Abstract
The function of the Saccharomyces cerevisiae Ppz1 phosphatase is controlled by its inhibitory subunit Hal3. Hal3 is a moonlighting protein, which associates with Cab3 to form a decarboxylase involved in the CoA biosynthetic pathway. Hal3 is composed by a conserved core PD region, required for both Ppz1 regulation and CoA biosynthesis, a long N‐terminal extension, and an acidic C‐terminal tail. Cab3 has a similar structure, but it is not a Ppz1 inhibitor. We show here that deletion or specific mutations in a short region of the N‐terminal extension of Hal3 compromise its function as a Ppz1 inhibitor in vivo and in vitro without negatively affecting its ability to interact with the phosphatase. This study defines a R‐K‐X(3)‐VTFS‐ sequence whose presence explains the unexpected ability of Cab3 (but not Hal3) to regulate Ppz1 function in Candida albicans. This sequence is conserved in a subset of fungi and it could serve to estimate the relevance of Hal3 or Cab3 proteins in regulating fungal Ppz enzymes. We also show that the removal of the motif moderately affects both Ppz1 intracellular relocalization and counteraction of toxicity in cells overexpressing the phosphatase. Thus, our work contributes to our understanding of the regulation of Ppz phosphatases, which are determinants for virulence in some pathogenic fungi.
Keywords: disordered regions, mutagenesis, protein phosphatase, regulatory subunit, yeasts
Mutagenesis of the N‐terminal extension of yeast Hal3 revealed a KR‐(X3)‐VTFS motif important for regulation of phosphatase Ppz1. A similar motif in Cab3, but not in Hal3, is found in most species of the CTG clade, explaining why Cab3, and not Hal3, regulates Ppz1 in C. albicans. Hal3 and Cab3 are components of PPCDC, a key enzyme in CoA biosynthesis. Thus, this motif determines the moonlighting nature of these proteins.

Abbreviations
- CoA
coenzyme A
- GFP
green fluorescent protein
- MAP kinase
mitogen‐activated protein kinase
- PP1c
catalytic subunit of protein phosphatase 1
- SGD
Saccharomyces Genome Database
Introduction
Protein phosphorylation is a widespread mechanism for regulating cellular functions. The phosphorylation status of a given protein is the result of the balanced action of protein kinases and protein phosphatases, often in response to intra or extracellular signalling cues. In addition to the well‐known role of protein kinases, the relevance of protein phosphatases in signal transmission has become evident in the last few years [1].
The Saccharomyces cerevisiae Ppz1 and Ppz2 Ser/Thr protein phosphatases define a subfamily of the PPP enzymes that are found only in fungi [2]. PPZ1 and PPZ2 are paralogous genes, with Ppz1 being the most functionally relevant isoform. Ppz1 is a 692‐residue protein composed of a C‐terminal catalytic domain (about 60% identical to Glc7, the catalytic subunit of genuine yeast PP1), and a long N‐terminal segment (~ 350 residues) [3]. Ppz1 plays diverse roles in the yeast cell, but it is crucial for the maintenance of monovalent cation homeostasis. This occurs by decreasing potassium influx by both high‐affinity Trk transporter‐dependent and ‐independent mechanisms, and by downregulating the expression of the ENA1 gene, encoding a Na+/K+‐ATPase induced by high salt and alkaline pH [4, 5, 6, 7, 8, 9]. Consequently, ppz1 mutants are hypertolerant to Na+ and Li+ cations, as well as sensitive to conditions or drugs affecting the integrity of the cell wall, such as high temperature or treatment with calcofluor white or caffeine [5, 10, 11, 12, 13].
It is known that excessive Ppz1 activity leads to a total blockage in the cell cycle at the G1‐S transition [11, 14, 15, 16, 17] and it has been demonstrated that PPZ1 is the gene for which the cell shows the lowest dosage tolerance limit [18], suggesting that Ppz1 is among the most toxic proteins in yeast when overexpressed [17, 19, 20, 21]. Consequently, Ppz1 activity must be finely regulated. In S. cerevisiae Ppz phosphatases are modulated by two regulatory subunits, Hal3 and Vhs3 [11, 22]. Both genes are paralogs, although the function of Hal3 as a Ppz regulator is more relevant in vivo than that of Vhs3.
Hal3 (also known as Sis2) was identified as a gene able to provide increased tolerance to salt stress [23], and to rescue cell cycle blockage in a Sit4‐deficient mutant when overexpressed [24]. Both effects were subsequently explained by the ability of Hal3 to bind to the catalytic C‐terminal domain of Ppz1 and inhibit its activity [11, 15]. In agreement with this regulatory role, deletion of HAL3 renders the cell hypersensitive to NaCl or LiCl, whereas overexpression of the gene provides tolerance to these cations, decreases tolerance to caffeine, and induces lysis in Slt2‐deficient cells. Overexpression of the HAL3 gene fully counteracts the toxic effects caused by high levels of Ppz1 [11, 15, 20], and it has been very recently demonstrated that this effect requires Hal3‐mediated relocalization of Ppz1 from the cell periphery to internal membranes [25].
In S. cerevisiae and other related fungi, Hal3 and Vhs3 are moonlighting proteins. At least one of them is required for the formation of an atypical heterotrimeric phosphopantothenoylcysteine decarboxylase (PPCDC) enzyme [26] and therefore, they are necessary in the biosynthesis of coenzyme A (CoA). This trimeric structure involves a third component, Cab3 (Ykl088w), which is also structurally related to Hal3 and Vhs3, in a way that the catalytic site is formed at the interface of one molecule of Hal3 or Vhs3 (providing a key catalytic His residue) and one of Cab3 (supplying a required Cys residue). It has been shown that Cab3 interacts with Ppz1 but does not regulate the phosphatase in vivo, nor inhibits the enzyme in vitro [26].
The Hal3 protein (562 amino acids) can be divided into three segments. (a) an N‐terminal region (about 250 residues), unrelated to other proteins outside yeasts, and for which a disordered structure is predicted; (b) a structured core region of similar size (named here PD region), which is responsible for the PPCDC activity and, hence, displays sequence identity with both bacterial and eukaryotic PPCDCs; and (c) a C‐terminal tail, of roughly 80 residues in length, very rich in acidic residues. Previous work determined that the conserved PD core of Hal3 is necessary for both PPCDC activity and the regulatory role on Ppz1 [27, 28, 29], whereas the acidic C‐terminal tail is dispensable for Hal3 function as a PPCDC, but is crucial for its role as a Ppz1 inhibitor [28]. Similarly, the N‐terminal domain, although not functional by itself, might contribute to stabilize the interaction between Hal3 and Cab3 when forming the PPCDC trimer. Interestingly, it was found that the PD and the N‐terminal regions were ineffective, by themselves, in inhibiting full‐length Ppz1 or its catalytic C‐terminal domain, but a polypeptide containing both regions, although very inefficient on the full‐length phosphatase, was able to substantially inhibit the activity of the catalytic Ppz1 C‐ter polypeptide [28]. This evidence hinted that the N‐terminal region of Hal3 might contribute to the function of the protein as a Ppz1 regulator.
The interest in the regulation of Ppz1 activity was increased upon reports that the phosphatase was a determinant for virulence in the pathogenic fungi Candida albicans [30] and Aspergillus fumigatus [31]. C. albicans encodes two genes resembling the Hal3/Vhs3/Cab3 triad. The protein encoded by orf19.7378 was designated as CaHal3 on the basis of its closer similarity to S. cerevisiae Hal3/Vhs3 proteins in the PD domain including the presence of the typical catalytic His in its conserved environment, whereas the product of orf19.3260 was more reminiscent of Cab3 and contained the conserved catalytic Cys [32]. CaHal3 and CaCab3 exhibit the characteristic three regions found in S. cerevisiae Hal3: a conserved PD, a long N‐terminal extension (relatively divergent), and an acidic C‐terminal tail. Remarkably, although both proteins behaved as their S. cerevisiae counterparts concerning PPCDC function, only CaCab3, but not CaHal3, was able to act as a Ppz1 inhibitor when expressed in S. cerevisiae [32]. This unexpected result was difficult to explain on the basis of the conservation of residues located at the PD region previously known to be relevant for Ppz1 regulation [27, 32].
These open questions prompted us to undertake a detailed structure–function analysis of the role of the N‐terminal region of S. cerevisiae Hal3 concerning its function as a regulator of the Ppz1 phosphatase. We demonstrate that this region plays an important role in the ability of Hal3 to inhibit Ppz1 without affecting its binding capacity to the phosphatase, Moreover, we have been able to pinpoint the relevant region to residues 90–105, where K90, R91 and the 95VTFS98 sequence are key determinants. We also show that this region is conserved in CaCab3 but not in CaHal3, and that their exchange abolishes the ability of CaCab3 to regulate S. cerevisiae Ppz1 function, thus providing a basis to explain the apparently contradictory behaviour of these proteins as Ppz1 regulators. Finally, the extension of this analysis to other fungi supports the notion that Cab3, and not Hal3, would be a moonlighting protein in most of the species belonging to the CTG clade and in a few other related families.
Results
Coarse mapping of the N‐terminal extension of Hal3 reveals a defined region required for Ppz1‐mediated function.
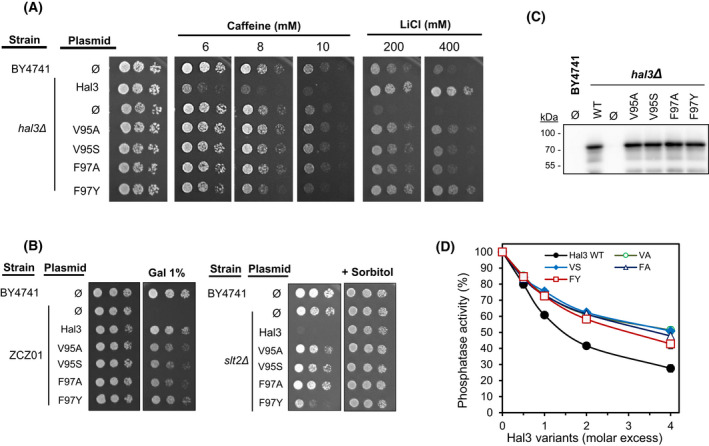
As an initial attempt to identify structural determinants in the nearly 250 residues of the N‐terminal Hal3 extension that could be relevant for its inhibitory function on the Ppz1 phosphatase, we created seven different versions bearing deletions of residues 39–51 (Δ39–51), 67–110 (and its subdeletions Δ67–93 and Δ94–110), 118–148 (Δ118–148), 209–227 (Δ209–227) and 248–256 (Δ248–256) (Fig. 1A). These specific regions were selected based on two criteria: (a) sequence conservation among different yeast species, and (b) the results of our recently reported chemical cross‐linking experiments [33], which suggested several interactions between the catalytic C‐terminal domain of Ppz1 and diverse regions within the N‐terminal Hal3 extension. These versions were cloned in plasmid pWS93, to allow multicopy expression from the ADH1 promoter, and were subsequently introduced into diverse yeast strains to test for the function of these Hal3 versions as Ppz phosphatase regulators. For comparison, we initially included in the study two previously characterized constructs producing a Hal3 version devoid of its N‐terminal domain with (PD‐Cter) or without (PD) the acidic C‐terminal tail [28]. Overexpression of native Hal3 confers higher sensitivity to caffeine and increased tolerance to toxic cations, such as LiCl. As shown in Fig. 1B, all three versions bearing total or partial deletions in the region comprising residues 67–110 did not decrease tolerance to caffeine and barely improved tolerance to LiCl, whereas the remaining constructs resulted in phenotypes very similar to those conferred by expression of native Hal3. A second test for Hal3 function was based in the ability of the regulator, when expressed in multicopy, to induce cell lysis in strains deficient in the MAP kinase Slt2. As mentioned in the Introduction, such behaviour derives from an increase in Trk1,2‐mediated influx attributed to the inhibition of Ppz1. We show in Fig. 1B (right panel) that all variants, with the exception of Δ67–110, Δ67–93 and Δ94–110, did block proliferation of the slt2Δ mutant. The constructs producing PD‐Cter (and particularly PD) showed a marked loss of function in all tests.
Fig. 1.

Coarse functional mapping of the S. cerevisiae Hal3 N‐terminal extension. (A) Schematic cartoon depicting the regions deleted in the Hal3 N‐terminal disordered domain. The right segment (in red) represents the start of the PD domain. (B) The wild‐type BY4741 strain and its hal3Δ derivative were transformed with the pWS93 vector (Ø) or the same plasmid bearing the native HAL3 gene or the indicated deletion variants. Cultured were spotted as described in material and methods on plates containing synthetic medium lacking uracil and different concentrations of caffeine or LiCl. The same constructs were introduced into cells lacking the Slt2 MAP kinase (slt2Δ, right panel). For slt2Δ cells 10% sorbitol, which acts as osmotic support, was added to the medium as positive growth control). (C) The indicated strains were transformed with pWS93‐based constructs bearing the different variants of Hal3. Protein extracts were prepared and resolved (40 μg) by SDS/PAGE. The expression levels of the various Hal3 versions were examined by immunoblot using anti‐HA antibodies. The experiment was repeated twice with different protein extracts and gave coincident results. The lower panel shows the relevant section of the gel stained with red ponceau to monitor loading and transfer efficiency. Ø denotes empty plasmid. (D) Strain ZCZ01, overexpressing PPZ1 from the strong GAL1‐10 promoter was transformed and cultures spotted as above. Induction of Ppz1 overexpression was achieved by inclusion of 1% galactose (GAL) in 2% raffinose containing plates. Pictures were taken in all cases after 3 days.
These results indicate that, because all phenotypes tested are related to the role of Hal3 as an inhibitory subunit of Ppz1, the region ranging from residue 67 to 110 must contain important determinants for such function. It can also be concluded that these determinants cannot be exclusively allocated to the 67–93 or 94–110 subregions, since both partial deletions perfectly mimic the complete one. However, we knew from previous work that, in this experimental setting, the levels of PD‐Cter and PD could be markedly lower than that of native Hal3, so there was the possibility that the inability of our full N‐terminal deletion variants to replace the functions of native Hal3 in overexpression assays could be due to lower amounts of protein. We, therefore, tested the levels of protein accumulation of native Hal3 and of all nine variants. As deduced from Fig. 1C, in contrast to the PC‐Cter and PD variants, which are produced clearly in lower amounts, all partial deletions are produced at levels similar to those of native Hal3, ruling out the possibility that the failure to mimic the function of native Hal3 could be due to insufficient levels of protein. The PC‐Cter and PD variants were not included in the subsequent experiments because their lower levels impede the proper interpretation of the obtained results.
We also took advantage of the ability of Hal3 to counteract the dramatic growth arrest suffered by cells overexpressing PPZ1. Fig. 1D illustrates that, while the strain ZCZ01 (which expresses Ppz1 from the strong GAL promoter) cannot grow when galactose is added to the medium, co‐expression with native HAL3 fully restores growth. The same beneficial effect is observed for all Hal3 variants with the exception, once more, of deletions in residues in the region spanning from 67 to 110.
We next investigated the ability of these variants to regulate Ppz1 activity in vitro. To this end, the different versions were expressed in E. coli and purified as GST‐fusions, the GST moiety removed, and the recombinant protein tested for Ppz1 inhibition. As shown in Fig. 2, the inhibitory capacity of the Δ67–93, Δ94–110 and, in particular, the Δ67–110 variant, was clearly poorer than that of native Hal3. The Δ39–51 and Δ118–148 deletions were indistinguishable from the full‐length Hal3 protein, whereas the versions with deletions affecting the segments proximal to the catalytic domain (Δ209–227 and Δ248–256) were slightly more effective as inhibitors than native Hal3. Remarkably, when the different Hal3 versions were tested with a recombinant version of Ppz1 devoid of the phosphatase N‐terminal extension, all Hal3 variants were equally powerful inhibitors of the phosphatase activity, suggesting a functional interaction between the 67–110 region of Hal3 and the N‐terminal extension of Ppz1.
Fig. 2.
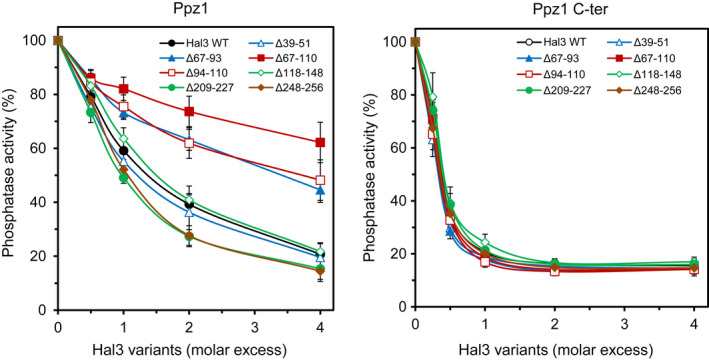
In vitro Ppz1 inhibitory activity of different Hal3 deletion variants. The Ppz1 phosphatase activity was measured as described in materials and methods using 2–10 pmols of full‐length Ppz1 or 5 pmols of the Ppz1 C‐terminal catalytic domain (Ppz1‐Cter), and pNPP as substrate. The phosphatase was pre‐incubated for 5 min at 30 °C with increasing amounts of native Hal3 or the indicated variants and the assay started by addition of the substrate. Values are means ± SE from at least four different assays and are expressed as the percentage of phosphatase activity relative to the control without inhibitor. At least two different preparations of the phosphatases and the inhibitors were used for these assays. The specific activities of Ppz1 and Ppz1‐Cter preparations were 5.05 ± 0.11 and 5.76 ± 0.09 nmol·min−1 × μg−1 respectively.
Fine mapping of the 67–110 region of Hal3
The results described above clearly identified the 67–110 segment as relevant for Hal3 function. To pinpoint the specific functional determinants within this region, we generated a set of six new smaller deletions (Fig. 3A) that were subjected to functional evaluation. As illustrated in Fig. 3B, only the deletion affecting residues 105–110 behaved as native Hal3 in both the caffeine and LiCl sensitivity tests. Interestingly, although the remaining deletions eliminated Hal3 function in the caffeine test, only the Δ90–98 and Δ97–110 changes markedly decreased the tolerance to LiCl in comparison with native Hal3, while all three deletions spanning the 67–91 region displayed a rather limited effect. When the effect of the deletions was tested in the strain deficient in SLT2, it could be observed (Fig. 3C) than all Hal3 variants, except the one bearing the Δ105‐110 deletion, were unable to abolish growth of the MAP kinase mutant. This pattern was similar to that observed for caffeine tolerance and suggests that structural elements important for both functions reside within residues 67–105. Interestingly, only the Δ97–110 deletion clearly failed to attenuate the toxicity derived from PPZ1 overexpression (with perhaps a minor effect of the Δ90–98 isoform). Immunoblot analyses of Hal3 and ZCZ01 cells demonstrated that the level of protein accumulation of all of these variants was as high as that of native Hal3 (Fig. 3D).
Fig. 3.

Functional relevance of the Hal3 67–110 N‐terminal region. (A) The sequence of the region analysed is shown. Specific deletions subjected to functional analysis are denoted by empty boxes. (B, C) Strains containing the indicated constructs were analysed as in Fig. 1. All pictures were taken after 3 days of incubation, except plates containing the slt2Δ strain (4 days). (D) Immunoblot analysis of diverse Hal3 deletions comprising residues 67–110. The indicated strains were transformed with pWS93‐based constructs bearing the different variants of Hal3. The expression levels of the various Hal3 versions were monitored by immunoblot using anti‐HA antibodies as in Fig. 1. Two independent sets of extracts were analysed with equivalent results. Ø denotes empty plasmid, WT, native Hal3. (E) The inhibitory capacity of the indicated Hal3 variants was determined using full‐length recombinant Ppz1 as in Fig. 2. Data are mean ± SE from five different assays using at least two different preparations of the phosphatase and the inhibitors.
The ability of the new versions to inhibit Ppz1 is presented in Fig. 3E. As observed, the Δ67–91 and the Δ105–110 deletions have little or no impact on the inhibitory capacity of Hal3. In contrast, deletion of regions 90–98 and 97–110 were less effective in inhibiting the activity of the phosphatase, while the other variants exhibited intermediate effects. These results confirmed that the relevant determinants for Ppz1 regulation present in the N‐terminal region of Hal3 could be restricted to a segment contained within residues 90–105.
The 90KRVPAVTFS98 region is crucial for regulation of Ppz1 function and activity
To further refine our search for specific Hal3 residues required for Ppz1 regulation, we introduced diverse mutations in the region encoding amino acids 90–105 (Fig. 4A). Phenotypic testing (Fig. 4B and C) indicates that mutations changing K90 and R91 to Glu or Gln virtually eliminated the ability of Hal3 to induce sensitivity to caffeine but barely affected its ability to confer tolerance to LiCl. Similarly, these mutations eliminated the deleterious effect of Hal3 expression in SLT2‐deficient cells. Interestingly, the Hal3 versions bearing mutations that change the 95VTFS98 and 96TFS98 amino acid sequences were largely ineffective in mimicking native Hal3 in all phenotypes tested. The combined mutation changing T96 and S98 to Ala had no effect on Hal3 function, but the change of these residues to the phosphomimetic amino acids Glu and Asp affected the behaviour of Hal3 in caffeine and in the slt2 background. Finally, the triple change of 102QQQ104 to Ala did not alter Hal3 properties for any of the phenotypes tested. These results indicated that the 95VTFS98 region was important for all Ppz1‐related Hal3 functions, while mutation of K90 and R91 showed selective effects depending on the test performed. In contrast, mutation of the triple Gln stretch had no effect. Immunoblot analysis (Fig. 4D) demonstrated that the level of all variants was at least as high as that of native Hal3. The phenotypic behaviour of the Hal3 variants was largely in agreement with their ability to inhibit Ppz1 in vitro. As shown in Fig. 4E, amino acid changes in the 95VTFS98 region strongly decreased the ability of Hal3 to inhibit Ppz1. Changes restricted to residues 96TFS98, those affecting K90 and R91, as well as the transformation of T96 and S98 in phosphomimetic residues, also resulted in partial loss of their inhibitory activity. In contrast, mutation of 102QQQ104 or T96 and S98 to Ala had virtually no effect on the in vitro inhibition of Ppz1 by Hal3.
Fig. 4.

Mapping of a functional region to Hal3 residues from 90 to 105. (A) Sequence of the region examined. Residues affected by mutations are underlined. (B, C) The phenotypic tests were performed as in Fig. 1. All pictures were taken after 3 days. (D) Immunoblot analysis of diverse changes comprising Hal3 residues 90–105. A representative experiment out of two is shown. See legend to Fig. 1 for details. (E) The ability of the different Hal3 variants to inhibit Ppz1 was tested as in Fig. 2. The mean ± SE from four different assays is shown, including at least two different preparations of Ppz1 and the Hal3 variants.
Because the 95VTFS98 and 96TFS98 mutations provided rather similar phenotypes, we wanted to know whether V95 had any relevance. Therefore, we converted V95 to Ala or Ser. As shown in Fig. 5, cells expressing these mutated forms of Hal3 did not behave as those carrying native Hal3 in the caffeine tolerance test nor when expressed in the slt2 background. They were also slightly less tolerant to LiCl and somewhat less effective than native Hal3 in protecting cells from strong overexpression of PPZ1 (Fig. 5B, left panel). We also changed F97 to Ala and Tyr. The rather conservative F97Y change had mild or no consequences on the phenotypes tested, while the mutation to Ala caused noticeable effects, with an intensity similar to that observed for the mutation of V95. The level of protein accumulation of the Hal3 variants were indistinguishable from those of the native protein (Fig. 5C). Curiously, all four HAL3 mutations yielded less powerful in vitro inhibitors of Ppz1 activity (Fig. 5D). Taken together, these results indicate that the 95VTFS98 region, and specifically residues V95 and F97 in the N‐terminal extension of Hal3, are important for the function of this protein as inhibitor of the Ppz1 protein phosphatase.
Fig. 5.
Identification of V95 and F97 as key residues in the N‐terminal extension of Hal3. (A, B) All plates were incubated for 3 days prior documentation. (C) Immunoblot analysis of the indicated variants. Two independent set of extracts were analysed with identical results. (D) Inhibition of Ppz1 is expressed as the mean ± SE from four different assays with at least two different preparations of the phosphatase and the inhibitors.
The loss of functional capacity of the Hal3 variants is not due to lack of interaction with Ppz1
The loss of the ability of the Hal3 variants to inhibit Ppz1 could be due to the inability of bound Hal3 to block the catalytic reaction or simply to the lack of binding to the phosphatase. To discern among these possibilities, we carried out an in vitro binding assay using recombinant GST‐Ppz1 bound to glutathione‐Sepharose beads and crude extracts from yeast expressing different HA‐tagged Hal3 variants. As shown in Fig. 6, most variants exhibited a binding capacity similar to that of native Hal3. Only the Δ67–110 version showed a slightly decreased interaction, but the change was not statistically significant. In contrast, variants displaying a notable incapacity to inhibit Ppz1, such as the Δ90–98 deletion or the quadruple 95VTFS98 mutation, demonstrated a clear increase in binding capacity. These results indicate that the loss of Hal3 inhibitory capacity caused by these amino acid changes is not due to their incapacity to associate to the phosphatase.
Fig. 6.

Evaluation of the physical interaction of different Hal3 versions with Ppz1. Equal amounts (6 μg) of GST‐tagged full‐length Ppz1 were immobilized on glutathione beads and used as bait to pull‐down the different variants of Hal3 expressed from the pWS93 plasmid in the strain IM021 (ppz1 hal3). After washing, beads were processed for SDS/PAGE (8% gels) and native Hal3 (WT) and its variants immunodetected by means of their HA‐tag as described in methods (upper panel). The membrane was stained prior transfer with red ponceau to assess similar recovery of the bait in the assay (middle panel). Lower panel. Quantification of the relative Hal3 vs Ppz1 amounts obtained by scanning and integration of immunoblots and recovered GST‐Ppz1 signals (upper and middle panels) from at least six experiments. Data are expressed as the mean ± SE. *P < 0.05, **P < 0.01 according to two‐tailed Student's test.
Mutation of potential Ser phosphorylation sites in the N terminus does not affect Hal3 function
According to diverse databases, Hal3 can be phosphorylated in vivo at least at 18 residues, almost without exception located within the first 250 residues. Eight of them, placed in a small cluster within residues 47 and 60 (S47, S50, S54, S55, S56, S57, T58 and S60) have been consistently identified in many proteome‐wide experiments (see SGD at https://www.yeastgenome.org/locus/S000001780/protein). With the objective to evaluate the potential role of these phosphorylation sites in the function of Hal3 we changed S47, S50, S54, S56 and S60 to both alanine and aspartic residues. These versions were cloned in the multicopy plasmid YEp195 and expressed from the native HAL3 promoter. However, tolerance tests to caffeine, LiCl or NaCl did not reveal any significant difference with the native Hal3 protein (not shown). We then also tested a double S54A S56A (and the corresponding phosphomimetic version), as well as the entire deletion of the 47–60 region, again with negative results (not shown). Therefore, we must conclude that this set of phosphorylatable residues plays no role in Hal3 function.
The ability of Hal3 N‐terminal variants to recruit overexpressed Ppz1 to internal membranes
We recently reported that the ability of Hal3 to rescue the growth defect associated with PPZ1 overexpression involved not only the capacity to inhibit the phosphatase activity but also the ability to remove Ppz1 from the cell periphery and relocate it to internal (mostly vacuolar) membranes. To test how the changes introduced in the N‐terminal extension of Hal3 could affect such ability, we transformed the MAC003 strain, a ZCZ01 derivative in which the PPZ1 coding sequence is followed by a C‐terminal GFP tag. As shown in Fig. 7 (A and B), transformation with the empty vector does not lead to changes in the subcellular localization of Ppz1, whereas the presence of native Hal3 results in recruitment of Ppz1 to internal membranes in 80% of the cells. It should be noted that this value is somewhat higher than that the previously reported [25], likely because Hal3 is expressed here from a more powerful promoter. The localization of the phosphatase in cells expressing the PD domain alone were indistinguishable from those transformed with the empty vector, whereas the same protein fragment containing the C‐terminal acidic tail (PD + CtD) provided a limited, but measurable capacity to internalize Ppz1 (about 28% of cells). Remarkably, cells expressing the Δ67–93, Δ67–110 or Δ94–110 variants showed a mild but significant difference when compared to cells expressing native Hal3, ranging from 67% to 71% of cells with internalized Ppz1. When these constructs were tested for their ability to suppress the growth defect of the MAC003 strain, we observed (Fig. 7C) that the cells expressing the Δ67–93, Δ67–110 or Δ94–110 versions exhibited a mild growth defect when compared with native Hal3, indicating partial compensation of Ppz1 toxicity. The version of Hal3 devoid of its N‐terminal extension (PD + CtD) exhibited a very strong growth defect, although still distinguishable from the complete lack of growth observed for the strain expressing only the PD domain or the empty plasmid. However, this later result must be taken with caution because of the above‐mentioned low levels of the PD + CtD and PD proteins.
Fig. 7.

The ability of Hal3 variants in recruiting overexpressed Ppz1 to internal membranes. (A) Micrographs of representative cultures of MAC003 cells (overexpressing C‐terminally GFP tagged Ppz1) bearing an empty pWS93 vector (Ø) or the same vector with native Hal3. Exposition was 3 and 1.5 s respectively. Bars represent 5 μm. (B) MAC003 cultures expressing the indicated Hal3 variants were monitored after 6 h of PPZ1 overexpression and the percentage of cells showing internalized Ppz1 was determined by blind counting of a range between 293 and 813 cells. Ns, not significant *P < 0.05; ***P < 0.001, compared with native Hal3 and determined by two‐tailed Student's test. (C) MAC003 cells were grown overnight on synthetic medium lacking uracil and then transferred to YP raffinose containing 2% galactose at initial OD600 of 0.004. Growth was monitored every 30 min. The mean ± SE from six independent cultures is shown. Ø, empty pWS93 vector.
Functional relevance of the N‐terminal region of C. albicans Cab3 for Ppz1 regulation
As mentioned in the Introduction, C. albicans encodes homologues of Hal3 and Cab3, both with large N‐terminal extensions, although only Cab3 could regulate Ppz1. We observed that CaCab3 and CaHal3 largely differ in the N‐terminal segment that we identified here to be relevant for Hal3 function in S. cerevisiae, and that CaCab3, but not CaHal3, includes the residues corresponding to R91 and the entire 95VTFS98 region (Fig. 8A). We then wondered whether these structural features could be a differential trait between CaCab3 and CaHal3 that may contribute to the distinct behaviour of both proteins with regard to Ppz1 regulation. To test this possibility, we replaced the 89–134 region of CaCab3 with the 73–90 region of CaHal3 and vice versa (Fig. 8A), introduced the hybrid constructs in wild‐type cells as well as hal3 and slt2 deletion mutants, and subjected these cells to phenotypic tests. Expression of CaCab3 had little impact on caffeine sensitivity but, as previously reported [32], it markedly improved tolerance to LiCl in a hal3Δ strain (Fig. 8B). We also observed a significant effect on growth in the slt2Δ background. Interestingly, these phenotypic traits are lost in the hybrid CaCab3 version carrying the CaHal373–90 region. However, introduction of the CaCab389–134 segment into CaHal3 resulted in a behaviour identical to that of native CaHal3 (or the empty vector). In any case, the lack of effect of the hybrid proteins cannot be attributed to a deficient expression, since immunoblot experiments demonstrated that their protein levels in the hal3 (Fig. 7C) or wild‐type strains (not shown) were similar or even higher than that of native S. cerevisiae Hal3 or CaCab3. These results indicate that the conserved region found relevant in S. cerevisiae for regulation of S. cerevisiae Ppz1 function is also important in CaCab3. However, it is not sufficient to transform CaHal3 into a functional regulator of ScPpz1, suggesting that additional determinants are needed.
Fig. 8.

Exchanging relevant N‐terminal regions between C. albicans CaCab3 and CaHal3. (A) Sequence comparison between the 60–111 region of S. cerevisiae Hal3 (NP_012998.1) with the corresponding regions of CaCab3 (XP_717950.1) and CaHal3 (XP_716619.1). Protein sequences were obtained from the NCBI protein databank and alignments were generated with Clustal omega. Residues found in this work to be relevant for the Hal3 regulatory function on Ppz1 are displayed in yellow background. The segments exchanged between CaCab3 and CaHal3 are highlighted in bold and the corresponding region in ScHal3 in italics. B) The different constructs were introduced into the wild‐type BY4741 strain and its hal3 and slt2 derivatives and plated on synthetic medium lacking uracil and supplemented with 8 mM caffeine or 200 mM LiCl (for the hal3 background) or in the presence or absence of 10% sorbitol (for the slt2 background). Plates were incubated in all cases for 3 days. C) The indicated constructs were introduced in the hal3 strain, protein extracts prepared and the level of expression of the native and hybrid proteins was assessed by immunoblot using anti‐HA antibodies. The lower panel shows red ponceau staining of the membrane. Two independent sets of extracts were analysed with similar results.
The N‐terminal traits identified in Hal3 as functionally relevant for Ppz1 regulation are conserved only in a subset of yeast species
The analysis of fungal genomes for the presence of Hal3 proteins displaying an N‐terminal extension preceding the PD domain revealed that this feature was, with very few exceptions, restricted to the order Saccharomycetales (see Discussion). However, even within this subset of species the overall level of identity in this region was rather poor. Therefore, based on our experimental data, we analysed the N‐terminal region of 83 Hal3/Vhs3 and Cab3‐like proteins from 35 species for the presence of the R/K‐R/K‐X3‐VTFS signature or degenerate variants (Fig. 9). Remarkably, all members of the Saccharomycetaceae (NCBI:txid4893) and Saccharomycodaceae (NCBI:txid34365) families examined present the canonical K‐R‐X3‐VTFS signature in Hal3 (and in Vhs3, when the HAL3 paralog exists), whereas in Cab3 only a limited version of the consensus, always lacking the basic residues, is found. In contrast, all members of the CTG‐clade examined, with the exception of Meyerozyma guilliermondii, consistently exhibit slightly degenerate versions of the [K‐X4‐V‐(ST)‐F and R/K‐X3‐V‐(ST)‐F] motifs in Cab3, but not in Hal3. Similar sequences in Cab3 are also found in the Phaffomycetaceae, Pichiaceae and Dipodascaceae families, in some cases with Val substituted by Ile.
Fig. 9.

Identification of the regulatory consensus sequence in diverse clades of the order Saccharomycetales. The phylogeny of the selected species was based in the alignment provided in reference [51] and represented using MEGA X software [52]. The identification of the consensus sequences was done by inspection of Clustal Omega alignments and search of FASTA files for degenerate combinations with in‐house developed software. Key consensus residues are denoted in bold. The anomalous sequence in P. kudriavzevii is shaded. An asterisk denotes that the sequence found in the database appears N‐terminally incomplete. (A), Phaffomycetaceae; (B), Pichiaceae; (C), Dipodascaceae.
Discussion
The fact that Ppz1 is a virulence determinant in some pathogenic fungi [30, 31], together with the unusual intolerance of S. cerevisiae for higher‐than‐normal levels of this phosphatase, has stirred the interest in its regulation and, in particular, in the role played by its inhibitory subunit Hal3. Previous work revealed that, although the isolated Hal3 N‐terminal segment had no function by itself, it could contribute to the regulation of Ppz1 [28]. We show here that, indeed, this region is vital for regulation of Ppz1. This is in keeping with the very recent evidence obtained by chemical cross‐linking showing that the vast majority of interactions detected between the catalytic C‐terminal domain of Ppz1 and Hal3 involve residues located within this N‐terminal extension [33].
Our results indicate an overall correlation between the failure of the diverse variants to mimic native Hal3 phenotypes in vivo and the loss of capacity to act as Ppz1 inhibitors in vitro. We also show that the inability to inhibit Ppz1 properly is not caused by defective physical interaction between both proteins, which is actually enhanced in some cases such as the Δ90–98 deletion or the mutations affecting the 95VTFS98 region. The irrelevance of the R/K‐R/K‐X3‐VTFS motif for the interaction between Ppz1 and Hal3 fits with the earlier observations that ScCab3 and CaHal3, both devoid of such motif, still interact with ScPpz1 or CaPpz1 [26, 32]. Therefore, our data indicate that this short region is somehow involved in the inhibitory mechanism. In this regard, it is suggestive that the differential in vitro effect on the inhibitory activity observed for the large Hal3 deletions on full‐length Ppz1 (Fig. 2A) is not detected when the catalytic C‐terminal domain is employed. In this case, all variants behave as strong inhibitors, indistinguishable from the native Hal3 protein. These results suggest that specific interactions between the N‐terminal domains of Ppz1 and Hal3 might take place, and that these interactions could modulate the capacity of Hal3 as Ppz1 inhibitor. This hypothesis would fit with the early notion that the N‐terminal extension of Ppz1 protects the phosphatase from excessive Hal3 inhibition [11].
We show here that the impact of some deletions or specific mutations in the Hal3 N‐terminal region is different depending on the phenotype tested. Thus, all three deletions involving the 67–91 region result in a very severe loss of function when cells are tested in caffeine, but only a mild effect when challenged with LiCl (Fig. 3B). Likewise, the T96E S98D variant behaves as native Hal3 in LiCl tolerance while it is largely inefficient when tested in media supplemented with caffeine. In fact, disparate phenotypes have been previously reported for specific mutations affecting Hal3's PD core [27, 34], or for the expression of the Schizosaccharomyces pombe SpHal3 protein (which lacks an N‐terminal extension) in budding yeast [35]. It is worth noting that it has been reported that specific deletions of the N‐terminal extension of S. cerevisiae Ppz1 display a differential phenotype‐specific behaviour [14]. This would argue in favour of a functional role of the N‐terminal extension of Hal3 in modulating Ppz1 function, as well as supporting a functional interplay between these N‐terminal moieties. In any case, we observe here a perfect phenotypic correlation between caffeine sensitivity and the growth test performed in the slt2Δ background. We interpret this as the result of the described impact of caffeine on the cell wall integrity pathway (see [36] and references therein).
Our experiments defined residues 90–105, highlighting K90 or R91 (or both), and the 95VTFS98 region as important functional determinants for the inhibition of the Ppz1 phosphatase by the Hal3 protein. We also selected for analysis the diverse Ser residues located within the 47–60 segment. There were several reasons for this choice. This region contains 5 of the 18 phosphorylatable residues experimentally found in Hal3 (of which 16 are located within the first 250 amino acids), and these residues consistently appear as phosphorylated in most public data sets. More importantly, it was recently reported that Hal3 S54 and S56 were specifically dephosphorylated in cells treated with acetic acid [37], suggesting that these phosphorylation sites might be responsive to alterations in the intracellular pH. Because of the known role of Ppz1 in monovalent cation homeostasis and intracellular pH maintenance [4, 5, 7, 21] we considered that this might be a hint for a role of this phosphosites in Ppz1 regulation. However, the independent mutation of any of these Ser residues to Ala (non‐phosphorylatable) or Asp (phosphomimetic), including the simultaneous change of S54 and S56, as well as the entire deletion of this region, did not alter the properties of Hal3. Therefore, we propose that this specific cluster plays no role in the regulation of Ppz1 in vivo.
It has been very recently demonstrated that the counteraction of the toxic effects of Ppz1 overexpression by Hal3 involves the relocation of the phosphatase from the cell periphery to internal membranes [25]. We show here that particular deletions at the N‐terminal extension of Hal3 that greatly impair both the ability to inhibit Ppz1 in vitro and its function in vivo, only moderately weakens their ability to relocate Ppz1 and to compensate the growth defect caused by an excess of Ppz1 (Fig. 7). This suggest that the functional region identified here has relatively little relevance concerning subcellular redistribution of overexpressed Ppz1. As mentioned above, the observation that the versions of Hal3 devoid of the entire N‐terminal region fail to promote Ppz1 internalization could be simply due to the markedly lower levels of these variants. This might also reflect a role of the N‐terminal region of Hal3 in the stabilization of the polypeptide. In any case, our results further confirm the relevance of intracellular redistribution in the neutralization of the deleterious effects derived from excess of Ppz1 activity.
The comparison of Hal3‐like fungal proteins indicates that an N‐terminal extension preceding the PD domain is found only in the genome of Saccharomycetes. Exceptions to this rule appear only in the Exobasidiomycetes and Ustilaginomycetes clades (see below). In other ascomycota classes, the protein is reduced to the PD domain and often lacks the acidic tail. Our work demonstrating the functional relevance of a specific signature present in the N‐terminal region in S. cerevisiae provides an explanation as to why the S. pombe SpHal3 protein, which lack the N‐terminal extension, or the C. neoformans CnHal3a or CnHal3b proteins (with a very short N‐terminal region) were ineffective in inhibiting their own Ppz enzymes or the S. cerevisiae Ppz1 [35, 38]. Similarly, the Hal3 protein from the Ustilaginomycete Ustilago maydis, which contains a very large N‐terminal extension but lacks the above‐mentioned signature, cannot inhibit the UmPpz1 or ScPpz1 phosphatases [39]. It must be stressed that the failure of these Hal3 proteins to inhibit their corresponding Ppz1 enzymes cannot be attributed to structural differences in the phosphatases, because all of them were potently inhibited by ScHal3 [32, 35, 38, 39]. Taken together with the results shown here concerning CaCab3 mutagenesis, these data reinforce the notion that the R/K‐R/K‐X3‐VTFS motif, or slightly degenerated versions of it, could be an important structural feature for the role of Hal3 as regulator of the Ppz1 phosphatase. Our comparative analysis also reveals two scenarios: (1) the regulatory consensus is present in Hal3 (but not in Cab3) in the Saccharomycetaceae and Saccharomycodaceae families; (2) it is found in Cab3 (but not in Hal3) not only in C. albicans and in some other pathogenic Candida species [40], but the vast majority of the members of the CTG‐clade, plus in the examples of the Phaffomycetaceae, Pichiaceae and Dipodascaceae families examined. The genomes of all these species encode both Hal3 and Cab3 proteins, so it is expected that the PPCDC enzyme will be a heterotrimer involving both proteins. Because our work associates the presence of the consensus with the ability to regulate Ppz1, it can be concluded that, in the first scenario, the moonlighting capacity corresponds to Hal3, whereas in the second case, Cab3 is the moonlighting protein.
Our results raise a key question: why is the R/K‐R/K‐X3‐VTFS motif important in regulating Ppz1 function? Although this sequence bears some similarity with the RVXF motif found in many PP1c regulatory subunits [41, 42], in our case the basic residues are more distant from the VXF core. In addition, the RVXF motif has been described as necessary for the interaction of the regulators with PP1c, whereas in our case it is dispensable for interaction. Therefore, our data do not support the notion that the R/K‐R/K‐X3‐VTFS motif is replacing the widespread RVXF motif, but that it would have a Ppz1‐specific role. In any case, our results show that the presence of a bulky residue at position F97 is important, as the mutation to Ala has a stronger effect than the change to Tyr (Fig. 5B). In contrast, in the case of V95, decreasing the size of the residue to Ala or its hydrophobicity (by mutation to Ser) resulted in a loss of function of similar potency. It is suggestive that this motif lies in a small segment for which a strong prediction for a disordered binding region has been observed (Fig. 10). Such regions have been reported to be extensively involved in regulatory and signalling functions [43]. It could be hypothesized that the interaction of these residues with the catalytic domain of Ppz1 may contribute to the appropriate positioning of Hal3 to effectively impair the catalytic reaction. In fact, our results showing that the Δ67–93, Δ94–110 and Δ67–110 variants could not block full‐length Ppz1 activity, but that they were as effective as the native Hal3 protein in inhibiting the C‐terminal Ppz1 catalytic domain, suggest that these interactions might involve residues of the Ppz1 N‐terminal extension. This extension has been also predicted as a disordered region, not only in S. cerevisiae but also in many other fungi [19] and, in the case of S. cerevisiae, to strongly determine the properties of the phosphatase [14, 19].
Fig. 10.

Prediction of intrinsically disordered (IDRs) and folding upon binding regions for the N‐terminal region of S. cerevisiae Hal3. Prediction of IDRs according the IUPred2 software is denoted in red, and prediction of disordered binding regions based in the Anchor2 software in blue. The 0.5 cut‐off (discontinuous line) corresponds to 5% false positive prediction on IDRs or ordered protein segments. The cartoon on the top depicts the position of the major deletions described in our work, as well as the location of the KR‐X3‐VTFS motif. The N‐terminal extension is shown in light blue.
Materials and methods
Yeast strains and growth conditions
Saccharomyces cerevisiae cells were grown at 28 °C in YP medium (10 g·L−1 yeast extract, 20 g·L−1 peptone) or in synthetic medium (SC) lacking the appropriate requirements when carrying plasmids for selection [44], supplemented with a carbon source (20 g·L−1) such as glucose (Glu, as in YPD), raffinose (Raff) or galactose (Gal), or as indicated. All yeast strains used in this work are listed in Table 1. Growth on plates was tested by spotting cells at an initial OD600 of 0.05, plus two 1/5 serial dilutions. Growth assays in liquid media were done as in [20], except that measures were made every 30 min in a Bioscreen C apparatus (Thermo Labsystems, Madrid, Spain), with shaking for 7 min before each measurement.
Table 1.
Yeast strains used in this work.
| Strain | Genotype | References |
|---|---|---|
| BY4741 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | [53] |
| BY4741 hal3Δ | BY4741 hal3::KANMx | [53] |
| BY4741 slt2Δ | BY4741 slt2::KANMx | [53] |
| ZCZ001 | BY4741 pGAL1:PPZ1: kanMX6 | [20] |
| MAC003 | BY4741 pGAL1:PPZ1‐EGFP:HISMX6 | [25] |
| IM021 | MAT a ura3‐52 leu2‐3, 112 trp1‐1 his4 can‐1r ppz1::KANMX hal3::LEU2 | [27] |
DNA techniques and plasmid constructions
Escherichia coli (strain DH5α) was used as standard plasmid DNA host and was grown in LB medium at 37 °C supplemented with 50 μg·mL−1 ampicillin when needed for plasmid selection. E. coli strain BL21 (DE3) RIL was used for heterologous protein expression as described below.
The diverse variants of S. cerevisiae Hal3 were made by 2‐step PCR, using pGEX‐6P‐1‐ScHal3 as template and the oligonucleotides described in Table S1. After the overlapping PCR step the PCR products and the pGEX‐6P‐1‐ScHal3 vector were cut at the indicated restriction sites and ligated (Table S2). These constructs were used to produce in E. coli the diverse protein variants of Hal3. The construction of vectors for expression in E. coli of GST‐fused full length Ppz1 and the Ppz1‐Cter (Δ1–344) catalytic moiety has been previously described [22]. For expression in yeast, the Hal3 variants generated in pGEX‐6P‐1 were digested with EcoRI and XhoI, and cloned into the high copy‐number plasmid pWS93, which provide a C‐terminal 3xHA tag [45], linearized with EcoRI and SalI. Construction of plasmid pWS93‐ScHAL3 was reported earlier [26].
The C. albicans hybrid proteins were generated from the pre‐existing CaHal3 and CaCab3 genes cloned in pGEX‐6P‐1 [32]. These constructs were used as templates to amplify overlapping fragments with the oligonucleotides pairs 5′ CaHal3 megaprimer/3′ CaHal3 megaprimer and 5′ CaCab3 Megaprimer/3′ CaCab3 Megaprimer, respectively. In a second step, the CaHal3 fragment was reamplified using primers pGEX 5′ sequencing Primer and 3′ CaHal3 + 80Bsu15I, whereas for the CaCab3 fragment the primers employed were pGEX 5′ sequencing Primer and 3′ CaCab3 + 100Bst1107I. Each pair of overlapping fragments were amplified with oligonucleotides pGEX 5′ sequencing Primer and 3′ CaHal3 + 80Bsu15I (for Cahal3) or 3′ CaCab3 + 100Bst1107I (for CaCab3). The former amplification fragment was inserted in pGEX‐CaHal3 by EcoRI/Bsu15I digestion, whereas the latter was cloned in pGEX‐CaCab3 by EcoRI/Bst1107I digestion. Once in pGEX‐6P‐1, both hybrid genes were transferred to pWS93 for expression in yeast by digestion with EcoRI/KspAI.
To perform point substitutions in HAL3 affecting phosphorylatable Ser in the 47–60 region, a Quickchange site‐directed mutagenesis method [46] was carried out using YEplac195 HAL3, which contains the HAL3 gene (2.4 Kbp) cloned at EcoRI/HindIII sites, as a template [27]. The amplification of the entire template was done with Q5 High‐Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA), at 1 min·kb−1 of extension time and final reaction volume of 50 μL, with the appropriate oligonucleotides described in Table S1. The PCR product was purified with NucleoSpin Gel and PCR Clean‐Up kit (Macherey‐Nagel, Düren, Germany) and was eluted in 30 μL. Finally, the PCR template was digested with DpnI (FastDigest, Thermo Scientific, Waltham, MA, USA) and 1/3 of the PCR product was used to transform E. coli.
Deletion of the 47–60 region was made by two parallel PCR amplifications using YEplac195 Hal3 as template and the pairs of oligonucleotides 5‐Hal3‐200 / 3‐Hal3delta47_60, and 5‐Hal3delta47_60 / 3‐Hal3+920. Both PCR product were diluted, mixed and used as template for a further amplification with the 5‐Hal3‐200 / 3‐Hal3+920 pair of primers. The resulting fragment (about 920 bp) was digested with XbaI and BamHI and cloned in these same sites in YEplac195 HAL3. Oligonucleotides employed are described in Table S1.
The construction of pWS93‐PD and pWS93‐PD + CtD can be found in [28].
Yeast protein extraction and immunoblot
The appropriate strains were grown to exponential phase in SC medium lacking uracil (10 mL, OD600 = 0.8) with glucose as carbon source (except for the ZCZ01 strain, which was grown on raffinose). Cells were collected by centrifugation at 4 °C, washed twice with cold water and frozen at −20 °C until use. Protein extraction, SDS/PAGE and immunoblot analysis were performed as in [34].
Fluorescence microscopy
MAC003 cells carrying the plasmids of interest were grown overnight to saturation in synthetic media with glucose lacking uracil. Then, cells were diluted to OD600 = 0.004 in fresh synthetic media without uracil containing 2% raffinose, instead of glucose, and growth resumed at 28 °C until OD600 = 0.6 was reached. At this point, galactose was added to the culture at a final concentration of 2%, to induce Ppz1 overexpression. Cell monitoring was performed using a μ‐Slide 8 Well (Ibidi GmbH, Gräfelfing, Germany) as described in [25] after 6 h of Ppz1 overexpression. Pictures were taken using a Nikon Eclipse TE‐2000E fluorescence microscope.
Recombinant expression of proteins in E. coli
All recombinant proteins were expressed in E. coli BL21 (DE3) RIL cells as described for Ppz1 in [47] using Terrific Broth (tryptone 12 g·L−1, 24 g·L−1 yeast extract, 4 mL·L−1 glycerol, 17 mm KH2PO4, 72 mm K2HPO4). Affinity‐purification and removal of the GST‐moiety by treatment with PreScission protease (GE Healthcare, Chicago, IL, USA) was done as in [34]. Quantification of the purified proteins was carried out by SDS/PAGE followed by Coomassie Blue staining and integration of the relevant bands with the Gel Analyser software using a bovine serum albumin calibration curve as reference.
Detection of Ppz1–Hal3 interactions.
The interaction between the diverse variants of Hal3 and Ppz1 was evaluated in vitro by incubating aliquots (∼ 100 μL) of the glutathione‐agarose beads containing 6 μg of GST‐Ppz1 with protein extracts (600 μg) prepared from strain IM021 (ppz1 hal3) transformed with plasmid pWS93 (negative control) and with the same plasmid carrying the diverse HA‐tagged version of Hal3 as in [28, 34]. Quantification of the level of interaction was done by determining the ratio between the immunoreactive Hal3 signal and the corresponding Ponceau Red stained phosphatase bait and taking the value for native ScHal3 as the unit. At least two different preparations of the phosphatase and of each Hal3 variant were used in the assays.
Assay of protein phosphatase activity.
The ability of the different versions of Hal3 to inhibit in vitro full‐length Ppz1 or its catalytic domain was evaluated using recombinant proteins produced in E. coli as GST fusions after removal of the GST moiety. To this end, the Ppz1 phosphatase activity was measured with 10 mm p‐nitrophenylphosphate (pNPP) as substrate and using 2–10 pmols of full‐length Ppz1 or 1–5 pmols of Ppz1‐Cter essentially as described previously [34, 48].
Other methods
To select the set of Hal3‐like proteins for N‐terminal comparisons, a BLASTP analysis was performed with both the entire sequence and residues 1–250 of S. cerevisiae Hal3 (NP_012998.1) at the NCBI non‐redundant protein database, excluding Saccharomyces cerevisiae (taxid:4932). The full sequences of fungal Hal3‐like proteins were aligned using the Clustal Omega algorithm of the EMBL‐EBI server [49]. Assignation to homologues of ScHal3/ScVhs3 or ScCab3 was done according to the p‐values of the BLASTP analysis followed by identification in Cab3 of the conserved Cys residue in the substrate recognition clamp as well as the canonical PXMNXXMW motif [26]. The prediction of intrinsically disordered regions (IDRs) was done with the iupred2 software (with the “short disordered regions” setting) and disordered binding regions were identified using the ANCHOR2 prediction algorithm [50].
Conflict of interest
The authors declare no conflict of interest.
Author contributions
JA and AC planned the experiments; CS performed most experiments; MA and DV performed some experiments and contributed with reagents. JA, AC, CS and MA analysed the data; JA wrote the paper, and all authors contributed to its edition.
Supporting information
Table S1. Oligonucleotides used in this work.
Table S2. PCR amplifications and cloning strategies.
Acknowledgements
The technical assistance of Montserrat Robledo is acknowledged. We thank J. Piñol for allowing access to microscopy facilities and L. Yenush for critical review of the manuscript. We also thank Prof I. Sa Correia (Univ. of Lisbon) for sharing with us information about acetic acid‐induced phosphorylation changes in Hal3 before publication. This research was funded by the Ministerio de Economía, Industria y Competitividad, Spain, grant number BFU2017‐82574‐P, to JA and AC. MA, CS and DV were recipients of PIF fellowships from the Universitat Autònoma de Barcelona, Spain. The funding source(s) was not involved in the study design, the collection, analysis and interpretation of data, the writing of the report or in the decision to submit the article for publication.
Data availability statement
The data that supports the findings of this study are available in Figures 1 to 10, and Table1 and in the supplementary material of this article. Any other data or aditional information are available from the corresponding author [joaquin.arino@uab.es] upon reasonable request.
References
- 1. Brautigan DL. Protein ser/Thr phosphatases–the ugly ducklings of cell signalling. FEBS J. 2013;280:324–5. [DOI] [PubMed] [Google Scholar]
- 2. Ariño J, Velázquez D, Casamayor A. Ser/Thr protein phosphatases in fungi: structure, regulation and function. Microb Cell. 2019;6:217–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Posas F, Casamayor A, Morral N, Ariño J. Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino‐terminal region. J Biol Chem. 1992;267:11734–40. [PubMed] [Google Scholar]
- 4. Yenush L, Merchan S, Holmes J, Serrano R. pH‐responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1‐related Ppz1 phosphatase. Mol Cell Biol. 2005;25:8683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Posas F, Camps M, Ariño J. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J Biol Chem. 1995;270:13036–41. [DOI] [PubMed] [Google Scholar]
- 6. Ruiz A, Yenush L, Ariño J. Regulation of ENA1 Na+‐ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot Cell. 2003;2:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yenush L, Mulet JM, Ariño J, Serrano R. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 2002;21:920–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruiz A, Ruiz MC, Sanchez‐Garrido MA, Arino J, Ramos J. The Ppz protein phosphatases regulate Trk‐independent potassium influx in yeast. FEBS Lett. 2004;578:58–62. [DOI] [PubMed] [Google Scholar]
- 9. Ruiz A, Ariño J. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot Cell. 2007;6:2175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee KS, Hines LK, Levin DE. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1‐related protein phosphatases function within the PKC1‐mediated pathway. Mol Cell Biol. 1993;13:5843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Nadal E, Clotet J, Posas F, Serrano R, Gomez N, Ariño J. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p ser/Thr protein phosphatase. Proc Natl Acad Sci USA. 1998;95:7357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merchan S, Bernal D, Serrano R, Yenush L. Response of the Saccharomyces cerevisiae Mpk1 mitogen‐activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot Cell. 2004;3:100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Posas F, Casamayor A, Ariño J. The PPZ protein phosphatases are involved in the maintenance of osmotic stability of yeast cells. FEBS Lett. 1993;318:282–6. [DOI] [PubMed] [Google Scholar]
- 14. Clotet J, Posas F, De Nadal E, Arino J. The NH2‐terminal extension of protein phosphatase PPZ1 has an essential functional role. J Biol Chem. 1996;271:26349–55. [DOI] [PubMed] [Google Scholar]
- 15. Clotet J, Garí E, Aldea M, Ariño J. The yeast Ser/Thr phosphatases Sit4 and Ppz1 play opposite roles in regulation of the cell cycle. Mol Cell Biol. 1999;19:2408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stevenson LF, Kennedy BK, Harlow E. A large‐scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci USA. 2001;98:3946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Velázquez D, Albacar M, Zhang C, Calafí C, López‐Malo M, Torres‐Torronteras J, et al. Yeast Ppz1 protein phosphatase toxicity involves the alteration of multiple cellular targets. Sci Rep. 2020;10:15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Makanae K, Kintaka R, Makino T, Kitano H, Moriya H. Identification of dosage‐sensitive genes in Saccharomyces cerevisiae using the genetic tug‐of‐war method. Genome Res. 2013;23:300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calafí C, López‐Malo M, Albacar M, Casamayor A, Ariño J. The N‐terminal region of yeast protein phosphatase Ppz1 is a determinant for its toxicity. Int J Mol Sci. 2020;21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calafí C, López‐Malo M, Velázquez D, Zhang C, Fernández‐Fernández J, Rodríguez‐Galán O, et al. Overexpression of budding yeast protein phosphatase Ppz1 impairs translation. Biochim Biophys Acta Mol cell Res. 2020;1867:118727. [DOI] [PubMed] [Google Scholar]
- 21. Albacar M, Sacka L, Calafí C, Velázquez D, Casamayor A, Ariño J, et al. The toxic effects of Ppz1 overexpression involve Nha1‐mediated deregulation of K+ and H+ homeostasis. J Fungi (Basel). 2021;7:1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruiz A, Muñoz I, Serrano R, Gonzalez A, Simon E, Arino J. Functional characterization of the Saccharomyces cerevisiae VHS3 gene: a regulatory subunit of the Ppz1 protein phosphatase with novel, phosphatase‐unrelated functions. J Biol Chem. 2004;279:34421–30. [DOI] [PubMed] [Google Scholar]
- 23. Ferrando A, Kron SJ, Rios G, Fink GR, Serrano R. Regulation of cation transport in Saccharomyces cerevisiae the salt tolerance gene HAL3 . Mol Cell Biol. 1995;15:5470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Di Como CJ, Bose R, Arndt KT. Overexpression of SIS2, which contains an extremely acidic region, increases the expression of SWI4, CLN1 and CLN2 in sit4 mutants. Genetics. 1995;139:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albacar M, Velázquez D, Casamayor A, Ariño J. The toxic effects of yeast Ppz1 phosphatase are counteracted by subcellular relocalization mediated by its regulatory subunit Hal3. FEBS Lett, in press. 2022;596:1556–66. [DOI] [PubMed] [Google Scholar]
- 26. Ruiz A, Gonzalez A, Munoz I, Serrano R, Abrie JA, Strauss E, et al. Moonlighting proteins Hal3 and Vhs3 form a heteromeric PPCDC with Ykl088w in yeast CoA biosynthesis. Nat Chem Biol. 2009;5:920–8. [DOI] [PubMed] [Google Scholar]
- 27. Munoz I, Ruiz A, Marquina M, Barcelo A, Albert A, Arino J. Functional characterization of the yeast Ppz1 phosphatase inhibitory subunit Hal3: a mutagenesis study. J Biol Chem. 2004;279:42619–27. [DOI] [PubMed] [Google Scholar]
- 28. Abrie JAA, González A, Strauss E, Ariño J. Functional mapping of the disparate activities of the yeast moonlighting protein Hal3. Biochem J. 2012;442:357–68. [DOI] [PubMed] [Google Scholar]
- 29. Abrie JA, Molero C, Ariño J, Strauss E. Complex stability and dynamic subunit interchange modulates the disparate activities of the yeast moonlighting proteins Hal3 and Vhs3. Sci Rep. 2015;5:15774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ádám C, Erdei É, Casado C, Kovács L, González A, Majoros L, et al. Protein phosphatase CaPpz1 is involved in cation homeostasis, cell wall integrity and virulence of Candida albicans . Microbiology. 2012;158:1258–67. [DOI] [PubMed] [Google Scholar]
- 31. Manfiolli AO, de Castro PA, Dos Reis TF, Dolan S, Doyle S, Jones G, et al. Aspergillus fumigatus protein phosphatase PpzA is involved in iron assimilation, secondary metabolite production, and virulence. Cell Microbiol. 2017;19:e12770. [DOI] [PubMed] [Google Scholar]
- 32. Petrényi K, Molero C, Kónya Z, Erdődi F, Ariño J, Dombrádi V. Analysis of two putative Candida albicans Phosphopantothenoylcysteine decarboxylase/protein phosphatase Z regulatory subunits reveals an unexpected distribution of functional roles. PLoS One. 2016;11:e0160965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Casamayor A, Velázquez D, Santolaria C, Albacar M, Rasmussen MI, Højrup P, et al. Comparative analysis of type 1 and type Z protein phosphatases reveals D615 as a key residue for Ppz1 regulation. Int J Mol Sci. 2022;23:1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santolaria C, Velázquez D, Strauss E, Ariño J. Mutations at the hydrophobic core affect Hal3 trimer stability, reducing its Ppz1 inhibitory capacity but not its PPCDC moonlighting function. Sci Rep. 2018;8:14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Molero C, Petrényi K, González A, Carmona M, Gelis S, Abrie JA, et al. The Schizosaccharomyces pombe fusion gene hal3 encodes three distinct activities. Mol Microbiol. 2013;90:367–82. [DOI] [PubMed] [Google Scholar]
- 36. Ruta LL, Farcasanu IC. Saccharomyces cerevisiae and caffeine implications on the eukaryotic cell. Nutrients. 2020;12:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerreiro JF, Mira NP, Santos AXS, Riezman H, Sá‐Correia I. Membrane phosphoproteomics of yeast early response to acetic acid: role of Hrk1 kinase and lipid biosynthetic pathways, in particular sphingolipids. Front Microbiol. 2017;8:1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang C, García‐Rodas R, Molero C, de Oliveira HC, Tabernero L, Reverter D, et al. Characterization of the atypical Ppz/Hal3 phosphatase system from the pathogenic fungus Cryptococcus neoformans . Mol Microbiol. 2019;111:898–917. [DOI] [PubMed] [Google Scholar]
- 39. Zhang C, de la Torre A, Pérez‐Martín J, Ariño J. Protein phosphatase Ppz1 is not regulated by a Hal3‐like protein in plant pathogen Ustilago maydis . Int J Mol Sci. 2019;20:3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szabó K, Miskei M, Farkas I, Dombrádi V. The phosphatome of opportunistic pathogen Candida species. Fungal Biol Rev. 2021;35:40–51. [Google Scholar]
- 41. Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase‐1. J Biol Chem. 2003;278:18817–23. [DOI] [PubMed] [Google Scholar]
- 42. Casamayor A, Ariño J. Controlling ser/Thr protein phosphatase PP1 activity and function through interaction with regulatory subunits. Adv Protein Chem Struct Biol. 2020;122:231–88. [DOI] [PubMed] [Google Scholar]
- 43. Yang J, Gao M, Xiong J, Su Z, Huang Y. Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding. Protein Sci. 2019;28:1952–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in yeast genetics: a cold Spring Harbor Laboratory course manual. Suffolk County, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 45. Song W, Carlson M. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 1998;17:5757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu H, Naismith JH. An efficient one‐step site‐directed deletion, insertion, single and multiple‐site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molero C, Casado C, Ariño J. The inhibitory mechanism of Hal3 on the yeast Ppz1 phosphatase: a mutagenesis analysis. Sci Rep. 2017;7:8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garcia‐Gimeno MA, Munoz I, Arino J, Sanz P. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J Biol Chem. 2003;278:47744–52. [DOI] [PubMed] [Google Scholar]
- 49. Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL‐EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Erdős G, Dosztányi Z. Analyzing protein disorder with IUPred2A. Curr Protoc Bioinformatics. 2020;70:e99. [DOI] [PubMed] [Google Scholar]
- 51. Mühlhausen S, Kollmar M. Molecular phylogeny of sequenced Saccharomycetes reveals polyphyly of the alternative yeast codon usage. Genome Biol Evol. 2014;6:3222–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotides used in this work.
Table S2. PCR amplifications and cloning strategies.
Data Availability Statement
The data that supports the findings of this study are available in Figures 1 to 10, and Table1 and in the supplementary material of this article. Any other data or aditional information are available from the corresponding author [joaquin.arino@uab.es] upon reasonable request.


