Abstract
MALDI imaging allows for the near-cellular profiling of proteoforms directly from microbial, plant, and mammalian samples. Despite detecting hundreds of proteoforms, identification of unknowns with only intact mass information remains a distinct challenge, even with high mass resolving power and mass accuracy. To this end, many supplementary methods have been used to create experimental databases for accurate mass matching, including bulk or spatially resolved bottom-up and/or top-down proteomics. Herein, we describe the application of 193 nm ultraviolet photodissociation (UVPD) for fragmentation of quadrupole isolated singly charged ubiquitin (m/z 8565) by MALDI-UVPD on a UHMR HF Orbitrap. This platform permitted the high-resolution accurate mass measurement of not just terminal fragments but also large internal fragments. The outlined workflow demonstrates the feasibility of top-down analyses of isolated MALDI protein ions and the potential toward more comprehensive characterization of proteoforms in MALDI imaging applications.
Graphical Abstract
INTRODUCTION
Matrix-assisted laser desorption/ionization (MALDI) is a powerful technique for generating low-charge-state biomolecules from droplet preparations, cultures, or biological tissues. Despite the popularity of MALDI for protein analyses, application and outcomes of intact protein mass spectrometry imaging (MSI) are limited due to the challenge of detecting and annotating protein ions. While time-of-flight (TOF) and Fourier transform ion cyclotron resonance (FTICR) are commonly used mass analyzers that have both been applied to analyze intact proteins,1 high-mass-resolving-power measurements by FTICR, and recently ultra-high-mass range (UHMR) Orbitrap,2,3 offer advantages over TOF for more precise identification of proteoforms, distinct forms of proteins potentially harboring multiple post-translational modifications (PTMs).4,5
However, assigning proteoforms solely based on accurate mass can be challenging regardless of the analyzer employed and often requires supplemental analyses.6 For example, top-down proteomics (TDP) can deliver accurate mass databases for peak annotation in MALDI; however, proteome coverage between methods may not significantly overlap due to different preparation requirements. Thus, direct fragmentation of ions generated by MALDI is appealing, especially for confident annotations of unknowns in MSI applications. However, traditional dissociation techniques exhibit a charge-state dependence and/or inefficient fragmentation with increased mass.
While MALDI in-source decay (ISD) delivered exciting results,7 the technique lacks a mass isolation step and thus proteoform selectivity, which can be problematic for complex mixtures. Commonly used activation methods (e.g., collision- or electron-based) are generally difficult to apply to singly charged protein ions generated by MALDI.8,9 Alternatively, photon-based methods such as ultraviolet photodissociation (UVPD) and infrared multiphoton photodissociation (IRMPD) have been demonstrated as viable TDP techniques.10,11 UVPD has also been suggested to be less sensitive to precursor charge state and thus has potential for MALDI applications,12,13 but UVPD has been almost exclusively applied for electrospray generated ions.14 First demonstrated three decades ago, 193 nm MALDI-UVPD of protein ions showed great promise.15 Herein, we report application of MALDI-UVPD to singly charged ubiquitin. Retrospectively, we have noted significantly improved peak assignments with the high mass resolving power and accuracy offered by the UHMR Q Exactive HF Orbitrap. This proof-of-principle study demonstrates the potential to further develop the platform to increase sensitivity and expand UVPD to proteoforms annotated within MALDI-MSI applications.
METHODS
An elevated pressure MALDI source from Spectroglyph (Kennewick, WA)16 was mounted on a Thermo Scientific Q Exactive HF Orbitrap MS upgraded with UHMR boards and operated under custom privilege licenses, as described in great depth elsewhere.2 Detailed notes for instrumental parameters and sample preparation are presented within the Supporting Information. Briefly, the spectrometer was operated under default parameters with several parameters specifically optimized for MALDI-UVPD. For all experiments, the source and instrument parameters remained the same: 500 MALDI laser shots were used per microscan, with 5 microscans coadded prior to averaging, and a total of 1500 transients were averaged. UVPD implementation was accomplished with a 193 nm excimer laser (ExciStar XS 200, Coherent, Santa Clara, CA) triggered externally from “IonGun” experiments,17 and one excimer laser pulse was used per microscan. Optics for alignment were housed within a light tight enclosure (Supplementary Figure S1), and the measured attenuation of laser pulse energies was 73% (Supplementary Figure S2); pulse energies noted throughout the text are the set energies within the laser control software.
Raw spectral data resultant from 1500 averages is located within Supporting Information for further inspection. Here, spectra were annotated and visualized in LcMsSpectator (v1.1.7158.24217);18 all ion types (a, b, c, x, y, z) and side chain fragment ions (d, v, w) were annotated with possible neutral losses of water or ammonia with a minimum signal-to-noise ratio (SNR) of 5, Pearson correlation of 0.8, and relative intensity threshold of 3. Heat maps of fragment ion types were plotted using results from Xtract deconvolution and UV-POSIT assignment.19 ClipsMS was used to evaluate internal fragmentation after Xtract processing in Freestyle (v.1.8 SP1).20 All fragment types were searched for a minimum of 20 amino acids. All analyses used a mass tolerance of 5 ppm.
RESULTS AND DISCUSSION
Previously, MALDI-UVPD of singly charged ubiquitin was completed by MALDI TOF/TOF, but despite averaging 30 000 experiments, the spectrum was poorly annotated.21 Indeed, high baseline from chemical noise is observed in the previous report,21 likely a result of spectral congestion or metastable decay. Here, MALDI-UVPD allowed for dissociation of ubiquitin with a conservative annotation of sequence coverage at 86.7% (SNR of 5) as shown for 3.0 mJ experiments (Figure 1A). As seen within the inset zooms (Figure 1B,C), the mass range (m/z 4000–8500) is dense in fragment ions, but the resolving power and increased time scale for detection afforded by UHMR HF Orbitrap permitted isotopic resolution throughout the spectrum (Supplementary Figure S3).
Figure 1.
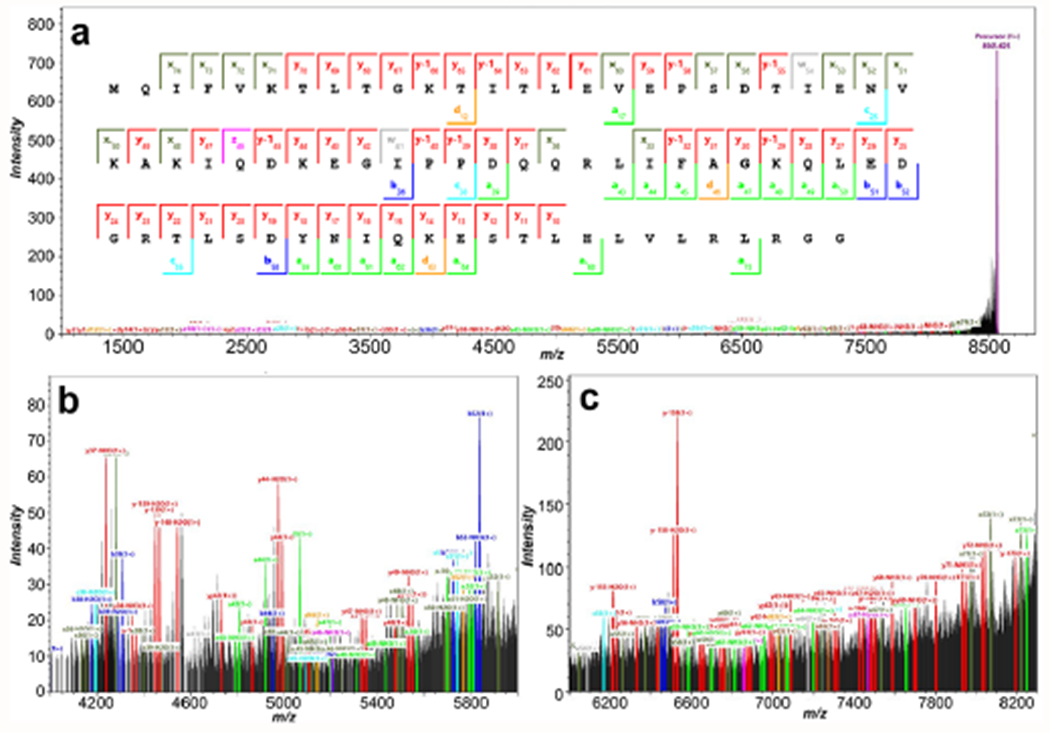
(a) The annotated coverage map within the average spectrum of 1500 single 3.0 mJ pulse MALDI-UVPD acquisitions for ubiquitin (SNR of 5, Pearson correlation of 0.8); where the precursor was identified at −1.48 ppm and ion types were manually validated. The overall sequence coverage was observed at 86.7%; however, this does not account for internal fragmentation pathways, and annotation was conservative with higher signal-to-noise and isotope correlation filters. A zoomed inset from m/z 4000 to 6000 is shown in (b), and an inset from m/z 6000 to 8300 is shown in (c) from the overall average spectrum.
Less conservative annotation of the spectrum using reduced thresholds (SNR of 3, Pearson correlation of 0.7) resulted in higher sequence coverage at 96% for the same 3.0 mJ experiment but is likely to also annotate isotopic noise. In contrast, using higher-energy collisional dissociation (HCD) with collision voltages less than 200 V, negligible fragmentation of the precursor was observed, and collisional energies greater than 200 V afforded up to 8% sequence coverage (Supplementary Figure S4). These current results demonstrate a breakthrough for intact protein MALDI-UVPD, with high levels of sequence coverage generated per unit time (Supplementary Figures S5 and S6), and high levels of coverage are obtained from only 100 averages (Supplementary Figure S6, analyzed area of 0.09 mm2 in 51.2 s).
Furthermore, these high-m/z regions (Figure 1B,C) contained nearly continuous series of isotopic peaks. Many cannot be matched to N-/C- terminal ions but are presumably overlapping internal fragments with variable neutral losses. Considering internal fragments, overfragmentation is a concern (Figure 2), occurring readily above 4.5 mJ with an observable decrease in fragment ion counts. Inspection for internal fragments (Supplementary Figure S7) also showed complete coverage above 3.5 mJ, but given the large number of theoretical fragments, false positives are likely, and our current analysis solely relied on mass accuracy. Several high-intensity fragment ions also formed hot spots (Figure 2), where b/y ions were at acidic or proline residues (y37 D/Q; y44 D/K; y39 P/D; y40 P/P; y55 D/T; y58 E/P; b52 D/G), which are well-recognized and consistent with recent statistical analysis of TDP data sets.22,23 Further investigation of the fragmentation mechanisms for singly charged protein ions may help us better annotate the unassigned fragments, where charge-state-dependent structures have been confirmed recently through simulations,24 and dependencies of MALDI-UVPD have not been explored for singly charged proteins. Regardless, we demonstrate UVPD can feasibly be accomplished for characterizing intact proteoforms desorbed and ionized by MALDI.
Figure 2.
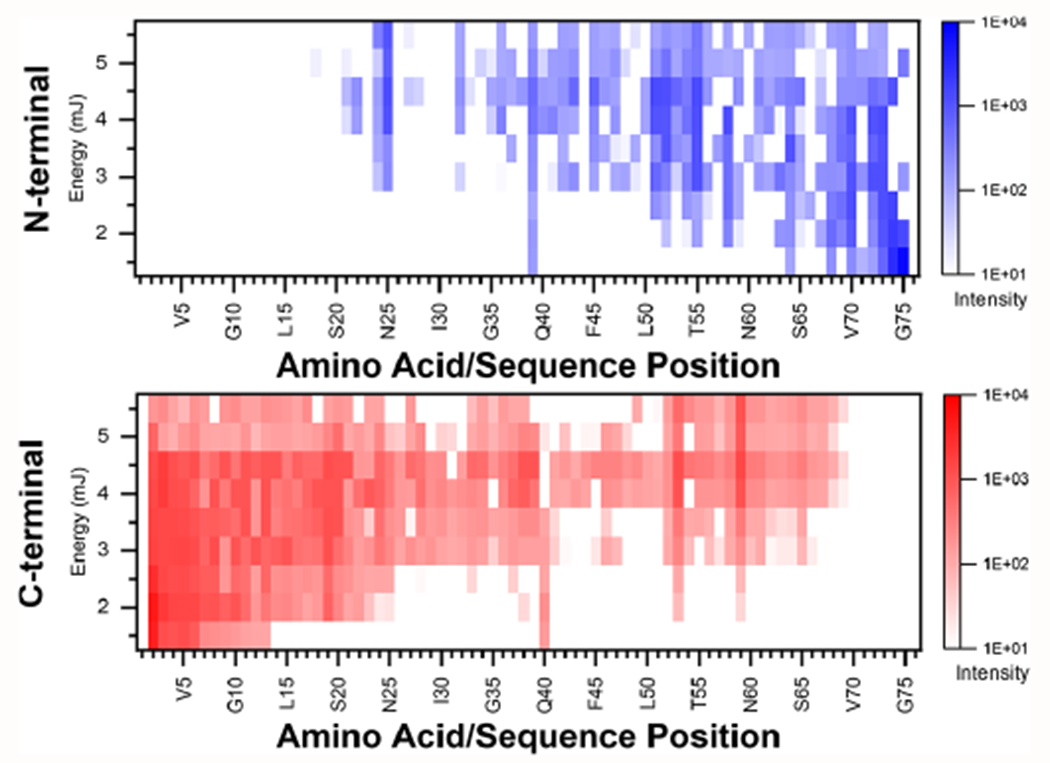
Heat map of fragment ion coverage where possible neutral losses of water or ammonia were investigated and plotted against the position within the sequence of ubiquitin. Fragment ion types (a, b, c or x, y, z) from the N- or C-terminus, respectively, were summed for all MALDI-UVPD experiments with 1500 averages from 1.5 to 5.5 mJ pulse energy. Distinct areas of hotspots can be identified within the heatmap signifying the relative intensity of the fragment ions within the UVPD experiments.
Distinct differences in proportions of all fragment ions produced from MALDI generated ions can also be noted (Figure 3) in comparison to 193 nm UVPD of low-charge-state ubiquitin produced through electrospray methods.25 As expected, for MALDI-UVPD above 3.0–4.0 mJ, decreases in ion type counts were observed for a/x and b/y fragment ions, and increased counts were noted for side chain fragment ions (d, v, w). Photodepletion of the precursor was also observed at only 32% for the 3.0 mJ experiments (Supplementary Figure S8), opening several avenues for advanced experiments such as inclusion of multiple laser shots per acquisition. However, advanced modulation of the ion packet may be needed as benefits can be limited.26
Figure 3.
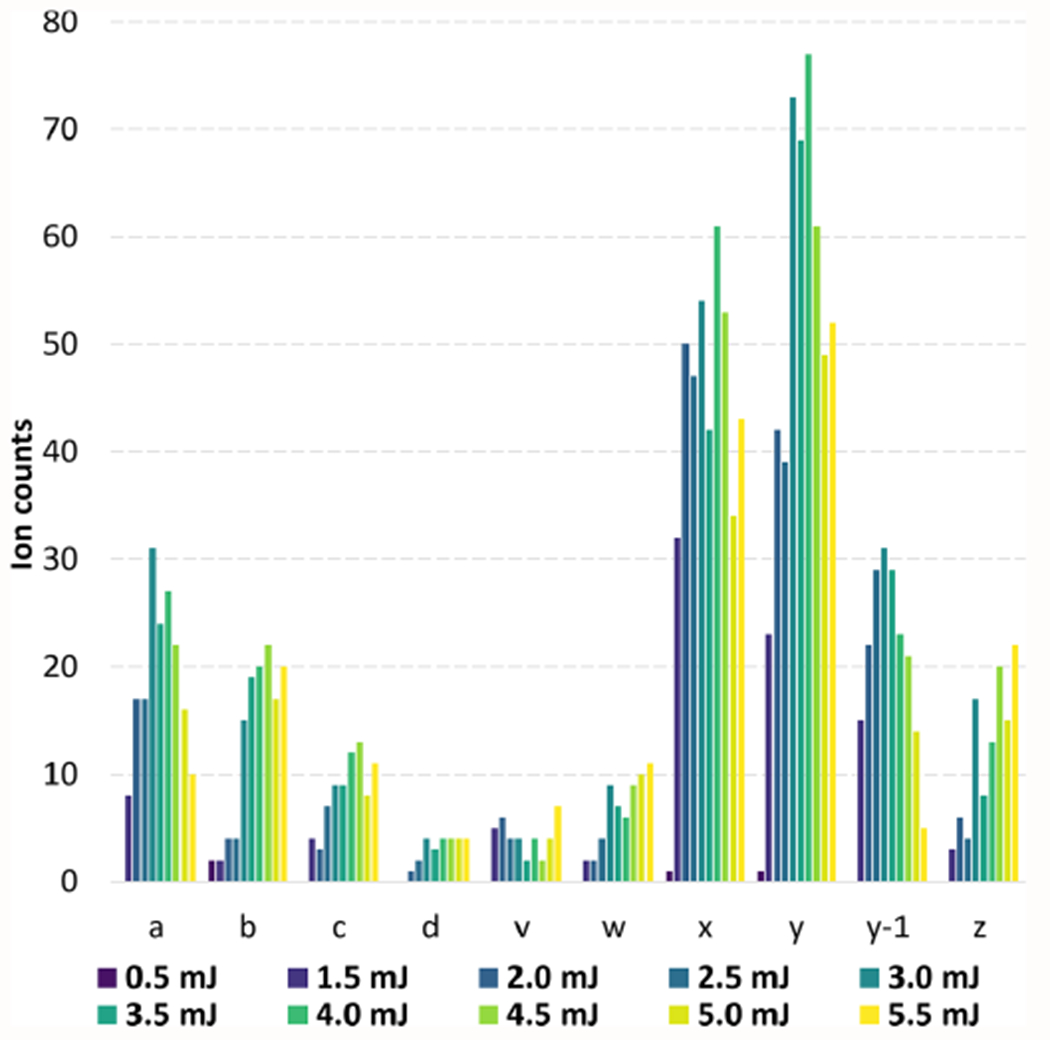
Annotation of spectra (SNR of 5, Pearson correlation of 0.8) for each laser pulse energy used for MALDI-UVPD experiments with 1500 averages. The distribution of the ubiquitin fragment type ions annotated was inspected and plotted. While side chain fragments (d, v, and w) increase with laser pulse energy and the same trend is observed for c/z type ions, a/x fragment ion counts reduce with higher fluence.
Ultimately, the scope and upper bounds of MALDI-UVPD are not currently known, and to date, our UHMR Q Exactive HF has not detected a proteoform larger than 17 kDa from tissue. Yet, these small proteoforms can have important and diverse biological functions (i.e., signaling, metabolism, etc.) and are difficult to detect and characterize by traditional methodologies including bottom-up proteomics and sequencing. Further development of the instrument will offer both increased sequence coverage and reduced experimental time, opening new avenues for automation within data-dependent fragmentation for MSI applications.27 Yet in the most basic form, broad utility for MALDI characterization of small proteoforms has been demonstrated with vast potential for integration into spatial proteomics.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Mikhail Belov at Spectroglyph, LLC, as well as Dr. Gordon Anderson and Chris Anderson at GAA Custom Electronics, LLC, for technical support for the MALDI source. The authors would also like to acknowledge Drs. Kyle Fort, Maria Reinhardt-Szyba, and Alexander Makarov of Thermo Fisher Scientific for technical guidance and licensing of the instrument. This research was funded by the National Institutes of Health (NIH) Common Fund, Human Biomolecular Atlas Program (HuBMAP) grant UG3CA256959-01, doi.org/10.46936/staf.proj.2020.51770/60000309 (L.P.T.), and a portion of this research was performed on project award doi.org/10.46936/intm.proj.2019.51159/60000152 (W.K.) from the Environmental Molecular Science Laboratory (EMSL), a Department of Energy (DOE) Office of Science User Facility sponsored by the Office of Biological and Environmental Research program under Contract No. DE-AC05-76RL01830.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/jasms.2c00302
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.2c00302.
Extended methods: sample preparation, MALDI-UVPD implementation, and operation. Supplementary Figure S1: Digital photographs of the aligned excimer beam and UVPD optics chain on the instrument. Supplementary Figure S2: Measured pulse energy and power of the 193 nm excimer beam after transmission through the optics. Supplementary Figure S3: Zoomed insets of the nonannotated averaged spectrum of 3.0 mJ experiments and the full isolation spectrum of singly charged ubiquitin. Supplementary Figure S4: Testing of HCD collision energy (CE) voltages for fragmentation of singly charged ubiquitin. Supplementary Figure S5: Tracking of sequence coverage through additional averaging of 2, 3, and 4 mJ MALDI-UVPD experiments. Supplementary Figure S6: Sequence coverage plots for 1, 2, 3, 4, and 5 mJ pulse energy experiments with the averaging of 100 microscans. Supplementary Figure S7: Internal fragment sequence coverage maps from 1500 microscan experiments from clipsMS. Supplementary Figure S8: Plot of photodepletion and sequence coverage for all experiments resultant from averaging of 1500 microscan events (PDF)
Supplementary MALDI-UVPD ubiquitin spectra (ZIP)
The authors declare no competing financial interest.
Contributor Information
Kevin J. Zemaitis, Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99352, United States
Mowei Zhou, Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99352, United States.
William Kew, Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99352, United States.
Ljiljana Paša-Tolić, Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99352, United States.
REFERENCES
- (1).Han J; Permentier H; Bischoff R; Groothuis G; Casini A; Horvatovich P Imaging of protein distribution in tissues using mass spectrometry: An interdisciplinary challenge. TrAC Trends in Analytical Chemistry 2019, 112, 13–28. [Google Scholar]
- (2).Zemaitis KJ; Veličković D; Kew W; Fort KL; Reinhardt-Szyba M; Pamreddy A; Ding Y; Kaushik D; Sharma K; Makarov AA; Zhou M; Paša-Tolić L Enhanced Spatial Mapping of Histone Proteoforms in Human Kidney Through MALDI-MSI by High-Field UHMR-Orbitrap Detection. Anal. Chem 2022, 94 (37), 12604–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zhou M; Fulcher JM; Zemaitis KJ; Degnan DJ; Liao Y-C; Veličković M; Veličković D; Bramer LM; Kew WR; Stacey G; Paša-Tolić L Discovery top-down proteomics in symbiotic soybean root nodules. Frontiers in Analytical Science 2022, 2, 1012707. [Google Scholar]
- (4).Smith LM; Kelleher NL; Linial M; Goodlett D; Langridge-Smith P; Ah Goo Y; Safford G; Bonilla L; Kruppa G; Zubarev R; Rontree J; Chamot-Rooke J; Garavelli J; Heck A; Loo J; Penque D; Hornshaw M; Hendrickson C; Pasa-Tolic L; Borchers C; Chan D; Young N; Agar J; Masselon C; Gross M; McLafferty F; Tsybin Y; Ge Y; Sanders I; Langridge J; Whitelegge J; Marshall A Proteoform: a single term describing protein complexity. Nat. Methods 2013, 10 (3), 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Smith LM; Kelleher NL Proteoforms as the next proteomics currency. Science 2018, 359 (6380), 1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ryan DJ; Spraggins JM; Caprioli RM Protein identification strategies in MALDI imaging mass spectrometry: a brief review. Curr. Opin. Chem. Biol 2019, 48, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).van der Burgt YEM; Kilgour DPA; Tsybin YO; Srzentić K; Fornelli L; Beck A; Wuhrer M; Nicolardi S Structural Analysis of Monoclonal Antibodies by Ultrahigh Resolution MALDI In-Source Decay FT-ICR Mass Spectrometry. Anal. Chem 2019, 91 (3), 2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mayer PM; Poon C The mechanisms of collisional activation of ions in mass spectrometry. Mass Spectrom. Rev 2009, 28 (4), 608–639. [DOI] [PubMed] [Google Scholar]
- (9).Lermyte F; Valkenborg D; Loo JA; Sobott F Radical solutions: Principles and application of electron-based dissociation in mass spectrometry-based analysis of protein structure. Mass Spectrom. Rev 2018, 37 (6), 750–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Shaw JB; Robinson EW; Paša-Tolić L Vacuum Ultraviolet Photodissociation and Fourier Transform-Ion Cyclotron Resonance (FT-ICR) Mass Spectrometry: Revisited. Anal. Chem 2016, 88 (6), 3019–3023. [DOI] [PubMed] [Google Scholar]
- (11).Floris F; van Agthoven M; Chiron L; Soulby AJ; Wootton CA; Lam YP; Barrow MP; Delsuc MA; O’Connor PB 2D FT-ICR MS of Calmodulin: A Top-Down and Bottom-Up Approach. J. Am. Soc. Mass Spectrom 2016, 27 (9), 1531–8. [DOI] [PubMed] [Google Scholar]
- (12).Holden DD; Sanders JD; Weisbrod CR; Mullen C; Schwartz JC; Brodbelt JS Implementation of Fragment Ion Protection (FIP) during Ultraviolet Photodissociation (UVPD) Mass Spectrometry. Anal. Chem 2018, 90 (14), 8583–8591. [DOI] [PubMed] [Google Scholar]
- (13).Dilillo M; de Graaf EL; Yadav A; Belov ME; McDonnell LA Ultraviolet Photodissociation of ESI- and MALDI-Generated Protein Ions on a Q-Exactive Mass Spectrometer. J. Proteome Res 2019, 18 (1), 557–564. [DOI] [PubMed] [Google Scholar]
- (14).Becher S; Wang H; Leeming MG; Donald WA; Heiles S Influence of protein ion charge state on 213 nm top-down UVPD. Analyst 2021, 146 (12), 3977–3987. [DOI] [PubMed] [Google Scholar]
- (15).Gimon-Kinsel ME; Kinsel GR; Edmondson RD; Russell DH Photodissociation of high molecular weight peptides and proteins in a two-stage linear time-of-flight mass spectrometer. J. Am. Soc. Mass Spectrom 1995, 6 (7), 578–587. [DOI] [PubMed] [Google Scholar]
- (16).Belov ME; Ellis SR; Dilillo M; Paine MRL; Danielson WF; Anderson GA; de Graaf EL; Eijkel GB; Heeren RMA; McDonnell LA Design and Performance of a Novel Interface for Combined Matrix-Assisted Laser Desorption Ionization at Elevated Pressure and Electrospray Ionization with Orbitrap Mass Spectrometry. Anal. Chem 2017, 89 (14), 7493–7501. [DOI] [PubMed] [Google Scholar]
- (17).Fort KL; Dyachenko A; Potel CM; Corradini E; Marino F; Barendregt A; Makarov AA; Scheltema RA; Heck AJR Implementation of Ultraviolet Photodissociation on a Benchtop Q Exactive Mass Spectrometer and Its Application to Phosphoproteomics. Anal. Chem 2016, 88 (4), 2303–2310. [DOI] [PubMed] [Google Scholar]
- (18).Park J; Piehowski PD; Wilkins C; Zhou M; Mendoza J; Fujimoto GM; Gibbons BC; Shaw JB; Shen Y; Shukla AK; Moore RJ; Liu T; Petyuk VA; Tolić N; Paša-Tolić L; Smith RD; Payne SH; Kim S Informed-Proteomics: open-source software package for top-down proteomics. Nat. Methods 2017, 14 (9), 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rosenberg J; Parker WR; Cammarata MB; Brodbelt JS UV-POSIT: Web-Based Tools for Rapid and Facile Structural Interpretation of Ultraviolet Photodissociation (UVPD) Mass Spectra. J. Am. Soc. Mass Spectrom 2018, 29 (6), 1323–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lantz C; Zenaidee MA; Wei B; Hemminger Z; Ogorzalek Loo RR; Loo JA ClipsMS: An Algorithm for Analyzing Internal Fragments Resulting from Top-Down Mass Spectrometry. J. Proteome Res 2021, 20 (4), 1928–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Moon JH; Shin YS; Cha HJ; Kim MS Photodissociation at 193 nm of some singly protonated peptides and proteins with m/z 2000–9000 using a tandem time-of-flight mass spectrometer equipped with a second source for delayed extraction/post-acceleration of product ions. Rapid Commun. Mass Spectrom 2007, 21 (3), 359–368. [DOI] [PubMed] [Google Scholar]
- (22).Haverland NA; Skinner OS; Fellers RT; Tariq AA; Early BP; LeDuc RD; Fornelli L; Compton PD; Kelleher NL Defining Gas-Phase Fragmentation Propensities of Intact Proteins During Native Top-Down Mass Spectrometry. Journal of The American Society for Mass Spectrometry 2017, 28 (6), 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Macias LA; Sipe SN; Santos IC; Bashyal A; Mehaffey MR; Brodbelt JS Influence of Primary Structure on Fragmentation of Native-Like Proteins by Ultraviolet Photodissociation. J. Am. Soc. Mass Spectrom 2021, 32 (12), 2860–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sever AIM; Konermann L Gas Phase Protein Folding Triggered by Proton Stripping Generates Inside-Out Structures: A Molecular Dynamics Simulation Study. J. Phys. Chem. B 2020, 124 (18), 3667–3677. [DOI] [PubMed] [Google Scholar]
- (25).Bashyal A; Sanders JD; Holden DD; Brodbelt JS Top-Down Analysis of Proteins in Low Charge States. J. Am. Soc. Mass Spectrom 2019, 30 (4), 704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dunham SD; Sanders JD; Holden DD; Brodbelt JS Improving the Center Section Sequence Coverage of Large Proteins Using Stepped-Fragment Ion Protection Ultraviolet Photodissociation. J. Am. Soc. Mass Spectrom 2022, 33, 446. [DOI] [PubMed] [Google Scholar]
- (27).Ellis SR; Paine MRL; Eijkel GB; Pauling JK; Husen P; Jervelund MW; Hermansson M; Ejsing CS; Heeren RMA Automated, parallel mass spectrometry imaging and structural identification of lipids. Nat. Methods 2018, 15 (7), 515–518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


