Abstract
A 16‐year‐old male was admitted to the hospital for weakness of both lower extremities. Magnetic resonance imaging revealed an intraspinal extramedullary subdural mass at the thoracic 9 level. Microscopically, the tumor cells were small to medium sized and round to ovoid in shape. They were distributed in diffuse sheets or showed nodular appearance. The nucleus of the tumor had mild‐to‐moderate atypia, with vesicular chromatin and prominent nucleoli. A smaller proportion of tumor cells demonstrated rhabdoid morphology. Focal myxoid stromal change was present, in which tumor cells exhibited spindle shapes. Approximately two mitoses were counted per 10 high‐power fields. No necrosis was observed. The tumor cells were focal positive for CD99; multifocal positive for WT1; diffuse positive for nestin, synaptophysin, and D2‐40; partial positive for GFAP; focal positive for desmin and SSTR2; and scattered positive for S‐100 protein. The Ki‐67 labeling index was approximately 20%. Genetic testing revealed CIC‐LEUTX gene fusion. Considering the patient's history, clinical data, pathological findings and genetic findings, we rendered a rare tumor named CIC‐rearranged sarcoma with CIC‐LEUTX gene fusion.
Keywords: CIC‐LEUTX fusion, immunohistochemistry, RNA‐based next generation sequencing, sarcoma, spinal tumor
INTRODUCTION
CIC‐rearranged sarcomas are classified as undifferentiated small round cell sarcomas and uncertain differentiation tumors in mesenchymal tumors of non‐meningeal epithelial origin in the 5th edition of the WHO Classification of Tumors of the Soft Tissue and Bone and Tumors of the Central Nervous System, respectively. 1 , 2 CIC‐rearranged sarcomas are most common with CIC‐DUX4 fusions 3 and other rare partner genes, including FOXO4, LEUTX, NUTM1, and NUTM2A. 4 , 5 , 6 , 7 Most tumors arise in the deep soft tissue of the trunk and extremities, followed by visceral location. Only two cases of CIC‐rearranged sarcoma located in the spinal cord have been reported so far, both of which were CIC‐DUX4 fusion. 8 , 9 However, the CIC‐rearranged sarcomas with CIC‐LEUTX fusion in the spinal cord have not been reported, and this is the first case.
CLINICAL SUMMARY
The patient, a 16‐year‐old male, was admitted to the hospital due to acute onset of weakness of both lower extremities for 20 days and aggravation for five days. The patient experienced bilateral lateral thigh soreness and discomfort without obvious predisposing factors 20 days previously, but it resolved spontaneously, followed by weakness of both lower extremities, which was not taken seriously. Then he gradually developed dragging while walking. Five days before presenting at the hospital, the patient was unable to walk or stand, which continued without relief. There was no obvious abnormality in cranial computed tomography (CT), cranial magnetic resonance imaging (MRI) and magnetic resonance angiography (MRA) and lumbar MRI, and no special treatment was given in other hospitals. For further diagnosis and treatment, the patient came to the outpatient department of the hospital and planned to be admitted to the neurology department with myelitis. On admission, MRI of the thoracic spine revealed mass isointensity on T1 and T2‐weighted images at the level of thoracic 9. The spinal cord was compressed, and the subdural space above and below the lesion was widened (Fig. 1A,B). The mass displayed obvious enhancement onT1‐weighted contrast‐enhanced images and was approximately 1.2 cm in diameter (Fig. 1C). He was then transferred to the department of neurosurgery for further surgical treatment. The thoracic 6–9 segments were opened by surgery, and the tumor tissue in the thoracic 9 thoracic canal was found. The tumor was located under the dura mater and related to dura mater closely. The tumor was rubbery and rich in blood supply. The tumor was closely adhered to the spinal cord on the left ventral side, and the boundary was unclear. The boundary was carefully separated to protect the spinal cord, and the tumor was then completely removed. The size of the tumor was approximately 1.2 × 1.2 × 1.5 cm. After the operation, the limb weakness and sensory disturbance were significantly improved, and the patient could walk independently. Two months after the operation, spinal canal radiotherapy and ifosfamide and liposome doxorubicin chemotherapy were performed. Postoperative follow up was uneventful, and the thoracic spine MRI revealed no tumor recurrence at five months.
Fig 1.
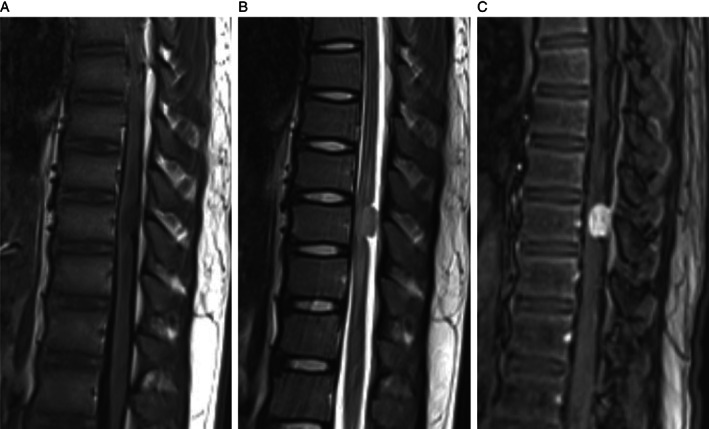
(A) T1‐weighted sequences reveal an isointense mass. (B) T2‐weighted sequences reveal an isointense mass. (C) The mass displays obvious enhancement in T1‐weighted contrast‐enhanced images.
PATHOLOGIC FINDINGS
On macroscopic examination, the lesion showed a pile of gray‐white tissue, measuring 1.5 × 1 × 0.4 cm in size. On hematoxylin and eosin (HE) staining, the case comprised solid proliferation with sheets of tumor cells and exhibited a nodular appearance with thick collagenous septa separating the tumor into compartments locally (Fig. 2A). The tumor cells were small to medium sized and round to ovoid in shape with clear or mildly eosinophilic scant cytoplasm. They were relatively uniform in appearance, but the nuclei showed pleomorphism focally. The nuclei showed coarse chromatin and prominent nucleoli (Fig. 2B). A smaller proportion of tumor cells demonstrated rhabdoid cells with eccentrically located nuclei and extensive eosinophilic cytoplasm (Fig. 2c). The tumor showed focal prominent myxoid stromal change with spindle‐shaped cells (Fig. 2D). The mitoses were approximately 2/10 high‐power fields, and no hemorrhage or necrosis was observed. In the tumor tissue adjacent to the dura, pigment granules could be observed. Antibodies used for immunohistochemical staining are listed with their hosts, clones, dilutions, and sources in Table 1. The tumor cells were focal positive for CD99 (Fig. 3A); multifocal positive for WT1 with unequal strength (Fig. 3B); diffuse positive for nestin, synaptophysin (Fig. 3C); and D2‐40 (Fig. 3D); partial positive for GFAP (Fig. 3E); focal positive for desmin and SSTR2; and scattered positive for S‐100 protein. The Ki‐67 labeling index was approximately 20% (Fig. 3F). Nuclear INI‐1 staining was retained (Fig. 3G), whereas STAT6, PR, EMA, CK, Olig2, PLAP, CD45, HMB45, Melan‐A, CD34, CD31, and ERG were negative.
Fig 2.
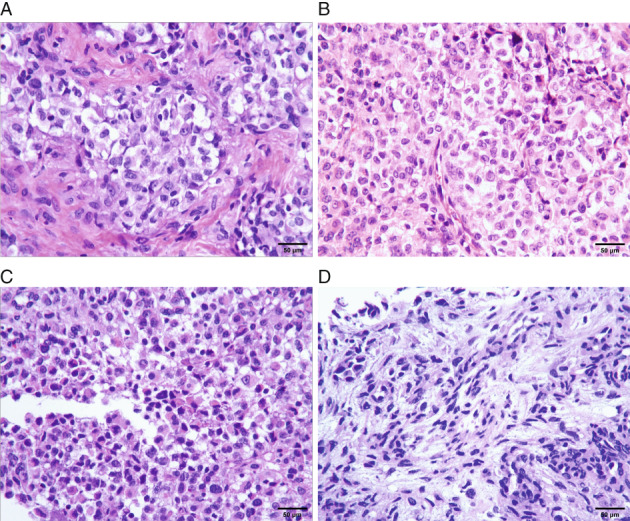
HE staining. (A, B) The tumor shows a nodular appearance or diffuse sheets. The tumor cells are relatively uniform in appearance, but the nuclei show pleomorphism focally. The nuclei shows coarse chromatin and prominent nucleoli. (C) A smaller proportion of tumor cells demonstrates rhabdoid cells with eccentrically located nuclei and extensive eosinophilic cytoplasm. (D) The tumor shows focal prominent myxoid stromal change with spindle‐shaped cells.
Table 1.
List of antibodies used for immunohistochemical staining
| Antibody | Host | Clone | Source | Dilution |
|---|---|---|---|---|
| CD99 | Mouse monoclonal | O13 | ZSGB, Beijing, China | Prediluted |
| WT1 | Mouse monoclonal | MX012 | Maxim, Fujian, Fuzhou, China | Prediluted |
| nestin | Rabbit monoclonal | EP287 | ZSGB, Beijing, China | Prediluted |
| synaptophysin | Rabbit monoclonal | EP158 | ZSGB, Beijing, China | 1:300 |
| D2‐40 | Mouse monoclonal | D2‐40 | ZSGB, Beijing, China | Prediluted |
| GFAP | Rabbit monoclonal | EP13 | ZSGB, Beijing, China | Prediluted |
| SSTR2 | Rabbit monoclonal | EP149 | ZSGB, Beijing, China | Prediluted |
| S‐100 protein | Mouse monoclonal | 4C4.9 | Celnovte, Henan, Zhengzhou, China | 1:300 |
| Ki‐67 | Mouse monoclonal | MIB‐1 | ZSGB, Beijing, China | 1:300 |
| INI‐1 | Mouse monoclonal | 25 | ZSGB, Beijing, China | Prediluted |
| STAT6 | Rabbit monoclonal | EP325 | ZSGB, Beijing, China | Prediluted |
| desmin | Rabbit monoclonal | EP15 | ZSGB, Beijing, China | Prediluted |
| PR | Mouse monoclonal | C4D10 | Celnovte, Henan, Zhengzhou, China | 1:400 |
| EMA | Mouse monoclonal | GP1.4 | ZSGB, Beijing, China | 1:100 |
| CK | Mouse monoclonal | AE1/AE3 | Celnovte, Henan, Zhengzhou, China | 1:400 |
| Olig2 | Rabbit monoclonal | EP112 | Celnovte, Henan, Zhengzhou, China | 1:800 |
| PLAP | Rabbit monoclonal | SP15 | ZSGB, Beijing, China | Prediluted |
| CD45 | Mouse monoclonal | 2B11&PD7/26 | Celnovte, Henan, Zhengzhou, China | 1:400 |
| HMB45 | Mouse monoclonal | HMB45 | Chang Dao, Shanghai, China | Prediluted |
| Melan‐A | Mouse monoclonal | A103 | Chang Dao, Shanghai, China | Prediluted |
| CD34 | Mouse monoclonal | QBEnd/10 | Celnovte, Henan, Zhengzhou, China | 1:300 |
| CD31 | Mouse monoclonal | MX032 | Maxim, Fujian, Fuzhou, China | Prediluted |
| ERG | Rabbit monoclonal | MXR004 | Maxim, Fujian, Fuzhou, China | Prediluted |
Fig 3.
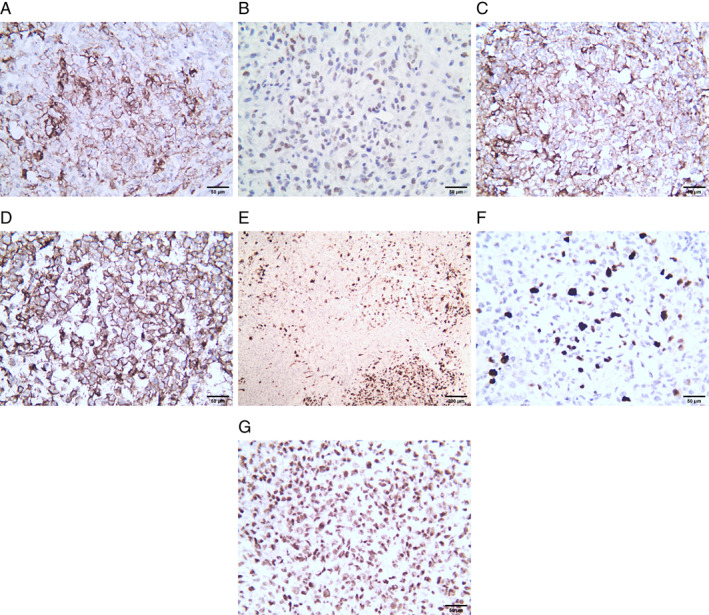
Immunohistochemical staining. (A) CD99 immunostaining is focal positive. (B) WT1 immunostaining is multifocal positive. (C) Synaptophysin immunostaining is diffuse positive. (D) D2‐40 immunostaining is diffuse positive. (E) GFAP immunostaining is partial positive. (F) The Ki‐67 labeling index is approximately 20%. (G) Nuclear INI‐1 staining is retained.
GENETIC FINDINGS
Fusion testing was performed using an RNA‐based next generation sequencing (NGS) assay at Genetron Health (Beijing). Gene test results showed that a fusion of CIC‐LEUTX genes located in exon 20 of the CIC gene and exon 3 of the LEUTX gene. The breakpoints were at chr19: 42799216 and chr19: 40276580 for CIC and LEUTX, respectively (Fig. 4). Using the same detection method, no mutation was found in IDH1/2, BRAF, TERT, H3F3A, HIST1H3B, and HIST1H3C. Chromosome 7 was not amplified, and chromosome 10 was not deleted. Chromosome 1p/19q was intact.
Fig 4.
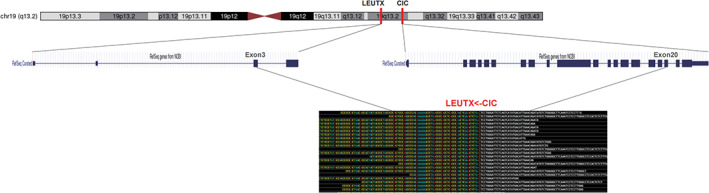
A fusion of CIC‐LEUTX genes located in exon 20 of the CIC gene and exon 3 of the LEUTX gene. The breakpoints are at chr19: 42799216 and chr19: 40276580 for CIC and LEUTX, respectively.
DISCUSSION
The first case of t(4;19)(q35;q13.1)‐associated sarcoma was reported by Richkind et al. 10 in 1996 in a 12‐year‐old boy who presented with an ankle soft tissue mass and synchronous lung metastases and died within 10 months. In 2006, Kawamura‐Saito et al. 11 also discovered two cases of CIC‐DUX4 fusion round cell sarcoma diagnosed as Ewing‐like sarcoma with chromosomal translocation t(4;19)(q35;q13). However, Choi et al. 12 did not recommend the appellation “Ewing‐like sarcoma” for CIC‐DUX4 sarcoma despite its superficial resemblance. First, the location of the tumor is different. Ewing sarcoma (ES) most often presents as a primary bone tumor, whereas CIC‐DUX4 sarcoma is most frequently reported in soft tissue tumors. Second, in terms of treatment and prognosis, unlike ES, which is sensitive to chemotherapy, CIC‐DUX4 sarcoma usually appears to quickly develop chemoresistance and has a more aggressive clinical course. In addition, the two types of tumors differ in histology, immunohistochemistry, and molecular characteristics. Specht et al. 13 also believed that the distinct gene signature and immunoprofile of CIC‐DUX4 sarcoma suggested a distinct pathogenesis from ES.
Most CIC‐rearranged sarcomas occur in the deep soft tissues of the limbs or trunk, followed by the head and neck, retroperitoneum, or pelvis; uncommonly in the viscera including kidney, gastrointestinal tract, or brain; and only rarely with primary osseous involvement. A case of primary cardiac CIC‐rearranged undifferentiated sarcoma in an infant was reported recently. 14 There is a wide age range but a predilection for young adults, and there is a slight male predominance. 3 CIC‐rearranged sarcomas are typically composed of diffuse sheets of undifferentiated round cells, displaying at least in part a lobulated growth pattern, divided by fibrous septa. A minor component of spindle or epithelioid/ rhabdoid cells may be seen. The tumor cells have relatively uniform cytomorphology but often reveal mild nuclear pleomorphism, with vesicular chromatin and prominent nucleoli. The cytoplasm is lightly eosinophilic or clear. Stromal myxoid change is a common finding, in which tumor cells exhibit reticular or pseudoacinar arrangements. Necrosis is common, and mitotic activity is brisk. Changes in histomorphology after therapy have been reported, included focal pleomorphism cells, eosinophilic cytoplasmic globules, nuclear pseudoinclusions and hyaline cartilage formations. 8 , 15 Immunohistochemically, these tumors usually show weak CD99 positivity and absent NKX2.2 expression. Hung et al. 16 evaluated the expression of ETV4 and WT1 in CIC‐rearranged sarcomas and found that the sensitivity and specificity of ETV4 expression were 90% and 95%, respectively. While the sensitivity and specificity of WT1 were 95% and 81%, respectively. It is concluded that diffuse ETV4, along with at least focal WT1 expression, is helpful to distinguish CIC‐rearranged sarcoma from ES and other histologic mimics. In addition, a small number of cases can express myogenic markers and cytokeratin, calretinin, neurofilament protein, S‐100 protein, ERG, FLI1, MUC‐4, and D2‐40, but there is no specificity. 3 , 4 , 9 , 13 , 15 The histomorphological and immunohistochemical findings in this case were similar to those described in the literature. However, pigment granules can be seen in the tumor tissue adjacent to the meninges, which need to be differentiated from melanoma, and the relevant melanoma markers were all negative, which excluded this diagnosis. In this case, GFAP was partially expressed in tumor cells, and there is no report that CIC‐rearranged sarcoma expresses GFAP. The tumor location was extramedullary, and glioma‐related genes were all negative. Therefore, gliomas were excluded, and it may be caused by a heterogeneous expression of tumor cells.
The CIC gene on 19 q13 encodes a transcriptional repressor with a high‐mobility group box. CIC‐DUX4 fusion is the most common form in 95% of CIC‐rearranged sarcomas, with DUX4 either on 4q35 or 10q26. 1 The DUX4 fusion to CIC upregulates PEA3 family genes, such as ETV1, ETV4, and ETV5 genes, which might play an important role in tumorigenesis. 11 There are several other rare non‐DUX4 partner genes, such as FOXO4, LEUTX, NUTM1, and NUTM2A. It is worth noting that in a report on angiosarcomas, three cases were found to have CIC rearrangement, and one of them had the CIC‐LEUTX fusion gene. 5 Still, these three cases lacked angiogenesis, and their histology was similar to CIC‐rearranged sarcoma. Only vascular‐related immunohistochemical markers CD31 and ERG were positive, and these cases were thus classified as angiosarcoma. It is more appropriate to classify them as CIC‐rearranged sarcomas based on histomorphological and molecular detection. Two other reports on CIC‐LEUTX fusion gene occurred in the central nervous system, but neither was a CIC‐rearranged sarcoma. One article included two cases of childhood gliomas: one case of anaplastic ganglioglioma and one case of anaplastic astrocytoma with epithelioid GBM features. 17 The other article was a case of an embryonal tumor of the central nervous system in a child. This case not only had the CIC‐LEUTX fusion gene but also had NBN germline mutations and TSC2 lineage mutations. 18 So far, there are only three reports on CIC‐LEUTX fusion tumors, with a total of four cases, and this case is the fifth case (Table 2). The LEUTX gene is located on 19q13. LEUTX plays an important role in embryonal genome activation, and its expression is mostly suppressed postnatally. The fusion with KAT6A was discovered earlier, and it was reported in acute myeloid leukemia. 19 Recent studies suggested that CIC‐LEUTX fusion mechanisms might be similar to those of CIC‐DUX4 fusion because both LEUTX and DUX4 belong to the same class of paired (PRD) homeobox genes. 5 Barresi et al. 20 recently reported a case of malignant epithelioid peripheral nerve sheath tumor in a child with BRD4‐LEUTX fusion. Increased levels of LEUTX transcripts in the tumor could be detected in this case, suggesting that the BRD4‐LEUTX fusion leads to LEUTX reactivation.
Table 2.
Reported cases of tumors with CIC‐LEUTX fusion
| Reference | Age/sex | Location | Immunohistoche‐mistry | Pathological diagnosis | Genetic alteration | Therapy | Follow up (Month) | Status |
|---|---|---|---|---|---|---|---|---|
| Huang et al., 2016 | 26/F | Thigh |
Positive:CD31, ERG |
Angiosarcomas | CIC‐LEUTX | NA |
10 |
Recured |
| Negative:CD99,S100,desmin | 33 | Died | ||||||
| Lake et al., 2020 | 19/F | Frontoparietal | Positive: synaptophysin, NSE | anaplastic ganglioglioma | CIC‐LEUTX |
GTR |
10 |
Progression Alive, progressive disease |
| RT + TMZ → STR + RT (CSI) + Carboplatin→cytoxan/doxo/ifos/etoposide | 56 | |||||||
| Lake et al., 2020 | 12/F | intraventricular | Positive: GFAP, synaptophysin, CD34 | anaplastic astrocytoma with epithelioid GBM features | CIC‐LEUTX | GTR + RT/TMZ | 3 | Alive, on therapy |
| Hu et al., 2020 | 2/M | left temporal lobe‐basal ganglia and left parietal lobe |
Positive: synaptophysin |
CNS embryonal tumor | CIC‐LEUTX, TCS2 and NBN |
GR |
1 |
Progression Alive |
| Negative: GFAP | (CTX + CBP + VCR/DDP + VP‐16), RT, and TSC2 targeted drug (everolimus) | 11 | ||||||
| This case | 16/M | Intraspinal extramedullary subdural | Positive:CD99, WT1, nestin, synaptophysin, D2‐40 | CIC‐rearranged sarcoma | CIC‐LEUTX | GRT + RT+ ifos/LD | 5 | Alive, on therapy |
CBP, carboplatin; CSI, craniospinal irradiation; CNS, central nervous system; CTX, cyclophosphamide; DDP, cisplatin; doxo, doxorubicin; GBM, glioblastoma; GR, gross resection; GTR, gross total resection; ifos, ifosfamide; LD, liposome doxorubicin; NA, not applicable; NSE, neuron‐specific enolase; RT, radiotherapy; STR, subtotal resection; TMZ, temozolomide; VP‐16, etoposide; VCR, vincristine.
Only six original articles and a total of nine case reports can be found concerning the CIC‐rearranged sarcoma located in the central nervous system. 6 , 8 , 9 , 14 , 21 , 22 As to the CIC‐rearranged sarcoma located in the spinal cord, only two case reports can be found. 8 , 9 Genetic testing revealed that both cases were CIC‐DUX4 fusions. The first case was a 15‐year‐old female with a T5–T6 epidural tumor, and the second case was a 23‐year‐old male with a C3–C5 intramedullary tumor. Our case was a 16‐year‐old male, and the tumor was located in the extramedullary subdural space at the T9 level. Because of the location of the tumors, the clinical manifestations of patients were different. The clinical symptoms were back pain, weakness in the extremities, and weakness in both lower extremities separately. Radiological findings also varied in the three patients. MRI revealed hypointensity and isointensity on T1‐weighted images in the second case and our case, respectively. After gadolinium administration, the tumors both displayed enhancement. On T2‐weighted images, MRI revealed hyperintensity in the other two cases and isointensity in our case. Tumor resection and radiation therapy were performed in all three patients. The first patient and our patient were also treated with chemotherapy. The first patient died of disease after 22 months. The second patient and our patient have survived without tumor progression for 10 and five months, respectively. Maximal safe tumor resection and adjuvant radiation therapy might play an essential role in the management of this rare tumor. 9 The other seven cases were located in the brain, and five of them were detected in CIC‐NUTM1 gene fusion. CIC‐NUTM1 fusion seems to exhibit significant tropism for the central nervous system.
CIC‐rearranged sarcomas should first be differentiated from ES. Although both are small round cell tumors and may have glycogen‐rich cytoplasm. CIC‐rearranged sarcomas showed significantly higher degrees of lobulation, nuclear pleomorphism, the prominence of the nucleoli, spindle cell elements, and myxoid changes. The CD99 expression was focal and heterogenous in the CIC‐rearranged sarcomas, whereas in the ES, its expression was diffuse and strong. 14 Yoshimoto et al. 23 found that CCND2 and MUC5AC are reliable biomarkers to distinguish CIC‐DUX4 sarcoma from ES. In addition, it should be differentiated from other small round cell tumors and epithelioid cell tumors, such as atypical teratoid/rhabdoid tumors, sarcomas with BCL‐6 interacting corepressor genetic alterations, alveolar rhabdomyosarcomas, poorly differentiated synovial sarcomas, lymphoma, desmoplastic small round cell tumors, carcinomas, and melanomas. There is clinicopathologic overlap between these tumors, and they can be initially identified by immunohistochemistry. The final diagnosis needs to rely on genetic testing. Fluorescence in situ hybridization analysis had a significant risk of false‐negative results, while NGS‐based diagnostic methods were more sensitive. 24 CIC‐rearranged sarcoma can be accurately classified by DNA methylation array analysis. 25 , 26
The prognosis of CIC‐rearranged sarcoma is aggressive in clinical course and is prone to metastasis, most often to lung, liver, brain, lymph nodes and bone. CIC‐rearranged sarcoma has a significantly worse prognosis than ES and has a poor response to chemotherapy for ES. 1 , 3 , 14 With the in‐depth study on the molecular mechanism of CIC‐rearranged sarcoma, targeted molecular therapy based on precision therapy has made progress. Oyama et al. 27 found that bortezomib and crizotinib can significantly suppress tumor cell growth in CIC‐DUX4 sarcoma in vitro. Yoshimoto et al. 23 found that palbociclib and trabectedin can block the growth of CIC‐DUX4 sarcoma in mice. The optimal treatment for patients with CIC‐rearranged sarcoma remains to be further clarified, and a large number of clinical trials and comprehensive analysis of multi‐institutional data is required.
DISCLOSURE
The authors declare no conflict of interest.
REFERENCES
- 1. WHO Classification of Tumours Editorial Board . WHO classification of tumours. In: Soft Tissue and Bone Tumours [M], 5th edn. Lyon: IARC Press, 2020. [Google Scholar]
- 2. Louis DN, Perry A, Wesseling P et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncology 2021; 23: 1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antonescu CR, Owosho AA, Zhang L et al. Sarcomas with CIC‐rearrangements are a distinct pathologic entity with aggressive outcome:A clinicopathologic and molecular study of 115 cases. Am J Surg Pathol 2017; 41: 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugita S, Arai Y, Tonooka A et al. A novel CIC‐FOXO4 gene fusion in undifferentiated small round cell sarcoma: A genetically distinct variant of Ewing‐like sarcoma. Am J Surg Pathol 2014; 38: 1571–1576. [DOI] [PubMed] [Google Scholar]
- 5. Huang SC, Zhang L, Sung YS et al. Recurrent CIC gene abnormalities in angiosarcomas: A molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 gene alterations. Am J Surg Pathol 2016; 40: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sturm D, Orr BA, Toprak UH et al. New brain tumor entities emerge from molecular classification of CNS‐PNETs. Cell 2016; 164: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sugita S, Arai Y, Aoyama T et al. NUTM2A‐CIC fusion small round cell sarcoma: A genetically distinct variant of CIC‐rearranged sarcoma. Hum Pathol 2017; 65: 225–230. [DOI] [PubMed] [Google Scholar]
- 8. Donahue JE, Yakirevich E, Zhong S et al. Primary spinal epidural CIC‐DUX4 undifferentiated sarcoma in a child. Pediatr Dev Pathol 2018; 21: 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamada S, Muto J, Leon JCAD et al. Primary spinal intramedullary Ewing‐like sarcoma harboring CIC‐DUX4 translocation: A similar cytological appearance as its soft tissue counterpart but no lobulation in association with desmoplastic stroma. Brain Tumor Pathol 2020; 37: 111–117. [DOI] [PubMed] [Google Scholar]
- 10. Richkind KE, Romansky SG, Finklestein JZ. T(4;19)(q35;ql3.1):A recurrent change in primitive mesenchymal tumors? Cancer Genet Cytogenet 1996; 87: 71–74. [DOI] [PubMed] [Google Scholar]
- 11. Kawamura‐Saito M, Yamazaki Y, Kaneko K et al. Fusion between CIC and DUX4 up‐regulates PEA3 family genes in Ewing‐like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet 2006; 15: 2125–2137. [DOI] [PubMed] [Google Scholar]
- 12. Choi E‐YK, Thomas DG, McHugh JB et al. Undifferentiated small round cell sarcoma with t(4;19)(q35;q31.1)CIC‐DUX4 fusion.A novel highly aggressive soft tissue tumor with distinctive histolopathology. Am J Surg Pathol 2013; 37: 1379–1386. [DOI] [PubMed] [Google Scholar]
- 13. Specht K, Sung Y‐S, Zhang L, Richter GHS, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC‐DUX4 positive round cell tumors compared to EWSR1‐rearranged Ewing sarcomas–further evidence towards distinct pathologic entities. Genes Chromosomes Cancer 2014; 53: 622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang M, Yang Y, Guan X, Yao X, Guo Y, He L. Primary cardiac CIC‐rearranged undifferentiated sarcoma in an infant. Pediatr Investig 2021; 5: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshida A, Goto K, Kodaira M et al. CIC‐rearranged sarcomas:A study of 20 cases and comparisons with Ewing sarcomas. Am J Surg Pathol 2016; 40: 313–323. [DOI] [PubMed] [Google Scholar]
- 16. Hung YP, Fletcher CD, Hornick JL. Evaluation of ETV4 and WT1 expression in CIC‐rearranged sarcomas and histologic mimics. Mod Pathol 2016; 29: 1324–1334. [DOI] [PubMed] [Google Scholar]
- 17. Lake JA, Donson AM, Prince E et al. Targeted fusion analysis can aid in the classification and treatment of pediatric glioma, ependymoma, and glioneuronal tumors. Pediatr Blood Cancer 2020; 67: e28028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu W, Wang J, Yuan L et al. Case report: A unique case of pediatric central nervous system embryonal tumor harboring the CIC–LEUTX fusion, germline NBN variant and somatic TSC2 mutation: Expanding the spectrum of CIC‐rearranged neoplasia. Front Oncol 2020; 10: 598970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chinen Y, Taki T, Tsutsumi Y et al. The leucine twenty homeobox (LEUTX) gene,which lacks a histone acetyltransferase domain, is fused to KAT6A in therapy‐related acute myeloid leukemia with t(8;19)(p11;q13). Genes Chromosomes Cancer 2014; 53: 299–308. [DOI] [PubMed] [Google Scholar]
- 20. Barresi S, Giovannoni I, Rossi S et al. A novel BRD4‐LEUTX fusion in a pediatric sarcoma with epithelioid morphology and diffuse S100 expression. Genes Chromosomes Cancer 2021; 60: 647–652. [DOI] [PubMed] [Google Scholar]
- 21. Bielle F, Zanello M, Guillemot D et al. Unusual primary cerebral localization of a CIC–DUX4 translocation tumor of the Ewing sarcoma family. Acta Neuropathol 2014; 128: 309–311. [DOI] [PubMed] [Google Scholar]
- 22. Loarer FL, Pissaloux D, Watson S et al. Clinicopathologic features of CIC‐NUTM1 sarcomas, a new molecular variant of the family of CIC‐fused sarcomas. Am J Surg Pathol 2019; 43: 268–276. [DOI] [PubMed] [Google Scholar]
- 23. Yoshimoto T, Tanaka M, Homme M et al. CIC‐DUX4 induces small round cell sarcomas distinct from Ewing sarcoma. Cancer Res 2017; 77: 2927–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sbaraglia M, Righi A, Gambarotti M, Tos APD. Ewing sarcoma and Ewing‐like tumors. Virchows Arch 2020; 476: 109–119. [DOI] [PubMed] [Google Scholar]
- 25. Koelsche C, Hartmann W, Schrimpf D et al. Array‐based DNA‐methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol 2018; 31: 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miele E, Vito RD, Ciolfi A et al. DNA methylation profiling for diagnosing undifferentiated sarcoma with capicua transcriptional receptor (CIC) alterations. Int J Mol Sci 2020; 21: 1818–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oyama R, Takahashi M, Yoshida A et al. Generation of novel patient‐derived CIC‐DUX4 sarcoma xenografts and cell lines. Sci Rep 2017; 7: 4712. [DOI] [PMC free article] [PubMed] [Google Scholar]


