Abstract
The processing of the Coronavirus polyproteins pp1a and pp1ab by the main protease Mpro to produce mature proteins is a crucial event in virus replication and a promising target for antiviral drug development. Mpro cleaves polyproteins in a defined order, but how Mpro and/or the polyproteins determine the order of cleavage remains enigmatic due to a lack of structural information about polyprotein-bound Mpro. Here, we present the cryo-EM structures of SARS-CoV-2 Mpro in an apo form and in complex with the nsp7-10 region of the pp1a polyprotein. The complex structure shows that Mpro interacts with only the recognition site residues between nsp9 and nsp10, without any association with the rest of the polyprotein. Comparison between the apo form and polyprotein-bound structures of Mpro highlights the flexible nature of the active site region of Mpro, which allows it to accommodate ten recognition sites found in the polyprotein. These observations suggest that the role of Mpro in selecting a preferred cleavage site is limited and underscores the roles of the structure, conformation, and/or dynamics of the polyproteins in determining the sequence of polyprotein cleavage by Mpro.
Keywords: SARS CoV-2, 3CL main protease (Mpro), polyprotein, proteolytic processing, cryogenic electron microscopy (cryo-EM)
Viruses use a wide variety of mechanisms for expressing genes, omitting several prerequisites for protein translation in eukaryotic host cells, such as having a 5′ cap and poly-A tail on each mRNA. Positive-sense single-stranded viral RNA can express multiple proteins from a single mRNA frame by producing a precursor polyprotein followed by proteolytic cleavage to separate the mature proteins (1). All retroviruses and the majority of RNA viruses employ this strategy of translating their RNA genomes into long polyprotein chains. These polyproteins comprise multiple recognition sites, which are cleaved by viral proteases to produce mature and functional proteins. Polyprotein processing by viral proteases is an essential step in virus replication (2), and the structural distinction of these proteases from host proteins makes them ideal targets for developing antiviral inhibitors.
SARS-CoV-2 is a member of the Coronaviridae family of viruses consisting of a long positive-sense single-stranded RNA genome of approximately 30 kb in length with a 5′ cap and a 3′ poly-A tail (1). Two-thirds of its RNA genome from the 5′-end encodes the pp1a and pp1b polyproteins. These polyproteins are subsequently processed into 16 individual nsps (nsp1-nsp16) to form the RNA genome replication and mRNA transcription complexes (Replication/Transcription Complex). Processing of these polyproteins is mediated by two viral proteases: papain-like protease (PLpro) and the main protease (Mpro), also known as 3C-like cysteine protease (3CLpro) (3).
Mpro, encoded by nsp5, is responsible for its own release from the polyprotein through autoproteolysis (also known as cis-cleavage), resulting in an ∼34 kDa protein. The active form of Mpro is a homodimer (∼70 kDa) that cleaves at ten different sites found between nsp6 and nsp16 to release mature nsp proteins. The Mpro recognition sites consist of a consensus sequence of Q↓ (S/A/G/N) at the P1↓P1' (↓ denotes the Mpro cleavage site) positions, which is highly conserved among different coronaviruses (4, 5). The identification and cleavage of different recognition sequences by Mpro has been studied through in vitro protease assays and X-ray crystallographic studies of Mpro in complex with recognition peptides. Changing the Mpro recognition sequence not only at the P1↓P1' positions but also at the flanking P2 or P2' positions affects the efficiency of Mpro cleavage (6, 7). X-ray crystal structures of Mpro in complex with recognition site peptides highlighted the crucial interactions for the substrate recognition and proteolysis mechanism of Mpro (8, 9, 10).
The various cleavage sites present on the viral polyproteins are not randomly digested during processing. Different intermediates from Mpro-mediated polyprotein processing were found during murine hepatitis virus infection (11, 12) and alphacoronavirus human CoV 229E (HCoV-229E) infection (5). Stepwise cleavage of the viral polyprotein was also observed in the SARS-CoV-1 and SARS-CoV-2 systems. Limited proteolysis and native mass spectrometry (native MS) studies of the SARS-CoV-1 polyprotein nsp7-10 region by Mpro revealed that among the three cleavage sites found in the nsp7-10 polyprotein, the nsp9/10 site is cleaved first, followed by cleavage at the nsp8/9 and nsp7/8/sites (13). Mpro-mediated processing of polyprotein nsp7-11 region in a stepwise manner is also observed in SARS CoV-2 by the hydrogen-deuterium exchange mass spectrometry (HDX MS) (14). However, the mechanism by which the polyprotein and/or Mpro determines the order of cleavage remains unclear in the absence of a structure of the complex between Mpro and the polyprotein substrate.
In this study, we investigated the structural basis of stepwise SARS-CoV-2 polyprotein cleavage by determining cryo-EM structures of Mpro in the absence and presence of a polyprotein substrate. Based on the structural findings and the results of the proteolysis assay, we propose that the polyprotein is the major determinant of the sequence of polyprotein cleavage by Mpro.
Results
SARS-CoV-2 polyprotein nsp7-10 processing by Mpro protease
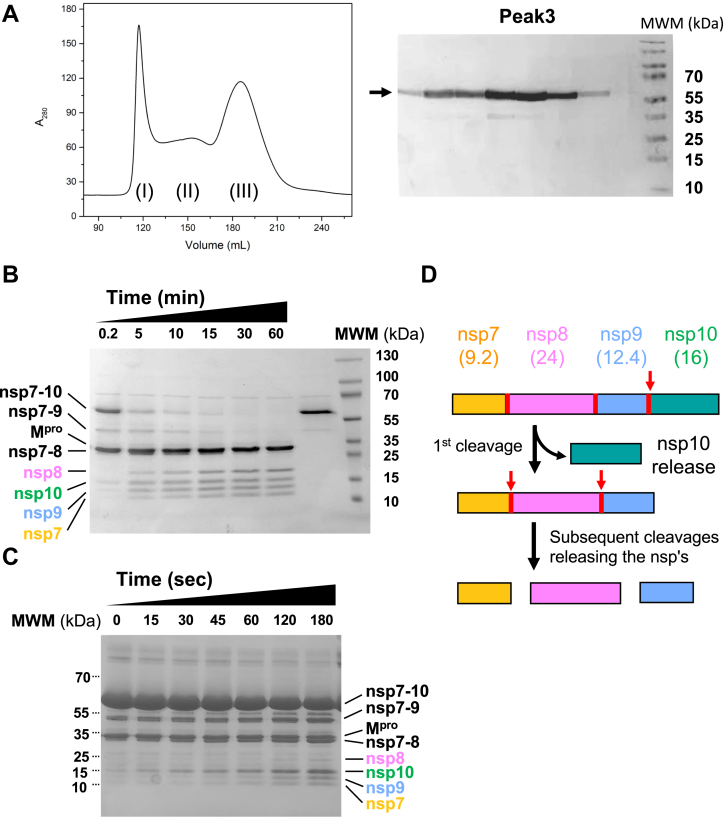
In this study, we used the nsp7-10 region as a representative polyprotein (∼58 kDa) instead of the full-length pp1a (∼490 kDa) or pp1a/b (∼794 kDa) viral polyproteins, since the nsp7-10 polyprotein can be prepared homogeneously by a bacterial expression system. The addition of ZnCl2 to bacterial growth medium during nsp7-10 polyprotein expression and the maintenance of a high salt concentration during purification (Fig. 1A) facilitated nsp7-10 polyprotein preparation, which is essential for proteolysis assays and structure studies. To confirm that the nsp7-10 polyprotein prepared in this study is processed by Mpro in a stepwise manner, as observed in previous studies (13, 14), we carried out a limited proteolysis assay. The proteolytic reaction was initiated by mixing equimolar amounts of nsp7-10 and Mpro, and all proteins, including cleaved products and intermediates, were analyzed by SDS‒PAGE (Figs. 1B and S8). Mpro first cleaved the nsp9/10 site, thus releasing nsp10 from the polyprotein (0.2 min), which simultaneously appeared with an intermediate nsp7-9 protein. As the reaction proceeded, another intermediate (nsp7-8) and other products (nsp7, nsp8 or nsp9) were released, and polyprotein processing was completed after 1 h under our experimental conditions. We also monitored the same cleavage reaction with polyprotein in excess (Mpro:nsp7-10 = 1:4) to further confirm that the nsp9/10 site was cleaved first (15–60 s, Fig. 1, C and D). This result was consistent with the results of previous studies (13, 14), indicating that the nsp7-10 polyprotein was suitable for the structural investigation of the polyprotein and Mpro complex.
Figure 1.
nsp7-10 polyprotein processing by Mpro.A, preparation of the nsp7-10 polyprotein. Different oligomers of nsp7-10 polyprotein (peaks I, II, and III) were separated by size-exclusion chromatography in the presence of 1 M NaCl in the purification buffer (left). Proteins in fraction III were analyzed by SDS‒PAGE (right). The molecular weight of the nsp7-10 polyprotein is 58.2 kDa. B, limited nsp7-10 polyprotein proteolysis by Mpro. Substrate (nsp7-10), products (nsp7, nsp8, nsp9, and nsp10), intermediates (nsp7-8, nsp7-9), and Mpro were separated by SDS‒PAGE and labeled. The time after mixing Mpro with nsp7-10 (1:1 ratio) is indicated above the lanes. C, limited nsp7-10 proteolysis assay under substrate excess conditions (Mpro:nsp7-10 = 1:4). D, schematic illustration of stepwise nsp7-10 polyprotein cleavage by Mpro. Nonstructural proteins within nsp7-10 and their molecular weights are indicated. Mpro recognition sites found between nsps are depicted as red lines, and the cleavage order is indicated by red arrows. Mpro, main protease.
Cryo-EM structure of Mpro in complex with the nsp7-10 polyprotein
Next, we determined the 3D structure of Mpro and the nsp7-10 polyprotein complex by cryo-EM single-particle reconstruction. To form a stable complex of Mpro and nsp7-10, the Cys145 residue of Mpro was replaced with Ala (Mpro-C145A, Fig. S1A), which eliminates the catalytic activity of the protease without influencing its substrate binding at the Mpro active site (15, 16). Mpro-C145A was mixed with nsp7-10 at a molar ratio of 1:2, and the complex was isolated through size-exclusion chromatography (Fig. S1B). Atomic resolution (greater than 3.0 Å) structure determination of macromolecules below 100 kDa, including Mpro (68 kDa as a dimer), by cryo-EM remains a challenging task (17). To achieve this goal, UltraAuFoil R1.2/1.3 grids were used to attain thinner ice on EM grids during vitrification and to minimize beam-induced particle motions during data collection. The reductions in background noise and particle motion improve the image quality, which contributes to the high-resolution cryo-EM structure determination of smaller macromolecules through optimized motion correction, particle picking, and alignment (17, 18). Cryo-EM micrographs were recorded using the conventional defocus-based method with a Titan Krios 300 kV microscope equipped with a Gatan K3 summit direct electron detector, and the structure was determined with an overall resolution of 2.49 Å as C2 symmetry (Figs. S2 and S3).
Mpro exclusively contacts the recognition site within the polyprotein
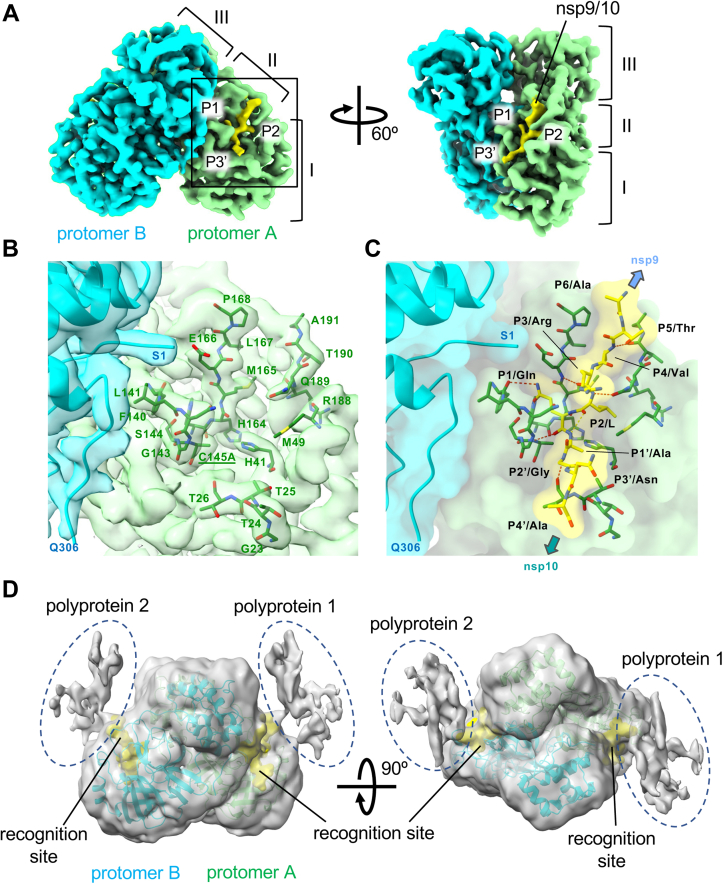
The cryo-EM map of the Mpro-C145A and nsp7-10 complex showed high-resolution features of the main and side chains of Mpro (domain I, residues 8–101; domain II, residues 102–184; domain III, residues 201–303) (Fig. 2, A and B). In sharp contrast, observation of the nsp7-10 polyprotein density map is limited to the Mpro recognition site (ten residues, from P6 to P4' positions), which is accommodated in the substrate binding cleft of both protomers of the Mpro dimer (Fig. 2, A and C and Movie S1). Mpro exclusively binds the polyprotein through the recognition site, from residues P1 to P6 on one side of the scissile bond and from residues P1' to P4' on the other (Fig. 2C). The cryo-EM density corresponding to the side chains of P4 to P2' residues from the recognition site of polyprotein was determined local resolutions between 2.4 ∼ 2.6 Å (Fig. S4B); thus, the amino acid sequence of these residues can be assigned to the nsp9/10 recognition site without ambiguity (amino acid sequence ATVRLQ↓AGNA, ↓ indicates a scissile bond) in comparison to the two other possible sites without ambiguity (nsp7/8, NRATL↓QAIAS; nsp8/9, SAVKL↓QNNEL) (Fig. S4A). The binding of the nsp9/10 recognition site at the Mpro active site is consistent with the stepwise Mpro-mediated polyprotein cleavage shown by the limited proteolysis assay (Fig. 1) and previously published native MS and HDX MS (13, 19).
Figure 2.
Cryo-EM structure of the nsp7-10 polyprotein bound Mpro-C145A.A, cryo-EM density map of the Mpro-C145A and nsp7-10 complex. Density maps corresponding to each protomer of the Mpro dimer, and the recognition site are indicated by color. Three domains of Mpro and the P1, P2, and P3' positions of the recognition site are indicated. B, a magnified view of the active center cleft of Mpro (boxed area of protomer A in panel A). The cryo-EM density map is partially transparent, and amino acid residues contacting the nsp9/10 region (<4 Å) are depicted as stick models and labeled. The density map and the model of nsp9/10 are omitted for clarity. C, a magnified view of the nsp9/10 and Mpro interaction. Structures of the nsp9/10 and Mpro residues participating in the nsp9/10 interaction are depicted as stick models with a partially transparent surface model of the Mpro and polyprotein complex. Locations of the nsp9 and nsp10 connecting to the nsp9/10 recognition site (stick model) are indicated. Orientation is the same as B. D, low path filtered cryo-EM density map of the Mpro and polyprotein complex (gray transparent overlayed with Mpro and nsp9/10 recognition site) shows the densities of the nsp7-10 polyprotein outside from the nsp9/10 region (dashed ovals). These densities are located above the active center of Mpro without any contact with Mpro. Mpro, main protease.
The cryo-EM density for the rest of the nsp7-10 polyprotein could not be traced; however, upon Gaussian filtering to the cryo-EM density map, fragmented densities likely corresponding to the main body of polyprotein became visible near the Mpro active site (Fig. 2D and Movie S2). The individually determined structures of nsp7, nsp8, nsp9, and nsp10 (20, 21, 22, 23) cannot be fitted to the map due to the lack of any 2D or 3D structural detail in such lower resolution and fragmented maps. We generated a series of cryo-EM maps by masking out the Mpro dimer density and focused refinement around the polyprotein but could not yield any stable reconstruction. The poorly defined density of the polyprotein in the complex with Mpro suggests that Mpro contacts the polyprotein primarily through recognition site residues.
The nsp9/10 site is accommodated in the active site cavity of Mpro extending along the substrate binding cleft, forming antiparallel β strands (Fig. 2C). Mpro interacts with the P4 to P4' position residues of the recognition site of the polyprotein substrate by forming hydrogen bonds with the main chain of the substrate. In addition, the P4(Val), P2(Leu), P1(Gln), P1' (Ala), and P3' (Asn) side chains of the substrate form hydrogen bonds and/or van der Waals (vdW) interactions with Mpro. The three residues (P4, P2, and P'1) of the N-terminal (or nonprime) side of the cleavage site sequence fit into the narrow cavity of the Mpro catalytic center. The glutamine at P1 rotated toward the oxyanion hole residues (Gly143 and Ser144) of Mpro is positioned in the manner usually accepted to stabilize the scissile bond residues in Mpro, and P2(Leu) is surrounded by the H41, M49, H164, M165, and Q189 residues of Mpro. The side chain of P3(Arg) is exposed on the surface, while the carbonyl oxygen interaction with Glu166 helps in its positioning close to the S1 site. The P4 to P6 residues of the recognition site are positioned near the S4 site, where the main chain at P4 makes H-bonding contacts with Thr190. In contrast, the C-terminal (prime) side of the sequence is surface exposed except P1', which is buried near the S1' subsite. The Asn residue at P3' aligns and interacts with the T24 and Thr25 residues. The binding of the nsp9/10 site at the Mpro active center determined by the cryo-EM study is nearly identical to that in the X-ray structure of Mpro-H41A in complex with an nsp9/10 recognition site peptide (PDB: 7DVY) (Fig. S4C) (10).
Cryo-EM structure of the apo form of Mpro
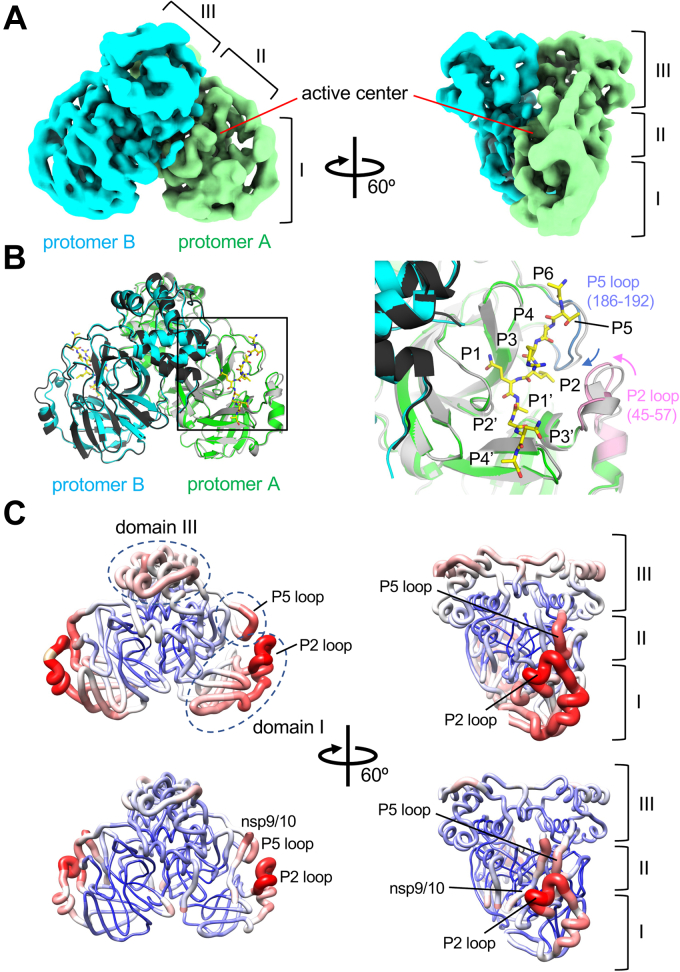
In addition to the Mpro-C145A and nsp7-10 polyprotein complex structure, we determined the cryo-EM structure of wild-type Mpro without polyprotein with an overall resolution of 3.4 Å (Figs. 3A and S5). The cryo-EM structures of Mpro in the absence and in the presence of polyprotein substrate are nearly identical (RMSD: 0.604 Å), except for noticeable differences at the active center near the P2 loop (residues T45 to L57) and P5 loop (residues from V186 to Q192) (Fig. 3B) (Fig. S7). Although substrate binding at the Mpro active center does not trigger any major conformational changes in Mpro, it does stabilize the catalytic center of the protease. Compared with the positions in the apo form, the P2 and P5 loops of Mpro move toward its active center in the presence of a polyprotein substrate, which reduces the B-factor of the P5 loop and domain I region (including the P2 loop) (Fig. 3C). The B-factor of domain III is also reduced in the Mpro/nsp7-10 complex, although domain III is not directly involved in the formation of the substrate binding cavity of Mpro. The flexible nature of the active center and domain III of Mpro may contribute to the cleavage of multiple recognition sites found in the pp1a and pp1ab polyproteins during Mpro-mediated polyprotein processing.
Figure 3.
Cryo-EM structure of wild-type Mpro.A, cryo-EM density map of Mpro. Density maps corresponding to each protomer of the Mpro dimer are colored and indicated. Three domains and the active center of Mpro are indicated. B, left, comparison of the Mpro structures in the apo form (gray and block) and in the nsp7-10 complex (green, cyan, and yellow); Right, a magnified view of the active center of the Mpro (boxed area of protomer A in the right panel). The flexible P2 (pink) and P5 loops (blue) around the substrate binding cleft move toward the nsp9/10 recognition site in the Mpro and polyprotein complex (indicated by pink and blue arrows). C, comparison of the B-factor distributions in the apo form (top) and in the nsp7-10 complex (bottom) of Mpro. Cartoon representation of the models with gradients of color (blue, white to red) and thickness (narrow to wide) reflecting the scale of the B factors (low to high). Domains and loops of the apo-form Mpro showing a higher B-factor compared with the Mpro and nsp7/10 complex are indicated by black dashed ovals. Mpro, main protease.
Discussion
Since the complete genome sequence of SARS-CoV-2 became available in early January 2020, extensive structural studies of Mpro have been conducted to understand the mechanism of the trans-proteolytic process by using peptides (8 ∼ 20 residues) containing recognition sequences as model substrates (9, 10, 24). However, there is a lack of structural information regarding the trans-proteolytic process performed by Mpro with polyprotein substrates. In this study, we determined the cryo-EM structure of the Mpro and polyprotein complex to shed light on the interactions of Mpro with the different recognition sites on the polyprotein. We revealed that Mpro associates with the recognition site residues but makes only limited contact with the rest of the polyprotein structure or sequences (Fig. 2D). The flexible nature of the active site of Mpro observed in the cryo-EM structure of Mpro without substrate provides an explanation for its capability to access multiple recognition sites within the polyprotein.
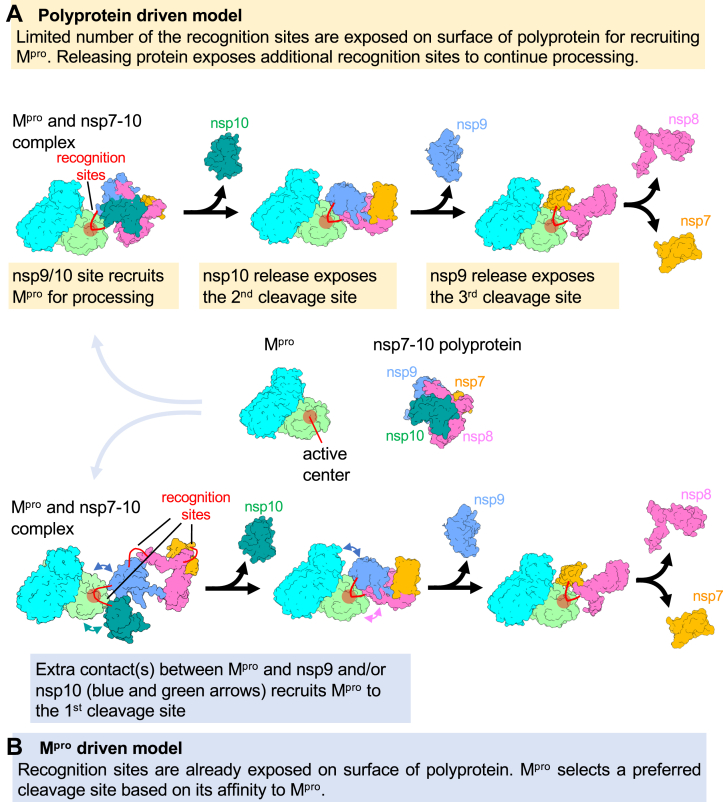
Based on the observations from this study and previously published studies (13, 19), we propose the following two models, which could explain the possible factors behind the stepwise polyprotein processing mechanism. In the “polyprotein-driven model” (Fig. 4A), a limited number of recognition sites are exposed on the surface of the polyprotein for recruiting Mpro at the primary cleavage sites. Cleavage at these sites and the release of cleaved proteins exposes the secondary and other recognition sites that recruit Mpro to continue stepwise processing. In this model, cleavage order is primarily determined by the polyprotein, and Mpro plays a limited role in determining the order of cleavage site selection.
Figure 4.
Two distinct models explaining the stepwise polyprotein processing by Mpro.A, polyprotein-driven model. B, Mpro-driven model. The different preprocessed nsps are colored separately and indicated and both the protomers of Mpro are colored cyan and light green with the active site region circled in red.
In the second “Mpro-driven model” (Fig. 4B), most recognition sites are exposed on the surface of the polyprotein, and Mpro selects a preferred cleavage site based on its affinity for the Mpro active site and/or additional contact between Mpro and the polyprotein beyond the recognition site. Since the recognition sequences are highly conserved, Mpro may need additional contact with the polyprotein beyond the recognition site to make energetically favored associations of Mpro with the primary cleavage sites over others. The cryo-EM structure of Mpro and the polyprotein complex (Fig. 2) and the results from the HDX MS study (14) support the “polyprotein-driven model” (Fig. 4A), as Mpro makes contacts only with the recognition site residues and does not interact with the polyprotein beyond it. Further structural evidence from a stable polyprotein substrate in apo form and complex with Mpro would be needed to determine the approach used by Mpro or polyproteins or both for stepwise processing.
The cryo-EM study provides additional valuable insight regarding the dynamics of the substrate binding region of Mpro (Fig. 3C). Conservation of the domain architecture in the cryo-EM structure of Mpro in comparison with the X-ray crystal structures of Mpro (Fig. S4C) confirms that cryo-EM could be used as an alternative method for investigating the structure of smaller proteins such as Mpro. The dynamic nature of polyproteins makes their crystallization difficult and arduous. The results from this study suggest that cryo-EM is a better-suited method for structural studies of multi-conformation and/or flexible proteins.
In summary, we provide insights into how SARS-CoV-2 Mpro recognizes and cleaves multiple sites on polyprotein substrates, and through limited interaction with Mpro, the polyprotein regulates the stepwise order of processing. This study describes the role of polyproteins in the sequential order of processing by Mpro and lays the foundation for future studies aimed at understanding the interactions of these proteins during coronavirus replication.
Experimental procedures
Cloning, protein expression, and purification
The plasmid pSUMO was used for producing WT and derivative of SARS-Co-V2 Mpro as described (16). Recombinant Mpro was expressed in E. coli BL21(DE3) cells and purified by Ni-affinity and size-exclusion chromatography (Fig. S1A). The final purified protein in buffer containing 20 mM HEPES (pH 7.5), 5% glycerol, 100 mM NaCl, and 0.5 mM DTT was stored at −80 °C for further use.
The codon-optimized gene encoding the nsp7-10 region of the SARS-CoV-2 polyprotein was synthesized commercially (GenScript) and cloned between the NdeI and XhoI sites of the pET15b vector, which introduces an N-terminal 6x His-tag. nsp7-10 was expressed in BL21(DE3) cells in LB growth medium containing 0.1 mM ZnCl2 along with ampicillin. The next day, 1000 ml of the medium was inoculated with 5 ml of overnight-grown culture. The protein overexpression was induced with 0.4 mM IPTG at OD600 0.6 to 0.7 and grown for 16 h at 16 °C after induction.
The cells were pelleted by centrifugation at 7000g for 20 min (4 °C) and resuspended in 100 ml of lysis buffer (20 mM Tris-HCl pH 8.0, 5% glycerol, 500 mM NaCl and 1 mM DTT). Cells were disrupted by sonication (60% amplitude, for 5 min; pulse 5 s on/10 s off), after which the lysate was cleared by centrifugation (39,000g, 30 min, 4 °C).
The polyprotein was purified after loading the supernatant on a HisTrap HP column equilibrated with buffer A (20 mM Tris pH 8.0, 500 mM NaCl, 5% glycerol, 1 mM DTT) and eluting the protein after washing with a gradient with buffer B (20 mM Tris pH 8.0, 200 mM NaCl, 5% glycerol, 300 mM imidazole, 1 mM DTT). Protein-containing fractions were diluted four times with buffer C (20 mM Tris pH 8.0, 5% glycerol, 1 mM DTT) and loaded on a HiTrap Q column. The protein was eluted with 50% buffer D (20 mM Tris pH 8.0, 1 M NaCl, 5% glycerol, 1 mM DTT) and loaded on Superdex 200 26/600 preequilibrated with buffer E (20 mM HEPES pH 7.5, 10% glycerol, 1 M NaCl and 5 mM DTT) (Fig. 1A).
Polyprotein processing assay
The processing of nsp7-10 polyprotein by the Mpro protease was monitored in an in vitro assay (Fig. 1, B and C). The reaction was set up in assay buffer (20 mM HEPES pH 7.5, 5% glycerol, 1 mM DTT, 100 mM NaCl) by incubating wild-type Mpro and nsp7-10 polyprotein at a molar ratio of 1:1 (Fig. 1B) or 1:4 (Fig. 1C) at room temperature. A total reaction volume of 150 μl was prepared, which included adding the polyprotein in the assay buffer followed by the addition of WT Mpro protease. From this reaction mixture, 10 μl aliquots were taken at regular time intervals starting from 0 to 60 min and mixed with Laemmli sample buffer to stop the reaction (four volumes, 1× final). The aliquots of the reaction mixtures were denatured (60 °C, 10 min) and analyzed by SDS‒PAGE.
Cryo-EM sample preparation, data collection, and processing
To prepare the Mpro and nsp7-10 complex for cryo-EM data collection, Mpro-C145A (5 mg/ml) and nsp7-10 polyprotein (9 mg/ml) were mixed (1:2 M ratio, 500 μl volume) and preincubated overnight at 4 °C in a solution containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 0.5 mM DTT, and 5% glycerol. The complex was loaded on a Superose 6 10/300 column (GE Healthcare, currently Cytiva) preequilibrated with 20 mM HEPES (pH 7.5), 100 mM NaCl, and 0.5 mM DTT buffer and isolated from a single peak (Fig. S1B). A 3.5 μl sample containing the complex (1 mg/ml) was applied to a glow-discharged UltraAuFoil grid (R1.2/1.3, mesh 300) (Electron Microscopy Sciences) and then blotted and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI) with 95% humidity at 4 °C. The cryo-EM grids for the apo-form wild-type Mpro were prepared as described above. The cryo-EM grids were imaged using a 300 keV Titan Krios (Thermo Fisher) microscope equipped with a K3 direct electron detector (Gatan) and controlled by the Latitude S (Gatan, Inc) software at the National Cancer Institute Cryo-EM Facility at Frederick. The defocus range for data collection was −1.0 to −2.5 μm, and the magnification was ×105,000 in electron counting mode (pixel size = 0.873 Å/pixel). Forty frames per movie were collected with a dose of 1.125 e−/Å2/frame, giving a total dose of 50 e−/Å2 (Table S1).
The Mpro-C145A-nsp710 polyprotein complex data were processed using cryoSPARC (25) (Figs. S2 and S3). A total of 5606 movies were collected, and the movies were aligned and dose-weighted using patch-motion correction. CTF fitting was performed with Patch-CTF estimation. Initially, ∼1000 particles were manually picked to generate particle templates followed by automated picking, resulting in a total of 418,049 particles subjected to 2D classification. After two rounds of 2D classification to remove junk particles, 361,048 particles were used to generate two ab initio models. Junk particles were removed, resulting in a dataset of 275,629 particles chosen for the 3D classification (heterogenous refinement). Poorly populated classes were removed, resulting in a dataset of 49,995 particles to generate the density map at 2.49 Å resolution. The particles were 3D autorefined without the mask and postprocessed (homogenous refinement). The cryo-EM data of the apo-form wild-type Mpro were processed by the same method as those of the Mpro-C145A and nsp7-10 complex and are outlined in Fig. S6 (Table S1).
Model building and refinement
To refine the Mpro-C145A and nsp7-10 polyprotein structure, the X-ray crystal structure of Mpro (PDB: 7LBN) (16) was manually fit into the cryo-EM density map using Chimera (26). The structure of nsp7-10 recognition sequence was manually built, and real-space refined by using Coot (27). The structure was real-space refined using Phenix (28) with secondary structure, Ramachandran, rotamer, and reference model restraints. The structure of apo-form wild-type Mpro was refined as described for the Mpro and nsp7-10 complex (Table S2). Figures were prepared by ChimeraX and PyMOL.
Data availability
The cryo-EM density maps and the refined models have been deposited in the Electron Microscopy DataBank (www.ebi.ac.uk/emdb/) and Protein Data Bank (www.rcsb.org) under accession numbers EMD-28162 and 8EIR (Mpro-C145A and nsp7-10) and EMD-28200 and 8EKE (wild-type Mpro).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Joyce Jose for providing a plasmid with the Mpro-encoding gene. We thank Cryo-EM facility at Penn State for the initial screening and optimization of the cryo-EM specimen preparations. We thank Boris Krichel at the University of Wisconsin-Madison, Ruchi Yadav, Xavi Ruiz, and Eddy Arnold at Rutgers University, Valentine Courouble and Patrick Griffin at Scripps Research Institute, Florida, for critical discussion of the manuscript.
Author contributions
M. N. and K. S. M. conceptualization; M. N. investigation; M. N., J.-P. A., and K. S. M. formal analysis; M. N., J.-P. A., and K. S. M. writing – review and editing; T. J. E. resources.
Funding and additional information
This research was supported in part by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. This work was supported by an NIH grant (R35 GM131860) to K. S. M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Contributor Information
Jean-Paul Armache, Email: jza449@psu.edu.
Katsuhiko S. Murakami, Email: kum14@psu.edu.
Supporting information
References
- 1.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta S.P. Elsevier/Academic Press; London: 2017. Viral Proteases and Their Inhibitors. [Google Scholar]
- 3.Gorkhali R., Koirala P., Rijal S., Mainali A., Baral A., Bhattarai H.K. Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinform. Biol. Insights. 2021;15 doi: 10.1177/11779322211025876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziebuhr J., Siddell S.G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 1999;73:177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., et al. SARS-CoV-2 M(pro) inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17:222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 7.Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antivir. Res. 2011;92:204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J., Worrall L.J., Vuckovic M., Rosell F.I., Gentile F., Ton A.T., et al. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat. Commun. 2020;11:5877. doi: 10.1038/s41467-020-19662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald E.A., Frey G., Namchuk M.N., Harrison S.C., Hinshaw S.M., Windsor I.W. Recognition of divergent viral substrates by the SARS-CoV-2 main protease. ACS Infect. Dis. 2021;7:2591–2595. doi: 10.1021/acsinfecdis.1c00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Zhu Y., Liu X., Jin Z., Duan Y., Zhang Q., et al. Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2117142119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bost A.G., Carnahan R.H., Lu X.T., Denison M.R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 2000;74:3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deming D.J., Graham R.L., Denison M.R., Baric R.S. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 2007;81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krichel B., Falke S., Hilgenfeld R., Redecke L., Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav R., Courouble V.V., Dey S.K., Harrison J., Timm J., Hopkins J.B., et al. Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro. Sci. Adv. 2022;8 doi: 10.1126/sciadv.add2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira J.C., Fadl S., Villanueva A.J., Rabeh W.M. Catalytic dyad residues His41 and Cys145 impact the catalytic activity and overall conformational fold of the main SARS-CoV-2 protease 3-chymotrypsin-like protease. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.692168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan A., Narwal M., Majowicz S.A., Varricchio C., Toner S.A., Ballatore C., et al. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022;5:169. doi: 10.1038/s42003-022-03090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzik M.A., Jr., Wu M., Lander G.C. High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Nat. Commun. 2019;10:1032. doi: 10.1038/s41467-019-08991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passmore L.A., Russo C.J. Specimen preparation for high-resolution cryo-EM. Methods Enzymol. 2016;579:51–86. doi: 10.1016/bs.mie.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yadav R., Courouble V.V., Dey S.K., Harrison J.J.E.K., Timm J., Hopkins J.B., et al. Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro. bioRxiv. 2022 doi: 10.1101/2022.05.27.493767. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konkolova E., Klima M., Nencka R., Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020;211 doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S., Chen H., Ye F., Chen Z., Yang F., Zheng Y., et al. Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design. Signal Transduct. Target. Ther. 2020;5:131. doi: 10.1038/s41392-020-00241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23 doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e3. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kneller D.W., Zhang Q., Coates L., Louis J.M., Kovalevsky A. Michaelis-like complex of SARS-CoV-2 main protease visualized by room-temperature X-ray crystallography. IUCrJ. 2021;8:973–979. doi: 10.1107/S2052252521010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 26.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Afonine P.V., Mustyakimov M., Grosse-Kunstleve R.W., Moriarty N.W., Langan P., Adams P.D. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps and the refined models have been deposited in the Electron Microscopy DataBank (www.ebi.ac.uk/emdb/) and Protein Data Bank (www.rcsb.org) under accession numbers EMD-28162 and 8EIR (Mpro-C145A and nsp7-10) and EMD-28200 and 8EKE (wild-type Mpro).