Abstract
Cytochrome P450 (CYP) is a heme-containing enzyme that catalyzes hydroxylation reactions with various substrate molecules. Steroid hydroxylases are particularly useful for effectively introducing hydroxyl groups into a wide range of steroids in the pharmaceutical industry. This study reports a newly identified CYP steroid hydroxylase (BaCYP106A6) from the bacterium Bacillus sp. and characterizes it using an in vitro enzyme assay and structural investigation. Bioconversion assays indicated that BaCYP106A1 catalyzes the hydroxylation of progesterone and androstenedione, whereas no or low conversion was observed with 11β-hydroxysteroids such as cortisol, corticosterone, dexamethasone, and prednisolone. In addition, the crystal structure of BaCYP106A6 was determined at a resolution of 2.8 Å to investigate the configuration of the substrate-binding site and understand substrate preference. This structural characterization and comparison with other bacterial steroid hydroxylase CYPs allowed us to identify a unique Arg295 residue that may serve as the key residue for substrate specificity and regioselectivity in BaCYP106A6. This observation provides valuable background for further protein engineering to design commercially useful CYP steroid hydroxylases with different substrate specificities.
Keywords: Crystal structure, cytochrome P450, steroid hydroxylase, X-ray crystallography
Introduction
Steroids are biological compounds that play key roles in the body, including controlling metabolism and the immune system, synthesizing muscle and bone, and maintaining homeostasis [1-3]. Steroid-based drugs are one of the most widely used clinical drugs to treat inflammation, rheumatoid arthritis, cancer, and allergic reactions and are also used as convulsants and contraceptives [4-7]. Since hydroxylated steroids generally exhibit higher biological activity than less polar steroids, research on hydroxylated steroid production methods has focused on and expanded from a chemical method that has disadvantages in terms of time and cost to an eco-friendly biocatalytic method using a bacteria-derived enzyme such as Cytochrome P450 (CYP) [5, 8].
CYP is a large family of heme-containing monooxygenases that catalyzes various reactions in secondary metabolite and natural product biosynthesis and xenobiotic metabolism in humans [9]. Plants have hundreds of CYPs per species, which are used to synthesize substrates, such as alkaloids, terpenes, and flavonoids [10]. Among them, microbial CYPs catalyze a broad spectrum of substrates and are more popular as biocatalysts industrially because of their ease of heterologous expression compared with mammalian and plant CYP [11-13]. Accordingly, the bioconversion of steroids using microbial CYPs has been addressed. For example, P450lun from Curvularia lunata was applied to produce cortisol from 11-deoxycortisol [14, 15]. CYP105A2 from Pseudonocardia autotrophica and CYP105A1 from Streptomyces griseolus were also used for transforming vitamin D3 to 1α,25-dihydroxy-vitamin D3, which can be used to treat vitamin D3 deficiency and metabolic disorder such as hypocalcemia, psoriasis, and osteoporosis [16-18].
The CYP106 and CYP109 families hydroxylate various types of steroids and terpenoids. For example, CYP106A1 from Bacillus megaterium DSM319 showed hydroxylation of a triterpene, 11-keto-β-boswellic acid (KBA), at the 7β- and 15α-positions using the whole cell system [19]. The well-studied BmCYP106A2 from B. megaterium ATCC 13368 also hydroxylated the 15β position of 3-oxo-Δ4-steroids such as progesterone, corticosterone, testosterone, and androstenedione, and 7β position of dehydroepiandrosterone (DHEA) and pregnenolone [20-22]. Additionally, CYP109A2, another CYP from B. megaterium DSM319, hydroxylated vitamin D3 resulting in 25-hydroxyvitamin D3 [23]. Recently, our group showed that BaCYP106A2 from Bacillus sp. PAMC 23377 converts 4-androstenedione or nandrolone into 15β-hydroxy-androstenedione or 7β,15β-dihydroxyandrostenedione, respectively [24].
Although these members are similar, with approximately 30% or greater sequence identity, the substrate preferences and hydration site regions of each CYP differ. This indicates that sequential alignment does not distinguish the characteristics of CYPs, and biochemical and structural investigations of each CYP are necessary to understand the detailed mechanisms of bacterial CYPs for steroid hydroxylation.
In the present study, we report the biochemical characterization and crystal structure of BaCYP106A6—at a 2.8-Å resolution—from multi-Bacillus species isolated from soil. Accordingly, the results provide valuable insights for further protein engineering to design commercially applicable CYP steroid hydroxylases with different substrate specificities.
Materials and Methods
Materials
The substrates were purchased from Sigma-Aldrich (Korea) and Tokyo Chemical Industry Co. Ltd. (Japan). All the enzymes, including Taq polymerase and restriction enzymes, were obtained from Takara Clontech (Korea). All other chemicals and solvents used were of the highest commercially available grade (ACS, HPLC grade; Fisher Scientific, Korea). Ampicillin, α-aminolevulinic acid, NADPH, catalase, glucose-6-phosphate dehydrogenase, and glucose-6-phosphate, the redox partners of spinach FDX and FDR, were purchased from Sigma-Aldrich. Isopropyl 1-thio-β-d-galactopyranoside (IPTG) and kanamycin were purchased from Duchefa Biochemie (Korea).
Over-expression and Purification of BaCYP106A6
BaCYP106A6-pET28a was introduced into Escherichia coli C41(DE3) and inoculated into Luria–Bertani (LB) medium supplemented with ampicillin (100 μg/ml). Thereafter, the seed culture was transferred to LB containing 0.5 mM FeCl3·6H2O, 1 mM 5-aminolevulinic acid hydrochloride (5-ALA), 100 μg/ml ampicillin at 37°C. The culture was induced with 0.5 mM IPTG when the optical density at 600 nm (OD600) reached 0.6–0.8. After 72 h of incubation at 20°C, the cells were harvested and suspended in 50 mM potassium buffer (pH 7.4). The cell extract was sonicated and centrifuged at 10,000 rpm for 20 min to obtain the soluble protein solution. Next, the protein solution was mixed with Co2+ resin solution and eluted with elution buffer (potassium phosphate buffer, pH 7.4) containing 20, 100, and 250 mM imidazole. The eluted fraction containing the protein was concentrated using a 30 K Amicon Ultra centrifugal filter (Millipore, Ireland).
Enzyme Analysis and Substrate-Binding Assay
The concentration of the purified BaCYP106A6 was calculated using a UV-visible spectrophotometer (Biochrome Libra, UK). The concentration of BaCYP106A6 was determined by CO-bound reduced difference spectroscopy using an extinction coefficient of 91 mM-1cm-1. CO difference spectroscopy measurements were performed according to the method of Omura and Sato [25]. Protein purity and molecular weight were analyzed using SDS-PAGE.
To determine the degree of steroid substrate-binding to a specific enzyme, spin-shift states were identified using a UV-visible spectrophotometer and tandem quartz cuvettes. The sample cuvette chambers were filled with a total volume of 1 ml in 50 mM potassium buffer (pH 7.4), including purified protein diluted to 1 μM and substrate by concentration, and the standards were prepared without substrate. The substrates were dissolved in DMSO at a storage concentration of 40 mM, diluted in the range of 0–500 μm and used for the assay. The UV-visible absorbance spectrum was measured between 350 and 500 nm until no spectral changes were observed. The equilibrium Kd of BaCYP106A6 was calculated using a quadratic equation by titrating the substrate concentration until saturation:
where [E] and [S] are the concentrations of the enzyme and substrate, respectively, Kd is the binding constant, ΔAmax is the maximal absorption shift, and ΔA is the peak-to-trough ratio [26]. The dissociation constant (Kd) of BaCYP106A6 was determined by titrating different concentrations of the substrates until saturation.
In vitro Biotransformation Assays of BaCYP106A6 Activity
For the in vitro assay, two steroid substrates were used: progesterone and androstenedione. A stock solution (100 mM) of the substrate was dissolved in DMSO and stored until use. The in vitro reaction was carried out in a total volume of 250 μl in 50 mM potassium buffer (pH 7.4) consisting of 10 μM BaCYP106A6, 25 μg FDX, 0.1 U FDR, 100 μM steroid substrates, 5 mM MgCl2, 100 μg/ml catalase, 1 mM NADPH, 1 U glucose-6-phosphate dehydrogenase (G6P-DH), and 10 mM glucose-6-phosphate. The reaction was initiated by adding 1 mM NADPH. Reaction mixtures were incubated at 30°C for 2 h, extracted twice with an equal volume of ethyl acetate, and then dried completely with nitrogen gas. Finally, the dried samples were dissolved in a 6:4 (acetonitrile/water) solution for HPLC analysis.
HPLC and NMR Analysis
HPLC analyses for product separation were performed using an Agilent 1100 series system (G1311A Quaternary pump, G1379A Solvent degasser, G1315B Diode array detector, and G1313A Standard autosampler; Agilent Technologies, USA). This device was connected to a reversed-phase C18 GP column (4.6 × 250 mm, 5 μm; Mightysil; Kanto Chemical, Japan), and the analysis temperature was maintained at 40°C. The mobile phase was mixed with two solvents, water (A) and acetonitrile (B), at a rate of 1 ml·min−1. The HPLC system started with acetonitrile and water at a ratio of 15:85, increased to 50:50 for 8 min, and then to 90:10 for 18 min. The ratio was maintained for 19 min, reduced to 15:85 for 21 min, and finally, ran for 25 min. To detect the substrate and product, the UV detector was set to 242 or 245 nm. Mass analysis was performed using quadrupole time-of-flight/electrospray ionization mass spectrometry in the positive ion (+) mode using ultra-performance liquid chromatography (SYNAPT G2‐S/ACUITY; Waters Corp., USA). The products isolated from the steroids were analyzed using a 700 MHz NMR spectrometer (Korea Basic Science Institute, Korea). For 1H, 13C NMR, HMBC, HSQC, COSY, and ROESY, 7.3 and 15 mg progesterone and androstenedione products, respectively, were dissolved in 1 ml CDCl3.
Crystallization and Data Collection
Initial crystallization screening was conducted using a TTP Labtech Mosquito LCP Crystallisation Robot (TTP Labtech, UK) with commercially available screening kits, such as MCSG1-4 (Molecular dimensions, UK), Index, and SaltRx (Hampton Research, USA). The sitting drop vapor-diffusion method was performed at 293 K in 96-well plates (Emerald Bio, , USA). A 200-nL protein solution and an equal volume of reservoir solution were mixed and equilibrated against 80 μl of reservoir solution. BaCYP106A6 crystals were grown within 2 days under 0.2 M ammonium citrate dibasic and 25% (w/v) PEG 3350 condition. To obtain larger crystals, crystallization conditions were optimized using the hanging drop vapor-diffusion method at 293 K in 24-well plates. Each drop, consisting of 1 μl of protein solution and 1 μl of reservoir solution, was equilibrated against 500 μl of reservoir solution. Optimized crystals appeared after 2 days under 0.18 M ammonium citrate dibasic and 22% (w/v) PEG 3350 condition at 293 K. Because of fragileness, a single BaCYP106A6 crystal was directly mounted without soaking in a cryoprotection agent. The 2.8 Å resolution of the diffraction dataset was obtained on a BL-5C beamline at the Pohang Accelerator Laboratory (Korea). The dataset containing 360 images with an oscillation range of 1° rotation was indexed, integrated, and scaled using the program HKL-2000 [27].
Structure Determination and Refinement
The crystal structure of BaCYP106A6 was determined by the molecular replacement method using the MOLREP program from the CCP4i suite. The crystal structure of CYP106A2 from B. megaterium (PDB code 5IKI; sequence identity, 65%) was used as the search model [28]. The Matthews coefficient calculation result predicted that two molecules are contained in the asymmetric unit, with a Matthews coefficient of 2.50 Å3Da-1 and solvent content of 50.79 %. Next, the initial model was iteratively rebuilt using Coot [29], refined REFMAC5 [30], and phenix.refine [31]. The final model had an Rcryst value of 0.22 and Rfree value of 0.27. Model quality was checked using MolProbity [32]. Detailed refinement statistics are presented in Table 1. All graphical structural representations were generated using PyMOL [33]. The coordinates and structural factors of BaCYP106A6 were deposited in the Protein Data Bank RCSB under accession code 8HG9.
Table 1.
X-ray diffraction data collection and refinement statistics.
| Data set | BaCYP106A6 |
|
| |
| X-ray source | BL-5C beamline |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a=53.335, b=98.997, c=92.99, α=γ=90, β=106.412 |
| Wavelength (Å) | 0.9794 |
| Resolution (Å) | 50.00–2.80 (2.85–2.80) |
| Total reflections | 142028 |
| Unique reflections | 22,227 (1,192) |
| Average I/σ (I) | 31.0 (5.0) |
| R merge a | 0.126 (0.471) |
| Redundancy | 6.4 (7.4) |
| Completeness (%) | 99.1 (97.0) |
|
| |
| Refinement | |
|
| |
| Resolution range (Å) | 37.59-2.80 (2.94-2.80) |
| No. of working set reflections | 22,146 (3,161) |
| No. of test set reflections | 1,011 (122) |
| No. of atoms | 6,309 |
| No. of water molecules | 58 |
| R cryst b | 0.22 (0.28) |
| R free c | 0.27 (0.39) |
| r.m.s. bond length (Å) | 0.013 |
| r.m.s. bond angle (°) | 1.667 |
| Average B value (Å2) (protein) | 42.28 |
| Average B value (Å2) (solvent) | 36.55 |
|
| |
| Ramachandran plot | |
|
| |
| Favored (%) | 96.08 |
| Allowed (%) | 3.13 |
| Outliers (%) | 0.78 |
aRmerge = Σ | <I> - I | /Σ<I>.
bRcryst = Σ | |Fo| - |Fc| | /Σ|Fo|.
cRfree calculated with 5% of all reflections excluded from refinement stages using high-resolution data.
Values in parentheses refer to the highest-resolution shells.
Cloning and Construction of Recombinant Plasmids
The gene for BaCYP106A6 was amplified by PCR using the genomic DNA of Bacillus sp. as the template. The PCR primers used were 5′- GGA TCC ATG TTG AAA GAA GTC ATT CC-3′ (BamHI site underlined) as the forward and 5′-CTC GAG TCC TTA CTT ATA CAC GTT CA-3′ (XhoI site underline) as the reverse primers. The fragment was digested with restriction enzymes and ligated into a pET28a (+) vector to construct BaCYP106A6-pET28a. Sequence analyses for cloning were performed by Macrogen Inc. (Korea).
Results and Discussion
Purifcation and Characterization of BaCYP106A6
To characterize BaCYP106A6, His-tagged BaCYP106A6 was successfully expressed and purified in a soluble form in E. coli host cells. Two-step purification yielded BaCYP106A6 with high purity (> 95%). SDS-PAGE analysis showed a single homogeneous band of purified proteins at the expected molecular weight of 47.1 kDa (Fig. S1A). Purified BaCYP106A6 induces a type I spin-shift in the resting state. UV-visible absorption spectroscopy of BaCYP106A6 revealed a significant heme Soret peak at 417 nm in the substrate-free oxidized form. This peak could be shifted to 448 nm in the reduced CO-bound form, which is a characteristic feature of the cysteine-thiolate-ligated heme of CYP in a Fe (II)-CO complex (Fig. S1B) [34-36].
Substrate-Binding and Steroid Assays Using BaCYP106A6
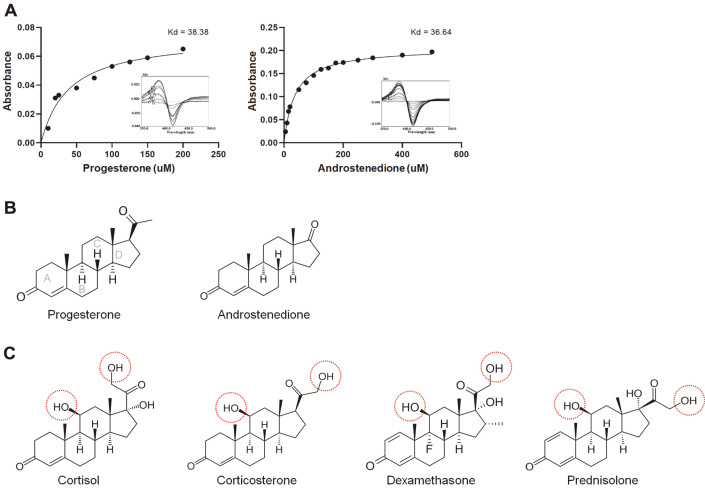
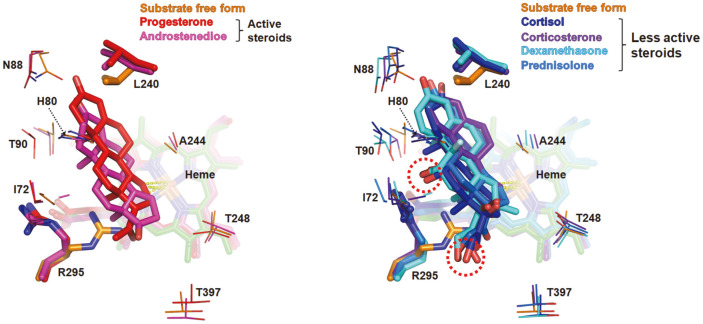
The binding of steroids to P450 causes a type I spectral shift due to the substitution of axial water molecules in the heme iron coordination sphere [20], revealing maximum and minimum spectral values of ~390 and ~420 nm, respectively. The dissociation constant (Kd) of CYP was determined by titrating different concentrations of substrates until saturation. Based on this characteristic, we conducted substrate-binding experiments with different steroidal substrates using differential spectroscopy before conducting in vitro conversions. As shown in Fig. 1A, the binding assay demonstrates that two of the six steroidal substrates could bind BaCYP106A6. The equilibria Kd of BaCYP106A6 for progesterone and androstenedione were 38.38 and 36.64 μM, respectively (Figs. 1A and 1B). In contrast, cortisol, corticosterone, dexamethasone, and prednisolone showed no active or minor type I spin states, even at high substrate concentrations (Fig. S2). Therefore, the tested steroids could be divided into two groups based on their affinity: active and less active steroids. Interestingly, the less active steroid group had a hydroxyl group at the 11β position, while the active steroid group interacting with BaCYP106A6 did not have a hydroxyl group at the corresponding site (Figs. 1B and 1C). Moreover, the less active steroid group had an additional methanol group at the acetyl group linked at the 17β position. Therefore, we hypothesized that the hydroxyl group and the side chain length at the 17β position are critical factors determining substrate affinity to BaCYP106A6.
Fig. 1. Steroid affinity test on BaCYP106A6.
(A) Type I shift spectra and Kd values using progesterone and 4- androstenedione. A total of six substrates were tested for BaCYP106A6 binding. Progesterone and androstenedione showed binding affinity, whereas corticosterone, cortisol, dexamethasone, and prednisolone did not show consistent data for affinity calculation. (B) Steroids with a BaCYP106A6-associated conversion rate > 20% were considered active steroids. (C) Steroids with a BaCYP106A6-associated conversion rate < 20% were considered less active steroids. The conversion rate of steroids was analyzed by HPLC after 15 min incubation at 20°C.
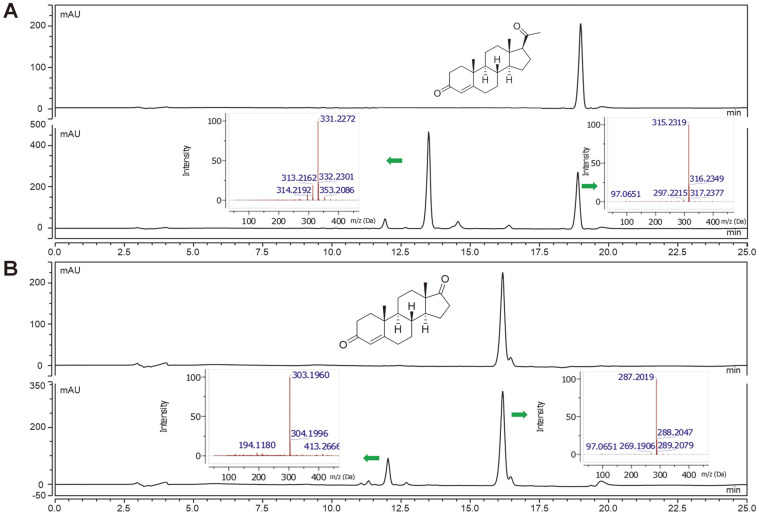
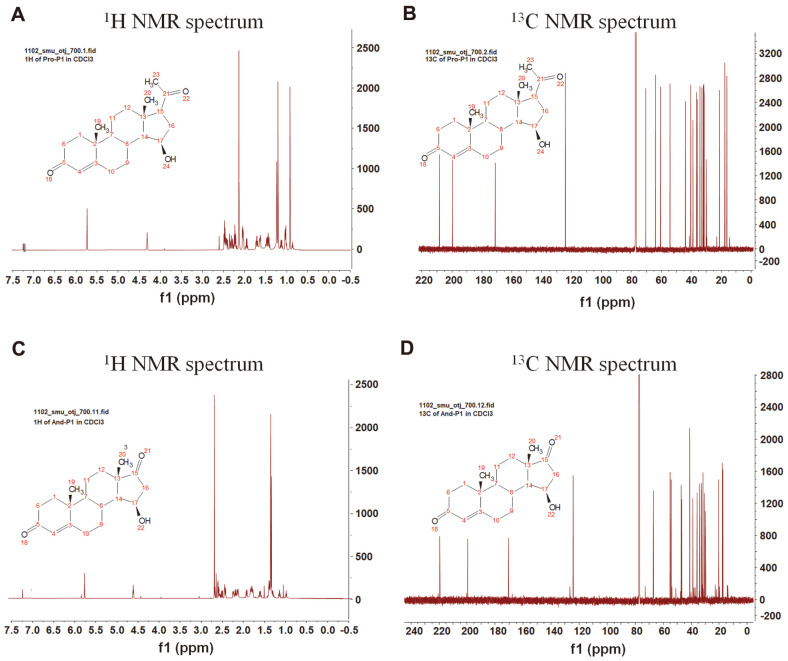
Next, we performed a bioconversion reaction of progesterone and androstenedione using recombinant BaCYP106A6. Before the analysis, we confirmed that FDX/FDR could be a redox partner of BaCYP106A6. As expected from the binding assay, the results indicated that BaCYP106A6 could catalyze the hydroxylation of these steroids. In the case of progesterone, after incubation with BaCYP106A6, the retention time at λmax:245 nm was shifted to ~13.41 min, which is equivalent to the mass for the molecular formula C21H30O4 for [M + H]+ m/z+ ~331.2273, confirming the monohydroxylated progesterone product (Fig. 2A). Similarly. The analysis of androstenedione from the reaction mixture showed monohydroxylated products as observed by mass [M + H]+ m/z+ ~303.1960 (Fig. 2B). Calculating the retention area showed that progesterone showed the highest conversion (~70%), while androstenedione showed a 30% conversion from the substrate after 15 min of incubation. Furthermore, NMR analysis of the chemical structures of the hydroxylated products of steroids indicated that the C15 region was the hydroxylation site by BaCYP106A6 with high regio- and stereoselectivity (Fig. 3). By comparing the NMR spectroscopic data reported in the literature, the structures of the products were determined to be 15β-hydroxy progesterone and 15β-hydroxy androstenedione [37, 38].
Fig. 2. In vitro HPLC and LC/MS analysis of BaCYP106A6.
The HPLC spectra of progesterone (A) and androstenedione (B) with their corresponding BaCYP106A6 catalyzed reaction mixture. Each substrate generated a monohydroxylated product. The masses of the substrate and monohydroxylated product are shown by the arrows.
Fig. 3. Nuclear magnetic resonance (NMR) analysis.
1H and 13C NMR analyses of hydroxylated products of progesterone (A, B) and androstenedione (C, D).
Structure Determination and Overall Structure of BaCYP106A6
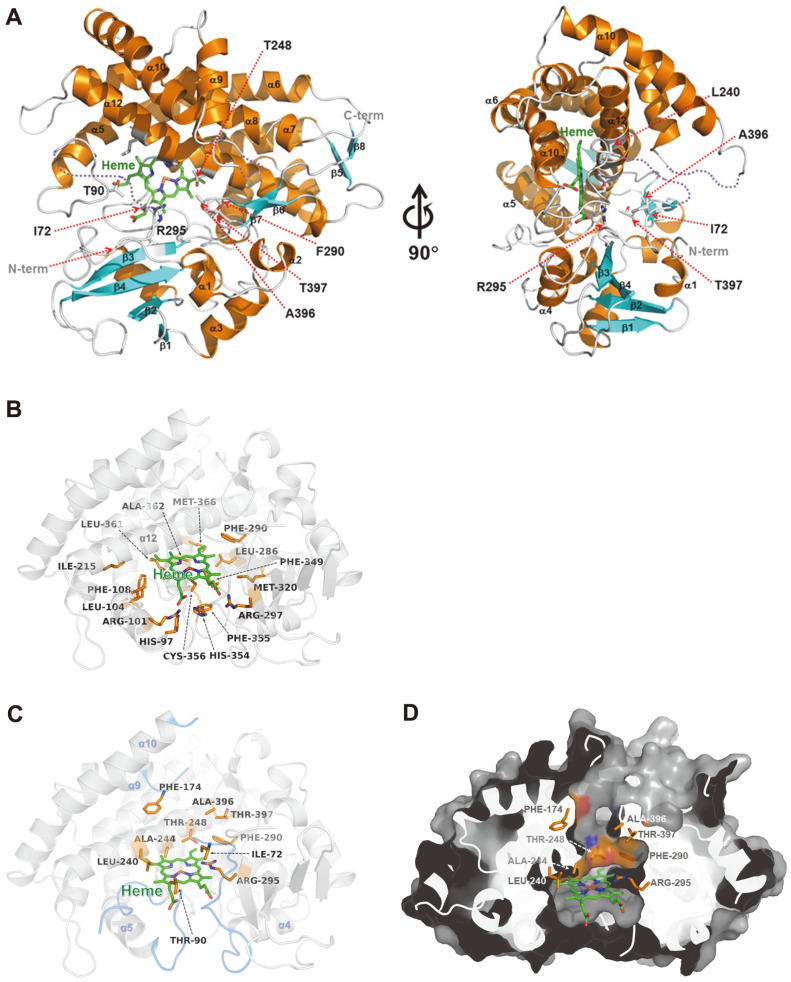
The crystal structure of BaCYP106A6 was determined at a 2.8-Å resolution using X-ray crystallography. The phase problem was solved using the crystal structure of CYP106A2 (PDB code:5IKI) via the molecular replacement method. Next, the coordinates were refined as the values for Rwork and Rfree of the final model were 22 and 27%, respectively (Table 1). Due to an ambiguous electron density map, all amino acids were built except for the four N-terminal residues and residues from the 74–84 and 178–181 regions. The final model of BaCYP106A6 contained two monomers in the asymmetric unit, with the heme cofactor located at the center of each monomer. The two monomers form 2-fold rotational symmetry with a small interface area, which seems too small to form a realistic dimeric interface, indicating that BaCYP106A6 exists as a monomer. Consistent with this, size-exclusion chromatography analysis indicated that BaCYP106A6 is a stable monomer in solution (Fig. S3).
The monomeric structure of BaCYP106A6 consists of 17 α-helices and eight β strands and exhibits a typical fold seen in CYP monooxygenases with a centered heme cofactor molecule (Fig. 4A). Structural analysis of BaCYP106A6 revealed a heme cofactor-binding site and putative substrate-binding residues. The heme molecule is tightly bound to the central region of the BaCYP106A6 structure and is surrounded by α5, α12, α14, and α16 helices. The Cys356 residue is involved in the coordination of heme iron. The carboxyl groups of heme interact with the side chains of His97, Arg101, Arg297, and His354. The nonpolar part of the heme is surrounded by hydrophobic residues Leu104, Phe108, Ile215, Leu286, Phe290, Met320, Phe349, Phe355, Leu361, Ala362, and Met366. These conserved heme-binding features are typical of CYP monooxygenases (Fig. 4B). The putative substrate-binding pocket is located directly above the heme molecule and consists of Ile72, His80, Asn88, Thr90, Leu240, Ala244, Thr248, Arg295, Ala396, and Thr397 (Figs. 4C and 4D).
Fig. 4. The overall structure and active site of BaCYP106A6.
(A) The overall structure of BaCYP106A6 is presented as a ribbon diagram in the front view (left) and a 90° rotated view (right). The α-helices and β-strands are colored orange and cyan, respectively. The bound heme molecules are represented by a stick model and colored green. (B) Heme-binding motif of BaCYP106A6. Hydrophobic residues surrounding the heme molecule and residues interacting with carboxyl groups of heme are presented as an orange stick model. The heme molecule is represented by a green stick model. (C) The putative substratebinding pocket of BaCYP106A6. Several specific residues comprising substrate-binding pockets are presented as orange stick models. Disordered regions of the α4–α5 and the α9–α10 loops are colored marine. (D) Surface representation of BaCYP106A6 shown in the same orientation as
Substrate Selectivity of BaCYP106A6
The binding mode of steroids was analyzed to better understand the substrate selectivity of BaCYP106A6. First, because the structure has unconnected residues between the 74–84 and 178–181 regions, the complete structure was generated by the SWISS modeling server based on the BaCYP106A6 structure as a template [39]. The steroids were superposed with abietic acid from the CYP106A2 structure (PDB:5IKI) and corticosterone from the CYP109E1 structure (PDB:5L91) in the putative binding pocket, and energy minimization was conducted using the YASARA Energy Minimization Server [40]. Energy minimization with a substrate-complexed structure avoids substrate hindrance or residual crashes between steroids and proteins. The results showed that the distances between the iron of heme and the C15 of progesterone and androstenedione were 4.3 and 3.8 Å, respectively, indicating that the orientation and distance of steroids are in a feasible conformation.
Compared with the substrate-free structure, the direction of the side chain of Leu240 and Arg295 was mainly changed among residues interacting with substrates in steroid-complexed structures. The side chain of Leu240 leans to the heme molecule in the substrate-free structure, and the side chain is pushed away by the C4 of the steroids upon steroid binding, generating a hydrophobic interaction. In the case of Arg295, its side chain protrudes inside the binding pocket and occupies a large volume of the substrate-binding pocket in a substrate-free structure (Fig. S4). However, in the steroid-binding mode, Arg295 was tilted opposite the active site (Fig. 5). These changes imply that Leu240 and Arg295 may be critical residues that recognize specific BaCYP106A6 substrates. Moreover, Arg295 was closely located to the hydroxyl group at the 11β position of the less active steroid group and was positioned in the hydrogen interaction range of 3–5 Å. Another possible interaction is between Arg295 and the 17β side chain. It is worth noting that we speculated that the hydroxyl group at 11β and/or the 17β position side chain were critical points distinguishing the types of steroids from the substrate-binding assay. Since only less active steroids have a hydroxyl group at the 11β position and this hydroxyl group is likely to interact with Arg295, we provisionally concluded that the interaction of Arg295 with negatively charged functional groups, such as the hydroxyl group at the 11β and/or 17β positions of less active steroids, may interfere with the less active steroids that are suited to the active site, resulting in low activity.
Fig. 5. Overlay of steroids as obtained by superposition onto BaCYP106A6.
The active (left) and less active steroids (right) occupy the substrate-binding pocket depicted in red and blue series colors. Energy-minimized structures and heme molecules are colored the same for each steroid. The hydroxyl groups at 11β and 17β of the less active steroids, marked with red dotted circles, are placed near an Arg295.
Comparison of Substrate-Binding Pocket between CYP106 and CYP109 Proteins
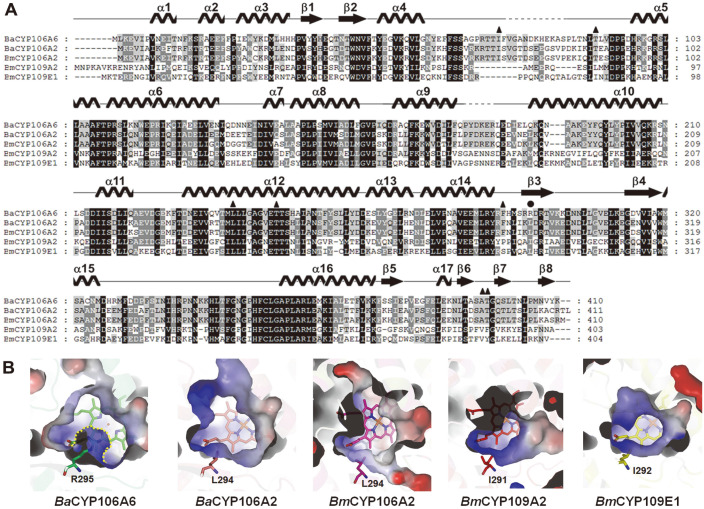
A DALI structural homology search revealed that the BaCYP106A6 structure showed high structural similarity (Z-score:54.1) to BmCYP106A2 from B. megaterium ATCC13368 (PDB code: 4YT3 and 5IKI), BmCYP109A2 from B. megaterium (PDB code 5OFQ), and BmCYP109E1 from B. megaterium (PDB code 5L92) (Table 2). Multiple sequence alignment indicated that the Arg295 residue is unique to the BaCYP106A6 sequence, and other CYP106 and CYP109 homologs do not have this positively charged residue at the corresponding position (Fig. 6A). The substrate-binding site of BaCYP106A6 is mainly composed of hydrophobic residues and interacts with the backbone of the steroids. However, the existence of Arg295 generates a unique environment in the substrate-binding site (e.g., changing the charge distribution and size of the substrate access channel) (Fig. 6B). Thus, it is thought that BaCYP106A6 may have a different substrate-binding mode and specificity controlled by Arg295 compared with those of other CYP106 and CYP109 proteins.
Table 2.
Structural homolog search results for BaCYP106A6 from a DALI search (DALI-Lite server).
| Protein | PDB code | DALI Z-score | UniProtKB code | Sequence % ID with BaCYP106A6 (aligned residue number) | Reference |
|---|---|---|---|---|---|
| Abietic acid-bound CYP106A2 (B. megaterium) | 5IKI | 54.1 | Q06069 | 65 (379/399) | [28] |
| CYP109A2 (B. megaterium) | 5OFQ | 45.2 | D5DF88 | 36 (373/387) | [23] |
| Corticosterone bound CYP109E1 (B. megaterium) | 5L91 | 45.1 | D5DKI8 | 42 (368/391) | [43] |
| Cytochrome P450 TbtJ1 (Thermobispora bispora) | 5VWS | 43.4 | D6Y4Z8 | 36 (367/376) | [44] |
| CYP109B1 (Bacillus subtilis) | 4RM4 | 43.3 | O34374 | 40 (351/364) | [45] |
| Mycinamicin bound Cytochrome P450 (Micromonospora griseorubida) | 2Y5N | 43,0 | Q59523 | 30 (370/400) | [46] |
| Cytochrome P450 MoxA (Nonomuraea recticatena) | 2Z36 | 42.5 | Q2L6S8 | 27 (373/404) | [47] |
| Cytochrome P450 OleP from (Streptomyces antibioticus) | 5MNV | 41.9 | Q59819 | 29 (367/397) | [48] |
Fig. 6. Multiple sequence alignment and binding mode of steroids.
(A) Multiple sequence alignment of BaCYP106A2 with other homolog CYPs, BaCYP106A2 (Bacillus sp. PAMC23377; PDB code 5XNT), BmCYP106A2 (B. megaterium; UniProtKB code: Q06069; PDB code 4YT3), BmCYP109A2 (B. megaterium; UniProtKB code: D5DF88; PDB cod 5OFQ), and BmCYP109E1 (B. megaterium; UniProtKB code: D5DKI8; PDB code 5L90). Secondary structures of BaCYP106A6 are shown above the aligned sequence. Multiple sequence alignment was conducted using ClustalX [41] and edited using GeneDoc. (B) The active site of CYPs. Electrostatic surfaces and charge distribution of the proteins were analyzed using the Adaptive Poisson–Boltzmann Solver [42]. Arg295 and the corresponding residues from CYPs are represented as different-colored sticks.
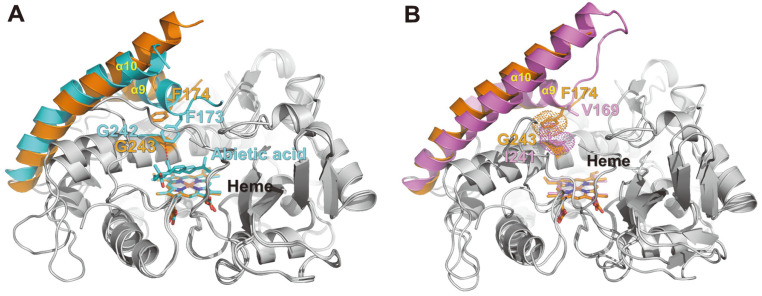
Besides the Arg295 residue, CYP106 and CYP109 proteins have several striking residue composition differences in the substrate-binding pocket. In BmCYP106A2 (PDB code 5IKI), Phe174, located in the α9–α10 loop region, forms hydrophobic interactions with the bound substrate (abietic acid). These interactions may induce α9–α10 loop and α9 helix movements inside the substrate-binding pocket. This Phe residue is highly conserved in the CYP106 protein sequences (Fig. 7A). On the other hand, in BmCYP109E1 (PDB code 5L90), this Phe is substituted for valine (V169). Previous site-directed mutagenesis experiments have shown that the V169A mutant of BmCYP109E1 almost lost its ability to produce 16β-hydroxytestosterone. Another residue is Gly243, which is located on the α12 helix of BaCYP106A6. Although CYP106 proteins have glycine at this position, CYP109 proteins have isoleucine. The I241A mutant of BmCYP109E1 was almost completely deprived of 16β-hydroxytestosterone production. Interestingly, in the BaCYP106A6 structure, the Phe174 side chain occupied the space of the Ile241 side chain in BmCYP109E1 (Fig. 7B). Taken together, these residues located in the substrate-binding pocket are crucial for distinguishing the substrate specificities of CYP106 and CYP109 proteins. In addition, there are sequence differences between the α4–α5 loop and α9–α10 loop regions. Although direct structural comparisons are impossible due to the partial disorder in these regions, these loop regions are also thought to be important for the substrate-binding and specificity difference between CYP106 and CYP109 proteins.
Fig. 7. Structural comparison of BaCYP106A6 with other CYPs.
(A) Superposition of BaCYP106A6 and abietic acid-bound BaCYP106A2 (PDB code 5IKI). The α9, α10 helices, loop regions of BaCYP106A6, and the corresponding regions of BaCYP106A2 are shown as ribbon diagrams colored orange and cyan, respectively. The phenylalanine residues of BaCYP106A6 and BaCYP106A2 are represented by stick models. Bound heme molecules and abietic acid are also represented by stick models. (B) Superposition of BaCYP106A6 and BmCYP109E1 (PDB code 5L90). BmCYP109E1 is colored violet. The residues Val169 and Ile241 of BmCYP109E1 are represented by violet stick models. Phe174 of BaCYP106A6 and Ile241 of BmCYP109E1 occupy the same position in each substrate-binding pocket.
In this study, we isolated the BaCYP106A6 enzyme for the first time and investigated its function and structure. Our goal was to characterize its hydroxylating activity and analyze its structure to provide new information on steroid hydroxylases. Based on the BaCYP106A6-associated bioconversion results, steroids can be divided into active steroids with high catalytic activity and less active steroids. The less active steroids have the hydroxyl group at 11β and the larger side chain at 17β of steroids in common. Furthermore, the crystal structure revealed that BaCYP106A6 has a unique Arg295 residue, which is part of the constituent residues of the substrate-binding site, and possibly interacts with these functional groups. Therefore, this residue is considered an important target for substrate reactivity. Although additional structural investigations of BaCYP106A6 complexed with active steroids and mutational studies are needed, this finding provides insight into the specificity and regioselectivity of BaCYP106A6 hydroxylation. In addition, we provide fundamental information for future mutational studies on altering the hydroxylation of steroids and changing product yield.
Biochemical and structural investigations revealed a substrate preference for BaCYP106A6. Furthermore, structural analyses and molecular alignment–energy minimization showed how BaCYP106A6 distinguishes specific steroids.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This research was part of the project titled “Development of potential antibiotic compounds using polar organism resources (20200610, KOPRI Grant PM23030),” funded by the Ministry of Oceans and Fisheries, Korea. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2019R1D1A3A03103903). We thank the Division of Magnetic Resonance, Korea Basic Science Institute, Ochang, Chungbuk, Korea, for NMR analyses.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
References
- 1.Grossman CJ. Interactions between the gonadal steroids and the immune system. Science. 1985;227:257–261. doi: 10.1126/science.3871252. [DOI] [PubMed] [Google Scholar]
- 2.Monostory K, Dvorak Z. Steroid regulation of drug-metabolizing cytochromes P450. Curr. Drug Metab. 2011;12:154–172. doi: 10.2174/138920011795016854. [DOI] [PubMed] [Google Scholar]
- 3.Carson JA, Manolagas SC. Effects of sex steroids on bones and muscles: Similarities, parallels, and putative interactions in health and disease. Bone. 2015;80:67–78. doi: 10.1016/j.bone.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS. Microbial conversion of steroid compounds: recent developments. Enzyme Microb. Technol. 2003;32:688–705. doi: 10.1016/S0141-0229(03)00029-2. [DOI] [Google Scholar]
- 5.Donova MV, Egorova OV. Microbial steroid transformations: Current state and prospects. Appl. Microbiol. Biotechnol. 2012;94:1423–1447. doi: 10.1007/s00253-012-4078-0. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Jiang Y, Guengerich XFP, Ma L, Li S, Zhang W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020;295:833–849. doi: 10.1016/S0021-9258(17)49939-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Peng Y, Zhao J, Li Q, Yu X, Acevedo-Rocha CG, et al. Bacterial cytochrome P450-catalyzed regio- and stereoselective steroid hydroxylation enabled by directed evolution and rational design. Bioresour. Bioprocess. 2020;7:2. doi: 10.1186/s40643-019-0290-4. [DOI] [Google Scholar]
- 8.Mahato SB, Garai S. Advances in microbial steroid biotransformation. Steroids. 1997;62:332–345. doi: 10.1016/S0039-128X(96)00251-6. [DOI] [PubMed] [Google Scholar]
- 9.Rudolf JD, Chang CY, Ma M, Shen B. Cytochromes P450 for natural product biosynthesis in: Streptomyces: sequence, structure, and function. Nat. Prod. Rep. 2017;34:1141–1172. doi: 10.1039/C7NP00034K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guengerich FP, Munro AW. Unusual cytochrome P450 enzymes and reactions. J. Biol. Chem. 2013;288:17065–17073. doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei K, Chen H. Global identification, structural analysis and expression characterization of cytochrome P450 monooxygenase superfamily in rice. BMC Genomics. 2018;19:35. doi: 10.1186/s12864-017-4425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Du L, Li F, Zhang X, Qu Z, Han L, et al. Mechanistic insights into interactions between bacterial class i P450 enzymes and redox partners. ACS Catal. 2018;8:9992–10003. doi: 10.1021/acscatal.8b02913. [DOI] [Google Scholar]
- 13.Ma B, Wang Q, Ikeda H, Zhang C. Hydroxylation of steroids by a microbial substrate-promiscuous P450 cytochrome (CYP105D7): key arginine residues for rational design. Appl. Environ. Microbiol. 2019;85:e01530–19. doi: 10.1128/AEM.01530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Sanga K ichiro, Chikaoka Y, Itagaki E. Purification and properties of cytochrome P-450 (P-450lun) catalyzing steroid 11β-hydroxylation in Curvularia lunata. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1993;1203:215–223. doi: 10.1016/0167-4838(93)90086-7. [DOI] [PubMed] [Google Scholar]
- 15.Sakaki T. Practical application of cytochrome P450. Biol. Pharm. Bull. 2012;35:844–849. doi: 10.1248/bpb.35.844. [DOI] [PubMed] [Google Scholar]
- 16.Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol. Rev. 1998;78:1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi K, Sugimoto H, Shinkyo R, Yamada M, Ikeda S, Ikushiro S, et al. Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1. Biochemistry. 2008;47:11964–11972. doi: 10.1021/bi801222d. [DOI] [PubMed] [Google Scholar]
- 18.Kawauchi H, Sasaki J, Adachi T, Hanada K, Beppu T, Horinouchi S. Cloning and nucleotide sequence of a bacterial cytochrome P-450VD25 gene encoding vitamin D-3 25-hydroxylase. BBA - Gene Struct. Expr. 1994;1219:179–183. doi: 10.1016/0167-4781(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 19.Brill E, Hannemann F, Zapp J, Brüning G, Jauch J, Bernhardt R. A new cytochrome P450 system from Bacillus megaterium DSM319 for the hydroxylation of 11-keto-β-boswellic acid (KBA) Appl. Microbiol. Biotechnol. 2014;98:1703–1717. doi: 10.1007/s00253-013-5029-0. [DOI] [PubMed] [Google Scholar]
- 20.Bleif S, Hannemann R, Zapp J, Hartmann D, Jauch J, Bernhardt R. A new Bacillus megaterium whole-cell catalyst for the hydroxylation of the pentacyclic triterpene 11-keto-β-boswellic acid (KBA) based on a recombinant cytochrome P450 system. Appl. Microbiol. Biotechnol. 2012;93:1135–1146. doi: 10.1007/s00253-011-3467-0. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz D, Zapp J, Bernhardt R. Hydroxylation of the triterpenoid dipterocarpol with CYP106A2 from Bacillus megaterium. FEBS J. 2012;279:1663–1674. doi: 10.1111/j.1742-4658.2012.08503.x. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz D, Zapp J, Bernhardt R. Steroid conversion with CYP106A2 - production of pharmaceutically interesting DHEA metabolites. Microb. Cell Fact. 2014;13:81. doi: 10.1186/1475-2859-13-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdulmughni A, Jóźwik IK, Brill E, Hannemann F, Thunnissen AMWH, Bernhardt R. Biochemical and structural characterization of CYP109A2, a vitamin D3 25-hydroxylase from Bacillus megaterium. FEBS J. 2017;284:3881–3894. doi: 10.1111/febs.14276. [DOI] [PubMed] [Google Scholar]
- 24.Kim KH, Lee CW, Dangi B, Park SH, Park H, Oh TJ, et al. Crystal structure and functional characterization of a cytochrome P450 (BaCYP106A2) from Bacillus sp. PAMC 23377. J. Microbiol. Biotechnol. 2017;27:1472–1482. doi: 10.4014/jmb.1706.06013. [DOI] [PubMed] [Google Scholar]
- 25.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. J. Biol. Chem. 1964;239:2370–2378. doi: 10.1016/S0021-9258(20)82244-3. [DOI] [PubMed] [Google Scholar]
- 26.Johnston JB, Ouellet H, de Montellano PR. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010;285:36352–36360. doi: 10.1074/jbc.M110.161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski MW, Otwinowski ZZ, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Janocha S, Carius Y, Hutter M, Lancaster CRD, Bernhardt R. Crystal structure of CYP106A2 in substrate-free and substratebound form. ChemBioChem. 2016;17:852–860. doi: 10.1002/cbic.201500524. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, et al. Macromolecular structure determination using Xrays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40:1–8. [Google Scholar]
- 34.Perera R, Sono M, Sigman JA, Pfister TD, Lut Y, Dawson JH. Neutral thiol as a proximal ligand to ferrous heme iron: Implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. USA. 2003;100:3641–3646. doi: 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean KJ, Warman AJ, Seward HE, Marshall KR, Girvan HM, Cheesman MR, et al. Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions. Biochemistry. 2006;45:8427–8443. doi: 10.1021/bi0601609. [DOI] [PubMed] [Google Scholar]
- 36.Zhong F, Lisi GP, Collins DP, Dawson JH, Pletneva EV. Redox-dependent stability, protonation, and reactivity of cysteinebound heme proteins. Proc. Natl. Acad. Sci. USA. 2014;111:E306–E315. doi: 10.1073/pnas.1317173111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zehentgruber D, Hannemann F, Bleif S, Bernhardt R, Lütz S. Towards preparative scale steroid hydroxylation with cytochrome P450 monooxygenase CYP106A2. ChemBioChem. 2010;11:713–721. doi: 10.1002/cbic.200900706. [DOI] [PubMed] [Google Scholar]
- 38.Kiss FM, Schmitz D, Zapp J, Dier TKF, Volmer DA, Bernhardt R. Comparison of CYP106A1 and CYP106A2 from Bacillus megaterium - identification of a novel 11-oxidase activity. Appl. Microbiol. Biotechnol. 2015;99:8495–8514. doi: 10.1007/s00253-015-6563-8. [DOI] [PubMed] [Google Scholar]
- 39.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77:114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jóźwik IK, Kiss FM, Gricman Ł, Abdulmughni A, Brill E, Zapp J, et al. Structural basis of steroid binding and oxidation by the cytochrome P450 CYP109E1 from Bacillus megaterium. FEBS J. 2016;283:4128–4148. doi: 10.1111/febs.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gober JG, Ghodge S V, Bogart JW, Wever WJ, Watkins RR, Brustad EM, et al. P450-mediated non-natural cyclopropanation of dehydroalanine-containing thiopeptides. ACS Chem. Biol. 2017;12:1726–1731. doi: 10.1021/acschembio.7b00358. [DOI] [PubMed] [Google Scholar]
- 45.Zhang A, Zhang T, Hall EA, Hutchinson S, Cryle MJ, Wong LL, et al. The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis. Mol. Biosyst. 2015;11:869–881. doi: 10.1039/C4MB00665H. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Tietz DR, Rutaganira FU, Kells PM, Anzai Y, Kato F, et al. Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions. J. Biol. Chem. 2012;287:37880–37890. doi: 10.1074/jbc.M112.410340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasutake Y, Imoto N, Fujii Y, Fujii T, Arisawa A, Tamura T. Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) Biochem. Biophys. Res. Commun. 2007;361:876–882. doi: 10.1016/j.bbrc.2007.07.062. [DOI] [PubMed] [Google Scholar]
- 48.Parisi G, Freda I, Exertier C, Cecchetti C, Gugole E, Cerutti G, et al. Dissecting the cytochrome p450 olep substrate specificity: evidence for a preferential substrate. Biomolecules. 2020;10:1411. doi: 10.3390/biom10101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.