Abstract
The switch from the normal quiescent vasculature to angiogenesis in tumors is induced by a variety of growth factors, released from cancer and stromal cells upon oxygen and nutrients deprivation. Vascular endothelial growth factor A (VEGF‐A) is a potent‐secreted mitogen and the only growth factor specific to endothelial cells that is observed almost ubiquitously at sites of angiogenesis. Expression of VEGF‐A in cancer cells is controlled through transcriptional and post‐transcriptional mechanisms. Post‐transcriptional regulation of VEGF‐A occurs at multiple levels, through the control of splicing, mRNA stability and translation rate, enabling a fine‐tuned expression and release of VEGF‐A. Mounting evidence is highlighting the important role played by microRNAs (miRNAs) in the control of VEGF‐A mRNA stability and translation in cancer. Moreover, non‐coding RNAs, as long non‐coding RNAs and circular RNAs, are emerging as crucial modulators of VEGF‐A‐targeting miRNAs, with consequent ability to modulate VEGF‐A expression. This review discusses the recent progress on the ncRNA‐related networks controlling VEGF‐A expression in cancer cells and provides insights into the complexity of VEGF‐A post‐transcriptional regulation.
Keywords: circRNA, microRNA, post‐transcriptional regulation, VEGF, VEGFA
Abbreviations
- bFGF
basic Fibroblast Growth Factor
- CAFs
Cancer‐Associated Fibroblasts
- circRNA
circular RNA
- CRC
colorectal cancer
- GBM
glioblastoma multiforme
- GC
gastric cancer
- HIF
Hypoxia‐Inducible Factor
- IL8
interleukin‐8
- lncRNA
long non‐coding RNA
- MALAT1
Metastasis‐Associated Lung Adenocarcinoma Transcript 1
- miRNA
microRNA
- MV
microvesicles
- OSCC
oral squamous cell carcinoma
- TAMs
Tumor‐Associated Macrophages
- TNF‐α
Tumor Necrosis Factor Alpha
- TTP
tristetraprolin
- VEGF‐A
Vascular Endothelial Growth Factor A
- VEGF
Vascular Endothelial Growth Factor
- VHL
von Hippel–Lindau
1. INTRODUCTION
Tumor progression includes an initial “avascular” stage when the tumors are small and dormant and a subsequent “vascular” stage, characterized by the development of a unique tumor vasculature necessary for the metabolic demand of tumors that have exceeded a certain size (usually 1–2 mm). The induction of this vasculature, termed “angiogenic switch,” is caused by a variety of soluble growth factors, as for example vascular endothelial growth factor (VEGF), tumor necrosis factor alpha (TNF‐α), basic fibroblast growth factor (bFGF) and interleukin‐8 (IL8), released by cancer cells and by other cell types infiltrated in the tumor microenvironment, such as cancer‐associated fibroblasts (CAFs) and macrophages (TAMs). 1 , 2 , 3 , 4 The timing of “angiogenic switch” depends on the tumor type and site, and represents a prerequisite for tumor spreading and metastatic dissemination.
Vascular endothelial growth factor A (VEGF‐A) is the key mediator of angiogenesis in cancer. 5 In physiological and pathological conditions, VEGF‐A affects both the development of new blood vessels (angiogenesis) and the survival of endothelial cells (vascular maintenance) by binding to the two tyrosine‐kinase receptors VEGFR1 and VEGFR2. 6 Moreover, VEGF‐A is known as a vascular permeability factor, based on its ability to induce vascular leakage. 7 , 8 The crucial role played by VEGF‐A/VEGFR system in angiogenesis has been evidenced by the study of knock‐out mouse models, which showed that loss of a single VEGFA allele leads to developmental deformities in the forming vasculature and embryonic death between days 11 and 12. Mice lacking either VEGFR‐1 or VEGFR‐2 die even earlier, between embryonic days 8.5 and 9.5. 6 The requirement of a very strict regulation of VEGF‐A level during embryonic development has been also highlighted by the analysis of the effect of VEGF‐A overexpression. Of note, even two‐ to three‐fold overexpression of VEGF‐A from its endogenous locus results in severe abnormalities in heart development and embryonic lethality at E12.5‐E14. 9
Importantly, VEGF‐induced tumor vasculature is structurally and functionally different from normal vasculature. Indeed, tumor blood vessels are irregularly shaped, tortuous, leaky, and hemorrhagic. Tumor blood flow is, therefore, suboptimal causing hypoxia and further VEGF production. Major stimuli responsible for VEGF‐A up‐regulation in cancer cells are low oxygen tension, oncogene activation, hormones, exposure to growth factors, and cytokines.
2. REGULATION OF VEGFA EXPRESSION
The need for finely tuned VEGF‐A expression is highlighted by a complex regulation at multiple levels, including transcriptional regulation, mRNA stabilization, alternative splicing, and translational regulation. Furthermore, even the availability of extracellular VEGF‐A is strictly controlled through the intervention of VEGF‐releasing proteases and soluble carrier molecules, both regulating VEGF gradient formation. 10 As mentioned above, among stimuli responsible for the transcriptional up‐regulation of the VEGFA gene, hypoxia received particular interest because of its role in cancer progression. 11 Specifically, hypoxia‐inducible factor (HIF) is stabilized by exposure to low oxygen tension, as a result of von Hippel–Lindau (VHL) protein destabilization, and transcriptionally activates the hypoxia response element (HRE) within VEGFA promoter. However, transcriptional regulation accounts for only a fraction of the VEGFA gene expression, which is strictly controlled by post‐transcriptional mechanisms. 12 Among post‐transcriptional regulatory layers, alternative splicing is a major mechanism that gives rise, depending on the inclusion/exclusion of exons 6–8, to several distinct isoforms of VEGFA, which differ in their expression patterns, affinity to receptors, biochemical and biological properties. VEGF121 (equivalent to murine VEGF120) and VEGF165 (equivalent to murine VEGF164) are the most abundant isoforms in human tumors. These splice variants differ by the presence or absence of exons 6a, 6b, and 7. Heparin‐binding VEGF isoforms (such as VEGF165) produce a branching network with narrow vessels, while non heparin‐binding isoforms (such as VEGF120) results in poorly branching, tortuous, and leaky vessels. 10
Moreover, the use of an alternative 3′ splice site in the last exon of VEGFA (exon 8) leads to the production of the same isoforms but differing in their six C‐terminal amino acid sequences, named VEGFXXXb, characterized by reduced angiogenic potential, due to their lower affinity to VEGF receptors. 13 VEGFXXXb isoforms are generally down‐regulated in human tumors as a result of the activation of splicing factors as SRSF1 (a.k.a. ASF/SF2). 14 Cooperation between SRSF1 and lncRNA MALAT1 (Metastasis‐Associated Lung Adenocarcinoma Transcript 1) has been also reported to favor VEGFXXX vs. VEGFXXXb in breast cancer cells. 15 Interestingly, the use of inhibitors of SRPK1, the kinase responsible for SRSF1 activation, has been recently proposed as powerful antiangiogenic strategy to be used for the treatment of various pathological conditions, included cancer. Inhibition of SRPK1, through the blocking of SRSF1 phosphorylation and activity, leads to the induction of VEGFXXXb isoforms and, consequently, significantly reduces the angiogenic potential of targeted cells. 16 , 17 , 18
mRNAs‐encoding VEGF‐A isoforms are generally unstable under normal oxygen and nutrient conditions. It has been shown that multiple regions in the VEGF mRNA cooperate both to ensure the rapid degradation of the mRNA under normoxic conditions and to allow stabilization of the mRNA in response to hypoxia. 11 Major factors responsible for destabilization of VEGF‐A mRNA are ARE‐binding proteins AUF1 and TTP (tristetraprolin), interacting with AU‐rich elements present in VEGF‐A 3′‐UTR. 12 AU‐rich elements within the VEGF‐A 3′‐UTR may, on the contrary, confer hypoxia‐dependent mRNA stability and enhanced translation rate, when involved in interaction with RNA‐binding proteins such as HuR (ELAVL1), PTBP1, and NF90 (ILF3). 12 , 19 , 20 Expression of VEGF‐A is also controlled at translation level by elements present in the long GC‐rich 5′‐UTR region of the gene, which contains three in‐frame alternative CUG start codons, one uORF element and two internal ribosome entry sites (IRES). These last enable efficient translation in the presence of stimuli/stresses (as for example hypoxia) while inhibiting cap‐dependent translation (reviewed in Refs 21 and 22).
3. CONTROL OF VEGF‐A EXPRESSION BY MICRORNAS
The long 3′‐UTR region of VEGF‐A emerged as a major regulator of mRNA stability and translation rate also due to the presence of several binding sites for microRNAs. MicroRNAs (miRNAs) are single‐stranded non‐coding RNAs of 18–25 nucleotides that predominantly act as translational repressors. Through sequence‐specific interaction with the 3′‐untranslated region (UTR) of target mRNA, miRNAs suppress gene expression via transcript degradation or inhibition of protein translation.
The first experimental evidence of the relevance of miRNAs in the regulation of VEGFA expression and in angiogenesis was obtained by the analysis of the phenotype of mice lacking miRNA‐processing enzyme Dicer. 23 Subsequently, a multitude of miRNAs has been reported to target directly the 3′‐UTR of VEGF‐A in various cell types; these include miR‐20, miR‐29b, miR‐93, miR‐126, miR‐150‐5p, miR‐190, miR‐195, miR‐200, miR‐203, miR‐205‐5p, miR‐206, miR‐361, miR‐497, miR‐503, miR‐613, and miR‐638 (reviewed in Ref 24 and reported in Refs 25, 26, 27, 28).
miRNAs have been shown to control VEGFA expression also in the endothelial cell context. 29 , 30 , 31 An example of this regulation is represented by some miRNAs of the miR‐15/107 family, 32 for example, miR‐16 and miR‐424, sharing the same seed sequence required for target recognition, and inhibiting the expression not only of VEGFA but also of VEGFR2 (reviewed in Ref 33). Interestingly, as VEGF‐A has been reported to exert both autocrine and intracrine signaling in the endothelium (reviewed in Ref 34), targeting of VEGFA 3′‐UTR by miRNAs might contribute to the lowering of VEGF‐A expression in endothelial cells and to the inhibition of these signaling pathways in the absence of angiogenic stimuli.
VEGF‐A‐targeting miRNAs are generally down‐regulated in tumors, contributing to the induction of proliferation, invasion, and angiogenesis. Interestingly, miRNAs can compete with VEGF‐A stabilizing RBPs by sharing common binding sites in the 3′‐UTR. A representative example is the antagonistic effect of miR‐200 family miRNAs and the protein HuR. 35 Of note, the evidence that many of the miRNAs that target VEGF‐A 3′‐UTR are also responsible for the negative regulation of other key players in angiogenesis, as for example HIF‐1α, highlighted that inhibition of angiogenesis in physiological conditions is based on a complex network of regulatory layers. The inhibitory activity of miRNAs on the expression of angiogenesis‐related genes is almost always lost in cancer cells, through both downregulation of these tumor suppressor miRNAs as well as the inhibition of their activity exerted by additional non‐coding RNAs as circular RNAs (circRNAs) and long non‐coding RNAs (lncRNAs) which are frequently upregulated and function as sponges to limit microRNAs’ function. These circRNA‐miRNA and lncRNA‐miRNA regulatory networks are discussed below.
4. CIRCULAR RNAS CONTROLLING VEGF‐A EXPRESSION
CircRNAs are covalently closed RNA loops that originate from back‐splicing events occurring mainly in protein‐coding genes. Owing to the closed continuous loop structure, circRNAs can escape exonuclease‐mediated degradation and are more stable than other RNA species. 36 CircRNAs may be overexpressed in cancer cells, contributing to the various features of malignancy, as proliferation, migration, and invasion, and are efficiently released by cancer cells into extracellular vesicles, acting as autocrine and paracrine mediators. 37 CircRNAs may exert at intracellular level various functions, the best known of which is miRNA sponging. 38 Through base‐pairing with a miRNA sequence, a circRNA may sequester the miRNA and block its inhibitory activity on target mRNAs. Therefore, the overexpression of a circRNA usually results in the upregulation of the target mRNAs of the sequestered miRNA. Other major activities of circRNAs are the enhancement of protein complexes formation and/or activity, and the regulation of transcription or translation. Intriguingly, a few circRNAs may be also translated into polypeptides, when presenting an epitranscriptomic m6A modification, needed for translation initiation. 37 With regard to VEGF‐A, a number of circRNAs have been functionally characterized as inducers of VEGF‐A protein expression, mainly through their ability to sequester miRNAs targeting the 3′‐UTR region of VEGF‐A (summarized in Table 1 and Figure 1). Interestingly, various miRNAs that are sponged by VEGF‐A‐inducing circRNAs are also down‐regulated in cancer cells, as, for example, members of the miR‐29 family, miR‐195, miR‐205‐5p, and miR‐613. miR‐29 family comprises three mature miRNAs, namely miR‐29a, miR‐29b, and miR‐29c, characterized by identical seed sequence (AGCACCA). The miR‐29‐3p arm represents the most abundant and functionally relevant arm of all three family members. miR‐29 is recognized as one of the critical miRNAs with tumor suppressor activity that play a role in cancer pathogenesis 39 ; it is down‐regulated in the vast majority of cancer types, where it regulates proliferation, apoptosis, epithelial‐mesenchymal transition (EMT), fibrosis and metastasis, by targeting key regulatory players in various oncogenic pathways. Of note, miR‐29 may be sponged by three circular RNAs, namely circ‐MYLK, circ_001971, and circ_0044366, all leading to the induction of VEGF‐A expression in various cancer types (Table 1). Circ‐MYLK promotes bladder cancer progression by activating VEGFA/VEGFR2 and downstream Ras/ERK signaling pathway. 40 Circ_001971 expression induces proliferation, invasion, and angiogenesis in colorectal cancer (CRC). 41 Circ_0044366 (termed circ29) is released in exosomes by gastric cancer (GC) cells and reaches endothelial cells, where it modulates VEGF‐A expression and VEGF‐A/VEGFR signaling by sequestering miR‐29a. 42 Also miR‐877 is potentially sponged by multiple circRNAs in cancer cells, enabling VEGF‐A induction. Specifically, circ‐RanGAP1‐mediated miR‐877–3p/VEGFA axis promotes GC progression. Circ‐RanGAP1 is upregulated in the plasma exosomes from GC patients and, interestingly, these exosomes are able to induce the invasive potential of cultured GC cells. 43 miR‐877–3p has been shown to be targeted by the sponging activity of circ_0001766 in oral squamous cell carcinoma (OSCC), thus enhancing cell proliferation. 44 A number of additional circRNA‐miRNA interactions impinging on the expression of VEGF‐A and angiogenesis have been identified in various cancer types and are listed in Table 1.
TABLE 1.
CircRNAs involved in the regulation of VEGF‐A expression in the indicated cancer types
| circRNA (host gene) | Sponged miRNA or other function | Cancer type | Refs |
|---|---|---|---|
| circSMARCA5 | Interacts with SRSF1 leading to enhanced VEGFxxx/VEGFxxxb ration | Glioblastoma | 47 |
| circRPL15 | miR‐146b‐3p | Glioma | 53 |
| circPVT1 | miR‐195 | Papillary thyroid carcinoma | 54 |
| circ_0001429 (MANBA) | miR‐205‐5p | Bladder cancer | 55 |
| circ_0056618 (SPOPL) | miR‐206 | Colorectal cancer | 56 |
| circMYLK | miR‐29a | Bladder cancer | 40 |
| circ_0044366 (a.k.a. circ29)(ATP5G1) | miR‐29a | Gastric cancer | 42 |
| circ_001971 (alias hsa_circ_0001060)(UXS1) | miR‐29c‐3p | Colorectal cancer | 41 |
| circRhoC | miR‐302e sponging and interaction with VEGFA protein | Ovarian cancer | 57 |
| circITGA7 | miR‐34a‐5p | Glioma | 58 |
| circ_0001178 (USP25) | miR‐382‐5p | Hepatocellular carcinoma | 59 |
| circAP2A2 | miR‐382‐5p | Hemangioma | 60 |
| circSCAF11 | miR‐421 sponging, leading to Sp1 increase and VEGFA transcriptional activation | Glioma | 46 |
| circMYOF | miR‐4,739 | Pancreatic ductal adenocarcinoma | 61 |
| circ_0023404 (RNF121) | miR‐5,047 | Cervical tumors | 62 |
| circ_0030998 (LAMP1) | miR‐567 | Colorectal cancer | 63 |
| circSHKBP1 (circ_0000936) | miR‐582‐3p sponging, leading to HuR protein induction and VEGFA mRNA stabilization | Gastric cancer | 45 |
| circCCT3 | miR‐613 | Colorectal cancer and pancreatic ductal adenocarcinoma | 64 |
| circASH2L | miR‐665 | Ovarian cancer | 65 |
| circ_0001766 (PDIA4) | miR‐877‐3p | Oral squanous cell carcinoma | 44 |
| circRanGAP1 | miR‐877‐3p | Gastric cancer | 43 |
| circPOK (Zbtb7a) | Interaction with ILF2/ILF3 complex and VEGFA mRNA stabilization | Mesenchymal tumors | 66 |
| circATXN1 | miR‐526b‐3p | Glioma | 67 |
FIGURE 1.
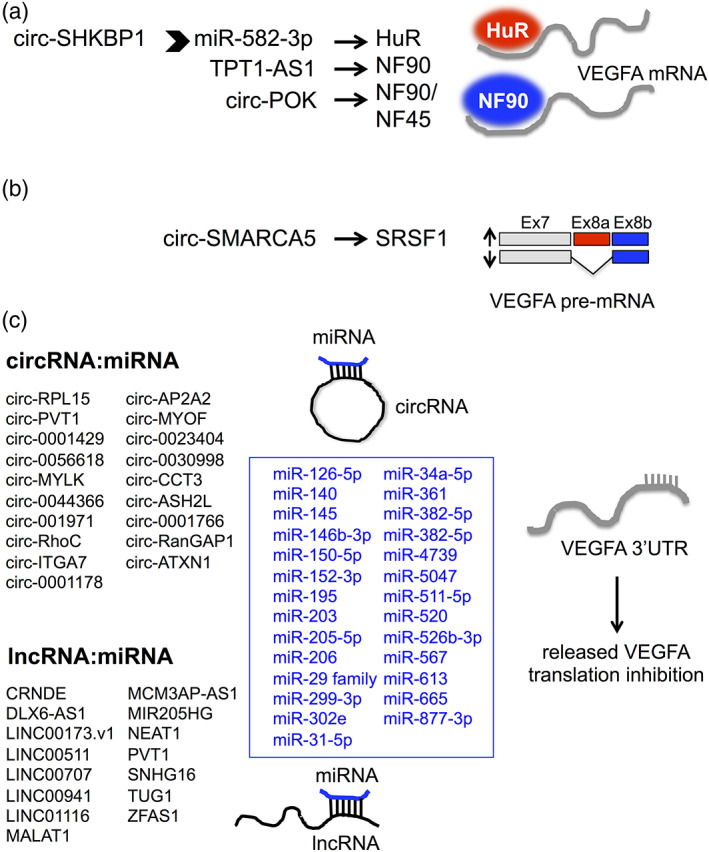
Different ncRNA‐driven post‐transcriptional mechanisms regulating VEGFA expression in cancer cells. (a) ncRNAs may induce the expression of VEGFA by regulating the expression (as in the case of the circ‐SHKBP1/miR‐582‐3p/HuR axis) or the activity (as in the case of TPT‐AS1/NF90 and circ‐POK/NF90 axis) of key proteins involved in VEGFA mRNA stabilization, such as HuR and NF90. (b) circ‐SMARCA5 impinges on the expression level of VEGFA pro‐ and anti‐angiogenic isoforms, favoring the former over the latter, by interacting with splicing factor SRSF1. (c) Summary of the so far reported circRNAs (upper list) and lncRNAs (lower list) able to sponge VEGFA‐targeting miRNAs (indicated in blue) in various cancer types
CircRNA may also affect VEGF‐A expression indirectly, for example, by targeting VEGF‐A regulatory proteins and impinging on different VEGF‐A regulatory layers. Interesting examples of these alternative mechanisms are represented by circ‐SHKBP1, circ‐SCAF11, circ‐SMARCA5, and circPOK. In the first case, circ‐SHKBP1 (circ_0000936) is able to sponge miR‐582‐3p in GC cells, leading to released expression of HuR protein, which in turn enhances VEGF‐A mRNA stability and expression. Of note, GC cells release circ‐SHKBP1 in the extracellular space and exosomal circ‐SHKBP1 contributes to enhance GC cell growth. 45 Circ‐SCAF11 is involved in VEGF‐A induction during the progression of glioma; circ‐SCAF11indeed is able to sponge miR‐421 and release the expression of its target Sp1, a key transcription factor responsible for VEGF‐A expression. 46 The last example refers to a circRNA, circ‐SMARCA5, which acts in advanced brain tumors. Circ‐SMARCA5 controls the ratio between pro‐ and anti‐angiogenic VEGF‐A isoforms in glioblastoma multiforme (GBM), a highly vascularized tumor, by interacting with SRSF1, a key splicing factor controlling the VEGFXXX/VEGFXXXb ratio. Of note, blood vascular microvessel density negatively correlated with the expression of circ‐SMARCA5 in GBM biopsies while positively correlated with that of SRSF1 and the VEGFXXX/VEGFXXXb ratio. 47 The last example is represented by circPOK, a proto‐oncogenic RNA in mesenchymal tumor progression, encoded by the Zbtb7a gene. CircPOK acts antithetically to its linear transcript counterpart (Pokemon), which acts as a tumor suppressor gene. CircPOK favors proliferation and angiogenesis by interacting with and activating the NF45/NF90 complex (encoded by ILF2 and ILF3 genes), leading to the activation of many pro‐angiogenic and growth factors, such as IL6 and VEGF‐A, through transcriptional and post‐transcriptional regulation, respectively. 66
5. LNCRNAS CONTROLLING VEGF‐A EXPRESSION
The inhibitory activity of miRNAs on VEGF‐A 3′‐UTR may be also regulated by long non‐coding RNAs (lncRNAs), which may function as sponges for miRNAs. Moreover, lncRNAs may impact on VEGF‐A abundance by regulating the activity of transcriptional or post‐transcriptional modulators of VEGF‐A expression (Table 2 and Figure 1). The most functionally relevant lncRNA:miRNA networks enclose miRNAs with a well‐established tumor suppressor activity, such as miR‐29 family members (sponged by LINC00511 and PVT1), miR‐195 (belonging to miR‐15/107 group and sponged by NEAT1 and DLX6‐AS1), miR‐34a‐5p (sponged by TUG1), and miR‐145 (sponged by MALAT1).
TABLE 2.
LncRNAs involved in the regulation of VEGF‐A expression in the indicated cancer types
| LncRNA | Sponged miRNA or other function | Cancer type | Refs |
|---|---|---|---|
| CamK‐A | Activation of NF‐kB and consequent VEGFA induction | Multiple tumors | 68 |
| CRNDE | miR‐203 | Hepatoblastoma | 69 |
| DLX6‐AS1 | miR‐195 | Bladder cancer | 70 |
| FLANC | Increases the half‐life of phosphorylated STAT3 to induce VEGFA expression | Colorectal cancer | 71 |
| LINC00173.v1 | miR‐511‐5p | Lung cancer | 72 |
| LINC00511 | miR‐29b‐3p | Pancreatic ductal adenocarcinoma | 73 |
| LINC00707 | miR‐382‐5p | Cervical cancer | 74 |
| LINC00941 | miR‐877‐3p | Non‐small cell lung cancer | 75 |
| LINC01116 | miR‐31‐5p | Glioma | 76 |
| MALAT1 | miR‐126‐5p | Colorectal cancer | 51 |
| MALAT1 | miR‐140 | Hepatocellular carcinoma | 77 |
| MALAT1 | miR‐145 | Breast cancer | 78 |
| MALAT1 | miR‐150‐5p | Osteosarcoma | 79 |
| MALAT1 | miR‐206 | Endothelial cells | 51 |
| MIR205HG | miR‐299‐3p | Melanoma | 80 |
| NEAT1 | miR‐126‐5p | Thyroid carcinoma | 81 |
| NEAT1 | miR‐195 | Sinonasal SCC | 82 |
| NEAT1 | miR‐205‐5p | Colorectal cancer | 83 |
| NEAT1 | miR‐361 | Hemangioma | 84 |
| PVT1 | miR‐152‐3p | Colorectal cancer | 85 |
| PVT1 | miR‐29c | Non‐small cell lung cancer | 86 |
| SNHG16 | miR‐520 | Lung cancer | 87 |
| TNK2‐AS1 | Interacts with STAT3 and elevate VEGFA expression | Lung cancer | 88 |
| TPT1‐AS1 | Interacts with NF90 and enhances VEGFA mRNA stability | Colorectal cancer | 89 |
| TUG1 | miR‐299‐3p | Renal cell carcinoma | 90 |
| TUG1 | miR‐34a‐5p | Hepatoblastoma | 91 |
| TUG1 | Sponges miR‐143‐5p enabling HIF‐1a expression and VEGFA upregulation | Osteosarcoma | 92 |
| ZFAS1 | miR‐150‐5p | Colorectal cancer | 93 |
The example of PVT1 is of particular interest as the PVT1 (Plasmacytoma Variant Translocation 1) gene locus encodes many ncRNA variants, including both linear and circular isoforms, the most abundant being the exon 2‐derived circPVT1 (reviewed in Refs 48 and 49) and it has been reported that linear PVT1 and circPVT1, both overexpressed in cancer cells, may positively regulate VEGF‐A expression.
Overexpression of PVT1 and circPVT1 in cancer cells may result from gene amplification and/or from promoter activation. Interestingly, different promoters drive expression of PVT1 and circPVT1, as a second promoter region present in the first intron of the gene regulates circPVT1 transcription. Upregulation of PVT1 and circPVT1 in cancer cells may be caused by c‐Myc transcription factor and by the TEAD/YAP/mutant p53 complex, respectively (reviewed in Refs 48 and 49). CircPVT1 could promote VEGFA expression by sponging miR‐195, thus contributing to the malignant progression of papillary thyroid carcinoma. Interestingly, also linear PVT1 had been previously reported to sequester miR‐195, despite its effect on VEGF‐A wasn't analyzed in that study. 48 Linear PVT1 induces VEGF‐A expression by sponging miR‐152‐3p and miR‐29c in colorectal cancer and non‐small cell lung cancer, respectively (Table 2).
Another lncRNA relevant in the cancer context, MALAT1, highly expressed in a variety of tumors following transactivation by oncogenic transcription factors such as HIF‐1 and YAP, 50 , 51 and implicated in various cancer‐related activities as proliferation, invasion, and angiogenesis, has been shown to control VEGF‐A expression by sponging a variety of miRNAs, as miR‐126‐5p, miR‐140, miR‐145, miR‐150‐5p in various cancer types and miR‐206 in endothelial cells. Interestingly, MALAT1 had been previously reported to control VEGF‐A splicing, favoring VEGFXXX vs VEGFXXXb isoforms. 15 This lncRNA thus emerges as a regulator of VEGF‐A acting at different post‐transcriptional levels. Several groups have shown that many lncRNAs contribute to tumor angiogenesis and progression, inducing VEGFA expression by sequestering distinct miRNAs in a wide variety of cancer types (summarized in Table 2). In addition to sponging of VEGFA‐targeting miRNAs, various lncRNAs may impact on VEGF‐A expression through the modulation of the expression and/or activity of VEGF‐A post‐transcriptional regulators. One such example is lncRNA TPT1‐AS1, a liver‐metastasis associated lncRNA in colorectal cancer. TPT1‐AS1 is upregulated in CRC and its high expression is associated with poor outcome. Functionally, TPT1‐AS1 interacts with NF90 protein thus favoring VEGFA mRNA stability, leading to enhanced angiogenic and metastatic potential of CRC cells. 89 Of note, TPT1‐AS1 and VEGFA mRNA are significantly correlated in a CRC cohort.
6. CONCLUSIONS
Mounting evidence has emerged in the last few years about the relevance of VEGFA post‐transcriptional regulation exerted by non‐coding RNAs in a wide variety of cancer types. These regulatory networks impinge on the selection of specific VEGFA isoforms, on their stability and translation rate. ncRNA‐based networks impinging on the angiogenic potential of cancer cells represent a powerful source of inspiration for the development of novel therapeutic approaches aimed at blocking the spreading of tumors. As the use of anti‐VEGF agents for the treatment of cancer has not been encouraging as hoped, mainly due to development of resistance, the possibility of exploiting ncRNA‐driven networks to block the angiogenic potential of cancer cells could present some advantages, compared to single‐target treatments such as the anti‐VEGF therapies. These include (a) the direct impact of some ncRNAs on the expression of VEGFA, therefore, acting upstream, with respect to current antiangiogenic therapies; (b) the ability of some ncRNAs to impinge, simultaneously, on the expression of various growth factors controlling angiogenesis, thus potentially overcoming the development of resistance; (c) the possibility of using cell‐derived microvesicles (MV) to deliver ncRNA molecules, efficiently and in targeted manner, to cancer cells. With regard to this last possibility, it has been recently reported that the MV‐delivered miR‐29a/c significantly suppresses VEGFA expression in GC cells, inhibiting vascular cell growth, metastasis, and tube formation. In vivo data proved that MVs function as a potential carrier of miRNAs for targeted therapy in gastric cancer. 52 Interestingly, the miR‐29 mimic MRG‐201 is currently being tested in phase I clinical trials via intradermal injection (Clinical-Trials.gov: NCT02603224) for fibrosis‐related pathologies and could represent a potential effective therapy for the treatment of cancers presenting miR‐29 downregulation/inactivation. ncRNA‐based therapeutics thus represents an attractive approach to target angiogenesis‐related networks for the treatment of cancer.
ACKNOWLEDGMENTS
I have no conflict of interest to declare. The author received funding from AIRC (Associazione Italiana per la Ricerca sul Cancro) under IG 2018—ID. 21434 project—P.I. Fontemaggi Giulia.
Fontemaggi G. Non‐coding RNA regulatory networks in post‐transcriptional regulation of VEGFA in cancer. IUBMB Life. 2023;75(1):30–39. 10.1002/iub.2620
REFERENCES
- 1. Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. [DOI] [PubMed] [Google Scholar]
- 2. Folkman J. Incipient angiogenesis. Jnci‐J Natl Cancer. 2000;I(92):94–95. [DOI] [PubMed] [Google Scholar]
- 3. McAllister SS, Weinberg RA. The tumour‐induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fukumura D, Xavier R, Sugiura T, et al. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94:715–725. [DOI] [PubMed] [Google Scholar]
- 5. Linderholm B, Tavelin B, Grankvist K, Henriksson R. Vascular endothelial growth factor is of high prognostic value in node‐negative breast carcinoma. J Clin Oncol. 1998;16:3121–3128. [DOI] [PubMed] [Google Scholar]
- 6. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. [DOI] [PubMed] [Google Scholar]
- 7. Senger DR, Galli SJ, Dvorak AM, Perruzzi CA, Harvey VS, Dvorak HF. Tumor‐cells secrete a vascular‐permeability factor that promotes accumulation of ascites‐fluid. Science. 1983;219:983–985. [DOI] [PubMed] [Google Scholar]
- 8. Bates DO, Curry FE. Vascular endothelial growth factor increases microvascular permeability via a Ca2+−dependent pathway. Am J Physiol‐Heart C. 1997;273:687–694. [DOI] [PubMed] [Google Scholar]
- 9. Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. [DOI] [PubMed] [Google Scholar]
- 10. Vempati P, Popel AS, Mac Gabhann F. Extracellular regulation of VEGF: Isoforms, proteolysis, and vascular patterning. Cytokine Growth F R. 2014;25:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dibbens JA, Miller DL, Damert A, Risau W, Vadas MA, Goodall GJ. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol Biol Cell. 1999;10:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arcondeguy T, Lacazette E, Millevoi S, Prats H, Touriol C. VEGF‐A mRNA processing, stability and translation: A paradigm for intricate regulation of gene expression at the post‐transcriptional level. Nucleic Acids Res. 2013;41:7997–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nowak DG, Woolard J, Amin EM, et al. Expression of pro‐ and anti‐angiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J Cell Sci. 2008;121:3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bates DO, Cui TG, Doughty JM, et al. VEGF(165)b, an inhibitory splice variant of vascular endothelial growth factor, is down‐regulated in renal cell carcinoma. Cancer Res. 2002;62:4123–4131. [PubMed] [Google Scholar]
- 15. Pruszko M, Milano E, Forcato M, et al. The mutant p53‐ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017;18:1331–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li QY, Zeng CY, Liu HZ, et al. Protein‐protein interaction inhibitor of SRPKs alters the splicing isoforms of VEGF and inhibits angiogenesis. Iscience. 2021;24:102423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gammons MV, Lucas R, Dean R, Coupland SE, Oltean S, Bates DO. Targeting SRPK1 to control VEGF‐mediated tumour angiogenesis in metastatic melanoma. Brit J Cancer. 2014;111:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hatcher JM, Wu GW, Zeng CY, et al. SRPKIN‐1: A covalent SRPK1/2 inhibitor that potently converts VEGF from pro‐angiogenic to anti‐angiogenic isoform. Cell Chem Biol. 2018;25:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan XHC, Steitz JA. Overexpression of HuR, a nuclear‐cytoplasmic shuttling protein, increases the in vivo stability of ARE‐containing mRNAs. EMBO J. 1998;17:3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vumbaca F, Phoenix KN, Rodriguez‐Pinto D, Han DK, Claffey KP. Double‐stranded RNA‐binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol. 2008;28:772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robichaud N, Sonenberg N, Ruggero D, Schneider RJ. Translational control in cancer. Csh Perspect Biol. 2019;11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G‐Quadruplex is essential for cap‐independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 23. Yang WJ, Yang DD, Na SQ, Sandusky GE, Zhang Q, Zhao GS. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Wang LY, Chen C, Chu XY. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol Cancer. 2018;17:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang JL, Zhang JX, Pang XC, et al. MiR‐205‐5p suppresses angiogenesis in gastric cancer by downregulating the expression of VEGFA and FGF1. Exp Cell Res. 2021;404:112579. [DOI] [PubMed] [Google Scholar]
- 26. Huang JJ, Wang X, Wen GB, Ren Y. miRNA‐205‐5p functions as a tumor suppressor by negatively regulating VEGFA and PI3K/Akt/mTOR signaling in renal carcinoma cells. Oncol Rep. 2019;42:1677–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stahlhut C, Suarez Y, Lu J, Mishima Y, Giraldez AJ. miR‐1 and miR‐206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development. 2012;139:4356–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanitz A, Imig J, Dziunycz PJ, et al. The expression levels of MicroRNA‐361‐5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. Plos One. 2012;7:e49568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suarez Y, Fernandez‐Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. [DOI] [PubMed] [Google Scholar]
- 30. Suarez Y, Fernandez‐Hernando C, Yu J, et al. Dicer‐dependent endothelial microRNAs are necessary for postnatal angiogenesis. P Natl Acad Sci USA. 2008;105:14082–14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of dicer and drosha for endothelial MicroRNA expression and angiogenesis. Circ Res. 2007;101:59–68. [DOI] [PubMed] [Google Scholar]
- 32. Turco C, Donzelli S, Fontemaggi G. miR‐15/107 microRNA Gene Group: Characteristics and functional implications in cancer. Front Cell Dev Biol. 2020;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chamorro‐Jorganes A, Araldi E, Suarez Y. MicroRNAs as pharmacological targets in endothelial cell function and dysfunction. Pharmacol Res. 2013;75:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Domigan CK, Ziyad S, Iruela‐Arispe ML. Canonical and noncanonical vascular endothelial growth factor pathways new developments in biology and signal transduction. Arterioscl Throm Vas. 2015;35:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang SH, Lu YC, Li X, et al. Antagonistic function of the RNA‐binding protein HuR and miR‐200b in post‐transcriptional regulation of vascular endothelial growth factor‐a expression and angiogenesis. J Biol Chem. 2013;288:4908–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 37. Fontemaggi G, Turco C, Esposito G, Di Agostino S. New molecular mechanisms and clinical impact of circRNAs in human cancer. Cancer. 2021;13:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 39. Kwon JJ, Factora TD, Dey S, Kota J. 1 a systematic review of miR‐29 in cancer. Mol Ther‐Oncolytics. 2019;12:173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhong ZY, Huang MG, Lv MX, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. [DOI] [PubMed] [Google Scholar]
- 41. Chen C, Huang ZG, Mo XY, et al. The circular RNA 001971/miR‐29c‐3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Canc Res. 2020;39:91. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Li S, Li JL, Zhang HY, et al. Gastric cancer derived exosomes mediate the delivery of circRNA to promote angiogenesis by targeting miR‐29a/VEGF axis in endothelial cells. Biochem Bioph Res Co. 2021;560:37–44. [DOI] [PubMed] [Google Scholar]
- 43. Lu J, Wang YH, Yoon C, et al. Circular RNA circ‐RanGAP1 regulates VEGFA expression by targeting miR‐877‐3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. [DOI] [PubMed] [Google Scholar]
- 44. Shao YX, Song YH, Xu SM, Li SY, Zhou HW. Expression profile of circular RNAs in Oral squamous cell carcinoma. Front Oncol. 2020;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie MY, Yu T, Jing XM, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR‐582‐3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meng Q, Li S, Liu Y, et al. Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR‐421/SP1/VEGFA Axis. Mol Ther‐Nucl Acids. 2019;17:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barbagallo D, Caponnetto A, Brex D, et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma Multiforme through the binding of SRSF1. Cancer. 2019;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Traversa D, Simonetti G, Tolomeo D, et al. Unravelling similarities and differences in the role of circular and linear PVT1 in cancer and human disease. Brit J Cancer. 2022;126:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Palcau AC, Canu V, Donzelli S, Strano S, Pulito C, Blandino G. CircPVT1: a pivotal circular node intersecting long non‐coding‐PVT1 and c‐MYC oncogenic signals. Mol Cancer. 2022;21:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shih CH, Chuang LL, Tsai MH, et al. Hypoxia‐induced MALAT1 promotes the proliferation and migration of breast cancer cells by sponging MiR‐3064‐5p. Front Oncol. 2021;11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun X, Luo LH, Li JR. LncRNA MALAT1 facilitates BM‐MSCs differentiation into endothelial cells via targeting miR‐206/VEGFA axis. Cell Cycle. 2020;19:3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang HY, Bai M, Deng T, et al. Cell‐derived microvesicles mediate the delivery of miR‐29a/c to suppress angiogenesis in gastric carcinoma. Cancer Lett. 2016;375:331–339. [DOI] [PubMed] [Google Scholar]
- 53. Wang B, Duan R, Li ZB, Wang L. Circ‐RPL15/miR‐146b‐3p/VEGFA feedback loop is responsible for triggering proliferation and migration in glioma. Eur Rev Med Pharmaco. 2020;24:6204–6210. [DOI] [PubMed] [Google Scholar]
- 54. Zeng LW, Yuan SF, Zhou PF, Gong JM, Kong XD, Wu M. Circular RNA Pvt1 oncogene (CircPVT1) promotes the progression of papillary thyroid carcinoma by activating the Wnt/beta‐catenin signaling pathway and modulating the ratio of microRNA‐195 (miR‐195) to vascular endothelial growth factor a (VEGFA) expression. Bioengineered. 2021;12:11795–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao WF, Zhao YG, Wang L, Huang XK. Circ0001429 regulates progression of bladder cancer through binding miR‐205‐5p and promoting VEGFA expression. Cancer Biomark. 2019;25:101–113. [DOI] [PubMed] [Google Scholar]
- 56. Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR‐206 and upregulating CXCR4 and VEGF‐A in colorectal cancer. Eur Rev Med Pharmaco. 2020;24:4190–4202. [DOI] [PubMed] [Google Scholar]
- 57. Wang LL, Zong ZH, Liu Y, Guan X, Chen S, Zhao Y. CircRhoC promotes tumorigenicity and progression in ovarian cancer by functioning as a miR‐302e sponge to positively regulate VEGFA. J Cell Mol Med. 2019;23:8472–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qi L, Wang WY, Zhao GF, et al. Circular RNA circitga7 accelerates glioma progression via miR‐34a‐5p/VEGFA axis. Aging‐Us. 2021;13:13138–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gao S, Hu W, Huang X, et al. Circ_0001178 regulates miR‐382/VEGFA axis to facilitate hepatocellular carcinoma progression. Cell Signal. 2020;72:109621. [DOI] [PubMed] [Google Scholar]
- 60. Yuan XQ, Xu YN, Wei ZQ, Ding Q. CircAP2A2 acts as a ceRNA to participate in infantile hemangiomas progression by sponging miR‐382‐5p via regulating the expression of VEGFA. J Clin Lab Anal. 2020;34:e23258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zheng DD, Huang XX, Peng JF, et al. CircMYOF triggers progression and facilitates glycolysis via the VEGFA/PI3K/AKT axis by absorbing miR‐4739 in pancreatic ductal adenocarcinoma. Cell Death Discov. 2021;7:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guo J, Chen MM, Ai GH, Mao WP, Li H, Zhou JH. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR‐5047. Biomed Pharmacother. 2019;115:108957. [DOI] [PubMed] [Google Scholar]
- 63. Jin LY, Han C, Zhai TY, Zhang XY, Chen C, Lian L. Circ_0030998 promotes tumor proliferation and angiogenesis by sponging miR‐567 to regulate VEGFA in colorectal cancer. Cell Death Discov. 2021;7:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li WL, Xu YQ, Wang XD, et al. circCCT3 modulates vascular endothelial growth factor a and Wnt signaling to enhance colorectal cancer metastasis through sponging miR‐613. DNA Cell Biol. 2020;39:118–125. [DOI] [PubMed] [Google Scholar]
- 65. Chen JX, Li XC, Yang L, Li MM, Zhang Y, Zhang JR. CircASH2L promotes ovarian cancer tumorigenesis, angiogenesis, and Lymphangiogenesis by regulating the miR‐665/VEGFA Axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guarnerio J, Zhang Y, Cheloni G, et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29:628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu XB, Shen SY, Zhu L, et al. SRSF10 inhibits biogenesis of circ‐ATXN1 to regulate glioma angiogenesis via miR‐526b‐3p/MMP2 pathway. J Exp Clin Canc Res. 2020;39:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sang LJ, Ju HQ, Liu GP, et al. LncRNA CamK‐A regulates Ca2+‐signaling‐mediated tumor microenvironment remodeling. Mol Cell. 2018;72:71–83. [DOI] [PubMed] [Google Scholar]
- 69. Chen LJ, Yuan MX, Ji CY, et al. Long non‐coding RNA CRNDE regulates angiogenesis in Hepatoblastoma by targeting the MiR‐203/VEGFA Axis. Pathobiology. 2020;87:161–170. [DOI] [PubMed] [Google Scholar]
- 70. Wang HB, Niu XB, Jiang HS, et al. Long non‐coding RNA DLX6‐AS1 facilitates bladder cancer progression through modulating miR‐195‐5p/VEGFA signaling pathway. Aging‐Us. 2020;12:16021–16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pichler M, Rodriguez‐Aguayo C, Nam SY, et al. Therapeutic potential of FLANC, a novel primate‐specific long non‐coding RNA in colorectal cancer. Gut. 2020;69:1818–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen JR, Liu AB, Wang ZH, et al. LINC00173.v1 promotes angiogenesis and progression of lung squamous cell carcinoma by sponging miR‐511‐5p to regulate VEGFA expression. Mol Cancer. 2020;19:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao XH, Liu YM, Li ZH, et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa‐miR‐29b‐3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guo H, Li J, Fan FR, Zhou P. LINC00707 regulates miR‐382‐5p/VEGFA pathway to enhance cervical cancer progression. J Immunol Res. 2021;2021:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ren MH, Chen S, Wang LG, Rui WX, Li P. LINC00941 promotes progression of non‐small cell lung cancer by sponging miR‐877‐3p to regulate VEGFA expression. Front Oncol. 2021;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ye JL, Zhu JL, Chen HR, et al. A novel lncRNA‐LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int J Cancer. 2020;146:248–261. [DOI] [PubMed] [Google Scholar]
- 77. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non‐coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR‐140. Am J Physiol‐Cell Ph. 2020;318:C649–C663. [DOI] [PubMed] [Google Scholar]
- 78. Huang XJ, Xia Y, He GF, et al. MALAT1 promotes angiogenesis of breast cancer. Oncol Rep. 2018;40:2683–2689. [DOI] [PubMed] [Google Scholar]
- 79. Vimalraj S, Subramanian R, Dhanasekaran A. LncRNA MALAT1 promotes tumor angiogenesis by regulating MicroRNA‐150‐5p/VEGFA signaling in osteosarcoma: In‐vitro and in‐vivo analyses. Front Oncol. 2021;11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guo JL, Gan Q, Gan CB, Zhang XN, Ma XP, Dong ML. LncRNA MIR205HG regulates melanomagenesis via the miR‐299‐3p/VEGFA axis. Aging‐Us. 2021;13:5297–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zeng WW, Lin Y, Lin H, Wu XM. Silencing NEAT1 suppresses thyroid carcinoma via miR‐126/NEAT1/VEGFA axis. Front Biosci‐Landmrk. 2020;25:564–576. [DOI] [PubMed] [Google Scholar]
- 82. Lu HL, Kang F. Down‐regulating NEAT1 inhibited the viability and vasculogenic mimicry formation of sinonasal squamous cell carcinoma cells via miR‐195‐5p/VEGFA axis. Biosci Rep. 2020;40:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liu HJ, Li AY, Sun ZC, Zhang JY, Xu H. Long non‐coding RNA NEAT1 promotes colorectal cancer progression by regulating miR‐205‐5p/VEGFA axis. Hum Cell. 2020;33:386–396. [DOI] [PubMed] [Google Scholar]
- 84. Yu XY, Liu XY, Wang R, Wang L. Long non‐coding RNA NEATI promotes the progression of hemangioma via the miR‐361‐5p/VEGFA pathway. Biochem Bioph Res Co. 2019;512:825–831. [DOI] [PubMed] [Google Scholar]
- 85. Lai SW, Chen MY, Bamodu OA, et al. Exosomal lncRNA PVT1/VEGFA axis promotes colon cancer metastasis and stemness by downregulation of tumor suppressor miR‐152‐3p. Oxid Med Cell Longev. 2021:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mao ZJ, Xu BT, He LX, Zhang GD. PVT1 promotes angiogenesis by regulating miR‐29c/vascular endothelial growth factor (VEGF) signaling pathway in non‐small‐cell lung cancer (NSCLC). Med Sci Monitor. 2019;25:5418–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen L, Qiu CH, Chen Y, Wang Y, Zhao JJ, Zhang M. LncRNA SNHG16 drives proliferation, migration, and invasion of lung cancer cell through modulation of miR‐520/VEGF axis. Eur Rev Med Pharmaco. 2020;24:9522–9531. [DOI] [PubMed] [Google Scholar]
- 88. Wang Y, Han DM, Pan LM, Sun J. The positive feedback between lncRNA TNK2‐AS1 and STAT3 enhances angiogenesis in non‐small cell lung cancer. Biochem Bioph Res Co. 2018;507:185–192. [DOI] [PubMed] [Google Scholar]
- 89. Zhang YY, Sun JY, Qi Y, et al. Long non‐coding RNA TPT1‐AS1 promotes angiogenesis and metastasis of colorectal cancer through TPT1‐AS1/NF90/VEGFA signaling pathway. Aging‐Us. 2020;12:6191–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li YS, Zheng D, Pan LT, et al. Knockdown of TUG1 by shRNA inhibited renal cell carcinoma formation by miR‐299‐3p/VEGF axis in vitro and in vivo. Eur J Pharmacol. 2019;860:172536. [DOI] [PubMed] [Google Scholar]
- 91. Dong R, Liu GB, Liu BH, et al. Targeting long non‐coding RNA‐TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016;7:e2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yu X, Hu L, Li SY, et al. Long non‐coding RNA taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF‐1 alpha via miR‐143‐5p. Cell Death Dis. 2019;10:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen XX, Zeng KX, Xu M, et al. SP1‐induced lncRNA‐ZFAS1 contributes to colorectal cancer progression via the miR‐150‐5p/VEGFA axis. Cell Death Dis. 2018;9:982. [DOI] [PMC free article] [PubMed] [Google Scholar]


