2 and Bfl-1, were significantly upregulated in zBBΔBRM/1G4 T cells (figure 4I). However, cell division, as measured by CellTrace Violet staining, did not differ between groups (online supplemental figure 10D). We further examined the expression patterns of CCR7 and CD45RO markers on TCR-T cells to determine their differentiation status. Flow cytometry analyses were performed either on unstimulated cells or on cells stimulated with irradiated-K562 SCT for 7-day intervals. We found a significant increase in the population of effector memory CD8 T cells (CD8 TEM, CCR7−CD45RO+) following the stimulation (online supplemental figure 10E). However, there was no significant difference among all groups in the frequency of the central memory T cells (CCR7+CD45RO+), suggesting that the incorporation of 4-1BB signaling domain in the CD3ζ does not significantly affect the differentiation of TCR-T cells. Collectively, these results suggest that the superior in vitro expansion of zBBΔBRM/1G4 T cells could be attributable to the additional 4-1BB signaling, which is likely related to upregulation of anti-apoptotic factors, rather than increased cell division.
zBBΔBRM/1G4 T cells exert superior antitumor effects in vivo
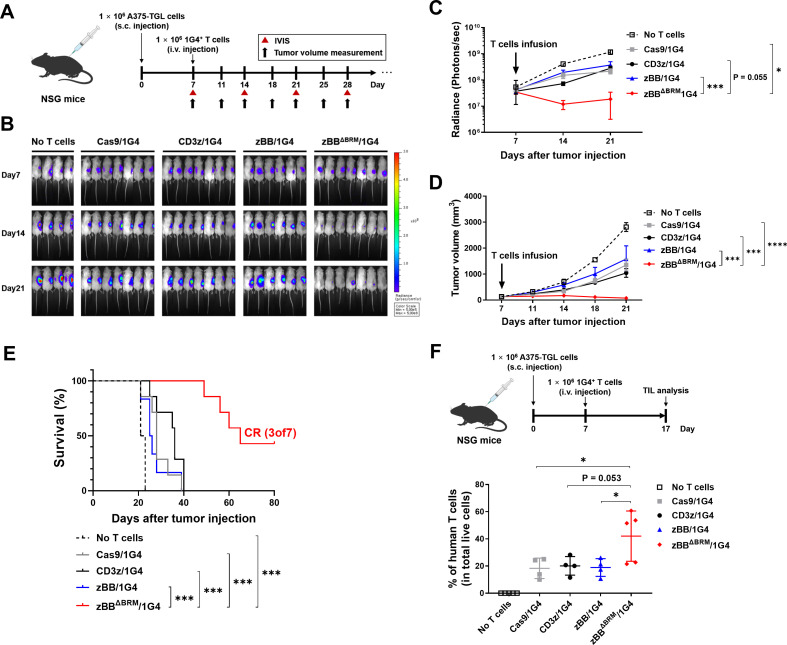
To determine whether the modified-CD3ζ-KI TCR-T cells enhance antitumor effects in vivo, we used a xenograft model, prepared by subcutaneously injecting A375-TGL cells into NSG mice. After allowing the A375-TGL cells to grow for 7 days, mice were intravenously injected with each of 1G4 TCR-T cells (figure 5A). Tumor regression was assessed by monitoring the reduction in tumor burden using an IVIS, and tumor size was measured using a digital caliper. Treatment with zBBΔBRM/1G4 T cells resulted in a dramatic reduction in tumor growth, whereas treatment with other 1G4 TCR-T cells mildly reduced tumor development (figure 5B–D, online supplemental figure 11A). zBBΔBRM/1G4 T cells prolonged mouse survival compared with controls and led to complete remission in three of the seven mice (figure 5E). Lastly, to determine the extent of in vivo expansion of TCR-T cells, we sacrificed mice on day 17 in the same xenograft setting as above (figure 5F, top). Assessment of the proportion of TILs showed enhanced longevity of zBBΔBRM/1G4 T cells (figure 5F, bottom and online supplemental figure 11B), potentially accounting for their superior antitumor effect in vivo.
Figure 5.
zBBΔBRM-KI TCR-T cells exert superior antitumor effects in vivo. (A) Experimental design showing subcutaneous infusion of NSG mice with A375 cells (day 0) and subsequent injection with 2.5×106 dLNGFR-positive T cells (1×106 1G4 TCR+ T cells; day 7), followed by monitoring of A375 tumor growth (ie, progression). (B) Representative in vivo bioluminescent imaging of the luciferase activity (n=8–9 mice/group). (C) Quantification of bioluminescence imaging data (total photon counts). Data have been presented as mean±SEM. The indicated p values, as determined using two-way analysis of variance (ANOVA) with Holm-Sidak’s correction for multiple comparisons, correspond to comparisons of the Cas9/1G4 T cell group with the zBBΔBRM/1G4 T-cell group. (D) Tumor volume of the A375-bearing NSG mice. Data have been presented as mean±SEM. The indicated p values, as determined using two-way ANOVA with Holm-Sidak’s correction for multiple comparisons, correspond to comparisons of control (Cas9), CD3z, or zBB group with the zBBΔBRM group, as indicated. (E) Kaplan-Meier curves showing overall survival of mice. Results are representative of one of two repeat experiments. Statistical significance was determined using Mantel-Cox test. (F) Experiments were conducted in the same manner as in (A) above, but with sacrifice of mice on day 17, followed by analysis of TILs. Top, experimental design; bottom, percentage of TILs (hCD3+ T cells among total live cells) in tumors. Data have been pooled from two independent experiments. *p<0.05, one-way ANOVA with Tukey’s multiple comparisons post hoc test (F). BRM, basic-rich motif; CR, complete remission; dLNGFR, delta low-affinity nerve growth factor receptor; i.v., intravenous; IVIS, in vivo imaging system; s.c., subcutaneous; TCR, T-cell receptor; TILs, tumor-infiltrating lymphocytes.
Discussion
A major breakthrough in the history of CAR-T cell therapy is the design of second-generation CAR-T incorporating a co-stimulatory signaling domain into the signaling domain of CAR, which has a dramatic impact on their in vivo persistence and proliferation. We therefore sought to recapitulate such functional improvements in TCR-T therapy by engineering the CD3ζ chain within the TCR complex. A prerequisite for successful second-generation TCR-T is that any modification should not impair the expression or native signaling (signal 1) of the transgenic TCR. However, we unexpectedly found that the modified protein formed by fusion of the 4-1BB signaling domain to the N-terminal CD3ζ ICD failed to form a TCR complex. While fusion of the same domain to the C-terminus led to partial recovery of the TCR, proximal signaling remained significantly compromised, as revealed by decreased phosphorylation of CD3ζ and ZAP-70. Notably, the positional preference of the modification was opposite in the case of CAR expression, favoring BBz over zBB, reflecting the marked structural differences between CAR (single polypeptide) and TCR (a complex of multiple polypeptides).
Through a series of mutational studies, we found that the BRM within the 4-1BB signaling domain negatively affected the formation and signaling of the TCR complex. Thus, mutation of lysine and arginine to serine or truncation of the entire BRM significantly improved the TCR expression and signaling. BRMs are found in the ICDs of many immune receptors, such as CD3ζ, CD3ε, and CD28, and have been shown to facilitate binding of the ICD to the negatively charged inner leaflet of the plasma membrane.45 46 49 50 In particular, lipid binding of CD3ζ through the BRM is thought to be responsible for sequestering key tyrosine residues in the immunoreceptor tyrosine-based activation motif, thus functioning as a safety control for resting T cells.45 Although the exact role of the BRM in 4-1BB has not yet been identified, it is conceivable that fusion of an intact 4-1BB ICD containing the BRM with CD3ζ may further reinforce the lipid association of the modified CD3ζ, resulting in impaired native TCR signaling. Consistent with this interpretation, TCR signaling was restored by removal of the BRM from 4-1BB. However, it should also be noted that mutations in wild-type-CD3ζ BRM have been reported to impair TCR signaling,45 possibly by disrupting the interactions with Lck,46 suggesting that an appropriate charge balance in the modified-CD3ζ is essential for optimal T-cell activation.
Our immunoprecipitation analyses revealed that both zBB and zBBΔBRM are able to recruit TRAF2, in addition to ZAP-70, via the TBM motif in the 4-1BB ICD. Furthermore, the resulting zBBΔBRM/1G4 T cells demonstrated superior proliferative capacity after repeated antigen stimulation in vitro. It is likely that the additional signaling through 4-1BB/TRAF2 is responsible for the enhanced T-cell survival, consistent with observations in 4-1BB-based second-generation CAR-T cells. However, a more in-depth analysis of the signaling of zBBΔBRM/1G4 T cells is required to further validate this hypothesis.
We showed that zBBΔBRM/1G4 T cells demonstrate superior in vivo proliferation as well as potent antitumor activity compared with conventional 1G4 TCR-T cells in A375 melanoma xenograft models. Our study demonstrates that engineering of second-generation TCR-T can be accomplished by modifying the signaling components of the TCR, unlike previous studies on TCR-T which have primarily focused on engineering target recognition by the TCR. Several approaches have been employed previously to enhance the antitumor activity of TCR-T cells, including the removal of inhibitory programmed cell death protein-1 through CRISPR/Cas9 gene editing51 and the co-expression of transforming growth factor β R2-41BB chimeric switch receptor.52 In the case of CAR-T cells, the incorporation of cytokine signaling (signal 3) has been shown to improve their effector function and proliferation.53 54 Additionally, genetic ablation or overexpression of certain transcription factors,55 and epigenetic regulators56 57 has been shown to promote T-cell stemness and resistance to exhaustion, which could be crucial for developing effective CAR-T therapies for solid tumors. Considering that these approaches successfully enhanced the therapeutic activity of second-generation CAR-T cells, our TCR-T platform may also synergize with these genetic modifications. It is also noteworthy that the application of our modified-CD3ζ is not limited to TCR-T therapy but could also be used in other ACT platforms, such as TIL and virus-specific T-cell therapy. To demonstrate our concept, we optimized the ICD of 4-1BB in the current study, but other co-stimulatory signaling domains, such as CD28, OX40, and inducible T cell costimulator (ICOS), could be employed in future studies. Overall, our strategy for improving the intracellular signaling of TCR-T cells may provide new insights into second-generation TCR-T engineering that could help provide effective immunotherapy against solid tumors.
Footnotes
Twitter: @SangJoonLah, @HyuncheolJung, @Hyeong-Ryeol Choi
SL and SK contributed equally.
Contributors: CHK, SL, SK and Y-HL designed the experiments. CHK, SL and CH wrote the manuscript. SL, SK and IK performed the experiments and analyzed the data. HK and HRC assisted with cloning experiments. HJ assisted with co-immunoprecipitation experiments. CHK is responsible for the overall content as a guarantor.
Funding: This work was supported by a grant from the Samsung Science & Technology Foundation (SRFC-MA1701-07, CHK).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
Human peripheral blood collection from healthy donors was performed according to an Institutional Review Board-approved protocol (IRB-18-071, KAIST). All animal studies were conducted under a protocol approved by and in compliance with policies set by the Institutional Animal Care and Use Committee, KAIST (IACUC-18-192).
References
- 1.June CH, Riddell SR, Schumacher TN. Adoptive cellular therapy: a race to the finish line. Sci Transl Med 2015;7:280ps7. 10.1126/scitranslmed.aaa3643 [DOI] [PubMed] [Google Scholar]
- 2.Sadelain M, Rivière I, Riddell S. Therapeutic T cell engineering. Nature 2017;545:423–31. 10.1038/nature22395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.June CH, O’Connor RS, Kawalekar OU, et al. Car T cell immunotherapy for human cancer. Science 2018;359:1361–5. 10.1126/science.aar6711 [DOI] [PubMed] [Google Scholar]
- 4.Sadelain M, Brentjens R, Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov 2013;3:388–98. 10.1158/2159-8290.CD-12-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson HJ, Rafiq S, Brentjens RJ. Driving CAR T-cells forward. Nat Rev Clin Oncol 2016;13:370–83. 10.1038/nrclinonc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross G, Waks T, Eshhar Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc Natl Acad Sci U S A 1989;86:10024–8. 10.1073/pnas.86.24.10024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eshhar Z, Waks T, Gross G, et al. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc Natl Acad Sci U S A 1993;90:720–4. 10.1073/pnas.90.2.720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haynes NM, Snook MB, Trapani JA, et al. Redirecting mouse CTL against colon carcinoma: superior signaling efficacy of single-chain variable domain chimeras containing TCR-zeta vs Fc epsilon RI-gamma. J Immunol 2001;166:182–7. 10.4049/jimmunol.166.1.182 [DOI] [PubMed] [Google Scholar]
- 9.Brocker T, Karjalainen K. Signals through T cell receptor-zeta chain alone are insufficient to prime resting T lymphocytes. J Exp Med 1995;181:1653–9. 10.1084/jem.181.5.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong MC, Latouche JB, Krause A, et al. Cancer patient T cells genetically targeted to prostate-specific membrane antigen specifically lysE prostate cancer cells and release cytokines in response to prostate-specific membrane antigen. Neoplasia 1999;1:123–7. 10.1038/sj.neo.7900018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher J, Brentjens RJ, Gunset G, et al. Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRzeta /CD28 receptor. Nat Biotechnol 2002;20:70–5. 10.1038/nbt0102-70 [DOI] [PubMed] [Google Scholar]
- 12.Savoldo B, Ramos CA, Liu E, et al. Cd28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. J Clin Invest 2011;121:1822–6.:46110. 10.1172/JCI46110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brentjens RJ, Davila ML, Riviere I, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 2013;5:177ra38. 10.1126/scitranslmed.3005930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter DL, Levine BL, Kalos M, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med 2011;365:725–33. 10.1056/NEJMoa1103849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. 10.1056/NEJMoa1407222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Call ME, Pyrdol J, Wiedmann M, et al. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 2002;111:967–79. 10.1016/s0092-8674(02)01194-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson LA, Heemskerk B, Powell DJ, et al. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J Immunol 2006;177:6548–59. 10.4049/jimmunol.177.9.6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbins PF, Li YF, El-Gamil M, et al. Single and dual amino acid substitutions in TCR cdrs can enhance antigen-specific T cell functions. J Immunol 2008;180:6116–31. 10.4049/jimmunol.180.9.6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt TM, Aggen DH, Stromnes IM, et al. Enhanced-affinity murine T-cell receptors for tumor/self-antigens can be safe in gene therapy despite surpassing the threshold for thymic selection. Blood 2013;122:348–56. 10.1182/blood-2013-01-478164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linnemann C, Mezzadra R, Schumacher TNM. TCR repertoires of intratumoral T-cell subsets. Immunol Rev 2014;257:72–82. 10.1111/imr.12140 [DOI] [PubMed] [Google Scholar]
- 21.Dash P, Fiore-Gartland AJ, Hertz T, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 2017;547:89–93. 10.1038/nature22383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helsen CW, Hammill JA, Lau VWC, et al. The chimeric TAC receptor co-opts the T cell receptor yielding robust anti-tumor activity without toxicity. Nat Commun 2018;9:3049. 10.1038/s41467-018-05395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Yang Z, Horan LH, et al. A novel antibody-TCR (abtcr) platform combines fab-based antigen recognition with gamma/delta-TCR signaling to facilitate T-cell cytotoxicity with low cytokine release. Cell Discov 2018;4:62. 10.1038/s41421-018-0066-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baeuerle PA, Ding J, Patel E, et al. Synthetic truc receptors engaging the complete T cell receptor for potent anti-tumor response. Nat Commun 2019;10:2087. 10.1038/s41467-019-10097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Liu G, Wang J, et al. Chimeric STAR receptors using TCR machinery mediate robust responses against solid tumors. Sci Transl Med 2021;13:586.:eabb5191. 10.1126/scitranslmed.abb5191 [DOI] [PubMed] [Google Scholar]
- 26.Miyao K, Terakura S, Okuno S, et al. Introduction of genetically modified cd3ζ improves proliferation and persistence of antigen-specific CTLs. Cancer Immunol Res 2018;6:733–44. 10.1158/2326-6066.CIR-17-0538 [DOI] [PubMed] [Google Scholar]
- 27.Matsui K, Boniface JJ, Reay PA, et al. Low affinity interaction of peptide-MHC complexes with T cell receptors. Science 1991;254:1788–91. 10.1126/science.1763329 [DOI] [PubMed] [Google Scholar]
- 28.Corr M, Slanetz AE, Boyd LF, et al. T cell receptor-MHC class I peptide interactions: affinity, kinetics, and specificity. Science 1994;265:946–9. 10.1126/science.8052850 [DOI] [PubMed] [Google Scholar]
- 29.Harding FA, McArthur JG, Gross JA, et al. CD28-mediated signalling co-stimulates murine T cells and prevents induction of anergy in T-cell clones. Nature 1992;356:607–9. 10.1038/356607a0 [DOI] [PubMed] [Google Scholar]
- 30.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol 2004;4:336–47. 10.1038/nri1349 [DOI] [PubMed] [Google Scholar]
- 31.Schwartz RH. T cell anergy. Annu Rev Immunol 2003;21:305–34. 10.1146/annurev.immunol.21.120601.141110 [DOI] [PubMed] [Google Scholar]
- 32.Fathman CG, Lineberry NB. Molecular mechanisms of CD4+ T-cell anergy. Nat Rev Immunol 2007;7:599–609. 10.1038/nri2131 [DOI] [PubMed] [Google Scholar]
- 33.Philip B, Kokalaki E, Mekkaoui L, et al. A highly compact epitope-based marker/suicide gene for easier and safer T-cell therapy. Blood 2014;124:1277–87. 10.1182/blood-2014-01-545020 [DOI] [PubMed] [Google Scholar]
- 34.Crosson SM, Dib P, Smith JK, et al. Helper-Free production of laboratory grade AAV and purification by iodixanol density gradient centrifugation. Mol Ther Methods Clin Dev 2018;10:1–7. 10.1016/j.omtm.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014;24:1012–9. 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen T, Yu YYL, Fremont DH. Preparation of stable single-chain trimers engineered with peptide, beta2 microglobulin, and MHC heavy chain. Curr Protoc Immunol 2009;Chapter 17:17. 10.1002/0471142735.im1705s87 [DOI] [PubMed] [Google Scholar]
- 37.Call ME, Schnell JR, Xu C, et al. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 2006;127:355–68. 10.1016/j.cell.2006.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Au-Yeung BB, Shah NH, Shen L, et al. ZAP-70 in signaling, biology, and disease. Annu Rev Immunol 2018;36:127–56. 10.1146/annurev-immunol-042617-053335 [DOI] [PubMed] [Google Scholar]
- 39.Jang IK, Lee ZH, Kim YJ, et al. Human 4-1BB (CD137) signals are mediated by TRAF2 and activate nuclear factor-kappa B. Biochem Biophys Res Commun 1998;242:613–20. 10.1006/bbrc.1997.8016 [DOI] [PubMed] [Google Scholar]
- 40.Zapata JM, Perez-Chacon G, Carr-Baena P, et al. CD137 (4-1BB) signalosome: complexity is a matter of trafs. Front Immunol 2018;9:2618. 10.3389/fimmu.2018.02618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapoport AP, Stadtmauer EA, Binder-Scholl GK, et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med 2015;21:914–21. 10.1038/nm.3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran I, Lowther DE, Dryer-Minnerly R, et al. Systemic and local immunity following adoptive transfer of NY-ESO-1 spear T cells in synovial sarcoma. J Immunother Cancer 2019;7:276. 10.1186/s40425-019-0762-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Z, Condomines M, van der Stegen SJC, et al. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell 2015;28:415–28. 10.1016/j.ccell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Qiu S, Chen J, et al. Chimeric antigen receptor designed to prevent ubiquitination and downregulation showed durable antitumor efficacy. Immunity 2020;53:456–70. 10.1016/j.immuni.2020.07.011 [DOI] [PubMed] [Google Scholar]
- 45.Zhang H, Cordoba S-P, Dushek O, et al. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci U S A 2011;108:19323–8. 10.1073/pnas.1108052108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Guo X, Shi X, et al. Ionic CD3-lck interaction regulates the initiation of T-cell receptor signaling. Proc Natl Acad Sci U S A 2017;114:E5891–9. 10.1073/pnas.1701990114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arch RH, Thompson CB. 4-1BB and ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kappab. Mol Cell Biol 1998;18:558–65. 10.1128/MCB.18.1.558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Philipson BI, O’Connor RS, May MJ, et al. 4-1BB costimulation promotes CAR T cell survival through noncanonical NF-κb signaling. Sci Signal 2020;13:625.:eaay8248. 10.1126/scisignal.aay8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol 2000;7:1023–6. 10.1038/80930 [DOI] [PubMed] [Google Scholar]
- 50.Xu C, Gagnon E, Call ME, et al. Regulation of T cell receptor activation by dynamic membrane binding of the cd3epsilon cytoplasmic tyrosine-based motif. Cell 2008;135:702–13. 10.1016/j.cell.2008.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stadtmauer EA, Fraietta JA, Davis MM, et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020;367:eaba7365. 10.1126/science.aba7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth TL, Li PJ, Blaeschke F, et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell 2020;181:728–44. 10.1016/j.cell.2020.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kagoya Y, Tanaka S, Guo T, et al. A novel chimeric antigen receptor containing A JAK-STAT signaling domain mediates superior antitumor effects. Nat Med 2018;24:352–9. 10.1038/nm.4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell M, Gottschalk S. Engineered cytokine signaling to improve CAR T cell effector function. Front Immunol 2021;12:684642. 10.3389/fimmu.2021.684642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan O, Giles JR, McDonald S, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 2019;571:211–8. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fraietta JA, Nobles CL, Sammons MA, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 2018;558:307–12. 10.1038/s41586-018-0178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prinzing B, Zebley CC, Petersen CT, et al. Deleting dnmt3a in car T cells prevents exhaustion and enhances antitumor activity. Sci Transl Med 2021;13:620.:eabh0272. 10.1126/scitranslmed.abh0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005519supp001.pdf (13.8MB, pdf)
Data Availability Statement
Data are available upon reasonable request.