Abstract
When ecosystems are under severe pressure or environments change, trophic position and intraspecific niche width may decrease or narrow, signalling that conservation action is required. In New Zealand, alpine and subalpine ecosystems have been extensively modified through farming since 19th‐century European settlement, with consequences for indigenous species such as the kea Nestor notabilis.
We investigated feather stable isotope values in the kea and predicted a lower trophic position in modern kea populations, to reflect reduced lowland habitat and a mixed diet with more plant material. We predicted that size and sex would influence trophic values in this sexually dimorphic species, with larger birds more likely to have a high protein diet.
We examined potential dietary changes in 68 museum collected kea from 1880s to 2000s, first recording accession details including provenance and sex and measuring culmen length. We used bulk carbon and nitrogen stable isotope analyses (BSIAs) of feathers and a further feather subset using compound‐specific stable isotope analyses of amino acids (CSIA‐AA) to obtain isotopic values and estimate trophic position.
BSIA showed δ15N values in kea feathers declined through time and could indicate that early century kea were highly omnivorous, with δ15N values on average higher than in modern kea. Variance in δ15N values was greater after 1950, driven by a few individuals. Few differences between males and females were evident, although females in the south region had lower δ15N values. There was a tendency for large male birds to have higher trophic values, perhaps reflecting dominant male bird behaviour noted in historical records. Nonetheless, CSIA‐AA performed on a subset of the data suggested that variation in BSIA is likely due to baseline changes rather than relative trophic position which may be more homogenous than these data indicate. Although there was more variability in modern kea, we suggest caution in interpretation.
Stable isotope data, particularly CSIA‐AA, from museum specimens can reveal potential change in ecological networks as well as sexually dimorphic feeding patterns within species. The data can reveal temporal and regional variation in species trophic position and changes in ecosystem integrity to inform conservation decision‐making.
Keywords: amino acids, CSIA‐AA, museum collections, niche width, omnivory, parrot, stable isotopes, trophic position
Kea parrots in New Zealand were hunted as sheep killers for more than 100 years, while forest and high country habitat were destroyed for farming. We analysed carbon and nitrogen isotope ‘fingerprints’ in feathers from museum collections to recreate past kea diets, and found that kea are generally omnivorous.
1. INTRODUCTION
Native species decline is frequently linked to human impacts on ecosystems, including land use change, accelerated climate change, pollution, invasive species impacts and direct exploitation of species through harvesting (Allentoft et al., 2014; Elliott & Kemp, 2004; Sandom et al., 2014). Species extinction is only one outcome of ecosystem damage; other impacts can include contracted species distributions, misaligned life histories or phenology, reduced foraging opportunities for individuals and genetic bottlenecks (Dussex et al., 2015). Species declines and extinctions are particularly evident in island ecosystems, where human colonisation is recent and endemic species, including ground nesting avifauna, are particularly vulnerable (Blackburn et al., 2004; Duncan & Blackburn, 2004). Nonetheless, impacts can be difficult to pinpoint because it can be challenging to reconstruct diet, movement or behaviour patterns for organisms that have been extirpated.
Natural history collections provide remarkable glimpses into such histories, and allow exploration of these impacts and their ongoing effects through time (Holmes et al., 2016; Roberts et al., 2016; Smith et al., 2021; Wehi et al., 2012). The painstaking work of curators, collectors and others has resulted in many thousands of skins and mounts available for scientific study. As human‐induced environmental impacts accelerate in the Anthropocene, there has been a quickening of interest in the use of these specimens, with the aim of increasing knowledge of past organisms and the ecosystems in which they lived, to aid current conservation decision‐making. For example, one recent study of genetic population structure based on collections of fossil bones indicates that post‐glacial contractions rather than human activity led to modern genetic bottlenecks in kea (Dussex et al., 2014). Another study based on bones from museum collections confirmed colonisation of modern lakes by a genetically, morphologically and ecologically distinct swan that replaced a predecessor extirpated by hunting (Rawlence et al., 2017).
Morphological and genetic studies are only the beginning, however; natural history collections may also be used for long‐term studies based on isotopic research (English et al., 2018; Hallworth et al., 2018; Wehi et al., 2012). Stable isotope data provide insights into both the trophic position of species, and shifts in foraging patterns (Chiaradia et al., 2010; Hobson et al., 1994). It is possible to track animals moving between isotopically different food sources that vary spatially and to construct longitudinal time series (e.g. Flockhart et al., 2017; Wassenaar & Hobson, 1998). Stable isotope values in inert tissues, such as the keratin in bird feathers, reliably records diet, habitat and geographical location information. Feathers, like other keratin‐based tissues such as nails and claws, are metabolically inert and therefore reflect diet during the period of their growth (Hobson & Clark, 1992).
Stable isotope analyses use ratios of light and heavy forms of carbon (12C/13C, represented as δ13C) and nitrogen (14N/15N, represented as δ15N) to determine likely dietary preferences and trophic position of consumers. Because the heavier 15N isotope is assimilated into tissues while the lighter 14N isotope is metabolised in a process called isotopic fractionation, consumers are 15N enriched compared to their food resource. As such, omnivores feeding at a high trophic position will usually have high bulk δ15N values compared with herbivores, although there are difficulties in assessing baseline isotope values among ecosystems. Both δ13C and δ15N vary in plants depending on the types of compounds obtained, the manner in which uptake occurs, or mycorrhizal associations (Katzenberg, 1989; Szpak, 2014). As well as with climatic variables such as mean annual precipitation, or in ecosystems where there is loss of biodiversity and hence a reduction in food chain length. In addition, variation may be linked to habitats associated with anthropogenic input such as fertilisers, and 20th‐century anthropogenic foods that may contain products derived from C4 plants (i.e. cane sugar or corn), leading to distinct δ13C values in tissues (e.g. Newsome et al., 2010). These bulk stable isotope values can thus be useful for identifying individual organisms that scavenge around human food sources. In addition to these baseline and variation issues in consumed foods, many generalist species consist of aggregations of specialised individuals (Bolnick et al., 2007). This may be seen in populations with intense competition (Kronfeld‐Schor & Dayan, 1999) and morphological character displacement (Losos, 2000), including sexually dimorphic species (Tomotani et al., 2021).
Recent development of compound‐specific stable isotope analysis of amino acids (CSIA‐AAs) can now more accurately assess trophic position by removing the uncertainty around baselines, where sampling of potential food items and other materials in the same spatio‐temporal context is limited, lacking or simply impossible. Indeed, CSIA‐AA offers a refinement of the bulk stable isotope approach by leveraging information from individual molecules. As building blocks of proteins, amino acids are a major source of nitrogen in feathers and play a fundamental role in all metabolic processes (O'Connell, 2017). As such, amino acid nitrogen stable isotopes (δ15NAA) now represent a powerful tool that can be applied to disentangle past (e.g. Tomotani et al., 2021) and present food web structures (Hetherington et al., 2017; McMahon et al., 2016; Sabadel et al., 2016). The rationale behind the CSIA‐AA approach is that some AA compounds, referred to as ‘trophic’ (e.g. glutamic acid), fractionate substantially while others, referred to as ‘source’ (e.g. phenylalanine), only fractionate a little, thus creating a nitrogen isotope baseline (Ohkouchi et al., 2017; Popp et al., 2007). Additionally, the difference between trophic and source AAs enables us to better constrain trophic positions and track changes in the δ15N baseline (Popp et al., 2007; Quillfeldt & Masello, 2020; Sabadel et al., 2019, 2020).
In countries such as New Zealand, where human arrival first occurred around 800 years ago, recent large‐scale human‐induced environmental change, such as the deforestation of high‐altitude regions, is well‐documented. Much of this ‘high country’ was converted from forest to farmland in the 19th century after European arrival (McGlone, 1983), leading to reduced alpine and subalpine biodiversity. The ‘alpine parrot’ or kea Nestor notabilis is now a threatened species with low and declining population numbers of around 5,000 individuals (Dussex et al., 2014) compared to more than 150,000 in the 19th century (Elliott & Kemp, 2004). Kea are large, sexually dimorphic and highly social parrots (Diamond & Bond, 1991). Their diet consists of invertebrates, fruit and seeds (Buller, 1882; Clark, 1970), and they play an important role in seed dispersion (Young et al., 2012). Kea nonetheless have a chequered reputation as both carnivores and scavengers and have been blamed for sheep killing (often described as ‘kea strike’) on high country farms, particularly in the late 19th and early 20th centuries (e.g. Benham, 1906; Reid, 2019).
In this study, we used museum specimens to examine kea diet from the late 19th century through to 2007. We first evaluated trophic position to identify any likely foraging differences based on culmen length, bill dimorphism of males and females, age and collection year. We additionally aimed to identify anthropogenic food sources in kea diet. We then compared niche space among northern and southern population groups and by sex.
2. MATERIALS AND METHODS
2.1. Natural history collections
We examined 84 kea specimens and skins held in five natural history collections, at Auckland Museum, Te Papa National Museum, Canterbury Museum and Otago Museum collections in New Zealand and Naturhistorisches Museum Wien (the Vienna Natural History Museum), Austria (Data S1, Wehi et al., 2022a, 2022b). Details were missing for some individuals, thus reducing the total number of individuals whose feathers were analysed for isotope values. However, we recorded all accession details for kea where these were available, including collection location and year, collector, weight, circumstances of collection and sex if previously determined. Collection dates ranged from 1877 through to 2007. Sampling was approved by all museums, the Komiti Taoka Tuku Iho, and via a wildlife authority application 52312‐DOA from the Department of Conservation.
2.2. Morphometrics, sex and location assignment
We age‐estimated the same birds based on eye ring coloration (where the presence of a yellow ring around the eye indicates the bird is immature) (Bond et al., 1991). We recorded culmen length, as the chord from the beak tip to the cere, using callipers, for all 84 birds we examined, with the exception of one individual with a severely twisted beak. Damage to the beak and other characteristics were recorded. Of the 70 wild birds, 48 had their sex noted in accession details (n = 14 female, n = 34 male). For 18 of the 22 remaining wild birds whose sex was not recorded, we estimated sex by fitting a predictive model to the data. Culmen length and age were predictor variables (see the Statistical analyses on bulk isotope analysis section). Sex was not able to be estimated for four of the 22 birds with unknown sex, because both culmen length and age were unknown. Capture locations for sampled specimens are marked on Figure 1.
FIGURE 1.
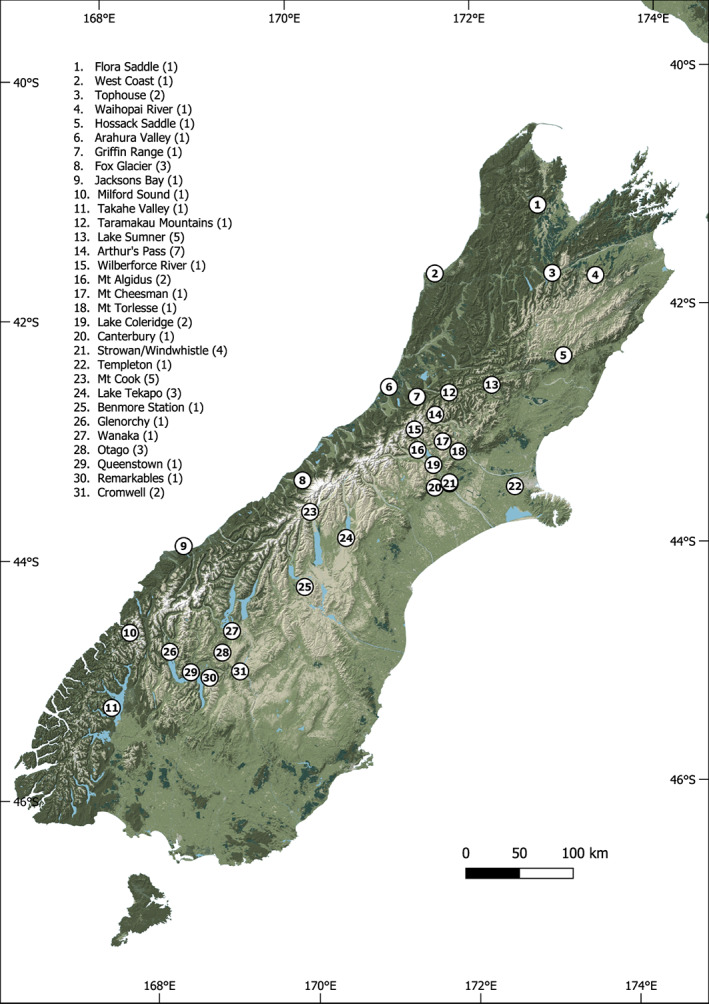
Capture locations in the South Island of New Zealand for kea from museum collections used in this study. Recorded locations were part of the provenance data attached to museum specimens. Kea specimens used for the north group in the Canterbury region were from locations 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 and 18. Birds in the south group from the Otago region were from locations 20, 21, 22, 25, 26, 27, 28, 29, 30. Birds from the northern area and West Coast of the South Island were excluded from these groupings
We examined the geographical distribution of capture locations and assigned wild caught birds by region as ‘north’ or ‘south’, based on geographical clumping of collection locations in the Canterbury (north) and Otago (south) regions (Figure 1), which also reflect underlying genetic patterns (Dussex et al., 2015). Twelve wild kea with locational data were unassigned, because their location fell substantially outside the main groupings or were ambiguous.
2.3. Feather sampling and bulk stable isotope analysis
For each of 68 individuals for whom most accession data were available, three feathers were removed from the upper middle back between the shoulder blades on each bird skin. Feathers are moulted annually but irregularly, and for adults, after fledging. Feathers (including duplicates from six birds to provide error estimates) were washed in 2:1 chloroform:methanol solution for 24 hr, agitated, rinsed and air‐dried in a fume cupboard for 48 hr, before the top 1 cm of the feather vane was finely clipped with scissors and weighed into tin capsules.
Bulk carbon and nitrogen isotopes (with a precision of ±0.1‰ for δ13C and ± 0.1‰ for δ15N) and total %C and %N content were measured on around 0.5 mg of feathers tightly wrapped in 6 × 4 mm tin capsules in duplicate. Carbon and nitrogen isotopes were measured using an Isoprime isotope ratio mass spectrometer (VG Micromass, UK), interfaced to an Eurovector EA elemental analyser (VG Micromass, UK) in continuous flow mode (EA‐IRMS). Samples were measured at the Stable Isotope Facility, GNS Science in Lower Hutt.
Results were expressed in conventional delta notation (d), defined as the part per mil (‰), according to the following equation:
where X represents 13C or 15N, and R sample and R standard are the fractions of heavy to light isotopes in the sample and standard respectively. The feather δ13C and δ15N values were determined and normalised using working reference materials (leucine, cane sugar, EDTA, caffeine, beet sugar) calibrated to international standards (IAEA‐N1, IAEA‐N2, IAEA‐CH6, IAEA‐CH7, IAEA‐S1 and IAEA‐S2) and were reported relative to Vienna PeeDee Belemnite for carbon (VPDB) and atmospheric N2 for nitrogen. Analytical precision of working standards and reference materials were <±0.1‰ for δ13C and ±0.1‰ for δ15N. Blanks were included to check for background interference. Reference materials and working standards were used to normalise values via a multi‐point (4 point) calibration curve which corrected for linearity and stretching. The C:N ratios are calculated using the atomic ratio of carbon to nitrogen abundance (C:N = [%C/%N] × [14/12)]).
2.4. Amino acid stable isotope analyses
Amino acid stable isotope analyses on feathers from 18 individuals were carried out in the Isotrace Lab at the University of Otago. Kea feather amino acids were extracted by hydrolysing each feather sample with 2 ml 6 M HCl at 110°C for 24 hr in a N2 atmosphere. An internal standard, norleucine (50 μl of 1 mg/ml solution), was added to monitor the wet chemistry and amino acid stable isotope values. Solutes were dried under a gentle flow of N2 at 60°C and subsequently converted into N‐acetylisopropyl ester derivatives following the protocol described in Sabadel et al. (2016), modified from Styring et al. (2012). Details of the derivatisation procedure are published in the supplement of Sabadel et al. (2016). Nitrogen stable isotopes of AA compounds were measured by gas chromatography/combustion/isotope ratio mass spectrometer (GC‐IRMS), using a Thermo Trace gas chromatograph, the GC combustion III interface and a Delta plus XP isotope ratio mass spectrometer (Thermo Fisher Scientific, UK). Two hundred nanolitre aliquots of derivatised AA were injected. The inlet was set at 270°C in splitless mode, carried by helium gas at 1.4 ml/min and compounds were separated on a VF‐35 ms column (0.32 mm ID and a 1.0 μm film thickness). The GC programme is published in the supplement of Sabadel et al. (2016). The oxidation reactor was set at 980°C, the reduction reactor at 650°C and a liquid N2 cold trap was employed after the reduction reactor. Samples were analysed along with AA standards of known isotopic composition. Eleven AAs (listed below) from each sample were measured with no peak co‐elutions. In order of elution: alanine (Ala), glycine (Gly), valine (Val), leucine (Leu), isoleucine (Ile), Threonine (Thr), serine (Ser), proline (Pro), asparagine + aspartic acid (Asx), glutamine + glutamic acid (Glx) and phenylalanine (Phe). Note that during the hydrolysis step, asparagine is converted to aspartic acid (hence the notation Asx) and glutamine is converted to glutamic acid (hence the notation Glx). Raw δ15NAA values were corrected by plotting the mean δ15NAA value of each individual AA standard measured on GC‐IRMS vs. their ‘real’ value measured on EA‐IRMS. Our R 2 was always >0.99. Raw δ15NAA values in our samples were then corrected using the fitting equation. The δ values were reported following the conventional method of expressing δ at natural abundance, in per mil (‰), relative to an international standard: atmospheric N2. Precision (1SD) of δ15NAA ranged from 0 to 1‰ with a mean of 0.4‰.
2.5. Trophic position estimations
Trophic position based on amino acid isotope values was calculated using the δ15NGlx and δ15NPhe relationship (TPGlx‐Phe, Equation 1) described in Chikaraishi et al. (2010):
| (1) |
where β is the isotopic difference between δ15NGlx and δ15NPhe values in the primary producers: δ15NGlx − δ15NPhe = −8.4‰ for terrestrial plants (Chikaraishi et al., 2010). The trophic discrimination factor, representing the difference in fractionation per trophic position of δ15NGlx and δ15NPhe (TDFGlx‐Phe), was 7.6‰, chosen in accordance with Chikaraishi et al. (2010), and recently confirmed as appropriate for New Zealand terrestrial birds by Tomotani et al. (2021).
2.6. Statistical analyses on Bulk Stable Isotope Analysis (BSIA)
All statistical analyses were carried out in R 4.0.2 (R Core Team, 2020). We first validated published estimates of morphological sex assignation using a logistic regression model with sex and culmen length as predictor variables. The ‘glm’ R function was used to fit the models. To do this, we trained the model for birds that had previously been assigned sex by museum staff (most likely resulting from inspection after dissection), and then made predictions for individuals that had not been assigned sex previously. The prediction accuracy, measured by fivefold cross‐validation using the caret r package (Kuhn, 2008), was estimated to be 76.9% (SD = 10.5%).
We used linear mixed effects modelling, with the ‘lmer’ function from the lme4 package (Bates et al., 2015), to investigate the association between multiple predictor variables and the outcome variables: δ13C and δ15N. For each of the δ13C and δ15N values, we fitted two models to the data. The first investigated a possible trend over time (as represented by sampling year), and the second focused on the effect of region (‘North’ and ‘South’; Figure 1) and habitat type (‘Wild’ and ‘Human’). In both models, other covariate effects were added to improve model accuracy and to control for potential confounding effects. A range of possible fixed effect interactions and random effects were assessed using the lmerTest package (Kuznetsova et al., 2017). The final models included no interactions and a single random effect: ‘Location’, describing the sample locations (Figure 1).
In the δ15N ‘time model’, Habitat Type (‘Wild’ and ‘Human’) and Age (‘Immature’ and ‘Mature’) effects were removed due to model instability caused by multicollinearity between the predictor variables. Equivalently, in the ‘region and habitat type model’, the Year variable was removed. The following equations were used in the models to investigate the relationship between the response (δ13C and δ15N) and predictor variables:
Time model:
Region/Habitat model:
In the above equations, YearS and CulmenS were modelled as continuous (scale) variables, and the remaining effects were two‐level categorical (factor) variables. YearS and CulmenS are standardised versions of Year and Culmen, used to improve model stability. Residual diagnostics were performed using the influence.ME r package (Nieuwenhuis et al., 2012) and showed that the models provided an acceptable fit to the data.
We examined the variance in two time periods using variance ratio tests (F tests), first, in birds from the late 19th century and before 1950, and second, in birds from after 1950, to investigate whether populations had become less specialised and intraspecific competition was stronger. Finally, we used the package siber (Jackson et al., 2011) to assess the possible existence of different bivariate isotopic niches associated with different regions (‘north’ and ‘south’) and with sex (‘female’ and ‘male’), where ellipses are used to identify the 95% CI (confidence intervals) for the four different combinations of region and sex.
3. RESULTS
3.1. Natural history collections
Of the 84 kea specimens that we examined, 77 were recorded as wild collected and seven had been captured and held in captivity (for unknown periods) before their death. Of the wild individuals, nine had neither an approximate collection location nor year recorded and were excluded. Of 68 kea with full or partial accession data, four were missing the year of collection, and two early specimens (1887 and 1905) did not have a location recorded. A further individual had a twisted beak that we were unable to measure. A small number had damaged beaks that nevertheless allowed a close length estimate. We therefore used the data from 61 wild individuals in the mixed effect model.
3.2. Morphological differences between males and females
Using the logistic regression model, adults with a culmen length >52.80 mm were classified as male, and adults with a culmen length <52.80 mm were classified as female; immature birds with a culmen length >45.36 mm were classified as males and immature birds with a culmen length <45.36 mm were classified as females. The cross‐validated prediction success rate of 76.9% provided confidence that morphological assignation of sex using this method resulted in a high level of accuracy. Of the individuals that we used for statistical analysis, the sex of only three birds (AV14648, AV3079 and NMW12.212) changed from an initial classification, out of 14 birds for whom we had estimated sex based on previous estimates of bill size ranges.
3.3. Dietary indicators
3.3.1. Bulk carbon and nitrogen stable isotope values
The linear mixed effects model investigating change over time (see Table S1 for model summary results) showed that kea relative trophic position, as estimated by comparing their δ15N values, has decreased since the late 19th century. However, it did not support differences in relative trophic position between the sexes (Figures 2 and 3a). The earliest feather samples had higher δ15N values than more recent feather samples, with an estimated mean δ15N of 6.5‰ (95% CI: 4.8‰, 8.2‰), considerably more than the estimated mean δ15N of 2.0‰ (95% CI: 0.2‰, 3.9‰) in modern kea (Figure 2). Culmen length was not a good predictor of relative trophic position, but the few kea with high δ15N feather values were all males (Figure 3a). A separate comparison of atomic C:N ratios with culmen length suggested a decrease in variance as culmen length increased (Figure 3b) and this result was confirmed when the variance of the lower quantile of culmen values was compared with the upper quantile (F 15,14 = 26.38, p < 0.0001) (Table S3). A plot of the data initially suggested that δ15N values variance might also reduce after 1950, but the ratio of these variances was not statistically significant (F 33,21 = 1.21, p = 0.66 (Table S4).
FIGURE 2.
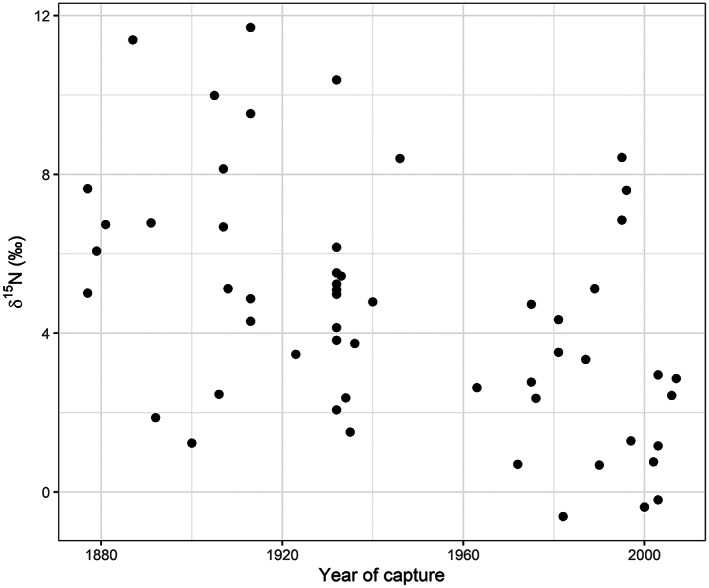
Relationship of bulk δ15N values from kea feathers and time, plotted by year
FIGURE 3.
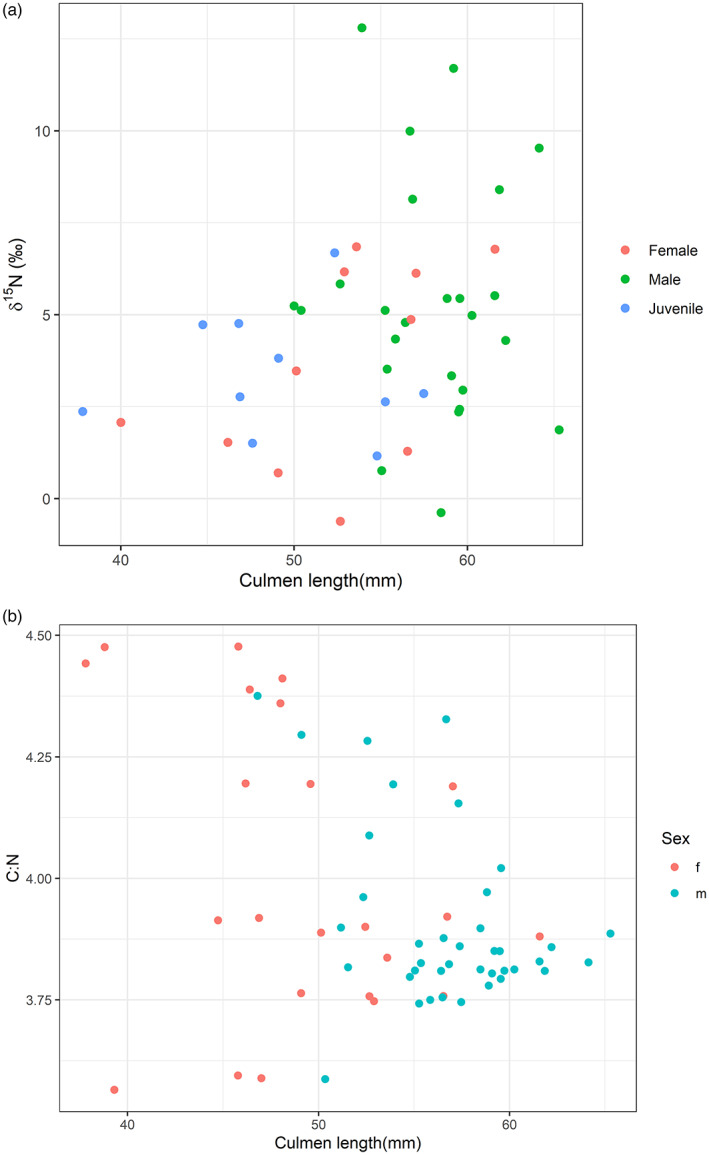
(a) Relationship between kea culmen length and bulk δ15N values for adult males, adult females and immature kea. (b) Relationship between kea culmen length and C:N bulk ratios estimated from kea feathers. C:N ratios become less heterogeneous as culmen length increases in male and female kea
Feather bulk δ13C values fell in the range of −24.0 to −22.0‰, with no apparent pattern in values through time (Figure S1). Nonetheless, a small number of individuals had δ13C values that fell outside this range; four birds—one from Arthurs Pass and three from Westland (two from Arahura Valley and one from Jacksons Bay)—had feather δ13C values around −20.0‰. Descriptive statistics suggest that δ13C variance increased after 1950, and the ratio of these variances was statistically significant (F 33,21 = 0.34, p = 0.006).
The linear mixed effects model with the primary focus on region and habitat effects (Table S2) identified a statistically significant association (p < 0.001) between bulk δ15N values and habitat type (‘human’ and ‘wild’). Wild habitat types have higher δ15N values (x̄ = 5.7‰, 95% CI: 4.3‰, 7.1‰) compared with human habitats (x̄ = 2.5‰, 95% CI: 0.9‰, 4.1‰). There was no statistical difference however between the north and south regions. An equivalent model was fitted with δ13C values as the dependent variable, and in this case, no statistically significant effects were identified.
Exploration of the possible existence of different isotopic niches associated with the assigned north and south regions and with sex showed a large degree of overlap in ellipses (Figure S2), indicating generally minor differences in isotopic niche for both δ13C and δ15N values with the four combinations possible. However, female kea in the south region did show a smaller standard ellipse area, representing the size of their isotopic niche, than all other groups (Figure 4).
FIGURE 4.
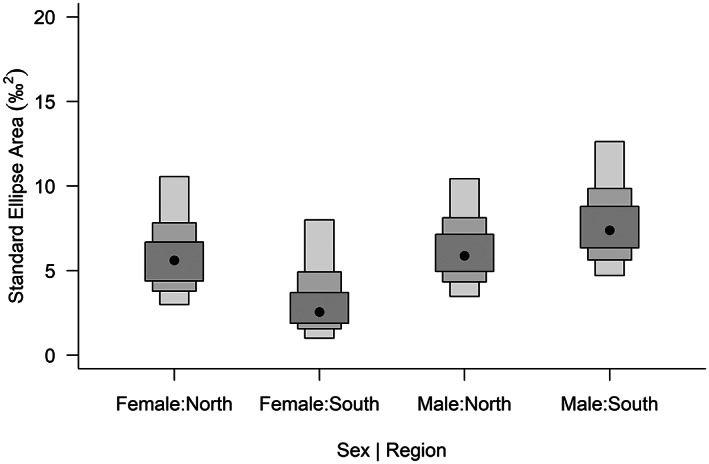
Isotopic niche representation of kea females and males in the North and South groups, showing the standard ellipse area. Black dots represent medians
3.3.2. Compound‐specific stable isotope values
Compound‐specific stable isotope of amino acid values ranged from −22.5‰ (δ15NThr) to 15.1‰ (δ15NPro) across the whole dataset. All AA groups (and individual AAs) are correlated with bulk δ15N values, except for threonine. Bulk δ15N correlated strongly with δ15NPhe values (F 1,16 = 28.6, p < 0.0001; Figure S3). Trophic position calculated using CSIA‐AA varied between 2 and 3 for all individuals analysed. Kea collected before 1950 had a more homogenous diet, mainly omnivorous, than kea collected more recently, with trophic position spanning completely herbivorous to completely carnivorous (Figure 5), but where trophic position did not vary significantly through time (Figure S4).
FIGURE 5.
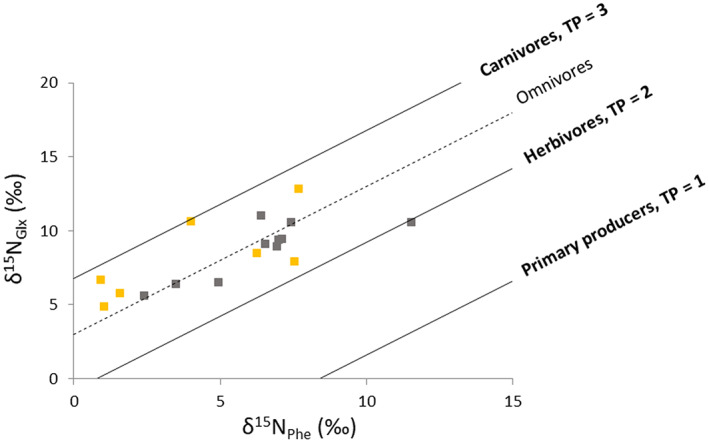
Cross‐plot for the δ15N values of trophic and source AAs, Glx and Phe, respectively, from kea feathers. Brown depicts ‘ancient’ kea (1887–1950) and yellow depicts ‘modern’ kea (1950–2007). Trophic isoclines with a slope of 1.0 and y‐intercept intervals of 7.6‰ represent different trophic positions (TPGlx‐Phe = 1, 2 and 3) according to equation 1 from Chikaraishi et al. (2010)
4. DISCUSSION
Both bulk and CSIA‐AA data support omnivory as the primary feeding classification for kea. Previous studies of modern kea have similarly indicated a diverse diet that is likely to be predominantly plant and insect based (Clark, 1970; Diamond & Bond, 1991; Greer et al., 2015), as might be expected from a parrot with a relatively unspecialised tongue (Kirk et al., 1993). Nonetheless, kea are also known as rubbish dump scavengers, and population biology research has notably occurred at high‐country refuse dumps and ski field carparks where kea congregate to scavenge food from humans (e.g. Bond & Diamond, 1992).
Culmen lengths assigned to females in this research were slightly larger than those assigned for wild female kea in previous field studies. Bond et al. (1991) estimated that 79% of birds with culmen lengths <43.9 mm were females. In this study, we estimated adult female culmens as < 52.80 mm (with males above this value), and for immature birds, we estimated female culmen size as <45.36 mm, with immature male culmens above this length. Bond et al. (1991) noted that the bills of captive individuals were often much larger than those seen in the field, suggesting diet plays a role in its growth. It is unclear what has led to the pattern observed in the data shown here, but it is possible that, because many of the birds in this study were collected in the late 19th and the first half of the 20th century, it reflects a different nutritional environment for kea. Further comparisons with modern wild kea culmen, and exploration of nutritional effects, could be beneficial to conservation planning and management.
The downward trend in the BSIA results through time, particularly feather δ15N values, could be related to a variety of processes, including baseline and/or trophic change. CSIA‐AA data add to this picture by indicating that trophic position is not a likely explanation. However, correlation of bulk δ15N with δ15NPhe values strongly suggests that changes in bulk δ15N values are driven by changes in the nitrogen baseline (Figure S3), which could result, for example, from the use of synthetic fertilisers, or because kea reside more often in native forested habitats with undisturbed soils (Stevenson et al., 2010). In this case, the change could be due to changes in feeding grounds or the food resources consumed or both, rather than change in relative trophic position.
The high bulk δ15N values for some large male kea (caught in 1887 at an unknown location (AV420), in 1913 near the Wilberforce River (AV83) and in 1932 at the Waihopai River (AV2332)) align with 19th‐ and early 20th‐century farmer reports of kea behaviour that record dominant male nuisance birds ‘leading’ a flock, and sheep attacks in some areas (Benham, 1906). However, we did not see high bulk δ15N values in individuals from other areas where this behaviour was recorded, such as in Central Otago (Reid, 2019). Similarly, although relative trophic position as represented by bulk δ15N values was more varied in more recently collected kea, this result was primarily driven by a few birds. In the CSIA‐AA analyses, two modern birds, an adult male from Arthur's Pass in 1976 and a young Mt Cook female from 1975, along with an immature male from Lake Coleridge killed in 1907, had the highest trophic position. Although field studies have recorded that juveniles in particular are curious with highly exploratory behaviour that might lead to foraging differences (Diamond & Bond, 1991), we did not observe differences between mature and immature birds in this study. Nonetheless, the atomic C:N ratios seem to indicate that smaller birds, including females, might have a less homogenous and perhaps more opportunistic diet than mature birds with larger bills.
We observed a large overlap in isotopic niche between the north and south kea (approximating the Canterbury and Otago regions; Figure 1). These regions roughly corresponded to the ‘north’ and ‘central/southern’ regions, with genetic separation between these populations, as described by Dussex et al. (2014, 2015). We excluded West Coast birds from this analysis, in particular because of the high rainfall and extensive rainforest in this region, which could lead to potential isotopic baseline differences. The niche overlap suggests a similar overall diet across these regions, although there may be differences in isotopic baselines that are currently undetected and are affecting these results. There were, nonetheless, some differences between modern birds associated with human locations, where there are frequent reports of anthropogenic feeding, and modern birds collected at locations where foraging is more likely to consist of wild foods. The low δ15N values for individuals at human influenced locations is difficult to interpret, because nitrogen in the basal food supply may be low, for example, in undisturbed podocarp forests where kea may also forage (Stevenson et al., 2010). Wet and cool environments may also reduce basal δ15N values (Rawlence et al., 2016). It is thus unclear how prevalent scavenging behaviour is among kea, but more studies could examine anthropogenic impacts on declining populations.
Overall, the niche widths seen here do not show general regional differences. However, the standard ellipse areas indicated that niche space varied for females in the Otago region, suggesting some sexually dimorphic feeding patterns, although we did not see this difference for females in the Canterbury region. High positive correlations between diet and phenotypic features can provide evidence for strong competition among individuals (Losos, 2000) or reflect morphological dimorphism that allows the sexes to avoid intraspecific competition for food (Tomotani et al., 2021). Benham (1906) suggested that sheep attacks varied regionally in the early 20th century, with adult male kea more likely to lead flock behaviour and attack sheep in Otago rather than Canterbury, but evidence is limited. Male kea also provision breeding females (Jarrett & Wilson, 1999), which might lead to more homogenous diets for these birds. Because kea appear to moult annually, but not systematically, it is impossible to disentangle seasonal factors using isotope analysis (monthly data shown in Table S5). However, we speculate that juvenile exploration of food sources, patterns of adult male dominance and possibly other behaviours might lead to potential differences in diet.
CSIA‐AA plays a particularly useful role in disentangling isotopic niche and is particularly suitable for use with museum collections where specimens may be collected from a range of time periods and locations. A limitation of BSIA is that it is impossible to distinguish between changes in baseline foods and foraging patterns without sampling of adequate materials to construct a baseline database, whereas with CSIA‐AA, these issues can be solved by examining δ15NPhe values for example. In this study, CSIA‐AA revealed that the bulk δ15N values observed for kea were in fact driven by a change in the nitrogen baseline and not by a trophic change suggesting caution in interpreting relative trophic position. These baseline issues are likely to become more acute as both anthropogenic influences on landscapes increase (Guiry et al., 2018) and weather patterns shift, creating change in weathering and erosion, rainfall and other environmental patterns that influence isotopic soil and plant nitrogen stable isotope values (Rawlence et al., 2016). Relative trophic positions calculated here using CSIA‐AA values suggest that kea diet in the more distant past was less variable across the population in comparison to modern kea where individual diets range between carnivorous and herbivorous (Figure 5). CSIA‐AA thus represents an additional level of precision that can enhance the quantity and quality of data derived from museums specimens and unlock new information on foraging ecology, behaviour and ecosystem changes.
Museum, herbaria and archaeological collections, and the ongoing collection of specimens, are critical to quantify environmental change (e.g. Robbirt et al., 2011). Conservation decision‐making has at times assumed that current realised niche has persisted over hundreds if not thousands of years, when this may not be the case now (Lyman, 1996), and, with ongoing and rapidly increasing environmental change, is unlikely to be the case in the future (e.g. Bond et al., 2019). Archaeological samples can likewise extend understanding of past environments and their changes, by thousands of years (Guiry et al., 2020; Ostrom et al., 2017). Many isotopic studies have now indicated that in addition to pinned specimens, skins and mounts, preserved museum samples such as fish can yield good quality, usable stable isotope values to reveal such changes despite having soaked in carbon or methyl‐based preservatives for many years (Chua et al., 2020; Durante et al., 2020).
Here, we have focused on kea skins to demonstrate the value of examining past trophic position and make headway into investigating changes (or otherwise) that might result from anthropogenic change. We have quantified relative trophic niche in both early century collected kea as well as modern birds, and examined foraging patterns for both males and females at different locations. These types of data provide a foundation that will help inform understanding of sexually dimorphic feeding patterns and can be used in evidence‐based conservation decision‐making, including diet optimisation or spatial planning. There is enormous possibility to learn more from museum collections, using a variety of tools including stable isotope analysis.
AUTHORS' CONTRIBUTIONS
P.M.W. conceived the idea and designed the methodology with K.M.R.; K.M.R., P.M.W. and A.J.M.S. interpreted bulk isotope analyses; A.J.M.S. conducted and interpreted all CSIA‐AA analyses; T.J. led the statistical analyses with input from P.M.W.; all authors contributed to the writing of the manuscript and approved the final publication.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Supporting information
Appendix S1 Supporting information
ACKNOWLEDGEMENTS
Funding from the Centre for Sustainability, University of Otago and Rutherford Discovery Fellowship funding 14‐LCR‐001 supported P.M.W. The Royal Society Marsden fund (19‐UOO‐212) supported A.J.M.S. Chris Garden created the distribution map and took the photo in the graphical abstract. Gretchen Brownstein helped with preliminary figures and data curation. The authors thank all the curators and museum staff who assisted with this project, especially Emma Burns, Kane Fleury, Paul Scofield, Brian Gill and Matt Rayner, Colin Miskelly and Anita Dornauf. Open access publishing facilitated by University of Otago, as part of the Wiley ‐ University of Otago agreement via the Council of Australian University Librarians.[Correction added on 30‐May‐22, after first online publication CAUL funding statement added].
Wehi, P. M. , Rogers, K. M. , Jowett, T. , & Sabadel, A. J. (2023). Interpreting past trophic ecology of a threatened alpine parrot, kea Nestor notabilis, from museum specimens. Journal of Animal Ecology, 92, 273–284. 10.1111/1365-2656.13742
Handling Editor: Beatriz Willink
DATA AVAILABILITY STATEMENT
Data available via the Dryad Digital Repository https://doi.org/10.5061/dryad.j3tx95xh7 (Wehi et al., 2022a). Supplementary information is available at https://doi.org/10.5281/zenodo.6508721 (Wehi et al., 2022b).
REFERENCES
- Allentoft, M. , Heller, R. , Oskam, C. L. , Lorenzen, E. D. , Hale, M. L. , Gilbert, M. T. , C. Jacomb, R. N. Holdaway , & Bunce M. (2014). Extinct New Zealand megafauna were not in decline before human colonization. Proceedings of the National Academy of Sciences of the United States of America, 11, 4922–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D. , Mächler, M. , Bolker, B. , & Walker, S. (2015). Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67(1), 1–48. [Google Scholar]
- Benham, W. B. (1906). Carnivorous habits of the New Zealand kea parrot. Nature, 1902(73), 559. [Google Scholar]
- Blackburn, T. M. , Cassey, P. , Duncan, R. P. , Evans, K. L. , & Gaston, K. J. (2004). Avian extinction and mammalian introductions on oceanic islands. Science, 305(5692), 1955–1958. [DOI] [PubMed] [Google Scholar]
- Bolnick, D. , Svanback, R. , Araujo, M. S. , & Persson, L. (2007). Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proceedings of the National Academy of Sciences of the United States of America, 104, 10075–10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, A. B. , & Diamond, J. (1992). Population estimates of kea in Arthur's Pass National Park. Notornis, 39(3), 151–160. [Google Scholar]
- Bond, A. B. , Wilson, K. J. , & Diamond, J. (1991). Sexual dimorphism in the kea Nestor notabilis . Emu, 91(1), 12–19. [Google Scholar]
- Bond, M. O. , Anderson, B. J. , Henare, T. H. A. , & Wehi, P. M. (2019). Effects of climatically shifting species distributions on biocultural relationships. People and Nature, 1(1), 87–102. [Google Scholar]
- Buller, W. L. (1882). Manual of the birds of New Zealand (Vol. 16). G. Didsbury, government printer. [Google Scholar]
- Chiaradia, A. , Forero, M. G. , Hobson, K. A. , & Cullen, J. M. (2010). Changes in diet and trophic position of a top predator 10 years after a mass mortality of a key prey. ICES Journal of Marine Science, 67(8), 1710–1720. [Google Scholar]
- Chikaraishi, Y. , Ogawa, N. O. , & Ohkouchi, N. (2010). Further evaluation of the trophic level estimation based on nitrogen isotopic composition of amino acids. In Ohkouchi N., Tayasu I., & Koba K. (Eds.), Earth, life, and isotopes (pp. 37–51). Kyoto University Press. [Google Scholar]
- Chua, K. W. J. , Liew, J. H. , Shin, K.‐H. , & Yeo, D. C. J. (2020). Effects of ethanol preservation and formalin fixation on amino acid stable isotope analysis (δ13C and δ15N) and its ecological applications. Limnology and Oceanography: Methods, 18(2), 77–88. [Google Scholar]
- Clark, C. M. H. (1970). Observations on populations, movements and food of the kea Nestor notabilis . Notornis, 17, 105–114. [Google Scholar]
- Diamond, J. , & Bond, A. B. (1991). Social behavior and the ontogeny of foraging in the kea (Nestor notabilis). Ethology, 88(2), 128–144. [Google Scholar]
- Duncan, R. P. , & Blackburn, T. M. (2004). Extinction and endemism in the New Zealand avifauna. Global Ecology and Biogeography, 13, 509–517. [Google Scholar]
- Durante, L. M. , Sabadel, A. J. M. , Frew, R. D. , Ingram, T. , & Wing, S. R. (2020). Effects of fixatives on stable isotopes of fish muscle tissue: Implications for trophic studies on preserved specimens. Ecological Applications, 2020, e02080. [DOI] [PubMed] [Google Scholar]
- Dussex, N. , Rawlence, N. J. , & Robertson, B. C. (2015). Ancient and contemporary DNA reveal a pre‐human decline but no population bottleneck associated with recent human persecution in the kea (Nestor notabilis). PLoS ONE, 10(2), e0118522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex, N. , Wegmann, D. , & Robertson, B. C. (2014). Postglacial expansion and not human influence best explains the population structure in the endangered kea (Nestor notabilis). Molecular Ecology, 23(9), 2193–2209. [DOI] [PubMed] [Google Scholar]
- Elliott, G. , & Kemp, J. (2004). Effect of hunting and predation on kea, and a method of monitoring kea populations. Results of kea research on the St. Arnaud range. Department of Conservation Science Internal Series, 181, 1–7. [Google Scholar]
- English, P. A. , Green, D. J. , & Nocera, J. J. (2018). Stable isotopes from museum specimens may provide evidence of long‐term change in the trophic ecology of a migratory aerial insectivore. Frontiers in Ecology and Evolution, 6, 14. [Google Scholar]
- Flockhart, D. T. , Brower, L. P. , Ramirez, M. I. , Hobson, K. A. , Wassenaar, L. I. , Altizer, S. , & Norris, D. R. (2017). Regional climate on the breeding grounds predicts variation in the natal origin of monarch butterflies overwintering in Mexico over 38 years. Global Change Biology, 23(7), 2565–2576. [DOI] [PubMed] [Google Scholar]
- Greer, A. L. , Horton, T. W. , & Nelson, X. J. (2015). Simple ways to calculate stable isotope discrimination factors and convert between tissue types. Methods in Ecology and Evolution, 6(11), 1341–1348. [Google Scholar]
- Guiry, E. J. , Beglane, F. , Szpak, P. , Schulting, R. , McCormick, F. , & Richards, M. P. (2018). Anthropogenic changes to the Holocene nitrogen cycle in Ireland. Science Advances, 4(6), eaas9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiry, E. J. , Orchard, T. J. , Royle, T. C. A. , Cheung, C. , & Yang, D. Y. (2020). Dietary plasticity and the extinction of the passenger pigeon (Ectopistes migratorius). Quaternary Science Reviews, 233, 106225. [Google Scholar]
- Hallworth, M. T. , Marra, P. P. , McFarland, K. P. , Zahendra, S. , & Studds, C. E. (2018). Tracking dragons: Stable isotopes reveal the annual cycle of a long‐distance migratory insect. Biology Letters, 14(12), 20180741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington, E. D. , Olson, R. J. , Drazen, J. C. , Lennert‐Cody, C. E. , Balance, L. T. , Kaufmann, R. S. , & Popp, B. N. (2017). Spatial food‐web structure in the eastern tropical Pacific Ocean based on compound‐specific nitrogen isotope analysis of amino acids. Limnology and Oceanography, 62(2), 541–560. [Google Scholar]
- Hobson, K. A. , & Clark, R. G. (1992). Assessing avian diets using stable isotopes I: Turnover of 13C in tissues. Condor, 94, 181–188. [Google Scholar]
- Hobson, K. A. , Piatt, J. F. , & Pitocchelli, J. (1994). Using stable isotopes to determine seabird trophic relationships. Journal of Animal Ecology, 63, 786–798. [Google Scholar]
- Holmes, M. W. , Hammond, T. T. , Wogan, G. O. U. , Walsh, R. E. , LaBarbera, K. , Wommack, E. A. , Martins, F. M. , Crawford, J. C. , Mack, K. L. , Bloch, L. M. , & Nachman, M. W. (2016). Natural history collections as windows on evolutionary processes. Molecular Ecology, 25, 864–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A. L. , Parnell, A. C. , Inger, R. , & Bearhop, S. (2011). Comparing isotopic niche widths among and within communities: SIBER—Stable Isotope Bayesian Ellipses in R. Journal of Animal Ecology, 80(3), 595–602. [DOI] [PubMed] [Google Scholar]
- Jarrett, M. I. , & Wilson, K. J. (1999). Seasonal and diurnal attendance of Kea (Nestor notabilis) at Halpin Creek rubbish dump, Arthur's Pass, New Zealand. Notornis, 46(2), 273–286. [Google Scholar]
- Katzenberg, M. A. (1989). Stable isotope analysis of archaeological faunal remains from southern Ontario. Journal of Archaeological Science, 16(3), 319–329. [Google Scholar]
- Kirk, E. J. , Powlesland, R. G. , & Cork, S. C. (1993). Anatomy of the mandibles, tongue and alimentary tract of kakapo, with some comparative information from kea and kaka. Notornis, 40(1), 55–63. [Google Scholar]
- Kronfeld‐Schor, N. , & Dayan, T. (1999). The dietary basis for temporal partitioning: Food habits of coexisting Acomys species. Oecologia, 121, 123–128. [DOI] [PubMed] [Google Scholar]
- Kuhn, M. (2008). Caret package. Journal of Statistical Software, 28(5), 17. [Google Scholar]
- Kuznetsova, A. , Brockhoff, P. B. , & Christensen, R. H. B. (2017). lmerTest PACKAGE: Tests in linear mixed effects models. Journal of Statistical Software, 82(13), 1–26. [Google Scholar]
- Losos, J. (2000). Ecological character displacement and the study of adaptation. Proceedings of the National Academy of Sciences of the United States of America, 97, 5693–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, R. L. (1996). Applied zooarchaeology: The relevance of faunal analysis to wildlife management. World Archaeology, 28(1), 110–125. [Google Scholar]
- McGlone, M. S. (1983). Polynesian deforestation of New Zealand: A preliminary synthesis. Archaeology in Oceania, 18(1), 11–25. [Google Scholar]
- McMahon, K. W. , Thorrold, S. R. , Houghton, L. A. , & Berumen, M. L. (2016). Tracing carbon flow through coral reef food webs using a compound‐specific stable isotope approach. Oecologia, 180(3), 809–821. [DOI] [PubMed] [Google Scholar]
- Newsome, S. D. , Ralls, K. , Van Horn Job, C. , Fogel, M. L. , & Cypher, B. L. (2010). Stable isotopes evaluate exploitation of anthropogenic foods by the endangered San Joaquin kit fox (Vulpes macrotis mutica). Journal of Mammalogy, 91(6), 1313–1321. [Google Scholar]
- Nieuwenhuis, R. , te Grotenhuis, M. , & Pelzer, B. (2012). influence.ME: Tools for detecting influential data in mixed effects models. The R Journal, 4(2), 38–47. [Google Scholar]
- O'Connell, T. (2017). ‘Trophic’ and ‘source’ amino acids in trophic estimation: A likely metabolic explanation. Oecologia, 184(2), 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkouchi, N. , Chikaraishi, Y. , Close, H. G. , Fry, B. , Larsen, T. , Madigan, D. J. , McCarthy, M. D. , McMahon, K. W. , Nagata, T. , Naitoa, Y. I. , Ogawa, N. O. , Popp, B. N. , Steffan, S. , Takano, Y. , Tayasu, I. , Wyatt, A. S. J. , Yamaguchi, Y. T. , & Yokoyama, Y. (2017). Advances in the application of amino acid nitrogen isotopic analysis in ecological and biogeochemical studies. Organic Geochemistry, 113, 150–174. [Google Scholar]
- Ostrom, P. H. , Wiley, A. E. , James, H. F. , Rossman, S. , Walker, W. A. , Zipkin, E. F. , & Chikaraishi, Y. (2017). Broadscale trophic shift in the pelagic North Pacific revealed by an oceanic seabird. The Royal Society, 284(1851), 20162436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp, B. N. , Graham, B. S. , Olson, R. J. , Hannides, C. C. , Lott, M. J. , López‐Ibarra, G. A. , Galván‐Magaña, F. , & Fry, B. (2007). Insight into the trophic ecology of yellowfin tuna, Thunnus albacares, from compound‐specific nitrogen isotope analysis of proteinaceous amino acids. Terrestrial Ecology, 1, 173–190. [Google Scholar]
- Quillfeldt, P. , & Masello, J. F. (2020). Compound‐specific stable isotope analyses in Falkland Islands seabirds reveal seasonal changes in trophic positions. BMC Ecology, 20(1), 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Retrieved from https://www.R‐project.org/ [Google Scholar]
- Rawlence, N. J. , Kardamaki, A. , Easton, L. J. , Tennyson, A. J. , Scofield, R. P. , & Waters, J. M. (2017). Ancient DNA and morphometric analysis reveal extinction and replacement of New Zealand's unique black swans. Proceedings of the Royal Society B: Biological Sciences, 284(1859), 20170876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlence, N. J. , Wood, J. R. , Bocherens, H. , & Rogers, K. M. (2016). Dietary interpretations for extinct megafauna using coprolites, intestinal contents and stable isotopes: Complimentary or contradictory? Quaternary Science Reviews, 142, 173–178. [Google Scholar]
- Reid, C. E. (2019). Understanding attacks by kea (Nestor notabilis), an endemic parrot, on sheep (Ovis aries) in the South Island high country (PhD thesis). Massey University. [Google Scholar]
- Robbirt, K. M. , Davy, A. J. , Hutchings, M. J. , & Roberts, D. L. (2011). Validation of biological collections as a source of phenological data for use in climate change studies: A case study with the orchid Ophrys sphegodes . Journal of Ecology, 99, 235–241. [Google Scholar]
- Roberts, D. L. , Taylor, L. , & Joppa, L. N. (2016). Threatened or data deficient: Assessing the conservation status of poorly known species. Diversity and Distributions, 22, 558–565. [Google Scholar]
- Sabadel, A. J. M. , Durante, L. M. , & Wing, S. R. (2020). Stable isotopes of amino acids from reef fishes uncover Suess and nitrogen enrichment effects on local ecosystems. Marine Ecology Progress Series, 647, 149–160. [Google Scholar]
- Sabadel, A. J. M. , Van Oostende, N. , Ward, B. B. , Woodward, E. M. , Van Hale, R. , & Frew, R. D. (2019). Characterization of particulate organic matter cycling during a summer North Atlantic phytoplankton bloom using amino acid C and N stable isotopes. Marine Chemistry, 214(2019), e103670. [Google Scholar]
- Sabadel, A. J. M. , Woodward, E. M. S. , Van Hale, R. , & Frew, R. D. (2016). Compound‐specific isotope analysis of amino acids: A tool to unravel complex symbiotic trophic relationships. Food Webs, 6, 9–18. [Google Scholar]
- Sandom, C. , Faurby, S. R. , Sandel, B. , & Svenning, J. C. (2014). Global late Quaternary megafauna extinctions linked to humans, not climate change. Proceedings of the Royal Society B: Biological Sciences, 281, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K. J. , Trueman, C. N. , France, C. A. M. , Sparks, J. P. , Brownlow, A. C. , Dähne, M. , Davison, N. J. , Guðmundsson, G. , Khidas, K. , Kitchener, A. C. , Langeveld, B. W. , Lesage, V. , Meijer, H. J. M. , Ososky, J. J. , Sabin, R. C. , Timmons, Z. L. , Víkingsson, G. A. , Wenzel, F. W. , & Peterson, M. J. (2021). Stable isotope analysis of specimens of opportunity reveals ocean‐scale site fidelity in an elusive whale species. Frontiers in Conservation Science, 2(2021), 13. [Google Scholar]
- Stevenson, B. A. , Parfitt, R. L. , Schipper, L. A. , Baisden, W. T. , & Mudge, P. (2010). Relationship between soil δ15N, C/N and N losses across land uses in New Zealand. Agriculture, Ecosystem and Environment, 139, 736–741. [Google Scholar]
- Styring, A. K. , Kuhl, A. , Knowles, T. D. , Fraser, R. A. , Bogaard, A. , & Evershed, R. P. (2012). Practical considerations in the determination of compound‐specific amino acid δ15N values in animal and plant tissues by gas chromatography‐combustion‐isotope ratio mass spectrometry, following derivatisation to their N‐acetylisopropyl esters. Rapid Communications in Mass Spectrometry, 26(19), 2328–2334. [DOI] [PubMed] [Google Scholar]
- Szpak, P. (2014). Complexities of nitrogen isotope biogeochemistry in plant‐soil systems: Implications for the study of ancient agricultural and animal management practices. Frontiers in Plant Science, 5, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomotani, B. M. , Salvador, R. B. , Sabadel, A. J. M. , Miskelly, C. M. , Brown, J. , Delgado, J. , Boussès, P. , Cherel, Y. , Waugh, S. M. , & Bury, S. J. (2021). Extreme bill dimorphism leads to different but overlapping isotopic niches and similar trophic positions in sexes of the charismatic extinct huia. Oecologia, 198, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar, L. I. , & Hobson, K. A. (1998). Natal origins of migratory monarch butterflies at wintering colonies in Mexico: New isotopic evidence. Proceedings of the National Academy of Sciences of the United States of America, 95(26), 15436–15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehi, P. M. , Rogers, K. , Jowett, T. , & Sabadel, A. J. M. (2022a). Interpreting past trophic ecology of a threatened species, kea (Nestor notabilis), from museum specimens. Dryad Digital Repository, 10.5061/dryad.j3tx95xh7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehi, P. , Rogers, K. , Jowett, T. , & Sabadel, A. (2022b). Interpreting past trophic ecology of a threatened species, kea (Nestor notabilis), from museum specimens. Zenodo, 10.5281/zenodo.6508721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehi, P. M. , Whaanga, H. , & Trewick, S. A. (2012). Artefacts, biology and bias in museum collection research. Molecular Ecology, 21, 3103–3109. [DOI] [PubMed] [Google Scholar]
- Young, L. M. , Kelly, D. , & Nelson, X. J. (2012). Alpine flora may depend on declining frugivorous parrot for seed dispersal. Biological Conservation, 147(1), 133–142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting information
Data Availability Statement
Data available via the Dryad Digital Repository https://doi.org/10.5061/dryad.j3tx95xh7 (Wehi et al., 2022a). Supplementary information is available at https://doi.org/10.5281/zenodo.6508721 (Wehi et al., 2022b).


