Abstract
Herpud1 is an ER-localized protein that contributes to endoplasmic reticulum (ER) homeostasis by participating in the ER-associated protein degradation pathway. The Nrf1 transcription factor is important in cellular stress pathways. We show that loss of Nrf1 function results in decreased Herpud1 expression in cells and liver tissues. Expression of Herpud1 increases in response to ER stress, but not in Nrf1 knockout cells. Transactivation studies show that Nrf1 acts through antioxidant response elements located in the Herpud1 promoter, and chromatin immunoprecipitation demonstrates that Herpud1 is a direct Nrf1 target gene. These results indicate that Nrf1 is a transcriptional activator of Herpud1 expression during ER stress, and they suggest Nrf1 is a key player in the regulation of the ER stress response in cells.
Keywords: Nrf1, ER stress, transcriptional regulation
INTRODUCTION
Endoplasmic reticulum (ER) stress occurs when aberrant or unfolded protein accumulates in the ER as a result of physiological or pathological processes (Walter and Ron, 2011; Hetz, 2012). ER stress has been suggested to contribute to the pathogenesis of various disorders including neurodegeneration, cancer, obesity, type II diabetes and fatty liver disease (Tsai and Weissman, 2010; Clarke et al., 2014; Lee and Ozcan, 2014; Wang and Kaufman, 2014a; Wang and Kaufman, 2014b). Cells respond to ER stress by activating the unfolded protein response (UPR) pathway. The UPR functions to restore ER homeostasis by inhibiting protein synthesis and inducing genes that promote protein folding and degradation. The ER-associated protein degradation (ERAD) pathway, a process in the UPR pathway, functions to eliminate terminally misfolded proteins from the ER by triggering retrograde transport of protein to the cytoplasm for proteasome-dependent degradation (Ruggiano et al., 2014).
Nuclear factor erythroid-derived 2-related factor 1 (Nrf1, also known as NFE2L1) is a member of the Cap-N-Collar (CNC) subfamily of bZIP transcription factors that includes Nrf2, p45NFE2 and Nrf3 (Andrews et al., 1993; Chan et al., 1993; Moi et al., 1994; Kobayashi et al., 1999). Nrf1 is an essential gene and is critical for the maintenance of cellular homeostasis. The global knockout of Nrf1 in mice leads to embryonic lethality, and conditional gene targeting of Nrf1 in various tissues indicates Nrf1 is important in the regulation against development of steatohepatitis and neurodegeneration in mice (Chan et al., 1998; Xu et al., 2005; Lee et al., 2011). Nrf1 has been shown to regulate the expression of genes via cis-active sequences known as the antioxidant response element (Venugopal and Jaiswal, 1996; Kwong et al., 1999; Myhrstad et al., 2001). In addition, Nrf1 is important in the proteotoxic stress response through its role in regulating expression of genes encoding the proteasome (Radhakrishnan et al., 2010; Lee et al., 2011; Lee et al., 2013). In response to proteasome inhibition, Nrf1 mediates recovery of proteasome function, and promotes survival of cancer cells (Radhakrishnan et al., 2010). Aside from cellular stress response, Nrf1 has been shown to regulate differentiation of osteoblast and odontoblast (Narayanan et al., 2004; Xing et al., 2007; Kim et al., 2010). Additionally, loss of Nrf1 function leads to genetic instability and the promotion of tumorigenesis (Xu et al., 2005; Oh et al., 2012).
Herpud1 (homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1) is an ER-resident membrane protein (Kokame et al., 2000). Herpud1 has been shown to facilitate ER-associated degradation by functioning as a shuttle factor that delivers ubiquitinated substrates to the proteasome for degradation (Okuda-Shimizu and Hendershot, 2007; Huang et al., 2014). Herpud1 has also been shown to be involved in regulating ubiquitination of Hrd1, a ubiquitin–ligase complex responsible for protein degradation in the ER (Kny et al., 2011). Expression of Herpud1 is induced in primary neurons by agents that induce ER dysfunction, or in response to proteasome inhibition (Miura et al., 2010). Furthermore, Herpud1 is essential for neuronal survival; knockdown of Herpud1 expression leads to enhance susceptibility to ER stress-induced apoptosis (Chan et al., 2004; Hori et al., 2004; Nogalska et al., 2006; Shinozaki et al., 2013).
In the current study we investigated the role of Nrf1 in the regulation of Herpud1 expression. We demonstrate that Nrf1 is necessary for basal expression and induction of Herpud1 during ER stress in human and mouse cells. Through reporter gene assays and chromatin immunoprecipitation studies, we show that Nrf1 directly activates Herpud1 expression via ARE-like response elements in the Herpud1 promoter. Our results identify Herpud1 as a direct Nrf1 target gene, and implicate Nrf1 as an important transcription factor in the regulation of the ERAD pathway and ER stress response.
MATERIALS AND METHODS
Reagents
Dulbecco's modified Eagle's medium (DMEM), streptomycin, penicillin and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA). Tunicamycin, thapsigargin, glucose oxidase, and menadione were purchased from Sigma-Aldrich (St. Louis, MO). Direct-zol RNA MiniPrep kit was purchased from ZymoResearch (Irvine, CA). Bradford assay kit and iScript Reverse Transcription Supermix were from BioRad (Hercules, CA). FastStart SYBR Green reagent mix for quantitative RT-PCR was purchased from Roche (Indianapolis, IN). BioT was purchased from Bioland Scientific (Paramount, CA). Dual Reporter Assay Kit was from Promega (Madison, WI). Tcf11 antibody and SimpleChIP Plus Enzymatic Chromatin IP Kit were purchased from Cell Signaling (Danvers, MA). Enhanced chemiluminescence substrate kit was from Pierce Biotechnology (Rockford, IL). Herpud1 antibody (ab56742) was purchased from Abcam (Cambridge, MA).
RNA Isolation and Quantitative Real-Time PCR
RNA was extracted from tissues and cells using Direct-zol RNA MiniPrep kit according to the manufacturer's recommendation. cDNA synthesis was done using iScript Reverse Transcription kit. Quantitative amplification of cDNA was performed in a Step One Plus PCR machine (Life technologies, Grand Island, NY) using FastStart SyBr Green reagent in duplicate 10-μl reactions. PCR cycling conditions consist of 95 °C for 15 min and 45 cycles of 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 45 s. RPLP0 was used as an endogenous control and relative expression is calculated with the equation 2(Ct target gene-Ct RPLP0).
Cells
HEK293T and spontaneously immortalized mouse embryonic fibroblast cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 100 units/ml penicillin. Cells were cultured at 37 °C in humidified 5% CO2 atmosphere.
Plasmids
A luciferase reporter constructs containing the mouse Herpud1 promoter was generated by PCR amplification of the mouse genomic DNA sequence using primers that spans the −3727/+3 region of the Herpud1 open reading frame, and cloned into the NheI and XhoI sites of the pGL3-basic vector. Point mutations in the ARE sites of the Herpud1 promoter were generated by inverse PCR.
Transfection and Luciferase Assays
Cells were seeded onto a 24-well plate and grown to approximately 70% confluence prior to transfection using BioT reagent according to the manufacturer's protocol. Cellular extracts were prepared 48 hours after transfection, and Firefly- and Renilla-luciferase activities measured using Dual Reporter Assay Kit.
Chromatin Immunoprecipitation Assay (ChIP)
Chromatin immunoprecipitation assay was done using SimpleChIP Plus Enzymatic Chromatin IP Kit according to manufacturer's protocol with minor modifications. HEK293 Cells were fixed with 1% formaldehyde and the cross-linking reaction stopped by addition of glycine to a final concentration of 125 mM. Cells were then washed, lysed, and then sonicated. The supernatant was pre-cleared with Protein-G beads and incubated overnight at 4°C with Nrf1-specific rabbit polyclonal antibody (Wang and Chan, 2006), or unrelated rabbit polyclonal antibody as control. The DNA-protein complexes were then washed and eluted. Cross-linking was reversed, and recovered DNA was subjected to quantitative PCR using primers that flank (-2977 to +132 nt) the Herpud1 gene promoter. PsmB6 and beta-tubulin promoters were used as positive and negative controls, respectively.
Western Blotting
Cells were lysed in cold RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1X Protease Inhibitor). Lysates were cleared by centrifugation for 15 min at 4 °C, and Bradford reagent was used to measure protein concentrations. An equal volume of 2 X SDS sample buffer (100 mM Tris, pH 6.8, 25% glycerol, 2% SDS, 0.01% bromphenol blue, 10% 2-mercaptoethanol) was added to cell lysates, and the mixture was boiled for 5 min. Proteins were electrophoresed on SDS-polyacrylamide gels and then transferred onto nitrocellulose membranes. Membranes were then blocked in 5% non-fat dry milk in TBS-T (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, and 0.05% Tween 20) at room temperature for one hour, and then incubated with the indicated primary antibodies overnight at 4 °C followed by a incubation with a horseradish peroxidase-conjugated secondary antibody. Antibody-antigen complexes on the blots were detected using chemiluminescent detection system.
Primers
| Forward | Reverse | |
|---|---|---|
| Herpud1 | ACTCCTCGCTGAGCAGATTT | CTCTGTCTGAACGGAAACCA |
| Nqo1 | GCATTGGCCACACTCCACCAG | ATGGCCCACAGAGAGGCCAAA |
| Herpud1 (ChIP) | GATTGGGCCACGTTGGGAGAG | CCCGCAATCTCTGCAACGACA |
| Herpud1 (Promoter) | CGGAGCTCTCAATATCCTCCGATG | CATGTCTGGCTAGGCGGC |
| Herpud1 (M1) | AACAATTGGGCCACGTTGGCAC | GCCCGGAGAAAGGCGGCGGAG |
RESULTS AND DISCUSSION
ER stress, but not oxidative stress, induces Herpud1 expression
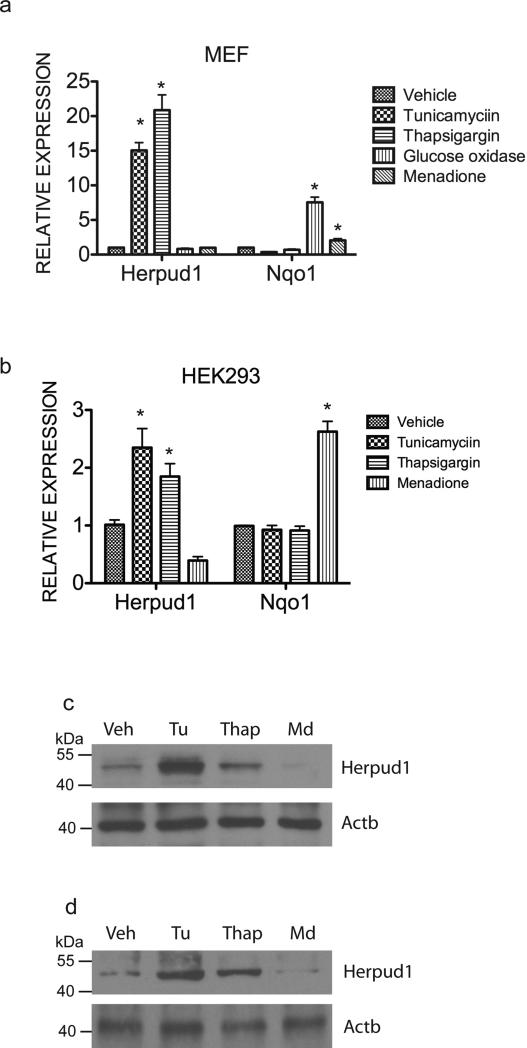
Due to a role of Nrf1 in antioxidant gene expression, we sought to investigate whether Herpud1 can be activated by oxidative stress. To do this, we analyzed Herpud1 expression in response to ER and oxidative stress stimuli. Both tunicamycin and thapsigargin treatment significantly increased the expression of Herpud1 mRNA in both HEK293T and MEF cells (Fig. 1a and b). Immunoblotting showed that Herpud1 protein levels were also increased significantly in tunicamycin- and thapsigargin-treated HEK293 and MEF cells relative to vehicle control cells (Fig. 1c and d). In contrast, no significant change in Herpud1 expression was observed in cells treated with menadione, or glucose oxidase to induce oxidative stress (Fig. 1a-c). As expected, induction of Nqo1 expression was observed in response to oxidative stress induction (Fig. 1a and b). These findings are consistent with previous results that demonstrated Herpud1 is associated with ER stress pathway.
Figure 1. Herpud1 expression is induced by ER stress, but not oxidative stress.
Expression levels of Herpud1 and Nqo1 mRNA examined by quantitative RT-PCR in (a) MEF and (b) HEK293 cells treated with tunicamycin (2μg/mL), thapsigargin (2μM), glucose oxidase (10mU) and menadione (2.5μM) for 24 hours. Expression levels were quantitated relative to endogenous RPLP0 levels as an internal reference, and calculated as 2(Ct test gene-Ct RPLP0). Data are shown as the means ± SEM of three independent experiments. *P < 0.05 compared to vehicle-treated cells. Western blotting of Herpud1 in (c) HEK293 and (d) MEF cells treated with tunicamycin (2μg/mL), thapsigargin (2μM) and menadione (2.5μM) for 24 hours. β-actin was used for loading control.
Constitutive and ER-stress induced expression of Herpud1 is Nrf1 dependent
To evaluate a possible role of Nrf1 in regulating Herpud1 expression, we first investigated the expression of Herpud1 in wild type and Nrf1 knockout MEF cells by RT–PCR. Compared to wild type MEF cells, Herpud1 expression was decreased in Nrf1 knockout cells (Fig. 2a). Treatment with tunicamycin resulted in up-regulation of Herpud1 in wild type cells. In contrast, Herpud1 induction by tunicamycin was blunted in Nrf1 KO cells (Fig. 2a). Consistent with these results, livers from mice with hepatocyte-specific disruption of Nrf1 also showed diminished basal and tunicamycin-induced expression of Herpud1 (Fig. 2b). Based on these findings, we conclude that Herpud1 expression is Nrf1-dependent.
Figure 2. Loss of Nrf1 results in decreased constitutive and ER-stress activated expression of Herpud1.
Herpud1 mRNA expression in (a) wild type and Nrfl1 knockout MEF cells, and (b) livers of control and Nrf1LKO mice compared by quantitative RT-PCR analysis. MEF cells were treated with tunicamycin for 24 hours prior to RNA isolation. Wild type and Nrf1 liver knockout mice were treated with 2mg/kg of tunicamycin for 48 hours prior to harvesting tissues for analysis. Expression levels were quantitated relative to endogenous RPLP0 levels as an internal reference, and calculated as 2(Ct test gene-Ct RPLP0). *P < 0.05 compared to vehicle treated cells or livers. #P < 0.05 compared to wild type cells or livers.
Herpud1 gene promoter is activated by Nrf1
We next determined whether Nrf1 regulates Herpud1 promoter activity. The Herpud1 regulatory region was cloned into a luciferase reporter vector and transient transfection studies were done. In a pattern similar to RT-PCR analysis, luciferase expression was markedly decreased in Nrf1 knockout cells, suggesting that Nrf1 activity is involved in the transcriptional control of Herpud1 (Fig. 3a). In support of this idea, an increase of the luciferase activity was observed in a dose-dependent manner when the Nrf1 expression vector was co-transfected with the reporter (Fig. 3a). Herpud1 promoter activity was increased by tunicamycin in both MEF (Fig. 3b) and HEK293 cells (Fig. 3c). In agreement with gene expression analysis, Herpud1 promoter was not activated by oxidative stress induced by glucose oxidase treatment (Fig. 3b). To investigate the ability of Nrf1 to potentiate ER stress activation of the Herpud1 promoter, wild type and Nrf1 knockout MEF cells transfected with the reporter plasmid were treated with tunicamycin. Herpud1 promoter activity was increased by tunicamycin treatment in wild type, but not in Nrf1 knockout cells (Fig. 3d). Together, these results demonstrate that ER stress increased Herpud1 promoter activity through Nrf1.
Figure 3. Transactivation of Herpud1 gene is Nrf1 dependent.
(a) Wild type and Nrf1 knockout MEF cells were transfected with a luciferase reporter plasmid (200ng/well) containingthe mouse Herpud1 gene promoter along with increasing concentrations of Nrf1 expression vector (50, 100, and 200ng/well). (b) MEF cells and (c) HEK293 cells transfected with Herpud1 reporter plasmid (2ng/well) were treated with tunicamycin, or glucose oxidase, and luciferase activity was analyzed after 24 hours after treatment. (d) Wild type and Nrf1 knockout MEF cells transfected with the Herpud1 reporter (200ng/well) were treated with tunicamycin, and luciferase activity was analyzed after 24 hours. Luciferase values were normalized to Renilla-luciferase activity from pRL-null transfection control plasmid (1ng/well). Histograms represent mean values ± SEM for 3 experiments each containing 3 replicates. *P<0.05 relative to vehicle (DMSO) control. #P < 0.05 compared to wild type cells. ##P<0.05 compared to Nrf1 mutant cells.
Nrf1 regulates ARE dependent activation of Herpud1 promoter
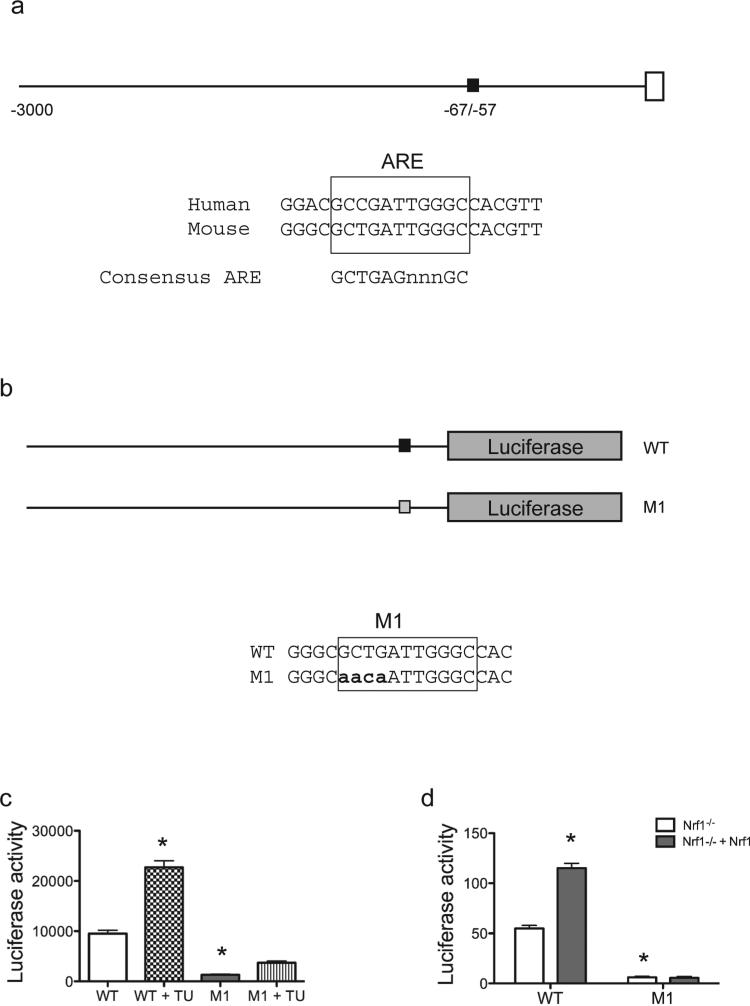
Transcription of Herpud1 has been shown to be regulated by ER stress-responsive cis-acting elements (ERSE-I, ERSE-II and C/EBP-ATF composite site) located at the proximal region of the Herpud1 promoter (Kokame et al., 2001; Ma and Hendershot, 2004; Yamamoto et al., 2004; Liang et al., 2006). Transcription factors found to transactivate Herpud1 at ERSE-I or ERSE-II include XBP-1, ATF6, and Luman. While ATF4 binds and activates through the C/EBP-ATF sequence, it is dispensable for ER-stress mediated induction of Herpud1 (Ma and Hendershot, 2004). We scanned the mouse Herpud1 promoter region for potential Nrf1 binding sites. Based on the GCTGAGnnnGC motif, an ARE-like motif showing good match to the consensus motif was identified between nucleotide −67/–57 of the promoter (Fig. 4a). To investigate the functional capacity of this site, point mutations were introduced into this ARE-like site in the Herpud1 promoter construct (Fig. 4b). Point mutations in the ARE site resulted in decreased expression, and induction by tunicamycin (Fig. 4c). Co-expression of Nrf1 was able to increase the activity of the wild-type Herpud1 promoter construct, but activation by Nrf1 was abolished in the mutant reporter construct (Fig. 4d). These results suggest that ARE-like site in the Herpud1 promoter is critical for Nrf1-mediated basal and ER-stress induced activation in MEF cells, and this data provides the first demonstration that an ARE also regulates the Herpud1 core promoter.
Figure 4. Nrf1 activates the Herpud1 promoter through ARE sites.
(a) Schematic representation of the proximal promoter sequence of the mouse Herpud1 gene. Black box indicates the ARE site, which is conserved in both mouse and human promoter. (b) Schematic representation of wild type (WT), mutant 1 (M1) luciferase vectors. Mutations introduced in the ARE site are in bold font shown below the WT sequence. (c) HEK293 cells were transiently transfected with the indicated constructs. To activate ER stress, cells were incubated in tunicamycin (2μg/mL) for 24 h, and luciferase activity was analyzed and normalized to Renilla luciferase. (d) Nrf1−/− MEF cells and Nrf1−/− MEF cells complemented with Nrf1 cDNA were transiently transfected with the indicated constructs. Luciferase activity was analyzed and normalized to Renilla luciferase. The data are presented as means ± SEM for 3 experiments each containing 3 replicates. *P value <0.05 relative to DMSO treated wild type cells.
Nrf1 binds the Herpud1 Promoter in vivo
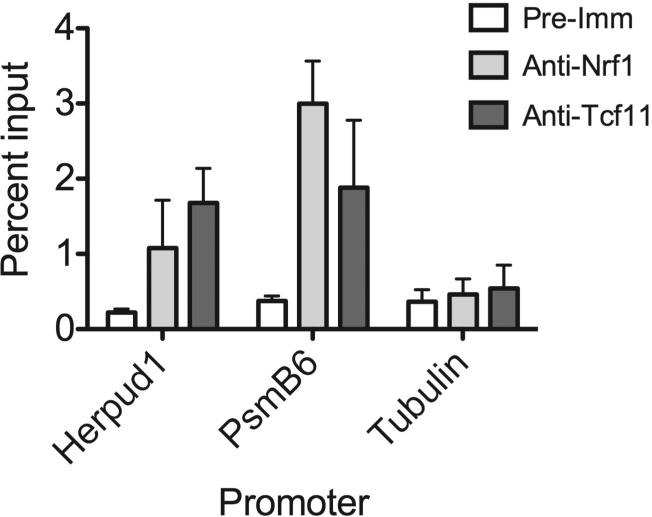
To directly assess Nrf1 binding to the Herpud1 gene promoter, we performed chromatin immunoprecipitation (ChIP) of endogenous Nrf1 in cells. Quantitative PCR analysis of ChIP samples showed Nrf1 co-immunoprecipiated with the Herpud1 promoter (Fig. 5). There was also significant amplification to the PsmB6 promoter, a previously published direct target gene of Nrf1 (Lee et al., 2011), in ChIP samples obtained using Nrf1-specific antibody. As expected, significant binding of Nrf1 on the tubulin gene promoter was not detected. These experiments suggest that Nrf1 is a direct mediator in controlling Herpud1 transcription.
Figure 5. Nrf1 binds the Herpud1 Promoter in vivo.
qPCR analysis were carried out on chromatin template precipitated using Nrf1-specific antibodies (anti-Nrf1 (Wang and Chan, 2006) and anti-Tcf11), or rabbit pre-immune IgG. Primers were targeted to the -500nt region of the Herpud1 promoter. Primers against PsmB6 and tubulin promoters were used as positive and negative controls, respectively. Histograms represent ChIP results expressed as the ratio to nonimmunoprecipitated chromatin (1%) used as an input control. The data are representative two independent experiments.
In summary, we show that: (i) Herpud1 is induced by ER, but not oxidative stress, (ii) disruption of Nrf1 function down regulates Herpud1 expression in mouse liver and cells, (iii) Nrf1 plays a role in basal and ER-stress induced activation of the Herpud1 promoter, (iv) Nrf1 activates through an ARE site in the promoter, and (v) Nrf1 binds the Herpud1 gene promoter. On the basis of these results, we conclude that Nrf1 is important for Herpud1 gene expression. Consistent with previously reported Herpud1 involvement in ERAD, it is expected that misfolded proteins in the ER would accumulate as a result of Nrf1 deficiency. However, the effect of Nrf1 knockout on degradation of model ERAD substrates is currently not known. Nonetheless, it is reasonable to postulate that reduction in Herpud1 function, in conjunction with reduced proteasome function that was previously demonstrated, contribute to ER stress in Nrf1 deficient cells. Previous Nrf1-knockout studies suggest a relationship between Nrf1 and ER stress (Lee et al., 2013). Disruption of the Nrf1 gene in mouse liver caused steatohepatitis, a disorder linked with high levels of ER stress (Xu et al., 2005; Zhang et al., 2011). Additionally, Nrf1 liver knockout mice show activation of the ER stress-signaling pathway (Lee et al., 2013). Because of the role of Herpud1 in ERAD, our data showing Herpud1 as a direct target of Nrf1 further links Nrf1 to protein quality control and ER stress response. As ER stress is associated with the development of multiple disorders including obesity, type II diabetes and cancer, further studies to elucidate where Nrf1 resides in the genetic cascade controlling expression of other genes in the ERAD and ER stress pathway would be of great interest.
Herpud1 is an ER-resident membrane protein.
Herpud1 plays a role in ER-associated degradation pathway and in maintaining ER homeostasis.
Herpud1 expression is down regulated in Nrf1 deficient cells.
Nrf1 transcription factor activates Herpud1 expression in response to ER stress.
ACKNOWLEDEMENTS
This research was supported by NIH grants CA091907 and NS065223.
Abbreviations
- ARE
antioxidant response element
- CNC
Cap and Collar
- ER
endoplasmic reticulum
- Nrf1
Nuclear factor erythroid-derived 2-related factor 1
- ERAD
ER-associated protein degradation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to disclose.
REFERENCES
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–8. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:11371–5. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. The EMBO journal. 1998;17:1779–87. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Fu W, Zhang P, Cheng A, Lee J, Kokame K, Mattson MP. Herp stabilizes neuronal Ca2+ homeostasis and mitochondrial function during endoplasmic reticulum stress. J Biol Chem. 2004;279:28733–43. doi: 10.1074/jbc.M404272200. [DOI] [PubMed] [Google Scholar]
- Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer Cell. 2014;25:563–73. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Hori O, Ichinoda F, Yamaguchi A, Tamatani T, Taniguchi M, Koyama Y, Katayama T, Tohyama M, Stern DM, Ozawa K, Kitao Y, Ogawa S. Role of Herp in the endoplasmic reticulum stress response. Genes Cells. 2004;9:457–69. doi: 10.1111/j.1356-9597.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- Huang CH, Chu YR, Ye Y, Chen X. Role of HERP and a HERP-related protein in HRD1-dependent protein degradation at the endoplasmic reticulum. J Biol Chem. 2014;289:4444–54. doi: 10.1074/jbc.M113.519561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Xing W, Wergedal J, Chan JY, Mohan S. Targeted disruption of nuclear factor erythroid-derived 2-like 1 in osteoblasts reduces bone size and bone formation in mice. Physiol Genomics. 2010;40:100–10. doi: 10.1152/physiolgenomics.00105.2009. [DOI] [PubMed] [Google Scholar]
- Kny M, Standera S, Hartmann-Petersen R, Kloetzel PM, Seeger M. Herp regulates Hrd1-mediated ubiquitylation in a ubiquitin-like domain-dependent manner. J Biol Chem. 2011;286:5151–6. doi: 10.1074/jbc.M110.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap'n' collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–52. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J Biol Chem. 2000;275:32846–53. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- Kokame K, Kato H, Miyata T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J Biol Chem. 2001;276:9199–205. doi: 10.1074/jbc.M010486200. [DOI] [PubMed] [Google Scholar]
- Kwong M, Kan YW, Chan JY. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. The Journal of biological chemistry. 1999;274:37491–8. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- Lee CS, Ho DV, Chan JY. Nuclear factor-erythroid 2-related factor 1 regulates expression of proteasome genes in hepatocytes and protects against endoplasmic reticulum stress and steatosis in mice. FEBS J. 2013;280:3609–20. doi: 10.1111/febs.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Lee C, Hu T, Nguyen JM, Zhang J, Martin MV, Vawter MP, Huang EJ, Chan JY. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc Natl Acad Sci U S A. 2011;108:8408–13. doi: 10.1073/pnas.1019209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ozcan U. Unfolded protein response signaling and metabolic diseases. J Biol Chem. 2014;289:1203–11. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Audas TE, Li Y, Cockram GP, Dean JD, Martyn AC, Kokame K, Lu R. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol Cell Biol. 2006;26:7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Herp is dually regulated by both the endoplasmic reticulum stress-specific branch of the unfolded protein response and a branch that is shared with other cellular stress pathways. J Biol Chem. 2004;279:13792–9. doi: 10.1074/jbc.M313724200. [DOI] [PubMed] [Google Scholar]
- Miura H, Hashida K, Sudo H, Awa Y, Takarada-Iemata M, Kokame K, Takahashi T, Matsumoto M, Kitao Y, Hori O. Deletion of Herp facilitates degradation of cytosolic proteins. Genes Cells. 2010;15:843–53. doi: 10.1111/j.1365-2443.2010.01422.x. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–30. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrstad MC, Husberg C, Murphy P, Nordstrom O, Blomhoff R, Moskaug JO, Kolsto AB. TCF11/Nrf1 overexpression increases the intracellular glutathione level and can transactivate the gamma-glutamylcysteine synthetase (GCS) heavy subunit promoter. Biochimica et biophysica acta. 2001;1517:212–9. doi: 10.1016/s0167-4781(00)00276-1. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Ramachandran A, Peterson MC, Hao J, Kolsto AB, Friedman AD, George A. The CCAAT enhancer-binding protein (C/EBP)beta and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. The Journal of biological chemistry. 2004;279:45423–32. doi: 10.1074/jbc.M405031200. [DOI] [PubMed] [Google Scholar]
- Nogalska A, Engel WK, McFerrin J, Kokame K, Komano H, Askanas V. Homocysteine-induced endoplasmic reticulum protein (Herp) is up-regulated in sporadic inclusion-body myositis and in endoplasmic reticulum stress-induced cultured human muscle fibers. J Neurochem. 2006;96:1491–9. doi: 10.1111/j.1471-4159.2006.03668.x. [DOI] [PubMed] [Google Scholar]
- Oh DH, Rigas D, Cho A, Chan JY. Deficiency in the nuclear-related factor erythroid 2 transcription factor (Nrf1) leads to genetic instability. FEBS J. 2012;279:4121–30. doi: 10.1111/febs.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–54. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–79. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki S, Chiba T, Kokame K, Miyata T, Kaneko E, Shimokado K. A deficiency of Herp, an endoplasmic reticulum stress protein, suppresses atherosclerosis in ApoE knockout mice by attenuating inflammatory responses. PLoS One. 2013;8:e75249. doi: 10.1371/journal.pone.0075249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Weissman AM. The Unfolded Protein Response, Degradation from Endoplasmic Reticulum and Cancer. Genes Cancer. 2010;1:764–778. doi: 10.1177/1947601910383011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14960–5. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014a;14:581–97. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- Wang S, Kaufman RJ. How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr Opin Lipidol. 2014b;25:125–32. doi: 10.1097/MOL.0000000000000056. [DOI] [PubMed] [Google Scholar]
- Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem. 2006;281:19676–87. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- Xing W, Singgih A, Kapoor A, Alarcon CM, Baylink DJ, Mohan S. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. The Journal of biological chemistry. 2007;282:22052–61. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4120–5. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136:343–50. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. The EMBO journal. 2011;30:1357–75. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]